Abstract
GPR37 and GPR37L1 are glia-enriched GPCRs that have been implicated in several neurological and neurodegenerative diseases. To gain insight into the potential molecular mechanisms by which GPR37 and GPR37L1 regulate cellular physiology, proteomic analyses of whole mouse brain tissue from wild-type (WT) versus GPR37/GPR37L1 double knockout (DKO) mice were performed in order to identify proteins regulated by the absence versus presence of these receptors (data are available via ProteomeXchange with identifier PXD015202). These analyses revealed a number of proteins that were significantly increased or decreased by the absence of GPR37 and GPR37L1. One of the most decreased proteins in the DKO versus WT brain tissue was S100A5, a calcium-binding protein, and the reduction of S100A5 expression in KO brain tissue was validated via Western blot. Co-expression of S100A5 with either GPR37 or GPR37L1 in HEK293T cells did not result in any change in S100A5 expression but did robustly increase secretion of S100A5. To dissect the mechanism by which S100A5 secretion was enhanced, cells co-expressing S100A5 with the receptors were treated with different pharmacological reagents. These studies revealed that calcium is essential for the secretion of S100A5 downstream of GPR37 and GPR37L1 signaling, as treatment with BAPTA-AM, an intracellular Ca2+ chelator, reduced S100A5 secretion from transfected HEK293T cells. Collectively these findings provide a panoramic view of proteomic changes resulting from loss of GPR37 and GPR37L1 and also impart mechanistic insight into the regulation of S100A5 by these receptors, thereby shedding light on the functions of GPR37 and GPR37L1 in brain tissue.
Keywords: S100A5, glia, calcium, secretion, astrocyte, oligodendrocyte, calcium-binding protein
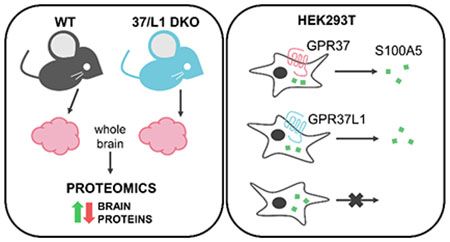
Graphical Abstract
INTRODUCTION
GPR37 and GPR37L1 are closely-related G protein-coupled receptors (GPCRs) that are highly expressed in the brain. These receptors exhibit differential expression in two distinct glial populations, with GPR37L1 being most highly expressed in astrocytes and GPR37 being enriched in oligodendrocytes.1,2,3,4,5,6,7,8,9 The first ligand reported for GPR37 was the invertebrate peptide known as head activator.10,11 Subsequently, the secreted vertebrate protein prosaposin and its active fragment prosaptide were reported to bind and activate both GPR37L1 and GPR37.12 Activation of GPR37 and/or GPR37L1 by prosaptide has been corroborated by several other groups,2,13,14,15 although several reports have also failed to detect significant activation of these receptors by prosaptide.16,17 Recent work by Liu et al. suggested a resolution of these seemingly disparate findings by providing evidence that prosaptide stimulation of GPR37 and GPR37L1 is dependent on cellular context, being readily observed in primary cells such as astrocytes but more difficult to observe in heterologous cells over-expressing the receptors.2 Additionally, the bioactive lipid neuroprotectin D1 has also recently been reported as a ligand for GPR37.13 Thus, despite the recent progress, more work is still needed to achieve a consensus as to the endogenous ligand(s) for GPR37 and GPR37L1.
Much of the current understanding of GPR37 and GPR37L1 function stems from studies that have focused on characterizing mice lacking the expression of one or both receptors. Knockout of GPR37 has been linked to altered dopamine signaling,18 precocious oligodendrocyte differentiation7 and increased susceptibility to demyelination,5 whereas knockout of GPR37L1 has been linked to precocious cerebellar development19 and enhanced seizure vulnerability.20 Moreover, mutations in GPR37L1 in humans have been associated with seizures and epilepsy.20,21 The link between GPR37L1 and changes in seizure susceptibility in both humans and mice is intriguing, given the predominant expression of GPR37L1 in astrocytes and the fact that most epilepsy-associated proteins are mainly expressed in neurons.22
The phenotypes observed in mice lacking GPR37 and GPR37L1, as well as the links between GPR37L1 variants and human disease, have led to interest in achieving a better understanding of the roles these receptors play in regulating cellular and molecular processes in the brain. Here we utilized a proteomic approach to study the effect of knocking out GPR37 and GPR37L1 on global protein expression in the mouse brain. Insights gained from these studies can lead to a better understanding of the processes and molecular mechanisms regulated by GPR37 and GPR37L1 and thereby help to guide future studies on these glia-enriched receptors.
MATERIALS AND METHODS
Animals
GPR37L1 knockout mice (Gpr37l1−/−) were obtained from the NIH Mutant Mouse Regional Centers (strain Gpr37l1tm1Lex, stock number 011709-UCD) and GPR37 knockout mice (Gpr37−/−) were obtained from Jackson Laboratories (strain Gpr37tm1Dgen, stock number 005806). The double knockout mouse (Gpr37−/− and Gpr37l1−/−) was made by crossing Gpr37−/− and Gpr37l1−/− mice, and genetic deletion of both genes was confirmed via DNA sequencing. All mice were maintained on a C57BL/6J background and housed on a 12-hour light/dark cycle with food and water ad libitum. All experiments were done in accordance with the guidelines of the Institutional Animal Care and Use Committee of Emory University.
Tissue preparation for proteomic analysis
Proteomic data was generated as stated in Smith et al. (2017).5 Brain lysates were prepared from 3-month-old WT (n=3) and GPR37L1/GPR37 double knockout mice (n=3) in urea buffer (8M urea, 100 mM NaHPO4, pH 8.5) with HALT protease and phosphatase inhibitor (Pierce) and processed at the Emory Proteomics Core. An aliquot of 100 μg from each sample was treated with 1 mM (final concentration) dithiothreitol (DTT) for 30 minutes followed by 5 mM (final concentration) iodoacetamide (IAA) for 30 minutes in the dark. Both steps were performed at room temperature. The protein mixtures were digested with 1:100 (w/w) lysyl endopeptidase (Wako) at 25°C for 2 hours. Trypsin (Promega) was then added at 1:50 (w/w) and digestion was allowed to proceed overnight. Resulting peptides were desalted with a Sep-Pak C18 column (Waters) and dried under vacuum.
Liquid chromatography coupled to mass spectrometry (LC-MS/MS)
Dried peptides were resuspended in peptide 100 μL of loading buffer (0.1% formic acid, 0.03% TFA, 1% acetonitrile). Peptide mixtures (2 μL) were separated on a self-packed C18 (1.9 μm Dr. Maisch, Germany) fused silica column (25 cm x 75 μm internal diameter (ID); New objective, Woburn, MA) by a Dionex Ultimate 3000 RSL CNano and monitored on a Fusion mass spectrometer (ThermoFisher Scientific, San Jose, CA). Elution was performed over a 140 minute gradient at a rate of 300nl/min with buffer B ranging from 3% to 99% (buffer A: 0.1% formic acid in water, buffer B: 0.1% formic acid in acetonitrile). The gradient starts off at 3% B and goes to 10% within 5 minutes, then from 10% to 40% within 100 minutes, then from 40% to 60% within 20 minutes, then from 60% to 99% in 5 minutes, and finally settling at 99% for the rest of the gradient. The mass spectrometer cycle was programmed to collect at the top speed for 3 second cycles. The spray voltage starts off at 2 kV and changes to 2.2 kV at 75 minutes. The MS scans (400-1500 m/z range, 200,000 AGC, 50 ms maximum ion time) were collected at a resolution of 120,000 at m/z 200 in profile mode. The higher energy dissociation (HCD) MS2 spectra (1.6 m/z isolation width, 0.5 m/z offset, 32% collision energy, 10,000 AGC target, 35 ms maximum ion time) were detected in the ion trap in centroid mode. Dynamic exclusion was set to exclude previous sequenced precursor ions for 30 seconds within a 10 ppm window. Precursor ions with +1, and +8 or higher charge states were excluded from sequencing.
Database search and label-free quantification
The Raw LC-MS/MS data was then analyzed via MaxQuant v1.5.4.1 using Andromeda search employing a target-decoy paradigm to control MS/MS peptide spectral match false discovery rate, and a database of contaminants plus the mouse Uniprot database (n=54,489 protein isoforms, downloaded April 2015). MaxQuant search parameters allowed up to 2 miscleavages, fully tryptic peptides only, and fixed modification of cysteine by carbamidomethylation (+57.0215 Da), plus variable oxidation of methionine (+15.9949 Da) and protein N-terminal acetylation (+42.0106 Da). Other search settings included a maximum peptide mass of 4,600 Da, a minimum peptide length of 7 residues, 0.05 Da tolerance for high resolution MS/MS scans. Co-fragmented peptide search was enabled to deconvolute multiplex spectra. The false discovery rate (FDR) for peptide spectral matches, proteins, and site decoy fraction were all set to 1 percent. The “match between runs” function was used to recover unsequenced precursors. Matches were matched within a 0.7 minute retention time match window and a 20 minute alignment. Quantitation of proteins was performed using LFQ with consideration of only razor and unique peptides. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE24 partner repository with the dataset identifier PXD015202.
Data processing and determination of differential protein abundance in GPR37/GPR37L1 double knockout vs. wild-type
Because data-dependent acquisition and label-free quantitation (LFQ) can miss individual sample measurements with a left-censoring (informative missingness) bias, the LFQ intensity quantitative data from MaxQuant for the n=6 samples were filtered to limit missingness, and then missing values were imputed as previously described. 25,26,27 Imputation of missing protein LFQ intensity values was performed using the imputation algorithm of Perseus27 as implemented in R and previously reported.25 Proteins were considered to have a detectable fold change if the fold change (FC) was a minimum of log2(1.286), 28.6% or more, represented by log2(DKO/WT) values of −0.36 and 0.36 or for downregulated and upregulated proteins, respectively. This threshold for fold change was determined as described in the below section. Significant differentially expressed proteins were then determined by additional filtering for t-test p values less than 0.05. In addition, any proteins that were sequenced and quantified on 1 unique or razor peptide maximum in any of the 6 samples were excluded.
