Abstract
Background/Aims
this experimental design was based on HIPK3 to explore the pathogenesis of ESCC.
Methods
RT-qPCR was used to detect the expression of CircHIPK3 and miR-599 in ESCC tissues and cell lines.CCK-8, colony formation, flow cytometry and transwell assay were used to detect the effects of CircHIPK3 and miR-599 on tumor cell proliferation, apoptosis and migration and invasion. Target gene prediction and screening, luciferase reporter assays were used to validate downstream target genes of CircHIPK3 and miR-599.mRNA and protein expression of c-MYC were detected by RT-qPCR and Western blotting. The tumor changes in mice were detected by in vivo experiments in nude mice.
Results
HIPK3 was highly expressed in ESCC tissues and cell lines. In addition, HIPK3 expression levels were associated with advanced TNM stage, lymph node metastasis and tumor size. Moreover, HIPK3 was significantly promoted cell proliferation and migration of ESCC cells. In addition, HIPK3 was able to inhibit miRNA-599 expression and up-regulate the expression level of c-MYC. Finally, the results of in vivo animal models confirmed that HIPK3 promoted ESCC progression by modulating the miR-599/c-MYC axis.
Conclusion
HIPK3 can regulate the proliferation of esophageal squamous cell carcinoma cells by regulating miR-599/c-MYC axis, thereby inhibiting the occurrence and development of esophageal squamous cell carcinoma.
Keywords: esophageal squamous cell carcinoma, HIPK3, miR-599, c-MYC, proliferation
Introduction
Esophageal cancer (EC) is one of the common malignant tumors in humans.16,39 It has a high incidence and a poor prognosis, and its regional differences are obvious.43 The primary histological type of primary EC is esophageal squamous cell carcinoma (ESCC).42 Esophageal squamous cell carcinoma is more common in China and other Eastern countries. Esophageal squamous cell carcinoma is more common in men over 50 years of age, and the incidence of men is much higher than that of women.4,31 Esophageal cancer mainly occurs in the middle and upper segments of the esophagus. Due to the anatomical location and the nature of the tumor, early esophageal cancer is not easy to be found, which affects the best time for treatment and belongs to refractory tumors. At present, patients with early and mid-stage esophageal cancer are treated with surgery or radiotherapy, supplemented by chemotherapy.21,35 However, after neoadjuvant chemotherapy for patients with locally advanced esophageal cancer of stage ⅡB and stage Ⅲ, the prognosis is significantly improved.11,28 Human esophageal cancer is a multi-step progression, and the process of esophageal epithelial malignancy develops from atypical hyperplasia, carcinoma in situ to advanced invasive carcinoma. The rapid advancement of molecular biology has proposed a new carcinogenic theory for the progression of esophageal cancer. The occurrence and development of tumors is a multi-gene, multi-stage, and multi-regulation process.17,19
Circular RNA (circ RNA) is a type of non-coding RNA widely found in mammals and is mainly involved in the regulation of genes in organisms.13,15 Most of circRNA is derived from the exon region of the gene, and a small portion is formed by intron splicing. Circ RNA is widely involved in the process of physiological and pathological regulation of humans.3,40 In tumor research, the mechanism by which circular RNA acts as a miRNA “cavernous body” to regulate downstream target genes has been widely reported.41 It has been found that a plurality of circular RNAs themselves contain at least one mi RNA binding site.8 Therefore, it can act as a “sponge” of RNA to adsorb mi RNA, thereby regulating the expression of downstream target genes inhibited by mi RNA through the mechanism of competitive endogenous RNAs (ce RNAs).38 The circular RNA HIPK3 (circ HIPK3) is highly expressed in the liver, brain, and lung.It mainly originates from the second exon of the gene HIPK3.The circular RNA of HIPK3 origin has three kinds of splicing bodies, circ HIPK3 and circ HIPK3.1.and circ HIPK3.2.18 While only circ HIPK3 is highly abundant and has significant functions in cells.37 However, the expression and regulation mechanism of circular RNAHIPK3 in ESCC has not been clarified.
miRNAs are endogenous non-coding single-stranded small RNA molecules 19 to 24 nucleotides in length in many organisms.32 miRNAs are involved in almost all tumor-related processes, including proliferation, differentiation, apoptosis, metastasis, and angiogenesis and so on.1,29 Many studies have shown that miRNA aberrant expression is closely related to the occurrence, development and prognosis of malignant tumors by regulating the expression of tumor suppressor genes and oncogenes.30 miR-599 is a class of mi RNA that promotes tumor cell proliferation.The oncogene c-MYC encodes a conserved protein molecule with a helix-loop-helix and leucine zipper structure, which is abnormally expressed in various cancer tissues and is closely related to cancer cell proliferation, tumor angiogenesis and cancer cell metastasis.6 As a transcriptional regulator, c-MYC can form heterodimers with other co-regulatory factors such as Max and regulate the expression of hundreds of genes.10 The study found that c-MYC regulation of non-coding miRNAs was an important component of the c-MYC regulatory network, which can alter the expression profile of miRNAs and promote cancer.2,20 However, there are few studies in the ESCC. In view of the above problems, this study aimed to explore the effect of HIPK3 on the proliferation and migration of ESCC by detecting the expression of HIPK3 in LSCC tissues and cells, and further reveal the mechanism by which HIPK3 regulated cell proliferation and migration. Defining the pathogenesis of esophageal squamous cell carcinoma may provide a more accurate and effective new therapeutic target for ESCC.
