Abstract
Background
The aim was to evaluate the association between the LINC00520 genetic polymorphisms and breast cancer (BC) susceptibility.
Methods
Nine single‐nucleotide polymorphisms (SNPs) on LINC00520 genotyping were performed in 504 BC patients and 505 cancer‐free controls in Chinese Han population to study the relationship between LINC00520 polymorphism and BC susceptibility. qRT‐PCR and luciferase tests were used to explore how rs12880540 affected the expression of LINC00520.
Results
The genotype GG (OR:3.58, 95%CI:1.32‐9.69) in rs8012083 increased the risk of triple‐negative BC. The genotype GG (OR:0.31, 95%CI:0.14‐0.69) in rs8012083, the genotype AA (OR:2.74, 95%CI:1.01‐7.42) in rs2152275, and genotype TG (OR:1.62, 95%CI:1.04‐2.52) in rs12880540 were associated with HER‐2 status. The dominant (OR:0.65, 95%CI:0.45‐0.95) and overdominant genetic model (OR:0.67, 95%CI:0.46‐0.98) consistently showed that rs11622641 T was significantly associated with lower risk of BC. Similarly, the recessive genetic model (OR:1.57, 95%CI:1.07‐2.30) of rs12880540 and the dominant (OR:1.62, 95%CI:1.24‐2.11) and overdominant (OR:1.56, 95%CI:1.19‐2.03) genetic model of rs2152278 may increase the risk of BC. The relative expression of LINC00520 increased linearly with the increase in the number of rs12880540 mutations. rs12880540 alleles were due to the interaction between LINC00520 and miR‐3122 at T, but the mutation of rs12880540 G > T had no effect on the binding ability of LINC00520 and miR‐3122.
Conclusion
A genetic variant of rs8012083 in LINC00520 may be used as a biomarker for triple‐negative BC after further evaluation of diagnostic tests. The genetic variant of LINC00520 was related to the susceptibility of BC, and rs12880540 might affect the corresponding mRNA expression of lncRNA LINC00520.
Keywords: breast cancer, LINC00520, lncRNA, single‐nucleotide polymorphisms, susceptibility
In our epidemiology study, nine single‐nucleotide polymorphisms (SNPs) in lncRNA LINC00520 were selected for genotyping among 504 breast cancer cases and 505 healthy controls. The 504 cases are all new breast cancer patients which were pathologically confirmed and the controls were from an epidemiological survey of cardiovascular disease in Henan Province with 20 000 participants and they were included without a history of cancer or other diseases. A genetic variant of rs8012083 in LINC00520 could be used as a biomarker for triple‐negative BC. The genetic variants of lncRNA LINC00520 were associated with the susceptibility of BC, and rs12880540 might affect the corresponding mRNA expression of lncRNA LINC00520.
1. INTRODUCTION
Breast cancer (BC) is the second most malignant tumor in the world, ranked after lung cancer. BC remains the leading cause of cancer deaths in Chinese women, placing a huge burden on public health.1 In China, new BC cases accounted for 12.2% of all new cancer cases and 9.6% of all deaths.2 With the Chinese population increasing and accelerated aging, the number of new BC cases has been continually rising in recent years and the age of onset tended to be younger.3 The high incidence and mortality of BC in China have become a public problem that seriously threatened women's physical and mental health.4 Finding the specific susceptibility biomarker and identifying individuals with higher risk of BC could be helpful to improve the early diagnostic rate of BC.
With the development of high‐throughput technology, emerging evidence demonstrated that genetic alterations, including single‐nucleotide polymorphisms (SNPs), insertions, deletions, and copy number variations, were associated with the risk of various diseases including BC.5, 6 Notably, SNPs, especially in irregular regions of protein‐coding and related with diseases, have been extensively studied and reported widely.7 The expression of the same lncRNA transcripts varies by different health conditions, age, tissues, even cells, which can indicate their potential as possible biomarkers and be predictive of diagnosis and prognosis of diseases.8, 9 Enormous studies have proven that lncRNAs were associated with tumorgenesis, progression, metastasis, and drug resistance, functioning as oncogenes, tumor suppressor genes, or both, involving in shaping multiple biological characteristics of cancer at transcriptional, posttranscriptional, and epigenetic levels.10, 11, 12 In addition, the abnormal expression levels of different lncRNAs with oncogenic and tumor suppressor functions have been frequently found in the development of BC.13 Particularly, adverse rules of miRNAs‐sponging lncRNAs, involved in ceRNA networks (ceRNETs), have been shown in BC.14 Eades discovered lincRNA‐ROR/miR‐145/ARF6 mRNA networks interacting with metastasis and prognosis in triple‐negative BC in 2015.15
Long intergenic nonprotein coding RNA 520 (LINC00520) is located at human chromosome 14q22.3. LINC00520, a highly conserved long‐chain noncoding RNA with a length of about 20kb, is widely expressed in various tissues. A study reported that LINC00520 was overexpressed in laryngeal squamous cell carcinoma tissue.16 The upregulation of LINC00520 expression might play an important biological role in the metastasis of laryngeal squamous cell carcinoma. The expression of LINC00520 was upregulated in primary nasopharyngeal carcinoma.17 In addition, the level of LINC00520 was significantly increased in renal cell carcinoma.18 LINC00520 promoted the migration of BC cells induced by Src, STAT3, and PI3K and played a functional role in BC.19 Although it was reported that the lncRNA LINC00520 was associated with BC, the complexity of its function was also determined by the complexity of the lncRNAs structure. However, the association among LINC00520 SNPs and the susceptibility of BC and its mechanism is not clear yet. In this study, the combined database, literature retrieval, and bioinformatics techniques were used to screen lncRNA and its SNP site associated with BC. The association between long noncoding RNA genetic variation and BC susceptibility was studied, as well as biological function was explored by the method of molecular epidemiology.
2. MATERIALS AND METHODS
2.1. Study subjects
A total of 1009 subjects were enrolled in the study, including 504 cases with BC and 505 age‐matched (±2 years) healthy controls (Table 1). Subjects were included in accordance with strict standards: (a) Patients with BC must have a definite pathological diagnosis of new cases; (b) The patients with BC were selected from the First Affiliated Hospital and the Third Affiliated Hospital of Zhengzhou University from 2013 to 2015; (c) Patients with BC have never received radiotherapy or chemotherapy or any other treatment before; (d) Except BC, there were no other diseases in all cases.
Table 1.
Basic characteristics of 504 breast cancer cases and 505 healthy controls
| Variables | Cases (%) | Controls (%) | P a | OR (95%CI) |
|---|---|---|---|---|
| n = 504 | n = 505 | |||
| Age (mean ± SD) | 48.00 ± 9.85 | 48.15 ± 9.61 | .906b | |
| Age at menarche (mean ± SD) | 14.21 ± 1.70 | 13.97 ± 1.75 | .030 b | |
| Menopause state | ||||
| Premenopause | 320 (63.5) | 294 (58.2) | 1.000 | |
| Postmenopause | 184 (36.5) | 211 (41.8) | .086 | 0.801 (0.622‐1.032) |
| Age at menopause | ||||
| ≤50 | 129 (70.1) | 146 (69.2) | 1.000 | |
| >50 | 55 (29.9) | 65 (30.8) | .844 | 0.958 (0.623‐1.472) |
| Number of pregnancies | ||||
| ≤1 | 53 (10.5) | 94 (18.6) | 1.000 | |
| 2 | 129 (25.6) | 177 (35.0) | .215 | 1.293 (0.861‐1.940) |
| 3 | 135 (26.8) | 128 (25.3) | .262 | 1.898 (0.620‐5.816) |
| ≥4 | 187 (37.1) | 106 (21.0) | .043 | 3.175 (1.037‐9.722) |
| Number of abortion | ||||
| ≤1 | 340 (67.5) | 345 (68.3) | 1.000 | |
| 2 | 105 (20.8) | 117 (23.2) | .545 | 0.911 (0.673‐1.233) |
| 3 | 35 (6.9) | 30 (5.9) | .735 | 0.913 (0.537‐1.551) |
| ≥4 | 24 (4.8) | 13 (2.6) | .308 | 1.444 (0.713‐2.926) |
| Breastfeeding | ||||
| No | 121 (24.0) | 94 (18.6) | 1.000 | |
| Yes | 383 (76.0) | 411 (81.4) | .037 | 0.724 (0.535‐0.980) |
| Family history | ||||
| No | 461 (91.5) | 481 (95.2) | 1.000 | |
| Yes | 43 (8.5) | 24 (4.8) | .017 | 1.869 (1.116‐3.130) |
| ER | ||||
| Negative | 149 (30.3) | |||
| Positive | 342 (69.7) | |||
| PR | ||||
| Negative | 191 (39.1) | |||
| Positive | 298 (60.9) | |||
| HER‐2 | ||||
| Negative | 136 (29.3) | |||
| Positive | 328 (70.7) | |||
Abbreviations: ER, estrogen receptor; HER‐2, human epidermal growth factor receptor‐2; PR, progesterone receptor.
