Abstract
The ImpX transporters of the Drug/Metabolite Transporter (DMT) superfamily were first proposed to transport riboflavin (vitamin B2) based on findings of a cis-regulatory RNA element responding to flavin mononucleotide (an FMN riboswitch). BdeHovibrio exovorous JSS has a homolog belonging to this superfamily. It has 10 TMSs and shows 30% identity to the previously characterized ImpX transporter from Fusobacterium nucleatum. However, the ImpX homolog is not regulated by an FMN-riboswitch. In order to test the putative function of the ImpX homolog from B. exovorous (BexImpX), we cloned and heterologously expressed its gene. We used functional complementation, growth inhibition experiments, direct uptake experiments and inhibition studies, suggesting a high degree of specificity for riboflavin uptake. The EC50 for growth with riboflavin was estimated to be in the range 0.5–1 μM, estimated from the half-maximal riboflavin concentration supporting the growth of a riboflavin auxotrophic E. coli strain, but the Khalf for riboflavin uptake was 20 μM. Transport experiments suggested that the energy source is the proton motive force, but that NaCl stimulates uptake. Thus, members of the ImpX family members are capable of riboflavin uptake, not only in riboflavin prototrophic species such as F. nucleatum, but also in the B2 auxotrophic species, B. exovorous.
Keywords: Riboflavin, ImpX transporter, Roseoflavin, FMN biosynthesis
Introduction
Bdellovibrios are predatory bacteria isolated from soil that can lyse and utilize the cytoplasmic constituents of other Gram-negative bacteria as nutrients, potentially providing an alternative approach to the biocontrol of human, animal and plant pathogens [Cao et al., 2012; Koval et al., 2013; McNeely et al., 2017]. Bdellovibrio is a genus of Gram-negative bacteria that belongs to the Oligoflexia class of Proteobacteria. Two known members of this genus, B. bacteriovorus and B. exovorus, are obligate predators of other Gram-negative bacteria. While the former species grows in the periplasmic space of the prey cell, the latter grows outside of the prey cell. These predators are ubiquitous in the environment and have been isolated from soil, compost, sewage, activated sludge and marine and terrestrial waters [Davidov et al., 2006a; Davidov et al., 2006b; Jurkevitch, 2006]. The complete genome sequences of both Bdellovibrio species are available, and this has allowed comparitive analyses of transport systems in these organisms [Heidari Tajabadi et al., 2017].
The water-soluble vitamin, riboflavin (RF; vitamin B2), is a precursor for two essential metabolic cofactors, flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD). Flavoproteins are universally important for biological processes, including energy metabolism, redox reactions, light emission, biosynthesis, and DNA repair [Garcia Costas et al., 2017; Liu et al., 2017; Plegaria et al., 2017; Stietz et al., 2017]. Riboflavin synthesis has been demonstrated in a wide range of microorganisms through a pathway that creates one riboflavin molecule from one molecule of GTP and two molecules of ribulose 5-phosphate [Fassbinder et al., 2000; Fischer and Bacher, 2010]. The enzymes involved in riboflavin synthesis (Fig. 1) are GTP cyclohydrolase I (RibA), 3,4-dihydroxy-2-butanone 4-phosphate synthase (RibB), pyrimidine deaminase/reductase (RibD), 6,7-dimethyl-8-ribityllumazine synthase (lumazine synthase, RibH) and riboflavin synthase (RibC) [Fischer and Bacher, 2010].
Fig. 1.
