SUMMARY
Pathological activation of TGF-β signaling is universal in fibrosis. Aberrant TGF-β signaling in conjunction with transdifferentiation of hepatic stellate cells (HSCs) into fibrogenic myofibroblasts plays a central role in liver fibrosis. Here we report that the DNA demethylase TET3 is anomalously upregulated in fibrotic livers in both humans and mice. We demonstrate that in human HSCs, TET3 promotes profibrotic gene expression by upregulation of multiple key TGF-β pathway genes, including TGFB1. TET3 binds to target gene promoters, inducing demethylation, which in turn facilitates chromatin remodeling and transcription. We also reveal a positive feedback loop between TGF-β1 and TET3 in both HSCs and hepatocytes. Furthermore, TET3 knockdown ameliorates liver fibrosis in mice. Our results uncover a TET3/TGF-β1 positive feedback loop as a crucial determinant of liver fibrosis and suggest that inhibiting TET3 may represent a therapeutic strategy for liver fibrosis and perhaps other fibrotic diseases.
In Brief
Xu et al. unmask a positive feedback loop between chromatin demethylase TET3 and TGF-β1 in stressed hepatocytes and stellate cells in humans and mice. Activation of this loop stimulates expression of fibrotic genes, whereas knockdown of TET3 reduces liver fibrosis in mice, suggesting a strategy for treating fibrosis.
Graphical Abstract
INTRODUCTION
Liver fibrosis is a major cause of morbidity and mortality worldwide. Fibrosis develops when the liver is injured because of chronic liver disease such as viral hepatitis, alcoholic fatty liver disease, and nonalcoholic fatty liver disease (NAFLD) or nonalcoholic steatohepatitis (NASH). To date, effective treatments to halt or reverse fibrosis are lacking. Understanding the signal transduction pathways responsible for the development and progression of fibrosis is key to prevention and cure.
A critical event in fibrogenesis is activation of hepatic stellate cells (HSCs), which are the primary source of extracellular matrix (ECM) proteins (Li et al., 2017; Tsuchida and Friedman, 2017). Quiescent HSCs control ECM turnover by releasing limited amounts of ECM molecules, matrix metalloproteinases, and their inhibitors. Liver injury activates and transforms HSCs into fibrogenic myofibroblasts that acquire the ability to produce α smooth muscle actin (α-SMA) and abundant ECM components, including type I collagen (COL1A1), fibronectin (FN1), and ECM remodeling enzymes such as thrombospondin 1 (TSP1) and tissue inhibitor of metalloproteinases-1 (TIMP1). HSC activation represents the major source of fibrogenic myofibroblasts irrespective of the underlying cause of liver damage (Henderson et al., 2013; Tsuchida and Friedman, 2017).
TGF-β is the master regulator of fibrosis (Meng et al., 2016; Murphy-Ullrich and Suto, 2018). The canonical TGF-β signaling components include TGF-β ligands, TGF-β receptor 2 (TGFBR2) and TGF-β receptor 1 (TGFBR1), and Smad proteins. TGF-β isoforms are secreted as latent precursors that need to be converted into biologically active forms by a variety of mechanisms in a cell-, tissue-, and/or disease-specific manner. Normally, only a small fraction of TGF-β is biologically active. Activated TGF-β binds to TGFBR2, which recruits and activates TGFBR1. TGFBR1 then phosphorylates Smad2 and Smad3, which form complexes with Smad4 and translocate into the nucleus to drive transcription of profibrotic molecules such as α-SMA, COL1A1, FN1, and TIMP1, thereby inducing myofibroblast activation and ECM deposition.
A pathological increase in TGF-β signaling is central to HSC activation, and HSCs themselves are the significant source of TGF-β1 and TSP1 (Breitkopf et al., 2005). Importantly, other hepatic cell types, including hepatocytes, also secrete TGF-β1, contributing critically to the profibrotic shift of HSCs (Benzoubir et al., 2013; Jee et al., 2015; Jeong et al., 2004; Tsuchida and Friedman, 2017). TSP1 acts as a primary regulator of TGF-β1 bioactivity in a number of fibrotic diseases (Li et al., 2017; Murphy-Ullrich and Suto, 2018). In addition to TSP1, TIMP1 has been shown to increase in hepatic fibrosis. Although transgenic TIMP1 overexpression promoted hepatic fibrosis in a carbon tetrachloride (CCl4)-induced liver fibrosis mouse model (Yoshiji et al., 2000), blocking TSP1-mediated TGF-β activation by a TSP1 antagonistic peptide attenuated dimethyl-nitrosamine-induced liver damage and fibrosis (Li et al., 2017; Murphy-Ullrich and Suto, 2018). Altogether, these studies highlight the importance of TGF-β1, TSP1, and TIMP1 in the genesis of liver fibrosis. However, how their expression is regulated remains incompletely understood.
The TET proteins belong to a novel class of DNA demethylases that oxidize 5-methylcytosine (5mC) to generate 5-hydroxymethylcytosine (5hmC), which is subsequently converted to unmethylated cytosine (An et al., 2017; Rasmussen and Helin, 2016; Wu and Zhang, 2017). Although much is known about the role of TETs in development and cancer, little is known of their function and mechanism in liver fibrosis, despite altered expressions of TETs having been noted in fibrotic liver diseases (Page et al., 2016). We have documented an increased expression of TET3 in uterine leiomyomas (uterine fibroids), benign tumors characterized by hyperplastic smooth muscle cells and excessive deposition of ECM from leiomyoma cells (Cao et al., 2019). In the present work, we uncover a positive feedback regulatory mechanism involving TET3 and TGF-β1 in both HSCs and hepatocytes as a critical determinant of liver fibrosis.
RESULTS AND DISCUSSION
TET3 Expression Increases in Fibrotic Livers
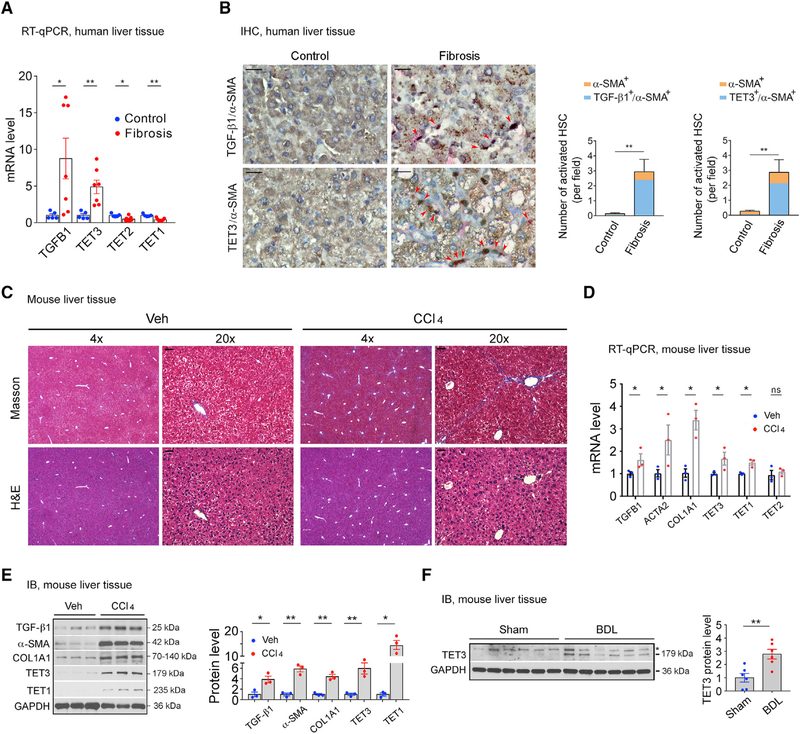
We analyzed TET expression in fibrotic and non-fibrotic livers. Liver tissues were obtained from patients with histories of chronic hepatitis B viral infection. TGF-β1 expression was significantly increased in fibrotic compared with non-fibrotic livers (Figure 1A). There was also increased expression of TET3 but decreased expression of TET2 and TET1 (Figure 1A). Next, we performed immunohistochemistry (IHC) analysis on human liver tissue sections (fibrosis with NASH versus normal control) revealing strong colocalizations of TGF-β1 and TET3 proteins with α-SMA-positive HSCs (Figure 1B), suggesting increased TET3 expression in activated HSCs. Next, we repetitively exposed mice to CCl4 to induce liver fibrosis (Liedtke et al., 2013). H&E and Masson’s trichrome staining assays confirmed fibrotic changes in the CCl4-treated livers (Figure 1C). qRT-PCR (Figure 1D) and immunoblotting (IB) (Figure 1E) analyses of liver tissues revealed increased expression of fibrotic markers TGF-β1 (TGFB1), α-SMA (ACTA2), and COL1A1 (COL1A1) in CCl4-treated compared with vehicle-treated livers. The expression of TET3 and TET1 (but not TET2) also increased in fibrotic livers (Figures 1D and 1E). An increase in TET3 expression was also observed in a bile duct ligation (BDL)-induced cholestatic liver fibrosis mouse model (Figure 1F; Cai et al., 2017). The consistent increase in TET3 expression in both human and mouse fibrotic livers prompted us to focus on investigating TET3 further in hepatic fibrosis.
