Abstract
In this study, the presence of Leishmania DNA and blood feeding sources in phlebotomine sand fly species commonly present in Sicily were investigated. A total of 1,866 female sand flies including 176 blood fed specimens were sampled over two seasons in five selected sites in Sicily (southern Italy). Sergentomyia minuta (n = 1,264) and Phlebotomus perniciousus (n = 594) were the most abundant species at all the sites, while three other species from the genus Phlebotomus (i.e., P. sergenti n = 4, P. perfiliewi n = 3 and P. neglectus n = 1) were only sporadically captured. Twenty-eight out of the 1,866 (1.5%) sand flies tested positive for Leishmania spp. Leishmania tarentolae DNA was identified in 26 specimens of S. minuta, while the DNA of Leishmania donovani complex was detected in a single specimen each of S. minuta and P. perniciosus. Interestingly, seven S. minuta specimens (0.4%) tested positive for reptilian Trypanosoma sp. Blood sources were successfully identified in 108 out of 176 blood fed females. Twenty-seven out of 82 blood sources identified in fed females of P. perniciosus were represented by blood of wild rabbit, S. minuta mainly fed on humans (16/25), while the sole P. sergenti fed specimen took a blood meal on rat. Other vertebrate hosts including horse, goat, pig, dog, chicken, cow, cat and donkey were recognized as blood sources for P. perniciosus and S. minuta, and, surprisingly, no reptilian blood was identified in blood-fed S. minuta specimens. Results of this study agree with the well-known role of P. perniciosus as vector of L. infantum in the western Mediterranean; also, vector feeding preferences herein described support the hypothesis on the involvement of lagomorphs as sylvatic reservoirs of Leishmania. The detection of L. donovani complex in S. minuta, together with the anthropophilic feeding-behaviour herein observed, warrants further research to clarify the capacity of this species in the transmission of pathogens to humans and other animals.
Introduction
Phlebotomine sand flies (Diptera: Phlebotominae) are insects of great interest in human and veterinary medicine. They are vectors of viral and bacterial pathogens and recognized as the main hematophagous arthropods proven to transmit protozoa of the genus Leishmania such as Leishmania infantum, the causative agent of canine leishmaniosis (CanL) in dogs and visceral (VL) or cutaneous (CL) leishmaniosis in humans in the Mediterranean area [1].
Sand flies are distributed throughout many regions of the world and their biodiversity and phenology have been investigated particularly in Leishmania endemic areas. In western Europe, only sand flies of the genus Phlebotomus are competent vectors for Leishmania transmission and P. perniciosus is the most widespread species [2].
Surveillance on phlebotomine sand fly vectors is pivotal to assess the risk for transmission of endemic Leishmania species, but also crucial to monitor the risk for introduction of new Leishmania species in non-endemic territories [2–3].
Several studies have investigated the presence of Leishmania DNA in phlebotomine sand flies, and the reported infection rate in P. perniciosus varied from 0.13% to 50% depending on the epidemiological context [4–5]. Although L. infantum is the only species widespread in Europe that causes illness, human cases due to non-indigenous Leishmania species are increasingly reported [6].
In Italy, imported leishmaniosis cases are mainly associated with international travellers and/or refugees coming from endemic zones, and consist of chronic forms diagnosed many months after entering the country [7–8]. Noteworthy, anthroponotic species such as Leishmania tropica and Leishmania donovani are included among the causative agents of imported human cases, and the risk of introduction and/or wider establishment of these species into Europe is higher since suitable sand fly vectors are present [3,6,9].
Sicily, one of the two major Italian islands, is located in the centre of the Mediterranean Sea. The island is characterized by a typical temperate climate, with mild and wet winters and hot dry summers. It is a well-known hyper-endemic region for CanL with a prevalence of infection up to 40% in dogs which are regarded as the main domestic reservoir hosts and source for human infection [10–11]. According to available entomological surveys on L. infantum vectors in Sicily, P. perniciosus has been recognized as the most abundant species, with P. neglectus and P. perfiliewi much less frequently detected [12–14]. The presence of P. sergenti has also been sporadically reported along the east side of the Island [12–14] implying the risk for L. tropica transmission. Although no autochthonous cutaneous leishmaniosis cases (ACL) caused by L. tropica have been detected in south-western Europe so far, Sicily is an area potentially susceptible to the introduction of this protozoan species due to its proximity to northern Morocco, an emerging ACL area [15], and to the large migratory flow of people between the two territories. As matter of fact, the flow of immigrants from endemic regions of North Africa and the Middle East, together with the rising number of reports of L. tropica human infections in Italy [6,7,16–17], could constitute the first step in its potential spread in Sicily and subsequently across other areas of southern Europe where the competent vector has been described [7].
Female sand flies are hematophagous insects and take blood meals on many vertebrate hosts as reptiles, birds, a variety of domestic and wild mammals and humans, as well showing an opportunistic feeding behaviour [1,4,18–19]. Interestingly, lagomorphs have been found to be the most frequent blood source for P. perniciosus in a large outbreak of human leishmaniosis in Madrid [20–21], and, xenodiagnostic studies demonstrated the role of hares and wild rabbits as reservoir hosts of L. infantum. Together these findings suggest the possible involvement of lagomorph species in the epidemiology of Leishmania infection [22–25]. Therefore, the identification of blood meal sources in wild-caught sand flies provides information on host-feeding patterns under natural conditions, which, in turn, results in data on potential reservoir hosts and essential knowledge for the establishment of efficient control strategies.
Given all of the above considerations, the aim of the present study was to investigate Leishmania infection rates and blood sources of phlebotomine sand fly species caught in Sicily over two transmission seasons.