Methods and Materials
Patient Samples and Cells
A total of 42 patients with postoperative ESCC and corresponding normal adjacent epithelial tissues (NATs, > 2 cm from tumor tissue) were selected from 2015 to 2017 in Zhengzhou, China. Informed consent was obtained from all patients. The study was approved by the Affiliated Cancer Hospital of Zhengzhou University Institutional Review Committee. The clinical pathological parameters and clinical features of all patients were shown in Table 1.
Table 1.
Correlation Between HIPK3 Expression and Clinicopathological Features in ESCC Patients
| Parameters | Group | N | HIPK3 Expression | P value | |
|---|---|---|---|---|---|
| High, n (%) | Low, n (%) | ||||
| Age (years) | ≤60 | 12 | 5(41.7) | 7(58.3) | 0.522 |
| >60 | 30 | 16(53.3) | 14(46.7) | ||
| Gender | Female | 11 | 6(54.5) | 5(45.5) | 0.656 |
| Male | 31 | 15(48.4) | 16(51.6) | ||
| Smoking | Ever and current | 23 | 13(56.5) | 10(43.5) | 0.331 |
| Never | 19 | 8(42.1) | 11(57.9) | ||
| Drinking | Ever and current | 26 | 14(53.8) | 12(46.2) | 0.218 |
| Never | 16 | 7(43.8) | 9(56.2) | ||
| Tumor size (cm) | >5 | 15 | 11(73.3) | 4(26.7) | 0.011 |
| ≤5 | 27 | 10(37.0) | 17(63.0) | ||
| Differentiation | Well or Moderate | 28 | 13(46.4) | 15(53.4) | 0.087 |
| Poor | 14 | 8(57.1) | 6(42.9) | ||
| TNM stage | A-IIB | 26 | 17(65.4) | 9(34.6) | 0.021 |
| IIIA | 16 | 4(25.0) | 12(75.0) | ||
| Lymph node metastasis | Positive Negative |
13 29 |
9 12 |
4 17 |
0.027 |
ESCC cell line (kyse-150, kyse-410, KYSE-510, ECA-109, EC-18 and TE-13) and normal immortalized cell line (NE1) were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). ESCC cell lines were subcultured in Eagle medium containing 10% fetal bovine serum.
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction
Total RNA was isolated from cells and tissues using TRIzol reagent in strict accordance with the instructions. Complementary DNA (cDNA) was then synthesized from total RNA using SuperScript III. The gene expression level of HIPK3 was determined by LightCyclerTM 480 system (Roche Diagnostics), and the small-nuclear RNA U48 was selected as an internal standard control. The qPCR assay was performed in a LightCyclerTM 480 system. The internal standard of miR-599 was U48.qRT-PCR analysis was performed using a SYBR Premix Ex Taq II kit (Takara, Otsu, Japan).The analysis was carried out using the 2−ΔΔCT method (Table 2).
Table 2.
The Oligonucleotides
| Sense | Antisense |
|---|---|
| c-MYC 5ʹ-CGAGTTTATTGTGCGAGCGAA −3’ | 5ʹ-CTGTGGCATCGCCAGGCC-3’ |
| U485ʹ-GAGGAGAACAAGAGAGAGAGAAATG-3 | 5ʹ-GCTGGGATGGAGACGCATGAGG-3 |
| GAPDH 5ʹ-CAAATGGAGCAGAGAGCCCT-3’ | 5ʹ-CCCCATGATACCGTGGGTAG-3’ |
Cell Transfection
miR-599 mime, miR-599 agomir and HIPK3 expressing vectors were purchased from Genecopia. In vitro cell experiments were transfected using Lipofectamine 2000 reagent (Invitrogen) and OptiMEM (Invitrogen).HIPK3 and the loader were first constructed to stably express TE-13 cells.Stable cell lines were then infected with miR-599 agomir or a negative control virus, followed by transfection using Lipofectamine 2000 reagent (Invitrogen) and OptiMEM (Invitrogen).
CCK-8 Method
Transfected cells were seeded into 96-well plates at a density of 1 x 103 cells per well. Three wells were replicated in each group and 10 μL/well of CCK8 was added to each well of cells for the last 2 hrs of incubation. Finally, the OD value of the cell liquid was measured by an enzyme-linked immunosorbent assay.
Colony Formation
The transfected cells (1 *103 cells/well) were placed in a 6-well plate to adhere to the cell wall within 24 hrs. After 24 hrs of exposure to ADR, the cells were washed and cultured in different media for 8 days.Finally, colonies were counted after fixation for 10 mins with 10% formaldehyde, and stained with 0.1% crystal violet for 10 min.Cell viability was plotted using GraphPad Prism 6 (GraphPadSofware, Inc., San Diego, CA, USA).
Apoptosis Detection
Cell apoptosis was determined by FITC Annexin V Apoptosis Detection Kit (BD, Franklin Lakes, NJ, USA). After treated with transfection reagents for 48 h, cells were washed twice with cold PBS and then 1 × 105 cells were re-suspended in 100 μL 1X binding buffer, then stained with 5 μL FITC Annexin V and 5μL PI for 10 min at room temperature (25 °C) in the dark. 400μL 1X binding buffer was added to each tube. The apoptosis cells were then detected by flow cytometry using fluorescence-activated cell-sorting (FACS) flow cytometer (BD Biosciences, San Jose, CA, USA).