Filtering of quantitative results (after imputation of missed quantitation)
We removed 581/4512 proteins from consideration that were sequenced and quantified on 1 unique or razor peptide maximum in any of the 6 samples. We determined the distribution of a true null 3×3 experiment of log2FC values. Each sample of 3 DKO and 3 WT measurements was converted to a null measurement by subtracting each of the log2 protein measurements from a paired measurement within the same genetic group, and then the null WT average log2 protein abundance was subtracted from the null DKO average, as previously described.29 The distribution histogram was fit to a Gaussian distribution per Dammer et al., 201529 where 1.96-fold of SD of the Gaussian fit to the histogram of the null 3931 measurements was log2(1.286), or a minimum FC of 28.6%. The counting of proteins beyond 1.96-fold(SD) corresponds to false positive quantitative measurements in a 3-sample vs. 3-sample comparison of WT null samples to DKO null samples that are expected to have no change. In the fit normal distribution, the relative area under the curve for the tails beyond this log2(FC) cutoff represents a probability of 0.05. At this FC cutoff, the null test produces 156/3931 (3.9%) of measurements that pass the FC criteria. This indicates that data filtered for minimum percent change 28.6% has been controlled to an empirical false discovery rate of less than 4%. Additional FDR calculations were performed in R via Perseus’ (v1.6.10.43) EdgeR implementation for determination of differential expression with FDR using default options of 0.04 dispersion factor, generalized linear modelling, and no additional normalization of LFQ abundances, non-log transformed. In addition, an R implementation of T test permutation with 10000 iterations for each protein using the independence test function of the R coin package run in R v3.5.2 generated permutation-based FDR values for all N=3931 proteins with at least 2 peptides quantified as log2(LFQ abundance). All protein measurements used were devoid of missing values following imputation as described. R code is available upon request.
Cell culture and cellular transfection
Human embryonic kidney cells (HEK293T cells) (ATCC) were maintained in complete media (DMEM 1X (Life Technologies) supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 1% penicillin/streptomycin (Quality Biological)) in a humid, 5% CO2, 37°C incubator. Cells were plated at 40% confluence the day prior to transfection. For standard transfections, 2 μg of each DNA construct was used for a total DNA amount of 4 μg per 10 cm plate. Transfections were done using Mirus TransIT-LT1 (Madison, WI) according to the manufacturer’s protocol. Following transfection, cells were incubated for 48 hours prior to harvesting. Constructs used included the following: empty vector, human GPR37L1 (untagged), human GPR37 (untagged), human S100A5-myc-DDK (Origene), human S100A4-myc-DDK (Origene), and human S100A6-myc-DDK (Origene).
Cell lysate preparation
Cells were lysed and harvested in low-salt harvest buffer (10 mM HEPES (pH 7.3), 50 mM NaCl, 5mM EDTA in ultrapure water) + HALT protease and phosphatase inhibitor (Thermo Fisher) (1% Triton X-100 was added to the low-salt harvest buffer if cells were solubilized). For solubilization, cells were rotated end-over-end at 4°C for at least 1 hour and then spun down at 15,000 RPM for 15 minutes to pellet. Laemmli sample buffer was added to cell lysate samples and left at room temperature overnight to prepare for Western blot analysis.
Western blot analyses
Protein samples were reduced and denatured in Laemmli buffer and analyzed via SDS-PAGE. Samples were loaded onto 4-20% Tris-glycine gels (BioRad) and then transferred to nitrocellulose membranes (BioRad). For analysis via chemiluminescence, membranes were blocked in 5% milk (5% non-fat milk in 50 mM NaCl, 10 mM HEPES, pH 7.3, 0.1% Tween-20 (Sigma) in ultrapure water) and then incubated with primary antibodies shaking overnight at 4°C. For analysis via infrared fluorescent Western detection, membranes were blocked with Odyssey Blocking Buffer (Licor) for 1 hour at room temperature and then incubated with primary antibodies shaking overnight at 4°C as per the manufacturer’s protocol. Membranes were then washed three times for 5 minutes each with wash buffer (1X PBS + 0.1% Tween-20) and then incubated with secondary antibodies that were diluted in blocking buffer. Membranes were again washed three times for 10 minutes each with wash buffer and were then imaged using the ChemiDoc Gel Imaging System. Protein quantification was done with ImageJ.
Secretion analyses
Transfected HEK293T cells were incubated in complete media (DMEM 1X + 10% FBS + 1% penicillin/streptomycin) for 48 hours and media was collected on the day of harvesting and HALT protease and phosphatase inhibitor (Thermo Scientific) was added to each media sample. Because the S100A protein constructs are all myc- and FLAG-tagged, myc-A/G agarose beads were used to pull down S100A proteins from the media. To pull down any S100A protein that may have been secreted into the media, each media sample was incubated with 60 μL (a non-saturated volume) of washed myc-A/G agarose bead slurry (Thermo Scientific) and rotated end-over-end at 4°C for 2 hours. Beads were then washed three times with low-salt harvest buffer and 2X Laemmli sample buffer was added to each bead sample and incubated at room temperature overnight prior to Western blot analysis.
For signaling studies, BAPTA-AM (Sigma-Aldrich), an intracellular calcium chelator, and prosaptide (TX14(A)) (Tocris Bioscience), a reported ligand for GPR37 and GPR37L1, were reconstituted to a stock concentration in the appropriate solvent. Cells were treated with compounds 24 hours after transfection and overnight prior to harvesting for protein and secretion analyses.
RNA extraction and cDNA conversion
Total mRNA was extracted from brain tissues and converted to cDNA for real-time quantitative polymerase chain reaction (RT-qPCR). Tissues were collected in 500 μL of TRIzol and dissociated by trituration. TRIzol mixtures were allowed to incubate at room temperature for 5 minutes. Following incubation, 100 μL of chloroform was added to each sample and tubes were shaken vigorously by hand for about 15 seconds to ensure thorough mixture. Samples were then allowed to incubate at room temperature for 5 minutes and then centrifuged at 15,000 rpm at 4°C for 15 minutes. The top, clear aqueous layer was collected into a new Eppendorf tube (about 200 μL in volume) and 250 μL of isopropyl alcohol was added to each sample and samples were inverted several times to ensure mixing. Samples were then incubated at room temperature for 10 minutes and centrifuged at 15,000 rpm at 4°C for 10 minutes to pellet the RNA. Supernatant was removed using a pipette and the RNA pellet was washed with 500 μL of 75% ethanol three times, spinning down at 15,000 rpm at 4°C for 5 minutes in between each wash. Following the final wash, the RNA pellet was allowed to dry on the benchtop at room temperature for 10-15 minutes or until most of the ethanol had evaporated. RNA pellets were then resuspended in RNAse-free water and incubated at 55°C for 10 minutes to rehydrate. RNA was stored at −80°C when not being used.
RNA was converted to cDNA via reverse transcription polymerase chain reaction (RT-PCR). Each RNA sample was diluted to 2-5 μg and incubated with 1 μL of 10 mM dNTPs, 0.5 μL of random primers (50 ng/μL), and 5.5 μL of DEPC dH2O at 65°C for 5 minutes. This RNA mixture was then incubated with 4 μL of 5X RT Buffer (Thermo Fisher), 2 μL of 0.1 M DTT, 1.5 μL of DEPC dH2O, 0.25 μL of RNaseOUT, and 0.25 μL of SuperScript III RT (Thermo Fisher) at room temperature for 10 minutes and then incubated at 42°C for 50 minutes.
Quantitative Polymerase Chain Reaction (qPCR)
cDNA made from the above protocol was used for quantitative analysis of mRNA expression in cell and tissue samples via the Bio-Rad Thermo Cycler C1000 Touch system using the DyNAmo ColorFlash SYBR Green qPCR kit (Thermo Fisher). Each reaction was performed in triplicate and the following thermal cycle protocol was used for all reactions: (1) 95°C for 7 mins, (2) 95°C for 10 seconds, (3) 60°C for 30 seconds, 40 cycles, steps 2-3, (4) melting curve obtained according to the instrument manufacturer. The following primers were made and purchased from Eurofins Genomics: GAPDH forward: 5’-ACC ACA GTC CAT GCC ATC AC-3’; GAPDH reverse: 5’-TCC ACC ACC CTG TTG CTG TA-3’; S100A5 forward: 5’-GCA AGC TGA CCC TGA GTA GG-3’; S100A5 reverse: 5’-CGC TGT TTT TGT CCA GGC TC-3’. Data were analyzed via the ΔΔCT method described previously30 and results are expressed as relative expression normalized to wild-type samples.
Antibodies used
PRDX6 (Cell Signaling Technologies) WB 1:1000; EBP (Invitrogen) WB 1:1000; S100A5 (Novus) WB 1:5000; GAPDH (Millipore) WB 1:5000; Flag-HRP (Invitrogen) WB 1:5000; GPR37L1 (Mab Technologies) WB 1:2000; GPR37 (Mab Technologies) WB 1:2000; IRDye 680LT goat anti-chicken (Licor) WB secondary antibody 1:2000; IRDye 800LT goat anti-rabbit (Licor) WB secondary antibody 1:2000.
S100A5 antibody validation
Lysates from HEK293T cells that were transfected with either S100A4-mycDDK (Origene), S100A5-mycDDK (Origene), or S100A6-mycDDK (Origene) were used for validation of S100A5 antibody specificity. Samples were prepared for Western blot as mentioned above and Western blot analysis was performed. Membranes were blotted with S100A5 (Novus, 1:5000) to check for specificity, and Flag-HRP (Invitrogen, 1:5000) to confirm that all three constructs were present in the samples (Supplemental Figure 4).
RESULTS
Proteomic analysis of global protein changes in the GPR37/GPR37L1 double knockout mouse brain vs. wild-type (WT) mouse brain
To gain insight into the potential physiological actions of GPR37 and GPR37L1 in the brain, mass spectrometry (MS)-based label-free proteomics was performed on whole mouse brain lysates. Because the seizure phenotype is most severe in the DKO mice, the proteomic screen was performed on wild-type (WT) and DKO mouse brains to assess whether the levels of certain proteins were altered by the loss of these two related GPCRs. Samples of WT and DKO whole brain lysates from adult (3 months old) mice were prepared in urea lysis buffer as previously described23 and submitted to the Emory Integrated Proteomics Core for proteomic analyses via liquid chromatography-mass spectrometry (LC/MS). In total, 4,512 unique proteins were identified in the proteomic analyses between the six total WT vs. DKO mouse brain lysates.