Materials and methods
Sand fly collection and identification
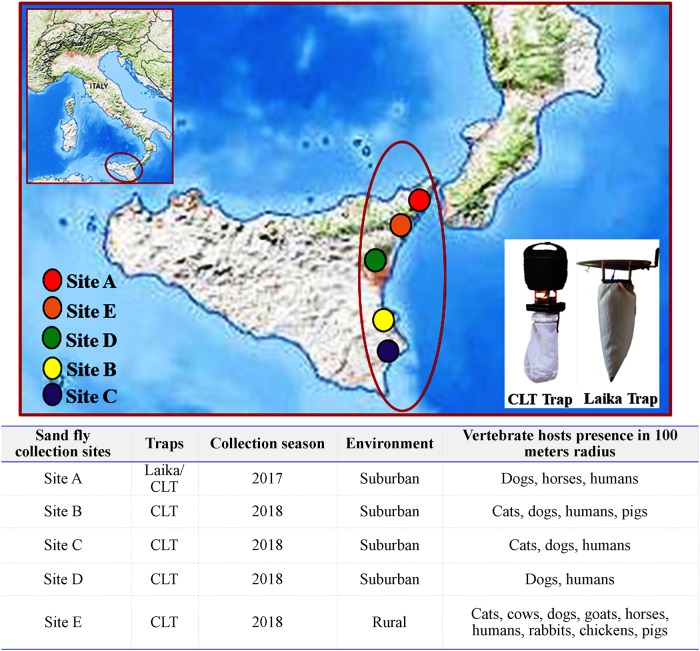
Sand flies in this study were sampled during two different entomologic surveys carried out in privately owned areas located in Sicily in 2017 and 2018 (Fig 1). In 2017, sand flies were captured in a suburban area (Site A. 38°13’59” N; 15°32’49” E; 263 m.a.s.l) nearby the didactic farm of the Department of Veterinary Science of the University of Messina using a classical light trap (CLT), equipped with a traditional incandescent lamp (12V, 8W) and five Laika 4.0 light traps with LED of different colours (Fig 1). Traps were placed from before sunset until sunrise for three consecutive nights each month from May to October [26]. In 2018, sand fly captures were performed in four different sites, i.e., in three shelters located in suburban areas of Syracuse (Site B. 37°04’54” N; 15°12’43” E; 69 m.a.s.l) (Site C. 37°04’30” N; 15°13’59” E; 416 m.a.s.l) and Catania (Site D. 37°36’45” N; 15°01’46” E; 948 m.a.s.l) and on a farm situated in a rural area of the municipality of Messina (Site E. 38°00’42” N; 15°25’18” E; 177 m.a.s.l). In each of the above sites, light traps (i.e., CLT) were placed from May to October and left working from before sunset until sunrise for two consecutive nights twice a month (Fig 1).
Fig 1. Geographical characteristics of sand fly sampling sites and presence of vertebrate hosts.
Access to the collection sites and sampling procedures were authorized by owners of the farms (Site A and Site E) and of the shelters (Sites B, C and D).
In the laboratory, sand flies were differentiated by sex with the aid of a stereomicroscope and thereafter processed for identification. Briefly, the head and posterior last tergites of females were dissected, cleared and slide-mounted for microscopic observation as described elsewhere [12], and identified to species level using morphological keys [27]. Females were further ranked as unfed (no visible blood in the abdomen), blood fed (presence of blood in the abdomen) and gravid (presence of eggs). Finally, the thorax and the abdomen of each specimen were individually transferred into 2 mL vials containing 70% ethanol and stored for DNA analysis.
DNA extraction and Leishmania spp. detection
Genomic DNA was extracted from portions (i.e., thorax and abdomen) of each sand fly specimen stored in 70% ethanol using the Citogene® Cell and Tissue kit (Citomed, Portugal) according to the manufacturer’s instructions. The DNA extracted was suspended in 30 μL of sterile water and stored at + 4°C until analysis.
Trypanosomatidae DNA amplification
The presence of Trypanosomatidae DNA in unfed, blood fed and gravid females was firstly screened using a one-step PCR protocol with a set of primers targeting sections of the ribosomal internal transcribed spacer (ITS-rDNA) [28]. For further molecular characterisation of ITS-rDNA positive samples, a two-step PCR protocol using a set of primers targeting conserved regions of the mitochondrial cytochrome b gene (cytB) was performed [29]. An additional one-step PCR with specific primers for small subunit ribosomal DNA gene (SSU-rDNA) partial amplification [30] was carried out, to characterise the samples where the presence of Trypanosoma spp. was suggested by BLAST analysis of cytB sequences (S1 Table).
Blood source identification
Identification of blood sources was conducted by the amplification of a 350 bp segment of the host mitochondrial cytB, using the modified vertebrate-universal specific primers (cytB1-F and cytB-2-R) on blood fed sand fly specimens [31]. The cytB PCR was carried out with 5 μL of extracted DNA in a final volume of 25 μL, using 12.5 μL of NZYTaq 2× Green Master Mix (Nyztech, Portugal) and 1.5 μL of each primer (10 pmol/μL). Amplification was performed as follows: one cycle at 94°C for 5 min, followed by 40 cycles consisting of denaturation at 94°C for 1 min, annealing at 55°C for 1 min and elongation at 72°C for 1 min, followed by final elongation at 72°C for 7 min [4] (S1 Table). Electrophoresis of PCR products was carried out in 1.5% agarose gel stained with 2.5 μL Greensafe premium® (Nzytech, Portugal), using a 100 bp DNA ladder as a molecular weight marker and final amplicons were visualized under UV light.
Sequence analysis
PCR products were purified and sequenced by Sanger’s method (StabVida, Portugal), using the same primers used for the PCR reactions. Nucleotide sequences obtained were examined using 4Peaks v1.8 (Nucleobytes, Netherlands) and analysed by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to find positional homologs. Obtained sequences were deposited at the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp/).
Phylogenetic analysis
Multiple sequence alignments of nucleotide datasets were performed using the iterative G-INS-I method as implemented in MAFFT v7 [32]. The obtained alignments were optimized via Gblocks [33], followed by their manual correction considering the encoding reading frame. Phylogenetic trees were constructed using the Maximum Likelihood method under the best-fitting evolutionary model (GTR + G +I; GTR—general time reversal, G—gamma distribution, I—proportion of invariant sites), selected based on the corrected Akaike information criterion, as suggested by Mega v6 [34]. The stability of the obtained tree topologies was assessed by bootstrapping with 1000 resamplings of the original sequence data. The generated trees were edited for display using FigTree v1.4.3 (available at http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analysis
Statistical analysis was performed only for data from sites where a large number of sand flies was captured. Variables were tested for normality of distribution using the Shapiro-Wilk test. As assumption of normality was not valid (P < 0.05), nonparametric analysis was carried out. Chi-square analysis was performed to evaluate the difference in host preference within and between sand fly species. A P value < 0.05 was considered as statistically significant. Data were analyzed using statistical software Prism v4.00 (GraphPad Software Ltd., USA).
Results
Sand fly identification and Leishmania/Trypanosoma infection rates
During the two entomological surveys, a total of 3,090 sand flies (n = 1,866 females and n = 1,224 males) were captured. Species and details on phlebotomine sand flies collected in each site are summarized in Table 1. Among the female sand flies, S. minuta was the most abundant species (n = 1,264; 67.7%); followed by four species from the genus Phlebotomus: P. perniciosus (n = 594; 31.8%); P. sergenti (n = 4; 0.2%); P. perfiliewi (n = 3; 0.2%) and P. neglectus (n = 1; 0.1%). One hundred seventy-six females were blood fed (9.4%) and 113 were gravid (6.1%) (Table 2).