Figure 1. TET3 Upregulation in Fibrotic Livers.
(A) qPCR of TGFB1 and TETs from non-fibrotic and fibrotic human liver tissues. Each data point represents an individual patient, with five to seven patients per group.
(B) IHC of liver tissue sections from patients with non-fibrotic and fibrotic disease co-stained for α-SMA (pink) and TGF-β1 (brown) or α-SMA (pink) and TET3 (brown), with nuclei counterstained in blue. Red arrowheads mark activated HSCs. Three sections per patient sample were counted, with four patients per group. Fields were picked randomly, with counting done in a blinded manner. Magnification, 20×; scale bars, 100 μm. Numbers of activated HSCs (α-SMA+) co-expressing TGF-β1 (TGF-β1+/α-SMA+) or TET3 (TET3+/α-SMA+) per field are presented.
(C) Masson’s trichrome and H&E staining on liver sections from mice treated with vehicle or CCl4. Magnifications of 4× and 20× are presented. Scale bars, 100 μm.
(D) qPCR of indicated genes from liver tissues isolated from mice treated with vehicle or CCl4. Each data point represents an individual mouse, with three mice per group.
(E) Immunoblotting (IB) of indicated proteins from liver tissues isolated from mice treated with vehicle or CCl4. Each data point represents an individual mouse, with three mice per group.
(F) IB of TET3 from liver tissues isolated from mice sham operated or with BDL. Each data point represents an individual mouse, with six mice per group. The asterisk indicates nonspecific protein bands.
All data are representative of at least two independent experiments. Error bars are mean with SEM. *p < 0.05 and **p < 0.01; significance by Student’s t test.
TET3 Promotes Expression of TGFB1 and Other TGF-β Pathway Genes in HSCs
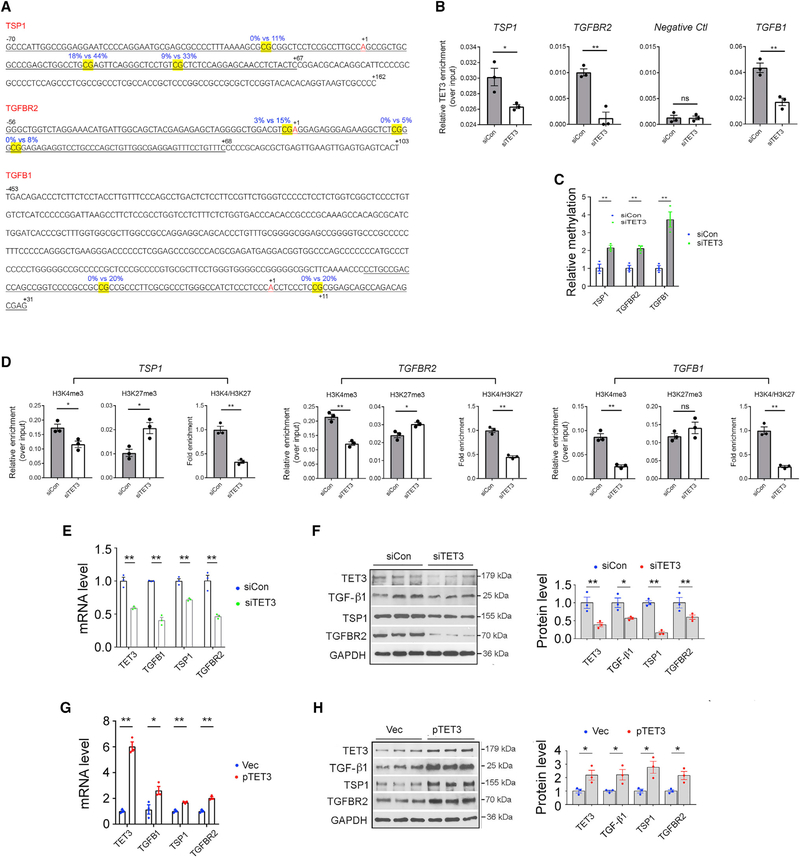
TET3 knockdown using a TET3-specific small interfering RNA (siRNA) (siTET3) induced epigenetic changes in human uterine leiomyoma cells by increasing methylation at three specific CpG sites in the critical transcriptional regulatory regions (CTRRs) of TSP1 and TGFBR2 (Figure 2A, yellow highlighted CpGs) (Cao et al., 2019). Altered methylation even at a single CpG site in transcriptional regulatory regions can profoundly affect transcription factor binding and gene expression (Ahrens et al., 2013; Kammel et al., 2016; Kim et al., 2003). Using chromatin immunoprecipitation coupled with qPCR (ChIP-qPCR) and quantitative methylation-specific PCR (QMSP), we showed that TET3 interacts with the CTRRs of TSP1 and TGFBR2, inducing demethylation at the specific CpGs, thereby facilitating chromatin remodeling and gene expression (Cao et al., 2019). To explore the possibility of a role of TET3 in hepatic fibrosis, we began to investigate whether TET3 regulates expression of TSP1 and TGFBR2 in HSCs, given that both liver fibrosis and uterine leiomyomas are fibrotic disorders and that HSCs and leiomyoma cells share the characteristics of ECM production induced by TGF-β signaling.
Figure 2. TET3-Dependent Epigenetic Regulation of TGF-β Pathway Genes.
(A) Sequences of CTRRs of TSP1, TGFBR2, and TGFB1. The differentially methylated CpGs are highlighted in yellow. Blue numbers mark percentage of methylation in siCon (left) or siTET3 (right) transfected leiomyoma cells as determined by genome-wide single-nucleotide-resolution methylation analysis. Black numbers indicate positions relative to the transcriptional start sites (+1). ChIP-qPCR amplified regions are underlined.
(B) LX-2 cells were transfected with siCon or siTET3, followed by ChIP-qPCR analysis at 48 h post-transfection. Data are presented as mean relative TET3 enrichment over input after normalization against preimmune IgG background signals.