The LFQ dataset was imputed using the imputation algorithm of Perseus to address missing values according to an assumption of informative missingness. 581 proteins from the original list were not considered due to unreliable, single peptide, measurement or high missingness of protein quantitation across the six measurements (Supplemental Material S1). Then, differentially expressed proteins between WT and DKO samples (n=3 each) were determined as described in the Methods under using filtering criteria specified in the section “Filtering of quantitative results” and are represented as either red or green dots on a volcano plot (Fig. 1A). Red points on the volcano plot represent proteins with significant negative fold change, and green points represent proteins with significant positive fold change. Empirical FDR based on the true null 3×3 experiment indicates an FDR of <4% without one-peptide identifications. Through determination of the distribution of a null experiment fit to a Gaussian distribution, data was filtered for a minimum percent change of 28.6% and a p value equal to or less than 0.05 (calculations for the null experiment and FDR can be found in Supplemental Material S2). Based on these criteria, the vast majority of proteins did not exhibit any significant changes in expression between WT and DKO brain tissues. However, out of over 4,500 proteins identified and quantified, a small handful of proteins was found to exhibit differential expression, with 78 proteins found to be significantly increased and 53 found to be significantly decreased (the full tables of significantly increased and decreased proteins can be found in Supplemental Material S3). These differentially-expressed proteins were then analyzed via GO-Elite “biological process,” “cellular component,” and “molecular function” categorical annotation using DAVID to identify the most enriched pathways and/or functions of either increased proteins (Fig. 1C) or increased and decreased proteins combined (Fig. 1D; the full list of proteins found for each ontology can be found in Supplemental Material S4). No ontologies were identified for the list of decreased proteins alone. A Z score of 1.96 or greater represented a functional enrichment or overrepresentation by Fisher’s Exact test that was significant within the given protein list. For increased proteins as well as the increased and decreased proteins combined, the only major enriched biological process was serine-type endopeptidase inhibitor activity (Z score=8.07 and 5.88, respectively). For “cellular component,” extracellular space and extracellular region were overrepresented with Z scores of 5.29 and 8.30 respectively for only increased proteins and Z scores of 4.37 and 7.27 respectively when analyzing both increased and decreased proteins together. For “molecular function,” the top 3 functional enrichments found when looking at only increased proteins were “negative regulation of hydrolase activity,” “steroid metabolic process,” and regulation of proteolysis with Z scores of 6.25, 5.03, and 4.02 respectively. Some other interesting associations were found for other molecular functions, such as glycerolipid metabolic process and response to hormone stimulus.
Figure 1. Imputation and analysis of proteomic data.
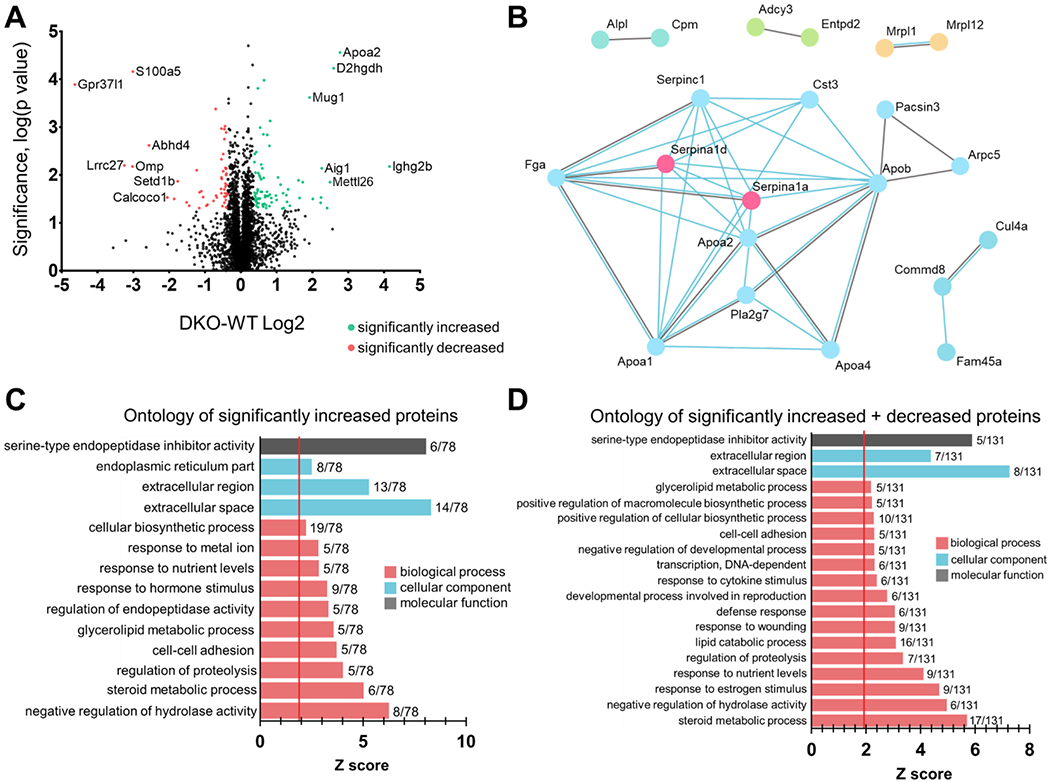
(A) Volcano plot depicting all protein hits from proteomic analyses comparing global protein changes between DKO and WT mouse brains. Whole mouse brain lysates were processed at the Emory Proteomics Core for analysis via mass spectrometry. The x-axis represents fold change (increased, positive Log2(DKO/WT) or decreased, negative Log2(DKO/WT)) and the y-axis represents −log10(p value), i.e. the bigger the −log10(p value), the more significant the data point is. The proteomic screen yielded only a small handful of proteins that exhibited both a significant and high fold change. Data points colored in red are those proteins that were significantly decreased (p≤0.05, DKO-WT Log2 ≤ −0.36) in DKO mouse brains compared to WT mouse brains, and data points colored in green are those that were significantly increased (p≤0.05, DKO-WT Log2 ≥ 0.36). Out of over 4,500 proteomic hits, only 78 were significantly increased (greater than or equal to +28.6% fold change) and only 53 were significantly decreased (less than or equal to −28.6% fold change) when both GPR37 and GPR37L1 were knocked out. WT, n=3; DKO, n=3. (B) A protein-protein interaction network between the 78 increased and 53 decreased proteins was generated via STRING analysis (string-db.org). This network identifies any protein-protein interactions determined experimentally (blue lines) or found in curated databases (gray lines). (C,D) Ontology graphs were generated each for the increased (C) and increased and decreased combined (D) lists of proteins.
When looking at ontologies for increased and decreased proteins lists together, the top 3 enrichments in “molecular function” were “steroid metabolic process” (Z score = 5.70), “negative regulation of hydrolase activity” (Z score = 4.97), and “response to estrogen stimulus” (Z score = 4.68). Interestingly, “glycerolipid metabolic process” appears on both ontology analyses for only increased proteins as well as both increased and decreased proteins lists combined. It should be noted that this direction of change is consistent with compensation for or responsiveness to accumulation of glycerolipids known to occur when saposins’ function is compromised at the lysosome, and a reported ligand for both GPR37L1 and GPR37 is prosaptide, an active fragment of prosaposin, which is known to be involved in lysosomal trafficking and biology. 12,31,32,33,34,35,36 Additionally, a protein-protein interaction network was generated for the list of proteins that were significantly differentially expressed (both up- and downregulated) (Fig. 1B). Only proteins that had an interaction with at least one other protein were displayed.
Proteome-wide changes in the GPR37/GPR37L1 double knockout mouse brain
The top decreased and increased proteins found in the proteomic analyses were used for further validation in in vitro studies (Tables 1 and 2). GPR37 and GPR37L1 have been reported to be expressed in distinct cell types and cell populations within the brain, with GPR37L1 being most highly expressed in astrocytes1,14,19,37 and GPR37 being most abundantly expressed in oligodendrocytes.1,7,38 Because of this cell-specific expression in the central nervous system, both the imputed lists for significantly increased and decreased proteins found via mass spec were analyzed for cell type expression to assess how knockout of both these receptors affected the cell type expression profile of proteins that were changed. Both lists were run against reported cell type-enriched expression databases via Differential Enrichment analysis of Proteomics data (DEP), including those by Sharma et al. (2015) and the Barres lab.9,23,25 Fisher’s Exact test was performed and heatmaps were prepared depicting −log(p-value). Interestingly, significantly upregulated proteins were found to be mostly expressed in neurons, microglia, and endothelial cells across the three different databases while significantly downregulated proteins were found enriched mostly in astrocytes and oligodendrocytes (Supplemental Figures 1, 2, 3). The lists of proteins that were found for each cell type by each database and their corresponding FET values are shown in Supplemental Material S5. Because GPR37 and GPR37L1 are most highly expressed in astrocytes and oligodendrocytes respectively, we focused on proteins that were significantly decreased (Table 1).
Table 1.
Top 7 most significantly decreased proteins in GPR37/GPR37L1 double knockout (DKO) vs. wild-type (WT) whole mouse brain identified via mass spectrometry
| Uniprot ID | Gene symbol | Percent decrease, LFQ signal | T-test p value ▲ | Traditional FDR (Benjamini-Hochberg) | Max single-sample Razor+Unique Peptide Counts | |
|---|---|---|---|---|---|---|
| DKO | WT | |||||
| P63084 | S100a5 | −88% | 6.94E-05 | 5.4% | 0 | 3 |
| Q99JG2 | Gpr37l1 | −96% | 1.27E-04 | 6.6% | 0 | 3 |
| P20917 | Mag | −38% | 4.16E-04 | 11.8% | 14 | 15 |
| P47199 | Cryz | −26% | 9.62E-04 | 18.2% | 8 | 10 |
| P52760 | Hrsp12 | −31% | 1.07E-03 | 18.2% | 8 | 9 |
| P21460 | Cst3 | −27% | 1.11E-03 | 18.2% | 5 | 5 |
| Q8VDK1–2 | Nit1 | −24% | 1.30E-03 | 18.2% | 9 | 9 |
Table 2.
Top 9 most significantly increased proteins in GPR37/GPR37L1 double knockout (DKO) vs. wild-type (WT) whole mouse brain identified via mass spectrometry
| Uniprot ID | Gene symbol | Percent decrease, LFQ signal | T-test p value ▲ | Traditional FDR (Benjamini-Hochberg) | Max single-sample Razor+Unique Peptide Counts | |
|---|---|---|---|---|---|---|
| DKO | WT | |||||
| P09813 | Apoa2 | 581% | 2.73E-05 | 5.3% | 2 | 3 |
| Q8CIM3 | D2hgdh | 501% | 5.85E-05 | 5.4% | 5 | 0 |
| Q91XF0 | Pnpo | 56% | 1.05E-04 | 6.6% | 4 | 3 |
| Q91V76 | 4931406C07Rik | 40% | 1.53E-04 | 6.6% | 8 | 8 |
| P28665 | Mug1 | 279% | 2.42E-04 | 9.2% | 16 | 7 |
| Q00623 | Apoa1 | 77% | 7.39E-04 | 18.2% | 14 | 14 |
| Q65CL1 | Ctnna3 | 46% | 1.02E-03 | 18.2% | 4 | 3 |
| Q00897 | Serpina1d | 55% | 1.40E-03 | 18.3% | 3 | 2 |
| Q9DB15 | Mrpl12 | 67% | 1.44E-03 | 18.3% | 3 | 3 |
The top 5 most significantly downregulated proteins included S100 calcium-binding protein A5 (S100A5), GPR37L1, myelin-associated glycoprotein (MAG), quinone oxidoreductase (CRYZ), and 2-iminobutanoate/2-iminopropanoate deaminase (HRSP12). We have previously published on the role of GPR37 knockout resulting in the downregulation of MAG.5 Western blot analyses were performed to either confirm or refute the other top hits based on the proteomic data.