Table 1. Total sand flies collected during the two entomological surveys.
| Site (Year) | Sand fly species | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sergentomyia minuta | Phlebotomus perniciosus | Phlebotomus sergenti | Phlebotomus perfiliewi | Phlebotomus neglectus | ||||||
| Females | Males | Females | Males | Females | Males | Females | Males | Females | Males | |
| Site A (2017) | 62 | 48 | 157 | 141 | 1 | 2 | ||||
| Site B (2018) | 154 | 99 | 54 | 81 | 2 | |||||
| Site C (2018) | 153 | 172 | 1 | 22 | 1 | |||||
| Site D (2018) | 248 | 162 | 9 | 78 | 3 | 5 | 2 | |||
| Site E (2018) | 647 | 144 | 373 | 239 | 3 | 27 | ||||
| Total | 1,264 | 625 | 594 | 561 | 4 | 5 | 3 | 1 | 33 | |
Table 2. Female sand flies sampled and molecularly analysed.
| Site (Year) | Female sand fly species | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sergentomyia minuta (n = 1,264) | Phlebotomus perniciosus (n = 594) | Phlebotomus sergenti (n = 4) | Phlebotomus perfiliewi (n = 3) | Phlebotomus neglectus (n = 1) | |||||||||||
| Unfed | Blood fed | Gravid | Unfed | Blood fed | Gravid | Unfed | Blood fed | Gravid | Unfed | Blood fed | Gravid | Unfed | Blood fed | Gravid | |
| Site A (2017) | 60 | 1 | 1 | 153 | 4 | 1 | |||||||||
| Site B (2018) | 121 | 33 | 43 | 6 | 5 | ||||||||||
| Site C (2018) | 137 | 11 | 5 | 1 | 1 | ||||||||||
| Site D (2018) | 239 | 4 | 5 | 7 | 2 | 2 | 1 | ||||||||
| Site E (2018) | 587 | 21 | 39 | 223 | 128 | 22 | 2 | 1 | |||||||
| Total | 1,144 | 37 | 83 | 427 | 138 | 29 | 3 | 1 | 2 | 1 | 1 | ||||
Leishmania DNA was detected in twenty-eight female sand flies out of 1,866 (1.5%). In detail, 26 out of 1,264 (2.1%) S. minuta specimens (n = 21 unfed; n = 4 blood-fed and n = 1 gravid) tested positive to Leishmania tarentolae DNA; whereas Leishmania donovani complex DNA was identified in a S. minuta unfed female (1/1264; 0.1%) and in a P. perniciosus blood-fed female (1/594; 0.2%). Seventeen out of the 26 L. tarentolae positive females (n = 13 unfed; n = 3 blood-fed and n = 1 gravid) were captured in the rural biotype (Site E), whereas 9 (n = 8 unfed and n = 1 blood-fed) were collected in periurban environments (Sites A-D). Both L. donovani complex positive females (n = 1 unfed and n = 1 blood-fed) were captured in the rural environment (Site E).
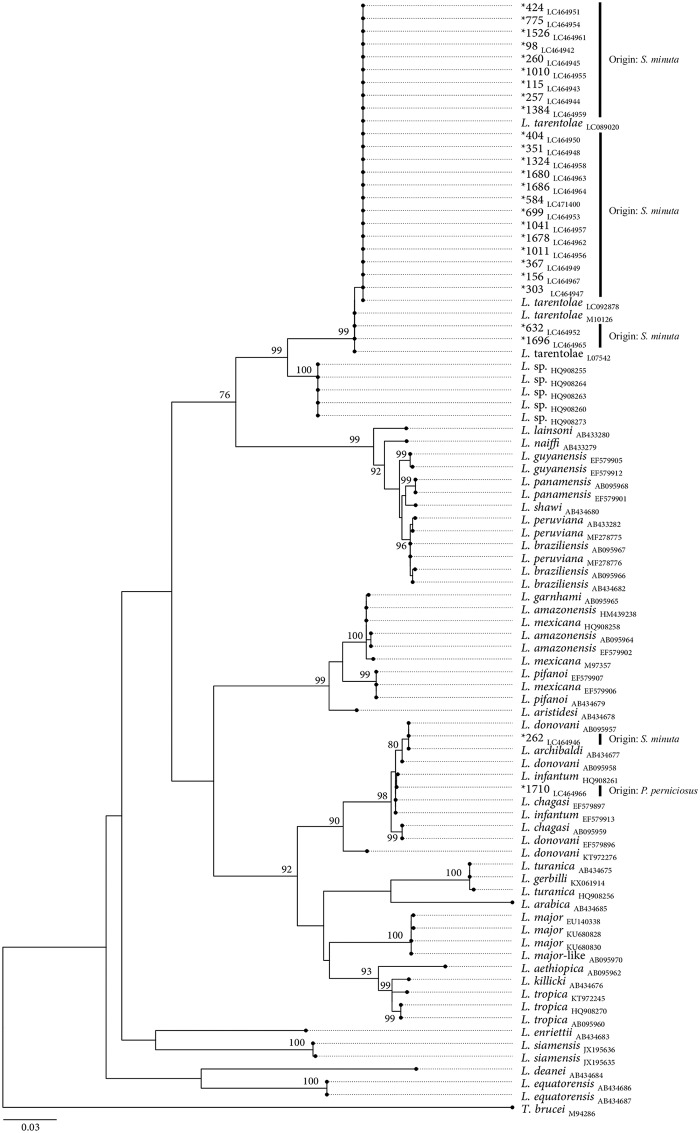
The 26 cytB sequences obtained from S. minuta revealed >99% sequence identity and 100% sequence coverage with reference sequences of L. tarentolae (accession number: LC092878). The cytB sequences obtained from S. minuta and P. perniciosus specimens, revealed > 99% identity and 100% sequence coverage with L. donovani complex sequences (accession numbers: CP022652; KX061917). All ctyB-rDNA obtained sequences were submitted to DDBJ (DDBJ Accession numbers: LC464942 to LC464968; LC471400). Based on phylogenetic analysis, 24 out of 26 obtained L. tarentolae sequences amplified from S. minuta, segregate together with L. tarentolae reference sequences in a monophyletic cluster, supported by a high bootstrap value (i.e., 99). Despite the heterogeneity of the sequences, phylogenetic analysis has not evidenced an unambiguous existence of different haplotypes. The two L. donovani complex sequences herein amplified from S. minuta and P. perniciosus extracts, segregate in the L. donovani complex cluster (Fig 2). Based on cytB phylogenetic tree, the sequences obtained from both specimens segregate independently. The existence of intra-groups was suggested within the L. donovani complex cluster, however none of them evidenced the monophyly of L. donovani/L. infanutm species.
Fig 2. Maximum likelihood phylogenetic tree based on unambiguous multiple-Leishmania cytB sequences alignment, using the GTR+G+I model of evolution.
