Abstract
Maintenance of glycolytic metabolism is postulated to be required for health of the spinal column. In the hypoxic tissues of the intervertebral disc and glycolytic cells of vertebral bone, glucose is metabolized into pyruvate for ATP generation and reduced to lactate to sustain redox balance. The rise in intracellular H+/lactate concentrations are balanced by plasma-membrane monocarboxylate transporters (MCTs). Using MCT4 null mice and human tissue samples, complimented with genetic and metabolic approaches, we determine that H+/lactate efflux is critical for maintenance of disc and vertebral bone health. Mechanistically, MCT4 maintains glycolytic and TCA cycle flux and intracellular pH homeostasis in the nucleus pulposus compartment of the disc, where HIF-1α directly activates an intronic enhancer in SLC16A3. Ultimately, our results provide support for research into lactate as a diagnostic biomarker for chronic, painful disc degeneration.
Keywords: Genetic Animals Models, Nutrition, Transcription Factors, Bone QCT/uCT
INTRODUCTION
For the past 20 years, low back pain (LBP) and neck pain have been the first and sixth leading causes of disability world-wide (1). Lifetime prevalence of LBP is 30.8% in industrialized countries with a peak incidence between the ages of 20–30 (2). Due to the staggering prevalence of LBP, the total US healthcare costs related to LBP are upwards of $625 billion dollars per year (3). Intervertebral disc degeneration is a major risk factor for chronic LBP and neck pain (4). Degeneration of the disc is characterized by a progressive loss of extracellular matrix content and intradiscal acidosis; both of which increase susceptibility to herniation, increased immune response and discogenic pain (5-10).
A renewed interest in cellular metabolism has highlighted the importance of bioenergetics in the pathogenesis of chronic skeletal disorders including disc degeneration, osteoporosis, and osteoarthritis (11-15). Discogenic back pain and associated disc degeneration are particularly related to pathological decreases in intracellular pH, perhaps secondary to metabolic dysregulation during the disease process (16,17). In fact, lactate, the end-product of glycolysis, has been considered a biomarker of disc degeneration since 1968 and was recently proposed as a biomarker of discogenic pain (18-22). Currently, researchers are using magnetic resonance spectroscopy (MRS) and chemical exchange saturation transfer MRI (qCEST-MRI) to measure intradiscal acidity as a diagnostic tool for identifying painful intervertebral discs in humans and a porcine model (21,22). Although quantifying intradiscal acidity is now an emerging diagnostic tool, it is still unclear how or why intradiscal acidity correlates with disc disease.
The avascular intervertebral disc is comprised of three distinct tissue compartments: an outer fibrocartilaginous annulus fibrosus (AF), surrounding an inner proteoglycan-rich NP, and cartilaginous endplates (CEP) at the junction with adjacent vertebral bodies. Due to a lack of local blood supply, NP cells manifest metabolic adaptations to their hypoxic, acidic, and nutrient-limiting environment (23-26). Although healthy NP cells survive at low pH, it has been shown that acidic environment (pH 6.2–6.5) causes increased expression of inflammatory cytokines IL-1β and IL-6 and elevated matrix catabolism mediated by matrix metalloproteinases (MMPs) (16,27). Acidic pH also elevates neurotrophic factors, nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF), through Acid-sensing ion channel-3 (ASIC-3) in NP cells (16).
Glycolytic cells employ a robust network of enzymes for intracellular pH regulation, many of which are controlled by the HIF-1α transcription factor. Previous studies demonstrated that NP cells buffer intracellular pH through bicarbonate recycling by the HIF-dependent and membrane-associated Carbonic Anhydrases 9/12, as well as through acid extrusion via Na+/H+ exchangers and Vacuolar-ATPases (17,28). Otherwise, the precise pH regulatory capabilities and the fate of H+/lactate have yet to be elucidated in glycolytic NP cells. One important hallmark of glycolytic cancer cells is H+-coupled lactate efflux driven by increased expression of monocarboxylate transporter 4 (MCT4) (29,30). Maintenance of H+/lactate export via MCT4 is thought to be required for both intracellular pH regulation and prolonged glycolysis. Intracellular lactic acidosis inhibits glycolytic enzyme activity and reciprocally blocks the NAD+ substrate production by lactate dehydrogenases (LDH) (31). In addition to MCT4, there are three MCTs capable of H+-coupled lactate transport with distinct tissue-specific function and localization (32,33). While MCT4 is up-regulated in glycolytic cell types for the purpose of H+/lactate efflux, MCT1 is a bi-directional transporter ubiquitously expressed throughout the body (34). The functions of MCT2/3 are not likely to be of consequence in the disc since MCT2 is considered a lactate importer with very low expression in human tissues, while MCT3 is restricted to the retinal pigment epithelium and choroid plexus (32,35). It is unknown if levels of MCTs are altered with disc degeneration, and if so, what the effect is on acid/base balance and metabolism.
In this study, we show that MCT4 expression is reduced in degenerated human discs and that loss of MCT4 expression leads to age-related disc degeneration and decreased vertebral bone quality in MCT4 knock-out mice. Using genetic and metabolic approaches, we showed that H+/lactate export is critical for maintaining glycolytic flux and blocking mitochondrial respiration in the NP compartment of the disc. Moreover, we demonstrate for the first time that HIF-1α directly activates an intronic enhancer in SLC16A3 under hypoxic conditions. Overall, this study underscores the importance of preserving metabolic homeostasis, through coordinated H+/lactate transport through MCT4, for disc and vertebral bone health.
MATERIALS AND METHODS
Human Sample Collection and Analyses
Human lumbar IVD tissues were obtained at surgery or post-mortem (PM) examination with informed consent of the patient or relatives (Sheffield Research Ethics Committee #09/H1308/70). Immunohistochemistry confirmed and localized expression of MCT1 (1:10, Santa Cruz, sc-50324) MCT4 (1:10, Santa Cruz, sc-50329), and EMMPRIN/CD147 (C-19) (1:10, Santa Cruz, sc-9754) and qRT-PCR confirmed mRNA expression of SLC16A1, SLC16A3, and BSG in 30 human IVDs: 2 PM and 28 surgical samples. See Supplemental Methods and Supplemental Table 1 for more details.
Mouse Study
All procedures regarding collection of animal tissues was performed as per approved protocols by Institutional Animal Care and Use Committee (IACUC) of the Thomas Jefferson University, in accordance with the IACUC’s relevant guidelines and regulations. MCT4+/− mice were purchased from Taconic Bioscience. Skeletally mature male mice at 8- and 14-months of age were analyzed for evidence of early-onset, age-related disc degeneration which is generally distinguishable between 6– 8 months of age. See Supplemental Methods.
Mouse Histological analysis
Caudal spines from 8 and 14-month old WT and MCT4 KO mice were harvested and fixed in 4% PFA for 24 hours and decalcified in 12.5% EDTA at 4oC for 15 days prior to paraffin embedding. Mid-coronal IVD sections (Ca5–8) were stained with Safranin O/Fast Green/Hematoxylin or Picrosirius red, then visualized using a light microscope (AxioImager 2, Carl Zeiss) or a polarizing microscope (Eclipse LV100 POL, Nikon). Histopathological scores were collected from n = 4 mice/genotype with 3 discs per mouse (total 12 discs/genotype) at 8 months; n = 8 mice/genotype with 3 discs per mouse (total 24 discs/genotype) at 14 months. Modified Thompson Grading was used to score NP and AF compartments (Table S2) and Boos Grading Scale was used to score the CEP compartment (5,36). Histological sections were incubated with antibody against KRT19 (1:3, DSHB, TROMA-III/supernatant), CA3 (1:150, Santa Cruz, sc-50715), MMP13 (1:200, Abcam, ab39012), Aggrecan (1:50, Millipore, AB1031), Collagen I (1:100, Abcam, ab34710), Collagen II (1:400, Fitzgerald, 70R-CR008), Collagen X (1:500, Abcam, ab58632), LDHA (1:200, Novus, NBP2–19320), CA9 (1:200, Novus, NB100–417), MCT1 (1:200, Santa Cruz), GLUT-1 (1:200, Abcam, ab40084) and ARGxx (1:200, Abcam, ab3773). See Supplemental Methods.
Width-to-height Aspect Ratio Measurement
Aspect ratio of NP was determined by width divided by height of the NP tissue measured from Safranin O/Fast Green staining images of mid-coronal tissue sections using ImageJ software (http://rsb.info.nih.gov/ij/). Data were collected from from n = 4 mice/genotype with 3 discs per mouse (total 12 discs/genotype) at 8 months; n = 8 mice/genotype with 3 discs per mouse (total 24 discs/genotype) at 14 months.
Immunofluorescence Microscopy
Mid-coronal disc tissue sections of 7 μm thickness were de-paraffinized and incubated in microwaved citrate buffer for 20 min, proteinase K for 10 min at room temperature, or Chondroitinase ABC for 30 min at 37 °C for antigen retrieval. Sections were blocked in 5% normal serum (Thermo Fisher Scientific, 10000 C) in PBS-T (0.4% Triton X-100 in PBS), and incubated with antibody against KRT19 (1:3, DSHB, TROMA-III/supernatant), CA3 (1:150, Santa Cruz, sc-50715), MMP13 (1:200, Abcam, ab39012), Aggrecan (1:50, Millipore, AB1031), Collagen I (1:100, Abcam, ab34710), Collagen II (1:400, Fitzgerald, 70R-CR008), Collagen X (1:500, Abcam, ab58632), LDHA (1:200, Novus, NBP2–19320), CA9 (1:200, Novus, NB100–417), MCT1 (1:200, Santa Cruz) in blocking buffer at 4 °C overnight. For GLUT-1 (1:200, Abcam, ab40084) and ARGxx (1:200, Abcam, ab3773) staining, Mouse on Mouse Kit (Vector laboratories, BMK2202) was used for blocking and primary antibody incubation. Tissue sections were thoroughly washed and incubated with Alexa Fluor®−594 (Ex: 591 nm, Em: 614 nm) conjugated secondary antibody (Jackson ImmunoResearch Lab, Inc.), at a dilution of 1:700 for 1 h at room temperature in dark. The sections were washed again with PBS-T (0.4% Triton X-100 in PBS) and mounted with ProLong® Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, P36934). All mounted slides were visualized with Axio Imager 2 (Carl Zeiss) using 5×/0.15 NAchroplan (Carl Zeiss) or 10×/0,3 EC Plan-Neofluar (Carl Zeiss) objectives, X-Cite® 120Q Excitation Light Source (Excelitas), AxioCam MRm camera (Carl Zeiss), and Zen2TM software (Carl Zeiss).
Micro-CT analysis
Micro-CT (μCT) scans (Bruker, Skyscan 1275) were performed on WT and MCT4 KO spines fixed with 4% PFA [77]. Lumbar spine segments incorporating L2–5 were scanned with an energy of 50 kVp, a current of 200 μA, at a resolution of 10 μm. 5 mice/ genotype were used. The cross sectional scans were analyzed to quantify changes in trabecular bone microarchitecture by first drawing a region of interest (ROI) that contoured the outer boundary of the trabecular bone throughout the entire caudal vertebral body, excluding the cortical bone. The 3D data sets were assessed for the following parameters using software supplied by the system manufacturer: trabecular bone volume (BV), total volume (TV), bone volume fraction (BV/ TV), trabecular thickness (Tb. Th), trabecular number (Tb. N), trabecular separation (Tb. Sp), connectivity density (Conn. Dens.), vertebral length and width. Tissue mineral density was calculated using a standard curve created with a mineral density calibration phantom pair (0.25 g/cm3 CaHA, 0.75 g/cm3 CaHA). Representative images of mineral density were created using ImageJ (NIH).
Isolation of NP cells, hypoxic culture, cell treatments
Rat NP cells were isolated as previously reported by our lab (24). Cells were maintained in Dulbecco’s Modification of Eagle’s Medium (DMEM) supplemented with 10% FBS and antibiotics. Cells were cultured in a Hypoxia Work Station (Invivo2 300, Ruskinn, UK) with a mixture of 1% O2, 5% CO2 and 94% N2. To investigate the effect of MCT inhibition, NP cells were treated with 1) a cocktail of MCT1 (ARC155858, 100 nM, Sigma) and MCT4 inhibitors (Parnell, KM, inventor; U.S. Patent, 15839539, Dec 2017; Publication #20180162822, May 2018; 30 μM) for 2 hours or 2) a dual MCT1 + MCT4 inhibitor (Syrosingopine, 10 μM, Sigma) for 2 hours (37) .
Microarray Analysis and Enriched Pathways
RNA was quantified on a Nanodrop ND-100 spectrophotometer, followed by RNA quality assessment analysis on an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA). Fragmented biotin labeled cDNA (from 100 ng of RNA) was synthesized using the GeneChip WT Plus kit according to ABI protocol (Thermo Fisher Scientific). Gene chips, Mouse Clariom S were hybridized with 2.5 μg fragmented and biotin-labeled cDNA in 100 μl of hybridization cocktail. Arrays were washed and stained with GeneChip hybridization wash & stain kit using Gene chip Fluidic Station 450. Chips were scanned on an Affymetrix Gene Chip Scanner 3000 7G, using Command Console Software. Quality Control of the experiment was performed by Expression Console Software v 1.4.1. Chp files were generated by sst-rma normalization from Affymetrix .cel file using Expression Console Software. Experimental group was compared with control group by using Transcriptome array console 4.0 and GeneSpring v14.5 software. The SST-RMA algorithm was used for data normalization. Only protein-coding genes were included in downstream analyses. The top 50% most expressed genes were kept and used for Significance Analysis of Microarrays (SAM). SAM was run at 5,000 permutations and the False Discovery Rate was set at 5%. DAVID was used to compute enriched pathways in the genes that are upregulated and the ones that are down-regulated. Gene ontology (GO) terms for biological process (BP), molecular function (MF) and cell compartment (CC) as well as KEGG pathways were examined. The genes that were included in the SAM analysis served as the background gene set. Analyses and visualizations were done in R version 3.3.0.