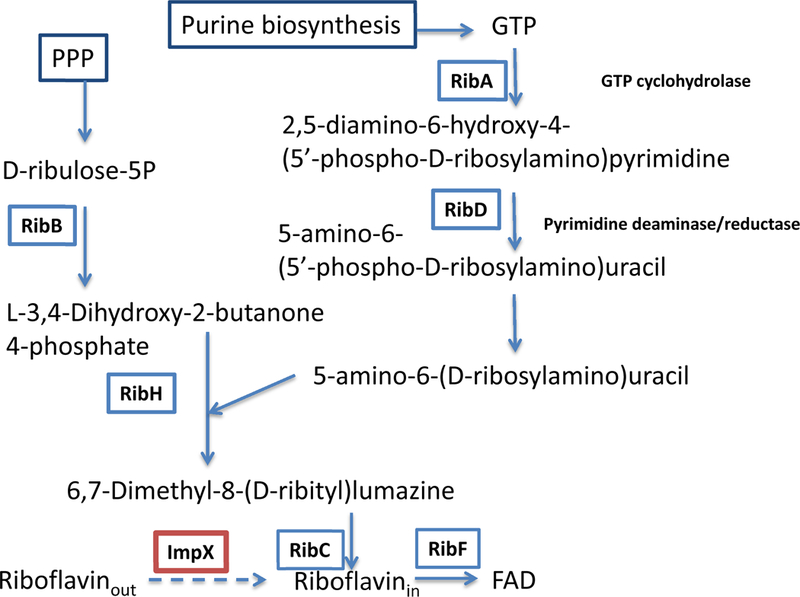
The pathway for riboflavin biosynthesis and transport in bacteria. PPP - pentose phosphate pathway.
The expression of riboflavin biosynthesis and transport genes in many bacteria is regulated by conserved cis-regulatory RNAs known as riboflavin nucleotide (RFN) elements that were first discovered in silico [Vitreschak et al., 2002] and subsequently identified as a class of FMN-sensing riboswitches that regulate gene expression by adopting alternative secondary structures after binding an FMN molecule [Mironov et al., 2002]. ribB expression is negatively regulated by an FMN-riboswitch in E. coli in the presence of FMN or the FMN-analogue, roseoflavin-mononucleotide [Pedrolli et al., 2015]. Roseoflavin, produced by Streptomyces davawensis, is supposed to suppress the activities of flavoproteins in E. coli [Langer et al., 2013] and the FMN riboswitch in B. subtilis [Jankowitsch et al., 2012; Ott et al., 2009].
The production of vitamins and exchange between microbes and hosts is important; exchange is mediated by efflux and uptake systems [Garcia-Angulo, 2017]. For example, the E. coli flavin efflux transporter YeeO has been characterized [McAnulty and Wood, 2014]. Additionally, riboflavin can be used as an electron shuttle for extracellular respiration. Previous analyses of bacterial genomes for genes potentially regulated by FMN riboswitches allowed prediction of multiple families of putative riboflavin uptake transporters [Sun and Rodionov, 2014; Vitreschak et al., 2002]. The functions of representative members of these predicted riboflavin transporter families have been confirmed experimentally [Gutierrez-Preciado et al., 2015; Rodionova et al., 2015]. The RibU transporters belong to the most broadly distributed class of ABC-type ECF-family riboflavin transporters in Gram-positive bacteria [Rodionov et al., 2009] and in T. maritima [Karpowich et al., 2016]. Two different types of ABC-type riboflavin transporters, RibXY (TCDB family 3.A.1.17.14) in Chloroflexi and Thermobaculum terrenum [Rodionova et al., 2015] and RfuABCD in spirochetes [Deka et al., 2013], are so far restricted to these taxonomic lineages. The PnuC-family riboflavin permease RibM (TC# 4.B.1.1.5) is found only in some Actinobacteria. However the phylogenetic distributions of two other riboflavin permeases, RibN (TC# 2.A.7.3.54) and RfnT (TC# 2.A.1.81.5), are restricted to several classes of Proteobacteria [Garcia Angulo et al., 2013]. A characterized RibZ permease TC# 2.A.1.3.72) has been identified in Firmicutes, but homologs are also present in Actinobacteria [Sun and Rodionov, 2014; Vitreschak et al., 2002]. Although ImpX (Integral Membrane Protein X) was originally identified in Fusobacterium nucleatum (a commensal-turned pathogen from the fusobacterial phylum) and Desulfitobacterium halfniense [Mironov et al., 2002], ImpX transporters regulated by FMN riboswitches were bioinformatically identified across the phyla of Firmicutes (e.g., in Bacillus clausii, Clostridium beijerinckii, Geobacillus sp. and Paenibacillus sp.) and Fusobacteria (in Ilyobacterpolytropus and Sebaldella termitidis), as well as in a single species from the proteobacterial phylum, Marinomonas sp. [Rodionova et al., 2015]. The involvement of ImpX from F. nucleatum in riboflavin uptake was confirmed by functional complementation [Gutierrez-Preciado et al., 2015].