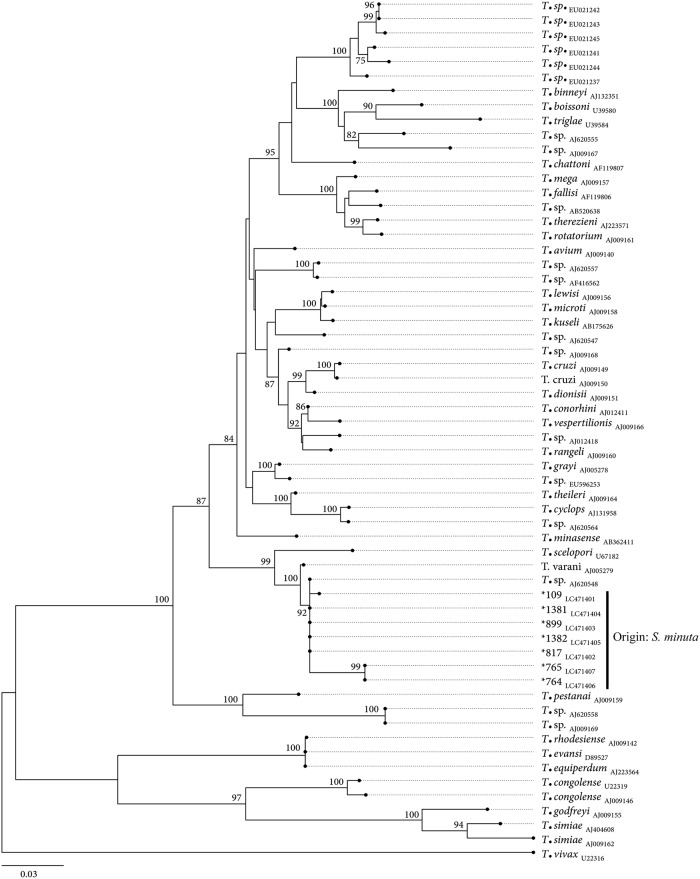
In addition to Leishmania, Trypanosoma DNA was detected in seven specimens of S. minuta, captured in rural Site E (n = 1) and periurban biotopes (Site A, n = 3; Site B, n = 1; Site C, n = 2). The 7 cytB sequences obtained revealed > 95% sequence identity and a sequence coverage > 80% with a reference sequence of Trypanosoma lewisi of Chinese origin (accession number: KR072974) [35]. The sequences were submitted to DDBJ (DDBJ Accession numbers: LC471393 to LC471399). The SSU-rDNA nucleotide sequences showed both 100% identity and coverage with the sequence of Trypanosoma sp. isolated from Gecko Tarentola annularis (accession number: AJ620548) [36]. The SSU-rDNA obtained sequences were submitted to DDBJ (DDBJ Accession numbers: LC471401 to LC471407). The phylogenetic tree shows that the obtained sequences share the same common ancestry of Trypanosoma varani (Accession number: AJ005279) forming together with Trypanosoma sp. isolated from Gecko a monophyletic cluster supported by a high bootstrap value (i.e., 92); whereas two sequences are different. These two sequences also share the same common ancestry of T. varani but segregate independently (Fig 3).
Fig 3. Maximum likelihood phylogenetic tree based on unambiguous multiple-Trypanosoma SSU-rDNA sequences alignment, using the GTR+G+I model of evolution.
At specific branch nodes bootstrap values (from 1000 random replicates of the original dataset) ≥ 75% are shown. The size bar indicates the number of nucleotide substitutions per site. The tree was rooted using as an outgroup a reference sequence of Trypanosoma brucei brucei (M94286). The sequences obtained in this study are identified with “*” and their respective accession numbers underscored.
At specific branch nodes bootstrap values (from 1000 random replicates of the original dataset) ≥ 75% are shown. The size bar indicates the number of nucleotide substitutions per site. The tree was rooted using as an outgroup a reference sequence of Trypanosoma vivax (U22316). The sequences obtained in this study are identified with “*” and their respective accession numbers underscored.
Identification of sand fly blood meals
The majority of blood-fed females were captured in site E (149/176; n = 128 P. perniciosus and n = 21 S. minuta) and underwent statistical analysis, whereas the remainder of 27 blood-fed specimens caught at the other sites (A, n = 4 P. perniciosus, n = 1 S. minuta; B, n = 6 P. perniciosus; C, n = 11 S. minuta; D, n = 4 S. minuta, n = 1 P. sergenti) were not statistically analysed (Table 2). Host mitochondrial cytB was successfully amplified from 169 out of 176 engorged females (96%), and the vertebrate hosts of 108 blood meals (64%) were distinctly identified (S2 Table).
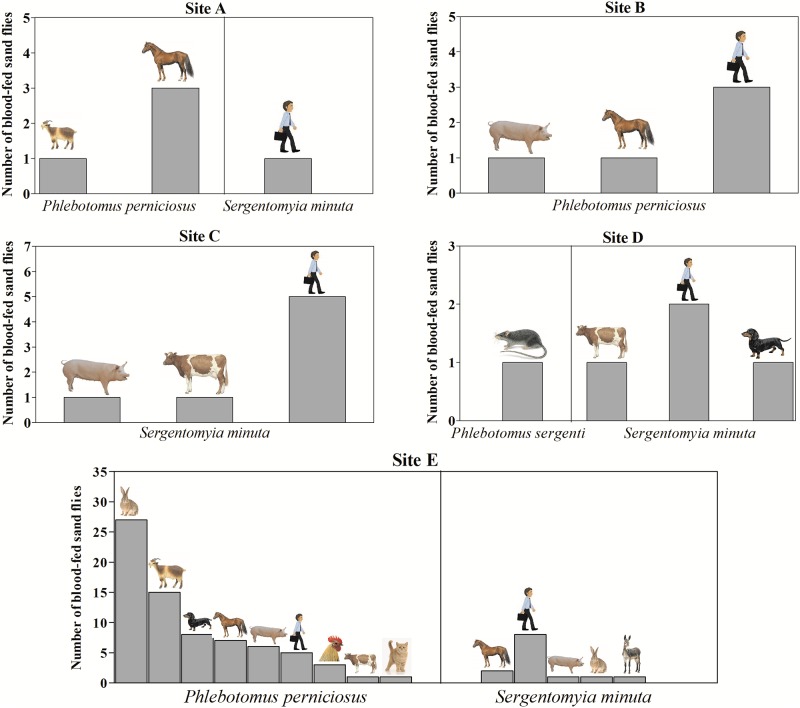
Wild rabbit (Oryctolagus cuniculus) (n = 28) and human (Homo sapiens) (n = 24) were the most frequent blood sources. Other vertebrate hosts also found were, i.e., goat (n = 16), horse (n = 13) pig (n = 9), dog (n = 9), chicken (n = 3), cow (n = 3), cat (n = 1), donkey (n = 1) and rat (n = 1). Blood sources for each sand fly species and site of capture are shown in Fig 4.
Fig 4. Vertebrate hosts identified per each sand fly species in selected sampling sites (A-E).