Cell Metabolomics and Data Analysis
NP cells were plated in 10cm plates at a density of 1 × 106 cells and cultured in DMEM with 10% dialyzed FBS (Sigma), 5mM D-glucose, and 4mM L-glutamine in hypoxic conditions until treated for MCT inhibitors for 2 hours. Cells were harvested in ammonium acetate (150 mM) on ice and immediately frozen in liquid nitrogen. Cell pellets were extracted with of 80% methanol (containing 2.5uM U13C succinate, and 5uM U13C citrate as internal standards). The supernatants transferred in GC sampling vial were dried under nitrogen flow, and a two-step derivatization with methoximation and silylation as reported (38). The derivatized metabolites were analyzed with gas chromatography time-of-flight mass spectrometry (GC-TOFMS premier, Waters, USA). The data was analyzed with Genedata Expressionist (Genedata, Basel, Switzerland) software. The output data was normalized to internal standards and the sum of all the metabolites within each sample. The metabolomics dataset comprised of 84 known metabolites that were standardized, i.e. subtracted the mean and divided by the standard deviation per metabolite, and was subsequently analyzed with multivariate statistics [PMID: 29654587] using R version 3.3.0. Metabolites with significant concentration differences were identified using SAM with 5,000 permutations and 5% FDR threshold. To determine the most enriched KEGG pathways, the P-value of each represented mouse metabolic pathway was calculated using hypergeometric test. See Supplemental Methods.
Stable Isotope Enrichment
NP cells were cultured and treated using the same protocol as Cell Metabolomics, however the DMEM was enriched with 2.5 mM [1,2-13C]-glucose as 50% of the glucose pool. A volume of 700 μl of cell culture media was used for ethyl chloroformate (ECF) derivatization following our previously published procedure with minor modifications (39). The derivatized metabolites were extracted with 300 μl of chloroform and 1μl was injected into the GC-MS system in splitless mode (GC-MS, Agilent, USA). The data was analyzed with MassHunter Quantitative Analysis software (Agilent, USA). All quantitative data is represented as mean ± SE, n=5 independent experiments.
Seahorse XF Analyzer Respiratory Assay
The Seahorse XF24 instrument was used to measure extracellular acidification rate (ECAR) and O2 consumption rate (OCR), as reported by Csordás et al (40). All quantitative data is represented as mean ± SE, n=3 independent experiments; 4–6 technical replicates per experiment. See Supplemental Methods.
Extracellular Lactate Measurement
Rat NP cells were plated in duplicate in a 6-well plate at 200,000 cells per well. Cells were incubated in hypoxia for 24 h prior to treatment with MCT inhibitors for 1 h. Lactate concentrations were measured in each sample using a clinical lactate measurement kit from ThermoFisher, complete with reagents and standard curve set.
Intracellular pH Measurement
Rat NP cells treated with MCT inhibitors for 1 h were plated in a 96-well plate at 5,000 cells per well. Intracellular pH was measured following the pHrodo Red AM Intracellular pH Indicator (ThermoFisher) protocol. A standard curve was calculated by clamping the intracellular pH with buffers at a pH of 4.5, 5.5, 6.5, and 7.5 after treatment with a 10 μM valinomycin/nigericin ionophore cocktail. All quantitative data is represented as mean ± SE, n=3 independent experiments; 4 technical replicates per experiment.
Transient Transfection and Dual Luciferase Assay
Cells were transferred to 48-well plates at a density of 2 × 104 cells/well 1 day before transfection. For each transfection, 250ng of plasmid (HRE-Luc (#26731) developed by Navdeep Chandel; pRL-TK (Promega) containing Renilla reniformis luciferase gene) were premixed with Lipofectamine 2000. Cells were harvested 48 h after transfection. The Dual-Luciferase Reporter Assay System (Promega) was used for sequential measurements of Firefly and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were performed with an Infinite 200 PRO plate reader (Tecan, Männedorf, Switzerland). See Supplemental Methods.
Immunofluorescence Microscopy
Mid-coronal disc tissue sections of 7 μm thickness were de-paraffinized and incubated in microwaved citrate buffer for 20 min, proteinase K for 10 min at room temperature, or Chondroitinase ABC for 30 min at 37 °C for antigen retrieval. Sections were blocked in 5% normal serum (Thermo Fisher Scientific, 10000 C) in PBS-T (0.4% Triton X-100 in PBS), and incubated with antibody against KRT19 (1:3, DSHB, TROMA-III/supernatant), CA3 (1:150, Santa Cruz, sc-50715), MMP13 (1:200, Abcam, ab39012), Aggrecan (1:50, Millipore, AB1031), Collagen I (1:100, Abcam, ab34710), Collagen II (1:400, Fitzgerald, 70R-CR008), Collagen X (1:500, Abcam, ab58632), LDHA (1:200, Novus, NBP2–19320), CA9 (1:200, Novus, NB100–417), MCT1 (1:200, Santa Cruz) in blocking buffer at 4 °C overnight. For GLUT-1 (1:200, Abcam, ab40084) and ARGxx (1:200, Abcam, ab3773) staining, Mouse on Mouse Kit (Vector laboratories, BMK2202) was used for blocking and primary antibody incubation. Tissue sections were thoroughly washed and incubated with Alexa Fluor®−594 (Ex: 591 nm, Em: 614 nm) conjugated secondary antibody (Jackson ImmunoResearch Lab, Inc.), at a dilution of 1:700 for 1 h at room temperature in dark. The sections were washed again with PBS-T (0.4% Triton X-100 in PBS) and mounted with ProLong® Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, P36934). All mounted slides were visualized with Axio Imager 2 (Carl Zeiss) using 5×/0.15 NAchroplan (Carl Zeiss) or 10×/0,3 EC Plan-Neofluar (Carl Zeiss) objectives, X-Cite® 120Q Excitation Light Source (Excelitas), AxioCam MRm camera (Carl Zeiss), and Zen2TM software (Carl Zeiss).
Protein extraction, Immunoprecipitation, and Western Blotting
Following hypoxic culture and HIF-1α knock-down, cells were washed on ice with ice-cold 1X PBS with protease inhibitor cocktail (Thermo Scientific). Cell were lysed with lysis buffer containing 1X protease inhibitor cocktail (Thermo Scientific), NaF (4 mM), Na3VO4 (20 mM), NaCl (150 mM), β- glycerophosphate (50 mM), and DTT (0.2 mM). Total protein was resolved on 10% SDS-polyacrylamide gels and transferred to PVDF membranes (Fisher Scientific). Membranes were blocked with 5% nonfat dry milk in TBST (50 mM Tris pH 7.6, 150 mM NaCl, 0.1% Tween 20) and incubated overnight at 4°C in 5% nonfat dry milk in TBST with anti-HIF-1α (1:500, R&D Systems, MAB1536); anti-MCT1 (1:500, Santa Cruz), anti-MCT4 (1:500, Alomone Labs, AMT-014), anti-CD147 (EMMPRIN G-19) (1:1000, Santa Cruz, sc-9757), or anti-β-tubulin (1:5000, DSHB, E7-C) antibodies. Specificity of all antibodies has been validated by the manufacturers using siRNA or negative control IgG. Immunolabeling was detected using ECL reagent (LAS4000, GE Life Sciences). Densitometric analysis was performed using ImageQuant TL (GE Life Sciences). All quantitative data is represented as mean ± SE, n ≥ 4 independent experiments.
Real Time RT-PCR Analysis
Total RNA was extracted from NP cells using RNAeasy mini columns (Qiagen). Purified DNA-free RNA was converted to cDNA using EcoDryTM Premix (Clontech). Equal amounts of template cDNA and gene- specific primers were incorporated into a SYBR Green master mixture (Applied Biosystems) and mRNA expression was quantified using the Step One Plus Real-time PCR System (Applied Biosystems). HPRT was used to normalize gene expression. Melting curves were analysed to verify the specificity of the RT- PCR and the absence of primer dimer formation. Each sample was assessed in duplicate and included a template- free control. All primers used were synthesized by Integrated DNA Technologies, Inc. All quantitative data is represented as mean ± SE, n ≥ 4 independent experiments.
Boinformatics Analysis
BigWig files from deposited ChIP-sequencing datasets (GSE101063, GSE120885) were uploaded and visualized using the using the UCSC Genome Browser (41,42) . Putative HRE consensus sequences (5′-[A/G]CGTG-3′) were determined using the JASPAR Core Database (http://jaspar.genereg.net/) with a relative score threshold of 0.9 (43). Multiz alignment of HRE motifs was performed using the Ensembl Lastz Database to analyze species conservation (http://www.ensembl.org/index.html).
Chromatin Immunoprecipitation
Rat NP cells were plated in 15-cm plates and cultured under normoxic or hypoxic conditions for 24 hours. ChIP assay was performed using ChIP-IT® high sensitivity kit (Active Motif, Carlsbad, CA) according to the manufacturer’s recommendations. Criteria for positive binding include ≥ 5 HIF-1α binding events/ 1,000 cells and binding efficiency 5-fold greater than negative control. Data is represented as mean ± SE, n ≥ 3 independent experiments. See Supplemental Methods.
Plasmid Cloning
Insert DNA was amplified from Human Mixed Genomic DNA (204ng) (Promega) by incubating with Q5 High-Fidelity DNA Polymerase (0.02 U/μl) (NEB), dNTPs (200 μM) (NEB) and primers (0.5 μM) (IDTDNA). PCR product was isolated from the rest of the sample using the QIAquick PCR Purification Kit (Qiagen) and product size confirmed on agarose gel. PCR product and recipient plasmid were restriction digested with Nhe1 and Xho1 enzymes (NEB) overnight. Products were run on an agarose gel and gel purification was performed to isolate DNA using QIAquick Gel Extraction Kit (NEB). DNA ligation was conducted to fuse the insert (0.06 pmol) and recipient plasmid (0.02 pmol) at a ratio of 1:3 recipient to donor using T4 DNA Ligase (NEB). Ligation reaction was transformed into DH5alpha cells and purified with Wizard Plus SV MiniPrep DNA Purification System (Promega). Final plasmid construct was verified by sequencing and analyzed using FinchTV software from Geospiza, Inc. (http://www.geospiza.com/finchtv/). See Supplemental Methods.
Statistical analysis
4–8 animals per strain (3 discs per animal) were used for all quantitative analysis. Quantitative parametric data are presented as mean ± SEM. Data distribution was checked with Shapiro-Wilk normality test, and Mann-Whitney test was used for non-normally distributed data. Differences between two groups were assessed by t-test. Comparisons among more than two groups were done by ANOVA. For histopathological analysis showing percent-degenerated-discs, χ2 test was used. Immunohistochemical analysis in human samples was presented as percentage immunopositivity and thus non-parametric, immunopositivity was compared across grades of degeneration using a Kruskall Wallis and Connover Ingman post hoc test, using Stats Direct. All in vitro experiments were performed in triplicate at minimum, and data are presented as mean ± S.E. Differences between groups were analyzed by the one-way and two-way ANOVA depending on the number of variables with appropriate post-hoc analyses (Sidak’s and Tukey’s multiple comparisons test) using Prism7 (Graphpad Software); p<0.05.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary files) or deposited on the GEO database (GSE130561).