(C) LX-2 cells were transfected with siCon or siTET3, followed by QMSP analysis 48 h later. Relative CTRR methylations of the indicated genes are shown.
(D) LX-2 cells were transfected with siCon or siTET3, followed by ChIP-qPCR analysis at 48 h post-transfection.
(E and F) qPCR (E) and IB (F) of indicated genes from LX-2 cells transfected with siCon or siTET3. RNA and protein were isolated at 48 h post-transfection.
(G and H) qPCR (G) and IB (H) of indicated genes from LX-2 cells transfected with empty vector (Vec) or a plasmid DNA expressing human TET3 (pTET3). RNA and protein were isolated at 48 h post-transfection.
All data are representative of three independent experiments. Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05 and **p < 0.01; significance by Student’s t test.
First, we tested whether the epigenetic regulation of TSP1 and TGFBR2 by TET3 could be recapitulated in human HSCs. To examine physical interactions between TET3 and the CTRRs of TGFB1 and TGFBR2, we used a TET3-specific antibody (Cao et al., 2019) to immunoprecipitate protein-DNA complexes from LX-2 cells (a human HSC line) transfected with siTET3 or siCon (a non-targeting control siRNA). qPCR was performed to amplify sequences of the CTRRs (Figure 2A, underlined) encompassing CpGs with altered methylation. Not surprisingly, binding of TET3 to the CTRRs was significantly reduced in TET3 knockdown versus control HSCs (Figure 2B). No binding of TET3 to a negative control region was detected irrespective of TET3 knockdown (Figure 2B). This region was shown not to be bound by TET3 in human cells by chromatin immunoprecipitation sequencing (ChIP-seq) (Deplus et al., 2013). Next, we tested effects of TET3 knockdown on methylation using QMSP. The QMSP primers were designed to specifically detect the CpGs with altered methylation (Figure 2A, yellow highlighted) (Cao et al., 2019). Compared with cells treated with siCon, cells treated with siTET3 had increased methylation at the CpGs (Figure 2C). To test whether TET3 knockdown alters chromatin structure, LX-2 cells were transfected with siCon or siTET3, followed by ChIP-qPCR, immunoprecipitating with antibodies specific for the H3K4me3 (active) or H3K27me3 (inactive) marks and amplifying parts of the CTRRs (Figure 2A, underlined). ChIP analysis showed that TET3 knockdown significantly decreased H3K4me3 (Figure 2D, left columns) and increased H3K27me3 (middle columns) association with the CTRRs of TSP1 and TGFBR2, such that the ratios of H3K4me3 to H3K27me3 decreased significantly (right columns). These results suggest that TET3 knockdown promotes a heterochromatin conformation, diminishing chromatin accessibility at the CTRRs in HSCs. Our results support the view that in HSCs, TET3 binds to the CTRRs of TSP1 and TGFBR2, inducing demethylation at specific CpGs, which in turn facilitates chromatin remodeling and transcription.
In light of the findings above demonstrating that the same epigenetic mechanism of TET3-mediated regulation of TSP1 and TGFBR2 occurs in both HSCs and leiomyoma cells, we conducted a careful inspection of our genome-wide methylation data (GEO: GSE117190; Cao et al., 2019) which revealed two CpG sites in the CTRR (−453 to +11) of TGFB1 (Kim et al., 1989) with increased methylation (0%−20%) in TET3-knockdown leiomyoma cells (Figure 2A). This CTRR enabled expression of chloramphenicol acetyltransferase (CAT) in multiple cell types and from different species; and importantly, the relative CAT activity paralleled the level of TGF-β1 mRNA in these cells (Kim et al., 1989). To determine whether TET3 regulates expression of TGFB1 in HSCs, ChIP-qPCR and QMSP experiments were carried out. TET3 knockdown in LX-2 cells led to decreased TET3 binding to the CTRR of TGFB1 (Figure 2B), increased methylation at the CpGs (Figure 2C, yellow highlighted), and reduced association of H3K4me3 with the CTRR of TGFB1 (Figure 2D). Furthermore, TET3 knockdown in LX-2 cells decreased expression of TGFB1, in addition to TSP1 and TGFBR2 at levels of both mRNA (Figure 2E) and protein (Figure 2F) and TET3 overexpression (from a human TET3 expression vector pTET3; Ko et al., 2013) increased their expression (Figures 2G and 2H). Collectively, our results show that in HSCs TET3 positively regulates expression of TGFB1, TSP1, and TGFBR2, likely in part by promoting demethylation at specific CpGs in the CTRRs.
TET3 and TGF-β1 Form a Positive Feedback Loop Promoting Profibrotic Gene Expression in HSCs
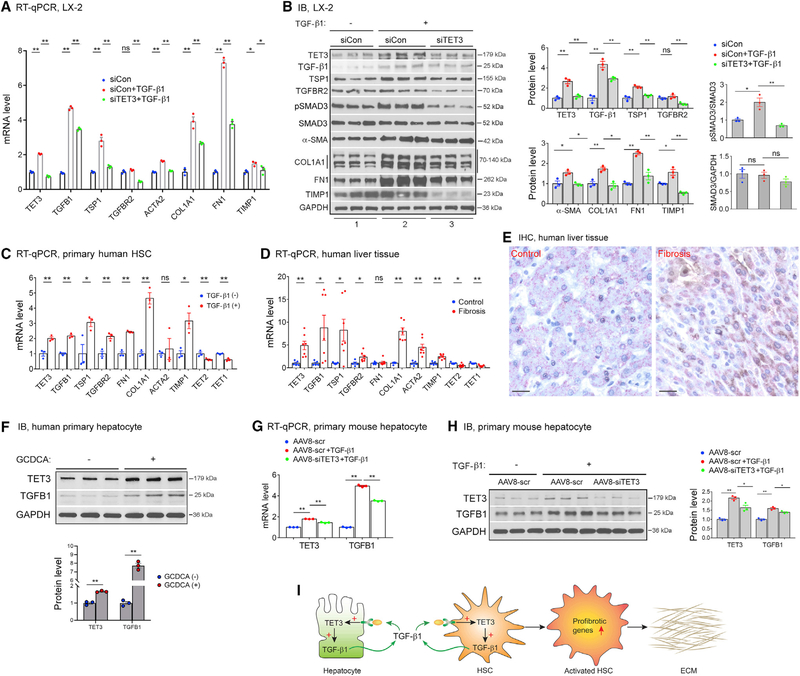
As TET3 enhances expression of multiple TGF-β pathway genes at multiple levels (i.e., the ligand TGF-β1, the receptor TGFBR2, and the activator TSP1) (Figure 2), we asked whether TET3 promotes TGF-β signaling and whether TET3 is required for increased TGF-β1-stimulated expression of profibrotic genes in HSCs. Thus, LX-2 cells were stimulated with TGF-β1 in the presence of TET3 siRNA knockdown. Intriguingly, TGF-β1 stimulation upregulated TET3 at levels of both mRNA (Figure 3A) and protein (Figure 3B). The expression of TGFB1, TSP1, and TGFBR2 also increased, with a concomitant increase in SMAD3 phosphorylation (marker for TGF-β signaling activation) and expression of α-SMA, COL1A1, FN1, and TIMP1 (markers for fibrotic activation) (Figures 3A and 3B). However, when cells were stimulated with TGF-β1 while TET3 was downregulated, the effects were significantly attenuated (Figures 3A and 3B). Taken together with the notion that TET3 positively regulates TGFB1 expression (Figure 2), we propose a positive feedback loop between TET3 and TGF-β1 that promotes TGF-β signaling and subsequent profibrotic gene expression in HSCs.
