Abstract
Purpose
The leucine zipper-EF-hand containing transmembrane protein 1 (LETM1) is a mitochondrial protein that has been associated with the occurrence and development of malignant tumors. Previous studies have shown that LETM1 expression is increased in several types of human cancer and is associated with a poor clinical outcome. However, the role of LETM1 in prostate cancer (PCa) has not yet been determined. In this study, we investigated the clinicopathological significance of LETM1 expression and its role in PCa progression.
Methods
We assessed the expression of LETM1 and genes related to cancer stemness, epithelial-mesenchymal transition (EMT), cell cycle, and PI3K/Akt signaling in 133 paraffin-embedded PCa tissue samples and cancer cells by using immunohistochemistry, immunofluorescence, and Western blotting.
Results
LETM1 expression was significantly increased in PCa, and it was positively correlated with Gleason score, pathologic tumor (pT) stage, clinical stage, and high microvessel density. Survival analysis showed that patients with PCa with a high level of LETM1 expression exhibited a low overall survival. Cox regression analysis indicated that LETM1 is an independent poor prognostic PCa factor. Additionally, the expression of LETM1 was correlated with cancer cell stemness-associated genes, EMT-related genes, cell cycle regulatory genes, and PI3K/Akt signaling gene expression in PCa. Furthermore, knocking down LETM1 expression down-regulated the expression of stemness-related proteins, while inhibiting tumor spheroid formation, EMT-like changes, cell proliferation, migration, and invasion in PCa cells. Importantly, the PI3K inhibitor LY294002 strongly inhibited the expression of LETM1, pPI3K-p85, and pAkt (Thr308, Ser473) in PCa cells.
Conclusion
These results indicate that LETM1 expression is associated with cancer cell stemness, promotes EMT-like changes and cell proliferation and is a potential prognostic biomarker for PCa.
Keywords: prostate cancer, leucine zipper-EF-hand containing transmembrane protein 1, epithelial-mesenchymal transition, prognosis
Introduction
Prostate cancer (PCa) is the second most prevalent cancer in men worldwide, accounting for an estimated 26,730 deaths in 2016.1,2 Metastasis is a high-risk complication and the major cause of death in patients with PCa.3,4 Androgen deprivation therapy remains the primary clinical treatment for patients in the early stages of PCa. As the disease develops into castration-resistant PCa, almost all patients with advanced PCa will experience recurrence and distal organ metastasis.5 Therefore, it is necessary to improve current therapeutic strategies and explore new molecular biomarkers for predicting the progression of PCa and for developing targeted therapies.
Cancer stem-like cells (CSCs) are a subpopulation of tumor cells with tumorigenic and metastatic capacities.6–8 Previous studies have shown that the proportion of stem cells present in tumors is closely related to disease progression. A high CSC count is associated with poor clinical outcome and metastasis.9 Several studies have confirmed that epithelial-mesenchymal transition (EMT) is associated with the emergence of drug resistance and acquisition of CSC-like properties.10,11 Inducing EMT creates a self-renewing state that enables redifferentiation to allow colonization and growth at distant metastatic sites.12,13 Many signals from the tumor microenvironment contribute to the induction of EMT and development prostate CSC phenotypes that, in turn, initiate a cascade of signals leading to metastasis.12 Conventional therapies have failed to eradicate carcinoma cells that have entered a CSC-like state via activation of EMT.14
Leucine zipper-EF-hand containing transmembrane protein 1 (LETM1) is a mitochondrial inner membrane protein that is conserved in eukaryotes from yeast to humans.15,16 The LETM1 gene was first identified in Wolf-Hirschhorn syndrome and encodes the human homolog of the yeast Mdm38p gene.17 LETM1-mediated mitochondrial biogenesis increases the glycolytic ATP supply, and the mitochondrial dynamics regulate stem cell fate.18,19 LETM1 is a biomarker associated with poor prognosis and tumor progression in patients with colorectal adenocarcinoma as well as breast, bladder, thyroid, and esophageal cancers.20–24 However, the clinical significance of LETM1, its function in tumorigenesis, and its regulation in PCa are not yet fully understood.
In this study, we investigated the clinical significance of LETM1 in PCa and evaluated the relationship between its expression and cancer stemness, cell proliferation, EMT-related genes, and the PI3K/Akt pathway.
Materials and Methods
Tissue Specimens
A total of 133 cases of PCa tissue samples and 14 cases of benign prostatic hyperplasia (BPH) tissue samples were obtained from patients who underwent prostate surgery and were stored in Xi’an Alena Biotechnology Ltd., Co. (Outdo Biotech) and Department of Pathology of Yanbian University Affiliate Hospital. No patient received preoperative chemotherapy or radiotherapy. Pathological parameters, including age, tumor grade, clinical stage, primary tumor (pT) stage, and lymph node metastasis were carefully reviewed in all prostate cancer patients. The written informed consent was obtained from each patient, and that their privacy would be maintained. This study was approved by the Human Ethics Committee and the Research Ethics Committee of Yanbian University, and conducted in accordance with the Declaration of Helsinki (No. YBUCM-81760531).
Cell Lines
LNCaP, PC3 and DU145 cell lines (purchased from American-Type Culture Collection, Manassas, USA) were maintained in RPMI-1640 with 10% heat-inactivated fetal bovine serum (FBS, Life Technologies, Grand Island, NY), 100 mg/mL penicillin G, and 50 mg/mL streptomycin (Life Technologies, Grand Island, NY) at 37°C in a humidified atmosphere containing 5% CO2. Cells were treated (for 48h unless otherwise specified) with LY24002 (Abcam, Cambridge, UK) according to the manufacturer’s instructions.