Cell Migration and Invasion Assay
Cell migration and invasion tests are shown in the literature.9
Luciferase Assay
The pmirGLO dual luciferase miRNA target expression vector was used in the 3ʹ-untranslated region (UTR) luciferase assay (Promega, Madison, WI, USA).Cells were seeded at a density of 4 x 105 cells/mL in 24-well plates.afterthat, cells were co-transfected with hsa-miR-599 mimics and wild-type or mutant target sequences using Lipofectamine 2000 (Invitrogen). Luciferase assay are shown in the literature.23
RNA Immunoprecipitation Assay (RIP)
RIP assay were performed using EZ-Magna RIP kit (Millipore, Billerica, MA, USA) and Argonaute 2 (Ago2) antidody (Abcam) to explore whether HIPK3 was existed in RNA induced silencing complex (RISC). Briefly, cells were lysed in RIP lysis buffer, followed by the incubation of protein A/G magnetic beads and antibody against IgG (Millipore) or Ago2 (Abcam). Then RNAs in magnetic beads-binding complexes were purified. At last, RT-qPCR assay was employed to measure the enrichment patterns of HIPK3 and miR-599 by IgG antibody or Ago2 antibody.
Western Blot
Western blot of c-MYC are shown in the literature.25 In vivo antitumor activity.
The study was conducted in strict accordance with the reorganization of the National Institutes of Health Laboratory Animal Care and Use Guidelines.The agreement was approved by the animal experiment ethics of the xxx hospital.BALB/c nude mice (4 weeks old, female) were maintained under pathogen free conditions.
Si-NC, si-HIPK3 or si-HIPK3/miR-599 cells were injected subcutaneously into the right flank of BALB/C nude mice (n=10/group).Tumor volume of nude mice was measured every 7 days after injection. All nude mice were sacrificed 4 weeks later and the tumors were dissected for qPCR or Western blot analysis experiments.
Statistical Method
The monitoring data were analyzed by SPSS19.0 statistical software. The data analysis results were expressed as mean±standard deviation (mean±SD). The data analysis between the two groups was performed by t test. One-way variance analysis (ANOVA) was used for data analysis among multiple groups, and LSD test was used for subsequent analysis.P <0.05 indicated that the difference was statistically significant.
Results
CircHIPK3 Was Up-Regulated in ESCC Tissues and Cell Lines
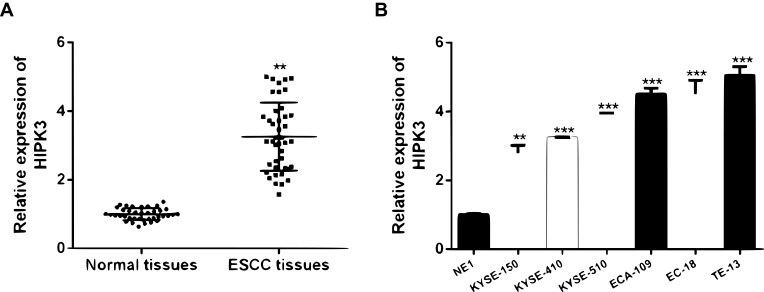
Firstly, the expression level of HIPK3 in ESCC was analyzed. It was found that HIPK3 was significantly up-regulated in cancer tissues compared with normal tissues (Figure 1A). Furthermore, similar to the tissue results, HIPK3 expression was significantly up-regulated in the ESCC cell line compared withNE1 cells (Figure 1B).
Figure 1.
HIPK3 was upregulated in ESCC tissues and cell lines. (A) The expression of HIPK3 in 42 pairsof ESCC tissues and in paired normal tissues. (B) The expression of HIPK3 in ESCC cell lines and NE1 cells. ** p <0.01, *** p <0.001.
Patients were divided into high expression group (n = 21) and low expression group (n = 21) using the median level (HIPK3) as the cutoff value. The results were shown in Table 1. The overexpression of HIPK3 was positively correlated with late TNM stage, lymph node metastasis and tumor size, but not with other parameters such as age, gender, alcohol
CircHIPK3 Promoted the Malignant Behaviors of ESCC Cells
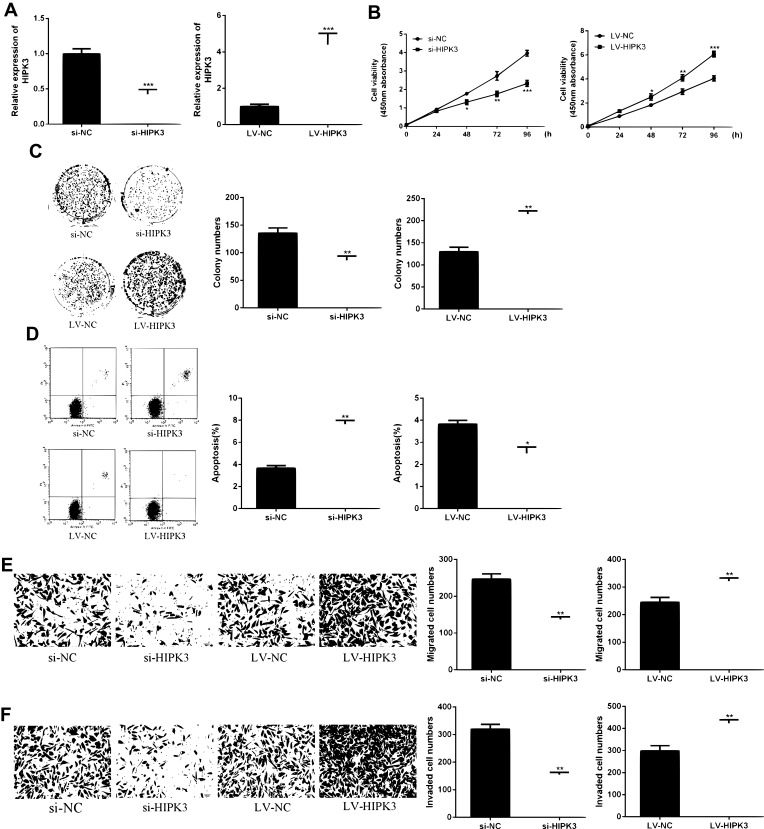
As shown in Figure 2A and B, HIPK3 expression was significantly changed after siRNA or LV-HIPK3 transfection, indicating a successful transfection.