In summary, ImpX transporters had been found previously only in bacteria that are capable of synthesizing riboflavin (B2 prototrophs). In this work, we analyze the distribution of ImpX homologs in an expanded set of bacterial genomes and report their identification in a few bacteria that do not synthesize riboflavin (B2 auxotrophs) including B. exovorus. We experimentally characterize the putative ImpX transporter from this organism.
Results
Bioinformatic analysis of ImpX transporters and FMN riboswitch regulons
Similarity searches identified orthologs of ImpX in bacterial genomes representing diverse taxonomic phyla of bacteria. These include Proteobacteria (including B. exovorous and Bacteriovorax marinus, another predatory bacterium from the Oligoflexia class, and 4 species of γ-proteobacteria), Fusobacteria (10 species), Actinobacteria (4 species), Firmicutes (9 species) and a single bacterium of the Chloroflexi phylum, Thermomicrobium roseum (Fig. 2). The presence of an FMN riboswitch upstream of the impX genes from Firmicutes, γ-proteobacteria and some Fusobacteria suggests that transporters with vitamin B2 specificity are conserved across the ImpX transporter family. Interestingly, the majority of bacterial genomes possessing an ImpX homolog also contain a complete set of riboflavin biosynthetic genes, suggesting that ImpX has a tendency to be found in B2 prototrophic microorganisms, possibly serving a scavenging functions or allowing riboflavin export. The only two B2 auxotrophic bacteria possessing ImpX transporters of those examined are B. exovorous and Streptobacillus moniliformis. The observed positive correlation between ImpX transporters and the presence of the B2 biosynthetic pathway suggests potential involvement of ImpX in vitamin B2 exchange between species in microbial communities, e.g., crossfeeding in biofilms.
Fig. 2.
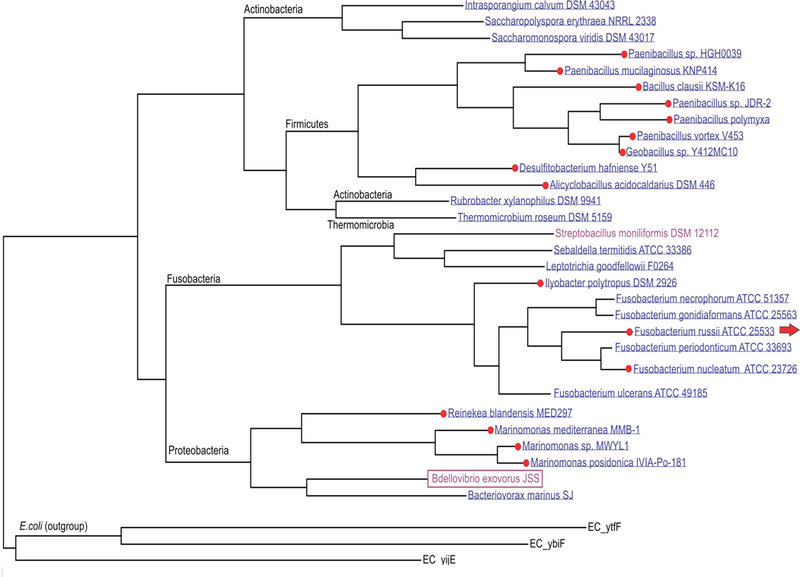
The maximum likelihood phylogenetic tree of ImpX proteins from Bdellovibrio exovorous and other bacterial species. Red dots show species that have an impX transporter gene controlled by an FMN riboswitch. The presence of a complete set of riboflavin biosynthetic genes in bacterial genomes is shown by blue (B2 prototrophs), while B2 auxotrophic species are shown in red. Three other members of the 10 TMS Drug/Metabolite Exporter (DME) family, YtfF, YbiF and YijE from E. coli, were used as an outgroup.