Western blot analysis revealed that there was a significant decrease of S100A5 across the single and double knockout mouse brains compared to WT mouse brain. Expression of S100A5 was found to be reduced in KO mouse brain tissue by approximately 75-80% (Fig. 2A, quantification Fig. 2B), a reduction that is in line with the proteomic data. Levels of S100A5 mRNA in DKO vs. WT mice were also analyzed. In line with the proteomic data and the Western blot data, mRNA levels of S100A5 were found by qPCR to be reduced about 75% in the DKO mouse brain. These data suggest that regulation of S100A5 by GPR37 and GPR37L1 occurs at least partially at the level of transcription.
Figure 2. S100A5 protein and mRNA levels are reduced in GPR37 knockout, GPR37L1 knockout, and GPR37/GPR37L1 double knockout mouse brains vs. WT mouse brain.
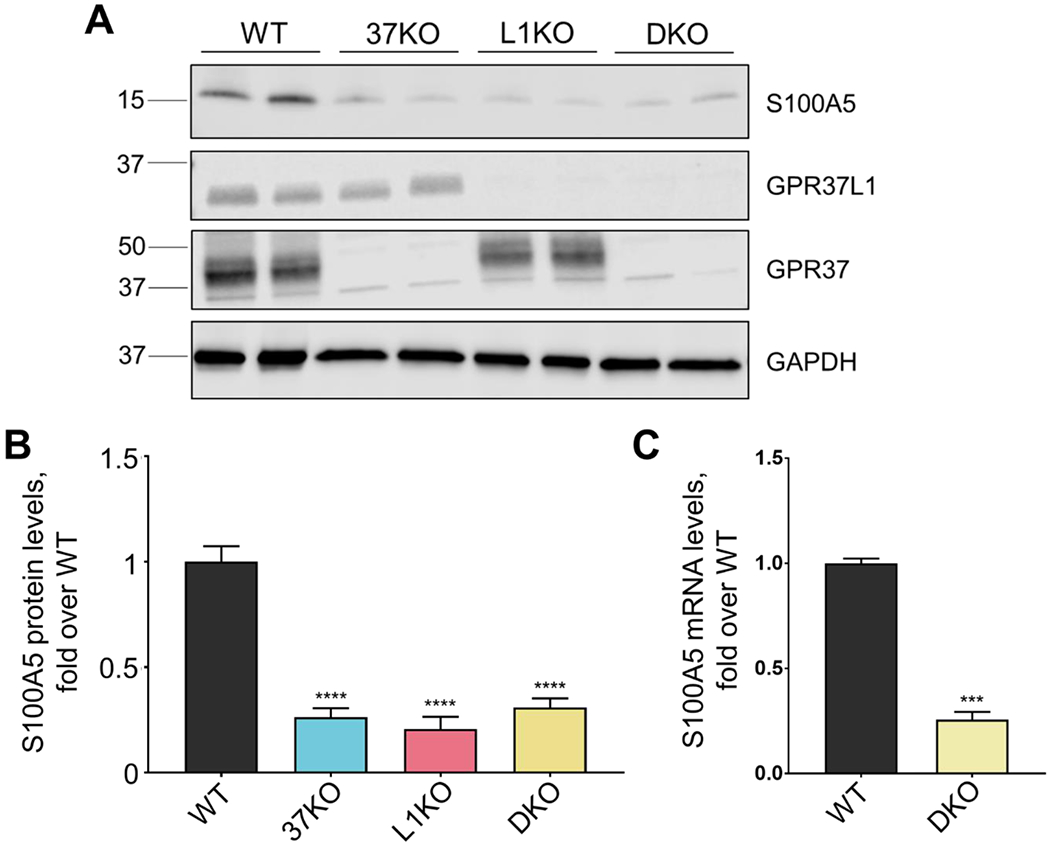
Representative Western blot depicting protein levels in whole mouse brain lysates. S100A5 is significantly downregulated in knockout mouse brains compared to wild-type mouse brains. (A) Western blots were quantified via ImageJ (B) (n=7, ****p<0.0001, bars represent SEM, one-way ANOVA, Dunnett’s post-hoc). (C) S100A5 mRNA levels in WT vs. double knockout mouse brain (n=4, ***p=0.0003, student’s t-test, bar represents SEM). All samples were normalized to GAPDH.
S100A5 secretion is regulated by GPR37 and GPR37L1 and is receptor-dependent
To further explore potential regulation of S100A5 by GPR37 and GPR37L1, we moved into a mammalian cell line. HEK293T cells were transfected with S100A5 in the absence and presence of either GPR37 or GPR37L1. As knockout of the receptors led to downregulation of S100A5 in vivo at the protein and transcript level, we assessed whether co-expression of S100A5 with either receptor might lead to upregulation of S100A5. However, there was little to no effect of the receptors on the total expression of S100A5 (Fig. 3A), an observation consistent with the aforementioned qPCR data suggesting that the effect of the receptors on S100A5 expression in vivo was mainly a result of transcriptional changes and not an effect on protein stability. Interestingly, though, we also analyzed media from transfected cells and found that S100A5 was only detectable in the media of cells in which S100A5 had been co-expressed with either GPR37 or GPR37L1. No detectable amounts of S100A5 were found in the condition in which S100A5 was expressed alone (Fig. 3B). While there appeared to be somewhat more S100A5 secreted from cells co-expressing S100A5 and GPR37 compared to cells co-expressing S100A5 and GPR37L1, statistical analyses revealed that the amount of S100A5 secreted from those two conditions was not significantly different. These data suggested that the presence of the receptors was able to induce secretion of S100A5 in transfected cells.
Figure 3. Co-expression of S100A5 with either GPR37 or GPR37L1 leads to robust secretion of S100A5 from HEK293T cells.
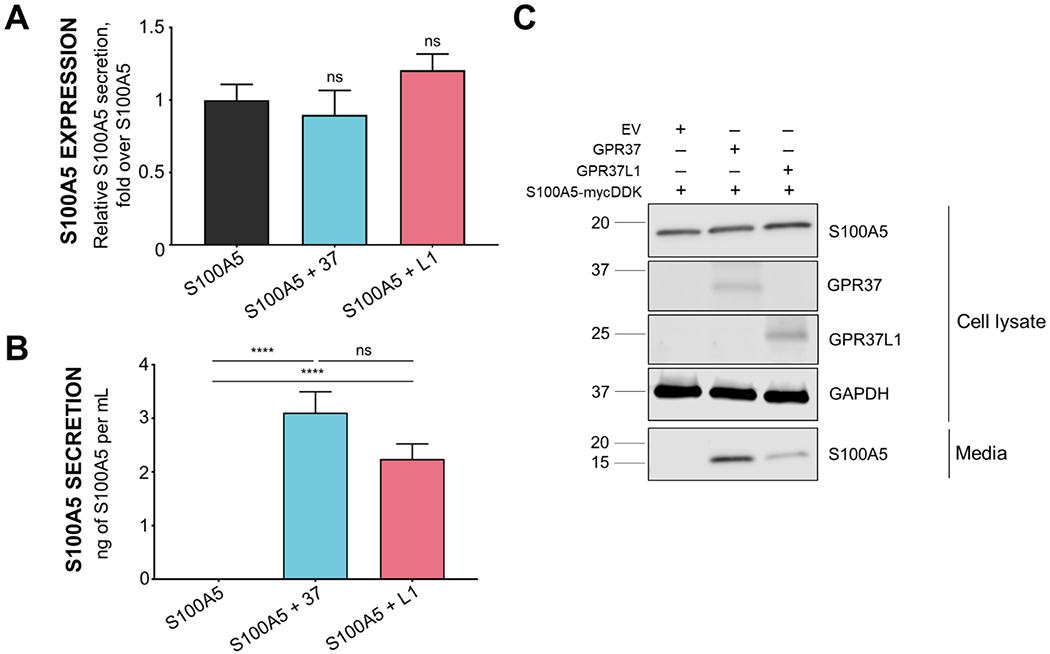
(A) Co-expression of S100A5 with either GPR37 or GPR37L1 in HEK293T cells did not significantly increase intracellular S100A5 expression (n=12 per condition, bars represent SEM, one-way ANOVA). (B) Co-expression of S100A5 with either GPR37 or GPR37L1 did increase S100A5 secretion compared to S100A5 expression alone (n=7, bars represent SEM one-way ANOVA, ****: p<0.0001). (C) Representative Western blot of cell lysates and media is shown on the right demonstrating expression of transfected proteins (cell lysate) and secretion (media) of S100A5 into the media. HEK293T cells were transfected to co-express S100A5 in the absence and presence of GRP37 or GPR37L1.
S100A proteins are known to be secreted, but almost nothing is known about the mechanisms regulating this secretion.39 Thus, we performed further studies to investigate the role of GPR37 and GPR37L1 in the regulation of the secretion of S100A5. To rule out that S100A5 secretion was indiscriminate or due to simple overexpression of the protein, cells were transfected with increasing amounts of the S100A5 construct in the absence of the receptors, and intracellular expression and extracellular secretion of S100A5 into the media were analyzed. Western blot analyses revealed that even at 6 μg of S100A5 construct transfected, three times the amount normally transfected into cells, there was no detectable secretion of S100A5 by the cells (Fig. 4A). Additionally, when the receptor constructs were transfected in increasing amounts (0-4μg) and co-expressed with a constant amount of S100A5 construct (2μg), there were receptor-dependent increases in the amount of secreted S100A5 (Fig. 4C–D), with receptor expression levels exhibiting a positive linear relationship to the amount of S100A5 secreted (Fig. 4B). There were no statistically significant differences in S100A5 secretion between the two conditions at any receptor concentration.
Figure 4. Receptor regulation of S100A5 secretion.