Two‐sided Chi‐square test, P < .05 was statistically different.
Student's t test.
The italic values represent P < .05.
The control was collected from the site of an epidemiological survey of cardiovascular disease involving more than 20 000 people in Henan Province during the same time. The healthy controls were matched with the BC group according to the age (±2 years), who were healthy without any history of cancer or any other chronic disease. None of the participants had any affinity.
All subjects volunteered to participate in the study and signed informed consent. The study has been approved by the Medical and Health Research Ethics Committee of Zhengzhou University.
2.2. Data collection
The data of the patients with BC were obtained through medical records of the hospital, including names, ages, nationality, place of origin, menarche age, menopause status, whether there was a history of breastfeeding and childbearing, cancer history of first‐degree relatives (parents, siblings, and children), second‐degree relatives (grandparents, uncles, aunts), along with estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor‐2 (Her‐2).
The data of controls were collected by face‐to‐face questionnaire designed by experts, asked by trained investigators. The content of the data was the same as BC cases. All the collected information was entered parallel with EpiData software. The whole process of investigation was carried out under strict quality control, including collection, collation, and entry of information.
2.3. DNA extraction
Peripheral blood (5 mL) was collected from each subject using an anticoagulation vessel containing ethylenediaminetetraacetic acid (EDTA) for DNA extraction and genotyping. The genomic DNA was extracted using genomic DNA extraction kit (DK601‐02 centrifugal column type) of Shanghai Lifefeng Biotech Co., Ltd and the operation process was carried out in accordance with the instructions strictly. All the extracted genomic DNA was labeled, measured, and finally stored in −80°C refrigerator for future use.
2.4. SNPs selection and genotyping
LncRNA database (http://www.lncrnadb.org/), NCBI database (https://www.ncbi.nlm.nih.gov/gene/), Ensembl database (http://asia.ensembl.org/Homo_sapiens/Info/Index), and 1000 Genomes Browser (http://browser.1000genomes.org/index.html) were used to determine the location of LINC00520. We selected the tagger SNPs in region of LINC00520 by lncRNASNP2 database (http://bioinfo.life.hust.edu.cn/lncRNASNP) based on Chinese Han population data of HapMap Project (HapMap Rel28, NCBI B36) with the criteria of a minor allele frequency (MAF) higher than 0.1 in Chinese Han population and the pairwise correlation was set as r 2 less than .8, which can reflect the degree of chain reaction between SNP. Secondly, we screened the SNP located in the Promoter Flanking Region and Enhancer regions of LINC00520. Then, lncRNASNP2 (http://bioinfo.life.hust.edu.cn/lncRNASNP) database was used to predict the potential miRNA binding sites of the selected SNPs. RNAfold website (http://rna.tbi.univie.ac.at/cgibin/RNAWebSuite/RNAfold.cgi) was utilized to predict the influence of the selected SNPs in LINC00520 on the secondary structure of LINC00520 (Table S1). Finally, nine SNPs (rs11622641, rs7157819, rs12880540, rs2152275, rs4144657, rs2152278, rs7142488, rs8008130, and rs8012083) in LINC00520 were selected for further research (Table S2).
In this study, rs11622641, rs7157819, rs12880540, and rs2152275 located in LINC00520 were genotyped by SNPscan. Because of the low score of other SNPs, SNPscan method was not suitable. Polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) technique was used for genotyping of rs4144657, rs2152278, and rs7142488 in LINC00520, while rs8008130 and rs8012083 using created restriction site PCR (CRS‐RFLP). While there was no suitable restriction endonuclease near the mutation site, we needed to change a base on the primer so that a new restriction site could be formed with the base of the mutation site, thus CRS‐RFLP method was used. Primers for PCR amplification were designed using primer 6.0 software. NCBI BLAST website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to evaluate the specificity of primers. Gradient PCR technique was used to standardize the conditions of DNA amplification and optimize the annealing temperature for the primers set. The suitable restriction enzymes were selected by WatCut website (http://watcut.uwaterloo.ca/template.php; Table S3). Moreover, approximately 5% of samples were randomly selected for Sanger sequencing, yielding a 100% concordance rate between the two methods.
2.5. Quantitative real‐time PCR analysis
The relative expression of LINC00520 was detected by the Eco Real‐Time PCR System (Illumina). The primers of LINC00520 and GAPDH were synthesized by TaKaRa Biotechnology Co. Ltd. The reaction conditions of qRT‐PCR were performed in strict accordance with the following instructions. All specific primers used in quantitative RT‐PCR were shown below, LINC00520 primer: 5′‐AAGCAGGACACAATTACAAC‐3′ and 5′‐GCAGGTCCGAGGTATTCGTC‐3′, GAPDH primer: 5′‐CGGAGTCAACGGATTTGGTCGTAT‐3′ and 5′‐AGCCTTCTTCATGGTGGTGAAGAC‐3′. All samples were independently measured in triplicate. Finally, the relative expression of the gene was the average of three replicates per sample. The relative expression of LINC00520 was calculated by 2−ΔΔCT method, and GAPDH was used as the internal reference gene (X ± SD).20
2.6. Cell culture, transfection, and luciferase reporter assay
To further examine whether rs12880540 could have impact on the binding of miR‐3122 and LINC00520, predicted by lncRNASNP2 database (Figure S1), 293T cell lines were obtained and grown in DMEM supplemented with 10% fetal bovine serum (GIBCO) in a humidified atmosphere of 5% CO2 at 37°C. All cell lines were never subcultured for more than 3 months and sequenced by DNA using the Applied Biosystems Amp F/STR Identifier kit and last performed in September 2018. The miR‐3122 mimics and negative control (NC) were synthesized in Genecreate. For transfection assays, 293T cells were seeded in 48‐well plates and simultaneously transfected with PGL3‐basic‐luc vector and miR‐3122 or NC mimics using Lipofectamine 3000 (Invitrogen). After 48 hours, cells were harvested, and Renilla luciferase/Firefly luciferase activities between different alleles were detected and analyzed according to the manufacturer's instruction (dual luciferase assay system, Promega). All experiments were performed independently in triplicate.
2.7. Quality control
Negative control was performed for each batch of PCR amplification to avoid false positive of sample contamination. About 5% of samples were randomly selected for sequencing to ensure the reliability of genotyping results. Figure S2 showed the PCR‐RFLP and CRS‐RFLP techniques, while the sequencing results were shown in Figure S3. The qRT‐PCR experiments were repeated three times independently to reduce the measurement error.
2.8. Statistical analysis
The sample size of the study (n = 460) was calculated using Power Analysis and Sample Size (PASS) software based on study power (0.9) and minimal alleles frequency (0.1). Student's t test was used for continuous normal distribution variables and Chi‐square test was used to classify variables. Hardy‐Weinberg equilibrium (HWE) was examined to compare the observed genotype frequencies of cancer‐free controls with the expected genotype frequencies by goodness‐of‐fit Chi‐square test. Odds ratios (ORs) and its 95% confidence intervals (CIs) were calculated to evaluate the association between SNPs and BC susceptibility adjusted for potential confounding factors, including age, age of menarche, age of menopause, menopausal status, pregnancy number, abortion number, breastfeeding history, and family history of BC in first‐degree relatives. MDR 2.0 software was used to analyze interaction between polymorphic sites in lncRNA gene and reproductive factors. SHEsis (http://analysis.bio-x.cn/myAnalysis.php) was conducted to calculate the difference of haplotype frequencies in both patients and controls. The input of data was carried out by EpiData software with double parallel entry. The statistical analysis was performed by SPSS 23.0 software.
3. RESULTS
3.1. Characteristics of the participants
The average age of BC cases was 48.00 years (±9.85), while that of the control group was 48.15 years (±9.61). No statistical differences were found in the distribution of age, menopause state, age at menopause, and number of abortion between the BC cases and cancer‐free controls (all P > .05). The mean age at menarche was 14.21 years (±1.70) in BC patients and 13.97 years (±1.75) in controls (P = .03). Breastfeeding was a protective factor (OR:0.73, 95%CI:0.54‐0.98), while carrying family history of tumor was a risk factor for BC cases (OR:1.87, 95%CI:1.12‐3.13). Approximately, half of the patients were positive in ER, PR, and Her‐2 (342 [69.7%], 298 [60.9%], and 328 [70.7%], respectively).