Figure 3. The TET3/TGF-β1 Positive Feedback Loop in HSCs and Hepatocytes.
(A and B) LX-2 cells were transfected with siCon or siTET3. 24 h later, cells were treated with TGF-β1 (5 ng/mL) (+) or vehicle (−), followed by RNA and protein extraction 24 h later. For pSmad3 and Smad3 analysis, protein was extracted 6 h after TGF-β1 stimulation. Expression of indicated genes was analyzed using qPCR (A) and IB (B). Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05 and **p < 0.01; significance by one-way ANOVA with Tukey post-test.
(C) qPCR of indicated genes from primary human HSCs stimulated with TGF-β1 (2 ng/mL) (+) or vehicle (−) for 24 h. Each data point represents an individual patient, with three patients per group. Error bars are mean with SEM. *p < 0.05 and **p < 0.01; significance by Student’s t test.
(D) qPCR of indicated genes from non-fibrotic and fibrotic human liver tissues. Each data point represents an individual patient, with five to seven patients per group. Error bars are mean with SEM. *p < 0.05 and **p < 0.01; significance by Student’s t test.
(E) Representative IHC of liver tissue sections from patients (same patient groups as in Figure 1B) with non-fibrotic and fibrotic disease co-stained for HepPar1 (pink) and TET3 (brown), with nuclei counterstained in blue. Magnification, 400×; bars, 100 μm. Three sections per patient sample were analyzed, with four patients per group.
(F) IB of TET3 and TGF-β1 from human primary hepatocytes treated with vehicle (−) or GCDCA (+) at a final concentration of 100 μM for 48 h. Error bars are mean with SEM of technical replicates (n = 3). **p < 0.01; significance by Student’s t test.
(G and H) Primary mouse hepatocytes were infected with AAV8-scr or AAV8-siTET3 for 24 h, followed by treatment with TGF-β1 (5 ng/mL) (+) orvehicle (−). RNA
(G) and protein (H) were extracted 48 h later. Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05 and **p < 0.01; significance by one-way ANOVA with Tukey post-test. All data are representative of at least two independent experiments.
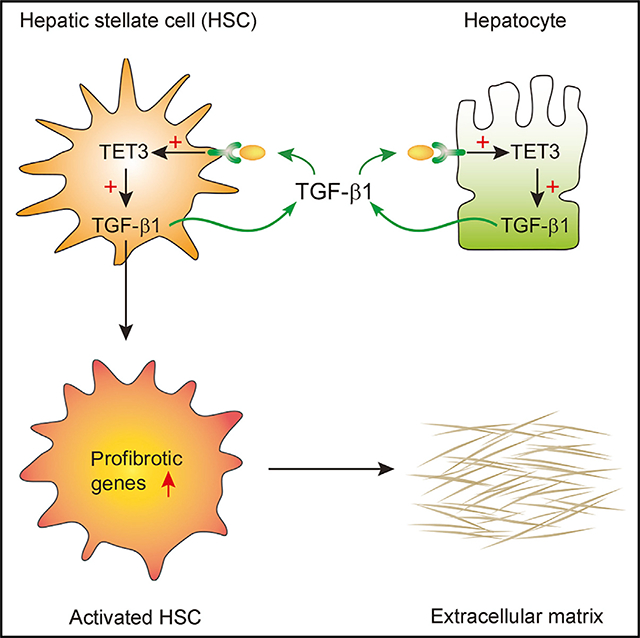
(I) A positive feedback model in liver fibrosis. Stressed hepatocytes upregulate TET3 leading to increased production of TGF-β1 from hepatocytes. TGF-β1 acts on both hepatocytes and HSCs to stimulate more TGF-β1 production via the TET3/TGF-β1 positive feedback mechanism. In HSCs, TET3 also promotes profibrotic gene expression and subsequent ECM production via increasing expression of multiple TGF-β pathway genes.
Next, we tested the regulatory pathway identified above in primary HSCs and human liver tissue samples. In line with findings from LX-2 cells, primary human HSCs treated with TGF-β1 showed increased expression of TET3, TGFB1, TSP1, TGFBR2, FN1, and COL1A1 (Figure 3C). Curiously, TGF-β1 stimulation decreased expression of TET2 and TET1 (Figure 3C). Remarkably, results derived from human liver tissue samples essentially mirrored those seen in primary human HSCs (Figure 3D). These results further support our conclusion that the TET3/TGF-β1 positive feedback loop promotes profibrotic gene expression in HSCs.
The TET3/TGF-β1 Loop Promotes TGF-β1 Production from Hepatocytes
Other hepatic cells, including hepatocytes, biliary epithelial cells, and immune cells, modulate HSC activation via production of cytokines and other signaling molecules (Tsuchida and Friedman, 2017). Damage to hepatocytes increases production of TGF-β1 from hepatocytes (Benzoubir et al., 2013; Jee et al., 2015; Jeong et al., 2004). To determine whether TET3 expression increases in hepatocytes under pathological conditions, human liver tissue sections (fibrosis with NASH versus normal control) were analyzed using IHC to colocalize TET3 with hepatocyte-specific antigen HepPar1. TET3 protein was detected in hepatocytes of fibrotic but not control livers; the TET3 staining was heterogeneous, with some hepatocytes showing predominantly nuclear and others showing predominantly cytoplasmic (Figure 3E). Heterogeneous subcellular localization of TET proteins has been previously documented (Huang et al., 2016; Mi et al., 2015; Zhang et al., 2014). Next, we treated isolated human primary hepatocytes with glycochenodeoxycholic acid (GCDCA) to induce hepatocyte stress (Cai et al., 2017). As seen in Figure 3F, TET3 protein increased in response to GCDCA treatment, with an expected increase in TGF-β1 as well, suggesting that stressed hepatocytes upregulate expression of both TET3 and TGF-β1. To determine whether the TET3/TGF-β1 loop plays a role in TGF-β1 production from hepatocytes, mouse primary hepatocytes were stimulated with TGF-β1 in combination with TET3 knockdown using adeno-associated virus (serotype 8) (AAV8)-based vectors. AAV8-mediated delivery has been shown to elicit high levels of liver-specific transgene expression in murine models (Lisowski et al., 2014; Zhang et al., 2018). The AAV8-siTET3 virus expresses siRNAs targeting mouse TET3. The AAV8-scr virus expresses non-targeting scrambled siRNAs. TGF-β1 stimulation increased expression of TET3 and TGF-β1 both at mRNA (Figure 3G) and protein (Figure 3H) levels, but the effects were attenuated when TET3 was downregulated (Figures 3G and 3H), supporting the notion that the TET3/TGF-β1 loop functions to promote TGF-β1 production from hepatocytes. Taken together, our results show that hepatic injury upregulates TET3 in both HSCs (Figure 1B) and hepatocytes (Figures 3E and 3F) and that the TET3/TGF-β1 loop acts to enhance TGF-β1 production from these cells. Therefore, we propose a model illustrated in Figure 3I. This model, however, does not exclude the possibility that TET3 regulates other genes and pathways in HSCs and hepatocytes which warrants future investigation.