B2 auxotrophic species must use a B2 transporter for uptake of this essential vitamin into the cell. However, B2 prototrophs can potentially use the ImpX transporter, not only for the uptake of exogenous B2 (when its concentration inside the cell is low), but also for the efflux of the vitamin when its intracellular concentration is high, due to the actvity of the biosynthetic pathway. In support of this hypothesis, many characterized members of the DME family are involved in multidrug or metabolite efflux such as the threonine/homoserine transporter, YbiF (TC# 2.A.7.3.6) [Livshits et al., 2003], and the cysteine exporter, YijE (TC# 2.A.7.3.26) [Yamamoto et al., 2015] of E. coli. Reconstruction of the riboflavin biosynthetic pathways in the two Bdellovibrio species with available genomes suggests that only B. bacteriovorus contains the complete set of B2 biosynthetic enzymes. It is a B2 prototroph without a riboflavin transporter. In contrast, the ribD, ribH and ribE genes are missing in B. exovorous JSS, which contains only homologs of ribBA and impX (gene locus tags, A11Q_1932 and A11Q_1837, respectively). Thus, the putative riboflavin transporter ImpX replaces the de novo biosynthetic pathway in this species (Fig.1).
Overexpression of ImpX in a ribC E. coli mutant complements the growth defect.
Taking into account the importance of vitamin B2 salvage for B. exovorus, the ImpX transporter from this organism (named BexImpX) was selected for experimental testing in vivo. To determine whether the identified candidate BexImpX protein is a riboflavin transporter, we cloned the impX gene from B. exovorus, inserted it into the chromosome of E. coli and evaluated its ability to complement the growth of a ΔribC E. coli strain. A low concentration of riboflavin in the medium did not support the growth of the ΔribC strain in M9 medium, reflecting the lack of a riboflavin transporter in E. coli (data not shown). This strain requires high concentrations of exogenous riboflavin to support growth. However, provision of BexImpX in this ΔribC strain allowed growth with 3 μM of riboflavin (Fig.3). These results indicate that ImpX facilitates the uptake of riboflavin. Some growth inhibition for ImpX overproducing cells appeared at 0.7mM of riboflavin in E. coli.
Fig. 3.

Growth of E. coli with varied concentrations of riboflavin (RF). The growth curves are marked purple for 0.7 mM RF with the ΔribC strain; other lines - ΔribC ImpX+, blue diamonds - 3μM RF; red squares - 10 μM RF; green triangles - 0.7 mM RF.
Inhibition of E. coli by roseoflavin in a BexImpX overproduction strain.
To support the inferred function of the riboflavin transporter, we tested the inhibition of E. coli growth by the riboflavin analog, roseoflavin, which targets flavin-dependent enzymes and FMN-riboswiches [Pedrolli et al., 2015]. The growth of E. coli wild type and the derivative strain expressing BexImpX was measured in 96-well plates using the plate-reader, Biotek Elx808 (Fig. 4). Growth of both strains in the presence of 20 μM roseoflavin was not affected, but at a higher concentration (240 μM), inhibition by roseoflavin was observed only for the BexImpX-producing strain. We conclude that roseoflavin was taken up by the BexImpX transporter and inhibited the growth of the impX+ strain.
Fig. 4.
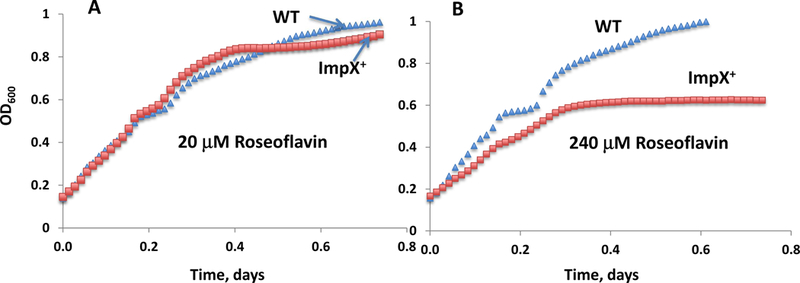
Growth inhibition of E. coli with roseoflavin at concentrations of 20 μM (A) and 240 μM (B); WT E. coli - blue triangles, isogenic impX+ strain of E. coli - red squares. The BW25113 wild type strain was used with conditions as specified under Materials and Methods.
Riboflavin uptake by ImpX in E. coli depends on the proton motive force.