Immunohistochemical (IHC) Analysis
Tissue sections on microscope slides were deparaffinized, hydrated, and treated with 3% H2O2 for 15 min to quench endogenous peroxidase activity. Sections were immersed in TE buffer (10 mM Tris and 1 mM EDTA, pH 9.3) for epitope retrieval in a microwave for 30 min. The slides were then incubated with 4% bovine serum albumin for 30 min to block nonspecific immunoreactivity. The sections were then incubated with primary antibodies for 60 min at room temperature. Antibodies used in present study were listed in Supplementary Table 1. Sections were then incubated with an anti mouse/rabbit antibody (Envision plus, Dako, Denmark, catalog: K801021-2) for 30 min at room temperature. The chromogen used was ImmPACT AEC Peroxidase Substrate (VECTOR Laboratories) for 20 mins. After reading and taking photographs of the slides, sections were then stripped one time used stripping buffer (20% SDS, 0.5M Tris, and mercaptoethanol) to removing the original antibody for one hour in a water bath at 56 °C to remove the original antibody and then for 10 mins in alcohol so that the sections could be restained. Omitting the primary antibody provided negative controls for immunostaining. All the primary antibody stained in the same blots, and in serial sections.
The double immunostaining procedure was performed using a two-step method with LETM1 antibody and anti-CD105 antibody (1:250, Abcam, Cambridge, UK, ab170943) to observe the relationship between the expression of LETM1 and microvessel density (MVD) in PCa. Primarily, for the LETM1 protocols, except that the chromogen with the 3, 3ʹ-diaminobenzidine (Dako) for 10 mins (FLEX20), all steps are the same. Then, subsequent staining of the same section was performed after incubating the samples with an antibody to CD105 by ImmPACT AEC Peroxidase Substrate for 20 mins.
Evaluation of the IHC Analysis
Two pathologists (LHP & YHX) who did not possess knowledge of the clinical data examined and scored all tissue specimens. IHC scores was measured, as follows: [IHC score 1], weak staining in <50% or moderate staining in <20% of carcinoma cells; [IHC score 2], weak staining in ≧50%, moderate staining in 20-50% or strong staining in <20% of carcinoma cells; [IHC score 3], moderate staining in ≧50% or strong staining in ≧20% of carcinoma cells. Cases with score 2 and 3 were regarded as positivity for each protein expression, the staining results were semi-quantitatively scored as negative and positive. In case of discrepancies, a final score was established by reassessment by both pathologists using a double-headed microscope.25
Western Blotting
Immuno-blot analysis was performed as previously described.4 Cells were collected and lysed with protease inhibitors. Equal amounts of proteins was loaded on SDS-PAGE gels and then transferred to a PVDF membrane. Membranes were blocked 2 h at RT with 5% skim milk (diluted in TBS), and then incubated with primary antibodies at 4 °C shaking for overnight. Membranes were then washed three times with TBST followed by incubation with horseradish peroxidase conjugated goat anti-rabbit/mouse IgG secondary antibody (1:500, BIOSS, Beijing, China) for 1h at room temperature. Finally, the membranes were washed three times with TBST, and the protein bands were detected using an ECL system (Merck) according to the manufacturer’s instructions.
Immunofluorescence (IF) Analysis
PC3 and DU145 cells were subcultured in a 6-well plate and incubated at 37 °C 5% CO2. After sample preparation by fixation, permeabilization, and blocking, the slides were incubated with primary antibody diluted in 3% BSA at 56 °C overnight. Following primary antibody incubation, the slides were then washed three times and incubated with conjugated secondary antibodies in 3% BSA for 1 h at RT. The slides were washed three times with PBST. The cells were counter stained with DAPI (Vector Laboratorise, Burlingame, CA, USA). Finally, the location of proteins in cell were observed in a confocal laser scanning microscope (Carl Zeiss, Thornwood, New York).26
Cell Transfection
LETM1 endoribonuclease-prepared short interfering RNAs (esiRNA) was designed and synthesized by Sigma-Aldrich (St. Louis, MO, USA). The sequence of LETM1 esiRNA was listed in Supplementary Table 2. Additionally, control esiRNA (control) was also used in this study. Transfections were performed using Lipofectamine 2000 (Invitrogen, Life Technologies, Grand Island, NY) according to the manufacturer’s instructions.
Tumor Sphere-Forming Assay
The PC3 and DU145 cells treated with or without LETM1 esiRNA for 48h were seeded in Corning ultra-low attachment 6-well plates in 2 mL of DMEM/F12 medium supplemented with 10 ng/mL each of human bFGF and human EGF and with 2% B27. After one week and two weeks, cell morphology was examined under light microscopy.
Migration and Invasion Assays
We used 24-well culture plate with an 8.0-µm transparent PET membrane separating the upper and lower halves of each well (Corning) to measure the migration ability and invasiveness of cells. For the invasion assay, we covered the upper side of the membrane with Matrigel (BD). The assays and counting of migrating or invading PC3 and DU145 cells were performed as described previously.26
Cell Cycle Experiment
We treated PC3 and DU145 with 200 pM LETM1 esiRNA for 48 h. Next, the cells were reseeded in new six-well plates and cultured. Then, the cells were fixed in 70% ethanol for 24 h at 4°C. Thereafter, the cells were washed with PBS, and then stained them with 10 µg/mL propidium iodide solution for 30 min. We analyzed the cell cycle profiled by flow cytometry.
Statistical Analysis
Correlations were examined using Pearson’s chi-square test as appropriate. Overall survival (OS) were determined using the Kaplan-Meier method and were compared using the Log rank test. The Cox proportional hazards model was used for multivariate analysis. The data are expressed as the mean ± standard deviation (SD). Comparisons between groups were analyzed using Student’s t-test. The statistical analysis was performed using SPSS 25.0 IBM Singapore Pte Ltd. (registration N 1975-01566-C) and GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). All tests were two sided, and P<0.05 was considered significant.