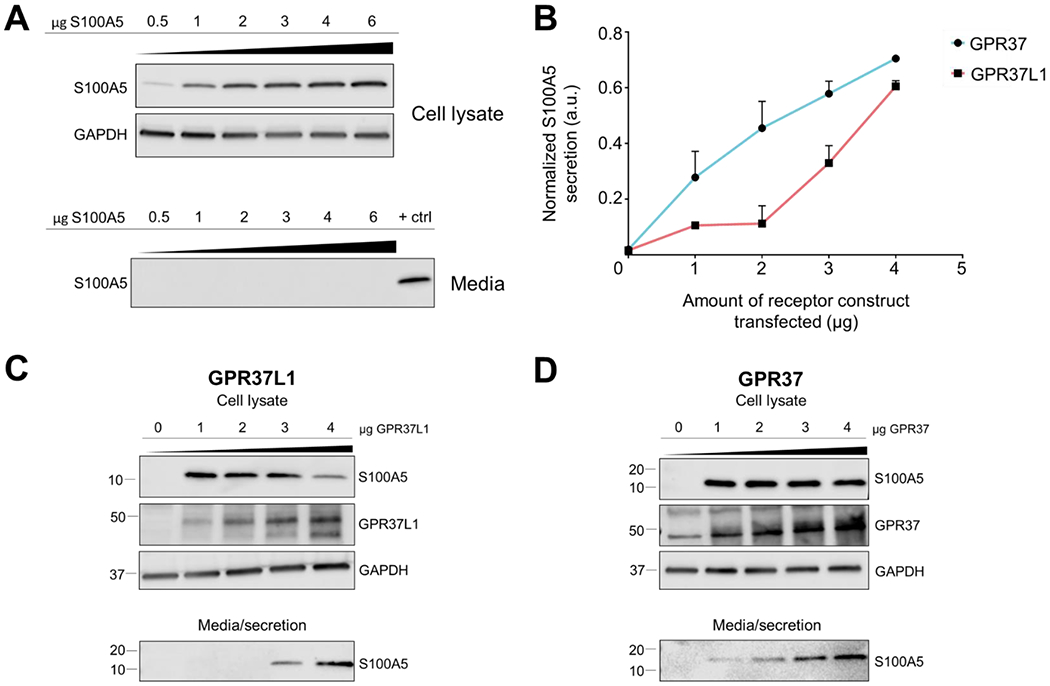
(A) To assess whether secretion of S100A5 can be induced by simply overexpressing the protein, increasing amounts of S100A5 plasmid (0.5 μg up to 6μg) were transfected into HEK293T cells. However, increasing S100A5 levels in HEK293T cells did not lead to detectable S100A5 secretion, suggesting that secretion of S100A5 is receptor-dependent. (B) Secretion of S100A5 increased according to the amount of receptor co-expressed (this panel shows a quantification of the data shown in panels C and D). (C) Representative Western blot depicting the effect of increasing GPR37L1 levels on S100A5 secretion (n=4). (D) Representative Western blot depicting effect of increasing GPR37 levels on S100A5 secretion (n=4). All samples were normalized to GAPDH. Bars represent SEM.
Calcium mediates S100A5 secretion downstream of GPR37L1 and GPR37 constitutive signaling
In further work, we investigated the mechanism by which GPR37 and GPR37L1 were able to enhance S100A5 secretion. HEK293T cells were transfected with S100A5 in the absence or presence of GPR37 or GPR37L1 and treated with increasing concentrations of BAPTA-AM, an intracellular Ca2+ chelator, or prosaptide (TX14A), a reported ligand for GPR37 and GPR37L1.2,12,15 Interestingly, when HEK293T cells co-expressing S100A5 with either receptor were treated with increasing concentrations of BAPTA-AM, there were significant reductions in S100A5 secretion (Fig. 5B, 5C). In contrast, treatment of transfected HEK293T cells with 100 nM prosaptide had no effect on S100A5 secretion (Fig. 5A).
Figure 5. Chelation of intracellular Ca2+ leads to decreased S100A5 secretion from HEK239T cells.
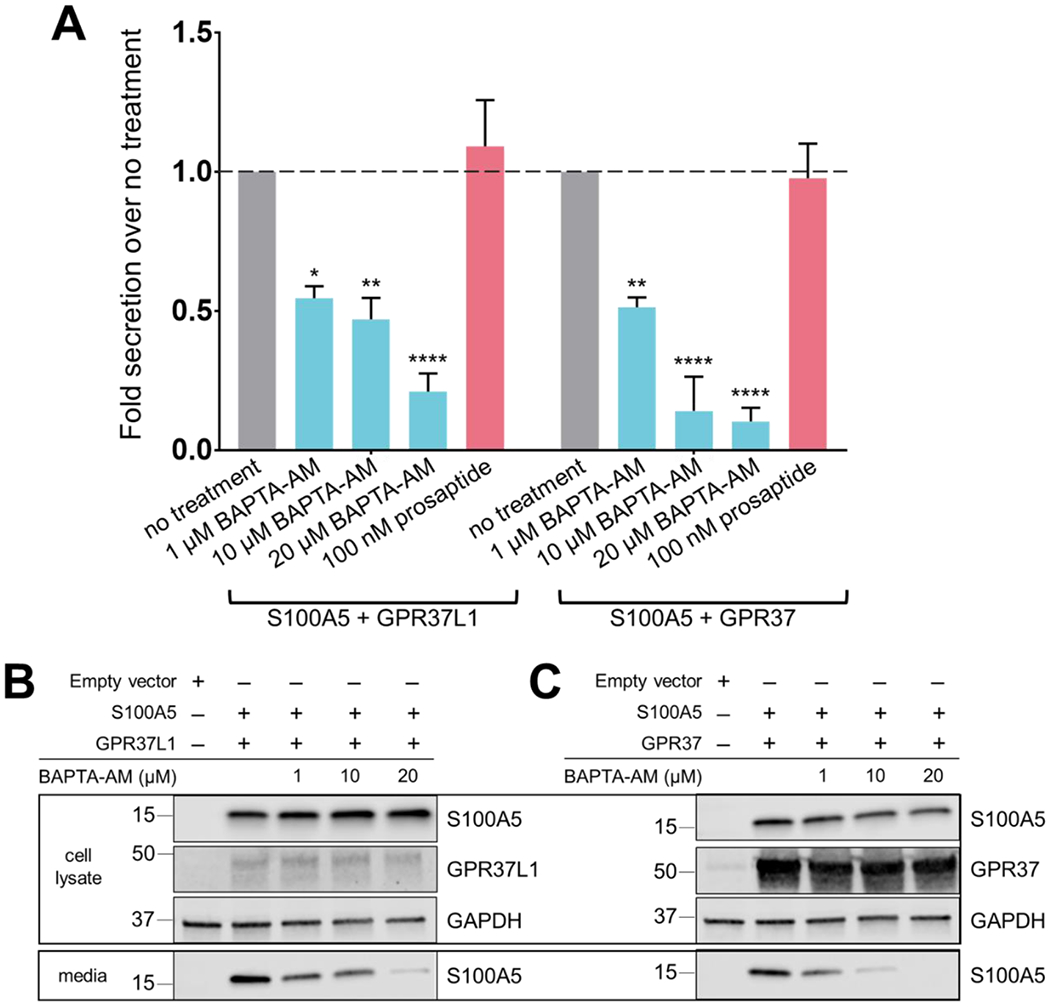
To elucidate the mechanism regulating S100A5 secretion, HEK293T cells were transfected with S100A5 and either GPR37 or GPR37L1. Cells were allowed to incubate for 24 hours following transfection and then treated with different pharmacological reagents. (A) Normalized quantification of S100A5 secretion when cells were treated with either BAPTA-AM or the putative GPR37L1 ligand prosaptide (TX 14A) compared to transfected cells that did not receive treatment. (B) Representative Western blots depicting effects of increasing concentrations of BAPTA-AM on S100A5 secretion in cells co-expressing S100A5 and GPR37L1. (C) Representative Western blot depicting effects of increasing concentrations of BAPTA-AM on S100A5 secretion in cells co-expressing S100A5 and GPR37. (n=3-4, bars represent SEM, two-way ANOVA within groups, Dunnett’s post hoc) (*p=0.0110; **p<0.006; ****p<0.0001)
Receptor-mediated secretion is restricted to homologous S100A proteins
To assess whether this GPR37- and GPR37L1-mediated S100A5 secretion was specific only to S100A5 or was general for other members of the S100A protein family, we performed the same experiments with S100A4 and S100A10. These two proteins were not found to be significantly changed in the proteome. S100A4 is a protein of the S100A protein family that is closely related to S100A5, sharing 53% protein sequence identity and 70% sequence similarity. S100A10 is more distally related, sharing only 33% protein sequence identity and 62% sequence similarity (positives, BLAST of human protein sequences). Additionally, on phylogenetic trees constructed by sequence alignments of the S100 protein family, S100A4 and S100A5 are grouped in the same subgroup, with S100A10 found in a distinct subgroup.39
HEK293T cells were transfected to express either S100A4 or S100A10 in the absence or presence of GPR37 or GPR37L1, and expression and secretion of the S100A proteins were analyzed. When S100A4 was co-expressed with either GPR37 or GPR37L1, S100A4 was detected in the media (Fig. 6A), whereas no detectable S100A4 was found when S100A4 was expressed alone. In contrast, co-expression of S100A10 with either receptor did not lead to any detectable secretion in the media (Fig. 6B). These data suggest that GPR37 and GPR37L1 can promote secretion of S100A proteins related to S100A5 but not all members of the S100A family.
Figure 6. Co-expression of homologous S100A proteins with either GPR37L1 or GPR37 also leads to secretion.
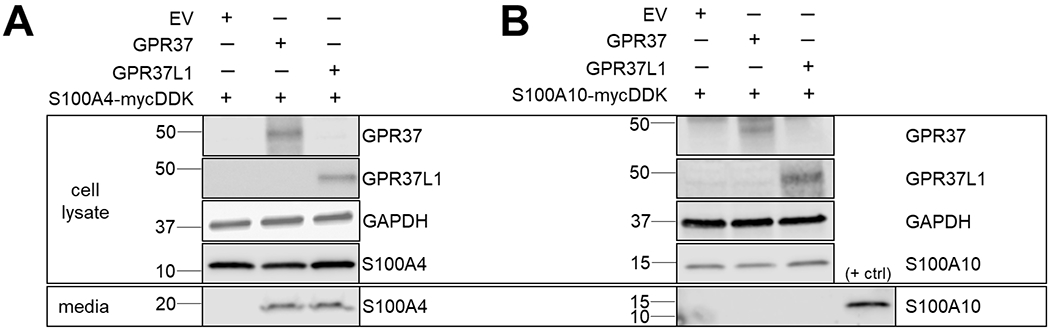
To explore whether GPR37- or GPR37L1-mediated secretion was specific only to S100A5 or general for other S100A protein family members, we examined whether co-expression of either GPR37 or GPR37L1 with S100A4 or S100A10 in HEK293T cells might lead to secretion of these proteins. (A) S100A4 co-expression with either receptor resulted in detectable secretion. (B) In contrast, S100A10 secretion was not observed upon co-expression with either receptor. The positive control shown here was a media sample containing secreted S100A5.