3.2. The associations between SNP genotypes and risk of BC
As indicated in Table 2, the genotype distributions of the nine SNPs in control group all conformed to HWE (P > .05). The relationship between SNPs and BC risk based on different genetic models is summarized in Table 2. When after adjusting the confounding factors in logistic regression analysis, the unconditional multiple logistic regression based on the dominant (OR:0.65, 95%CI:0.45‐0.95) and overdominant genetic model (OR:0.67, 95%CI:0.46‐0.98) consistently showed that rs11622641 T was significantly associated with lower risk of BC. Similarly, the recessive genetic model (OR:1.57, 95%CI:1.07‐2.30) also showed that the individuals with 12880540 G had increased risk of BC. The dominant (OR:1.62, 95%CI:1.24‐2.11) and overdominant (OR:1.56, 95%CI:1.19‐2.03) genetic model also revealed that rs2152278 T could increase the risk of BC.
Table 2.
Association between nine SNPs of LINC00520 and breast cancer susceptibility
| SNPs genetic model | Genotype | Cases (%) | Controls (%) | P a | P b | OR (95%CI) | P c |
|---|---|---|---|---|---|---|---|
| n = 504 | n = 505 | ||||||
| rs11622641 | .030 | .995 | |||||
| Codominant | CC | 445 (88.3) | 417 (82.6) | Reference | 1.000 | ||
| CT | 57 (11.3) | 84 (16.6) | .014 | 0.666 (0.453‐0.979) | .039 | ||
| TT | 2 (0.4) | 4 (0.8) | .439 | 0.359 (0.056‐2.289) | .278 | ||
| Dominant | CT + TT/CC | 0.650 (0.445‐0.949) | .026 | ||||
| Recessive | TT/CC + CT | 0.375 (0.059‐2.393) | .299 | ||||
| Overdominant | CT/CC + TT | 0.670 (0.455‐0.984) | .041 | ||||
| rs7157819 | .556 | .821 | |||||
| Codominant | CC | 375 (74.4) | 391 (77.4) | Reference | 1.000 | ||
| CT | 119 (23.6) | 105 (20.8) | .288 | 1.054 (0.768‐1.445) | .746 | ||
| TT | 10 (2.0) | 9 (1.8) | .819 | 0.971 (0.372‐1.445) | .952 | ||
| Dominant | CT + TT/CC | 1.047 (0.770‐1.423) | .770 | ||||
| Recessive | TT/CC + CT | 0.959 (0.369‐2.492) | .931 | ||||
| Overdominant | CT/CC + TT | 1.054 (0.769‐1.445) | .742 | ||||
| rs12880540 | .069 | .460 | |||||
| Codominant | TT | 208 (41.3) | 218 (43.2) | Reference | 1.000 | ||
| TG | 222 (44.0) | 237 (46.9) | .893 | 1.025 (0.776‐1.354) | .863 | ||
| GG | 74 (14.7) | 50 (9.9) | .041 | 1.551 (1.034‐2.328) | .034 | ||
| Dominant | TG + GG/TT | 1.098 (0.843‐1.429) | .488 | ||||
| Recessive | GG/ TG + TT | 1.566 (1.069‐2.295) | .021 | ||||
| Overdominant | TG/GG + TT | 0.948 (0.730‐1.232) | .690 | ||||
| rs2152275 | .886 | .680 | |||||
| Codominant | TT | 271 (53.8) | 265 (52.5) | Reference | 1.000 | ||
| TA | 199 (39.5) | 207 (41.0) | .646 | 1.001 (0.763‐1.315) | .992 | ||
| AA | 34 (6.7) | 33 (6.5) | .997 | 1.128 (0.658‐1.933) | .661 | ||
| Dominant | TA + AA/TT | 1.018 (0.784‐1.322) | .892 | ||||
| Recessive | AA/TA + TT | 1.127 (0.667‐1.906) | .655 | ||||
| Overdominant | TA/AA + TT | 0.988 (0.758‐1.289) | .930 | ||||
| rs8008130 | .702 | .644 | |||||
| Codominant | CC | 418 (82.9) | 427 (84.6) | Reference | 1.000 | ||
| CA | 79 (15.7) | 73 (14.5) | .598 | 1.165 (0.810‐1.674) | .411 | ||
| AA | 7 (1.4) | 5 (1.0) | .575 | 1.448 (0.410‐5.111) | .565 | ||
| Dominant | CA + AA/CC | 1.182 (0.831‐1.681) | .352 | ||||
| Recessive | AA/CA + CC | 1.418 (0.403‐4.997) | .586 | ||||
| Overdominant | CA/AA + CC | 0.862 (0.600‐1.239) | .424 | ||||
| rs4144657 | .252 | .122 | |||||
| Codominant | CC | 147 (29.2) | 126 (25.0) | Reference | 1.000 | ||
| CT | 267 (53.0) | 275 (54.5) | .235 | 0.859 (0.632‐1.168) | .333 | ||
| TT | 90 (17.9) | 104 (20.6) | .133 | 0.756 (0.511‐1.117) | .160 | ||
| Dominant | CT + TT/CC | 1.204 (0.897‐1.615) | .216 | ||||
| Recessive | TT/CC + CT | 0.836 (0.599‐1.167) | .292 | ||||
| Overdominant | CT/CC + TT | 0.964 (0.742‐1.253) | .785 | ||||
| rs2152278 | .001 | .080 | |||||
| Codominant | GG | 180 (35.7) | 238 (47.1) | Reference | 1.000 | ||
| GT | 250 (49.6) | 194 (38.4) | <.001 | 1.700 (1.278‐2.261) | <.001 | ||
| TT | 74 (14.7) | 73 (14.5) | .148 | 1.398 (0.939‐2.080) | .099 | ||
| Dominant | GT + TT/GG | 1.619 (1.239‐2.114) | <.001 | ||||
| Recessive | TT/GT + GG | 1.062 (0.735‐1.535) | .749 | ||||
| Overdominant | GT/TT + GG | 1.556 (1.194‐2.026) | .001 | ||||
| rs8012083 | .104 | .522 | |||||
| Codominant | AA | 288 (57.1) | 311 (61.6) | Reference | 1.000 | ||
| AG | 185 (36.7) | 176 (34.9) | .351 | 1.127 (0.855‐1.485) | .398 | ||
| GG | 31 (6.2) | 18 (3.6) | .053 | 1.666 (0.886‐3.133) | .113 | ||
| Dominant | AG + GG/AA | 1.179 (0.904‐1.539) | .224 | ||||
| Recessive | GG/AG + AA | 1.593 (0.855‐2.970) | .143 | ||||
| Overdominant | AG + GG + AA | 1.085 (0.826‐1.424) | .559 | ||||
| rs7142488 | .364 | .981 | |||||
| Codominant | TT | 410 (81.3) | 415 (82.2) | Reference | 1.000 | ||
| TC | 93 (18.5) | 86 (17.0) | .621 | 1.157 (0.823‐1.626) | .402 | ||
| CC | 1 (0.2) | 4 (0.8) | .374 | 0.241 (0.026‐2.209) | .208 | ||
| Dominant | TC + CC/TT | 1.111 (0.794‐1.555) | .539 | ||||
| Recessive | CC/TC + TT | 0.235 (0.026‐2.146) | .199 | ||||
| Overdominant | TC/CC + TT | 1.166 (0.830‐1.639) | .375 |
P value of Chi‐square test for comparison of genotype frequency between case and control groups.
P value of Hardy‐Weinberg test in control population.
P values adjusted for age, menarche age, menopausal status, number of pregnancies, number of abortions, history of breastfeeding, and family history of breast cancer in first‐degree relatives in logistic regression analysis.
The italic values represent P < .05.
3.3. Stratified analysis of SNP genotypes and BC risk
We also examined the effect of SNPs on BC susceptibility in different subgroups of demographic factors (Table 3). CT + TT in rs11622641 was a protective genotype of BC susceptibility in females with age less than or equal to 50 years (OR:0.55, 95%CI:0.33‐0.92), age at menarche less than 14 years (OR:0.55, 95%CI:0.34‐0.88), number of pregnancy more than twice (OR:0.60, 95%CI:0.36‐0.98), and number of abortion less than twice (OR:0.64, 95%CI:0.43‐0.94). BC risk increased in individuals carrying rs7157819 CT + TT and having had two or more abortions (OR:4.19; 95%CI:1.14‐9.49). Subjects at greater risk, who had a younger menopause age (≤50 years), carried variant genotypes TG + GG of rs12880540 (OR:1.80, 95%CI:1.06‐3.07). For CT + TT of rs4144657, the risk of BC was decreased in population who had no menopause (OR:0.65, 95%CI:0.44‐0.95) or without a family history of BC (OR:0.23, 95%CI:0.06‐0.87). The genotype GT + GG of rs2152278 increased the risk of BC in population who was less than 50 years (OR:1.63, 95%CI:1.16‐2.29), age at menarche less than 14 years (OR:1.52, 95%CI:1.10‐2.11), no menopause (OR:1.67, 95%CI:1.18‐2.36).