RESULTS
Reduced MCT4 Expression is Associated with Degeneration of Human Discs
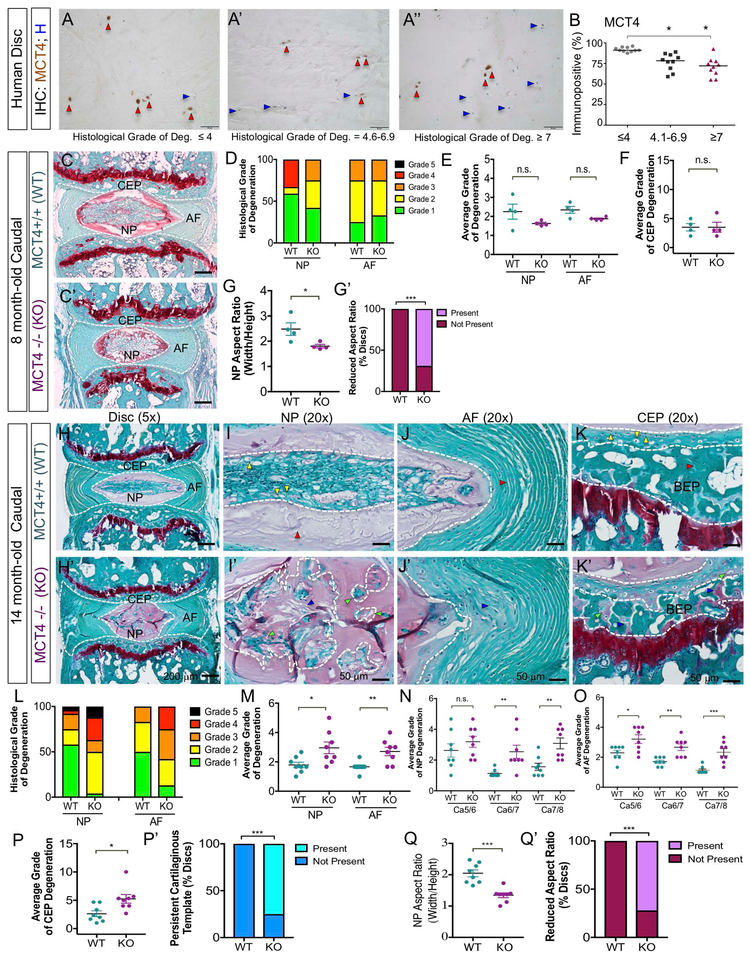
We first assessed whether expression levels of MCT1, MCT4, and CD147, an ancillary protein necessary for MCT trafficking and function, were correlated with human disc degeneration (see tissue sample details, Table S1). The degree of histological degeneration was determined via hematoxylin and eosin staining using previously published criteria (44). The majority of healthy, moderately-degenerate, severely degenerate, and immune cell-infiltrated intervertebral discs expressed SLC16A1 (MCT1), SLC16A3 (MCT4), and BSG (CD147) mRNA (Fig. S1A-C). Although SLC16A3 mRNA levels were consistent between grades of degeneration (Fig. S1B), the percent of cells expressing MCT4 protein significantly decreased in moderately- and severely- degenerate NP tissue (Fig.1A-B). The percent of cells expressing MCT1 and CD147 protein, however, were not affected by the grade of degeneration (Fig. S1D-G). Lack of HRP-positive staining in negative control samples confirms antibody specificities (Fig. S1H-J). These results underscore that loss of MCT4 expression, but not MCT1, correlates with the degenerative process in human discs.
Figure 1: Loss of MCT4 Causes Age-related Intervertebral Disc Degeneration.
A-A") Representative immunohistochemical images showing MCT4-positive cells (red arrows) and MCT4-negative cells (blue arrows) in healthy (A), mid-degenerate (A'), and severe degenerate (A") human samples (Scale bar = 50 μm). B) Percent MCT4 immunopositivity in healthy, moderately-, and severely- degenerate human discs. C, C”) Safranin-O/Fast Green staining of the intervertebral disc in 8 mo WT and global MCT4 KO mice (Scale bar = 200 μm). D) Histological grade of degeneration using modified-Thompson grading scale. E) Average grade of NP and AF degeneration. F) Average grade of CEP degeneration using modified-Boos scale. G, G’) Average aspect ratio of NP compartment and percent of discs with abnormally reduced aspect ratio. (n= 3 discs/ animal; 4 animals/ genotype;). H, H') Safranin-O/Fast Green staining of the disc in 14 mo WT and MCT4 KO mouse disc (Scale bar= 200 μm). (I, I’) High magnification view of NP compartment shows vacuolated cells (yellow arrows) and proteoglycan-rich matrix (red arrows) in WT animals and NP cell fragmentation (green arrows) and fibrotic matrix deposition (blue arrows) in MCT4 KO animals. (J,J’) High magnification view of AF compartment shows organized lamellae (red arrows) in WT animals and disorganized, mucinous lamellae (blue arrows) in MCT4 KO animals. (K, K’) High magnification view of CEP/BEP compartment shows articular chondrocytic cells (yellow arrows) and organized, ossified matrix (red arrows) in WT animals and hypertrophic chondrocytes (green arrows) and cartilaginous templates (blue arrows) in MCT4 KO animals. (Scale bars= 50 μm). L) Histological grade of degeneration using modified-Thompson grading scale. M-O) Average modified-Thompson grade of NP and AF degeneration on average and by disc level. P, P’) Average grade of CEP degeneration using modified-Boos scale and percent of discs showing persistent cartilaginous template in CEP/BEP region. Q, Q') Average aspect ratio of NP compartment and percent of discs with abnormally reduced aspect ratio in the NP compartment. (n=3 discs/ animal; 8 animals/ genotype). Statistical analysis on non-parametric human data was performed by Kruskall Wallis and Connover Ingman post hoc test * p-value ≤ 0.05. Statistical analysis on mouse data: t-test, One-way ANOVA. n.s.= not significant; *, p-value ≤ 0.05; **, p-value ≤ 0.01; ***, p-value ≤ 0.001.
Loss of MCT4 Drives Age-related Intervertebral Disc Degeneration in Mice
In mice, MCT4 is robustly expressed in the NP compartment compared to the AF and CEP, whereas MCT1 expression was pronounced in the CEP compartment (Fig. S2A-B’). Western blot analysis demonstrated the expression of MCT4 in NP tissue (Fig. S2C). To determine the role of MCT4 in maintenance of disc health, we examined the spinal phenotype of MCT4 knock-out (MCT4 KO) mice. MCT4 KO mice were born with normal Mendelian ratios (45) and spines did not show any gross phenotype. This is in contrast to the phenotype of global MCT1 KO mice, which are embryonically lethal (46). We observed that the discs of 8-month old skeletally mature MCT4 KO mice did not exhibit overt histological disc degeneration in NP, AF, or CEP compartments in the caudal or lumbar spine (Fig. 1C, C’; Fig. S3A-B). Quantitative histology using modified-Thompson grading (Table S2) showed that the relative distribution of grades of NP and AF degeneration were similar between WT and MCT4 KO animals (Fig. 1D) (5,36). Likewise, the average grades of NP, AF, and CEP degeneration were comparable between genotypes (Fig. 1E, F). On the other hand, the width/height aspect ratio of the NP compartment, measured and averaged over 10 cross-sectional slices, was significantly decreased in the majority of MCT4 KO discs. While this 2D measurement is prone to artifacts due to section angle and depth, the aspect ratio changes between genotypes were highly reproducible and therefore could provide some insights into volumetric changes in the NP tissue compartment. This was evidenced by a smaller and rounder NP compartment observed in histological analysis (Fig. 1G, G’).
To delineate if the effects of MCT4 loss on disc phenotype were more pronounced with age, we analyzed the spines of 14-month-old MCT4 KO mice. At this age, all disc tissue compartments in KO mice were degenerated in the caudal but not lumbar spine (Fig. 1H, H’; Fig. S3C-D). Due to differences in biomechanical environment between lumbar and caudal discs in mice, we often find that caudal discs are more susceptible to genetic and biological instigators of disc degeneration (5,9). The NP cells in WT animals were organized in a horizontal cell band bounded by organized Safranin-O stained matrix (Fig. 1I). On the other hand, NP cells in KO mice were organized in a honeycomb pattern separated with interterritorial matrix (Fig. 1I’). The AF compartment in KO animals lost the typical lamellar organization seen in WT animals (Fig. 1J, J’). The CEP cells in KO mice displayed hypertrophic morphology and Safranin-O positive cartilaginous remnants persisted in the bony endplate (BEP) (Fig. 1K, K’).
Modified-Thompson grading of histological sections showed that grade distributions for caudal NP and AF compartments were different between WT and KO animals (Fig. 1L). Importantly, the average Thompson grades of NP and AF degeneration were significantly higher in KO mice (Fig. 1M). Likewise, the level-by-level average grades for NP and AF were also different between the genotypes, indicating increased incidence of degeneration (Fig. 1N, O). Similarly, average grades of EP degeneration were higher in MCT4 KO mice, implying degeneration of both CEP and BEP (Fig. 1P). Quantification of incidence of cartilaginous remnants in the BEP showed that u75% KO discs presented this feature (Fig. 1P’). Moreover, the NP aspect ratio showed further reduction compared to 8-month old animals, and u70% of MCT4 KO discs were affected (Fig. 1Q, Q’).
Loss of MCT4 Compromises Vertebral Bone Quality
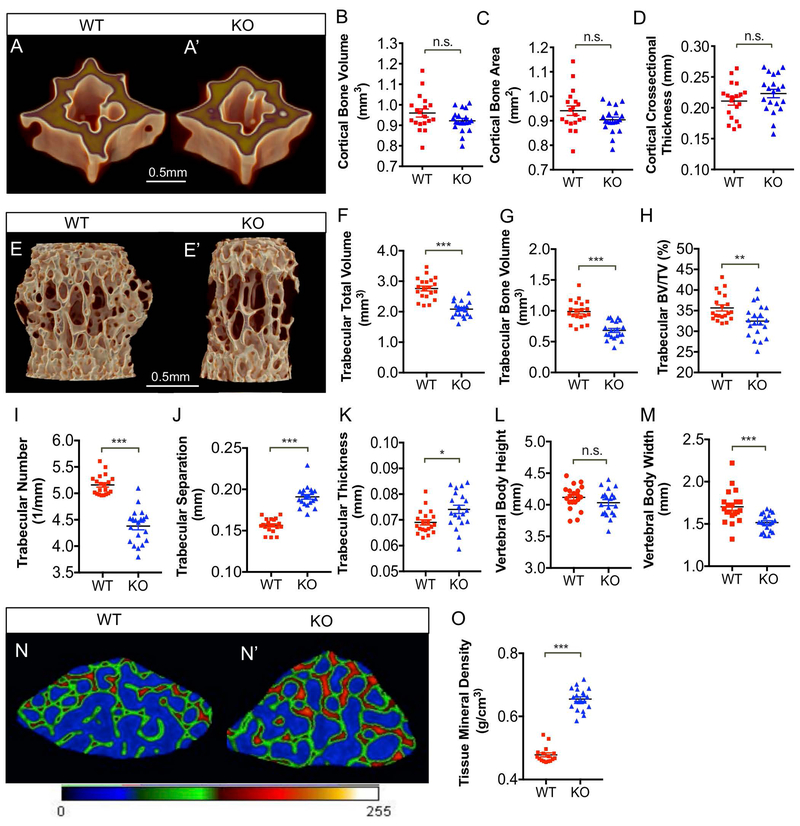
Recent studies have elucidated the role of glucose metabolism in maintaining bone homeostasis. It has been shown that glucose metabolism is required for osteoblast and osteoclast differentiation, as well as for cell-type specific processes including matrix deposition and resorption (47). In order to assess the impact of MCT4 deletion on vertebral bone health, we compared the cortical and trabecular bone morphology in the caudal and lumbar vertebrae from 14-month-old WT and MCT4 KO mice using micro-computed tomography (μCT). Reconstructed images from MCT4 KO mice showed no change in cortical bone volume, bone area, or cross-sectional thickness (Fig. 2A-D). On the other hand, the MCT4 KO mice clearly evidenced impaired trabecular bone quality (Fig. 3E, E’). The trabecular bone had reduced total volume, bone volume, and bone volume fraction in KO mice (Fig. 2F-G). Additionally, the trabeculae in MCT4 KO vertebrae decreased in number, however trabecular separation and thickness increased (Fig. 2I-K). Overall, KO mice had similar vertebral body height as WT animals, however vertebral body width was significantly reduced, likely contributing to the decreased trabecular bone volume (Fig. 2L, M). Based on the results, we postulate that the increased tissue mineral density in KO mice was compensatory for the alterations in trabecular number and separation (Fig. 2N-O). Taken together, μCT analysis showed that MCT4 KO vertebrae had inferior trabecular bone quality due to their reduced size and trabecular number, increased trabecular separation, and increased mineral density.
Figure 2: Loss of MCT4 Compromises Vertebral Bone Quality.
A, A') μCT reconstruction of cortical bone from vertebral bodies of 14 mo WT and MCT4 KO mice (Scale bar = 0.5 mm). B-D) Analysis of cortical bone showing no significant difference in cortical bone volume (B), cortical bone area (C), or cortical cross-sectional thickness (D). E, E') μCT reconstruction of trabecular bone from vertebral bodies of 14 mo WT and MCT4 KO mice (Scale bar = 0.5 mm). F-H) Analysis of trabecular bone volume showing significant differences in trabecular total volume (F), trabecular bone volume (G), and trabecular bone volume fraction (H). I-K) Analysis of trabecular bone morphology showing trabecular spacing (I), trabecular number (J), and trabeculae thickness (K) in WT and MCT4 KO mice. L, M) Average vertebral body height and width in WT and MCT4 KO animals. N, N’) Representative cross-section of vertebral body trabecular bone showing increased tissue mineral density (red colored bone vs. green colored bone) in WT and MCT4 KO mice. O) Quantification of trabecular tissue mineral density using ImageJ. (n= 4 vertebrae/aminal; 5 animals/ genotype). Statistical Analysis: t-test. n.s.= not significant; *, p-value ≤ 0.05; **, p-value ≤ 0.01; ***, p-value ≤ 0.001.