The E. coli wild type strain and the derivative strain producing BexImpX were assayed for riboflavin uptake, measured in M9 media containing 20 mM mannose in the presence and absence of Na+. The BexImpX-dependent uptake of riboflavin was observed during a 2–15 minute incubation, but no uptake was observed for the wild type E. coli strain. To determine the sodium or proton dependency, we measured riboflavin uptake in the BexImpX strain of E. coli in the presence and absence of 0–150 mM NaCl and 0–100 μM cyanide carbonyl m-chlorophenyl hydrazone (CCCP) (Fig.5A and 5B). Vitamin B2 uptake was most efficient in an M9 salts solution or in 0.1 X PBS buffer supplemented with 10 mM D-mannose. Under these conditions, an inhibitory effect in the presence of 25 μM CCCP was observed. The inhibitory effect on riboflavin uptake (35 μM) by nicotinamide or thiamine pyrophosphate at 0.9mM was less then 5%. The data obtained for the uptake using RF titration (Fig. 5C) was analyzed by Graph Pad software (Prizm 6).
Fig. 5.

Determination of riboflavin (RF) uptake using the fluorescence method. A. Dependency on the CCCP concentration. The increase in fluorescence after 4 minutes of uptake was measured using 0–25 μM CCCP. B. The dependency of the uptake of RF on the NaCl concentration. The increase in fluorescence after 4 minutes of uptake was measured with 0–150 mM NaCl. C. The dependency of uptake on the RF concentration (0–60 μM) giving a calculated Khalf = 20 μM. The increase in fluorescence after 4 and 8 minutes was measured as described in the text, normalized to a standardized OD600. FI is fluorescence intensity (the FI background for E. coli cells was subtracted).
Conclusion
In this communication, we provide evidence that an ImpX homologue from B. exovorous (BexImpX), which is distantly related to the previously characterized ImpX from F. nucleatum (Fig. 3), is probably a riboflavin-specific transporter. This suggests that close BexImpX homologues and probably all other ImpX family proteins shown in Fig. 2, have the same specificity.
To support the inferred functional relevance of the vitamin B2 transporter, we experimentally demonstrated the riboflavin transport activity of BexImpX heterologously expressed in E. coli, by showing that this transporter allows the growth of an E. coli ΔribC mutant auxotroph at low riboflavin concentrations. The overproduction of the ImpX transporter in E. coli showed a mild toxic effect at high concentrations of riboflavin. ImpX also mediates roseoflavin-dependent inhibition of E. coli growth. The transport assays revealed an activating effect of sodium at high concentrations. The transporter is probably a proton and/or sodium ion dependent pmf-driven system, as it was shown to be strongly inhibited by CCCP at a concentration of 25 μM (Figs. 5B and 5C). The dependency of uptake on RF concentrations of 0–60 μM using the fluorescence method yielded the 20 μM Khalf value for ImpX-mediated uptake (Fig. 5C).
Materials and Methods
Bacterial strains, plasmids, media and culture conditions
Bacterial strains and plasmids used in this study are listed in Table 1.
Table 1-.
Strains and plasmids used in this study.
| Strain or Plasmid | Relevant characteristics |
Source |
|---|---|---|
| Bdellovibrio exovorous JSS | Wild type | ATCC BAA-2330 |
| Caulobacter crescentus CB15N | Wild type | SDSU, Dr. Shapiro |
| Escherichia coli BW25113 | Wild type | In house |
| E. coli BSV13 | ΔribC | CGSC |
| Plasmid PKD13 | Amp+ Kan+ Temp- | In house |
| Plasmid PCP20 | Amp+ Kan+ Temp- | In house |
Initial studies on the growth of B. exovorous strain JSS (ATCC BAA-2330) were conducted in co-cultures in 1/10-strength yeast extract-peptone medium supplemented with calcium [Gordon et al., 1993]. For maintenance, co-cultures contained 1 ml of a stationary phase culture of B. exovorous strain JSS and 4 ml of a 24 h peptone-yeast extract (PYE) broth culture of Caulobacter crescentus CB2A [Koval et al., 2013]. This mixture was incubated in 125 ml flasks with 20 ml of defined HM buffer. The co-culture was incubated at 30°C with shaking at 150 rpm for 24–48 h. Cultures were transferred to sterile screw-capped tubes and kept at 4°C for up to 1 month. For long-term storage, a 24–48 h culture of strain B. exovorous JSS cells was frozen in the presence of 25 % (w/v) glycerol at −80°C [Koval et al., 2013].
