Abstract
Catheters are commonly inserted life supporting devices. Because serious complications can arise from malpositioned catheters, X-ray images are used to assess the position of a catheter immediately after placement. Previous computer vision approaches to detect catheters on X-ray images were either rule-based or only capable of processing a limited number or type of catheters projecting over the chest. With the resurgence of deep learning, supervised training approaches are beginning to show promising results. However, dense annotation maps are required, and the work of a human annotator is difficult to scale. In this work, we propose an automatic approach for detection of catheters and tubes on pediatric X-ray images. We propose a simple way of synthesizing catheters on X-ray images to generate a training dataset by exploiting the fact that catheters are essentially tubular structures with various cross sectional profiles. Further, we develop a UNet-style segmentation network with a recurrent module that can process inputs at multiple scales and iteratively refine the detection result. By training on adult chest X-rays, the proposed network exhibits promising detection results on pediatric chest/abdomen X-rays in terms of both precision and recall, with Fβ = 0.8. The approach described in this work may contribute to the development of clinical systems to detect and assess the placement of catheters on X-ray images. This may provide a solution to triage and prioritize X-ray images with potentially malpositioned catheters for a radiologist’s urgent review and help automate radiology reporting.
Keywords: X-ray, Catheter detection, Multi-scale, Deep learning, Recurrent network, Pediatric
Introduction
Catheters and tubes, including endotracheal tubes (ETTs), umbilical arterial catheters (UACs), umbilical venous catheters (UVCs), and nasogastric tubes (NGTs), are commonly used in the management of critically ill or very low birth weight neonates [8]. For example, ETTs assist in ventilation of the lungs and may prevent aspiration, umbilical catheters may be used for administration of fluids or medications and for blood sampling, and NGTs may be used for nutritional support, aspiration of gastric contents, or decompression of the gastrointestinal tract in critically ill neonates [3]. Because catheters and tubes (all referred as catheters in the following for simplicity) are typically placed without real-time image guidance, they are frequently malpositioned [12, 15], and serious complications can arise as a result [3]. Thus, the position of a catheter is usually assessed using X-ray imaging immediately following placement [3].
Paediatric radiologists are trained to accurately accomplish the task of detecting catheters on X-ray images and assessing placement with a low diagnostic error rate [10]. However, availability of expertise may be limited or delayed due to high image volumes. An automatic approach is desired to flag X-rays which may have a malpositioned catheter so that they can be immediately reviewed by a clinician or radiologist, thus promoting safer use of catheters. Since the location of a catheter impacts clinical decision making, we believe detection of catheters is a critical first step towards a fully automatic catheter placement evaluation system.
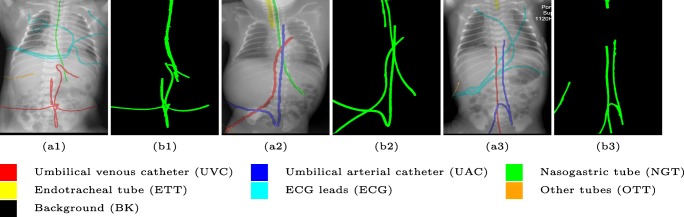
Automated catheter detection on pediatric X-ray is a challenging task. Although most catheters have a radiopaque strip to facilitate detection, the strip may become less apparent depending on the projection angle. Catheters maybe confused by other similar linear structures like ECG leads and anatomy including ribs. Additionally, portions of catheters can be occluded by anatomical structures given that radiographs are a 2D projection of a 3D structure. For example, when a NGT is placed within the oesophagus, the catheter itself becomes less apparent due to the high density of the adjacent vertebrae. Finally, the number and type of catheters that could possibly appear in pediatric X-rays are unknown a priori. The catheters may be intertwined with each other thus making simple line tracing methods fail. Figure 1 provides three sample pediatric X-ray images with some common catheters highlighted in different colors.
Fig. 1.
Detection of catheters is challenging on pediatric X-ray images. The number of catheters is not known prior to interpretation and they can be partially occluded by the body. ECG electrode leads and other unidentified catheters also serve as sources of confusion. (a1), (a2), and (a3) show the original pediatric X-rays, with the potential catheters, wires and lines (including ECG wires and other unidentified catheters) highlighted in different colors. (b1), (b2), and (b3) show the detected catheters by our proposed method
Previous methods have heavily relied on primitive low level cues and made superficial assumptions of catheter appearance and position. They all focused on catheters in chest X-rays without considering the presence of umbilical catheters. The template matching based region growing method would fail in our case due to the higher geometric complexity of the catheter.