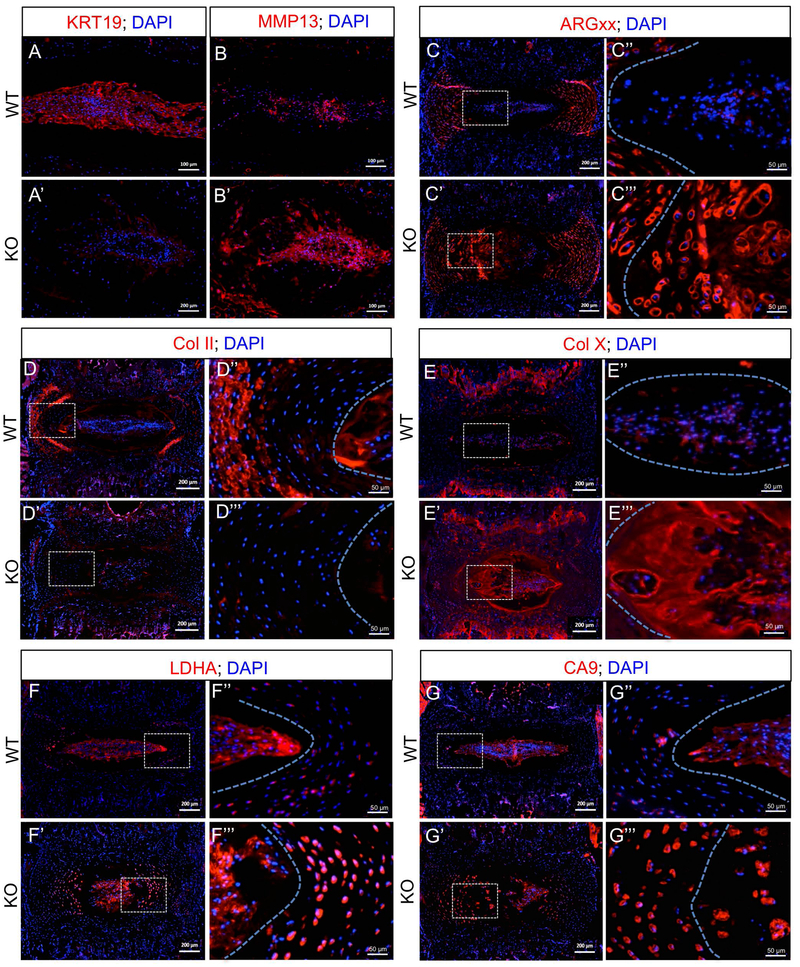
Figure 3: MCT4 Expression is Important for Maintenance of NP Cell Phenotype and Extracellular Matrix Composition.
Representative immunostaining showing expression of important proteins in the intervertebral disc in 14 mo WT and MCT4 KO mice. A, A’) NP phenotypic marker- KRT19 (Scale bar= 100 μm). B, B’) Matrix catabolic enzyme- MMP13 (Scale bar= 100 μm). C-E"') Extracellular matrix molecules: C-C’’’) ARGxx, (D-D’’’) Collagen II, and (E-E’’’) Collagen X imaged at 10X and 20X (Left scale bars= 200 μm; Right scale bars= 50 μm). F-G’’’) pH regulatory enzymes: F-F"') LDHA and (G-G’’’) CA9 imaged at 10X and 20X (Left scale bars= 200 μm; Right scale bars= 50 μm). Dotted lines were drawn to demarcate different tissue compartments within the disc. All staining was performed using at least 3 animals/genotype corresponding to Grade 2.
MCT4 Expression is Important for Maintenance of NP Cell Phenotype and Extracellular Matrix Composition
One of the hallmarks of disc degeneration is the altered phenotype of NP cells from notochordal to chondrocyte-like cells. These alterations coincide with decreased expression of NP phenotypic makers along with changes in extracellular matrix composition (5,48-50). MCT4 KO mice had decreased expression of NP phenotypic marker, KRT19, while GLUT1 and CA3 expression levels were unchanged (Fig 3A, A’; Fig. S4A-B’). The expression of extracellular matrix catabolic enzyme, MMP13, a classic marker of chondrocyte hypertrophy and disc degeneration, was increased in the NP compartment of KO mice (Fig. 3B, B’). We also analyzed expression of major disc extracellular matrix molecules and products of their catabolism. Although WT and KO animals had similar levels of expression of aggrecan and chondroitin sulfate (Fig. S4C, D’), there was a prominent increase in cleaved aggrecan neoepitope, ARGxx, in the NP and AF compartments of KO animals, suggesting elevated aggrecanase activity (Fig. 3C-C’’’). While collagen I expression was similar in the AF compartment, collagen II expression decreased in NP and AF compartments of KO mice (Fig. 3D-D’’’; Fig. S4E, E’). Collagen X, which shows some expression in the aging disc, showed a dramatic increase in the NP of MCT4 KO mice (Fig. 3E- E’’’). These results suggest that NP cells in MCT4 KO mice displayed a phenotype resembling hypertrophic chondrocytes.
Since NP cells under acidic conditions up-regulate expression of MMPs and decrease proteoglycan production, we assessed the levels pH regulatory enzymes in the KO mice (27). While the NP cells in MCT4 KO mice maintained high expression of LDHA and CA9, the inner AF cells showed elevated levels of both proteins (Fig. 3F-G’’’). Of note, inner AF cells have lower baseline expression of LDHA and CA9 than NP cells and may be more susceptible to fluctuations in intracellular pH. This is attributed to the fact that many pH regulatory enzymes, including LDHA and CA9, are transcriptional targets of HIF-1 - a master regulator of NP cell viability (17,51). CA9, in particular, regulates intracellular pH through a bicarbonate recycling mechanism that generates an abundance of H+ ions in the pericellular matrix through the process of CO2 hydration. It is possible that increased extracellular acidity, resulting from elevated CA9 expression, may contribute to the matrix catabolism observed in MCT4 KO mice. Lastly, the loss of MCT4 expression was not compensated by an increase in MCT1 expression in the disc (Fig. S4F-F’’’). Collectively, our results imply that loss of H+/lactate transport function in MCT4 KO mice results in significant alterations in NP phenotype and extracellular matrix composition of the disc.
Loss of MCT4 Leads to Diminished Transcriptional Program in vivo
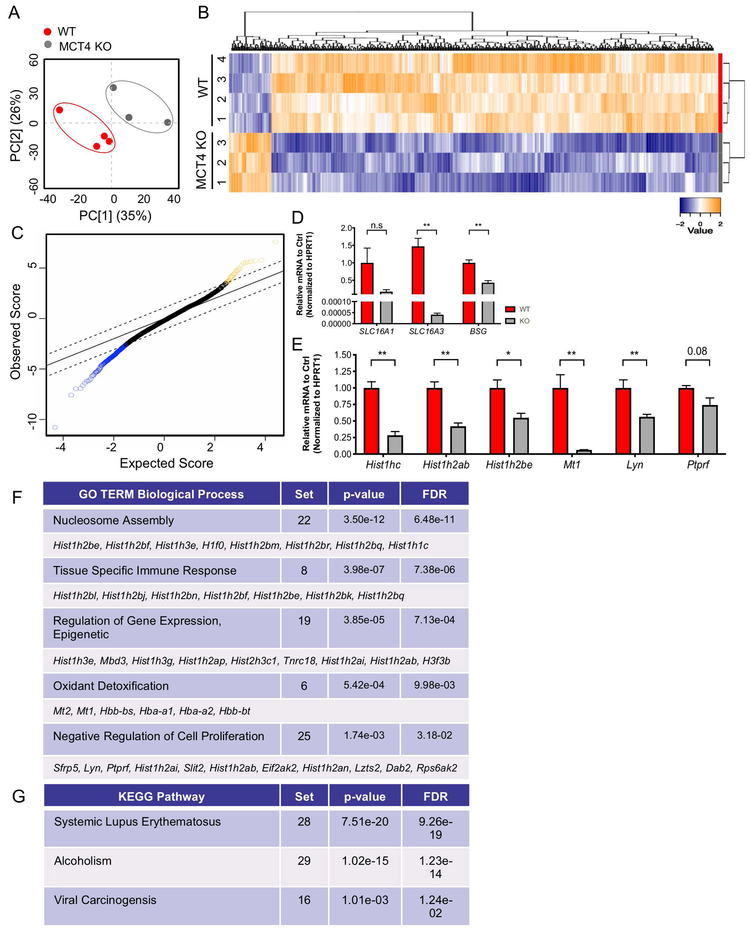
To get broader insights into the mechanism of disc degeneration instigated by loss of MCT4, we performed microarray analysis on RNA isolated from NP tissue from 14-month-old WT and MCT4 KO mice (GSE130561). The transcriptomic profiles of NP tissue from WT and MCT4 KO mice clustered distinctly along principal components (Fig. 4A). Analysis of differentially expressed genes (DEG) uncovered 337 down-regulated and 30 up-regulated genes with FDR ≤ 5%. These DEGs are represented in a heatmap with hierarchical clustering and significance analysis of microarrays (SAM) plot to demonstrate relative expression levels and significance scores (Fig. 4B, C). To assess levels of MCT and CD147 mRNA expression and to confirm the accuracy of the DEG list, we performed qRT-PCR analysis (Fig. 4D, E). As expected, levels of SLC16A3 and BSG were significantly lower in the MCT4 KO animals, however SLC16A1 levels were unchanged (Fig. 4D). We then confirmed the down-regulation of a set of genes that were identified from the SAM plot using qRT-PCR methods, including histone genes Hist1hc, Hist1h2ab, Hist1h2be as well as Mt1, Lyn, and Ptprf (Fig. 4E).
Figure 4: Loss of MCT4 Leads to Diminished Transcriptional Program.
A) Principal component analysis (PCA) of transcriptomic profiles from the NP compartment of 14 mo WT and MCT4 KO animals used in Clariom S Mouse Microarray. B). Heatmap and hierarchical clustering of SST-RMA normalized values of u350 differentially expressed protein-coding genes (DEGs) in WT and MCT4 KO samples with FDR ≤ 5%. C) The top 50% most expressed genes were used for Significance Analysis of Microarrays (SAM). SAM was run at 5,000 permutations and the False Discovery Rate was set at 5%. D) qRT-PCR analysis of SLC16A1, SLC16A3, and BSG mRNA expression in MCT4 KO mice. E) qRT-PCR analysis confirming decreased expression of DEGs identified from SAM plot. F, G) DAVID was used to compute enriched pathways in the genes that are upregulated. Gene ontology (GO) terms for biological process (BP) and KEGG pathways were examined. Top u10 DEGs involved in each process are shown. The number of DEGs from the multivariate analysis involved in each biological process shown as “Set”.
To identify enriched molecular pathways among the DEGs, we used DAVID Bioinformatics Resource (Fig. 4F, G) (Table S3). The enriched biological processes from the down-regulated DEG list were mainly involved in transcriptional reprogramming: i.e. nucleosome assembly, regulation of epigenetic gene expression, and negative regulation of cell proliferation (Fig. 4F). Additional enriched processes included tissue specific immune response and oxidant detoxification (Fig. 4F). The enriched KEGG pathways, on the other hand, included Systemic Lupus Erythematosus and Viral Carcinogenesis, disorders of the immune system, and alcoholism, a metabolic disorder (Fig. 4G); their enrichment within the DEG set was driven by the histone genes (Table S3). There were no enriched biological processes from the up-regulated DEG list, possibly because of the small number of genes. Overall, these microarray data underscored that long-term dysregulation of H+/lactate transport can have effects on epigenetic processes beyond those commonly associated with pH.
It is unclear if decreased transcriptional programming is directly or indirectly linked to lactate transport. In order to gain perspective on this question, we analyzed previously deposited RNAseq data from human pancreatic cell lines treated with MCT4 specific RNAi (52) (GSE63231). The down-regulated DEGs with a fold-change ≥ 2.5 showed very similar enriched biological processes as our dataset. The second and fifth highest enriched biological process gene ontology terms from the dataset deposited by Baek et al. were negative regulation of epigenetic gene expression (P-value = 3.3E-4) and nucleosome assembly (P-value = 8.8E-4), respectively. While not entirely conclusive, this does support the notion that MCT4 silencing may directly affect nuclear transcriptional processes.
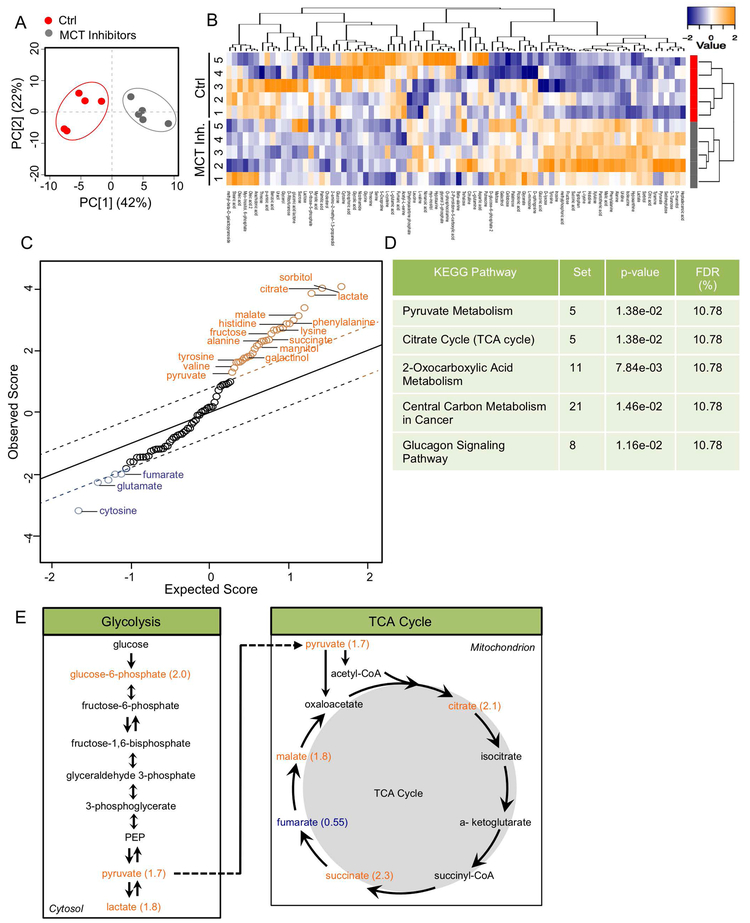
Inhibition of H+/lactate Efflux Causes an Increase in Pyruvate Metabolism and TCA cycle Metabolites
Loss of MCT4 caused major pathological changes in the intervertebral disc and vertebral bone in mice. While microarray analysis revealed that MCT4 deletion reduced transcriptional programming in aged mice, the initial drivers of disc degeneration caused by blocking H+/lactate efflux remained unclear. To investigate the acute metabolic changes following MCT4 inhibition, we performed metabolomic analysis on NP cells treated with an MCT inhibitors for 2 hours. For the following experiments, we inhibited MCT4 with a highly-specific pharmacological inhibitor, supplemented with a specific MCT1 inhibitor, ARC155858, to account for any lactate/H+ transport in the in vitro culture system (MCT Inh.). This experimental set-up allows us to delineate immediate metabolic changes that occur due to lactate accumulation before cells undergo adaptive changes. Control and MCT-inhibited cells clustered distinctly along principal components (Fig. 5A), a distinction that was further confirmed with hierarchical clustering (Fig. 5B). Interestingly, hierarchical clustering of all measured metabolites identified several groups with either increased or decreased concentration after MCT inhibition (Fig 5B). A SAM plot was used to identify and represent metabolites with significantly different concentrations between the two groups (FDR ≤ 5%). 15 metabolites were found to have increased concentration while 3 metabolites decreased in concentration after blocking H+/lactate efflux in hypoxic NP cells (Fig. 5C). Noteworthy, KEGG pathway enrichment analysis showed that pyruvate metabolism and citrate cycle (TCA cycle) pathways were over-represented in the panel of metabolites with significantly different concentrations (Fig. 5D). Accordingly, we hypothesize that MCT inhibition led to reprogramming of glucose metabolism in response to intracellular lactate accumulation (Fig. 5E). This reprogramming is evidenced by a u2-fold increase in pyruvate and TCA cycle intermediates citrate, succinate, and malate (Fig. 5E).