Inhibition of TET3 Attenuates Hepatic Fibrosis
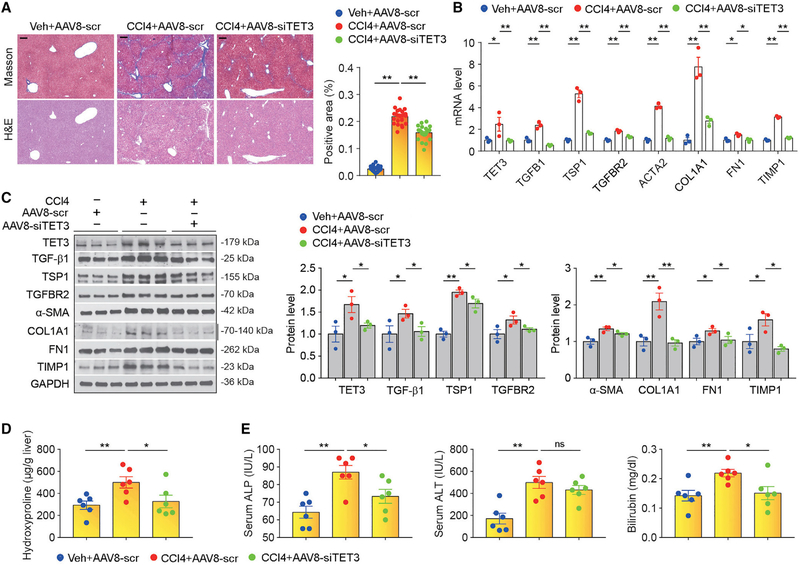
As proof of principle, we tested the model (Figure 3I) using the CCl4 mouse model of liver fibrosis. Mice were administrated with vehicle (mineral oil) plus AAV8-scr (group 1), CCl4 plus AAV8-scr (group 2), or CCl4 plus AAV8-siTET3 (group 3). Vehicle or CCl4 was injected intraperitoneally (i.p.) twice a week; AAV8-scr or AAV8-siTET3 was injected via tail vein once a week. Five weeks following the initial injection, mice were sacrificed, and blood and tissue samples were collected. As shown in Figure 4A, group 1 mice did not develop liver fibrosis, group 2 mice developed liver fibrosis, and group 3 mice developed liver fibrosis but significantly less than group 2 mice. Gene expression analysis revealed significantly increased expression of key pathway genes TET3, TGFB1, TSP1, TGFBR2, ACTA2, COL1A1, FN1, and TIMP1 in liver tissues from group 2 mice compared with group 1 mice at levels of both mRNA and protein (Figures 4B and 4C). The increase in expression of key pathway genes was significantly attenuated in liver tissues from group 3 mice compared with group 2 mice (Figures 4B and 4C). We also examined liver tissue hydroxyproline, which is a unique amino acid in collagen molecules and an important biomarker of liver fibrosis (Lee et al., 2005). The hydroxyproline content was significantly increased in group 2 mice compared with group 1 mice, but the increase was abolished in group 3 mice (Figure 4D). Blood chemistry results showed elevated alkaline phosphatase (ALP), alanine transaminase (ALT), and bilirubin in group 2 mice compared with group 1 mice, suggesting impaired liver function (Figure 4E). These markers, except for serum ALT, were reduced in group 3 mice (Figure 4E). Since AAV8 targets mainly hepatocytes (Davidoff et al., 2005; Lisowski et al., 2014; Rezvani et al., 2016), our results provide strong in vivo evidence supporting cross-talk between hepatocytes and HSCs via the TET3/ TGF-β loop in regulation of liver fibrosis (Figure 3I). Future studies aimed at developing strategies that allow simultaneous targeting of both hepatocytes and HSCs may prove more potent in therapy.
Figure 4. TET3 Inhibition Reduces Liver Fibrosis.
(A) Masson and H&E staining on liver sections from mice treated with vehicle plus AAV8-scr, CCl4 plus AAV8-scr, or CCl4 plus AAV8-siTET3. Magnification, 4×; scale bars, 500 μm. Quantification of fibrosis from liver tissue sections was performed using ImageJ. Four sections per mouse (six mice per group) were counted. Fields were picked randomly with counting done in a blinded manner. Error bars are mean with SEM. **p < 0.01; significance by one-way ANOVA with Tukey post-test.
(B and C) qPCR (B) and IB (C) of indicated genes from liver tissues isolated from mice treated as in (A). RNA and protein extracted from liver tissues pooled from six animals per group were analyzed. Data are representative of three independent experiments. Error bars are mean with SEM of technical replicates (n = 3). *p < 0.05 and **p < 0.01; significance by one-way ANOVA with Tukey post-test.
(D) Hydroxyproline contents from liver tissues isolated from mice treated as in (A). Each data point represents an individual mouse, with six mice in each group.
(E) Serum ALP, ALT and bilirubin from mice treated as in (A). Each data point represents an individual mouse, with six mice in each group. *p < 0.05 and **p < 0.01; significance by one-way ANOVA with Tukey post-test.
We believe that the molecular and functional interaction between TET3 and the TGF-β signaling pathway characterized in this report represents a critical mechanism in liver fibrosis. As aberrant TGF-β activation is a universal mechanism of fibrosis, our discovery of the TET3/TGF-β mechanism may have broad and far-reaching implications for fibrotic disease control.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yingqun Huang (yingqun.huang@yale.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patient liver tissue samples
Fibrotic and non-fibrotic liver tissues were obtained from 12 patients who underwent liver cancer resection in Shengjing Hospital of China Medical University between May 2018 and September 2018. Ethics approval for this study was obtained from the Institutional Review Board at China Medical University, and written informed consent was obtained from each patient. All patients had a history of hepatitis B virus infection. Among the 12 patients, 7 (4 males and 3 females, age 41–53 years old) were diagnosed with liver cancer with concurrent liver fibrosis (fibrosis group) and the other 5 (3 males and 2 females, age 35–47 years old) were diagnosed with liver cancer without liver fibrosis (control group). Fibrotic liver tissues were collected from cancer-free background livers of the fibrosis group; non-fibrotic liver tissues were collected from cancer-free background livers of the control group. Tissues were snap frozen in liquid nitrogen and stored at −80°C until later use for RT-qPCR analysis.
Archived deidentified paraffin embedded human liver tissue blocks (fibrosis with non-alcoholic steatohepatitis and normal control) were obtained from the Yale Pathology Tissue Services.
Mouse
All animal work was conducted in accordance with the guidelines of the Yale University Institutional Animal Care and Use Committee. All mice used in the study were male. C57BL/6 mice were purchased from Charles River (Cat# 027) and housed at 22°C-24°C with a 12 h light/12 h dark cycle with regular chow (Harlan Teklad no. 2018, 18% calories from fat). Livers from cholestatic BDL mice (The Jackson Laboratories, Cat# 000664) were used for this study, which have been described in our previous publication (Cai et al., 2017).
Mouse and human primary hepatocytes
Mouse primary hepatocytes were prepared as previously described (Zhang et al., 2018). Cells were maintained in complete culture medium (Williams’ Medium [GIBCO,12551] supplemented with 5% FBS, 10 mM HEPES buffer [GIBCO, 15630–080], 2 mM L-Glutamine [GIBCO, 25030–081], 1% SPA [GIBCO, 15240–062], 4 mg/L insulin [GIBCO, 12585–014] and 1 μM dexamethasone [Sigma, D4902]). For TET3 knockdown experiments, cells seeded in 12-well plates were infected with AAV8-scr or AAV8-siTET3 at 6,000 gc/cell at 3 h after seeding. Medium was changed the next day, and cells were stimulated with TGF-β1 at a final concentration of 5 ng/ml in insulin- and FBS-free culture medium, followed by RNA and protein extraction 48 h later.