Table 3.
Stratified analysis of nine SNPs in LINC00520 and genetic susceptibility to breast cancer
| rs11622641 (CT + TT/CC) | rs7157819 (CT + TT/CC) | rs12880540 (TG + GG/TT) | rs2152275 (TA + AA/TT) | |||||
|---|---|---|---|---|---|---|---|---|
| OR (95%CI)a | P a | OR (95%CI)a | P a | OR (95%CI)a | P a | OR (95%CI)a | P a | |
| Age (year) | ||||||||
| ≤50 | 0.549 (0.327‐0.923) | .024 | 1.084 (0.731‐1.606) | .689 | 1.029 (0.738‐1.433) | .868 | 0.944 (0.678‐1.316) | .735 |
| >50 | 0.743 (0.419‐1.315) | .307 | 1.155 (0.690‐1.934) | .583 | 1.215 (0.770‐1.915) | .403 | 1.174 (0.754‐1.827) | .478 |
| Age at menarche (year) | ||||||||
| ≤14 | 0.549 (0.342‐0.880) | .013 | 1.137 (0.778‐1.661) | .506 | 1.173 (0.848‐1.622) | .334 | 0.891 (0.646‐1.229) | .482 |
| >14 | 0.976 (0.503‐1.892) | .943 | 0.902 (0.525‐1.549) | .708 | 1.024 (0.640‐1.639) | .920 | 1.488 (0.931‐2.380) | .097 |
| Age at menopause (year) | ||||||||
| ≤50 | 0.764 (0.378‐1.541) | .452 | 1.149 (0.640‐2.063) | .642 | 1.800 (1.055‐3.070) | .031 | 1.148 (0.693‐1.900) | .592 |
| >50 | 0.461 (0.171‐1.248) | .127 | 0.892 (0.352‐2.258) | .809 | 1.104 (0.489‐2.492) | .811 | 1.501 (0.655‐3.439) | .337 |
| Menopause state | ||||||||
| Premenopause | 0.604 (0.354‐1.032) | .065 | 1.142 (0.757‐1.722) | .527 | 0.932 (0.661‐1.313) | .686 | 0.902 (0.640‐1.272) | .557 |
| Postmenopause | 0.696 (0.401‐1.206) | .196 | 1.023 (0.634‐1.650) | .927 | 1.449 (0.941‐2.231) | .092 | 1.249 (0.823‐1.897) | .296 |
| Number of pregnancies | ||||||||
| ≤2 | 0.785 (0.441‐1.397) | .411 | 0.941 (0.565‐1.568) | .816 | 1.289 (0.855‐1.942) | .225 | 1.195 (0.786‐1.818) | .405 |
| >2 | 0.596 (0.363‐0.980) | .041 | 1.130 (0.755‐1.691) | .551 | 0.952 (0.670‐1.352) | .783 | 0.963 (0.677‐1.370) | .836 |
| Number of abortions | ||||||||
| ≤2 | 0.637 (0.434‐0.935) | .021 | 0.925 (0.668‐1.282) | .641 | 1.061 (0.808‐1.393) | .672 | 1.126 (0.851‐1.490) | .407 |
| >2 | 0.777 (0.173‐3.495) | .742 | 4.192 (1.135‐9.486) | .032 | 1.609 (0.672‐3.852) | .286 | 0.591 (0.250‐1.396) | .231 |
| Breastfeeding | ||||||||
| No | 0.518 (0.211‐1.270) | .151 | 1.321 (0.690‐2.527) | .401 | 1.357 (0.742‐2.484) | .322 | 1.008 (0.564‐1.803) | .979 |
| Yes | 0.719 (0.469‐1.100) | .128 | 1.022 (0.718‐1.456) | .902 | 1.078 (0.797‐1.457) | .627 | 1.027 (0.764‐1.380) | .860 |
| Family history of breast cancer | ||||||||
| No | 0.705 (0.472‐1.052) | .087 | 1.015 (0737‐1.398) | .927 | 1.050 (0.796‐1.384) | .732 | 1.007 (0.767‐1.321) | .961 |
| Yes | 0.370 (0.095‐1.440) | .152 | 1.016 (0.272‐3.793) | .981 | 1.967 (0.673‐5.753) | .216 | 1.146 (0.388‐3.386) | .805 |
| rs8008130 (CA + AA/CC) | rs4144657 (CT + TT/CC) | rs2152278 (GT + TT/GG) | rs8012083 (AG + GG/AA) | rs7142488 (CT + TT/CC) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI)a | P a | OR (95%CI)a | P a | OR (95%CI)a | P a | OR (95%CI)a | P a | OR (95%CI)a | P a | |
| Age (year) | ||||||||||
| ≤50 | 1.070 (0.680‐1.684) | .771 | 0.709 (0.491‐1.024) | .067 | 1.630 (1.162‐2.285) | .005 | 1.212 (0.866‐1.695) | .262 | 1.248 (0.826‐1.885) | .293 |
| >50 | 1.466 (0.823‐2.612) | .194 | 1.284 (0.768‐2.145) | .341 | 1.471 (0.935‐2.315) | .095 | 1.218 (0.776‐1.909) | .391 | 0.878 (0.479‐1.610) | .673 |
| Age at menarche (year) | ||||||||||
| ≤14 | 1.462 (0.944‐2.264) | .089 | 0.829 (0.581‐1.184) | .303 | 1.520 (1.096‐2.108) | .012 | 1.260 (0.912‐1.741) | .161 | 1.104 (0.732‐1.665) | .637 |
| >14 | 0.872 (0.473‐1.605) | .660 | 0.832 (0.483‐1.432) | .506 | 1.909 (1.185‐3.074) | .008 | 1.001 (0.616‐1.627) | .997 | 0.904 (0.492‐1.659) | .744 |
| Age at menopause (year) | ||||||||||
| ≤50 | 1.314 (0.690‐2.506) | .406 | 1.156 (0.640‐2.090) | .631 | 1.385 (0.820‐2.338) | .224 | 0.928 (0.555‐1.551) | .775 | 0.765 (0.374‐1.565) | .463 |
| >50 | 1.767 (0.571‐5.472) | .324 | 2.382 (0.915‐6.201) | .076 | 1.243 (0.550‐2.812) | .601 | 1.092 (0.493‐2.418) | .828 | 0.794 (0.264‐2.387) | .682 |
| Menopause state | ||||||||||
| Premenopause | 1.085 (0.677‐1.738) | .736 | 0.650 (0.444‐0.951) | .027 | 1.669 (1.179‐2.363) | .004 | 1.330 (0.938‐1.887) | .110 | 1.355 (0.888‐2.067) | .159 |
| Postmenopause | 1.424 (0.828‐2.448) | .201 | 1.415 (0.866‐2.312) | .165 | 1.452 (0.943‐2.234) | .090 | 0.988 (0.647‐1.507) | .954 | 0.773 (0.432‐1.382) | .385 |
| Number of pregnancies | ||||||||||
| ≤2 | 0.867 (0.487‐1.545) | .629 | 1.028 (0.654‐1.614) | .906 | 1.707 (1.129‐2.581) | .011 | 1.332 (0.886‐2.002) | .168 | 1.131 (0.689‐1.856) | .625 |
| >2 | 1.494 (0.940‐2.375) | .090 | 0.745 (0.503‐1.104) | .142 | 1.608 (1.130‐2.288) | .008 | 1.107 (0.780‐1.572) | .569 | 1.126 (0.707‐1.796) | .617 |
| Number of abortions | ||||||||||
| ≤2 | 1.105 (0.766‐1.594) | .592 | 0.850 (0.626‐1.154) | .296 | 1.579 (1.198‐2.080) | .001 | 1.243 (0.945‐1.636) | .120 | 1.098 (0.772‐1.562) | .603 |
| >2 | 1.616 (0.509‐5.135) | .415 | 0.617 (0.243‐1.571) | .311 | 3.249 (1.289‐8.188) | .012 | 0.958 (0.397‐2.315) | .924 | 1.055 (0.368‐3.022) | .921 |
| Breastfeeding | ||||||||||
| No | 0.598 (0.262‐1.367) | .223 | 0.674 (0.354‐1.285) | .229 | 2.180 (1.178‐4.035) | .013 | 1.344 (0.744‐2.426) | .327 | 1.052 (0.492‐2.251) | .896 |
| Yes | 1.421 (0.957‐2.108) | .084 | 0.891 (0.638‐1.245) | .500 | 1.503 (1.115‐2.027) | .008 | 1.150 (0.852‐1.554) | .360 | 1.090 (0.747‐1.590) | .656 |
| Family history of breast cancer | ||||||||||
| No | 1.161 (0.806‐1.672) | .422 | 0.896 (0.659‐1.218) | .483 | 1.643 (1.244‐2.170) | <.001 | 1.135 (0.861‐1.498) | .369 | 1.080 (0.763‐1.528) | .665 |
| Yes | 2.196 (0.400‐12.052) | .365 | 0.230 (0.061‐0.871) | .030 | 1.158 (0.399‐3.363) | .788 | 3.074 (0.967‐9.771) | .057 | 2.835 (0.487‐16.498) | .246 |
P values adjusted for age, menarche age, menopausal status, number of pregnancies, number of abortions, history of breastfeeding, and family history of breast cancer in first‐degree relatives in logistic regression analysis.