Machine learning, especially deep learning, has recently received significant attention in the medical imaging community due to its demonstrated potential to complement image interpretation and augment image representation and classification. For example, super human performance has been achieved in organ segmentation in adult chest X-rays [5] and an algorithm is able to denoise low dose computed tomography with improved overall sharpness [29]. Here, we formulated catheter detection as a binary supervised segmentation task where the catheters are the foreground class and the remainder of the image is the background class. However, a large pixel level accurate annotation dataset is required for current supervised segmentation methods to work properly.
To alleviate this annotation problem, we proposed to use X-ray images with simulated catheters by exploiting the fact that catheters are essentially tubular objects with various cross sectional profiles. To be more specific, a synthetic 2D projection of a catheter is generated by first simulating a horizontal catheter profile and then using it as a brush tip to draw along a B-spline path. This generated catheter is then composited with an X-ray image serving as the training data. Another contribution of this work is a segmentation network that can inherently take into account multi-scale information. This network adopts a UNet-style form and contains a recurrent module that can process inputs iteratively in increasing scales.1 This setup draws the network’s attention to line structures and we have empirically shown that by iterating through the scale space of the input image, higher recall is achieved at a fixed precision level as compared to using a single scale. As far as we know, this is the first work that tried to detect all commonly seen catheters on pediatric X-ray images. Details about the methods are discussed in “Methodology”. Three sample detection results are shown in Fig. 1.
Related Works
There have been limited prior publications regarding automated catheter detection on X-ray images in general. In this section, we not only review catheter detection methods but also provide a brief overview of elongated structure detection as a broader concept.
- Catheter detection
Kao et al. [13] proposed a rule-based system to detect ETTs on pediatric chest X-rays. It was based on the presumption that ETTs usually have the highest intensity and are continuous in the cervical region. This system is sensitive to the positioning of the neonates and it is possible to confuse an ETT with a NGT when the assumptions no longer hold. Sheng et al. [22] proposed a method for identification of ETTs, NGTs and feeding tubes together in adult chest X-rays. The detection was based on the Hough transform assuming that catheters in small rectangular areas are straight. Their algorithm has high computation complexity and would fail if the catheter forms a loop or there are other sources of confusion present, e.g. ECG electrode leads or, ribs. Keller et al. [14] proposed a semi-automated system to detect catheters in chest X-rays with users supplying initial points for catheter tracking. Line tracing was accomplished by template matching of catheter profiles. Follow-up works have been conducted in [20, 21] where the initial seed for line tracing was automatically selected in the cervical region by detecting parallel lines. This approach is not suitable for umbilical catheters since their starting points are not well determined. Mercan et al. [19] proposed a patch-based neural network to detect chest tubes and a curve fitting approach to connect discontinued detected line segments. A very recent work used a fully convolutional neural network for detection of peripherally inserted central catheter (PICC) tip position on adult chest X-ray images [16]. A similar approach was taken by Ambrosini et al. [2] to detect catheters on X-ray fluoroscopy but using a UNet-style [18] network. All the supervised segmentation methods require humans to manually annotate catheter locations for supervised training.
- Elongated structure detection
One of the most common elongated structures in medical imaging is a blood vessel. Its detection has been researched in many imaging modalities, such as in retinal fundus imaging [17] and angiography [9]. The methods used in the literature have evolved from hard coded rule based methods into machine learning based methods. In the early days, researchers tried to devise metrics to measure the “vesselness” directly from feature sources like Hessian matrix [9] and co-occurrence matrix [26]. Later on, rather than relying on a single feature, researchers started to aggregate features from multiple sources, such as ridge based [24], wavelets [23] and then employed a supervised learning method on top to delineate the decision boundary between the vessel and non-vessel. Most recent progress was achieved by supervised deep learning where features were directly learnt from images without the intervention of domain expertise [17]. Since blood vessels are of various diameters by nature, multi-scale approaches have also been explored in the literature [30].