Results
LETM1 Expression is Associated with Unfavorable Clinicopathological Features and Poor Prognosis in PCa
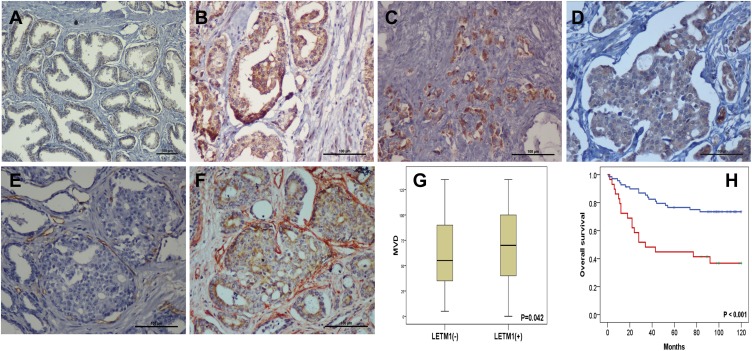
The expression of LETM1 in specimens from 14 cases of BPH and 133 cases of PCa was determined through IHC analysis. Our results showed that 3 out of 14 BPH cases (21.4%) and 95 out of 133 PCa cases (71.4%) positively stained for LETM1 (Figure 1A–D and Supplementary Table 3). LETM1 expression was mainly detected in the cytoplasm of prostate epithelial cells (Figure 1A–D). Chi-square analysis showed that high LETM1 expression was significantly associated with Gleason score (P = 0.001), pT stage (P = 0.005), and clinical stage (P = 0.006) (Table 1). CD105 (endoglin) was used as an endothelial cell marker, and its expression was observed in the new capillary blood vessels around cancer cells. Double IHC staining for LETM1 and CD105 in PCa tissues showed that microvessel density (MVD) was higher in LETM1-positive cases (67.68 ± 11.245) than in LETM1-negative cases (54.35 ± 10.291, P = 0.042) (Figure 1E–G). These results indicated that LETM1 expression might contribute to angiogenesis and tumor progression in PCa.
Figure 1.
LETM1 expression is associated with poor outcome in prostate cancer (PCa). Tissue Microarray (TMA) analysis immunohistochemistry staining of LETM1 was performed to investigate LETM1 expression in human PCa tissues; 133 cases of formalin-fixed primary PCa tissues and 14 benign prostatic hyperplasia (BPH) tissue samples were investigated. (A) Immunohistochemical analysis of LETM1 expression in BPH. (B) LETM1 expression in highly differentiated PCa tissues. (C) LETM1 expression in poorly differentiated PCa tissues. (D) LETM1 expression in lymphatic invasion PCa (original magnification 200×). Tumor angiogenesis is a key factor for LETM1 in promoting tumor progression. CD105 staining in LETM1-negative (E) and -positive (F) PCa samples. LETM1 (brown reaction product) in the cancer cell cytoplasm, and CD105 (red reaction product) in vascular endothelium (original magnification 200×). (G) Expression of LETM1 in PCa was significantly associated with increased MVD. (H) Kaplan-Meier analysis was used to test for a significant association between LETM1 expression and overall survival (OS) in PCa patients. Two types of OS outcomes were compared in the plot: a LETM1-positive group (red solid line) and a LETM1-negative group (blue solid line). The OS time in months is indicated on the x-axis, and the y-axis shows the probability of OS.
Table 1.
Comparison of Clinicopathologic Characteristics According to LETM1 Expression in Prostate Cancer
| Variable | n | LETM1(−) n(%) |
LETM1(+) n(%) |
χ2 | R | P |
|---|---|---|---|---|---|---|
| Age (years) | 1.266 | 0.092 | 0.260 | |||
| ≦65 | 52 | 18(34.6) | 34(65.4) | |||
| >65 | 81 | 20(24.7) | 61(75.3) | |||
| Gleason | 13.849 | 0.216 | 0.001 | |||
| Gleason4–6 | 10 | 8(80.0) | 2(20.0) | |||
| Gleason7–8 | 101 | 25(24.8) | 76(75.2) | |||
| Gleason9–10 | 22 | 5(22.7) | 17(77.3) | |||
| pT stage | 7.809 | 0.243 | 0.005 | |||
| T1 | 55 | 24(43.6) | 31(56.4) | |||
| T2 | 36 | 8(22.2) | 28(77.8) | |||
| T3 | 31 | 5(16.1) | 26(83.9) | |||
| T4 | 11 | 1(9.1) | 10(90.9) | |||
| Lymph node metastasis | 0.203 | 0.039 | 0.652 | |||
| Negative | 123 | 36(29.3) | 87(70.7) | |||
| Positive | 10 | 2(20.0) | 8(80.0) | |||
| Clinical stage | 7.650 | 0.241 | 0.006 | |||
| I | 52 | 23(44.2) | 29(55.8) | |||
| II | 38 | 9(23.7) | 29(76.3) | |||
| III | 30 | 5(16.7) | 25(83.3) | |||
| IV | 13 | 1(7.7) | 12(92.3) | |||
| CD68 | 0.330 | 0.048 | 0.566 | |||
| Negative | 59 | 19(32.2) | 40(67.8) | |||
| Positive | 74 | 19(25.7) | 55(74.3) | |||
| LC3A | 1.080 | 0.085 | 0.299 | |||
| Negative | 66 | 21(31.8) | 45(68.2) | |||
| Positive | 67 | 17(25.4) | 50(74.6) |
To assess the association between LETM1 expression and the survival of patients with PCa, Kaplan–Meier curves with a Log rank test were used to determine the overall survival (OS). The expression of LETM1 was significantly associated with poor OS in patients with PCa (P < 0.001) (Figure 1H). Furthermore, univariate and multivariate Cox regression analyses demonstrated that lymph node metastasis (both P < 0.001) and LETM1 expression in PCa (P < 0.001 and P = 0.006, respectively) were significant poor prognostic factors of OS. (Supplementary Table 4). These results indicated that LETM1 expression is a potential biomarker for poor PCa prognosis.