Figure 2.
CircHIPK3 promoted proliferation and metastasis of ESCC cells. (A) HIPK3 levels in si-HIPK3 or LV-HIPK3 transfected cells. (B) Effect of HIPK3 on cell viability. (C) Effect of HIPK3 on cell proliferation. (D) Effect of HIPK3 on apoptosis. (E, F) Effect of HIPK3 on cell migration and invasion.*p <0.05, ** p <0.01, *** p <0.001.
As shown in Figure 2C, compared with the control group, the proliferation rate of the HIPK3 overexpression group was significantly enhanced in the ESCC cells, and the cell proliferation rate of the HIPK3 silenced group was significantly decreased. As shown by Figure 2D, the number of cell colonies in the HIPK3 overexpression group was significantly increased in ESCC cells, and the number of cell colonies in the HIPK3 silenced group was significantly reduced compared with the control group. As shown in Figure 2E, compared with the control group, the apoptosis rate of the HIPK3 overexpression group was significantly decreased in ESCC cells, and the apoptosis rate of the HIPK3 silenced group was significantly increased. As shown by Figure 2E and F, the number of cell migration and invasion in the HIPK3 overexpression group was significantly increased in ESCC cells compared with the control group, and the number of cell migration and invasion in the HIPK3 silenced group was significantly reduced. In summary, the above results indicated that circHIPK3 exerted a carcinogenic effect and promoted ESCC cell growth and metastasis.
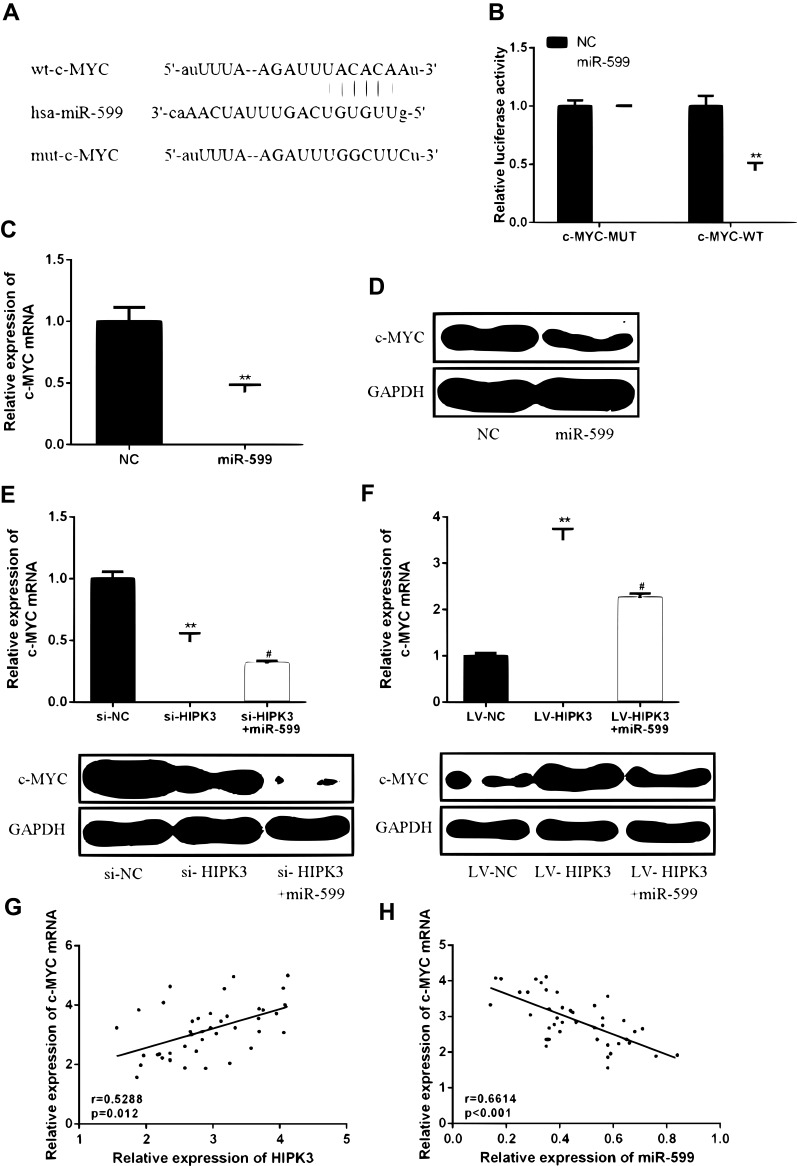
miR-599 Was the Target Gene of CircHIPK3
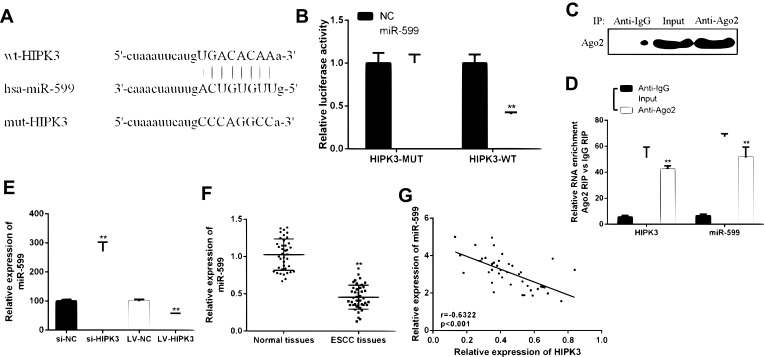
In order to determine the underlying mechanism of action of CircHIPK3 in ESCC, it was predicted that miR-599 was identified as a potential target for CircHIPK3 (Figure 3A) by bioinformatics. The WT-HIPK3 or mutant (Mut)-HIPK3 luciferase reporter plasmid for luciferase reporter assay in TE-13 cells was used to verify the predicted results. As shown in Figure 3B, ectopic expression of miR-599 significantly inhibited luciferase activity of WT-HIPK3, but had no significant effect on luciferase activity of Mut-HIPK3. To further test whether HIPK3 and miR-599 are associated through miRNA ribonucleoprotein complexes, Ago2 antibody was used to do the RNA immunoprecipitation experiment. The RT-PCR results showed that HIPK3 and miR-599 were preferentially enriched in the Ago2-containing miRNAs compared with the control IgG immune precipitates (Figure 3C and D). In addition, as shown in Figure 3E, HIPK3 knockdown can significantly increase the expression level of miR-599 in TE-13 cells. HIPK3 knockdown can significantly up-regulate the expression level of miR-599 in TE-13 cells, while HIPK3 overexpression can significantly down-regulate the expression level of miR-599 in TE-13 cells.