Methodology
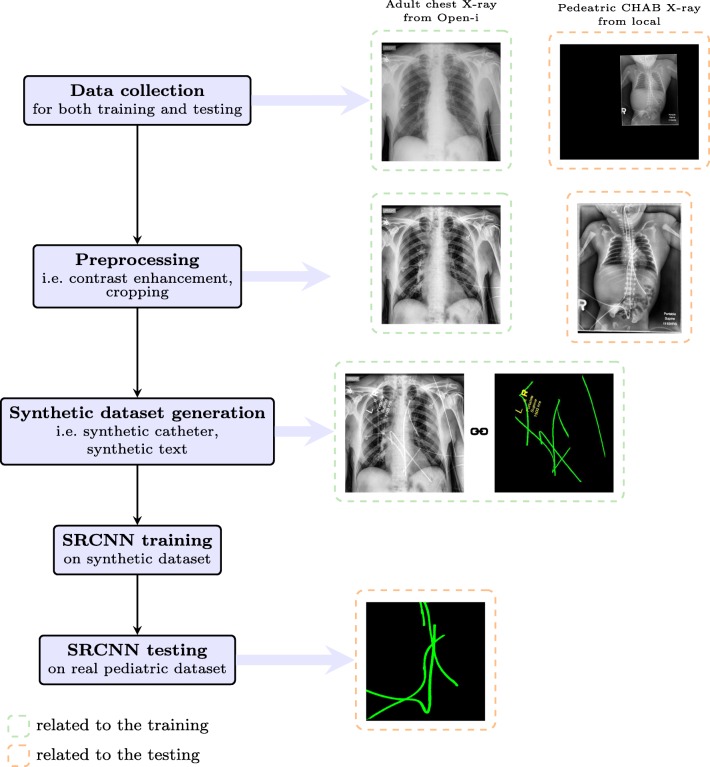
The whole catheter detection system consists of five steps, i.e., data collection, preprocessing, synthetic catheter generation, training and testing. A schematic overview can be found in Fig. 2.
Fig. 2.
An overview of the catheter detection pipeline (better viewed in color, best viewed in digital version)
Data Collection
The training dataset comes from the Open-i dataset [7] from the National Institutes of Health (NIH) which contains 7,471 adult chest X-rays. We randomly selected 2,515 frontal view images and generated synthetic catheters on them.
The test dataset is collected locally and only contains frontal chest-abdominal X-rays from patients < 4 weeks old. This is the most common radiograph obtained to confirm placement of catheters such as UACs and UVCs in neonates. Currently, the test set has 35 fully labeled images with different catheter types with sample images previously shown in Fig. 1. The annotation was carried out by a medical student with GIMP image processing software supervised by a pediatric radiologist. All the annotated catheters (lines excluding ECG leads) are treated as the same class in the detection.
Preprocessing
The X-ray images are of various contrast due to different acquisition protocols. Rather than making the network learn a contrast invariant feature, we normalized the contrast of the input X-rays before sending them for training by using contrast limited adaptive histogram equalization [31] as was done in other works. Moreover, zero-padding is used to account for the size variance of the patient in the X-ray image, therefore we cropped the body part of the locally collected image as shown in Fig. 2.
Synthetic Catheter Generation
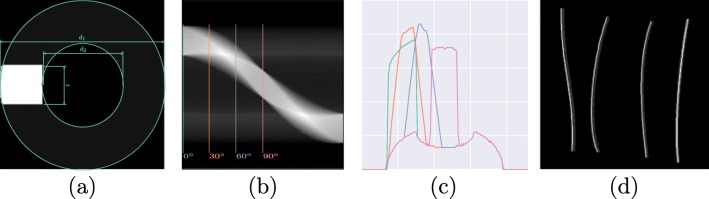
Catheters are essentially tubular objects, a portion of which is made of radiopaque material with a higher attenuation component designed for ease of detection. Figure 3a shows a simplified cross sectional profile. This profile would work for both NGTs and ETTs, as the only difference lies in the catheter width. Using a parallel beam geometry, the projected sinogram is obtained and shown in Fig. 3b. Figure 3c and d are the sampled projection profile at 0∘,30∘,60∘,90∘ and the synthetic catheters drawn with the corresponding profile. Note that the profile used to draw has to be resampled to accommodate the input image size. There are five parameters that are used to parameterize the simulated catheter: inner and outer catheter widths, d1 and d2; attenuation coefficients of the catheter and radioopaque material, c1 and c2; and the thickness of the strip, t. A similar approach is used for UVCs and UACs but with a profile of dual edges. The tracing of the catheter was simulated using a B-spline with control points randomly generated on the image. De Boor’s algorithm [6] was employed for the generation and the generated line was then rasterized with Xiaolin Wu’s antialiasing line drawing algorithm [27]. Implementation details can be found in “Implementation Details”.