LETM1 Expression Is Associated with Cancer Stemness in PCa
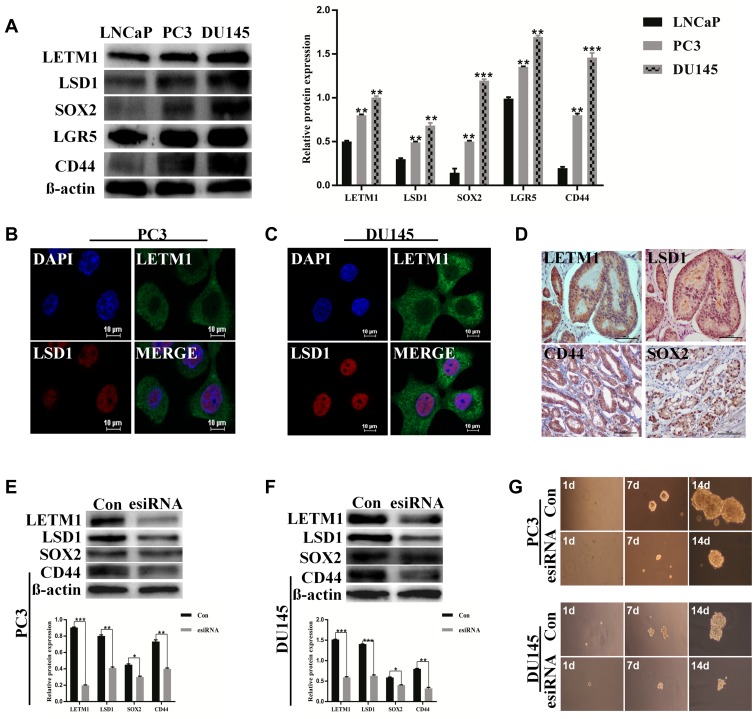
We examined the protein levels of LETM1 and stemness-related proteins in PCa cell lines using Western blotting. PC3 and DU145 cell lines have higher metastatic potential than LNCaP cells. As a result, stemness-related proteins such as LSD1, SOX2, LGR5, and CD44 were co-upregulated with LETM1 in DU145 and PC3 cells but not in LNCaP cells (Figure 2A) (all P < 0.01). Furthermore, LETM1-positive cell population markedly overlapped with the LSD1-positive cell population in PC3 and DU145 cells (Figure 2B and C), and LETM1/LSD1 co-localized in the same PCa tissues (Figure 2D). The expression of LETM1 and LSD1, SOX2, and CD44 in PCa tissue serial sections are shown in Figure 2D. To further verify the relationship between LETM1 and stemness-related proteins, we blocked LETM1 expression using LETM1-specific esiRNA in PCa cells. Knocking down LETM1 expression dramatically decreased the expression of cancer stemness-related proteins such as LSD1, SOX2, and CD44 in PCa cells (Figure 2E and F). Tumor spheroid formation assays showed that in LETM1-silenced PC3 and DU145 cells, tumor spheroid sizes at 1, 7, and 14 days were lower than those in the control group (Figure 2G). These results indicate that LETM1 activation regulates plasticity in PCa.
Figure 2.
LETM1 promotes prostate cancer (PCa) stem cell-like properties. (A) Western blot analysis for the protein levels of LETM1, LSD1, SOX2, LGR5, and CD44 in PCa cells. β-actin was used as a loading control. (B) and (C) Immunofluorescence staining for LETM1 and LSD1 in PC3 and DU145 cells (original magnification 600×). Blue color corresponds to DAPI staining; green to LETM1; red to LSD1; double labeling to merge. (D) Immunohistochemical staining for LETM1, LSD1, SOX2, and CD44 in the PCa tissue (original magnification 200×). Protein expression of LETM1, LSD1, SOX2, and CD44 after LETM1 knockdown compared with control in PC3 (E) and DU145 (F) cells. (G) Sphere-formation assay of PC3 and DU145 cells with LETM1-knockdown. *P < 0.05, **P < 0.01, ***P < 0.001 versus control.
LETM1 Expression Correlates with Cancer Epithelial Cell EMT-Like Changes in PCa
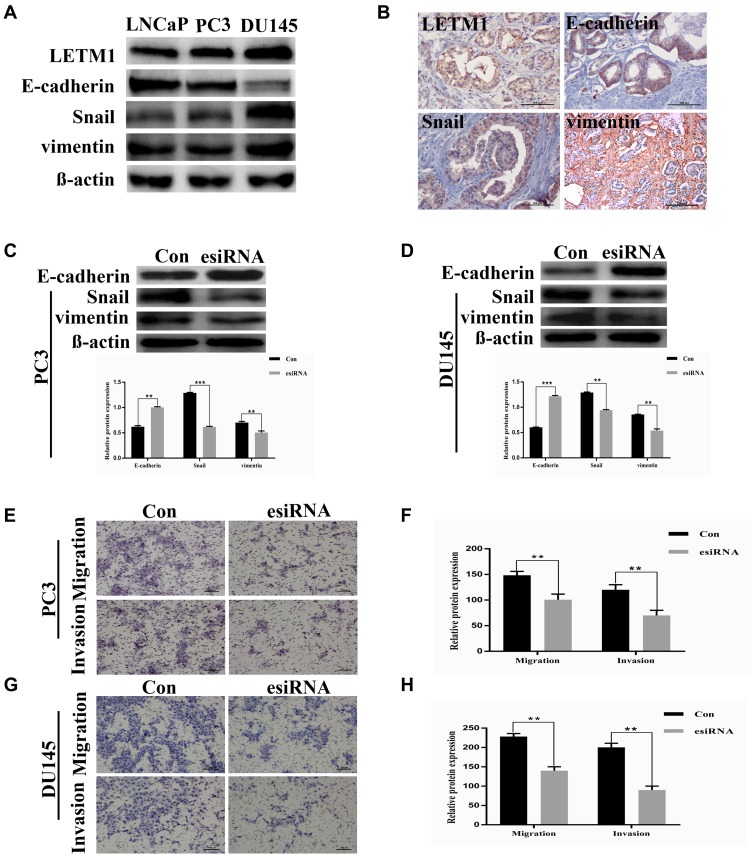
To investigate the correlation between LETM1 and EMT, protein expression levels of molecular markers including E-cadherin, vimentin, and Snail were analyzed in PCa cells and tissue serial sections through Western blotting and IHC staining. Our results confirmed that expression of LETM1, Snail, and vimentin was higher in poorly differentiated DU145 cells than in more differentiated LNCaP cells, and E-cadherin was down-regulated in DU145 cells (Figure 3A). LETM1 expression was significantly correlated with levels of Snail (P = 0.029) and vimentin (P = 0.025) in PCa tissues (Figure 3B and Table 2). Moreover, LETM1 knockdown in PCa cells resulted in significantly higher E-cadherin (PC3: P < 0.01; DU145: P < 0.001) expression and lower Snail (PC3: P < 0.001; DU145: P < 0.01) and vimentin (all P < 0.01) expression than those in the control group (Figure 3C and D).
Figure 3.
