Abstract
Sharing radiologic image annotations among multiple institutions is important in many clinical scenarios; however, interoperability is prevented because different vendors’ PACS store annotations in non-standardized formats that lack semantic interoperability. Our goal was to develop software to automate the conversion of image annotations in a commercial PACS to the Annotation and Image Markup (AIM) standardized format and demonstrate the utility of this conversion for automated matching of lesion measurements across time points for cancer lesion tracking. We created a software module in Java to parse the DICOM presentation state (DICOM-PS) objects (that contain the image annotations) for imaging studies exported from a commercial PACS (GE Centricity v3.x). Our software identifies line annotations encoded within the DICOM-PS objects and exports the annotations in the AIM format. A separate Python script processes the AIM annotation files to match line measurements (on lesions) across time points by tracking the 3D coordinates of annotated lesions. To validate the interoperability of our approach, we exported annotations from Centricity PACS into ePAD (http://epad.stanford.edu) (Rubin et al., Transl Oncol 7(1):23–35, 2014), a freely available AIM-compliant workstation, and the lesion measurement annotations were correctly linked by ePAD across sequential imaging studies. As quantitative imaging becomes more prevalent in radiology, interoperability of image annotations gains increasing importance. Our work demonstrates that image annotations in a vendor system lacking standard semantics can be automatically converted to a standardized metadata format such as AIM, enabling interoperability and potentially facilitating large-scale analysis of image annotations and the generation of high-quality labels for deep learning initiatives. This effort could be extended for use with other vendors’ PACS.
Keywords: Lesion tracking, Annotation and Image Markup (AIM), Data mining, Supervised training, Deep learning
Introduction
The ability to access radiologic image annotations is important in many clinical and research contexts, particularly in clinical oncology where index tumor measurements are tracked across sequential imaging studies to evaluate treatment response. For example, commonly upon an initial cancer diagnosis, an individual patient may transfer care from a primary health center to a tertiary care center or begin receiving care at multiple clinical sites that require coordination. At the population level, there is frequently a need to pool many patients’ medical data for use in cancer registries such as the Cancer Imaging Archive for radiomics research [1], during clinical drug trials, by big data applications, and for data sharing efforts such as the RSNA Image Share initiative [2].
In each of these scenarios, there is a need to easily access the lesion image annotation data; however, these efforts are often thwarted by the tendency of commercial PACS vendors to encode these annotations in formats lacking semantic standardization, such as DICOM presentation state objects (DICOM-PS) or in various DICOM structured reporting templates (DICOM-SR). While DICOM itself represents a standard format for encoding digital medical imaging and related data (International Organization of Standards 12052), visual annotations encoded within DICOM-PS objects lack semantic standardization. That is, due to the variability in how annotations are encoded in DICOM-PS by commercial PACS, these annotations are not readily interpretable by external systems. Our primary goal was to create a software tool that would enable line annotations created in a widely used commercial PACS (GE Centricity version 3.x) to be extracted and converted to the Annotation and Image Markup (AIM) open standard [3, 4] or the newly established AIM/DICOM-SR object for these annotations (TID-1500) [5, 6].
Once image annotations are in AIM format, lesion data may be processed by AIM-compliant tools such as ePAD, an open source, AIM-compliant imaging informatics platform [7] that can perform quantitative lesion tracking automatically. To utilize this functionality, lesion annotations must have matching names across sequential studies. However, when a user creates and saves line annotations in Centricity 3.x, the annotations are named in sequential order by creation time; for example, the first line annotation created is automatically named “lesion 1,” and the second is named “lesion 2.” Although commercial PACS including GE Centricity allow users to create text box annotations that may be placed near line annotations to manually label lesions, this practice may not be consistently followed and could be problematic when multiple annotations exist on the same image. Therefore, a secondary goal of our work was to create a method to match lesions across sequential studies and rename them accordingly, augmenting the image annotations in vendor systems that lack this information.