Fig. 3.
Simulation of catheters in 2D. a Simulated cross section profile. b Projection profile from 0∘ to 180∘. c: Projection profile sampled at 0∘,30∘,60∘,90∘. d Simulated catheter trace in 2D with the corresponding profile in (c)
Text Mask Generation
Our initial experiments showed that training with just synthetic lines would cause confusion for radiopaque markers (letters) which may occasionally be noted on radiographs and also share line like structures. Therefore, we explicitly created another class for text so that its misclassification can be penalized independently. For the sake of simplicity, we cropped the common text from the pediatric X-rays and randomly scaled and merged with the adult chest X-ray.
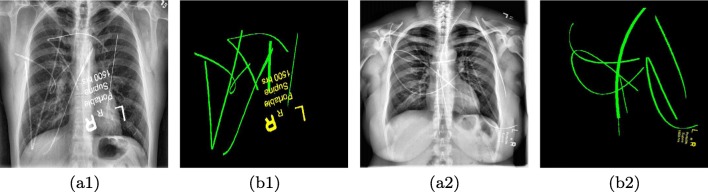
The generated catheter and text are then added to the adult chest X-ray with a weight sampled in the range of 0.15 to 0.35. Figure 4 shows two samples from our synthetic dataset.
Fig. 4.
Exemplar training image pairs for the proposed catheter detection network. (a1) and (a2) An adult chest X-ray with synthetic catheters overlaid on the image. (b1) and (b2) The annotation mask used for supervised training (best viewed in digital version)
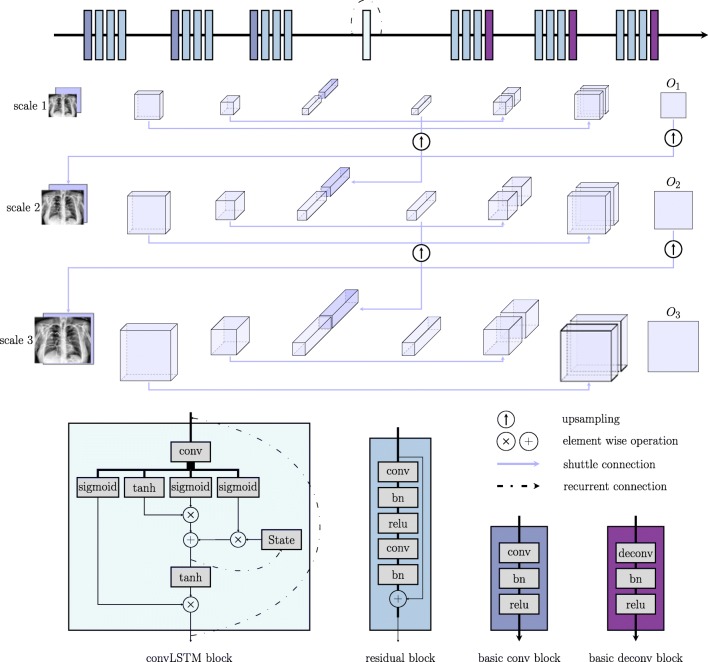
Network Architecture
Given an input image, the network has to learn to assign each pixel to one of three classes: background (cbg), catheter (ccatheter), and text (ctext). A scale recurrent neural network [25] was employed for this task. It is comprised of an encoder-decoder architecture with shuttle connections and recurrent modules. The encoder progressively increases the number of feature channels and decreases the spatial size (height, width) of the feature map to achieve a certain degree of translation invariance and save memory. The decoder in turn performs an inverse operation to gradually recover the size of the input. During the encoding and decoding process, every single pixel in the final output feature map contains information computed from a large portion of the image hence encodes the global information. The shuttle connection directly communicates lower level features to the higher level so that the network can make final predictions based on a fusion of both local and global cues. The network is fully convolutional thus can accept images of different scales. Input of increasing scale was sent to the network at different time steps. The recurrent module takes the form of convolutional long-short term memory (convLSTM) [28]. It takes concatenated inputs from the current and previous scale. To maintain size compatibility, we upscaled the feature maps from the previous scale with strided convolution.