Figure 5: Inhibition of H+/lactate efflux causes an increase in pyruvate metabolism and TCA cycle metabolites.
A) PCA of metabolomic profiles from Ctrl and MCT-inhibited NP cells used for GC-MS analysis. B). Heatmap and hierarchical clustering of z-score normalized concentrations of u90 metabolites. C) Metabolites with significant concentration differences were identified using SAM with 5,000 permutations and 5% FDR threshold. D) Metaboanalyst Functional Annotation analysis of metabolites using KEGG pathway terms; p-value of each represented mouse metabolic pathway was calculated using hypergeometric test. E) Schematic of pyruvate metabolism via TCA cycle denoting fold-change of affected metabolites. (n= 5 independent experiments, 5 samples/condition).
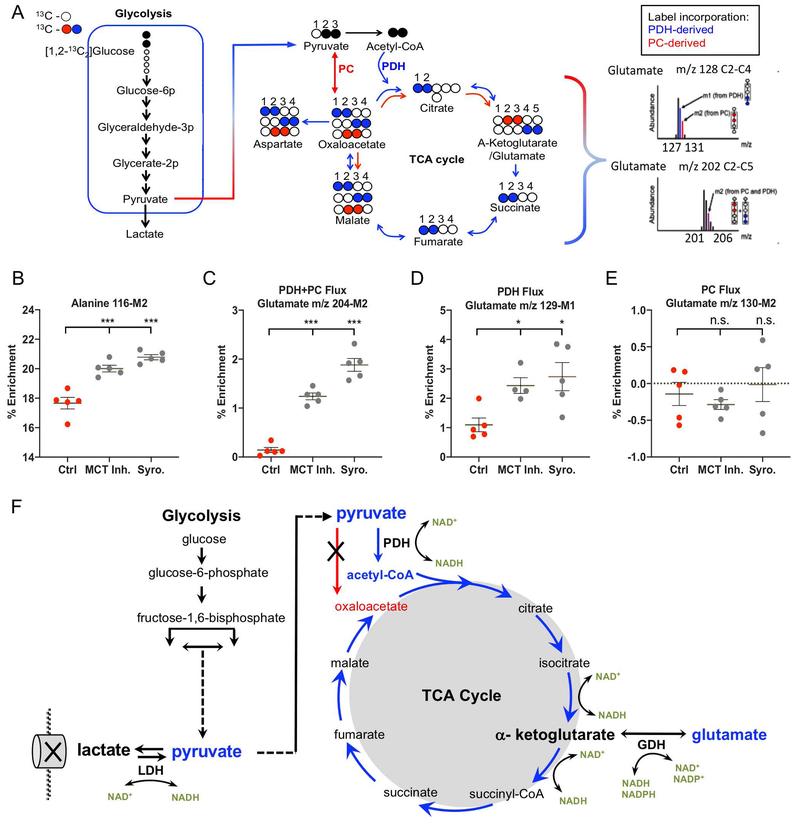
Inhibition of H+/lactate Efflux Causes an Increase in PDH Flux into the TCA Cycle
To gain further insights into this metabolic reprogramming, NP cells were cultured in media containing 5mM glucose enriched with 50% [1,2-13C]-glucose. After 24 hours of hypoxic preconditioning with 13C glucose, the cells were treated with MCT Inh. and a highly specific MCT1+4 dual inhibitor, Syrosingopine (Syro) for 2 hours. Figure 6A shows the framework for the tracer analysis using [1,2-13C]-glucose and details how TCA cycle flux was monitored with glutamate enrichments. First, we detected a significant increase in Alanine 116-M2 after MCT inhibition, reflecting elevated levels of intracellular M2 pyruvate, in agreement with metabolite analysis from Figure 5 (Fig. 6B). We then assessed the balance between pyruvate flux into the TCA cycle via pyruvate dehydrogenase (PDH) or pyruvate carboxylase (PC) using glutamate 13C enrichments. We first noted that MCT inhibition lead to a significant increase in glutamate 204-M2 enrichment (Fig. 6C). Glutamate m/z 204-M2 signifies the presence of tracer in the entire carbon backbone of glutamate C2-C5 and may reflect 13C incorporation at positions C3 and C4, denoting flux through PC, and/or 13C incorporation at positions C4 and C5, denoting flux through PDH. To parse out the relative contributions of PC and PDH flux into TCA cycle, we monitored the 3-carbon glutamate fragment (C2-C4) via glutamate m/z 128 (m/z 128-M0, 129-M1, 130-M2). Glutamate m/z 128 will have one 13C carbon if flux entered the TCA cycle via PDH (Glutamate m/z 129-M1), and two 13C carbons (at C3 and C4) if pyruvate flux entered the TCA via PC (Glutamate m/z 130-M2). Accordingly, MCT inhibition caused a significant increase in glutamate 129-M1 enrichment, implying an increase in PDH flux (Fig. 6D). On the other hand, there was no detectable glutamate 130-M2, suggesting no PC flux with or without MCT inhibition (Fig. 6E). These findings show that MCT inhibition in NP cells resulted in increased TCA cycle flux through PDH. These results are in agreement with the metabolic profile of MCT inhibited cells (Fig. 5), and suggest that TCA flux was directed from α-ketoglutarate towards glutamate production, in order to balance the redox ratio necessary for maintaining glycolytic and TCA cycle reactions (Fig. 6F).
Figure 6: Inhibition of H+/lactate efflux causes an increase in PDH flux into the TCA cycle resulting in glutamate enrichment.
A) Framework for metabolic flux analysis in NP cells cultured with [[1,2-13C]-glucose for 24 hours in Hx before a 2 hour treatment with MCT Inh and Syro. TCA cycle flux was monitored via glutamate isotopomers. [2,3-13C]-pyruvate can enter the TCA cycle through pyruvate dehydrogenase (PDH) or pyruvate carboxylase (PC). Glutamate 204-M2 reflects the presence of 13C at either the 3rd and 4th carbon position, or the 4th carbon and 5th carbons, and reflects the overall TCA cycle flux from either pyruvate entering from PC or PDH. Glutamate m/z 129-M1 reflects the presence of 13C at the 4th carbon position via PDH flux. Glutamate m/z 130-M2 reflects the presence of 13C at the 3rd and 4th carbon position, indicating pyruvate entering the oxaloacetate pool through PC. B) % Enrichment of alanine m/z 116-M2, indicating intracellular M2 pyruvate. C) PDH + PC flux measured by Glutamate m/z 204-M2. D) PDH flux measure by Glutamate m/z 129-M1. E) PC flux measure by Glutamate m/z 130-M2. (E) Schematic of the metabolic profile with tracer data showing pyruvate flux through PDH and increased flux through GDH to maintain NAD+ regeneration. n= 5 metabolic samples/condition. Statistical analysis: One-way ANOVA. n.s.= not significant; *, p-value ≤ 0.05; **, p-value ≤ 0.01; ***, p-value ≤ 0.001.
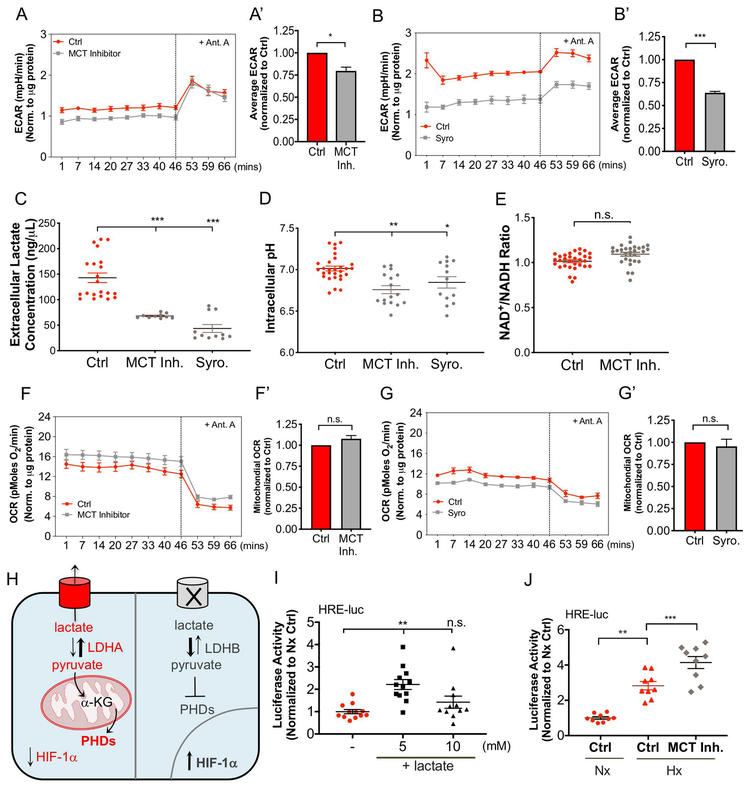
Inhibition of H+/lactate Efflux Decreases Extracellular Acidification Rate without Increasing Mitochondrial Oxygen Consumption Rate
To discern if acute inhibition of H+/lactate efflux is linked to a concomitant change in bioenergetics, we measured metrics of NP cell metabolism using a Seahorse Flux Analyzer. After short-term MCT4 inhibition with both the MCT Inh. cocktail (Fig. 7A, A’) and Syrosingopine (Syro) (Fig. 7B, B’), NP cells modestly decreased extracellular acidification rate (ECAR) over-time (Fig. 7A, B) and on average (Fig. 7A’, B’). While ECAR is generally considered a surrogate measure of glycolytic flux, this decrease is likely a reflection of H+/lactate transport inhibition. Accordingly, a considerable 50–75% decrease in extracellular lactate concentration was observed with both inhibitors, as expected (Fig. 7C). Of note, a substantial reduction in extracellular lactate/H+ efflux only correlated with a 25–40% decrease in ECAR, suggesting that pericellular CO2 hydration by CA9/12 contributes to the majority of extracellular proton production in NP cells (17). Given that CA9/12 is an intracellular pH regulator, it is not surprising that blocking H+/lactate efflux only caused a minor decrease in intracellular pH (Fig. 7D). To further elucidate the changes in NP cell bioenergetics, we measured the NAD+/NADH ratios after blocking H+/lactate export in NP cells. This ratio is primarily maintained during glycolysis by LDH activity, via the reduction of pyruvate to lactate, as well as in the TCA cycle by GDH activity, via the reduction of α-ketoglutarate to glutamate. Interestingly, MCT inhibition did not significantly alter the NAD+/NADH ratio, likely due to steady TCA cycle flux and glutamate production (Fig. 7E). Intriguingly, MCT inhibition did not cause a significant increase in mitochondrial oxygen consumption rate (OCR) with either mechanism of inhibition (Fig. 7F- G’), despite our results suggesting an increase in TCA cycle flux. This suggests that TCA cycle and electron transport chain may be uniquely uncoupled in NP cells.
Figure 7: Inhibition of H+/lactate Efflux Decreases ECAR without Increasing Mitochondrial OCR; Resulting intracellular lactate accumulation increases HIF-1α activity.