Figure 3.
HIPK3 directly targeted miR-599 in ESCC. (A) Bioinformatics analysis revealed predicted binding sites between HIPK3 and miR-599. (B) Relative luciferase activity in TE-13 cells co-transfected with wild-type (HIPK3-WT) or mutant reporter plasmid (HIPK3-mut) and miR-599 mimic or negative control. (C) Cellular lysates from ESCC cells were used for RIP with an Ago2 antibody. The Ago2 protein level was detected by Western blot, (D) and the relative expression of HIPK3 and miR-599 in the immunoprecipitate was measured by RT-PCR. (E) The miR-599 expression in ESCC tissues and matched normal tissues. (F) Expression of miR-599 in the presence of si-HIPK3 or LV-HIPK3. (G) The relationship between HIPK3 and miR-599 in the ESCC organization.(** P <0.01).
Next the expression pattern of miR-599 in ESCC was analyzed. As shown in Figure 3F, the expression level of miR-599 was significantly down-regulated in ESCC tissues compared with that in the normal tissues. In addition, it was found that there was a direct negative correlation between circHIPK3 expression and miR-599 levels in ESCC tissues (Figure 3G). In summary, these results indicated that circHIPK3 may exert its biological function through miR-599.
CircHIPK3 Promoted the Malignant Behaviors of ESCC Cells by Inhibiting miR-599
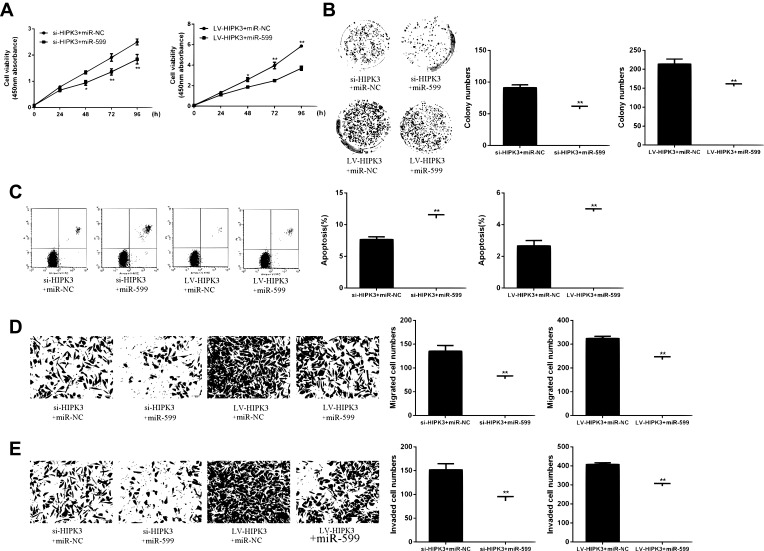
Next, whether HIPK3 exerts its biological effects by inhibiting miR-599was explored. As shown in Figure 4A, the cell proliferation rate of miR-559 mimetic + siHIPK3 group was significantly lower than that in HIPK3 silencing group. The cell proliferation rate of miR-559 mimetic + Lv HIPK3 group was significantly higher than that in HIPK3 overexpression group. As shown in Figure 4B, the number of cell colonies in the miR-559 mimetic + siHIPK3 group was significantly lower than that in the HIPK3 silencing group, while the number of cell colonies in the miR-559 mimetic + Lv HIPK3 group was significantly higher than that in the HIPK3 overexpression group. As shown in Figure 4C, the apoptosis rate of miR-559 mimetic + siHIPK3 group was significantly higher than that of HIPK3 silenced group, and the apoptosis rate of miR-559 mimetic + Lv HIPK3 group was significantly lower compared with that in HIPK3 overexpression group. As shown by Figure 4D and E, the miR-559 mimetic + siHIPK3 group had significantly increased cell migration and invasion compared withthat in HIPK3 silencing group. Cell migration and invasion of he miR-559 mimetic + Lv HIPK3 group were significantly reduced compared with that in the HIPK3 overexpressing group. In summary, these results indicated that HIPK3 exerted a biological role in ESCC cells via sponge miR-599.