The italic values represent P < .05.
3.4. Receptor status and the genotypes of nine SNPs
As shown in Table 4, only the genotype TG (OR:1.62, 95%CI:1.04‐2.52) in rs12880540, the genotype AA (OR:2.74, 95%CI:1.01‐7.42) in rs2152275, and GG (OR:0.31, 95%CI:0.14‐0.69) in rs8012083 were associated with HER‐2 status. There was no discovery between SNPs and ER and PR.
Table 4.
Association analysis between three SNPs in LINC00520 and ER, PR, and Her‐2 receptor status in breast cancer patients
| SNPs Genotype | ER | P a | OR (95%CI)a | PR | P a | OR (95%CI)a | Her‐2 | P a | OR (95%CI)a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative (n = 151) | Positive (n = 341) | Negative (n = 192) | Positive (n = 297) | Negative (n = 138) | Positive (n = 333) | |||||||
| rs11622641 | ||||||||||||
| CC | 135 | 299 | 1 | 171 | 260 | 1.000 | 120 | 295 | 1 | |||
| CT | 16 | 40 | .912 | 1.036 (0.553‐1.939) | 21 | 35 | .789 | 1.083 (0.603‐1.948) | 18 | 36 | .572 | 0.832 (0.439‐1.576) |
| TT | 0 | 2 | 1.000 | 0 | 0 | 2 | 1.000 | NC | 0 | 2 | 1.000 | 0 |
| CT + TT | 16 | 42 | .741 | 1.110 (0.597‐2.067) | 21 | 37 | .619 | 1.159 (0.649‐2.069) | 18 | 38 | .724 | 0.892 (0.475‐1.678) |
| rs7157819 | ||||||||||||
| CC | 116 | 252 | 1 | 141 | 225 | 1 | 108 | 249 | 1 | |||
| CT | 32 | 82 | .604 | 1.133 (0.706‐1.819) | 48 | 65 | .344 | 0.809 (0.521‐1.255) | 26 | 81 | .333 | 1.285 (0.773‐2.134) |
| TT | 2 | 7 | .987 | 0.988 (0.248‐3.943) | 3 | 7 | .557 | 0.515 (0.379‐6.052) | 4 | 3 | .099 | 0.273 (0.059‐1.275) |
| CT + TT | 35 | 89 | .626 | 1.121 (0.709‐1.772) | 51 | 72 | .454 | 0.849 (0.554‐1.301) | 30 | 84 | .578 | 1.148 (0.706‐4.866) |
| rs12880540 | ||||||||||||
| TT | 63 | 140 | 1 | 78 | 123 | 1 | 68 | 132 | 1 | |||
| TG | 65 | 151 | .679 | 1.094 (0.715‐1.673) | 81 | 134 | .674 | 1.090 (0.729‐1.632) | 52 | 154 | .034 | 1.617 (1.037‐2.522) |
| GG | 23 | 50 | .689 | 0.887 (0.492‐1.598) | 32 | 40 | .324 | 0.756 (0.434‐1.318) | 18 | 47 | .556 | 1.210 (0.641‐2.283) |
| TG + GG | 88 | 201 | .855 | 1.038 (0.698‐1.542) | 114 | 174 | .972 | 0.993 (0.682‐1.447) | 70 | 201 | .051 | 1.508 (0.998‐2.278) |
| rs2152275 | ||||||||||||
| TT | 77 | 186 | 1 | 103 | 158 | 1 | 79 | 172 | 1 | |||
| TA | 66 | 129 | .267 | 0.795 (0.530‐1.192) | 75 | 119 | .869 | 1.033 (0.701‐0.524) | 54 | 132 | .535 | 1.143 (0.749‐1.743) |
| AA | 8 | 26 | .583 | 1.267 (0.544‐2.956) | 14 | 20 | .805 | 0.911 (0.435‐1.908) | 5 | 29 | .048 | 2.735 (1.008‐7.418) |
| TA + AA | 74 | 155 | .403 | 0.846 (0.573‐1.251) | 89 | 139 | .942 | 1.014 (0.699‐1.470) | 59 | 161 | .241 | 1.277 (0.849‐1.920) |
| rs8008130 | ||||||||||||
| AA | 128 | 281 | 1 | 161 | 245 | 1 | 114 | 279 | 1 | |||
| AG | 22 | 54 | .819 | 1.066 (0.615‐1.850) | 29 | 47 | .856 | 0.953 (0.568‐1.600) | 23 | 48 | .506 | 0.827 (0.473‐1.447) |
| GG | 1 | 6 | .329 | 2.919 (0.340‐9.021) | 2 | 5 | .444 | 1.930 (0.359‐9.379) | 1 | 6 | .331 | 2.937 (0.335‐5.729) |
| AG + GG | 23 | 60 | .614 | 1.148 (0.672‐1.960) | 31 | 52 | .962 | 1.012 (0.614‐1.668) | 24 | 54 | .737 | 1.097 (0.638‐1.887) |
| rs4144657 | ||||||||||||
| CC | 45 | 96 | 1 | 58 | 83 | 1 | 43 | 94 | 1 | |||
| CT | 78 | 183 | .903 | 1.029 (0.653‐1.621) | 101 | 157 | .703 | 1.087 (0.707‐1.673) | 74 | 174 | .808 | 1.059 (0.666‐1.686) |
| TT | 28 | 62 | .855 | 0.947 (0.530‐1.693) | 33 | 57 | .653 | 1.136 (0.651‐1.982) | 21 | 65 | .309 | 1.381 (0.741‐2.573) |
| CT + TT | 106 | 245 | .976 | 1.007 (0.653‐1.551) | 134 | 214 | .650 | 1.100 (0.729‐1.659) | 95 | 239 | .585 | 1.132 (0.726‐1.764) |
| rs2152278 | ||||||||||||
| GG | 63 | 114 | 1 | 71 | 105 | 1 | 45 | 127 | 1 | |||
| GT | 69 | 174 | .125 | 1.395 (0.912‐2.135) | 97 | 144 | .822 | 1.048 (0.698‐1.574) | 75 | 154 | .105 | 0.689 (0.439‐1.081) |
| TT | 19 | 53 | .176 | 1.532 (0.826‐2.841) | 24 | 48 | .297 | 1.367 (0.760‐2.458) | 18 | 52 | .978 | 0.991 (0.519‐1.892) |
| GT + TT | 88 | 227 | .085 | 1.425 (0.952‐2.133) | 121 | 192 | .591 | 1.112 (0.755‐1.639) | 93 | 206 | .186 | 0.748 (0.486‐1.151) |
| rs8012083 | ||||||||||||
| AA | 78 | 205 | 1 | 109 | 171 | 1 | 76 | 194 | 1 | |||
| AG | 61 | 119 | .243 | 0.783 (0.519‐1.181) | 69 | 111 | .632 | 1.101 (0.743‐1.633) | 47 | 127 | .639 | 1.110 (0.717‐1.719) |
| GG | 12 | 17 | .141 | 0.551 (0.249‐1.218) | 14 | 15 | .261 | 0.639 (0.292‐1.396) | 15 | 12 | .005 | 0.305 (0.135‐0.693) |
| AG + GG | 73 | 136 | .139 | 0.744 (0.503‐1.100) | 83 | 126 | .925 | 1.018 (0.700‐1.481) | 62 | 139 | .640 | 0.907 (0.603‐1.364) |
| rs7142488 | ||||||||||||
| TT | 120 | 280 | 154 | 244 | 110 | 272 | ||||||
| TC | 31 | 60 | .626 | 0.884 (0.539‐1.451) | 38 | 52 | .539 | 0.861 (0.535‐1.387) | 28 | 60 | .843 | 0.949 (0.568‐1.586) |
| CC | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 |
| TC + CC | 31 | 61 | .672 | 0.899 (0.548‐1.410) | 38 | 53 | .587 | 0.877 (0.545‐1.410) | 28 | 61 | .899 | 0.967 (0.580‐1.614) |
P values adjusted for age, menarche age, menopausal status, number of pregnancies, number of abortions, history of breastfeeding, and family history of breast cancer in first‐degree relatives in logistic regression analysis.