Identification of blood source was successfully achieved in two out of the four engorged S. minuta specimens positive for L. tarentolae DNA, as being human (Homo sapiens) and donkey (Equus asinus); whereas the host mitochondrial cytB partial gene could not be amplified from the blood-fed P. perniciosus positive to L. donovani complex DNA.
Wild rabbits (27/82; 32.9%) represented the most preferred mammal species for P. perniciosus, while S. minuta mainly fed on humans (16/25; 64%). Results of the statistical analysis on frequency of blood sources for P. perniciosus and S. minuta from site E are summarized in S3 Table, where a statistically significant higher frequency of blood meal on rabbit by P. perniciosus compared to S. minuta (χ2 = 183.1; P = 0.01), and on human by S. minuta compared to P. perniciosus (χ2 = 27.7; P < 0.001), was detected.
Discussion
Our study aimed to assess the presence of natural Leishmania infection in wild-caught sand fly species common in Sicily, Southern Italy; additionally, the source of blood meals of fed females was also determined to gain evidence on sand fly feeding habits and to allow the identification of potential/alternative reservoir hosts for Leishmania.
Four out of five sand fly species analysed in this study are proven vectors of human leishmaniosis. Among the genus Phlebotomus, P. perniciosus was the most abundant species, which is in agreement with other entomological surveys conducted in Southern Italy [13,29,37–38], and the presence of L. donovani complex DNA in P. perniciosus herein reported confirms the role of this species in the maintenance and spread of leishmaniosis in the Mediterranean area. Only a single P. perniciosus (0.2%; n = 594) tested positive for L. donovani complex. To the best of our knowledge, the sole data on the prevalence infection for L. infantum in P. perniciosus in Sicily came from a study conducted in the city of Catania where a higher infection rate (i.e., 11%; 8/72) has been detected [13]. This difference may be related to the high endemicity of leishmaniosis in the area of Catania [11,13] and suggest that the risk of transmission of Leishmania can be high even in urban areas.
Phlebotomus neglectus and P. perfiliewi specimens were occasionally collected in suburban and rural environments, and P. sergenti, the vector able to transmit L. tropica, was also sporadically collected, with its presence limited to the eastern side of the Island [12–14].
The detection of L. donovani complex DNA in a S. minuta unfed female herein recorded, spurs to better investigate the potential role of this species in the circulation and eventually transmission of the parasite [39]. The positivity of this sand fly species may be correlated to its feeding behaviour and the large availability of mammalian reservoir hosts positive to Leishmania in endemic foci [38]. Although Sergentomyia species mainly feed on cold-blooded vertebrates, its sporadic/opportunistic anthropophilic feeding behaviour has been already suggested [4] and, accordingly, a higher human blood preference has been herein statistically demonstrated. Scientific evidence on the competence of S. minuta in transmitting Leishmania spp. to warm-blooded mammals is not available so far; however, it has recently been demonstrated that L. donovani, L. infantum and L. major are not able to develop late-stage infections in Sergentomyia schwetzi sand fly species [40].Sergentomyia spp. are widespread Mediterranean sand fly species, and proven vectors of reptile Leishmania species (e.g., L. tarentolae) non-pathogenic to humans. The positivity of S. minuta for L. tarentolae herein recorded (2.1%; 26/1264) confirms that this species feeds on cold-blooded animals, as this protozoan parasite is widespread in Gekkonidae species [41].
In Italy, the presence of Trypanosoma spp. in sand flies has been little reported. In particular, Trypanosoma platydactyli was described in S. minuta [42], and the infection by a Trypanosoma belonging to Trypanosoma theileri group with very high homology to other trypanosomes detected in European cervids was recently reported in Phlebotomus perfiliewi [43]. In the Mediterranean area, Trypanosoma nabiasi, a rabbit trypanosome, and its co-infection with L. infantum was found in P. perniciosus female sand flies caught in the context of human leishmaniosis outbreak in Madrid [44], and natural infection of sand flies by trypanosomes of lizards, amphibians, birds and rodents has been already reported mainly from the American continent and Asia [45–48]. Despite Trypanosoma spp. being protozoan parasites transmitted by hematophagous insects [49–50] the potential of phlebotomine sand flies in transmitting trypanosomes is still unclear. The findings herein obtained, suggest that S. minuta might potentially be a vector of both Leishmania and Trypanosoma parasites in southern Italy. Nevertheless, the detection of the trypanosomatid DNA is not sufficient to incriminate the species as a competent vector since it may simply originate from a blood meal without undergoing further development and/or transmission. The detection of L. donovani complex DNA and, in addition, the detection of reptilian Trypanosoma DNA within S. minuta indicates that more attention is required when identifying parasitic organisms by PCR within sand fly vectors in areas where leishmaniosis is endemic. As a matter of fact, it is important to highlight that, when Trypanosoma and Leishmania species are present in the same geographical area, mixed infections could appear within the same host and/or vectors representing a challenge to their diagnosis [51–53].
In regard to the detection of L. donovani complex sequences in both P. perniciosus and S. minuta, a species-specific identification cannot be concluded based on cytB. In fact, the monophyly of L. infantum and L. donovani species has not been consistently shown [29].
Concerning the identification of blood meal sources in this study, several vertebrate hosts have been recognized as blood sources in the most abundant sand fly species (P. perniciosus, S. minuta), confirming the opportunistic feeding behaviour of these hematophagous insects [4,18–19].
Knowledge of the host preferences of sand flies under natural conditions is essential to understand how host choice and blood-feeding behaviour of sand flies influence their vector capacity in leishmaniosis foci. Even though Canis lupus familiaris is regarded as the main domestic reservoir host for zoonotic leishmaniosis, it seems not to be the preferred species on which they feed [4,18]. The results from the current study show that despite three out of the five selected sites being shelters with high an availability of dogs (i.e., from 300 to 500 dogs per site), canine DNA was not detected in engorged P. perniciosus specimens caught in any of them. This finding could however be biased by the broad use of insecticides and repellents on dogs as preventive measures against sand fly bites and Leishmania transmission. In this study, during the sand fly sampling period, about a third of all dogs hosted in the three shelters were treated with actives repellent against sand flies.
Of the total engorged females investigated in this study 85% were caught on a farm located in a rural area far away from human settlements. There a large variety of vertebrate hosts were available, with dogs and rabbits (either domestic or sylvatic) being the most abundant species. At this site, wild rabbits represented the most preferred mammalian species for P. perniciosus, which agrees with what has already been reported in Spain [20–22]. In fact, high levels of anti-P. perniciosus saliva antibodies in wild rabbits suggested their exposure and attractiveness to sand flies [54]. The large availability of lagomorphs could contribute to the maintenance of high density of P. perniciosus in areas where these mammal species are abundant [54] and, the role of lagomorphs in sustaining sylvatic Leishmania cycle independently from the domestic one has been strongly suggested [22,25].