Figure 4.
(A) HIPK3 promoted cell growth and metastasis by inhibiting miR-599. (B) Cell viability assay. (C) Cell proliferation assay. (D) Apoptosis assay. (E, F) Cell migration and invasion assays.*p <0.05, ** p <0.01.
HIPK3 Increased c-MYC Expression by Inhibiting miR-559
In order to determine the specific mechanism of action of HIPK3 and miR-559 in ESCC progression, it was predicted that c-MYC was identified as a potential target for miR-559 (Figure 5A) by bioinformatics. As shown in Figure 5B, ectopic expression of c-MYC significantly inhibited the luciferase activity of WT-miR-559, but had no significant effect on the luciferase activity of mut-miR-559.Furthermore, in TE-13 cells, miR-559 mimics were able to significantly reduce c-MYC mRNA and protein expression compared with the control group (Figure 5C and D).These data indicated that c-MYC was a direct target of miR-559.
Figure 5.
HIPK3 increased c-MYC expression levels by inhibiting miR-599. (A) Bioinformatics analysis revealed predicted binding sites between c-MYC and miR-599. (B) Luciferase activity was detected in cells co-transfected with miR-599 and a luciferase reporter containing c-MYC or mutant forms. (C) c-MYC mRNA expression levels in TC-13 cells. (D) c-MYC protein expression levels in TC-13 cells. (E, F) Changes in expression of c-MYC protein in TE-13 cells. (G) A positive correlation between HIPK3 and c-MYC expression in ESCC tissues. (H) Negative correlation between c-MYC and miR-599 in ESCC organization.(#P <0.05, **P <0.01).
Next, whether HIPK3 promotes the development of ESCC by regulating the miR-559/c-MYC axiswas further studied. As shown in Figure 5E and F, HIPK3 knockdown inhibited c-MYC expression levels, while miR-559 mimics further increased c-MYC expression levels. In contrast, overexpression of HIPK3 significantly enhanced c-MYC expression levels, while miR-559 mimics further inhibited HIPK3-induced c-MYC upregulation. As shown in Figure 5G and H, c-MYC expression was positively correlated with HIPK3 expression and negatively correlated with miR-559 expression. These results indicated that HIPK3 can increase c-MYC expression levels by inhibiting miR-559.
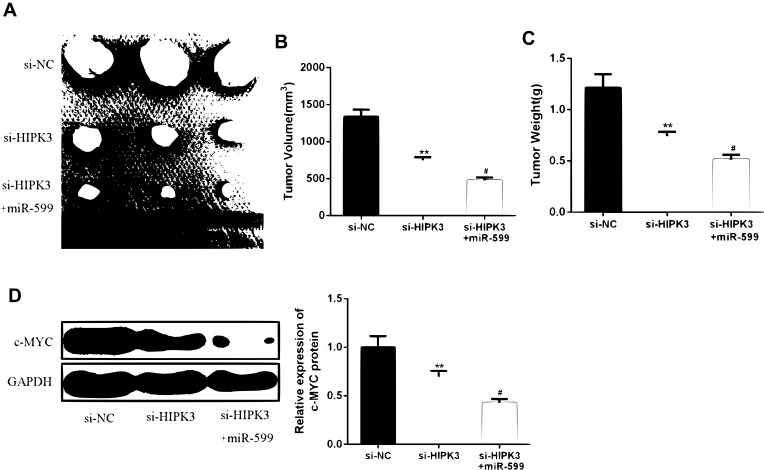
HIPK3 Promoted ESCC Progression in vivo by Modulating miR-599/c-MYC Axis
In order to further determine the effect of HIPK3 on ESCC progression in vivo, ESCC cells transfected with si-HIPK3 or si-HIPK3/miR-599 agomir were injected subcutaneously into nude mice. As shown in Figue 6A–C, the tumor size and tumor weight of the si-HIPK3 group were significantly lower compared with that in the control group. In the miR-599 agomir treatment group, the tumor growth of si-HIPK3 transfected cells was further inhibited. Furthermore, as shown in Figure 6D, HIPK3 silencing significantly inhibited c-MYC expression, while miR-599 agomir treatment further reduced c-MYC expression levels in tumors. These results indicated that HIPK3 promoted ESCC progression in vivo by inhibiting the miR-599/c-MYC axis.
Figure 6.
HIPK3 promoted ESCC progression in vivo by modulating the miR-599/c-MYC axis. (AC) Tumor size and tumor weight. (D) Expression changes of c-MYC protein in each group. (#P <0.05, **P <0.01).