Figure 5 provides a general overview of this architecture. Residual block was used to facilitate the training process. Both the skip connection and the residual block [11] benefits training by making the gradient propagate more easily through the network.
Fig. 5.
Overview of the network architecture. Note that for the last deconv block, bn and relu were replaced with a softmax layer to get a multi-channel likelihood map
Training Objective
The output of the network is a multi-channel feature map with the number of channels equal to the number of predicted classes. We normalized the feature maps with a softmax function so that each channel of the map can be interpreted as the likelihood of belonging to each class. Cross entropy (CE) loss was used to measure the difference between the output and the groundtruth. Loss at each scale was aggregated together as the final optimization objective, which can be expressed mathematically as:
| 1 |
where Oi is the output of the network at scale i and Gi is the corresponding groundtruth label map. m is the number of the scale and was chosen as 3 in this work. w is the weights to balance the unequal distribution of {cbg,ccatheter,ctext} and was chosen as 1, 40, and 80 respectively.
Experiment Setup
Implementation Details
The images from the Open-i dataset all have a width of 512. This size was found to be sufficiently large to discriminate different catheters. For NGTs and ETTs, d1 and d2 were selected as 160 and 80 while c1 and c2 were set as 0.1 and 1, and t was set to be 30 pixels. Note that in the current implementation, the size of d2 was not varied to cope with the width difference of NGTs and ETTs. For UACs and UVCs, only one projection profile at 0∘ was selected. The generated catheter width is 9, 9, and 6 pixels for NGTs, ETTs, UACs, and UVCs respectively, to accommodate image size. During training, the training image pairs were augmented with rotation (in the range of [− 60∘, 60∘]), horizontal flipping, and scale changes (in the range of [0.5, 1.1]) to generate random training image on the fly. Due to the scale change, the augmented images were cropped or padded to the size of 512 × 512. The testing images collected locally were all resized to a width of 480 and denoised using BM3D [4] with σ = 0.1.
The segmentation network was trained on the Cedar cluster of Compute Canada with a P100 GPU. Adam optimizer [35] with β1 = 0.9 and β2 = 0.999 was used for the optimization with a initial learning rate of 0.0001. The learning rate decayed by 0.1 every 10 epochs. The network was trained to convergence after 50 epochs. All training parameters were initialized with numbers drawn from a Gaussian distribution (0, 0.02). Batch size was chosen to be 2 due to the constraint of the GPU memory size. Since we have fixed the size of the network, the complexity of the network can be also regarded as O(N) where N is the number of pixels in the input X-ray image. In our experiment, the testing of each image can be accomplished in less than 1s when deployed on a single GPU.
Evaluation Metrics
In the evaluation, pixels of background and text were treated as the negative class and pixels of catheter were treated as the positive class. Since the class is highly imbalanced, precision and recall were computed with each expressed mathematically as:
| 2 |
where TP,TN,FP,and FN represents the number of true positives, true negatives, false positives, and false negatives respectively. The threshold for computing the precision and recall curve was sampled within the range of 0 to 255 at an interval of 30.
Another measure we used for the evaluation is the weighted harmonic mean of precision and recall (or Fβ-measure) which is defined as:
| 3 |
where β2 is a weighting term and was set as 0.3 to weight precision more than recall as in [1]. The threshold in calculating the corresponding precision and recall was an image dependent value defined as:
| 4 |
where, W,H are the width and height of the obtained catheter likelihood map Ok (assuming at the k-th channel of the network output).
Experiments
No prior method is applicable to detect all the catheters of interest, therefore we only compared our method with another deep learning approach which used fcn8s [18] for PICC line tip detection [16]. Further, in order to demonstrate the effectiveness of the recurrent module, we trained another network termed w/oR with the recurrent module removed under the exact same settings. This network resembles the typical UNet-style network used in [2] .