The results obtained in the current study show a different blood feeding pattern of S. minuta compared to P. perniciosus, suggesting that the two sand fly species do not prefer the same vertebrate host. It is well-known that S. minuta feeds on cold-blooded reptiles; noteworthy, reptile DNA was not amplified in engorged females herein analysed, and only DNA of warm-blooded vertebrates was amplified. Interestingly, S. minuta caught in site E mainly fed on humans, though the presence of this vertebrate host inside the farm is limited to a few hours during the day, and no inhabitations were present nearby.
Conclusions
The current survey describes Leishmania infection rate in P. perniciosus and S. minuta and documents, for the first time in Sicily, Trypanosoma DNA presence in S. minuta unfed females.
The findings herein reported, highlight the well-known role of P. perniciosus as competent vector of L. infantum in western Mediterranean area; also, the identification of blood meal sources suggests the importance of wild rabbits in the maintenance of P. perniciosus and their potential role as sylvatic reservoirs of Leishmania.
The role of wild animals in the epidemiology of leishmaniosis as well as that of S. minuta as a vector of Leishmania spp. to humans is worthy of future investigations to achieve efficient control strategies.
Supporting information
aTrypanossomatidae kinetoplast DNA. bHost mitochondrial DNA. cPCR product was previously diluted 1:50 in nuclease-free water. Abbreviations: cytB, cytochrome b; ITS-rDNA, ribosomal internal transcribed spacer DNA; SSU-rRNA, small subunit ribosomal DNA; bp, base pars.
(DOCX)
A: LC469805; LC469817; LC469821; LC469832; LC469834; LC469836; LC469839; LC469842-44; LC469846-48; LC469850-51; LC469853-56; LC469863; LC469865; LC469871; LC469875-76; LC469878; LC469882; LC469887. B: LC469829; LC469837; LC469840-41; LC469852; LC469857; LC469859-62; LC469867; LC469872; LC469874; LC469880-81; LC469891-93; LC469896; LC469898; LC469900-03. C: LC469807-10; LC469813-16; LC469818-20; LC469822-26. D: LC469806; LC469811; LC469827-28; LC469830; LC469833; LC469835; LC469869; LC469877; LC469884, LC469890; LC469897; LC469908. E: LC469838; LC469845; LC469849; LC469858; LC469868; LC469870; LC469873; LC469883; LC469899. F: LC469885-86; LC469888-89; LC469905; LC469907; LC469910-12. G: LC469812; LC469906; LC469909. H: LC469831; LC469894; LC469904. I: LC469866. L: LC469879. M: LC469895.
(DOCX)
(DOCX)
Data Availability
Obtained sequences were deposited at the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp/), and the following Accession numbers were assigned: LC464942-LC464968; LC469805-LC469912; LC471393-LC471407 (http://getentry.ddbj.nig.ac.jp).
Funding Statement
Sand fly collection in 2018 season has been partially funded by Bayer Animal Health. The founder did not play any role in the study design, data collection and analysis, decision to publish and preparation of the manuscript. Molecular analysis on sand flies were partially founded by the grant Research and Mobility no.015063 awarded by the University of Messina. Dr Carla Maia has been awarded with Investigator Starting Grant F/01302/2015, and André Pereira received PhD grant SFRH/BD/116516/2016. Bayer Animal Health provided support in the form of salaries for authors [MP], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.”
References
- 1.Maroli M, Feliciangeli MD, Bichaud L, Charrel RN, Gradoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med Vet Entomol. 2013; 27(2):123–47. 10.1111/j.1365-2915.2012.01034.x [DOI] [PubMed] [Google Scholar]
- 2.Alten B, Maia C, Afonso MO, Campino L, Jiménez M, González E, et al. Seasonal Dynamics of Phlebotomine Sand Fly Species Proven Vectors of Mediterranean Leishmaniasis Caused by Leishmania infantum. PLoS Negl Trop Dis. 2016; 10(2):e0004458 10.1371/journal.pntd.0004458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoniou M, Gramiccia M, Molina R, Dvorak V, Volf P. The role of indigenous phlebotomine sandflies and mammals in the spreading of leishmaniasis agents in the Mediterranean region. Eurosurveillance. 2013; 18(30):20540 10.2807/1560-7917.es2013.18.30.20540 [DOI] [PubMed] [Google Scholar]
- 4.Maia C, Parreira R, Cristóvão JM, Freitas FB, Afonso MO, Campino L. Molecular detection of Leishmania DNA and identification of blood meals in wild caught phlebotomine sand flies (Diptera: Psychodidae) from southern Portugal. Parasit Vectors. 2015; 8:173 10.1186/s13071-015-0787-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.González E, Alvares A, Ruiz S, Molina R, Jiménez M. Detection of high Leishmania infantum loads in Phlebotomus perniciosus captured in the leishmaniasis focus of southwestern Madrid region (Spain) by real time PCR. Acta Trop. 2017; 171:68–73. 10.1016/j.actatropica.2017.03.023 [DOI] [PubMed] [Google Scholar]
- 6.Dujardin JC, Campino L, Cañavate C, Dedet JP, Gradoni L, Soteriadou K, et al. Spread of vector-borne diseases and neglect of Leishmaniasis, Europe. Emerg Infect Dis. 2008; 14(7):1013–8. 10.3201/eid1407.071589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Muccio T, Scalone A, Bruno A, Marangi M, Grande R, Armignacco O, et al. Epidemiology of imported Leishmaniasis in Italy: Implications for a European endemic country. PLoS One. 2015; 10(7):e0134885 10.1371/journal.pone.0134885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gramiccia M, Scalone A, Di Muccio T, Orsini S, Fiorentino E, Gradoni L. The burden of visceral leishmaniasis in Italy from 1982 to 2012: a retrospective analysis of the multi-annual epidemic that occurred from 1989 to 2009. Euro Surveill. 2013; 18(29):20535 [PubMed] [Google Scholar]
- 9.Vaselek S, Volf P. Experimental infection of Phlebotomus perniciosus and Phlebotomus tobbi with different Leishmania tropica strains. Int J Parasitol. 2019; 49(11):831–5. 10.1016/j.ijpara.2019.05.009 [DOI] [PubMed] [Google Scholar]