A- B’) Seahorse Flux analysis of extracellular acidification rate (ECAR) over time (A, B) and on average (A’, B’) after MCT inhibition with an MCT inhibitor cocktail (MCT Inh) (A, A’) and Syrosingopine (Syro) (B, B’). C) MCT inhibition with MCT Inh. and Syro decreases extracellular lactate concentrations. D) Intracellular pH measurement after MCT inhibition with MCT Inh. and Syro. E) NAD+/NADH ratio after MCT inhibition. F-G’) Seahorse Flux analysis of oxygen consumption rate (OCR) over time (F, G) and on average (F’, G’) after MCT inhibition with MCT Inh (F, F’) and Syro (G, G’). H) Schematic of HIF-1α degradation vs. stabilization depending on status of PHD function. I, J) 5 mM exogenous lactate (I) and MCT inhibition (J) increase HIF activity as measured by dual luciferase assay. n= 4 independent experiments; 4-6 replicates/experiment. Statistical analysis: t-test, One-way ANOVA. n.s.= not significant; *, p-value ≤ 0.05; **, p-value ≤ 0.01; ***, p-value ≤ 0.001.
A Regulatory Feedback Loop Between HIF-1α and MCT4 is Mediated by Intracellular Lactate Levels
Intracellular lactate can act as a hypoxia mimetic factor by competitively inhibiting prolyl hydroxylase (PHD) function (53-55). This mechanism involves the conversion of lactate to pyruvate, which is routed into the TCA cycle, generating metabolic intermediates, namely succinate, fumarate, and oxaloacetate, which are structurally but not functionally similar to α-ketoglutarate; a substrate required for PHD-mediated degradation of HIF-1α (Fig. 7H). Accordingly, addition of exogenous lactate under normoxia increased HIF-1α activity in NP cells (Fig. 7I). This response was maximal at a 5mM dose and showed only a trend of increased activity at 10mM. Further support of this hypothesis was the observation that MCT inhibition increased HIF-1α activity in NP cells using an HRE-luciferase reporter (Fig. 7J).
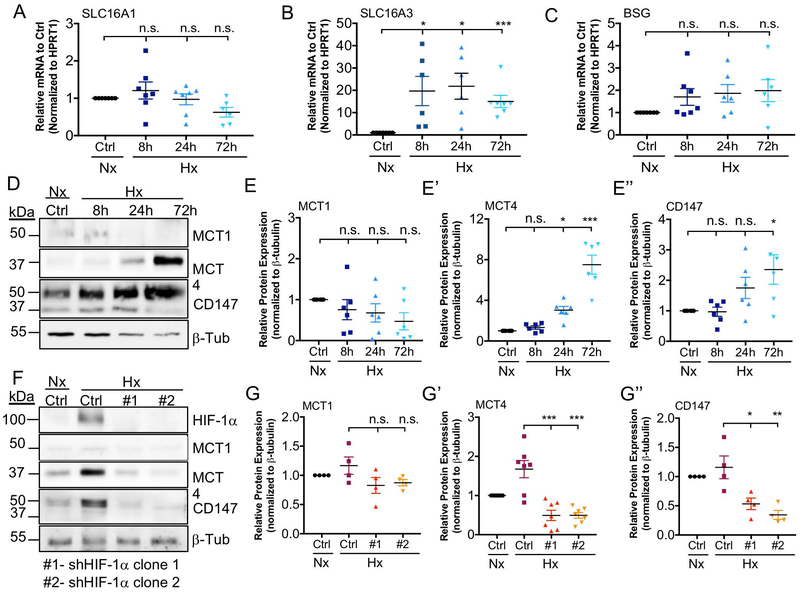
Since previous reports indicate that SLC16A3 (MCT4) is a hypoxia and HIF-1 responsive gene, we first investigated the effect of oxygen tension on SLC16A1 (MCT1), SLC16A3 (MCT4), and BSG (CD147) mRNA and protein levels in NP cells. SLC16A3, but not SLC16A1 or BSG showed significant hypoxic induction as early as 8 hours and persisted until 72 hours (Fig. 8A-C). MCT4 (Fig. 8D, E’) showed hypoxic-induction by 24 hours; interestingly, CD147 (Fig. 8D, E’’) followed a similar trend of increased levels which reached significance by 72 hours. Overall, it was evident that MCT4 and CD147 levels were sensitive to changes in oxygen tension and their expression was up-regulated under hypoxic conditions. To delineate the relationship between HIF-1α and MCT4, we transduced rat NP cells with lentiviruses expressing two distinct HIF-1α shRNA sequences. This silencing approach resulted in a robust knock-down of HIF-1α (Fig. 8F). MCT4 and CD147, but not MCT1, protein levels were significantly reduced by HIF-1α silencing under hypoxic conditions (Fig. 8F-G’’). Taken together, these results demonstrate a positive regulatory loop between HIF-1α and MCT4. Specifically, HIF-1α maintains MCT4 levels in hypoxia, and intracellular lactate may in turn modulate HIF-1α activity in NP cells.
Figure 8: MCT4 Expression is Hypoxia-inducible and HIF-dependent.
A-C) qRT-PCR analysis of SLC16A1 (A), SLC16A3 (B), and BSG (C) mRNA expression in rat NP cells cultured under hypoxia (1% O2) for up to 72 h. D) Representative Western blot analysis of MCT1, MCT4, and CD147 protein expression in rat NP cells cultured under hypoxia for up to 72 h. E-E’’) Densitometric analysis of MCT1 (E), MCT4 (E’), and CD147 (E’’) from Western blot experiment shown in (D). F) Representative Western blot analysis of HIF-1α, MCT1, MCT4, and CD147 protein expression in rat NP cells after silencing HIF-1α with two independent shRNAs. G-G’’) Densitometric analysis of Western blot showed in (F) above. (n=4 independent experiments). Statistical analysis: One-way ANOVA. n.s.= not significant; *, p-value ≤ 0.05; **, p-value ≤ 0.01; ***, p-value ≤ 0.001.
Transcriptional Activation of SLC16A3 (MCT4) in Hypoxia is Controlled by a HIF-1-dependent Intronic Enhancer
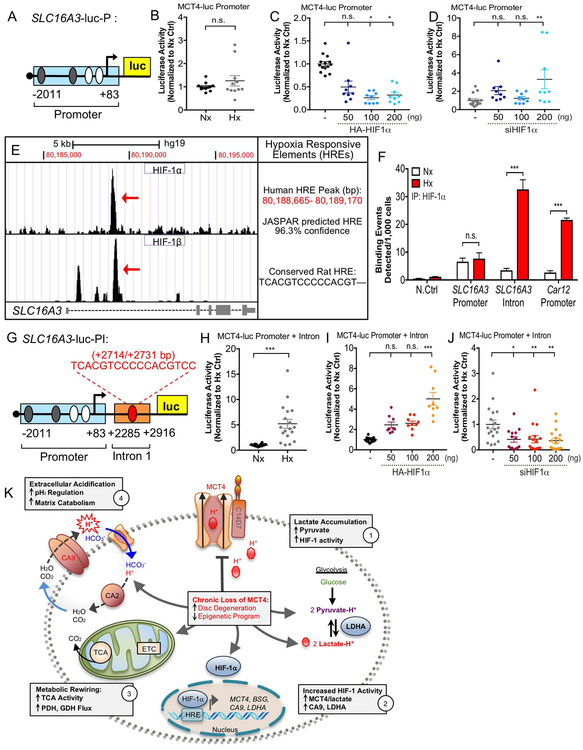
To gain mechanistic insights into HIF-1α regulation of SLC16A3 gene expression, we investigated the HIF-1α binding to hypoxia responsive elements (HREs) in the SLC16A3 promoter. Surprisingly, a luciferase reporter plasmid containing the 2kb proximal SLC16A3 promoter (SLC16A3-luc-P), with known HIF-1α binding sites (29), was insensitive to oxygen tension and did not show an anticipated response to HIF-1 modulation in NP cells, suggesting these sites were not functionally relevant in the disc (Fig. 9A-D). To discern additional putative binding sites, we examined HIF-1 binding enrichment along SLC16A3 from two deposited ChIP-sequencing datasets (41,56,57) (GSE101063, GSE120885). A shared 505bp peak of HIF-1α/β binding enrichment was found in the first intron of SLC16A3 between Chr17: 80,188,665–80,189,170 on Hg19 (Fig. 9E). Transcription factor binding analysis using JASPAR database identified a putative HRE site in this region with 96.3% confidence (43) (Fig. 9E). Importantly, the putative binding site in the human SLC16A3 intron was conserved in the rat SLC16A3 gene, as confirmed by Multiz alignment (Fig. 9E). Genomic ChIP showed that, in rat NP cells, HIF-1α binds to the intronic HRE site in hypoxia, and minimally binds the promoter HRE site in normoxia or hypoxia (Fig. 9F) (Table S4). HIF-1α binding enrichment along the SLC16A3 intronic site was similar to Car12, a known HIF-1 target gene in NP cells (Fig. 9F) (58). In order to validate the functionality of the intronic HRE for SLC16A3 transcription, we cloned the 505bp intronic binding region, containing the intronic HRE, downstream of the 2kb promoter (SLC16A3-luc-PI) (Fig. 9G). This reporter/SLC16A3-luc-PI was hypoxia-inducible in NP cells (Fig. 9H). Importantly, HIF-1α overexpression and silencing in NP cells resulted in increased and decreased activity of SLC16A3-luc-PI, respectively (Fig. 9I, J). Together, these data confirm that hypoxic induction of SLC16A3 transcription is mediated by HIF-1 binding to an intronic enhancer in NP cells. Given the enrichment of HIF-1 binding at this site in RCC cells, we postulate that it may be important for transcriptional activation of SLC16A3 in hypoxic tumors as in NP cells.
Figure 9: Transcriptional Activation of SLC16A3 in Hypoxia is Controlled by a HIF-1-dependent Intronic Enhancer.
A) Schematic of luciferase reporter plasmid (SLC16A3-luc-P) containing 2kb proximal promoter of SLC16a3 with predicted HREs. HREs in white are previously shown in literature. B-D). SLC16A3-luc-P is not hypoxia-inducible (B) or appropriately HIF-dependent (C, D) in NP cells as determined by dual luciferase assay. E) Analysis of ChIP-sequencing datasets identified novel 500 bp HIF-1 binding region in SLC16a3 intron. JASPAR analysis identified putative HRE site within this region with 96.3% confidence score. Multiz alignment confirmed HRE is conserved in the rat gene. F) Genomic ChIP assay showing HIF-1α binding enrichment along a negative control site, SLC16A3 promoter HRE, SLC16A3 intronic HRE, and positive control HRE on Car12. G) Schematic of SLC16A3-luc-PI plasmid construct with the putative enhancer cloned downstream of the 2 kb proximal promoter. H-J). SLC16A3-luc-PI is hypoxia-inducible (H) and HIF-dependent (I, J) in NP cells. (n=3 independent experiments; 3 technical replicates/experiment). Statistical analysis: t-test, One-way ANOVA. n.s.= not significant; *, p-value ≤ 0.05; **, p-value ≤ 0.01; ***, p-value ≤ 0.001. K) Schematic showing interplay between HIF-1, MCT4 and CA for pH regulation in NP cells. Metabolic consequences of decreased or loss of MCT4 function on NP cell health are depicted.
DISCUSSION
For the past two decades, several groups have attempted to understand the metabolic complexity and pH adaptations of disc cells in their physiologically hypoxic niche (59). It has been proposed that disturbances in the niche conditions and cellular metabolism underscore the molecular basis of age-dependent disc degeneration (59,60). In this study, we showed that MCT4 levels were decreased in degenerated human discs and that loss of MCT4 function contributes to disc degeneration in a mouse model. Mechanistically, blocking H+/lactate efflux may lead to disc degeneration through the dysregulation of multiple pathways. On one hand, loss of MCT4 correlated with decreased nucleosome assembly and epigenetic programming in degenerated NP tissue, whereas inhibition of MCT4 function instigated a metabolic rewiring of pyruvate through TCA cycle flux. This was particularly striking considering that NP cells are characterized by stabilized expression of HIF-1α, a master regulator of glycolytic metabolism. We also established a functional loop between HIF-1 and MCT4, whereby MCT4 expression is regulated by a HIF-1α-dependent intronic enhancer and HIF-1α activity is modulated by lactate. Our results show, for the first time, the critical importance of maintaining lactate transport function for the health and integrity of the intervertebral disc.
Previous studies have reported that age-related degenerative changes in the mouse disc are pronounced between 18–24 months (50,61). Here, we observed that loss of MCT4 increased degeneration of NP, AF, and CEP compartments in mice, characterized by gross histological alterations in cell phenotype and extracellular matrix integrity by 14 months of age. Whereas, at 8 months, decreased width-height aspect ratio in absence of overt morphological changes suggested an initial phase of degenerative processes. Such changes in disc volume and shape have been previously correlated with disc degeneration (62). In addition to alterations in the NP and AF compartments, a persistent cartilaginous template was also noted in the CEP/BEP. It is not unreasonable to think that these particular pathoanatomical changes are associated with chronic, long term metabolic stress. A smaller NP compartment can be due to decreased glucose utilization and subsequent production of anabolic intermediates. Meanwhile, CEP degeneration instigates nutrient deprivation and lactate accumulation, as it is the compartment through which metabolites and waste products diffuse (63,64). Collectively, these results imply that the phenotype in MCT4 KO mice resulted from cumulative postnatal degenerative processes, driven by metabolic constraints, rather than altered developmental effects. It is important to note that since the healthy intervertebral disc is an isolated, avascular, and immune privileged compartment, and circulating systemic factors have little effect on the compartment, the disc phenotype observed in these mice can be considered an intrinsic tissue response (9,65).