LETM1 expression correlates with cancer epithelial cells epithelial-mesenchymal transition (EMT) in prostate cancer (PCa). (A) Western blotting analysis for the protein expression of LETM1, E-cadherin, Snail, and vimentin in PCa cell lines. β-actin was used as a loading control. (B) Tissue microarray immunohistochemistry staining of PCa for LETM1, and EMT markers E-cadherin, Snail, and vimentin (original magnification 200×). (C) and (D) Effects of silencing LETM1 in PC3 and DU145 cells on E-cadherin, Snail, and vimentin levels. (E–H) PC3 and DU145 cells’ migration and invasion ability was investigated by transwell and Matrigel invasion assays after LETM1 knockdown. **P < 0.01, ***P < 0.001 versus control.
Table 2.
The Association Between Protein Expression of LETM1 and the Factors of EMT in Prostate Cancer
| Variable | n | LETM1(−) n(%) | LETM1(+) n(%) | χ2 | R | P |
|---|---|---|---|---|---|---|
| E-cadherin | 0.933 | 0.079 | 0.334 | |||
| Negative | 34 | 8(23.5) | 26(76.5) | |||
| Positive | 99 | 30(30.3) | 69(69.7) | |||
| Snail | 4.753 | 0.176 | 0.029 | |||
| Negative | 109 | 35(32.1) | 74(67.9) | |||
| Positive | 24 | 3(12.5) | 21(87.5) | |||
| Vimentin | 5.054 | 0.184 | 0.025 | |||
| Negative | 39 | 16(41.0) | 23(59.0) | |||
| Positive | 94 | 22(23.4) | 72(76.6) |
To further confirm the effects of LETM1 expression on cell migration and invasion of PCa cells, a transwell assay was performed. Our results showed that silencing LETM1 expression dramatically inhibited PC3 (Figure 3E and F) and DU145 (Figure 3G and H) cell migration and invasion. Collectively, our data indicate that LETM1 contributes to metastatic progression of cancer cells by promoting EMT in PCa.
LETM1 Is Associated with Cancer Cell Proliferation in PCa
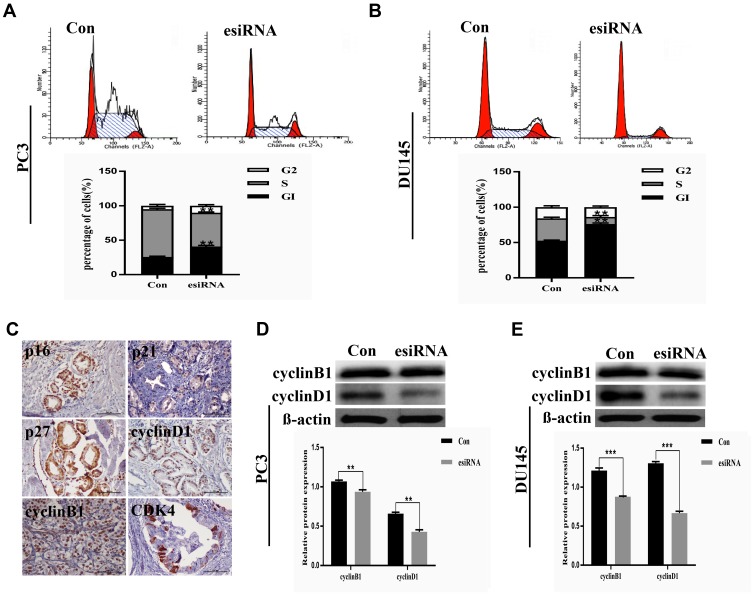
To investigate the function of LETM1 in the PCa cell cycle, we performed flow cytometry analysis on LETM1-silenced cells. Knockdown of LETM1 resulted in higher G0/G1-phase population and lower S-phase subpopulation in PC3 (Figure 4A) and DU145 (Figure 4B) cells than in the controls, but no significant changes were observed in the G2/M-phase subpopulations. To determine the association of LETM1 with the expression of cell cycle regulator genes (p16, p21, p27, cyclin B1, cyclin D1, and CDK4), we conducted IHC analysis of PCa tissues (Figure 4C). Our results showed that LETM1 expression was positively correlated with the expression of cell cycle regulators such as cyclin D1 (P < 0.001) and cyclin B1 (P < 0.001) (Table 3). After blocking LETM1 expression with LETM1-specific esiRNA, a dramatic decrease in cyclin D1 and cyclin B1 expression was observed in PC3 and DU145 cells (Figure 4D and E). Data indicate that LETM1 regulates cell cycle molecules and promotes proliferation in PCa cells.
Figure 4.
LETM1 positively regulates cell proliferation of PCa cells. Representative histograms of the cell cycle distribution in PC3 (A) and DU145 (B) cells after being transfected with LETM1 esiRNA were compared with the control using flow cytometry. The bar graph represents relative cell populations in different cell cycle phases such as G0/G1, S, and G2/M. (C) Immunohistochemical staining of cell cycle genes including p16, p21, p27, cyclin D1, cyclin B1, and CDK4 in PCa tissues (original magnification 200×). Protein expression of cyclin D1 and cyclin B1 after LETM1 knockdown compared with control in PC3 (D) and DU145 (E) cells. **P < 0.01, ***P < 0.001 versus control.
Table 3.