DISCUSSION
Previous findings revealed that the GPR37 single knockout (37KO), the GPR37L1 single knockout (L1KO), and the GPR37/GPR37L1 double knockout (DKO) mice exhibit increased susceptibility to induced seizures, with the DKO mice exhibiting the most pronounced susceptibility.20 Our proteomic analyses revealed that the loss of both GPR37 and GPR37L1 in the mouse brain led to the global increase and decrease of a small handful of proteins. Ontology analyses of significantly increased and decreased proteins combined revealed that proteins are involved in several processes such as “response to estrogen stimulus,” “steroid metabolic processes,” “defense response,” and “response to cytokine stimulation.” “Defense response” and “response to cytokine stimulation” are intriguing as it suggests that a subset of significantly differentially expressed proteins may be involved in controlling inflammatory responses. For “response to estrogen stimulus” and “steroid metabolic processes,” the steroid and estrogen connections may be interesting avenues for further investigation, given the genetic links between GPR37L1 and epilepsy20,21 and the vast literature on the role of sex steroids in modulating seizure vulnerability in both humans and rodent models.40,41,42,43,44,45,46
Another interesting finding was that the majority of the decreased proteins were found to be enriched in astrocytes and oligodendrocytes (the two cell types in which GPR37 and GPR37L1 are enriched) while many of the increased proteins were found to be enriched mainly in neurons, microglia, and endothelial cells. It is possible that knockout of both receptors leads to the decrease of this handful of proteins in the respective native cell types, astrocytes and oligodendrocytes, and subsequent deficits in the function of astrocytes and/or oligodendrocytes then lead to upregulation of certain proteins found in other cell types. One of the most decreased proteins from the proteomic analyses, S100A5, was found via Western blot to be reduced about 75-80% in single and double knockout mouse brain lysates. Moreover, mRNA transcripts of S100A5 were reduced by about 75% in the double knockout mouse brain, suggesting transcriptional down-regulation of S100A5 upon loss of GPR37 and GPR37L1. In parallel in vitro studies, we found that S100A5 levels were not affected by co-expression with GPR37 or GPR37L1, but surprisingly we observed robust secretion of S100A5 into the media upon co-expression with either receptor. S100A5 is a brain-enriched EF-hand motif-containing calcium-binding protein that can bind Ca2+, Zn2+, and Cu2+ 47 and has been reported to be expressed in astrocytes,48,49 the cell type in which GPR37L1 is most abundantly expressed.
Little is known about the biological function of S100A5, but there has been a significant amount of work on other members of the S100A family. For example, the closely-related S100A4 was found to be upregulated in astrocytes in models of traumatic brain injury (TBI) and kainic acid excitotoxicity, and is also known to be secreted to act as a neuroprotective and neurotrophic factor.50 Additionally, S100A6, another member of the S100A protein family that is highly homologous to S100A5, has also been reported to be secreted and regulated by pathological states.51,52,53 Over the years, a number of reports have described roles for S100 calcium-binding proteins in the regulation of ion channels and neuronal excitability. In a study done by Kubista et al., extracellular application of either S100B or S100A1 to Helix neurons led to hyperpolarization of the membrane resting potential and inhibited spontaneous discharge activity of action potentials due to effects on potassium channels.54 Several other studies also describe effects of manipulation of extracellular S100 proteins (with anti-serum or antibodies against S100 proteins, or treatment with S100 proteins) on neuronal activity. 55,56,57,58 Yet another study showed that secretion of S100B from astrocytes was dependent on presynaptic release of neurotransmitter.59 Taken together, these studies provide evidence that S100 proteins can be secreted in response to neuronal activity and imply an extracellular modulatory function of S100 proteins on neuronal excitability. Therefore, it is conceivable that alterations in GPR37- or GPR37L1-mediated regulation of S100A5 could lead to altered neuronal activity and/or excitability and thereby contribute to the seizure susceptibility phenotype observed in mice lacking these receptors.
The molecular mechanisms by which S100A proteins are secreted are completely unclear. Serotonin receptor agonists, glutamate, lipopolysaccharide, changes in extracellular Ca2+ and K+, and metabolic stress have all been reported to stimulate release of S100 proteins from astrocytes.60,61,62,63 Some studies suggest that secretion of S100 proteins may be passive or involve direct interaction with the plasma membrane.64,65 Our work provides additional mechanistic insight into the secretion of S100A proteins, as we found that co-expression with GPR37 or GPR37L1 promoted the secretion of both S100A5 and S100A4. As these S100A proteins bind calcium, and as GPR37 and GPR37L1 can increase calcium levels downstream of G protein coupling,10,11,13 we also assessed whether intracellular Ca2+ levels can affect secretion of S100A5. We found that treatment of transfected HEK293T cells with an intracellular Ca2+ chelator, BAPTA-AM, dramatically reduced secretion of S100A5. In further studies, it would be interesting to explore whether changes in intracellular Ca2+ levels result in specific conformational changes in S100A5 that promote secretion or conversely exert general effects on the secretion machinery. Several studies show that S100 calcium-binding proteins exposed to high intracellular Ca2+ levels can bind Ca2+ and subsequently adopt a conformation that exposes hydrophobic sites that then enables them to bind target proteins or membranes.66,67 Interestingly, S100 proteins have also been reported to be associated with synaptosomes, including several recent proteomic studies that reveal the presence of S100 proteins in the synaptosome.68,69,70 Several other studies show that S100 proteins are able to form tight complexes with binding sites in synaptosomal particulate fractions in both time- and temperature-dependent manners,71,72,73 and are shown to also localize to the synaptosome in mouse brain cortex.74,75 Additionally, two studies that investigated the localization of S100 proteins in synaptosomal fractions showed that divalent cations such as Ca2+ and Mg2+ were necessary for S100 proteins to interact directly with synaptosomes.73,76 While these studies highlight the role of S100 proteins in the neuronal synaptosome, it is possible that the binding of Ca2+ can affect the function and/or secretion of S100 proteins from glial cells as well.
We also assessed the effect of prosaptide, a reported endogenous ligand for GPR37 and GPR37L1, on S100A5 secretion, but found no effect of prosaptide treatment. This may be due to several factors, including that GPR37 and GPR37L1 have both been reported to exhibit high levels of constitutive activity in transfected cells.5,17 Moreover, recent work has revealed that treatment with prosaptide elicits robust GPR37 and GPR37L1 signaling in native cell types such as astrocytes but not in HEK293T cells when the receptors are overexpressed.2 Further work is needed on GPR37 and GPR37L1 to determine why their signaling activity is so dependent on cellular context and also to resolve the question of whether prosaptide and/or neuroprotectin D113 are both endogenous agonists for these receptors.
CONCLUSIONS
There is a growing body of evidence for dysregulation of GPR37 and GPR37L1 contributing to neurological disease, and thus a better understanding of the normal function of these two receptors is needed. In this study, we report proteomic data analyzing global protein changes that occur in the GPR37/GPR37L1 double knockout mouse brain compared to wild-type mouse brain. We validated S100A5 as being one of the most down-regulated proteins and also found that GPR37 and GPR37L1 promote S100A5 secretion in a Ca2+-dependent manner. These proteomic analyses raise many interesting questions for future studies and provide molecular insight into how loss of function of the glia-enriched receptors GPR37 and GPR37L1 can lead to altered seizure vulnerability and other deleterious effects.
Supplementary Material
Supplemental Figure S1. Differential expression analysis of Proteomics data (DEP) with Barres Lab database
Supplemental Figure S2. Differential Expression analysis of Proteomics data (DEP) with Sharma et al. (2015) database
Supplemental Figure S3. Differential Expression analysis of Proteomics data (DEP) with Sharma-Barres combination database with endothelial cells
Supplemental Figure S4. Antibody validation for detection of S100A5
Supplemental Material S1. (xlsx) Imputed WT vs. DKO whole proteome
Supplemental Material S2. (xlsx) Null experiment calculations for imputation of proteomic data
Supplemental Material S3. (xlsx) Complete lists of significantly increased and decreased proteins
Supplemental Material S4. (xlsx) GO ontology input and Z scores
Supplemental Material S5. (xlsx) FET heatmap values of brain cell types (all databases)
Supplemental Figure S5. Uncropped Western blots of Figure 2A (S100A5 protein and mRNA levels are reduced in GPR37 knockout, GPR37L1 knockout, and GPR37/GPR37L1 double knockout mouse brains vs. WT mouse brain
Supplemental Figure S6. Uncropped Western blots of Figure 3C (Co-expression of S100A5 with either GPR37 or GPR37L1 leads to robust secretion of S100A5 from HEK293T cells)
Supplemental Figure S7. Uncropped Western blots of Figure 4A (Receptor regulation of S100A5 secretion)
Supplemental Figure S8. Uncropped Western blots of Figure 4C (Receptor regulation of S100A5 secretion)
Supplemental Figure S9. Uncropped Western blots of Figure 4D (Receptor regulation of S100A5 secretion)
Supplemental Figure S10. Uncropped Western blots of Figure 5B (Chelation of intracellular Ca2+ leads to decreased S100A5 secretion from HEK239T cells)
Supplemental Figure S11. Uncropped Western blots of Figure 5C (Chelation of intracellular Ca2+ leads to decreased S100A5 secretion from HEK239T cells)
Supplemental Figure S12. Uncropped Western blots of Figure 6A (Co-expression of homologous S100A proteins with either GPR37L1 or GPR37 also leads to secretion)
Supplemental Figure S13. Uncropped Western blots of Figure 6B (Co-expression of homologous S100A proteins with either GPR37L1 or GPR37 also leads to secretion)
ACKNOWLEDGEMENTS
We would like to acknowledge the Emory Proteomics Core, one of the Emory Core Facilities, for assisting with the proteomic analyses described here. This work was funded by NIH grants R21-NS91986, R21-NS106323, and R01-NS088413, and T.T.N. was supported by NIH training grant T32-GM008602.