Results and Discussion
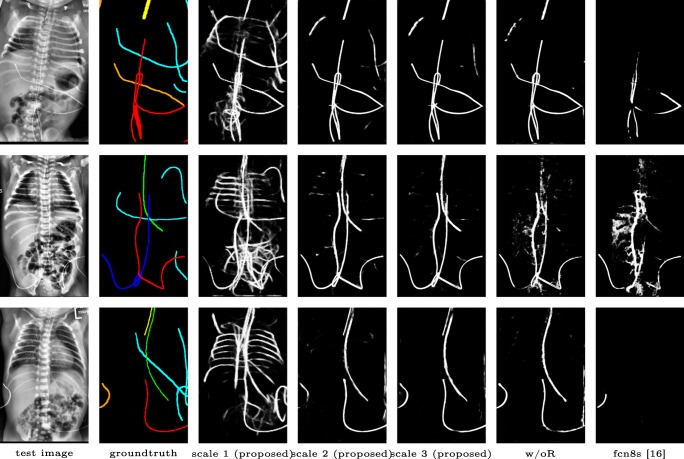
Qualitative visual examples of the raw catheter likelihood maps obtained directly from the network without any postprocessing are shown in Fig. 6. It can be seen that the proposed network at the highest scale (scale 3) achieves the best visual appearance as compared to the other methods. The maps from the proposed network at scale 2 and scale 3 look much cleaner than w/oR and fcn8s. We would attribute this to the iterative refinement of the detection results by using the recurrent module. When comparing results from the proposed network at different scales, we can see that the likelihood map from the smallest scale contains almost all line-like structures, including not only catheters but also ribs and ECG leads. This is because catheters, ribs, and ECG leads look similar at a smaller scale. These irrelevant line-like structures are gradually filtered out in higher scales because catheters, especially UVCs and UACs, begin to appear as two parallel edges whereas ribs and ECG leads continue to appear as a single solid line. One way of interpreting the immediate outputs at the two lower scales is by treating them as attention maps. The network draws its attention sequentially to the candidate line regions so that better feature representations can be learnt.
Fig. 6.
Raw catheter likelihood maps for different networks on test images: proposed, w/oR, and fcn8s (best viewed in digital version). The outputs at scale 1 and 2 can be treated as soft attention maps that the network has gradually learnt in detecting the interested catheter
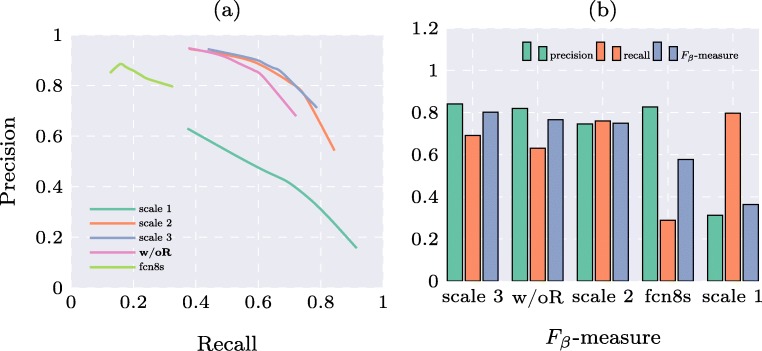
Precision and recall curves are shown in Fig. 7a and Fβ-measures computed from adaptive threshold are shown in Fig. 7b. Note that before computing these quantitative measures, the obtained binary map underwent morphological operations to filter out small irregular regions. It can be seen that the proposed method achieves higher precision and recall than fcn8s [16] (previously used for PICC line tip detection [16]) and w/oR (which resembles the typical UNet-style network used in [18]). The results at lowest scale have the highest recall but lowest precision. The results at the two higher scales achieve approximately the same performance. The reason we believe is that even though there are some improvements in the raw likelihood map, the middle scale has already detected all major parts of the catheter. The local refinement is too small to be manifested in the quantitative measures. Nonetheless, the Fβ-measure for the proposed method at the highest scale ranks the first among the comparators.
Fig. 7.
Quantitative results for different methods on the pediatric X-ray test set. a Precision and recall curves. bFβ-measures (methods ordered according to the value of the Fβ-measure)
Limitations
Catheters are represented as thin lines of just a few pixels wide on X-ray images. Due to a certain degree of error inherent to manually annotating catheters on the images, a slight pixel shift in the groundtruth annotation could adversely impact the quantitative results. In the future, we believe our method could provide assistance to annotators by providing initial annotations of potential lines for subsequent manual review.