Correlation of LETM1 Expression with Cell Cycle Genes Expression in Prostate Cancer
| Variable | n | LETM1(−) n(%) | LETM1(+) n(%) | χ2 | R | P |
|---|---|---|---|---|---|---|
| p16 | 1.943 | 0.117 | 0.163 | |||
| Negative | 13 | 6(46.2) | 7(53.8) | |||
| Positive | 120 | 32(26.7) | 88(73.3) | |||
| p21 | 0.001 | 0.003 | 0.973 | |||
| Negative | 48 | 14(29.2) | 34(70.8) | |||
| Positive | 85 | 24(28.2) | 61(71.8) | |||
| p27 | 0.033 | 0.015 | 0.857 | |||
| Negative | 8 | 2(25.0) | 6(75.0) | |||
| Positive | 125 | 36(28.8) | 89(71.2) | |||
| cyclinD1 | 17.803 | 0.355 | <0.001 | |||
| Negative | 31 | 17(54.8) | 14(45.2) | |||
| Positive | 102 | 21(20.6) | 81(79.4) | |||
| cyclinB1 | 23.356 | 0.400 | <0.001 | |||
| Negative | 80 | 33(41.3) | 47(58.7) | |||
| Positive | 53 | 5(9.4) | 48(90.6) | |||
| CDK4 | 0.048 | 0.019 | 0.826 | |||
| Negative | 100 | 29(29.0) | 71(71.0) | |||
| Positive | 33 | 9(27.3) | 24(72.7) |
Inhibiting PI3K/Akt Signaling Decreases the Expression of LETM1 in PCa Cells
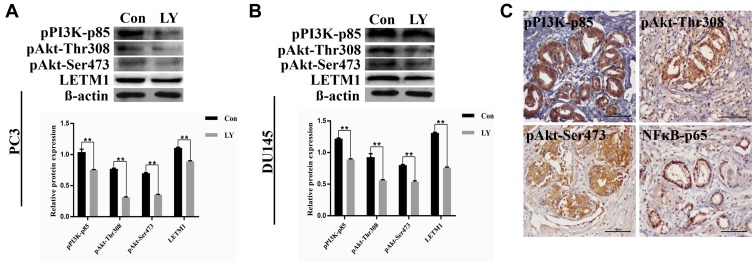
To investigate whether PI3K/Akt signaling was responsible for LETM1-mediated PCa progression, we blocked PI3K signaling using LY294002 in PCa cells. Our results showed that 20 μM LY294002 (a PI3K inhibitor) significantly inhibited LETM1, pPI3K-p85, and pAkt (Thr308, Ser473) expression in both PC3 (Figure 5A) and DU145 (Figure 5B) cells. In addition, IHC studies showed that LETM1 expression was significantly associated with the expression of pPI3K-p85 (P < 0.001) and pAkt-Thr308 (P < 0.001) in PCa tissues (Figure 5C and Table 4). These results indicate that the LETM1 gene is downstream of PI3K/Akt signaling and that PI3K signaling regulates LETM1 expression.
Figure 5.
LETM1 increases PCa tumorigenesis through the PI3K/Akt signaling pathway. (A) and (B) Western blotting analysis of pPI3K-p85, pAkt-Thr308, pAkt-Ser473, and LETM1 after treatment with the PI3K inhibitor LY294002 in PC3 and DU145 cells. (C) Immunohistochemical staining of pPI3K-p85, pAkt-Thr308, pAkt-Ser473, and NFκB-p65 in PCa tissues (original magnification 200×). **P < 0.01 versus control.
Table 4.
The Correlation of LETM1 Expression with PI3K/Akt/NF-Кb Signaling Genes Expression in Prostate Cancer
| Variable | n | LETM1(−) n(%) | LETM1(+) n(%) | χ2 | R | P |
|---|---|---|---|---|---|---|
| pPI3K-p85 | 35.784 | 0.480 | <0.001 | |||
| Negative | 71 | 34(47.9) | 37(52.1) | |||
| Positive | 62 | 4(6.45) | 58(93.5) | |||
| pAkt-Thr308 | 28.388 | 0.457 | <0.001 | |||
| Negative | 19 | 14(73.7) | 5(26.3) | |||
| Positive | 114 | 24(21.1) | 90(78.9) | |||
| pAkt-Ser473 | 1.104 | 0.090 | 0.293 | |||
| Negative | 28 | 6(21.4) | 22(78.6) | |||
| Positive | 105 | 32(30.5) | 73(69.5) | |||
| NFκB-p65 | 1.225 | 0.092 | 0.268 | |||
| Negative | 49 | 12(24.5) | 37(75.5) | |||
| Positive | 84 | 26(31.0) | 58(69.0) |
Discussion
LETM1 has many diverse functions including calcium or potassium/proton antiport, maintaining mitochondrial morphology, and facilitating mitochondrial translation.17 The expression level of LETM1 has been correlated with various human diseases, including cancer.17 Recent studies have shown that LETM1 regulates tumorigenesis, tumor progression, and cancer stemness.20,24 However, to our knowledge, this is the first study that investigated the relationship between LETM1 expression and cancer cell stemness and EMT-related proteins in PCa and clarified the significance of LETM1 expression in PCa tissues.
Recent studies have shown that high LETM1 expression is associated with poor prognosis and clinicopathological parameters of various human cancers.20–24 In spite of the difference in organs and cancer types, our findings are highly consistent with previous studies in which LETM1 expression was significantly higher in PCa than in BPH tissue. Moreover, the level of LETM1 expression was significantly associated with advanced grade, pT, and clinical stages. Subsequently, survival analysis showed that LETM1 overexpression was associated with poor PCa prognosis. In addition, various studies have shown that angiogenesis is crucial for the prognosis, metastasis, and progression of cancer.4,27 We found that the MVD was significantly higher in the LETM1-positive group than in the negative group. We speculate that LETM1 plays an important role in tumor initiation and cancer progression and indicates poor PCa prognosis.
PCa cells express several stem cell biomarkers, such as LSD1, SOX2, CD44, and CD133.28–32 LSD1 and its inhibitor have been reported to target CSC markers in several cancers.33,34 Sehrawat et al showed that LSD1 promotes PCa cell survival by activating gene networks that regulate the stem cell phenotype.35 SOX2 is a fundamental regulator of stem cell survival and pluripotency that promotes highly aggressive tumor phenotypes. CD44, a cell-surface glycoprotein, has been suggested to be a potential biomarker of prostate stem cells.28 Our previous study showed that LETM1 may serve as a novel cancer stemness gene for the prognostic evaluation of CRA and ESCC.20,24 In this study, stemness-related proteins were found to be co-upregulated with LETM1 in PCa cells. Furthermore, LETM1 knockdown significantly down-regulated CD44, SOX2, and LSD1 expression and the clonogenic ability of PCa cells. These results indicate that LETM1 may play an important role in promoting stemness properties in PCa cells, thereby increasing tumor progression, including tumor relapse and chemoresistance.