The C. crescentus strain used in this study, listed in Table 1, was obtained from Dr. L. Shapiro (Stanford University, Stanford, California, USA). C. crescentus was maintained on PYE medium. The E. coli ΔribC strain was obtained from the CGSC collection.
All of the constructs were confirmed by DNA sequencing using the primers listed in Table 2. To see if expression of the B. exovorous impX gene can restore growth of E. coil ribC mutant cells in the presence of riboflavin, a constitutive promoter driven impX gene was moved into the E. coli chromosome. First, the impX gene was amplified from B. exovorous ATCC BAA-2330 DNA using the primers Bex-Bam-R and Bex-Kpn-F (listed in Table 2). The resulting fragment was digested with KpnI and BamHI and cloned into the same sites of pKD13-rrnBT:Ptet, yielding the plasmid pKDT_Ptet-impX, in which impX expression is under the control of the tet promoter (Ptet). Present in this plasmid, the fragment “kmr:rrnBT:Ptet-impX” was PCR amplified, gel purified and then electroporated into wild type BW25113 cells expressing the λ-Red recombinase. The cells were incubated with shaking at 37°C for 1 hour and then applied onto LB + Km agar plates. The Kmr colonies were verified for the “kmr:rrnBT:Ptet-impX”substitution for the 67 bp intS/yfdG intergenic region between the 117th and 51st nucleotides relative to the start codon of yfdG by colony PCR (using primers kt and intS-ver-R) and subsequently by sequencing. This yielded strain BW25123_impX.
Table 2-.
Oligonucleotides used in this study
| Primer name | Sequence (5’ to 3’) |
|---|---|
| Bex-Bam-R | ATGGATCCTTAATTTGTATGACGGCGTGAAAGCTG |
| Bex-Kpn-F | TTAGGTACCATGGGTTTTATCTTTATTATTTTGGG |
| Bex-ver-R | TCAGGATAAGAAATAAGGAATACG |
| intS-Bex-P2 | GATAGTTGTTAAGGTCGCTCACTCCACCTTCTCATCAAGCCAGTCCGCCCTTAATTTGTATGACGGCGTGAAAGC |
| intS-ver-R | TCCAAGTCTTAATCGATCGATACTTG |
| Kt | CGGCCACAGTCGATGAATCC |
| rib-ver-R | TGATATTCAGCTCTGGCAGGTCGTG |
| ribC-ver-F | TGATATACTTCTGCACGTGAACAC |
| ribC1-P1 | ATGTTTACGGGGATTGTACAGGGCACCGCAAAACTGGTGTCGATTGACGAGTGTAGGCTGGAGCTGCTTC |
| ribC2-P2 | TCAGGCTTCTGTGCCTGGTTGATTCATGGCATTTTCTCGTGCCGCCAGCACATTCCGGGGATCCGTCGACCTG |
The chromosomal region carrying “kmr:rmBT:Ptet-impX” in BW25123_impX was transferred into E. coli ribC deletion mutant cells by P1 transduction. The selection medium was LB+Km+riboflavin. A P1 transductant was purified and confirmed by PCR, yielding E. coli ΔribC_impX that was used for the growth assays.
Riboflavin growth experiment and roseoflavin inhibition of the E. coli ImpX+ strain.
To compare growth of strains E. coli ΔribC and E. coli ΔribC_impX, minimal medium M9 with a carbon source (0.5% glucose) and a nitrogen source (20 mM NH4Cl) was used. The cells of E. coli ΔribC and ΔribC_impX were grown overnight in LB, supplemented with 0.7 mM riboflavin. The overnight cultures were washed twice using M9 salts. The strains inoculated into M9 for growth without washing are presented in Fig. 3.
The strains of E. coli: ΔribC ImpX+ and ΔribC (as control) were inoculated into M9 medium with different concentrations of riboflavin: 0, 0.03 μM, 0.27 μM and 26.5 μM. The cultures were started at OD600 = 0.02, and growth was measured by following the increase in the absorbance at 600 nm.
For the experiment with the riboflavin analogue inhibitor, roseoflavin, the E. coli wild type strains with and without impX were grown overnight and inoculated into LB medium at different concentrations of roseoflavin. The starting absorbance was OD600 = 0.005.