Discussion
Esophageal cancer is a common malignant tumor of the digestive system that originates in the esophageal mucosal epithelial tissue.14 Esophageal adenocarcinoma (esophageal adenocarcinoma) occurring from submucosal or cardiac glands of the esophagus is the main pathological type of esophageal cancer in Western countries. Esophageal squamous cell carcinoma (ESCC), which originates from esophageal squamous cells, has always been the main pathological type of esophageal adenocarcinoma.36 At the molecular biology level, the genetic susceptibility of esophageal cancer is related to the activation and expression of oncogenes and the loss or inactivation of tumor suppressor genes.19,33 At present, the specific pathogenesis of esophageal cancer is still unclear. Molecular biology research on esophageal cancer, searching for early diagnostic markers of esophageal cancer and related factors and mechanisms of invasion and metastasis have become a hot topic in this field.
Circular RNAs (circRNAs) exhibiting different properties than linear RNA.26 The current study found that circular RNA can play a role in miRNA molecular sponge and regulate gene transcription.22 CircRNA plays an important role in diseases.27 Studies have shown that the circular RNA cir-ITCH is lowly expressed in esophageal squamous cell carcinoma. As sponges of mir-7, mR-17 and mir-214, cir-ITCH can increase the expression level of ITCH, promote ubiquitination, and thus inhibit esophageal squamous cell carcinoma.34 Circ HIPK3 is another up-and-coming circular RNA that has received increasing attention. In normal cells, its abundance is comparable to or even higher than its linear RNA.18 In tumor research, it was found that circ HIPK3 can promote the proliferation of osteosarcoma.37 The results of this study showed that the expression of HIPK3 in cancer tissues and cells was significantly lower than that in normal adjacent normal tissues and normal cells (P<0.05).Moreover, overexpression of HIPK3 was positively correlated with advanced TNM stage and tumor size, but not with other parameters such as age, gender, alcohol consumption, smoking history or differentiation status.HIPK3 silencing inhibited cell viability, promoted apoptosis and reduced cell migration and invasiveness, while HIPK3 overexpression can increase cell viability, inhibit cell apoptosis and increase cell migration and invasion. In summary, HIPK3 can be used as a potential molecular target for the treatment of ESCC to control the development of tumors by inhibiting its expression.
As a target for circ RNA, mi RNA is the most widely studied class of non-coding RNAs.5 It can regulate cell proliferation, differentiation and apoptosis by degrading target mRNA or inhibiting its translation, and participate in processes such as individual development, body metabolism, tumorigenesis and development.32 Studies had found that overexpression of miR-599 inhibited the proliferation of breast cancer and lung cancer (!!! INVALID CITATION !!!).We screened miR-599 as a target gene for HIPK3 through a database. It regulated its expression by targeting the 3ʹUTR of the miR-199a-3p gene. HIPK3 knockdown can up-regulate the expression level of miR-599, and vice versa. The expression level of miR-599 was down-regulated in ESCC tissues compared with that in normal tissues, and an inverse correlation between circHIPK3 expression and miR-599 levels was observed in ESCC tissues.
c-MYC is a proto-oncogene of MYC family cells, which expresses a nuclear protein and participates in the regulation of other genes by binding DNA.24 The expression of c-MYC protein was higher in proliferating cells, which decreased with cell differentiation and lost the ability of promoting proliferation. When Down regulation is expressed, cells are induced to differentiate into terminal cells.7 c-MYC target-genes, which mediate these functions of c-MYC, represent a complex network of protein- and non-coding RNAs, including numerous miRNAs. For example, c-MYC directly regulates expression of the miR-17-92 cluster, miR-34a, miR-15a/16-1 and miR-9. Moreover, the expression and activity of c-MYC itself are under the control of miRNAs.12 Here, for the first time, we identified MYC as a target of miR-599 and showed that the miR-599-MYC axis regulates cell proliferation and migration in ESCC. circHIPK3 regulates the proliferation and migration of ESCC cells by serving as a miR-599 sponge and thus modulates MYC expression. Further understanding of the circHIPK3- miR-599- MYC axis may provide a novel therapeutic strategy for ESCC in the future.
Conclusion
HIPK3 inhibited ESCC proliferation and invasion and metastasis by regulating miR-599/c-MYC axis, suggesting that HIPK3 may be a potential therapeutic target for ESCC. It provided experimental basis for the study of clinical prognosis of the tumor and further targeted intervention therapy.
Ethical Statement and Consent for Publication
Written informed consent was obtained from all patients. The study was approved by the Affiliated Cancer Hospital of Zhengzhou University Institutional Review Committee.
Disclosure
The authors declare that they have no competing interests in this work.