The italic values representP < .05.
3.5. Molecular typing of BC and the genotypes of nine SNPs
Table 5 showed the association between nine SNPs of LINC00520 and the different molecular typing states of BC. The genotypic GG (OR:3.58, 95%CI:1.32‐9.69) in rs8012083 increased the risk of triple‐negative BC.
Table 5.
Association analysis between three SNPs in LINC00520 and molecular typing of breast cancer
| SNPs | Luminal | HER‐2 | Triple‐negative | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes (n = 344) | No (n = 127) | P a | OR (95%CI)a | Yes (n = 78) | No (n = 393) | P a | OR (95%CI)a | Yes (n = 49) | No (n = 422) | P a | OR (95%CI)a | |
| rs11622641 | ||||||||||||
| CC | 302 | 113 | 1.000 | 71 | 344 | 1.000 | 42 | 373 | 1.000 | |||
| CT | 40 | 14 | .973 | 0.989 (0.511‐1.915) | 7 | 47 | .532 | 0.764 (0.328‐1.777) | 7 | 47 | .384 | 1.486 (0.610‐3.623) |
| TT | 2 | 0 | 1.000 | 0 | 0 | 2 | 1.000 | 0 | 0 | 2 | 1.000 | 0 |
| CT + TT | 42 | 14 | .867 | 1.058 (0.549‐2.036) | 7 | 49 | .450 | 0.723 (0.312‐1.676) | 7 | 49 | .460 | 1.395 (0.576‐3380) |
| rs7157819 | ||||||||||||
| CC | 260 | 97 | 1.000 | 60 | 297 | 1.000 | 37 | 320 | 1.000 | |||
| CT | 78 | 29 | .765 | 0.927 (0.564‐1.525) | 17 | 90 | .953 | 0.982 (0.540‐1.786) | 12 | 95 | .653 | 1.178 (0.577‐2.407) |
| TT | 6 | 1 | .564 | 1.878 (0.221‐5.991) | 1 | 6 | .924 | 0.901 (0.104‐7.775) | 0 | 7 | 1.000 | 0 |
| CT + TT | 84 | 30 | .867 | 0.959 (0.588‐1.565) | 18 | 96 | .939 | 0.977 (0.544‐1.756) | 12 | 102 | .776 | 1.109 (0.544‐2.262) |
| rs12880540 | ||||||||||||
| TT | 146 | 54 | 1.000 | 33 | 167 | 1.000 | 21 | 179 | 1.000 | |||
| TG | 151 | 55 | .877 | 1.036 (0.662‐1.622) | 33 | 173 | .914 | 0.971 (0.570‐1.656) | 22 | 184 | .931 | 0.972 (0.507‐1.863) |
| GG | 47 | 18 | .614 | 0.848 (0.447‐1.609) | 12 | 53 | .625 | 1.203 (0.574‐2.521) | 6 | 59 | .874 | 1.083 (0.407‐2.876) |
| TG + GG | 198 | 73 | .955 | 0.988 (0.649‐1.503) | 45 | 226 | .926 | 1.024 (0.623‐1.684) | 28 | 243 | .986 | 0.994 (0.538‐1.838) |
| rs2152275 | ||||||||||||
| TT | 186 | 65 | 1.000 | 42 | 209 | 1.000 | 23 | 228 | 1.000 | |||
| TA | 132 | 54 | .471 | 0.853 (0.554‐1.314) | 31 | 155 | .958 | 0.986 (0.590‐1.649) | 23 | 163 | .257 | 1.439 (0.767‐2.699) |
| AA | 26 | 8 | .850 | 1.086 (0.463‐2.549) | 5 | 29 | .773 | 0.861 (0.312‐2.376) | 3 | 31 | .890 | 1.095 (0.304‐3.948) |
| TA + AA | 158 | 62 | .558 | 0.883 (0.583‐1.338) | 36 | 184 | .894 | 0.967 (0.591‐1.583) | 26 | 194 | .292 | 1.388 (0.755‐2.553) |
| rs8008130 | ||||||||||||
| AA | 284 | 109 | 1.000 | 69 | 324 | 1.000 | 40 | 353 | 1.000 | |||
| AG | 54 | 17 | .663 | 1.144 (0.625‐2.092) | 9 | 62 | .360 | 0.701 (0.328‐1.498) | 8 | 63 | .636 | 1.224 (0.530‐2.824) |
| GG | 6 | 1 | .367 | 2.694 (0.312‐3.239) | 0 | 7 | .999 | 0 | 1 | 6 | .884 | 1.181 (0.126‐2.109) |
| AG + GG | 60 | 18 | .490 | 1.229 (0.685‐2.204) | 9 | 69 | .220 | 0.625 (0.294‐1.326) | 9 | 69 | .626 | 1.219 (0.550‐2.701) |
| rs4144657 | ||||||||||||
| CC | 96 | 41 | 1.000 | 22 | 115 | 1.000 | 19 | 118 | 1.000 | |||
| CT | 186 | 62 | .389 | 1.234 (0.765‐1.989) | 39 | 209 | .934 | 0.976 (0.546‐1.743) | 23 | 225 | .251 | 0.675 (0.345‐1.321) |
| TT | 62 | 24 | .973 | 1.011 (0.550‐1.858) | 17 | 69 | .420 | 1.340 (0.657‐2.731) | 7 | 79 | .289 | 0.603 (0.237‐1.535) |
| CT + TT | 248 | 86 | .496 | 1.170 (0.745‐1.838) | 56 | 278 | .826 | 1.064 (0.614‐1.842) | 30 | 304 | .193 | 0.656 (0.348‐1.237) |
| rs2152278 | ||||||||||||
| GG | 117 | 55 | 1.000 | 33 | 139 | 1.000 | 22 | 150 | 1.000 | |||
| GT | 173 | 56 | .134 | 1.412 (0.900‐2.217) | 33 | 196 | .205 | 0.706 (0.412‐1.210) | 23 | 206 | .536 | 0.817 (0.429‐1.552) |
| TT | 51 | 16 | .199 | 1.537 (0.798‐2.959) | 12 | 58 | .701 | 0.865 (0.413‐1.812) | 4 | 66 | .158 | 0.445 (0.145‐1.367) |
| GT + TT | 227 | 72 | .094 | 1.440 (0.940‐2.206) | 45 | 54 | .247 | 0.742 (0.448‐1.229) | 27 | 272 | .308 | 0.725 (0.391‐1.344) |
| rs8012083 | ||||||||||||
| AA | 203 | 67 | 1.000 | 43 | 227 | 1.000 | 24 | 246 | 1.000 | |||
| AG | 125 | 49 | .626 | 0.896 (0.577‐1.391) | 31 | 143 | .678 | 1.115 (0.667‐1.863) | 18 | 156 | .817 | 1.082 (0.557‐2.100) |
| GG | 26 | 11 | .080 | 0.476 (0.207‐1.094) | 4 | 23 | .950 | 0.965 (0.314‐2.962) | 7 | 20 | .012 | 3.575 (1.318‐9.698) |
| AG + GG | 141 | 60 | .340 | 0.816 (0.538‐1.239) | 35 | 166 | .719 | 1.095 (0.667‐1.797) | 25 | 176 | .324 | 1.359 (0.739‐2.498) |
| rs7142488 | ||||||||||||
| TT | 281 | 101 | 1.000 | 63 | 319 | 1.000 | 38 | 344 | 1.000 | |||
| TC | 62 | 26 | .749 | 0.918 (0.542‐1.553) | 15 | 73 | .905 | 1.039 (0.552‐1.956) | 11 | 77 | .737 | 1.135 (0.542‐2.378) |
| CC | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 |
| TC + CC | 63 | 26 | .793 | 0.932 (0.551‐1.576) | 15 | 74 | .934 | 1.027 (0.546‐1.932) | 11 | 78 | .765 | 1.120 (0.535‐2.345) |
P values adjusted for age, menarche age, menopausal status, number of pregnancies, number of abortions, history of breastfeeding, and family history of breast cancer in first‐degree relatives in logistic regression analysis.
The italic values representP < .05.
3.6. Analysis of haplotype
The haplotype analysis was conducted to evaluate the combined effect of four LINC00520 SNPs. As shown in Table 6, Crs7157819Trs12880540Ars2152275Trs11622641 was the common haplotype in cases and controls with a frequency of 0.05% and 0.078%, respectively, and could decrease BC risk by 37% (OR:0.63, 95%CI:0.44‐0.91). However, other results showed that no haplotype was associated with BC risk with P < .05.