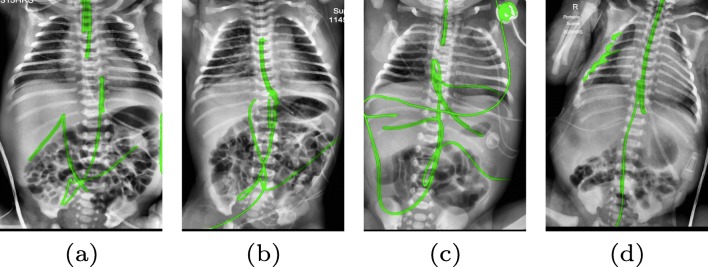
There are certain situations where our proposed method would fail. Figure 8a and b show a partially detected NGT. This mostly likely resulted from the decreased visibility of the radiopaque strip. Figure 8a also shows another failure situation where the inferior portion of the UVC is occluded by the abdomen. Figure 8c shows the case of a falsely detected unidentified line and Fig. 8d shows part of the lateral aspect of the rib cage falsely identified as a catheter. All the failure cases can be partially explained by the domain variance of the training and testing data. All the training images are on the adult chest X-ray images; thus, the network does not know how to handle the case where a catheter is projected over the abdomen. False positives on case (c) are all objects extraneous to the patient that cannot be found on the adult training images. There are not enough negative samples to train the segmentation network to differentiate these objects with the catheters of interest . A possible solution to mitigate this is to directly synthesize catheters on pediatric X-rays to avoid the domain shift. This is an area where further work remains.
Fig. 8.
Typical failure cases. a and b partially detected NGT possibly due to its similarity to ECG leads. Occlusion of UVC. c Confusion with other lines on the X-ray image. d Confusion with the lateral aspect of the rib cage (best viewed in digital version)
Conclusion
In this work, we have proposed the first deep learning system for catheter detection on pediatric chest/abdomen X-rays. The system is capable of detecting umbilical catheters, NGTs and ETTs in less than 1s when deployed on a single GPU. The system was trained on a dataset with synthetic catheters and has achieved promising detection results with Fβ = 0.8. The proposed scale recurrent network is able to utilize multi-scale information and learn to gradually draw its attention from general line structures to the catheters of interest. Further, our approach of synthesizing catheters on X-ray images by exploiting the fact that catheters are essentially tubular structures with various cross sectional profiles can lead to the efficient development of training datasets. Although we have experimented with only pediatric X-rays, we believe the methodology should be also applicable to adult X-rays provided the profile is carefully designed with consideration given to the large variation of catheter and wire types. The approach described in this work may contribute to the development of a system to detect and assess the placement of catheters on X-ray images, thus providing a solution to triage and prioritize X-ray images which have potentially malpositioned catheters for a radiologist’s urgent review. In the future, an automated catheter placement evaluation system may also be used to prepopulate draft radiology reports with text describing the catheters and tubes present on an X-ray image. This may increase the efficiency of radiology reporting. Computer-aided catheter placement evaluation, as an emerging area of research, is still in its infancy. It is our hope that this study can be beneficial towards building a general catheter placement evaluation system.
Footnotes
Our code is available at https://github.com/xinario/catheter_detection.githttps://github.com/xinario/catheter_detection.git.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Achanta R, Hemami S, Estrada F, Susstrunk S: Frequency-tuned salient region detection.. In: IEEE conference on computer vision and pattern recognition, 2009. cvpr 2009., IEEE, 2009, pp 1597–1604
- 2.Ambrosini P, Ruijters D, Niessen WJ, Moelker A, van Walsum T: Fully automatic and real-time catheter segmentation in x-ray fluoroscopy.. In: International conference on medical image computing and computer-assisted intervention, Springer, 2017, pp 577–585
- 3.Concepcion NDP, Laya BF, Lee EY. Current updates in catheters, tubes and drains in the pediatric chest: A practical evaluation approach. Eur J Radiol. 2017;95:409–417. doi: 10.1016/j.ejrad.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Dabov K, Foi A, Katkovnik V, Egiazarian K. Image denoising by sparse 3-d transform-domain collaborative filtering. IEEE Trans Image Process. 2007;16(8):2080–2095. doi: 10.1109/TIP.2007.901238. [DOI] [PubMed] [Google Scholar]