Several studies have confirmed that EMT-inducing factors promote EMT and confer CSC-like properties.13,36,37 Interrupting or reversing EMT inhibits the invasion and migration of laryngeal cancer cells.38 Xu et al showed that knocking down LETM1 markedly decreases the migration and invasion of renal cell carcinoma cells.39 In the present study, LETM1 expression was correlated with EMT-related genes in PCa tissues. In addition, knocking down LETM1 up-regulated the expression of the epithelial cell marker E-cadherin while down-regulating the expression of mesenchymal and transcription factor markers vimentin and Snail in PCa cells. LETM1 knockdown down-regulated the migration and invasiveness of the PCa cells. This finding suggested that LETM1 might be involved in PCa cell invasion and migration via activation of the EMT.
Cell cycle progression promotes cell proliferation, which in turn leads to clonal expansion of cells during tumor development.40 Cyclin D1 is an essential protein in the G1-phase of tumor cells and is highly expressed in actively proliferating non-small cell lung cancer cells.41 Cyclin B1, a crucial cell cycle checkpoint protein, promotes mitosis, and the overexpression of cyclin B1 may contribute to uncontrolled cell proliferation.42,43 In the present study, overexpression of LETM1 was associated with high expression of cyclin B1 and cyclin D1 in PCa tissues. Furthermore, LETM1 knockdown led to decreased S-phase subpopulations and increased G0/G1-phase subpopulations in PCa cells. Our findings are consistent with those of a recent study in which silencing LETM1 expression resulted in HeLa cell cycle arrest and influenced autophagy.16 Further, knocking down LETM1 markedly inhibits bladder cancer cell proliferation.22 This suggests that the expression of LETM1 leads to unrestrained PCa cell proliferation.
Recent studies have shown that the CSC-like phenotype is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer.44,45 By inhibiting the C-terminal modulator protein, LETM1 promotes activation of Akt/PKB,46 and overexpression of LETM1 may increase the activation of Akt in thyroid cancer cells.23 In the present study, we found a significant positive correlation between the expression levels of LETM1 and pPI3K/pAkt. Moreover, treatment with the PI3K-specific inhibitor LY294002 significantly down-regulated LETM1 expression in PCa cells. These results indicate that PI3K signaling is upstream of LETM1 signaling and regulates the stemness of PCa. Nevertheless, further studies are required to elucidate the specific mechanism.
Conclusion
In conclusion, LETM1 plays a key role in PCa progression by inducing EMT-like changes and increasing the proliferation capacity of cancer cells. Moreover, the PI3K/Akt pathway plays an essential role in the up-regulation of LETM1. Thus, LETM1 has the potential for being used as a therapeutic target and prognostic biomarker for PCa.
Funding Statement
This work was supported by grants from the National Natural Science Funds of China (81760531) and Jilin Provincial Education Department Project (JJKH20191138KJ).
Author Contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 3.Huang S, Wa Q, Pan J, et al. Transcriptional downregulation of miR-133b by REST promotes prostate cancer metastasis to bone via activating TGF-β signaling. Cell Death Dis. 2018;9(7):779–793. doi: 10.1038/s41419-018-0807-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ni WD, Yang ZT, Cui CA, Cui Y, Fang LY, Xuan YH. Tenascin-C is a potential cancer-associated fibroblasts marker and predicts poor prognosis in prostate cancer. Biochem Biophys Res Commun. 2017;486(3):607–612. doi: 10.1016/j.bbrc.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 5.Qin Y, Dey A, Purayil HT, Daaka Y. Maintenance of androgen receptor inactivation by S-nitrosylation. Cancer Res. 2013;73(22):6690–6699. doi: 10.1158/0008-5472.CAN-13-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124(6):1111–1115. doi: 10.1016/j.cell.2006.03.011 [DOI] [PubMed] [Google Scholar]
- 7.Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell. 2007;1(3):241–242. doi: 10.1016/j.stem.2007.08.012 [DOI] [PubMed] [Google Scholar]
- 8.Vermeulen L, De Sousa E Melo F, et al. Wnt activity defnes colon cancer stem cells and is regulated by the microenvironment. Elife. 2010;12(5):468–476. [DOI] [PubMed] [Google Scholar]
- 9.Steffensen KD, Alvero AB, Yang Y, et al. Prevalence of epithelial ovarian cancer stem cells correlates with recurrence in early-stage ovarian cancer. J Oncol. 2011;2011:620523–620534. doi: 10.1155/2011/620523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Li H, Ren G. Epithelial-mesenchymal transition and drug resistance in breast cancer (Review). Int J Oncol. 2015;47(3):840–848. doi: 10.3892/ijo.2015.3084 [DOI] [PubMed] [Google Scholar]
- 11.Li J, Gong X, Jiang R, et al. Fisetin inhibited growth and metastasis of triple-negative breast cancer by reversing epithelial-to-mesenchymal transition via PTEN/Akt/GSK3β signal pathway. Front Pharmacol. 2018;9:772–785. doi: 10.3389/fphar.2018.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani SA, Guo W, Liao MJ, et al. DNMT1 regulates epithelial-mesenchymal transition and cancer stem cells, which promotes prostate cancer metastasis. Neoplasia. 2016;18(9):553–566. doi: 10.1016/j.neo.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326(5949):144–147. doi: 10.1126/science.1175145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doonan PJ, Chandramoorthy HC, Hoffman NE, et al. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. FASEB J. 2014;28(11):4936–4949. doi: 10.1096/fsb2.v28.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piao L, Li Y, Kim SJ, et al. Association of LETM1 and MRPL36 contributes to the regulation of mitochondrial ATP production and necrotic cell death. Cancer Res. 2009;69(8):3397–3404. doi: 10.1158/0008-5472.CAN-08-3235 [DOI] [PubMed] [Google Scholar]
- 18.Tan DQ, Suda T. Reactive oxygen species and mitochondrial homeostasis as regulators of stem cell fate and function. Antioxid Redox Signal. 2018;29(2):149–168. doi: 10.1089/ars.2017.7273 [DOI] [PubMed] [Google Scholar]