In MCT4 KO mice there was an increase in degradation of the NP extracellular matrix- a known NP cellular response to acidity (6,27). Reduced H+/lactate transport resulted in increased levels of MMP13, as well as fragmented aggrecan neoepitope, ARGxx, implying enhanced proteolytic cleavage of matrix in KO mice. Additionally, decreased expression of collagen II and a concomitant increase in collagen X in conjunction with MMP13, implied a transition of notochordal NP towards a hypertrophic chondrocytic phenotype. This matrix transition has been recently noted in a mouse model of spontaneous age-related disc degeneration as well as during human disc degeneration (5,66,67). Furthermore, NP cells in MCT4 KO animals maintained expression of HIF-1 targets LDHA and CA9, both of which are integral components of NP cells glycolytic and pH regulatory systems. In addition to NP cells, AF cells displayed extracellular matrix alterations and prominent changes in pH regulatory proteins in response to MCT4 deletion. This was evident from decreased collagen II expression and robust induction in LDHA and CA9 expression. We ascertain that elevated LDHA and CA9 in MCT4 KO animals may be compensatory and, in fact, instigated by increased intracellular lactate concentrations. The generation of intracellular lactate/H+ via LDHA may be necessary for maintaining HIF levels, meanwhile CA9 may subsequently balance intracellular pH affected by lactate/H+ production. This illustrates the possibility that disc degeneration may be associated with elevated levels of intracellular lactate as well as increased extracellular acidification. Our studies of MCT4 KO mice clearly recapitulate the major pathoanatomical hallmarks of human disc degeneration, including loss of NP cell phenotype, acquisition of hypertrophic chondrocyte-like phenotype, and compromised matrix integrity (68,69). Based on our mouse studies and observation that human disc degeneration is associated with selective loss of MCT4 protein, we hypothesize that loss of MCT4 expression contributes to the cascade of events that occur during human disc degeneration.
Intervertebral disc degeneration and low back pain are correlated with the health and integrity of the spinal motion segment in its entirety, including the vertebral bodies. Decreased vertebral bone volume and impaired trabecular architecture in MCT4 KO mice clearly suggested that H+/lactate efflux is necessary for maintenance of vertebral trabecular bone quality. One interpretation of these results is that glucose metabolism is affected in the resident trabecular bone cells: the osteoblasts and osteoclasts. While the effect of lactate accumulation in these cells has not been studied in detail, it is known that they rely on glucose uptake for cell differentiation and bone formation (47,70). In fact, recent studies showed that osteoblasts preferentially use glycolysis for ATP production, even under aerobic conditions, mimicking the Warburg effect (71,72). It is therefore possible that loss of MCT4 may directly affect the metabolic profile of osteoblasts and interfere with proper trabecular bone formation and turnover in the vertebrae. Of note, unlike trabecular bone, the lack of phenotype in cortical bone may reflect differences in metabolic status between these bone compartments. The increased trabecular bone mineral density seen in the MCT4 KO mice is likely to be secondary to and compensatory for defects in bone volume and trabecular morphology.
Microarray studies were performed on MCT4 KO mice to understand how chronic accumulation of lactate affects the broader transcriptional program of NP cells which may lead to age-dependent disc degeneration. We found that the enriched biological processes in KO mice were driven by three major groups of genes concerned with nucleosome assembly and regulation of epigenetic gene expression; driven by histone genes, oxidant detoxification; driven by metallothioneins and hemoglobin genes, and negative regulation of cell proliferation; driven by Wnt-related genes and DNA-specific binding proteins. Our data is in agreement with MCT4 knock-down studies in human pancreatic cell lines, which showed DEGs involved with a similar down-regulation of epigenetic programming (52). These aforementioned changes raise an interesting question: Can H+/lactate transport directly affect gene transcription? Recent studies have demonstrated that, in fact, many essential glycolytic and mitochondrial enzymes “moonlight” in the nucleus where they are involved in activities such as DNA binding or the generation of intermediates that regulate gene transcription (73-76). Of particular interest for this study, phosphorylation of Tyr238 on LDH has been shown to promote nuclear translocation. In this scenario, locally produced lactate inhibits HDACs and increases gene transcription (77,78). Growing evidence thus suggests that metabolic intermediates and byproducts, such as lactate, can directly link cellular metabolism to functions such as cell proliferation and survival through altered gene transcription. Therefore, it is possible that the loss of MCT4 contributes to disc degeneration through a cumulative effect on the transcriptional program in NP cells.
We also delineated how disruption of H+/lactate efflux may initiate disc degeneration over time by instigating metabolic stress. Metabolomics analysis demonstrated that MCT inhibition increased the concentration of metabolites involved in pyruvate metabolism and the TCA cycle, meanwhile stable glucose isotope labeling confirmed an increase in intracellular M2 pyruvate (via M2 alanine enrichment) and TCA cycle flux through PDH. The fact that acute inhibition of a plasma membrane-associated ion transporter was capable of rewiring NP cell metabolism was striking. Such metabolic rewiring has only been observed in NP cells following decreased HIF-1α levels, which otherwise has stabilized expression and controls glycolysis (17,24,79). We hypothesize that MCT4 inhibition led to an increase in TCA cycle flux for two reasons. First, cytotoxic acidosis from intracellular H+/lactate accumulation may be prevented by lactate reconversion into pyruvate. Second, reducing equivalents generated from the increased TCA cycle flux and glutamate production may be required to maintain the redox state which is disturbed by feedback inhibition of lactate on LDH activity. The fact that acute MCT inhibition does not alter NAD+/NADH ratio speaks directly to this point. Taken together, we surmise that the metabolic plasticity of NP cells enabled them to withstand short-term MCT inhibition by up-regulating TCA cycle flux and maintaining redox ratios, however, we clearly show that long-term loss of MCT4 function causes disc degeneration. The increased expression of LDH and CA9 noted in AF cells in MCT4 KO mice supports the notion that long-term loss of MCT4 in vivo instigates cellular stress through altered intracellular pH and disturbed redox status.
In addition, our results provide the first evidence that lactate is an important signaling molecule in NP cells. In addition to modulating transcriptional program and instigating metabolic rewiring, we demonstrate that lactate can function as a hypoxia mimetic factor in NP cells. This occurs through the functional competition of lactate and pyruvate and other TCA metabolites such as succinate, fumarate and oxaloacetate with α-ketoglutarate, a cofactor necessary for HIF-1α hydroxylation and subsequent degradation by PHDs (53,54). Our results clearly show a dynamic relationship whereby alterations in lactate levels can modulate HIF-1α transcriptional activity using a luciferase reporter containing known hypoxia responsive elements. This may explain the elevation of HIF-1 targets, LDHA and CA9, in the discs of MCT4 KO mice.
Lastly, we show the first evidence to suggest a positive feedback loop exists between hypoxia-inducible MCT4 function and HIF-1α stability in NP cells. To show this, we provide a mechanism of MCT4 transcriptional-activation through a novel hypoxia and HIF-1 sensitive intronic enhancer which constitutes one arm of the HIF-1-MCT4 regulatory loop. Taken together, our studies clearly demonstrate that maintenance of proper H+/lactate transport in cells of the intervertebral disc and vertebrae is required for health of the spinal column with age.
Supplementary Material
Acknowledgements:
This work is supported by grants from the National Institutes of Health R01 AR055655, AR064733, AR074813 (MVR), R01 EY012042 (NJP), and a Sheffield Hallam University Vice Chancellors Scholarship (CLM). Elizabeth Silagi is supported by T32 AR052273. Stable Isotope and Metabolomics Core Facility of the Diabetes Research and Training Center (DRTC) of the Albert Einstein College of Medicine is supported by NIH/NCI grant (P60DK020541).
Footnotes
DISCLOSURES
The authors declare that they have no conflicts of interest or disclosures with the contents of this article.
REFERENCES
- 1.Murray CJL. The State of US Health, 1990–2010. JAMA. 2013. August 14;310(6):591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maher C, Underwood M, Buchbinder R. Non-specific low back pain. Lancet. 2017;389:736–47. [DOI] [PubMed] [Google Scholar]
- 3.Vos T, Barber RM, Bell B, Bertozzi-Villa A, Biryukov S, Murray CJL, Al E Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:743–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livshits G, Popham M, Malkin I, Sambrook PN, MacGregor AJ, Spector T, Williams FMK. Lumbar disc degeneration and genetic factors are the main risk factors for low back pain in women: The UK Twin Spine Study. Ann. Rheum. Dis 2011;70:1740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi H, Tessier S, Silagi ES, Kyada R, Yousefi F, Pleshko N, Shapiro IM, Risbud MV. A novel mouse model of intervertebral disc degeneration shows altered cell fate and matrix homeostasis. Matrix Biol. 2018;70:102–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silagi ES, Shapiro IM, Risbud M V. Glycosaminoglycan synthesis in the nucleus pulposus: Dysregulation and the pathogenesis of disc disease. Matrix Biol. 2018. March 1;71–72:368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Maitre CL, Pockert AP, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem. Soc. Trans 2007;35(Pt 4):652–5. [DOI] [PubMed] [Google Scholar]
- 8.Nachemson A Intradiscal Measurements of pH in Patients with Lumbar Rhizopathies Acta Orthop. Scand Taylor & Francis; 1969. January 8;40(1):23–42. [DOI] [PubMed] [Google Scholar]
- 9.Gorth DJ, Shapiro IM, Risbud MV. Transgenic mice overexpressing human TNF-α experience early onset spontaneous intervertebral disc herniation in the absence of overt degeneration. Cell Death Dis. 2018;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat. Rev. Rheumatol 2014;10(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merceron C, Mangiavini L, Robling A, Wilson TL, Giaccia AJ, Shapiro IM, Schipani E, Risbud M V. Loss of HIF-1alpha in the notochord results in cell death and complete disappearance of the nucleus pulposus. PLoS One. 2014;9(10):e110768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman R, Patil P, Tisherman R St., Croix C, Niedernhofer L, Robbins P, Ambrosio F, Van Houten B, Sowa G, Vo N. Age-dependent changes in intervertebral disc cell mitochondria and bioenergetics. Eur. Cells Mater 2018;36:171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee WC, Guntur AR, Long F, Rosen CJ. Energy metabolism of the osteoblast: Implications for osteoporosis. Endocr. Rev 2017;38:255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, Long F. WNT-LRP5 signaling induces warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 2013;17:745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida T, Kubota S, Aoyama E, Takigawa M. Impaired glycolytic metabolism causes chondrocyte hypertrophy-like changes via promotion of phospho-Smad1/5/8 translocation into nucleus. Osteoarthr. Cartil 2013;21:700–9. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert HTJ, Hodson N, Baird P, Richardson SM, Hoyland JA. Acidic pH promotes intervertebral disc degeneration: Acid-sensing ion channel −3 as a potential therapeutic target. Sci. Rep 2016. November 17;6:37360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silagi ES, Schoepflin ZR, Seifert EL, Merceron C, Schipani E, Shapiro IM, Risbud MV. Bicarbonate Recycling by HIF-1–Dependent Carbonic Anhydrase Isoforms 9 and 12 Is Critical in Maintaining Intracellular pH and Viability of Nucleus Pulposus Cells. J. Bone Miner. Res 2018;33(2):338–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diamant B, Karlsson J, Nachemson A. Correlation between lactate levels and pH in discs of patients with lumbar rhizopathies Experientia. Birkhäuser-Verlag; 1968. December;24(12):1195–6. [DOI] [PubMed] [Google Scholar]
- 19.Bartels EM, Fairbank JC, Winlove CP, Urban JP. Oxygen and lactate concentrations measured in vivo in the intervertebral discs of patients with scoliosis and back pain. Spine (Phila. Pa. 1976). 1998;23(1):1–7; discussion 8. [DOI] [PubMed] [Google Scholar]
- 20.Keshari KR, Lotz JC, Link TM, Hu S, Majumdar S, Kurhanewicz J. Lactic acid and proteoglycans as metabolic markers for discogenic back pain. Spine (Phila. Pa. 1976). 2008;33:312–7. [DOI] [PubMed] [Google Scholar]
- 21.Gornet MG, Peacock J, Claude J, Schranck FW, Copay AG, Eastlack RK, Benz R, Olshen A, Lotz JC. Magnetic resonance spectroscopy (MRS) can identify painful lumbar discs and may facilitate improved clinical outcomes of lumbar surgeries for discogenic pain. Eur. Spine J 2019;28:674–87. [DOI] [PubMed] [Google Scholar]