Human primary hepatocytes were obtained through the Liver Tissue Cell Distribution System, which was funded by NIH contract N01-DK-7–0004/HHSN267200700004C. Cells were maintained in complete culture medium (HMM medium [LONZA, CC-3197] supplemented with 1% SPA [GIBCO, 15240–062], 0.1 μM insulin [GIBCO, 12585–014] and 0.1 μM dexamethasone [Sigma, D4902]). To induce hepatocyte stress, the hepatocytes were incubated with glycochenodeoxycholic acid (GCDCA) (Sigma-Aldrich, G0759) at a final concentration of 100 μM for 48 h, followed by protein extraction.
METHOD DETAILS
RNA extraction and RT-qPCR assays
Total RNA was extracted from cultured cells or liver tissue samples using PureLink RNA Mini Kit (Ambion, 12183018A). 0.8 μg of total RNA was reverse transcribed to cDNA in a reaction volume of 10 μL using PrimeScript RT Reagent Kit (TAKARA, RR037A). Quantitative real-time PCR reactions were carried out using iQSYBRGreen (Bio-Rad) in a Bio-Rad iCycler. Gene expression levels were normalized against housekeeping genes HPRT1 and RPLP0. The specific PCR primers for human and mouse were summarized in Table S1.
Immunoblotting analysis
For liver tissue samples, 5–10 mg of tissues were homogenized by sonication (Qsonica, Model Q125) in 150 μL of 2xSDS-sample buffer. Samples were immediately heated at 100°C for 3 min with occasional vertexing. Then, 100 μL of 2xSDS-sample buffer was added to each sample and the 250 μL of tissue lysate were further homogenized by sonication and heated again at 100°C for 3–5 min with vertexing once every15 s. Samples (further diluted at 1:1 with 2xSDS-sample buffer before loading) were loaded 5–10 μL per well onto a 4%−15% gradient SDS gel, followed by western blot analysis. ImageJ was used to quantify the protein bands.
Histopathology Analysis
Mouse liver samples were collected and fixed at 4°C with 4% of phosphate buffered paraformaldehyde (Baker, 2106–01) for 24 h. Fixed tissue specimens were embedded in paraffin and cut into 4 mm-thick tissue sections for histopathological analysis and immunohistochemistry. Archived deidentified paraffin embedded human liver tissue sections (fibrosis with non-alcoholic steatohepatitis and normal control) were obtained from the Yale Pathology Tissue Services.
IHC
Paraffin sections were used to perform IHC using the EnVision G2 Doublestain System (K5361, DAKO, DAB+/Permanent Red). Briefly, paraffin slides were dewaxed in p-xylene (Sigma, 185566–1L) overnight, followed by sequential washes in 100%, 90%, 80%, and 70% ethanol and two times of ddH2O for 5 min each. At the end of the final wash, slides were immediately placed in boiling sodium citrate buffer (10 mM, pH 6.0) for 20 min for antigen retrieval. After gentle wash for 5 min, 3 times with 1xPBS, 200 μL of Dual Endogenous Enzyme Block was added to cover the specimen and incubation at RT was carried out for 5 min, followed by 2 times of PBS wash, 5 min each. 200 μL of primary antibody (TGF-β1 at 1:50 dilution, ab92486; TET3 at 1:50 dilution, GENE Tex, 121453; HepPar1 at 1:100 dilution, Millipore Sigma, 264M-94) or negative control IgG was added and incubation was carried out at RT for 1 h. The slides were washed for 5 min, 3 times with PBS. 200 μL of polymer/HRP was added for 1 h, followed by addition of DAB working solution (1:50 DAB chromogen/DAB buffer) and incubation for 5–15 min. Subsequently, the slides were sequentially incubated with double block for 3 min, primary antibody against α-SMA (diluted at 1:400, ab 293182), Mouse/Rabbit (Link), and polyer/AP for 30 min, and permanent red working solution for 5–20 min. Next, slides were incubated with hematoxylin for 1 min and then 0.037 mmol/L NH4OH for 2–5 min, followed by wash twice with ddH2O. After that, the slides were washed with 70%, 80%, 90% ethanol, and 100% ethanol and p-xylene for 5 min each. After air dry, the slides were mounted with aqueous mounting media (Immu-Mount from Thermo). IHC images were captured using confocal microscopy (Leica SP5).
Cell culture, transfection, and TGF-β1 treatment
LX-2 human hepatic stellate cells (SCC064, Sigma-Aldrich) were maintained in DMEM (GIBCO, 11965–092) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO, 26140–079) and 1% PS (penicillin and streptomycin) at 37°C in 5% humidified CO2 tissue culture incubator. Generally, cells were plated and transfected in a 24-well plate scale. For siRNA transfection, siTET3 or siCon (Cao et al., 2019) was mixed with 25 μL of OPTI-MEM (GIBCO, 31985–070) by gentle pipetting. In parallel, 1 μL of Lipofectamine 3000 (ThermoFisher Scientific, L3000015) was mixed with 25 μL of OPTI-MEM. Then, the contents from the two tubes were mixed by gentle pipetting. After incubation for 5 min at room temperature (RT), the resulting 50 μL solution was used to resuspend cell pellet (5 × 104 cells). After incubation at RT for 10 min, 450 μL of growth medium was added and the cell suspension was transferred into one well of a 24-well plate. For transfection with TET3-expression plasmid pTET3 (pcDNA-Flag-Tet3, Addgene plasmid #60940, Wang and Zhang, 2014) or control vector, 5 × 104 cells per well were seeded in a 24-well plate the night before transfection. The next day, 0.75 μg of pTET3 (or Vec) were mixed with 1 μL of P3000 in 25 μL of OPTI-MEM by gentle pipetting. In parallel, 0.5 μL of Lipofectamine 3000 were mixed with 25 μL of OPTI-MEM. The contents from the two tubes were combined by gentle pipetting and incubated for 5 min at RT. The resulting 50 μL of transfection solution was added to each well of cells seeded in a 24-well plate the night before and from which the growth medium had been removed. After 6 h of incubation in a tissue culture incubator, 500 μL of growth medium were added to each well of cells. For TGF-β1 (Sigma, T7039) treatment experiments, transfected cells were incubated in 500 μL of serum-free DMEM containing 5 ng/ml TGF-β1 (diluted with 4 mM HCl) or HCl (as control) for 24 h or 48 h, as indicated.
Genomic DNA extraction
Genomic DNA (gDNA) was isolated from LX-2 cells grown in 24-well plates using Quick-gDNA MicroPrep (Zymo, D3021) according to the manufacturer’s instructions.
QMSP
Genomic DNA was extracted from LX-2 cells in 24-well plates using Quick-gDNA MicroPrep (Zymo Research Corporation, Irvine, CA; D3021). For bisulfite treatment using EZ DNA Methylation-Gold Kit (Zymo, D5006), 90–500 ng of gDNA was used per column and 20–40 μL of elution buffer was used to elute DNA from each column. qPCR was performed in a 15 μL reaction containing 5 μL of eluted DNA using iQSYBRGreen (Bio-Rad, Hercules, CA; 1708880) in a Bio-Rad iCycler. PCR was conducted by initial denaturation at 95°C for 5 min, followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Two sets of PCR primers were designed: one for unmethylated and one for methylated DNA sequences. The PCR primers for methylated DNA were used at a final concentration of 0.6 μM in each PCR reaction. The relative levels of methylated versus unmethylated DNA sequences are presented. The primers used for QMSP are summarized in Table S1.