ABBREVIATIONS:
- GPR37L1
G protein-coupled receptor 37 like-1
- GPR37
G protein-coupled receptor 37
- WT
wild-type
- L1KO
GPR37L1-knockout
- 37KO
GPR37-knockout
- DKO
GPR37L1/GPR37 double knockout
- MS
mass spectrometry
REFERENCES
- 1.Cahoy JD; Emery B; Kaushal A; Foo LC; Zamanian JL; Christopherson KS; Xing Y; Lubischer JL; Krieg PA; Krupenko SA; Thompson WJ; Barres BA A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci 2008, 28, 264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu B, Mosienko V, Vaccari Cardoso B, Prokudina D; Huetelman M; Teschemacher AG; Kasparov S, Glio- and neuro-protection by prosaposin is mediated by orphan G-protein coupled receptors GPR37L1 and GPR37. Glia, 2018, 66, 2414–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marazziti D; Golini E; Gallo A; Lombardi MS; Matteoni R; Tocchini-Valentini GP Cloning of GPR37, a gene located on chromosome 7 encoding a putative G-protein-coupled receptor, from a human frontal brain EST library. Genomics, 1997, 45, 68–77. [DOI] [PubMed] [Google Scholar]
- 4.Marazziti D; Gallo A, Golini E; Matteoni R; Tocchini-Valentini GP Molecular cloning and chromosomal localization of the mouse Gpr37 gene encoding an orphan G protein-coupled peptide receptor expressed in brain and testis. Genomics, 1998, 53, 315–24. [DOI] [PubMed] [Google Scholar]
- 5.Smith BM; Giddens MM; Neil J; Owino S; Nguyen TT; Duong D; Li F; Hall RA Mice lacking Gpr37 exhibit decreased expression of the myelin-associated glycoprotein MAG and increase susceptibility to demyelination. Neuroscience, 2017, 358, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valdenaire O; Giller T; Breu V; Ardati A; Schweizer A; Richards JG A new family of orphan G protein-coupled receptors predominantly expressed in the brain. FEBS Lett, 1998, 424, 193–196. [DOI] [PubMed] [Google Scholar]
- 7.Yang HJ; Vainshtein A; Maik-Rachline G; Pelas E G protein-coupled receptor 37 is a negative regulator of oligodendrocyte differentiation and myelination. Nat. Commun, 2016, 7, 10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng Z; Kui S; Hla K; Li Y A novel endothelin receptor type-B-like gene enriched in the brain. Biochem. Biophys. Res. Commun, 1997, 233, 559–567. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y; Chen K; Sloan SA; Bennett ML; Scholze AR; O’Keeffe S, Phatnani HP; Guarnieri P; Caneda C; Ruderisch N; Deng S; Liddelow SA; Zhang C; Daneman R; Maniatis T; Barres BA; Wu JQ An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci, 2014, 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rezgaoui M; Susens U; Ignatov A; Geldreblom M; Glassmeier G; Franke I; Urny J; Imai Y; Takahashi R; Schaller HC The neuropeptide head activator is a high-affinity ligand for the orphan G-protein-coupled receptor GPR37. J. Cell. Sci, 2006, 119, 542–9. [DOI] [PubMed] [Google Scholar]
- 11.Gandia J; Fernandez-Duenas V; Morato X; Caltabiano G; Gonzalez-Muniz R; Pardo L; Stagljar I; Ciruela F The Parkinson’s disease-associated GPR37 receptor-mediated cytotoxicity is controlled by its intracellular cysteine-rich domain. J. Neurochem, 2013, 125, 25–35. [DOI] [PubMed] [Google Scholar]
- 12.Meyer RC; Giddens MM; Schaefer SA; Hall RA GPR37 and GPR37L1 are receptors for the neuroprotective and glioprotective factors prosaptide and prosaposin. Proc. Natl. Acad. Sci. U.S.A, 2013, 110, 9529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bang S; Xie YK; Zhang ZJ; Wang Z; Xu ZZ; Ji RR GPR37 regulates macrophage phagocytosis and resolution of inflammatory pain. J. Clin. Invest, 2018, 128, 3568–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolly S; Bazargani N; Quiroga AC; Pringle NP; Attwell D; Richardson WD; Li H G protein-coupled receptor 37-like 1 modulates astrocyte glutamate transporters and neuronal NMDA receptors and is neuroprotective in the ischemia. Glia, 2018, 66, 47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundius EG; Vukojevic V; Hertz E; Stroth N; Cederlund A; Hiraiwa M; Terenius L; Svenningsson P GPR37 protein trafficking to the plasma membrane regulated by prosaposin and GM1 gangliosides promotes cell viability. J. Biol. Chem, 2014, 289, 4660–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southern C; Cook JM; Neetoo-Isseljee Z; Taylor DL; Kettleborough CA; Merritt A; Bassoni DL; Raab WJ; Quinn E; Wehrmann TS; Davenport AP; Brown AJ; Green A; Wigglesworth MJ; Rees S Screening β-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors. J. Biomol. Screen, 2013, 18, 599–609. [DOI] [PubMed] [Google Scholar]
- 17.Coleman JL; Ngo T; Schmidt J; Mrad N; Liew CK; Jones NM; Graham RM; Smith NJ Metalloproteinase cleavage of the N terminus of the orphan G protein-coupled receptor GPR37L1 reduces its constitutive activity. Sci Signal. 2013, 9, ra36. [DOI] [PubMed] [Google Scholar]
- 18.Marazziti D; Golini E; Mandillo S; Magrelli A; Witke W; Matteoni R; Tocchini-Valentini GP Altered dopamine signaling and MPTP resistance in mice lacking the Parkinson’s disease-associated GPR37/parkin-associated endothelin-like receptor. Proc. Natl. Acad. Sci. U.S.A, 2004, 101, 10189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marazziti D; Di Pietro C; Golini E; Mandillo S; La Sala G; Matteoni R; Tocchini-Valentini GP Precocious cerebellum development and improved motor functions in mice lacking the astrocyte cilium-, patched 1-associated Gpr37l1 receptor. Proc. Natl. Acad. Sci. U.S.A, 2013, 110, 16486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giddens MM; Wong JC; Schroeder JP; Farrow EG; Smith BM; Owino S; Soden SE; Meyer RC; Saunders C; LePichon JB; Weinshenker D; Escayg A; Hall RA GPR37L1 modulates seizure susceptibility: Evidence from mouse studies and analyses of a human GPR37L1 variant. Neurobiol Dis., 2017, 106, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dershem R; Raghu, Metpally RPR; Jeffreys K; Krishnamurthy S; Carey DJ; Hershfinkel M; Robishaw JD; Breitweiser GE Rare variant pathogenicity triage and inclusion of synonymous variants improves analysis of disease associations. bioRxiv. 2018, 272955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J; Lin ZJ; Liu L; Xu HQ; Shi YW; Yi YH; He N; Liao WP Epilepsy-associated genes. Seizure, 2017, 44, 11–20. [DOI] [PubMed] [Google Scholar]
- 23.Seyfried NT; Dammer EB; Swarup V; Nandakumar D; Duong DM; Yin L; Deng Q; Nguyen T; Hales CM; Wingo T; Glass J; Gearing M; Thambisetty M; Troncoso JC; Geschwind DH; Lah JJ; Levey AI A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s Disease. Cell Syst., 2017, 4, 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaino JA The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res, 2019, 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai J; Johnson ECB; Dammer EB; Duong DM; Gearing M, Lah JJ; Levey AI; Wingo TS; Seyfried NT. Effects of APOE genotype on brain proteomic network and cell type changes in Alzheimer’s Disease. Front Mol Neurosci., 2018, 11, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karpievitch YV; Dabney AR; Smith RD Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinf., 2012, 13, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T, Mann M; Cox J The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods, 13, 731–740. [DOI] [PubMed] [Google Scholar]
- 28.Sharma K; Schmitt S; Bergner CG; Tyanova S; Kannaiyan N; Manrique-Hoyos N; Kongi K; Cantuti L; Hanisch U; Philips M; Rossner MJ; Mann M; Simons M Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci, 2015, 18, 1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammer EB; Lee AK; Duong DM; Gearing M; Lah JJ; Levey AI; Seyfried NT Quantitative phosphoproteomics of Alzheimer’s disease reveals cross-talk between kinases and small heat shock proteins. Proteomics, 2015, 15, 508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao X; Huang X; Zhou Z; Lin X An improvement of the 2^(-delta-delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath, 2013, 3, 71–85. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X; Sun L; Bastos de Oliveira F; Qi X, Brown WJ; Smolka MB; Sun Y; Hu F Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol, 2015, 201, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X; Sun L; Bracko O; Choi JW; Jia Y; Nana AL; Brady OA; Hernandez JCC; Nishimura N; Seeley WW; Hu F Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat. Commun, 2017, 25, 15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kollmann K; Damme M; Markmann S; Morelle W; Schweizer M; Hermans-Borgmeyer I; Rochert AK; Pohl S; Lubke T; Michalski JC; Kakela R; Walkley SU; Braulke T Lysosomal dysfunction causes neurodegeneration in mucolipidosis II ‘knock-in’ mice. Brain. 2012, 135, 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishimoto Y; Hiraiwa M; O’Brien JS Saposins: structure, function, distribution, and molecular genetics. J Lipid Res., 1992, 33, 1255–1267. [PubMed] [Google Scholar]
- 35.Kolter T; Sandhoff K Lysosomal degradation of membrane lipids. FEBS Lett., 2010, 584, 1700–1712. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y; Grabowski GA Saposin C is required for normal resistance of acid beta-glucosidase to proteolytic degradation. J Biol Chem., 2003, 34, 31918–23. [DOI] [PubMed] [Google Scholar]
- 37.Di Pietro C, Marazziti D; La Sala G; Abbaszadeh Z; Golini E; Matteoni R; Tocchini-Valentini GP (2017). Primary cilia in the murine cerebellum and in mutant models of medulloblastoma. Cell Mol. Neurobiol, 2017, 37, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai Y; Soda M; Hatakeyama S; Akagi T; Hashikawa T; Nakayama KI; Takahashi R CHIP is associated with Parkin, a gene responsible for familial Parkinson’s Disease, and enhances its ubiquitin ligase activity. Mol. Cell, 2002, 10, 55–67. [DOI] [PubMed] [Google Scholar]
- 39.Marenholz I; Heizmann CW; Fritz G S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun, 2004, 322, 1111–1122. [DOI] [PubMed] [Google Scholar]
- 40.Iacobas DA; Iacobas S; Nebieridze N; Velisek L; Veliskova J Estrogen protects neurotransmission transcriptome during status epilepticus. Front. Neurosci, 2018, 12, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihara Y; Itoh K; Tanaka M; Tsuji M; Kawamoto T; Kawato S; Vogel CFA; Yamazaki T Potential of 17β-estradiol synthesis in the brain and elongation of seizure latency through dietary supplementation with docohexaenoic acid. Sci. Rep, 2017, 7, 6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mostacci B; Eposto R; Lello S; Bisulli F; Licchetta L; Tinuper P Estrogen-related seizure exacerbation following hormone therapy for assisted reproduction in women with epilepsy. Seizure, 2018, 61, 200–202. [DOI] [PubMed] [Google Scholar]