- 22.Bez M, Zhou Z, Sheyn D, Tawackoli W, Giaconi JC, Shapiro G, Ben David S, Gazit Z, Pelled G, Li D, Gazit D. Molecular pain markers correlate with pH-sensitive MRI signal in a pig model of disc degeneration. Sci. Rep 2018;26:17363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita N, Chiba K, Shapiro IM, Risbud M V. HIF-1alpha and HIF-2alpha degradation is differentially regulated in nucleus pulposus cells of the intervertebral disc. J Bone Min. Res 2012;27(2):401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Risbud MV., Guttapalli A, Stokes DG, Hawkins D, Danielson KG, Schaer TP, Albert TJ, Shapiro IM. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J. Cell. Biochem 2006;98(1):152–9. [DOI] [PubMed] [Google Scholar]
- 25.Pan H, Strickland A, Madhu V, Johnson ZI, Chand SN, Brody JR, Fertala A, Zheng Z, Shapiro IM, Risbud MV. RNA binding protein HuR regulates extracellular matrix gene expression and pH homeostasis independent of controlling HIF-1α signaling in nucleus pulposus cells. Matrix Biol. 2018;77:23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi H, Merceron C, Mangiavini L, Seifert EL, Schipani E, Shapiro IM, Risbud MV. Hypoxia promotes noncanonical autophagy in nucleus pulposus cells independent of MTOR and HIF1A signaling. Autophagy. 2016;12(9):1631–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razaq S, Wilkins RJ, Urban JPG. The effect of extracellular pH on matrix turnover by cells of the bovine nucleus pulposus. Eur. Spine J 2003;12(4):341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Razaq S, Urban JP, Wilkins RJ. Regulation of intracellular pH by bovine intervertebral disc cells. Cell. Physiol. Biochem 2000;10(1–2):109–15. [DOI] [PubMed] [Google Scholar]
- 29.Ullah MS, Davies AJ, Halestrap AP. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1α-dependent mechanism. J. Biol. Chem 2006;281(14):9030–7. [DOI] [PubMed] [Google Scholar]
- 30.Chiche J, Ricci J-E, Pouysségur J. Tumor hypoxia and metabolism -- towards novel anticancer approaches. Ann. Endocrinol. (Paris). 2013. May;74(2):111–4. [DOI] [PubMed] [Google Scholar]
- 31.Parks SK, Chiche J, Pouysségur J. Disrupting proton dynamics and energy metabolism for cancer therapy Nat. Rev. Cancer Nature Publishing Group; 2013;13(9):611–23. [DOI] [PubMed] [Google Scholar]
- 32.Adijanto J, Philp NJ. The SLC16A family of monocarboxylate transporters (MCTs)-physiology and function in cellular metabolism, pH homeostasis, and fluid transport. Curr Top Membr. 2012;70:275–311. [DOI] [PubMed] [Google Scholar]
- 33.Philp NJ, Yoon H, Grollman EF. Monocarboxylate transporter MCT1 is located in the apical membrane and MCT3 in the basal membrane of rat RPE. Am. J. Physiol. Integr. Comp. Physiol 1998;274:R1824–8. [DOI] [PubMed] [Google Scholar]
- 34.Dimmer KS, Friedrich B, Lang F, Deitmer JW, Bröer S. The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem. J 2000;350 Pt 1:219–27. [PMC free article] [PubMed] [Google Scholar]
- 35.Philp NJ, Yoon H, Lombardi L. Mouse MCT3 gene is expressed preferentially in retinal pigment and choroid plexus epithelia. Am. J. Physiol. Physiol 2001;280:C1319–26. [DOI] [PubMed] [Google Scholar]
- 36.Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo award in basic science. Spine (Phila. Pa. 1976) 2002;27:2631–44. [DOI] [PubMed] [Google Scholar]
- 37.Benjamin D, Robay D, Hindupur SK, Pohlmann J, Colombi M, El-Shemerly MY, Maira S-M, Moroni C, Lane HA, Hall MN. Dual Inhibition of the Lactate Transporters MCT1 and MCT4 Is Synthetic Lethal with Metformin due to NAD+ Depletion in Cancer Cells. Cell Rep. 2018. December 11;25(11):3047–3058.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res 2009;8(10):4844–50. [DOI] [PubMed] [Google Scholar]
- 39.Qiu Y, Su M, Liu Y, Chen M, Gu J, Zhang J, Jia W. Application of ethyl chloroformate derivatization for gas chromatography-mass spectrometry based metabonomic profiling. Anal Chim Acta. 2007;583(2):277–83. [DOI] [PubMed] [Google Scholar]
- 40.Csordás G, Golenár T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la F Perez S, Bogorad R, Koteliansky V, Adijanto J, Mootha VK, Hajnóczky G. MICU1 Controls Both the Threshold and Cooperrative Activation of the Mitochondrial Ca2+ Uniporter. Cell Metab. 2013;17(6):976–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James Kent W, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, Van Der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F, Mathelier A. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;4:D260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sive JI, Baird P, Jeziorsk M, Watkins A, Hoyland JA, Freemont AJ. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs Mol. Pathol BMJ Publishing Group; 2002. April;55(2):91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bisetto S, Curry J, Philp NJ, Martinez-Outschoorn UE, Whitaker-Menezes D, Zhan T, Snyder CM, Tuluc M, Wilski NA. Monocarboxylate Transporter 4 (MCT4) Knockout Mice Have Attenuated 4NQO Induced Carcinogenesis; A Role for MCT4 in Driving Oral Squamous Cell Cancer. Front. Oncol 2018;8:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatel B, Bendahan D, Hourdé C, Pellerin L, Lengacher S, Magistretti P, Le Fur Y, Vilmen C, Bernard M, Messonnier LA. Role of MCT1 and CAII in skeletal muscle pH homeostasis, energetics, and function: In vivo insights from MCT1 haploinsufficient mice. FASEB J. 2017;31:2562–75. [DOI] [PubMed] [Google Scholar]
- 47.Karner CM, Long F. Glucose metabolism in bone. Bone. 2018;115:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Risbud MV., Schoepflin ZR, Mwale F, Kandel RA, Grad S, Iatridis JC, Sakai D, Hoyland JA. Defining the phenotype of young healthy nucleus pulposus cells: Recommendations of the spine research interest group at the 2014 annual ORS meeting. J. Orthop. Res 2015;33(3):283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silagi ES, Batista P, Shapiro IM, Risbud MV. Expression of Carbonic Anhydrase III, a Nucleus Pulposus Phenotypic Marker, is Hypoxia-responsive and Confers Protection from Oxidative Stress-induced Cell Death. Sci. Rep 2018;8(1):4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novais EJ, Diekman BO, Shapiro IM, Risbud MV. p16 Ink4a deletion in cells of the intervertebral disc affects their matrix homeostasis and senescence associated secretory phenotype without altering onset of senescence. Matrix Biol. 2019;doi: 10.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoepflin ZR, Silagi ES, Shapiro IM, Risbud MV. PHD3 is a transcriptional coactivator of HIF-1a in nucleus pulposus cells independent of the PKM2-JMJD5 axis. FASEB J. 2017;31(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baek GH, Tse YF, Hu Z, Cox D, Buboltz N, McCue P, Yeo CJ, White MA, DeBerardinis RJ, Knudsen ES, Witkiewicz AK. MCT4 Defines a Glycolytic Subtype of Pancreatic Cancer with Poor Prognosis and Unique Metabolic Dependencies. Cell Rep. 2014;24:2233–49. [DOI] [PubMed] [Google Scholar]
- 53.De Saedeleer CJ, Copetti T, Porporato PE, Verrax J, Feron O, Sonveaux P. Lactate Activates HIF-1 in Oxidative but Not in Warburg-Phenotype Human Tumor Cells. Bai Y, editor. PLoS One. 2012. October 17;7(10):e46571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu H, Forbes RA, Verma A. Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J. Biol. Chem 2002;277(26):23111–5. [DOI] [PubMed] [Google Scholar]
- 55.Lee DC, Sohn HA, Park Z-YY, Oh S, Kang YK, Lee K-MM, Kang M, Jang YJ, Yang S-JJ, Hong YK, Noh H, Kim J-AA, Kim DMJ, Bae K-HH, Kim DMJ, Chung SJ, Yoo HS, Yu D-YY, Park KC, Yeom Y Il. A lactate-induced response to hypoxia Cell. Elsevier Inc; 2015. April 23;161(3):595–609. [DOI] [PubMed] [Google Scholar]
- 56.Grampp S, Schmid V, Salama R, Lauer V, Kranz F, Platt JL, Smythies J, Choudhry H, Goppelt-Struebe M, Ratcliffe PJ, Mole DR, Schödel J. Multiple renal cancer susceptibility polymorphisms modulate the HIF pathway. PLoS Genet. 2017;13:e1006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smythies JA, Sun M, Masson N, Salama R, Simpson PD, Murray E, Neumann V, Cockman ME, Choudhry H, Ratcliffe PJ, Mole DR. Inherent DNA‐binding specificities of the HIF‐1α and HIF‐2α transcription factors in chromatin. EMBO Rep. 2019;20:e46401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silagi ES, Schoepflin ZR, Seifert EL, Merceron C, Schipani E, Shapiro IM, Risbud M V. Bicarbonate Recycling by HIF-1-Dependent Carbonic Anhydrase Isoforms 9 and 12 Is Critical in Maintaining Intracellular pH and Viability of Nucleus Pulposus Cells. J. Bone Miner. Res 2018. February;33(2):338–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Risbud M V, Schipani E, Shapiro IM. Hypoxic regulation of nucleus pulposus cell survival: from niche to notch. Am. J. Pathol 2010. April;176(4):1577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang CZ, Li H, Tao YQ, Zhou XP, Yang ZR, Li FC, Chen QX. The relationship between low pH in intervertebral discs and low back pain: A systematic review. Arch. Med. Sci 2012;8:952–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez-Garcia O, Matsuzaki T, Olmer M, Masuda K, Lotz MK. Age-related reduction in the expression of FOXO transcription factors and correlations with intervertebral disc degeneration. J. Orthop. Res 2017;35:2682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfirrmann CWA, Metzdorf A, Elfering A, Hodler J, Boos N. Effect of aging and degeneraton on disc volume and shape: A quantitative study in asymptomatic volunteers. J. Orthop. Res 2006;24:1086–94. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez AG, Slichter CK, Acosta FL, Rodriguez-Soto AE, Burghardt AJ, Majumdar S, Lotz JC. Human disc nucleus properties and vertebral endplate permeability. Spine (Phila. Pa. 1976). 2011;36:512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shirazi-Adl A, Taheri M, Urban JPG. Analysis of cell viability in intervertebral disc: Effect of endplate permeability on cell population. J. Biomech 2010;43:1330–6. [DOI] [PubMed] [Google Scholar]
- 65.Gorth DJ, Shapiro IM, Risbud M V. A new understanding of the role of IL‐1 in age‐related intervertebral disc degeneration in a murine model. J. Bone Miner. Res 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Xiong C, Kudelko M, Li Y, Wang C, Wong YL, Tam V, Rai MF, Cheverud J, Lawson HA, Sandell L, Chan WCW, Cheah KSE, Sham PC, Chan D. Early onset of disc degeneration in SM/J mice is associated with changes in ion transport systems and fibrotic events. Matrix Biol. 2018. September;70:123–39. [DOI] [PubMed] [Google Scholar]
- 67.Hristova GI, Jarzem P, Ouellet JA, Roughley PJ, Epure LM, Antoniou J, Mwale F. Calcification in human intervertebral disc degeneration and scoliosis. J. Orthop. Res 2011;29:1888–95. [DOI] [PubMed] [Google Scholar]
- 68.Roberts S, Evans H, Trivedi J, Menage J. Histology and pathology of the human intervertebral disc. J. Bone Joint Surg. Am 2006;88 Suppl 2:10–4. [DOI] [PubMed] [Google Scholar]
- 69.Thompson JP, Schechter MT, Adams ME, Tsang IK, Bishop PB, Pearce RH. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine (Phila. Pa. 1976) 1990;15:411–5. [DOI] [PubMed] [Google Scholar]
- 70.Lee SY, Abel ED, Long F. Glucose metabolism induced by Bmp signaling is essential for murine skeletal development. Nat. Commun 2018;9:4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Esen E, Lee SY, Wice BM, Long F. PTH Promotes Bone Anabolism by Stimulating Aerobic Glycolysis via IGF Signaling. J. Bone Miner. Res 2015;30:2137. [DOI] [PubMed] [Google Scholar]
- 72.Esen E, Long F. Aerobic glycolysis in osteoblasts. Curr. Osteoporos. Rep 2014;12:433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boukouris AE, Zervopoulos SD, Michelakis ED. Metabolic Enzymes Moonlighting in the Nucleus: Metabolic Regulation of Gene Transcription. Trends Biochem. Sci 2016;41:712–30. [DOI] [PubMed] [Google Scholar]
- 74.Castello A, Hentze MW, Preiss T. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol. Metab 2015;26:746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao X, Lin S-H, Ren F, Li J-T, Chen J-J, Yao C-B, Yang H-B, Jiang S-X, Yan G-Q, Wang D, Wang Y, Liu Y, Cai Z, Xu Y-Y, Chen J, Yu W, Yang P-Y, Lei Q-Y. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun 2016. December 30;7(1):11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao S, Torres A, Henry RA, Trefely S, Wallace M, Lee J V., Carrer A, Sengupta A, Campbell SL, Kuo Y-M, Frey AJ, Meurs N, Viola JM, Blair IA, Weljie AM, Metallo CM, Snyder NW, Andrews AJ, Wellen KE. ATP-Citrate Lyase Controls a Glucose-to-Acetate Metabolic Switch. Cell Rep. 2016. October 18;17(4):1037–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castonguay Z, Auger C, Thomas SC, Chahma M, Appanna VD. Nuclear lactate dehydrogenase modulates histone modification in human hepatocytes. Biochem. Biophys. Res. Commun 2014;454:172–7. [DOI] [PubMed] [Google Scholar]
- 78.Latham T, MacKay L, Sproul D, Karim M, Culley J, Harrison DJ, Hayward L, Langridge-Smith P, Gilbert N, Ramsahoye BH. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012;40:4794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Agrawal A, Guttapalli A, Narayan S, Albert TJ, Shapiro IM, Risbud MV. Normoxic stabilization of HIF-1alpha drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk. Am. J. Physiol. Cell Physiol 2007;293(2):C621–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary files) or deposited on the GEO database (GSE130561).