Table 6.
Haplotype analysis of four SNPs in LINC00520
| Gene | Haplotypea | Cases (%) | Controls (%) | χ2 | P | OR (95%CI) |
|---|---|---|---|---|---|---|
| LINC00520 | C G A C | 174.34 (0.173) | 164.71 (0.163) | 0.349 | .554 | 1.073 (0.849‐1.356) |
| C G T C | 53.43 (0.053) | 47.25 (0.047) | 0.409 | .522 | 1.140 (0.763‐1.704) | |
| C T A C | 29.54 (0.029) | 19.89 (0.020) | 1.948 | .163 | 1.502 (0.845‐2.671) | |
| C T A T | 50.10 (0.050) | 77.68 (0.077) | 6.316 | .012 | 0.627 (0.435‐0.905) | |
| C T T C | 556.93 (0.553) | 572.14 (0.566) | 0.418 | .518 | 0.943 (0.790‐1.126) | |
| T G T C | 128.34 (0.127) | 112.65 (0.112) | 1.193 | .275 | 1.162 (0.887‐1.522) |
Abbreviation: SNPs, single‐nucleotide polymorphisms.
The sequence of SNP locus is rs7157819, rs12880540, rs2152275, and rs11622641.
The italic values representP < .05.
3.7. Gene‐reproductive factor interaction analysis
We further explored the interaction between functional SNPs and other factors on BC risk using multifactor dimensionality reduction (MDR) analysis (Table 7). The optimal gene‐environment interactive model with testing balance accuracy (TBA) of 0.70 and cross‐validation consistency (CVC) of 7/10 revealed that the number of pregnancies more than twice with number of abortions more than twice carrying rs2152278 T allele had 5.24 times higher risk of BC over those without above characteristics.
Table 7.
Interaction analyses between the functional SNPs and environmental factors associated with breast cancer by MDR
| Model | TBA | CVC | χ2 | P | OR (95%CI) |
|---|---|---|---|---|---|
| Number of pregnancies | 0.596 | 10/10 | 31.413 | <.001 | 2.049 (1.592‐2.637) |
| Number of pregnancies, number of abortions | 0.688 | 10/10 | 143.543 | <.001 | 4.935 (3.774‐6.454) |
| Number of pregnancies, number of abortions, rs2152278 | 0.696 | 10/10 | 154.784 | <.001 | 5.239 (4.006‐6.853) |
The best model was selected as the one with the maximum testing balance accuracy and maximum cross‐validation consistency.
In this study, the best interaction model was the three‐factor model including number of pregnancies‐number of abortions‐LINC00520 rs2152278.
Abbreviations: CVC, cross‐validation consistency; MDR, multifactor dimensionality reduction; SNPs, single‐nucleotide polymorphisms; TBA, testing balance accuracy.
3.8. Functional correlation of rs12880540 genotypes with the relative expression of LINC00520 in plasma
As shown by qRT‐PCR analyses (Figure 1), the average relative expression of LINC00520 in the plasma of individuals with rs12880540 TG (1.61 ± 0.35, n = 44) and rs12880540 GG (2.25 ± 0.29, n = 22) was both higher (both P < .0029) than those only with rs12880540 TT (1.03 ± 0.48, n = 40). Furthermore, with the increase of C genotype, LINC00520 expression appeared to be elevated.
Figure 1.
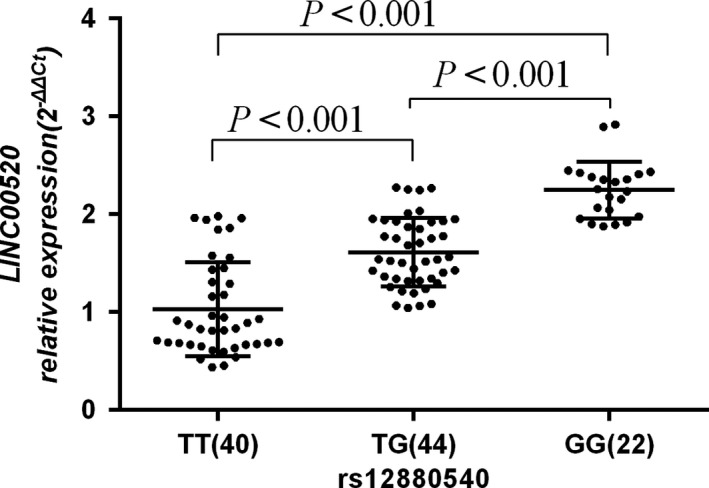
The relative expression of LINC00520 with different rs12880540 genotypes in plasma. The relative mRNA expression of LINC00520 levels in blood plasma from 106 cancer‐free controls was significantly higher in the TG (1.61 ± 0.35) and GG genotypes (2.25 ± 0.29) than the TT genotype (1.03 ± 0.48) (all P < .001)
3.9. Effect of rs12880540 mutation on the binding of miR‐3122 and LINC00520
As the database predicted, rs12880540 G > T mutation could gain/loss a binding site for miR‐3122 and LINC00520, which might regulate the expression of LINC00520 (Figure 2). To investigate the effect of rs12880540 on the binding of miR‐3122 and LINC00520, we constructed luciferase reporter containing part of LINC00520 covering rs12880540 G > T allele co‐transfected with miRNA mimics in 293T cells, measured and calculated the relatively luciferase activity. Luciferase reporter assays displayed that the luciferase activity of the construct with the risk rs12880540 G allele decreased when transfected with miR‐3122 mimics and there was no change when transfected with NC, which not indicated that rs12880540 G would affect the combination of miR‐3122 and LINC00520.
Figure 2.
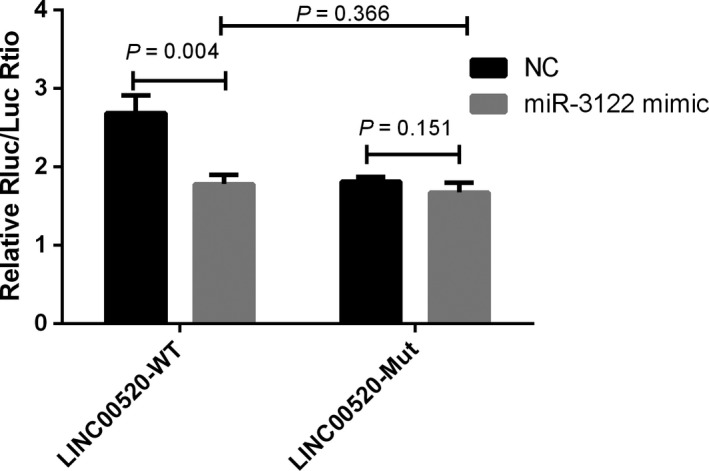
The effect of rs12880540 on the binding of miR‐3122 and LINC00520 revealed by luciferase reporter assays
4. DISCUSSION
So far as we know, no association between genetic variation of LINC00520 and BC susceptibility has been explored. In the study, a case‐control research was conducted to analyze the association between LINC00520 genetic susceptibility and the risk of BC. After adjusted by age, menarche age, menopausal status, number of pregnancies, number of miscarriages, history of breastfeeding, and family history of BC, the study found that the variation of rs11622641 allele was statistically associated with a reduced risk of BC. The variation of rs12880540 and rs2152278 alleles increased the risk of BC. The results showed that TG, rs2152275 genotype TG, and rs8012083 genotype AA of rs12880540 genotype GG were significantly correlated with the positive status of Her‐2 receptor. Haplotype analysis showed that LINC00520 Crs7157819 Trs12880540 Ars2152275 Trs11622641 haplotype could reduce the risk of BC. The homozygous mutant GG at rs8012083 site could significantly increase the risk of triple‐negative BC. A genetic variant of rs8012083 in LINC00520 could be used as a biomarker for triple‐negative BC. However, the mechanism of its influence on the susceptibility to triple‐negative BC needed to be further studied.