ChIP-qPCR
Experiments were performed in a 10-cm plate scale using Chromatin Prep Module Kit (Thermo, catalog number 26156) according to the manufacturer’s instructions. Agarose beads were used to pre-bind overnight with antibodies against TET3 (Millipore Sigma, ABE290), H3K4me3 (Cell signaling, C42D8) and H3K27me3 (Cell signaling, C36B11). Preimmune IgG was used as a negative control. LX-2 cells were cross-linked with 1% formaldehyde at RT for 10 min and the reaction was stopped by 1x glycine. ChIPs were carried out overnight at 4°C. Primers (Table S1) for the specific promoter regions of TGFB1, TSP1, and TGFBR2 were used to amplify input and ChIP-purified DNA. The relative enrichments of the indicated DNA regions were calculated using the Percent Input Method according to the manufacturer’s instructions and were normalized to % input. Data are presented after normalization against background IgG signals.
Mouse liver fibrosis induction and treatment
To induce liver fibrosis, mice at 8 weeks of age were intraperitoneally (i.p.) injected with vehicle (mineral oil, Sigma, M5310) or 10% CCl4 (Sigma, 319961) dissolved in mineral oil at 0.6 ml/kg twice a week for 4 weeks. For siRNA treatment studies, 8 wk-old mice were randomly divided into three groups: vehicle plus AAV8-scr (group 1), CCl4 plus AAV8-scr (group 2), and CCl4 plus AAV8-siTET3 (group 3). AAV8-siTET3 (Applied Biological Materials Inc., iAAV04929608) allows the expression of siRNAs specific for mouse TET3 from adeno-associated virus (serotype 8)-based vectors. AAV8-scr (Applied Biological Materials Inc., iAAV01508) expresses non-targeting scrambled siRNAs as a negative control. Both viruses were resuspended in PBS/0.05% sorbital buffer. Vehicle or CCl4 was injected i.p. twice a week; AAV8-scr or AAV8-siTET3 at 2 × 1010 gc/mouse was injected via tail vein once a week. Five weeks following the initial injection, mice were sacrificed for blood and tissue collection.
Hydroxyproline and blood chemistry
Hydroxyproline content of liver tissue (10 mg) was assessed using Hydroxyproline Assay Kit (Sigma-Aldrich, MAK008–1KT) according to the manufacture’s protocol. Serum bilirubin concentration was determined in a 96-well plate scale using Bilirubin Assay kit (Sigma-Aldrich, MAK126) according to the manufacture’s protocol. Briefly, 50 μL of Calibrator and 50 μL of ddH2O were transferred into two separate wells in a 96-well plate, followed by addition of 200 μL of ddH2O into each well for a final volume of 250 μl. Then, 50 μL of serum sample was added to each well, followed by addition of freshly prepared 200 μL of Working Reagent and incubation at RT for 10 min. Absorbance was measured at 530 nm (A530) in a multi-model plate reader (Molecular Device, Filter Max F5). Serum ALP and ALT were measured by the Department of Laboratory Medicine, Yale-New Haven Hospital.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Methods
The number of independent experiments and the statistical analysis for each figure are indicated in the legends. All statistical analyses were performed using GraphPad Prism version 8 for Windows (GraphPad Software, La Jolla California USA, https://www.graphpad.com) and are presented as mean ± SEM. Two-tailed Student’s t tests (or as otherwise indicated) were used to compare means between groups. p < 0.05 was considered significant.
DATA AND CODE AVAILABILITY
This study did not generate datasets or code.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-TET3 (Immunoblotting) | GeneTex | Cat# GTX121453 |
| Anti-TET1 (Immunoblotting) | GeneTex | Cat# GTX124207 |
| Anti-TET3 (ChIP) | Millipore Sigma | Cat# ABE290 |
| Anti-TGF-β1 | Abcam | Cat# ab92486 |
| Anti-TGFBR2 | Abcam | Cat# ab186838 |
| Anti-TSP1 | Abcam | Cat# ab85762 |
| Anti-SMAD3 | Cell Signaling | Cat# C67H9 |
| Anti-phospho-SMAD3 | Cell Signaling | Cat# C25A9 |
| Anti-COL1A1 | Santa Cruz | Cat# sc-293182 |
| Anti-FN1 | Abcam | Cat# ab194395 |
| Anti-α-SMA | Abcam | Cat# ab5694 |
| Anti-TIMP1 | Cell Signaling | Cat# 8946s |
| Anti-GAPDH | Abcam | Cat# ab128915 |
| Anti-HepPar1 | Millipore Sigma | Cat# 264M-94 |
| Anti-H3K4m3 (ChIP) | Cell Signaling | Cat# C42D8 |
| Anti-H3K27m3 (ChIP) | Cell Signaling | Cat# C36B11 |
| Anti-rabbit IgG, HRP-linked | Cell Signaling | Cat# 7074 |
| Anti-mouse IgG, HRP-linked | Cell Signaling | Cat# 7076 |
| Biological Samples | ||
| Human liver tissue blocks | Yale Pathology Tissue Services | https://medicine.yale.edu/pathology/ypts/ |
| Human primary hepatocytes and HSCs | Liver Tissue Cell Distribution System | N01-DK-7-0004/HHSN267200700004C |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human TGF-β1 | Sigma-Aldrich | Cat# T7039 |
| Glycochenodeoxycholic acid (GCDCA) | Sigma-Aldrich | Cat# G0759 |
| Dexamethasone | Sigma-Aldrich | Cat# D4902 |
| Insulin | GIBCO | Cat# 12585-014 |
| Williams Medium | GIBCO | Cat# 12551 |
| Carbon tetrachloride (CCl4) | Sigma-Aldrich | Cat# 319961 |
| Mineral oil | Sigma-Aldrich | Cat# M5310 |
| Critical Commercial Assays | ||
| EnVision G2 Doublestain System | DAKO | Cat# K5361 |
| Hydroxyproline Assay Kit | Sigma-Aldrich | Cat# MAK008-1KT |
| Bilirubin Assay Kit | Sigma-Aldrich | Cat# MAK126 |
| Pierce Agarose ChIP Kit | Thermo Scientific | Cat# 26156 |
| PrimeScript RT Reagent Kit | TaKaRa | RR037A |
| SYBR Green PCR Master Mix | Bio-Rad | 172-5124 |
| PureLink RNA Mini Kit | Ambion | 12183018A |
| Quick-gDNA MicroPrep | Zymo Research | D3021 |
| EZ DNA Methylation-Gold Kit | Zymo Research | D5006 |
| Experimental Models: Cell Lines | ||
| Human: LX-2 cells | Sigma-Aldrich | Cat# SCC064 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57B/6 (CCl4 model) | Charles River | Cat# 027 |
| Mouse: C57B/6J (BDL model) | The Jackson Laboratory | Cat# 000664 |
| Oligonucleotides | ||
| Primers for qPCR, ChIP, and QMSP see Table S1 | This paper | N/A |
| AAV8-siTET3: a cocktail of siRNAs targeting specifically to four different regions of mouse TET3: | Applied Biological Materials | Cat# iAAV04929608 |
| 5′-CGGTACCATCTCCTATTTCTCAGAGGGAG-3′ | ||
| 5′-GCCCACAAGGACCAACATAACCTCTACAA-3′ | ||
| 5′-CCTCTCCCTTTGCTCAGAGTTCCAGTTGC-3′ | ||
| 5′-GCCCTTGAGCTCCAACGAGAAGCTATTTG-3′ | ||
| AAV8-scr: a scrambled siRNAtargeting the following sequence: 5′-GGGTGAACTCACGTCAGAA-3′ | Applied Biological Materials | Cat# iAAV01500 |
| siTET3 (siRNA specific for human TET3) | Ambion (Cao et al., 2019) | Cat# 4392420/s47239 |
| siCon (nontargeting control siRNA) | Ambion (Cao et al., 2019) | Cat# AM4611 |
| Recombinant DNA | ||
| Plasmid: pcDNA-FLAG-Tet3 | Addgene | Cat# 60940 |
| Plasmid: pFLAG-CMV-2 (control vector) | Sigma-Aldrich | Cat# E7033 |
| Software and Algorithms | ||
| ImageJ | https://imagej.nih.gov/ij/ | |
| Prism 8 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
Highlights.