- 19.Khacho M, Clark A, Svoboda DS, et al. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19(2):232–247. doi: 10.1016/j.stem.2016.04.015 [DOI] [PubMed] [Google Scholar]
- 20.Piao L, Feng Y, Yang Z, et al. LETM1 is a potential cancer stem-like cell marker and predicts poor prognosis in colorectal adenocarcinoma. Pathol Res Pract. 2019;5:152437. doi: 10.1016/j.prp.2019.152437 [DOI] [PubMed] [Google Scholar]
- 21.Li N, Zheng Y, Xuan C, et al. LETM1 overexpression is correlated with the clinical features and survival outcome of breast cancer. Int J Clin Exp Pathol. 2015;8(10):12893–12900. [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B, Zhang J, Zhang X, et al. Suppression of LETM1 by siRNA inhibits cell proliferation and invasion of bladder cancer cells. Oncol Rep. 2017;38(5):2935–2940. doi: 10.3892/or.2017.5959 [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Lee WK, Seol MY, et al. Coupling of LETM1 up-regulation with oxidative phosphorylation and platelet-derived growth factor receptor signaling via YAP1 transactivation. Oncotarget. 2016;7(41):66728–66739. doi: 10.18632/oncotarget.11456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Ni W, Cui C, et al. Identification of LETM1 as a marker of cancer stem-like cells and predictor of poor prognosis in esophageal squamous cell carcinoma. Hum Pathol. 2018;8177(18):30258–30262. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Yeo S, Yin Y, et al. Tenascin-C, a prognostic determinant of esophageal squamous cell carcinoma. PLoS One. 2016;11(1):e0145807. doi: 10.1371/journal.pone.0145807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Zhang C, Qi W, et al. GLI1 promotes cancer stemness through intracellular signaling pathway PI3K/Akt/NFκB in colorectal adenocarcinoma. Exp Cell Res. 2018;373(1–2):145–154. doi: 10.1016/j.yexcr.2018.10.006 [DOI] [PubMed] [Google Scholar]
- 27.Yuri P, Hendri AZ, Danarto R. Association between tumor-associated macrophages and microvessel density on prostate cancer progression. Prostate Int. 2015;3(3):93–98. doi: 10.1158/0008-5472.CAN-11-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patrawala L, Calhoun T, Schneider-broussard R, et al. Highly purifified CD44þ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25(12):1696–1708. doi: 10.1038/sj.onc.1209327 [DOI] [PubMed] [Google Scholar]
- 29.Ugolkov AV, Eisengart LJ, Luan C, et al. Expression analysis of putative stem cell markers in human benign and malignant prostate. Prostate. 2011;71(1):18–25. doi: 10.1002/pros.21217 [DOI] [PubMed] [Google Scholar]
- 30.Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells. 2008;26(8):1931–1938. doi: 10.1634/stemcells.2007-1002 [DOI] [PubMed] [Google Scholar]
- 31.Kalantari E, Asgari M, Nikpanah S, et al. Co-expression of putative cancer stem cell markers CD44 and CD133 in prostate carcinomas. Pathol Oncol Res. 2017;23(4):793–802. doi: 10.1007/s12253-016-0169-z [DOI] [PubMed] [Google Scholar]
- 32.Ketscher A, Jilg CA, Willmann D, et al. LSD1 controls metastasis of androgen-independent prostate cancer cells through PXN and LPAR6. Oncogenesis. 2014;3(10):e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Lu F, Ren Q, et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71(23):7238–7249. doi: 10.1158/0008-5472.CAN-11-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Lu F, Wang J, et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 2013;5(2):445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sehrawat A, Gao L, Wang Y, et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci USA. 2018;115(18):4179–4188. doi: 10.1073/pnas.1719168115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morel AP, Lièvre M, Thomas C, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3(8):e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang R, Zhang C, Liu G, et al. MicroRNA-126 inhibits proliferation, migration, invasion and EMT in osteosarcoma by targeting ZEB1. J Cell Biochem. 2017;118(11):3765–3774. doi: 10.1002/jcb.v118.11 [DOI] [PubMed] [Google Scholar]
- 38.Mao W, Sun Y, Zhang H, et al. A combined modality of carboplatin and photodynamic therapy suppresses epithelial-mesenchymal transition and matrix metalloproteinase-2 (MMP-2)/MMP-9 expression in HEp-2 human laryngeal cancer cells via ROS-mediated inhibition of MEK/ERK signalling pathway. Lasers Med Sci. 2016;31(8):1697–1705. doi: 10.1007/s10103-016-2040-6 [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Huang B, Saiyang L, et al. Knockdown of LETM1 inhibits proliferation and metastasis of human renal cell carcinoma cells. Oncol Lett. 2018;16(5):6377–6382. doi: 10.3892/ol.2018.9449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong ZQ, Song MM, He Y, et al. Knockdown of Ezrin by RNA interference reverses malignant behavior of human pancreatic cancer cells in vitro. Asian Pac J Cancer Prev. 2012;13(8):3781–3789. doi: 10.7314/APJCP.2012.13.8.3781 [DOI] [PubMed] [Google Scholar]
- 41.Chikara S, Lindsey K, Dhillon H, et al. Enterolactone induces G1-phase cell cycle arrest in nonsmall cell lung cancer cells by downregulating cyclins and cyclin-dependent kinases. Nutr Cancer. 2017;69(4):652–662. doi: 10.1080/01635581.2017.1296169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Cai J, Zheng Z, et al. A novel ER-microtubule-binding protein, ERLIN2, stabilizes Cyclin B1 and regulates cell cycle progression. Cell Discov. 2015;1:15024–15041. doi: 10.1038/celldisc.2015.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki T, Urano T, Miki Y, et al. Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 2007;98(5):644–651. doi: 10.1111/j.1349-7006.2007.00444.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang L, Wang Z, Liu C, et al. TrkB prometes laryngeal cancer metastasis via activation PI3K/Akt pathway. Oncotarget. 2017;8(65):108726–108737. doi: 10.18632/oncotarget.21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang L, Graham PH, Hao J, et al. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013;4:e875. doi: 10.1038/cddis.2013.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J, Li Y, Kim SH, et al. New players in high fat diet-induced obesity: LETM1 and CTMP. Metabolism. 2014;63(3):318–327. doi: 10.1016/j.metabol.2013.10.012 [DOI] [PubMed] [Google Scholar]