- 43.Sarfi M; Elahdadi Salmani M; Goudarzi I; Lashkar Boluki T; Abrari K Evaluating the role of astrocytes on β-estradiol effect on seizures of Pilocarpine epileptic model. Eur. J. Pharmacol, 2017, 15, 32–38. [DOI] [PubMed] [Google Scholar]
- 44.Sato SM; Woolley CS Acute inhibition of neurosteroid estrogen synthesis suppresses status epilepticus in an animal model. Elife., 2016, 15, e12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramanujam B; Arora A; Malhotra V; Dash D; Mehta S; Tripathi M A case of recurrent status epilepticus and successful management with progesterone. Epileptic Disord., 2016, 18, 101–5. [DOI] [PubMed] [Google Scholar]
- 46.Wu YV; Burnham WM The anti-seizure effects of IV 5α-dihydroprogesterone on amygdala-kindled seizures in rats. Epilepsy Res., 2018, 146, 132–136. [DOI] [PubMed] [Google Scholar]
- 47.Schafer BW; Fritschy JM; Murmann P; Troxler H; Durussel I; Heizmann CW; Cox JA Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. J Biol. Chem, 2000, 275, 30623–30. [DOI] [PubMed] [Google Scholar]
- 48.Camby I; Nagy N; Lopes MB; Schafer BW; Maurage CA; Ruchoux MM; Murmann P; Pochet R; Heizmann CW; Brotchi J; Salmon I; Kiss R; Decaestecker C Supratentorial pilocytic astrocytomas, astrocytomas, anaplastic astrocytomas and glioblastomas are characterized by a differential expression of S100 proteins. Brain Pathol., 1999, 9, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camby I; Lefranc F; Titeca G; Neuci S; Fastrez M; Dedecken L; Schafer BW; Brotchi J; Heizmann CW; Pochet R; Salmon I; Kiss R; Decaestecker C Differential expression of S100 calcium-binding proteins characterizes distinct clinical entities in both WHO grade II and III astrocytic tumors. Neuropathol. Appl. Neurobiol, 2000, 26, 76–90. [DOI] [PubMed] [Google Scholar]
- 50.Dmytriyeva O; Panktratova S; Owczarek S; Sonn K; Soroka V; Ridley CM; Marsolais A; Lopez-Hoyos M; Ambartsumian N; Lukanidin E; Bock E; Berezin V; Kiryushko D The metastasis-promoting S100A4 protein confers neuroprotection in brain injury. Nat. Commun, 2012, 3, 1197. [DOI] [PubMed] [Google Scholar]
- 51.Bartowska K; Swiatek I; Aniszewska A; Jurewicz E; Turlejski K; Filipek A; Djavadian RL Stress-dependent changes in the CacyBP/SIP interacting protein S100A6 in the mouse brain. PLoS One, 2016, 12, e0169760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donato R; Sorci G; Giambanco I S100A6 protein: functional roles. Cell Mol. Life Science, 2017, 74, 2749–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lesniak W; Wilanowski T; Filipek A S100A6 – focus on recent developments. Biol. Chem, 2017, 398, 1087–1094. [DOI] [PubMed] [Google Scholar]
- 54.Kubista H; Donato R; Hermann A S100 calcium binding protein affects neuronal electrical discharge activity by modulation of potassium currents. Neuroscience, 1999, 90, 493–508. [DOI] [PubMed] [Google Scholar]
- 55.Melani R; Rebaudo R; Balestrino M; Cupello A; Haglid K; Hyden H Involvement of S-100 protein in anoxic long-term potentiation. Brain Res., 1999, 840, 171–174. [DOI] [PubMed] [Google Scholar]
- 56.Nikitin VP; Kozyrev SA; Shevelkin AV The effects of antibodies against proteins of the S100 group on neuron plasticity in sensitized and non-sensitized snails. Neurosci. and Behav.l Physiol, 2002, 32, 25–31. [DOI] [PubMed] [Google Scholar]
- 57.Nishiyama H; Knopfel T; Endo S; Itohara S Glial protein S100B modulates long-term neuronal synaptic plasticity. PNAS., 2001, 99, 4037–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rebaudo R; Melani R; Balestrino M; Cupello A; Haglid K; Hyden H Antiserum against S-100 protein prevents long-term potentiation through a cAMP-related mechanism. Neurochem. Res, 2000, 25, 541–545. [DOI] [PubMed] [Google Scholar]
- 59.Sakatani S; Seto-Ohshima A; Shinohara Y; Yamamoto Y; Yamamoto H; Itohara S; Hirase H Neural-activity-dependent release of S100B from astrocytes enhances kainite-induced gamma oscillations in vivo. J Neurosci., 2008, 28, 10928–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davey GE; Murmann P; Heizmann CW Intracellular Ca2+ and Zn2+ levels regulate the alternative cell density-dependent secretion of S100B in human glioblastoma cells. J. Biol. Chem, 2001, 276, 30819–30826. [DOI] [PubMed] [Google Scholar]
- 61.Gerlach R; Demel G; Konig HG; Gross U; Prehn JH; Raabe A; Seifert V; Kogel D Active secretion of S100B from astrocytes during metabolic stress. Neuroscience, 2006, 141, 1697–701. [DOI] [PubMed] [Google Scholar]
- 62.Guerra MC; Tortorelli LS; Galland F; Da Re C; Negri E; Engelke DS; Rodrigues L; Leite MC; Goncalves CA Lipopolysaccharide modulates astrocytic S100B secretion: a study in cerebrospinal fluid and astrocyte cultures from rats. J. Neuroinflammation, 2011, 8, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tramontina AC; Tramontina F; Bobermin LD; Zanotto C; Souza DF; Leite MC; Nardin P; Gottfried C; Goncalves CA Secretion of S100B, an astrocyte-derived neurotrophic protein, is stimulated by fluoxetine via a mechanism independent of serotonin. Prog Neuropsychpharmacol. Biol. Psychiatry, 2008, 32, 1580–3. [DOI] [PubMed] [Google Scholar]
- 64.Kathir KM; Ibrahim K; Rajalingam D; Prudovsky I; Yu C; Kumar TK S100A13-lipid interactions-role in the non-classical release of the acidic fibroblast growth factor. Biochim. Biophys. Acta, 2007, 1768, 3080–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zolese G; Tangorra A; Curatola G; Giambanco I; Donato R Interaction of S-100b protein with cardiolipin vesicles as monitored by electron spin resonance, pyrene fluorescence and circular dichroism. Cell Calcium, 1988, 9, 149–157. [DOI] [PubMed] [Google Scholar]
- 66.Bhattachara S; Bunick CG; Chazin WJ Target selectivity in EF-hand calcium binding proteins. Biochim. Biophys. Acta, 2004, 1742, 69–79. [DOI] [PubMed] [Google Scholar]
- 67.Zimmer DB; Weber DJ The calcium-dependent interaction of S100B with its target protein targets. Cardiovasc. Psychiatry Neurol, 2010, doi: 10.1155/2010/728052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Velasquez E; Nogueira FCS; Velasquez I; Schmitt A; Falkai P; Domont GB; Martins-de-Souza D Synaptosomal proteome of the orbitofrontal cortex from Schizophrenia patients using quantitative label-free and iTRAQ-based shotgun proteomics. J. Proteome Res, 2017, 16, 4481–4494. [DOI] [PubMed] [Google Scholar]
- 69.Eshraghi M; Gombar R; De Repentigny Y; Vacratsis PO; Kothary R Pathologic alterations in the proteome of synaptosomes from a mouse model of Spinal Muscular Atrophy. J. Proteome Res, 2019, 18, 3042–3051. [DOI] [PubMed] [Google Scholar]
- 70.Kong D; Tian X; Li Y; Zhang S; Cheng Y; Huo L; Ma H; Yang Z; Ren L; Zhang M; Zhang W Revealing the inhibitory effect of ginseng on mitochondrial respiration through synaptosomal proteomics. Proteomics, 2018, 18, p.e1700354. [DOI] [PubMed] [Google Scholar]
- 71.Donato R The specific interaction of S-100 protein with synaptosomal particulate fraction. Evidence for the formation of a tight complex between S-100 and its binding sites. J Neurochem., 1981, 36, 532–7. [DOI] [PubMed] [Google Scholar]
- 72.Curatola G; Mazzanti L; Ferretti G; Donato R S-100 protein-induced changes in the physical state of synaptosomal particulate fractions as monitored by spin labels. Arch Biochem Biophys., 1985, 240, 435–45. [DOI] [PubMed] [Google Scholar]
- 73.Donato R; Michetti F Specific binding sites for S-100 protein in isolated brain nuclei. J Neurochem., 36, 1698–1705. [DOI] [PubMed] [Google Scholar]
- 74.Starostina MV; Nikolaenkova AA; Malup TK; Korochkin LI; Sviridov SM Quantitative assay of S-100 protein in mouse brain cortex synaptosomes. Cell Mol Neurobiol., 13, 677–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Starostina MV; Sviridov SM Quantitative determination of neurospecific protein S-100 in mouse brain cortical synaptosomes. Biokhimiia, 46, 2030–42. [PubMed] [Google Scholar]
- 76.Cocchia D; Pansera F; Palumbo A; Donato R Immunocytochemical localization of S-100 protein binding sites in synaptosomal fractions and subfractions. Cell Mol. Neurobiol, 1982, 2, 265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Differential expression analysis of Proteomics data (DEP) with Barres Lab database
Supplemental Figure S2. Differential Expression analysis of Proteomics data (DEP) with Sharma et al. (2015) database
Supplemental Figure S3. Differential Expression analysis of Proteomics data (DEP) with Sharma-Barres combination database with endothelial cells
Supplemental Figure S4. Antibody validation for detection of S100A5
Supplemental Material S1. (xlsx) Imputed WT vs. DKO whole proteome
Supplemental Material S2. (xlsx) Null experiment calculations for imputation of proteomic data
Supplemental Material S3. (xlsx) Complete lists of significantly increased and decreased proteins
Supplemental Material S4. (xlsx) GO ontology input and Z scores
Supplemental Material S5. (xlsx) FET heatmap values of brain cell types (all databases)
Supplemental Figure S5. Uncropped Western blots of Figure 2A (S100A5 protein and mRNA levels are reduced in GPR37 knockout, GPR37L1 knockout, and GPR37/GPR37L1 double knockout mouse brains vs. WT mouse brain
Supplemental Figure S6. Uncropped Western blots of Figure 3C (Co-expression of S100A5 with either GPR37 or GPR37L1 leads to robust secretion of S100A5 from HEK293T cells)
Supplemental Figure S7. Uncropped Western blots of Figure 4A (Receptor regulation of S100A5 secretion)
Supplemental Figure S8. Uncropped Western blots of Figure 4C (Receptor regulation of S100A5 secretion)
Supplemental Figure S9. Uncropped Western blots of Figure 4D (Receptor regulation of S100A5 secretion)
Supplemental Figure S10. Uncropped Western blots of Figure 5B (Chelation of intracellular Ca2+ leads to decreased S100A5 secretion from HEK239T cells)
Supplemental Figure S11. Uncropped Western blots of Figure 5C (Chelation of intracellular Ca2+ leads to decreased S100A5 secretion from HEK239T cells)
Supplemental Figure S12. Uncropped Western blots of Figure 6A (Co-expression of homologous S100A proteins with either GPR37L1 or GPR37 also leads to secretion)
Supplemental Figure S13. Uncropped Western blots of Figure 6B (Co-expression of homologous S100A proteins with either GPR37L1 or GPR37 also leads to secretion)