DNA demethylase TET3 is increased in stressed hepatocytes and stellate cells (HSCs)
TET3 upregulates expression of TGF-β pathway genes including TGFB1 via demethylation
TGF-β1 stimulates TET3 expression, forming a positive feedback loop with TET3
The TET3/TGF-β1 loop promotes HSC activation and extracellular matrix production
ACKNOWLEDGMENTS
We thank Kathy Harry at the Yale Liver Center for isolation of primary mouse hepatocytes, Yalai Bai and David Rimm at Yale Pathology Tissue Services for providing human liver tissue samples, and Dr. Yi Zhang for the gift of plasmid pcDNA-Flag-Tet3. Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health of the United States under award R01DK119386 to Y.H. and NIDDK P30-34989 for the Yale Liver Center.
Footnotes
DECLARATION OF INTERESTS
All authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.12.092.
REFERENCES
- Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, et al. (2013). DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 18, 296–302. [DOI] [PubMed] [Google Scholar]
- An J, Rao A, and Ko M (2017). TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med 49, e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzoubir N, Lejamtel C, Battaglia S, Testoni B, Benassi B, Gondeau C, Perrin-Cocon L, Desterke C, Thiers V, Samuel D, et al. (2013). HCV core-mediated activation of latent TGF-β via thrombospondin drives the crosstalk between hepatocytes and stromal environment. J. Hepatol 59, 1160–1168. [DOI] [PubMed] [Google Scholar]
- Breitkopf K, Sawitza I, Westhoff JH, Wickert L, Dooley S, and Gressner AM (2005). Thrombospondin 1 acts as a strong promoter of transforming growth factor beta effects via two distinct mechanisms in hepatic stellate cells. Gut 54, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SY, Ouyang X, Chen Y, Soroka CJ, Wang J, Mennone A, Wang Y, Mehal WZ, Jain D, and Boyer JL (2017). Bile acids initiate cholestatic liver injury by triggering a hepatocyte-specific inflammatory response. JCI Insight 2, e90780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T, Jiang Y, Wang Z, Zhang N, Al-Hendy A, Mamillapalli R, Kallen AN, Kodaman P, Taylor HS, Li D, and Huang Y (2019). H19 lncRNA identified as a master regulator of genes that drive uterine leiomyomas. Oncogene 38, 5356–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff AM, Gray JT, Ng CY, Zhang Y, Zhou J, Spence Y, Bakar Y, and Nathwani AC (2005). Comparison of the ability of adeno-associated viral vectors pseudotyped with serotype 2, 5, and 8 capsid proteins to mediate efficient transduction of the liver in murine and nonhuman primate models. Mol. Ther 11, 875–888. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. (2013). TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, et al. (2013). Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med 19, 1617–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang G, Liang Z, Yang Y, Cui L, and Liu CY (2016). Loss of nuclear localization of TET2 in colorectal cancer. Clin. Epigenetics 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee MH, Hong KY, Park JH, Lee JS, Kim HS, Lee SH, and Jang SK (2015). New mechanism of hepatic fibrogenesis: hepatitis C virus infection induces transforming growth factor β1 production through glucose-regulated protein 94. J. Virol 90, 3044–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WI, Do SH, Yun HS, Song BJ, Kim SJ, Kwak WJ, Yoo SE, Park HY, and Jeong KS (2004). Hypoxia potentiates transforming growth factor-beta expression of hepatocyte during the cirrhotic condition in rat liver. Liver Int. 24, 658–668. [DOI] [PubMed] [Google Scholar]
- Kammel A, Saussenthaler S, Jähnert M, Jonas W, Stirm L, Hoeflich A, Staiger H, Fritsche A, Häring HU, Joost HG, et al. (2016). Early hypermethylation of hepatic Igfbp2 results in its reduced expression preceding fatty liver in mice. Hum. Mol. Genet 25, 2588–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Glick A, Sporn MB, and Roberts AB (1989). Characterization of the promoter region of the human transforming growth factor-beta 1 gene. J. Biol. Chem 264, 402–408. [PubMed] [Google Scholar]
- Kim J, Kollhoff A, Bergmann A, and Stubbs L (2003). Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum. Mol. Genet 12, 233–245. [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al. (2013). Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Shun CT, Chiou LL, Chen CH, Huang GT, and Sheu JC (2005). Hydroxyproline content of needle biopsies as an objective measure of liver fibrosis: emphasis on sampling variability. J. Gastroenterol. Hepatol 20, 1109–1114. [DOI] [PubMed] [Google Scholar]
- Li Y, Turpin CP, and Wang S (2017). Role of thrombospondin 1 in liver diseases. Hepatol. Res 47, 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C, Luedde T, Sauerbruch T, Scholten D, Streetz K, Tacke F, Tolba R, Trautwein C, Trebicka J, and Weiskirchen R (2013). Experimental liver fibrosis research: update on animal models, legal issues and translational aspects. Fibrogenesis Tissue Repair 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, Nygaard S, Grompe M, Alexander IE, and Kay MA (2014). Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 506, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XM, Nikolic-Paterson DJ, and Lan HY (2016). TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol 12, 325–338. [DOI] [PubMed] [Google Scholar]
- Mi Y, Gao X, Dai J, Ma Y, Xu L, and Jin W (2015). A novel function of TET2 in CNS: sustaining neuronal survival. Int. J. Mol. Sci 16, 21846–21857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy-Ullrich JE, and Suto MJ (2018). Thrombospondin-1 regulation of latent TGF-β activation: a therapeutic target for fibrotic disease. Matrix Biol. 68–69, 28–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A, Paoli P, Moran Salvador E, White S, French J, and Mann J (2016). Hepatic stellate cell transdifferentiation involves genome-wide remodeling of the DNA methylation landscape. J. Hepatol 64, 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, and Helin K (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani M, Espafñol-Surñer R, Malato Y, Dumont L, Grimm AA, Kienle E, Bindman JG, Wiedtke E, Hsu BY, Naqvi SJ, et al. (2016). In vivo hepatic reprogramming of myofibroblasts with AAV vectors as a therapeutic strategy for liver fibrosis. Cell Stem Cell 18, 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, and Friedman SL (2017). Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol 14, 397–411. [DOI] [PubMed] [Google Scholar]
- Wang Y, and Zhang Y (2014). Regulation of TET protein stability by calpains. Cell Rep. 6, 278–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, and Zhang Y (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet 18, 517–534. [DOI] [PubMed] [Google Scholar]
- Yoshiji H, Kuriyama S, Miyamoto Y, Thorgeirsson UP, Gomez DE, Kawata M, Yoshii J, Ikenaka Y, Noguchi R, Tsujinoue H, et al. (2000). Tissue inhibitor of metalloproteinases-1 promotes liver fibrosis development in a transgenic mouse model. Hepatology 32, 1248–1254. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, and Wong J (2014). Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT). J. Biol. Chem 289, 5986–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Geng T, Wang Z, Zhang R, Cao T, Camporez JP, Cai SY, Liu Y, Dandolo L, Shulman GI, et al. (2018). Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight 3, 120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or code.