Bioinformatic analyses
A search for B2 regulatory elements (FMN riboswitches) for the analysis and prediction of candidate transporters was conducted as described [Rodionova et al., 2015; Sun and Rodionov, 2014]. A maximum likelihood phylogenetic tree was constructed with the PhyML program, and an alignment was generated using the Muscle programs [Edgar, 2004; Lefort et al., 2017]. The operon co-localization of impX with the riboflavin biosynthetic genes is marked by an arrow; taxonomic groups are indicated in black font. The system analysis of homologues for riboflavin genes was produced using the PubSEED platform as described in [Magnusdottir et al., 2015].
Riboflavin uptake measurements
The E. coli wild type and the derivative strain expressing BeximpX were grown to OD600 equal to 1, and the cells were collected and washed with M9. Riboflavin uptake was measured in M9 salts or 0.1XPBS buffer or 40 mM Tris-HCl buffer supplemented with 20 mM mannose containing 0.04 mM riboflavin and 10 mM MgSO4. The effects of the different concentrations of NaCl or carbonyl cyanide m-chlorophenyl hydrazone (CCCP) on BexImpX-mediated uptake were tested (Fig 5A–B). Following uptake experiments at 25°C for various periods of time, cells were collected and washed with PBS buffer. For cell extraction of riboflavin, 0.2ml of 5% perchloric acid was added followed by incubation on ice for 10 minutes with periodic vortexing. After centrifugation, 0.05 ml of supernatant was added to 0.2 ml K2HPO4 on black plates, and the fluorescence was measured. The total concentration of FMN and riboflavin in the E. coli cells were detected by fluorescence as described in [Munro and Noble, 1999].
Acknowledgements:
This research was supported by the US National Institutes of Health (NIH) grant GM077402. D.A.R. was supported by the Russian Academy of Sciences via the program ‘Molecular and Cellular Biology’.
Footnotes
Statement of Ethics: No animal or human subjects were involved in this study, which is why there are no ethical conflicts to disclose.
Disclosure Statement: The authors declare that they have no conflicts of interest.
References
- Cao H, He S, Wang H, Hou S, Lu L, Yang X: Bdellovibrios, potential biocontrol bacteria against pathogenic Aeromonas hydrophila. Vet Microbiol 2012;154:413–418. [DOI] [PubMed] [Google Scholar]
- Davidov Y, Friedjung A, Jurkevitch E: Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like organisms. Environ Microbiol 2006a;8:1667–1673. [DOI] [PubMed] [Google Scholar]
- Davidov Y, Huchon D, Koval SF, Jurkevitch E: A new alpha-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol 2006b;8:2179–2188. [DOI] [PubMed] [Google Scholar]
- Deka RK, Brautigam CA, Biddy BA, Liu WZ, Norgard MV: Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. MBio 2013;4:e00615–00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Res 2004;32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassbinder F, Kist M, Bereswill S: Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS Microbiol Lett 2000;191:191–197. [DOI] [PubMed] [Google Scholar]
- Fischer M, Bacher A: Biosynthesis of Riboflavin. EcoSal Plus 2010;4. [DOI] [PubMed] [Google Scholar]
- Garcia Angulo VA, Bonomi HR, Posadas DM, Serer MI, Torres AG, Zorreguieta A, Goldbaum FA: Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium leguminosarum and other proteobacteria. J Bacteriol 2013;195:4611–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Costas AM, Poudel S, Miller AF, Schut GJ, Ledbetter RN, Fixen KR, Seefeldt LC, Adams MWW, Harwood CS, Boyd ES, Peters JW: Defining Electron Bifurcation in the Electron- Transferring Flavoprotein Family. J Bacteriol 2017;199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Angulo VA: Overlapping riboflavin supply pathways in bacteria. Crit Rev Microbiol 2017;43:196–209. [DOI] [PubMed] [Google Scholar]