References
- 1.Anokyedanso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10:5783–5794. doi: 10.2147/OTT.S150678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Yin D, Li L, Deng Y, Tian W. Screening aberrant methylation profile in esophageal squamous cell carcinoma for Kazakhs in Xinjiang area of China. Mol Biol Rep. 2015;42(2):457–464. doi: 10.1007/s11033-014-3788-z [DOI] [PubMed] [Google Scholar]
- 5.Cheng AM, Byrom M, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmore Jake E, Issa Ghayas C, Lemieux Madeleine E, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo L, Zheng L, Zhao Y, Wang Q. Profiling and bioinformatic analyses indicate differential circRNA and miRNA/isomiR expression and interactions. Biomed Res Int. 2018;2018:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guy JB, Espenel S, Vallard A, et al. Evaluation of the cell invasion and migration process: a comparison of the video microscope-based scratch wound assay and the boyden chamber assay. J Visual Exp Jove. 2017;2017:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeguchi M, Kohno Y, Kihara K, et al. Neoadjuvant chemotherapy for clinical Stage II and III thoracic esophageal squamous cell carcinoma with curative esophagectomy. J Cancer Ther. 2015;06(15):1207–1213. doi: 10.4236/jct.2015.615131 [DOI] [Google Scholar]
- 12.Jackstadt R, Hermeking H. MicroRNAs as regulators and mediators of c-MYC function. Biochim Biophys Acta. 2015;1849(5):544–553. doi: 10.1016/j.bbagrm.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 13.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji Y, Du X, Tian Y, et al. A Phase II study of S-1 with concurrent radiotherapy in elderly patients with esophageal cancer. Oncotarget. 2017;8(47):83022–83029. doi: 10.18632/oncotarget.v8i47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Xu WW, Han L, et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017a;36(28):3986–4000. doi: 10.1038/onc.2017.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Zhang X, Li N, Liu Q, Chen D. miR-30b inhibits cancer cell growth, migration, and invasion by targeting homeobox A1 in esophageal cancer. Biochem Biophys Res Commun. 2017b;485(2):506–512. doi: 10.1016/j.bbrc.2017.02.016 [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017c;18(9):1646–1659. doi: 10.15252/embr.201643581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long L, Pang X, Lei F, et al. SLC52A3 expression is activated by NF-κB p65/Rel-B and serves as a prognostic biomarker in esophageal cancer. Cell Mol Life Sci. 2018;75(14):2643–2661. doi: 10.1007/s00018-018-2757-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Young JJ, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–256. doi: 10.1038/ncb2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X, Zhao K, Guo W, et al. Salvage lymphadenectomy versus salvage radiotherapy/chemoradiotherapy for recurrence in cervical lymph node after curative resection of esophageal squamous cell carcinoma. Ann Surg Oncol. 2015;22(2):624–629. doi: 10.1245/s10434-014-4008-8 [DOI] [PubMed] [Google Scholar]
- 22.Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2016;18(5):780–788. [DOI] [PubMed] [Google Scholar]
- 23.Miyakawa H, Iguchi T. Comparative luciferase assay for establishing reliable in vitro screening system of juvenile hormone agonists. J Appl Toxicol. 2017;37(9):1082. doi: 10.1002/jat.v37.9 [DOI] [PubMed] [Google Scholar]
- 24.O’Donnell KA, Al E. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- 25.O’donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- 26.Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21. doi: 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen L, Wang H, Dong B, et al. Possible prediction of the response of esophageal squamous cell carcinoma to neoadjuvant chemotherapy based on gene expression profiling. Oncotarget. 2016;7(4):4531–4541. doi: 10.18632/oncotarget.6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21(6):1469–1477. doi: 10.1111/ejn.2005.21.issue-6 [DOI] [PubMed] [Google Scholar]
- 31.Teng H, Li X, Liu X, Wu J, Zhang J. The absence of human papillomavirus in esophageal squamous cell carcinoma in East China. Int J Clin Exp Pathol. 2014;7(7):4184. [PMC free article] [PubMed] [Google Scholar]
- 32.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17∼92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vrana D, Matzenauer M, Aujesky R, et al. Potential predictive role of MicroRNAs in the neoadjuvant treatment of esophageal cancer. Anticancer Res. 2017;37(2):403–412. doi: 10.21873/anticanres [DOI] [PubMed] [Google Scholar]
- 34.Wan L, Zhang L, Fan K, Cheng Z, Sun Q, Wang J. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Zhang W, Liu X, et al. Prognosis of esophageal squamous cell carcinoma patients with preoperative radiotherapy: comparison of different cancer staging systems. Thorac Cancer. 2014;5(3):204–210. doi: 10.1111/tca.2014.5.issue-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong CH, Ma BBY, Hui CWC, Tao Q, Chan ATC. Preclinical evaluation of afatinib (BIBW2992) in esophageal squamous cell carcinoma (ESCC). Am J Cancer Res. 2015;5(12):3588–3599. [PMC free article] [PubMed] [Google Scholar]
- 37.Xiaolong M, Kunpeng Z, Chunlin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi: 10.7150/jca.24619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong D, Dang Y, Lin P, et al. A circRNA–miRNA–mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220. doi: 10.1186/s12967-018-1593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu WW, Li B, Zhao JF, et al. IGF2 induces CD133 expression in esophageal cancer cells to promote cancer stemness. Cancer Lett. 2018;425:88–100. doi: 10.1016/j.canlet.2018.03.039 [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Jiang L, Sun D, Hou J, Ji Z. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 41.Zhang P, Meng X, Chen H, et al. PlantCircNet: a database for plant circRNA–miRNA–mRNA regulatory networks. Database. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, Zhou W, Xue L, Zhang W, Zhan Q. Nicotine activates YAP1 through nAChRs mediated signaling in esophageal squamous cell cancer (ESCC). PLoS One. 2014;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziqiang T. Regional differences of methylation of metallothionein-3 gene in tissues of esophageal cancer. Tumori. 2009;1:1137–1139. [Google Scholar]