- 10.Otranto D, Napoli E, Latrofa MS, Annoscia G, Tarallo VD, Greco G, et al. Feline and canine leishmaniosis and other vector-borne diseases in the Aeolian Islands: Pathogen and vector circulation in a confined environment. Vet Parasitol. 2017; 236:144–51. 10.1016/j.vetpar.2017.01.019 [DOI] [PubMed] [Google Scholar]
- 11.Brianti E, Napoli E, Gaglio G, Falsone L, Giannetto S, Solari Basano F, et al. Field Evaluation of Two Different Treatment Approaches and Their Ability to Control Fleas and Prevent Canine Leishmaniosis in a Highly Endemic Area. PLoS Negl Trop Dis. 2016; 10(9):e0004987 10.1371/journal.pntd.0004987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaglio G, Brianti E, Napoli E, Falsone L, Dantas-Torres F, Tarallo VD, et al. Effect of night time-intervals, height of traps and lunar phases on sand fly collection in a highly endemic area for canine leishmaniasis. Acta Trop. 2014; 133:73–7. 10.1016/j.actatropica.2014.02.008 [DOI] [PubMed] [Google Scholar]
- 13.Lisi O, D’Urso V, Vaccalluzzo V, Bongiorno G, Khoury C, Severini F, et al. Persistence of phlebotomine Leishmania vectors in urban sites of Catania (Sicily, Italy). Parasit Vectors. 2014; 7:560 10.1186/s13071-014-0560-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Urso V, Ruta F, Khoury C, Bianchi R, Depaquit J, Maroli M. About the presence of Phlebotomus sergenti Parrot, 1917 (Diptera: Psychodidae) in Eastern Sicily, Italy. Parasite. 2004; 1(3):279–83. [DOI] [PubMed] [Google Scholar]
- 15.El Alem MMM, Hakkour M, Hmamouch A, Halhali M, Delouane B, Habbari K, et al. Risk factors and prediction analysis of cutaneous leishmaniasis due to Leishmania tropica in Southwestern Morocco. Infect Genet Evol. 2018; 61:84–91. 10.1016/j.meegid.2018.03.017 [DOI] [PubMed] [Google Scholar]
- 16.Antinori S, Gianelli E, Calattini S, Longhi E, Gramiccia M, Corbellino M. Cutaneous leishmaniasis: an increasing threat for travellers. Clin Microbiol Infect. 2005; 11(5):343–6. 10.1111/j.1469-0691.2004.01046.x [DOI] [PubMed] [Google Scholar]
- 17.Gramiccia M, Di Muccio T, Marinucci M. Parasite identification in the surveillance of imported leishmaniasis cases in Italy. Parassitologia. 2004; 46: 207–10. [PubMed] [Google Scholar]
- 18.Latrofa MS, Iatta R, Dantas-Torres F, Annoscia G, Gabrielli S, Pombi M, et al. Detection of Leishmania infantum DNA in phlebotomine sand flies from an area where canine leishmaniosis is endemic in southern Italy. Vet Parasitol. 2018; 253:39–42. 10.1016/j.vetpar.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 19.Rossi E, Bongiorno G, Ciolli E, Di Muccio T, Scalone A, Gramiccia M, et al. Seasonal phenology, host-blood feeding preferences and natural Leishmania infection of Phlebotomus perniciosus (Diptera, Psychodidae) in a high-endemic focus of canine leishmaniasis in Rome province, Italy. Acta Trop. 2008; 105(2):158–65. 10.1016/j.actatropica.2007.10.005 [DOI] [PubMed] [Google Scholar]
- 20.González E, Jiménez M, Hernández S, Martín-Martín I, Molina R. Phlebotomine sand fly survey in the focus of leishmaniasis in Madrid, Spain (2012–2014): seasonal dynamics, Leishmania infantum infection rates and blood meal preferences. Parasit Vectors. 2017; 10:368 10.1186/s13071-017-2309-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González E, Gállego M, Molina R, Abras A, Alcover MM, Ballart C, et al. Identification of blood meals in field captured sand flies by a PCR-RFLP approach based on cytochrome b gene. Acta Trop. 2015; 152:96–102. 10.1016/j.actatropica.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 22.Jiménez M, González E, Martín-Martín I, Hernández S, Molina R. Could wild rabbits (Oryctolagus cuniculus) be reservoirs for Leishmania infantum in the focus of Madrid, Spain? Vet Parasitol. 2014; 202(3–4):296–300. 10.1016/j.vetpar.2014.03.027 [DOI] [PubMed] [Google Scholar]
- 23.Carrillo E, Moreno J, Cruz I. What is responsible for a large and unusual outbreak of leishmaniasis in Madrid? Trends Parasitol. 2013; 29(12):579–80. 10.1016/j.pt.2013.10.007 [DOI] [PubMed] [Google Scholar]
- 24.Jiménez M, González E, Iriso A, Marco E, Alegret A, Fúster F, et al. Detection of Leishmania infantum and identification of blood meals in Phlebotomus perniciosus from a focus of human leishmaniasis in Madrid, Spain. Parasitol Res. 2013; 112(7):2453–9. 10.1007/s00436-013-3406-3 [DOI] [PubMed] [Google Scholar]
- 25.Molina R, Jiménez MI, Cruz I, Iriso A, Martín-Martín I, Sevillano O, et al. The hare (Lepus granatensis) as potential sylvatic reservoir of Leishmania infantum in Spain. Vet Parasitol. 2012; 190(1–2):268–71. 10.1016/j.vetpar.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 26.Gaglio G, Napoli E, Arfuso F, Abbate JM, Giannetto S, Brianti E. Do Different LED Colours Influence Sand Fly Collection by Light Trap in the Mediterranean? Biomed Res Int. 2018; 2018:6432637 10.1155/2018/6432637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dantas-Torres F, Tarallo VD, Otranto D. Morphological keys for the identification of Italian phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae). Parasit Vectors. 2014; 7:479 10.1186/s13071-014-0479-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Tai NO, Osman OF, El Fari M, Presber W, Schönian G. Genetic heterogeneity of ribosomal internal transcribed spacer (its) in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms (sscp) and sequencing. Trans Royal Soc Trop Med Hyg. 2000; 94:1–5. [DOI] [PubMed] [Google Scholar]
- 29.Pereira A, Parreira R, Cristóvão JM, Castelli G, Bruno F, Vitale F, et al. Phylogenetic insights on Leishmania detected in cats as revealed by nucleotide sequence analysis of multiple genetic markers. Infect Genet Evol. 2020; 77:104069 10.1016/j.meegid.2019.104069 [DOI] [PubMed] [Google Scholar]
- 30.Kato H, Gomez EA, Cáceres AG, Vargas F, Mimori T, Yamamoto K, et al. Natural infections of man-biting sand flies by Leishmania and Trypanosoma species in the northern Peruvian Andes. Vector Borne Zoonotic Dis. 2011; 11(5):515–21. 10.1089/vbz.2010.0138 [DOI] [PubMed] [Google Scholar]