Materials and Methods
Data Extraction
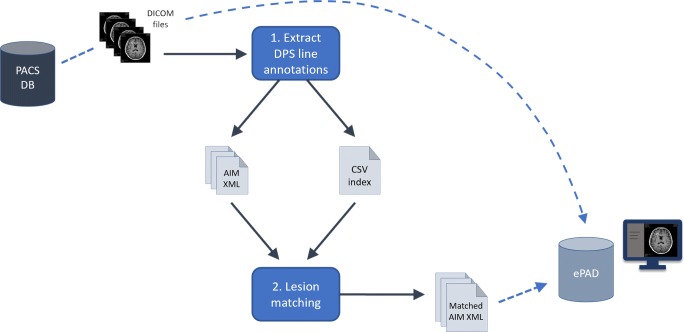
DICOM files including images and their accompanying DICOM-PS files, which contain the image annotations, were first exported from Centricity PACS for subsequent processing in our workflow (Fig. 1).
Fig. 1.
Schematic demonstrating the overall process of exporting DICOM files from the PACS database, performing lesion annotation extraction from the DICOM-PS objects followed by longitudinal lesion matching, and uploading the DICOM and matched annotation AIM XML files into ePAD. DB database, DPS DICOM presentation state objects, CSV comma-separated values file
Software
We created two software modules: the first module exports the line annotations in AIM XML format and the second module matches the same lesion across sequential imaging studies. With respect to this second task, a design assumption needed to be made since annotations in current vendor systems do not provide labels for lesions that can unambiguously track them. Therefore, for any given patient, we assume the same lesions are measured on each sequential study with no new lesions manifesting after the initial (baseline) study. In practice, consistent lesion measurement would need to be confirmed.
The first module was written in Java to leverage the AIM application programming interface (API) [7] and PixelMed library of DICOM tools (http://dclunie.com/pixelmed). The module is invoked with a parameter specifying the destination directory, which is then recursively scanned for DICOM files. Each DICOM file is sequentially parsed to identify line annotations encoded within DICOM-PS files in the Centricity convention. Using the AIM API for Java, these line annotations are converted to AIM format and exported as XML files that may then be transferred into an AIM-compliant PACS such as ePAD for viewing. Additionally, the module exports a CSV index file listing the parameters (e.g., patient ID, study accession number, line point coordinates, and corresponding image DICOM unique identifier) for each line annotation. This CSV index file facilitates access to these annotation data by other applications, including for potential conversion to other formats aside from AIM, using standard dataframe functionality such as the Pandas library in Python.
The second software module, created in Python, scans a provided destination directory to locate and load the CSV annotation index file created by the first script. For each patient, the algorithm matches lesion measurements across successive imaging studies using the annotations’ three-dimensional coordinates (Table 1). The Centricity PACS (and many other commercial PACS) does not enable the user to manually set or modify an annotation’s name, which is stored in the annotation’s metadata, so that the same lesion can be readily tracked on longitudinal studies. Thus, our algorithm reassigns lesion labels by looking at the patient’s baseline study and renaming the lesions (“lesion 1,” “lesion 2,” etc.) in order of the smallest to largest X, Y, and Z geographical body locations. In order to track the same lesions across sequential studies, on each follow-up study, the 3D location of each lesion is compared to that of each lesion on the baseline study to find a match that minimizes a spatial deviation penalty value. After this matching process completes, new AIM XML files are generated with the updated lesion names that match across sequential studies.
Table 1.
Pseudocode of the lesion tracking algorithm
| Lesion tracking pseudocode | |
|---|---|
| For the patient’s oldest (baseline) study, reorder/rename all lesions sequentially by smallest X, Y, and Z coordinate values (e.g., lesion 1, lesion 2, … lesion n) | |
| For each lesion annotation on the patient’s follow-up studies: | |
| Calculate total penalty values by summing the total positional difference as compared with each baseline lesion’s annotation X, Y, and Z values | |
| Match to the baseline lesion yielding the smallest total penalty value | |
| Rename the annotation so the new name corresponds with that of the matched lesion from the baseline study |
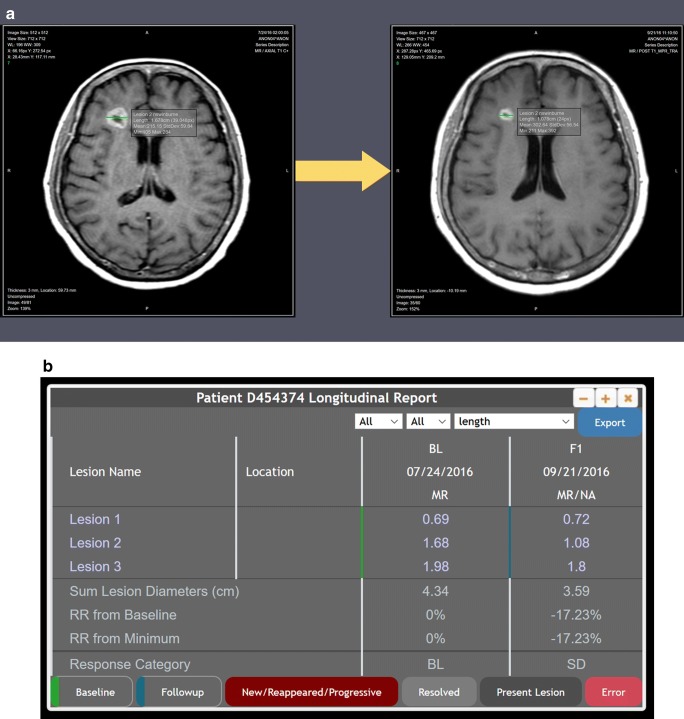
When the DICOM images and matched AIM XML files are uploaded into ePAD, pre-existing lesion tracking functionality in ePAD [8] automatically links matched lesions and provides quantitative metrics (e.g., change in lesion size over time) (Fig. 2).