Previous studies have shown that SNP could affect the level of genes expression through allelic changes, thus participating in the carcinogenesis of tumor. Compared with rs619586 A allele, the expression of MALAT1 in individuals carrying rs619586 G allele was lower, which might be related to the decreased susceptibility to BC.20 The G risk gene of rs6983267 site of lncRNA CCAT2 could increase the expression of CCAT2, which was related to the risk of colorectal cancer.21 The results showed that the genotype GG at rs12880540 site significantly increased the risk of BC compared with the genotype TT. The relative expression of LINC00520 showed a linear trend with the increase in the number of rs12880540 mutations. It was suggested that LINC00520 gene polymorphism could increase the risk of BC and affect the expression of LINC00520 in plasma, which played an important role in the occurrence and development of BC, consistent with the results of the increased expression of LINC00520 in BC.19
The SNP mutations could change the activity of lncRNAs transcriptional regulatory region and affect the expression of lncRNAs by changing the secondary structure of lncRNAs to obtain or lose the binding site of miRNAs, thus affecting the development, metastasis, and prognosis of tumor.22, 23, 24 The biological function of the secondary structure and the miRNA binding ability of the SNPs of the LINC00520 were predicted using the lncRNASNP2 database (http://bioinfo.life.hust.edu.cn/lncRNASNP) and the RNAfold website (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The predicted results showed that the secondary structure of the LINC00520 has changed after the mutation of the rs12880540 allele G to T, while the two binding sites of the miR‐92a‐2‐5p and the miR‐4648 and the four binding sites of the miR‐3122, the miR‐3913‐5p, the miR‐4259, and the miR‐4425 were lost. The results of double luciferase reporter gene experiment showed that LINC00520 and miR‐3122 interact with each other because of T, but the variation of rs12880540 G > T had little effect on the binding ability of LINC00520 and miR‐3122. Therefore, the effect of rs12880540 G > T on BC susceptibility may not be due to its change in binding ability of LINC00520 to miR‐3122.
BC is a disease with extremely complex pathogenesis. It is generally believed that BC is the result of the interaction of many factors, such as personal lifestyle, environment, genetic, and reproductive factors.25 So far, studies have found individual or family history of smoking, drinking, obesity, menarche at earlier age, late menopausal, infertile, multiple miscarriages, genetic factors BC or ovarian cancer, genetic mutations, endogenous hormone exposure, and exogenous hormone intake (oral contraceptive and hormone replacement therapy), as well as other environmental and reproductive factors as risk factors for BC, while breastfeeding and physical activity were well‐known protective factors.26, 27, 28, 29 The results of this study showed that there was obvious interaction between the number of pregnancies, the number of miscarriages, and rs2152278. However, the mechanism of its interaction needs to be further studied.
For the first time, we found the association between LINC00520 gene polymorphism and BC susceptibility. The following were advantages of this study. First of all, all the cases included in this study were new cases of BC with definite pathological diagnosis, which was beneficial to the control of current cases‐new cases bias. Secondly, the health control randomly selected from the community chronic disease survey of 20 000 people in Henan Province was used in this study. The age matching (±2 years old) between the case and the control made basic characteristics of the two groups similar, which was helpful to reduce confusion bias. Finally, 5% of the DNA samples were randomly selected for repeated experiments, and the different genotypes of each SNP were randomly selected for sequencing, so as to verify the consistency of the experimental results.
Inevitably, the study also has some limitations. The number of samples was 1009 and this study only aimed at the Chinese Han population, leading to the limited representativeness and universality of this study. Therefore, a larger sample size is needed to verify the experimental results of this study. Secondly, this study only aimed at the Chinese Han population. Due to ethnic or regional differences, the results of this study need to be verified by other regions and ethnic groups. Finally, because of the lack of relevant research of the LINC00520 and adequate reference and related biological function theory support, the research on the function of the LINC00520 gene only has a preliminary foundation study, which needs further research.
In conclusion, a genetic variant of rs8012083 in LINC00520 could be used as a biomarker for triple‐negative BC after further evaluation of diagnostic tests. The genetic variant of LINC00520 was related to the susceptibility of BC, and rs12880540 might affect the corresponding mRNA expression of lncRNA LINC00520.
CONFLICT OF INTERESTS
None declared.
Supporting information
ACKNOWLEDGMENTS
The authors thank the participants, the coordinators, and administrators for their supports during the study.
Guo Q, Xu L, Peng R, et al. Characterization of lncRNA LINC00520 and functional polymorphisms associated with breast cancer susceptibility in Chinese Han population. Cancer Med. 2020;9:2252–2268. 10.1002/cam4.2893
Qiaoyun Guo and Linping Xu contributed equally to this work and should be considered as co‐first authors
Funding information
This work was supported by the Major Science and Technology Project of Hainan Province (161100311400), the National Natural Science Foundation of China (U1604168), Medical Science and Technology Key Projects of Henan Province and Zhengzhou (201602295 and 20150374).
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Zuo T, Zeng H, Zhang S, He J. National cancer incidence and mortality in China, 2012. Chin J Cancer Res. 2016;28(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐132. [DOI] [PubMed] [Google Scholar]
- 4. Fan L, Strasser‐Weippl K, Li J‐J, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279‐e289. [DOI] [PubMed] [Google Scholar]
- 5. Kumaran M, Ghosh S, Joy AA, et al. Fine‐mapping of a novel premenopausal breast cancer susceptibility locus at Chr4q31.22 in Caucasian women and validation in African and Chinese women. Int J Cancer. 2020;146(5):1219‐1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Law PJ, Timofeeva M, Fernandez‐Rozadilla C, et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat Commun. 2019;10(1):2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng J, Huang X, Tan W, et al. Pancreatic cancer risk variant in LINC00673 creates a miR‐1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48(7):747‐757. [DOI] [PubMed] [Google Scholar]
- 8. Santosh B, Varshney A, Yadava PK. Non‐coding RNAs: biological functions and applications. Cell Biochem Funct. 2015;33(1):14‐22. [DOI] [PubMed] [Google Scholar]
- 9. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhartiya D, Scaria V. Genomic variations in non‐coding RNAs: structure, function and regulation. Genomics. 2016;107(2–3):59‐68. [DOI] [PubMed] [Google Scholar]
- 11. Pegueroles C, Gabaldon T. Secondary structure impacts patterns of selection in human lncRNAs. BMC Biol. 2016;14:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang X, Zhang W, Shao Z. Association between long non‐coding RNA polymorphisms and cancer risk: a meta‐analysis. Biosci Rep. 2018;38(4). 10.1042/BSR20180365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cerk S, Schwarzenbacher D, Adiprasito J, et al. Current status of long non‐coding RNAs in human breast cancer. Int J Mol Sci. 2016;17(9):1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gabriel E, Yong‐Shu Z, Qing‐Lin L, Ji‐Xiang X, Yuan Y, Qun Z. Long non‐coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5(2):134‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly‐Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080‐10100. [DOI] [PubMed] [Google Scholar]
- 16. Wu YY, Gao W, Zhang YL, et al. Expression and clinical significance of long non‐coding RNA LINC00520 in laryngeal squamous cell carcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2018;32(2):91‐95. [DOI] [PubMed] [Google Scholar]
- 17. Xie T, Pi G, Yang B, et al. Long non‐coding RNA 520 is a negative prognostic biomarker and exhibits pro‐oncogenic function in nasopharyngeal carcinoma carcinogenesis through regulation of miR‐26b‐3p/USP39 axis. Gene. 2019;707:44‐52. [DOI] [PubMed] [Google Scholar]
- 18. Chen B, Wang C, Zhang J, Zhou Y, Hu W, Guo T. New insights into long noncoding RNAs and pseudogenes in prognosis of renal cell carcinoma. Cancer Cell Int. 2018;18:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henry WS, Hendrickson DG, Beca F, et al. LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer. Oncotarget. 2016;7(50):81981‐81994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng R, Luo C, Guo Q, et al. Association analyses of genetic variants in long non‐coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene. 2018;642:241‐248. [DOI] [PubMed] [Google Scholar]
- 21. Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23(9):1446‐1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Minotti L, Agnoletto C, Baldassari F, Corra F, Volinia S. SNPs and somatic mutation on long non‐coding RNA: new frontier in the cancer studies? High Throughput. 2018;7(4):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sabarinathan R, Tafer H, Seemann SE, Hofacker IL, Stadler PF, Gorodkin J. The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Res. 2013;41(Web Server issue):W475‐W479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354‐361. [DOI] [PubMed] [Google Scholar]
- 25. Milne RL, Gaudet MM, Spurdle AB, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case‐control study. Breast Cancer Res. 2010;12(6):R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Islami F, Liu Y, Jemal A, et al. Breastfeeding and breast cancer risk by receptor status–a systematic review and meta‐analysis. Ann Oncol. 2015;26(12):2398‐2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Collaborative Group on Hormonal Factors in Breast Cancer . Menarche, menopause, and breast cancer risk: individual participant meta‐analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol. 2012;13(11):1141‐1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yanhua C, Geater A, You J, et al. Reproductive variables and risk of breast malignant and benign tumours in Yunnan province. China. Asian Pac J Cancer Prev. 2012;13(5):2179‐2184. [DOI] [PubMed] [Google Scholar]
- 29. Zhang Q, Liu LY, Wang F, Mu K, Yu ZG. The changes in female physical and childbearing characteristics in China and potential association with risk of breast cancer. BMC Public Health. 2012;12:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