- 31.Svobodová M, Alten B, Zídková L, Dvorák V, Hlavacková J, Mysková J, et al. Cutaneous leishmaniasis caused by Leishmania infantum transmitted by Phlebotomus tobbi. Int J Parasitol. 2009; 39(2):251–6. 10.1016/j.ijpara.2008.06.016 [DOI] [PubMed] [Google Scholar]
- 32.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013; 30(4):772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000; 17(4): 540–52. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013; 30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin RH, Lai DH, Zheng LL, Wu J, Lukeš J, Hide G, et al. Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen. Parasit Vectors. 2015; 8:665 10.1186/s13071-015-1281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol. 2004; 34(12):1393–404. 10.1016/j.ijpara.2004.08.011 [DOI] [PubMed] [Google Scholar]
- 37.Foglia Manzillo V, Gizzarelli M, Vitale F, Montagnaro S, Torina A, Sotera S, et al. Serological and entomological survey of canine leishmaniasis in Lampedusa island, Italy. BMC Vet Res. 2018; 14(1):286 10.1186/s12917-018-1606-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira S, Pita-Pereira D, Araujo-Pereira T, Britto C, Costa-Rego T, Ferrolho J, et al. First molecular detection of Leishmania infantum in Sergentomyia minuta (Diptera, Psychodidae) in Alentejo, southern Portugal. Acta Trop. 2017; 174:45–8 10.1016/j.actatropica.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 39.Maia C, Depaquit J. Can Sergentomyia (Diptera, Psychodidae) play a role in the transmission of mammal-infecting Leishmania? Parasite. 2016; 23:55 10.1051/parasite/2016062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadlova J, Dvorak V, Seblova V, Warburg A, Votypka J, Volf P. Sergentomyia schwetzi is not a competent vector for Leishmania donovani and other Leishmania species pathogenic to humans. Parasit Vectors. 2013; 6:186 10.1186/1756-3305-6-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pozio E, Gramiccia M, Gradoni L, Maroli M. Hemoflagellates in Cyrtodactylus kotschyi (Steindachner, 1870) (Reptilia, Gekkonidae) in Italy. Acta Trop. 1983; 40(4):399–400. [PubMed] [Google Scholar]
- 42.Gramiccia M, Gradoni L, Maroli M. Caractérisation enzymatique de Trypanosoma platydactyli CATOUlLLARD, 1909 isolé de Sergentomyia minuta minuta RONDANI, 1843 en Italie. Ann Parasitol Hum Comp. 1989; 64(2):154–6. French [Google Scholar]
- 43.Calzolari M, Rugna G, Clementi E, Carra E, Pinna M, Bergamini F, et al. Isolation of a Trypanosome Related to Trypanosoma theileri (Kinetoplastea: Trypanosomatidae) from Phlebotomus perfiliewi (Diptera: Psychodidae). Biomed Res Int. 2018; 2597074 10.1155/2018/2597074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González E, Molina R, Jiménez M. Rabbit trypanosome detection in Phlebotomus perniciosus sand flies from the leishmaniasis outbreak in Madrid, Spain. Acta Trop. 2018; 187: 201–6. 10.1016/j.actatropica.2018.08.011 [DOI] [PubMed] [Google Scholar]
- 45.Srisuton P, Phumee A, Sunantaraporn S, Boonserm R, Sor-Suwan S, Brownell N, et al. Detection of Leishmania and Trypanosoma DNA in Field-Caught sand flies from endemic and non-endemic areas of Leishmaniasis in southern Thailand. Insects. 2019; 10(8):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phumee A, Tawatsin A, Thavara U, Pengsakul T, Thammapalo S, Depaquit J, et al. Detection of an unknown Trypanosoma DNA in a Phlebotomus stantoni (Diptera: Psychodidae) collected from southern Thailand and records of new sand flies with reinstatement of Sergentomyia hivernus Raynal & Gaschen, 1935 (Diptera: Psychodidae). J Med Entomol. 2017; 54(2):429–34. 10.1093/jme/tjw161 [DOI] [PubMed] [Google Scholar]
- 47.Ferreira TS, Minuzzi-Souza TTC, Andrade AJ, de Coehlo TO, de Rocha DA, Obara MT, et al. Molecular detection of Trypanosoma sp. and Blastocrithidia sp. (Trypanosomatidae) in phlebotomine sand flies (Psychodidae) in the Federal District of Brazil. Rev Soc Bras Med Trop. 2015; 48(6):776–9. 10.1590/0037-8682-0076-2015 [DOI] [PubMed] [Google Scholar]
- 48.Kato H, Uezato H, Sato H, Bhutto AM, Soomro FR, Baloch JH, et al. Natural infection of sand fly Phlebotomus kazeruni by Trypanosoma species in Pakistan. Parasit Vectors. 2010; 3:10 10.1186/1756-3305-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton PB, Gibson WC, Stevens JR. Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomial RNA and protein-coding gene phylogenies. Mol Phylogenet Evol. 2007; 44(1):15–25. 10.1016/j.ympev.2007.03.023 [DOI] [PubMed] [Google Scholar]
- 50.Desser SS. The blood parasites of anurans from Costa Rica with reflections on the taxonomy of their trypanosomes. J Parsitol. 2001; 87(1):152–60. [DOI] [PubMed] [Google Scholar]
- 51.Bastrenta B, Mita N, Buitrago R, Vargas F, Flores M, Machane M, et al. Human mixed infections of Leishmania spp. and Leishmania-Trypanosoma cruzi in a sub Andean Bolivian area: identification by polymerase chain reaction/hybridization and isoenzyme. Mem Inst Oswaldo Cruz. 2003; 98(2):255–64. 10.1590/s0074-02762003000200015 [DOI] [PubMed] [Google Scholar]
- 52.De Araújo VAL, Boité MC, Cupolillo E, Jansen AM, Roque ALR. Mixed infection in the anteater Tamandua tetradactyla (Mammalia: Pilosa) from Pará State, Brazil: Trypanosoma cruzi, T. rangeli and Leishmania infantum. Parasitology. 2013; 140(4): 455–60. 10.1017/S0031182012001886 [DOI] [PubMed] [Google Scholar]
- 53.Díaz-Sáez V, Merino-Espinosa G, Morales-Yuste M, Corpas-López V, Pratlong F, Morillas-Márquez F, et al. High rates of Leishmania infantum and Trypanosoma nabiasi infection in wild rabbits (Oryctolagus cuniculus) in sympatric and syntrophic conditions in an endemic canine leishmaniasis area: epidemiological consequences. Vet Parasitol. 2014; 202(3–4):119–27. 10.1016/j.vetpar.2014.03.029 [DOI] [PubMed] [Google Scholar]
- 54.Martín-Martín I, Molina R, Rohoušová I, Drahota J, Volf P, Jiménez M. High levels of anti-Phlebotomus perniciosus saliva antibodies in different vertebrate hosts from the re-emerging leishmaniosis focus in Madrid, Spain. Vet Parasitol. 2014; 202(3–4):207–16. 10.1016/j.vetpar.2014.02.045 [DOI] [PubMed] [Google Scholar]