- Gordon RF, Stein MA, Diedrich DL: Heat shock-induced axenic growth of Bdellovibrio bacteriovorus. J Bacteriol 1993;175:2157–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Preciado A, Torres AG, Merino E, Bonomi HR, Goldbaum FA, Garcia-Angulo VA: Extensive Identification of Bacterial Riboflavin Transporters and Their Distribution across Bacterial Species. PLoS One 2015;10:e0126124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari Tajabadi F, Medrano-Soto A, Ahmadzadeh M, Salehi Jouzani G, Saier MH Jr.: Comparative Analyses of Transport Proteins Encoded within the Genomes of Bdellovibrio bacteriovorus HD100 and Bdellovibrio exovorus JSS. Journal of molecular microbiology and biotechnology 2017;27:332–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowitsch F, Schwarz J, Ruckert C, Gust B, Szczepanowski R, Blom J, Pelzer S, Kalinowski J, Mack M: Genome sequence of the bacterium Streptomyces davawensis JCM 4913 and heterologous production of the unique antibiotic roseoflavin. J Bacteriol 2012;194:6818–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkevitch E: Isolation and classification of bdellovibrio and like organisms. Curr Protoc Microbiol 2006;Chapter 7:Unit 7B 1. [DOI] [PubMed] [Google Scholar]
- Karpowich NK, Song J, Wang DN: An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin. J Mol Biol 2016;428:3118–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E: Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int J Syst Evol Microbiol 2013;63:146–151. [DOI] [PubMed] [Google Scholar]
- Langer S, Hashimoto M, Hobl B, Mathes T, Mack M: Flavoproteins are potential targets for the antibiotic roseoflavin in Escherichia coli. J Bacteriol 2013;195:4037–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V, Longueville JE, Gascuel O: SMS: Smart Model Selection in PhyML. Mol Biol Evol 2017;34:2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LK, Becker DF, Tanner JJ: Structure, function, and mechanism of proline utilization A (PutA). Arch Biochem Biophys 2017;632:142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits VA, Zakataeva NP, Aleshin VV, Vitushkina MV: Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res Microbiol 2003;154:123–135. [DOI] [PubMed] [Google Scholar]
- Magnusdottir S, Ravcheev D, de Crecy-Lagard V, Thiele I: Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet 2015;6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnulty MJ, Wood TK: YeeO from Escherichia coli exports flavins. Bioengineered 2014;5:386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely D, Chanyi RM, Dooley JS, Moore JE, Koval SF: Biocontrol of Burkholderia cepacia complex bacteria and bacterial phytopathogens by Bdellovibrio bacteriovorus. Can J Microbiol 2017;63:350–358. [DOI] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E: Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 2002;111:747–756. [DOI] [PubMed] [Google Scholar]
- Munro AW, Noble MA: Fluorescence analysis of flavoproteins. Methods Mol Biol 1999;131:25–48. [DOI] [PubMed] [Google Scholar]
- Ott E, Stolz J, Lehmann M, Mack M: The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol 2009;6:276–280. [DOI] [PubMed] [Google Scholar]
- Pedrolli D, Langer S, Hobl B, Schwarz J, Hashimoto M, Mack M: The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J 2015;282:3230–3242. [DOI] [PubMed] [Google Scholar]
- Plegaria JS, Sutter M, Ferlez B, Aussignargues C, Niklas J, Poluektov OG, Fromwiller C, TerAvest M, Utschig LM, Tiede DM, Kerfeld CA: Structural and Functional Characterization of a Short-Chain Flavodoxin Associated with a Noncanonical 1,2-Propanediol Utilization Bacterial Microcompartment. Biochemistry 2017;56:5679–5690. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, Eitinger T: A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 2009;191:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionova IA, Li X, Plymale AE, Motamedchaboki K, Konopka AE, Romine MF, Fredrickson JK, Osterman AL, Rodionov DA: Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ Microbiol Rep 2015;7:204–210. [DOI] [PubMed] [Google Scholar]
- Stietz MS, Lopez C, Osifo O, Tolmasky ME, Cardona ST: Evaluation of the electron transfer flavoprotein as an antibacterial target in Burkholderia cenocepacia. Can J Microbiol 2017;63:857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun EI, Rodionov DA: Computational analysis of riboswitch-based regulation. Biochim Biophys Acta 2014;1839:900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS: Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 2002;30:3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Nonaka G, Ozawa T, Takumi K, Ishihama A: Induction of the Escherichia coli yijE gene expression by cystine. Biosci Biotechnol Biochem 2015;79:218–222. [DOI] [PubMed] [Google Scholar]