Fig. 2.
Following lesion labeling and matching by our algorithm operating on semantically non-standard image annotations extracted from the commercial PACS, the lesion annotations from different time points are given unique labels that are correctly matched on the longitudinal studies and displayed as expected in ePAD (a), including baseline (left) and two-month follow-up (right) brain MRIs. Matched lesions are automatically linked in ePAD by its processing of the AIM annotations, enabling quantitative longitudinal lesion tracking (b). BL baseline, F1 follow-up exam #1, RR response rate, SD stable disease
Evaluation
Validation of the software modules was performed by exporting anonymized DICOM data containing brain MRI images and line annotations (2 sequential studies for each of 5 patients) from GE Centricity and running the software modules on the exported data, manually evaluating whether the AIM lesion annotation XML files were exported correctly and that lesions were matched appropriately. The test DICOM data included sequential studies for a given patient in which corresponding lesion names were named discordantly, e.g., “lesion 1” on the baseline study corresponded with “lesion 2” on the follow-up examination, and vice versa. After running the two software modules, the DICOM image files and final matched AIM XML files were uploaded into ePAD to evaluate whether the PACS’ quantitative data displayed correctly.
Availability
Custom code related to the conversion of lesion annotations from Centricity DICOM-PS to AIM format will be made available to academic institutions upon request.
Results
In our test data, line annotations were matched and renamed correctly. In particular, the algorithm correctly handled studies in which there were multiple lesions on the same axial image, and our coordinate-based approach to lesion matching performed as expected. When the images and annotations were uploaded into ePAD, the annotations on longitudinal studies were automatically linked, generating quantitative lesion tracking tables that display change in size over time using ePAD’s existing lesion tracking functionality (Fig. 2).
Discussion
Image annotations are routinely created by radiologists when they review and interpret images, commonly measuring lesions such as malignant tumors. These annotations are important for applications that need to know the location of lesions (e.g., deep learning models trained on lesions for detection or segmentation) and for applications that track lesion size over time, such as for response evaluation criteria in solid tumors (RECIST) reporting [9]. We have shown that it is possible to convert semantically non-standard image annotations in a commercial PACS into an open standard such as AIM and then use them for applications such as automated lesion tracking.
One of the challenges of applying machine learning to radiologic data is the need for large-scale image labeling data for use in either supervised or unsupervised machine learning. In contrast to efforts like ImageNet [10], which comprises millions of labeled non-medical images, radiologic image labeling is more difficult to crowdsource on a large scale due to the expertise needed for radiologic image interpretation. Meanwhile, the clinical PACS contains petabytes of historical image data that have been carefully reviewed by radiologists, many of which have lesion annotations that could be leveraged for machine learning. The software tools we created could help address the challenge of accessing historical image annotations and using them within a deep learning data pipeline for the large-scale generation of high-quality image labels [11].
The first software module we created extracts the Centricity formatted line annotations and converts them to the AIM standardized format. Recently, AIM has been harmonized with DICOM-SR [5, 6], which will allow AIM to be converted to DICOM-SR when the tooling for doing so becomes available in DICOM software libraries. ePAD already has the capability of importing AIM/DICOM-SR objects and converting them to AIM. While our effort was limited in scope to a single commercial vendor PACS, this exportation functionality could be extended for annotations created in other commercial PACS, facilitating the sharing or pooling of image annotation data across institutions.
The second software module we created matches lesion annotation names by minimizing a 3D spatial deviation penalty, and we showed it works successfully within the constraints described. We acknowledge that our current approach to labeling lesions consistently on longitudinal imaging studies may not be robust in all cases, and the number of patients we used to test our software is too small to thoroughly evaluate this; for example, a given lesion may disappear or new lesions could first manifest after the baseline study. Indeed, this lesion matching task may itself be best addressed with a deep learning approach that can handle the potential ambiguities inherent in matching lesions across studies and identifying new—or newly absent—lesions. Such future work could be built on top of our current work to provide more robust results.
The significance of our work is that we have shown that our software modules unlock line annotations created in a widely used commercial PACS for potential use in clinical and research workflows. We believe our approach and code could be extended to produce similar results with other vendor platforms. In the future, if vendors adopt AIM/DICOM-SR, this would provide a more direct path to semantic interoperability of image annotations and would facilitate the mining of these data that our software accomplishes. For existing DICOM imaging data repositories, our tools provide an immediate solution for accessing historical annotations in a vendor PACS, potentially improving patient care through imaging interoperability and enabling further steps into the era of quantitative imaging.
Funding
This work was supported in part by grants from the National Cancer Institute, National Institutes of Health, U01CA142555, 1U01CA190214, and 1U01CA187947.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nathaniel C. Swinburne, Email: swinburn@mskcc.org
Daniel L. Rubin, Email: dlrubin@stanford.edu
References
- 1.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The CANCER Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J Digit Imaging. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langer SG, Tellis W, Carr C, Daly M, Erickson BJ, Mendelson D, Moore S, Perry J, Shastri K, Warnock M, Zhu W. The RSNA Image Sharing Network. J Digit Imaging. 2015;28(1):53–61. doi: 10.1007/s10278-014-9714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Channin DS, Mongkolwat P, Kleper V, Rubin DL. The Annotation and Image Mark-up Project. Radiology. 2009;253(3):590–592. doi: 10.1148/radiol.2533090135. [DOI] [PubMed] [Google Scholar]
- 4.Channin DS, Mongkolwat P, Kleper V, Sepukar K, Rubin DL. The caBIG™ Annotation and Image Markup Project. J Digit Imaging. 2010;23(2):217–225. doi: 10.1007/s10278-009-9193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DICOM Standards Committee: DICOM PS3.21 2018e - Transformations between DICOM and other Representations; A.6 AIM v4 to DICOM TID 1500 Mapping 2018 [Internet]. [cited 2018 Oct 16]. Available from: http://dicom.nema.org/medical/dicom/current/output/chtml/part21/PS3.21.html. Accessed 16 Oct 2018
- 6.DICOM Standards Committee - Working Group 8 - Structured Reporting: Digital Imaging and Communications in Medicine (DICOM), Sup 200 - Transformation of NCI Annotation and Image Markup (AIM) and DICOM SR Measurement Templates. 2017. Available from: ftp://medical.nema.org/medical/dicom/final/sup200_ft_AIM_DICOMSRTID1500.pdf. Accessed 16 Oct 2018
- 7.Rubin DL, Willrett D, O’Connor MJ, Hage C, Kurtz C, Moreira DA. Automated Tracking of Quantitative Assessments of Tumor Burden in Clinical Trials. Transl Oncol. 2014;7(1):23–35. doi: 10.1593/tlo.13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abajian AC, Levy M, Rubin DL. Informatics in radiology: improving clinical work flow through an AIM database: a sample web-based lesion tracking application. Radiogr Rev Publ Radiol Soc N Am Inc. 2012;32(5):1543–1552. doi: 10.1148/rg.325115752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 11.Yan K, Wang X, Lu L, Summers RM: Deep Lesion: Automated Deep Mining, Categorization and Detection of Significant Radiology Image Findings using Large-Scale Clinical Lesion Annotations. ArXiv171001766 Cs [Internet]. 2017 Oct 4 [cited 2018 Aug 20]; Available from: http://arxiv.org/abs/1710.01766. Accessed 16 Oct 2018