- 5.Dai W, Doyle J, Liang X, Zhang H, Dong N, Li Y, Xing EP Scan: Structure correcting adversarial network for chest x-rays organ segmentation, 2017. arXiv:170308770
- 6.De Boor C, De Boor C, Mathématicien EU, De Boor C, De Boor C. A practical guide to splines. New York: Springer-Verlag; 1978. [Google Scholar]
- 7.Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, Thoma GR, McDonald CJ. Preparing a collection of radiology examinations for distribution and retrieval. J Am Med Inform Assoc. 2015;23(2):304–310. doi: 10.1093/jamia/ocv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn D, Kinoshita H, Livingstone V, Dempsey EM. Optimal line and tube placement in very preterm neonates: An audit of practice. Children. 2017;4(11):99. doi: 10.3390/children4110099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frangi AF, Niessen WJ, Vincken KL, Viergever MA: Multiscale vessel enhancement filtering.. In: International conference on medical image computing and computer-assisted intervention, Springer, 1998, pp 130–137
- 10.Fuentealba I, Taylor GA. Diagnostic errors with inserted tubes, lines and catheters in children. Pediatr Radiol. 2012;42(11):1305–1315. doi: 10.1007/s00247-012-2462-7. [DOI] [PubMed] [Google Scholar]
- 11.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition.. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp 770–778
- 12.Hoellering AB, Koorts PJ, Cartwright DW, Davies MW. Determination of umbilical venous catheter tip position with radiograph. Pediatr Crit Care Med. 2014;15(1):56–61. doi: 10.1097/PCC.0b013e31829f5efa. [DOI] [PubMed] [Google Scholar]
- 13.Kao EF, Jaw TS, Li CW, Chou MC, Liu GC. Automated detection of endotracheal tubes in paediatric chest radiographs. Comput Methods Programs Biomed. 2015;118(1):1–10. doi: 10.1016/j.cmpb.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Keller BM, Reeves AP, Cham MD, Henschke CI, Yankelevitz DF: Semi-automated location identification of catheters in digital chest radiographs.. In: Medical imaging 2007: Computer-aided diagnosis, international society for optics and photonics, vol 6514, 2007, p 65141O
- 15.Kieran EA, Laffan EE, Odonnell CP: Estimating umbilical catheter insertion depth in newborns using weight or body measurement: a randomised trial. Archives of Disease in Childhood-Fetal and Neonatal Edition pp fetalneonatal–2014, 2015 [DOI] [PubMed]
- 16.Lee H, Mansouri M, Tajmir S, Lev M H: A deep-learning system for fully-automated peripherally inserted central catheter (picc) tip detection. Journal of digital imaging, 1–10, 2010 [DOI] [PMC free article] [PubMed]
- 17.Liskowski P, Krawiec K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans Med Imaging. 2016;35(11):2369–2380. doi: 10.1109/TMI.2016.2546227. [DOI] [PubMed] [Google Scholar]
- 18.Long J, Shelhamer E, Darrell T: Fully convolutional networks for semantic segmentation.. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp 3431–3440 [DOI] [PubMed]
- 19.Mercan CA, Celebi MS. An approach for chest tube detection in chest radiographs. IET Image Process. 2013;8(2):122–129. doi: 10.1049/iet-ipr.2013.0239. [DOI] [Google Scholar]
- 20.Ramakrishna B, Brown M, Goldin J, Cagnon C, Enzmann D: Catheter detection and classification on chest radiographs: an automated prototype computer-aided detection (cad) system for radiologists.. In: Medical imaging 2011: Computer-aided diagnosis, international society for optics and photonics, vol 7963, 2011, p 796333
- 21.Ramakrishna B, Brown M, Goldin J, Cagnon C, Enzmann D: An improved automatic computer aided tube detection and labeling system on chest radiographs.. In: Medical imaging 2012: Computer-aided diagnosis, international society for optics and photonics, vol 8315, 2012, p 83150R
- 22.Sheng C, Li L, Pei W. Automatic detection of supporting device positioning in intensive care unit radiography. Int J Med Rob Comput Assisted Surg. 2009;5(3):332–340. doi: 10.1002/rcs.265. [DOI] [PubMed] [Google Scholar]
- 23.Soares JV, Leandro JJ, Cesar RM, Jelinek HF, Cree MJ. Retinal vessel segmentation using the 2-d gabor wavelet and supervised classification. IEEE Trans Med Imaging. 2006;25(9):1214–1222. doi: 10.1109/TMI.2006.879967. [DOI] [PubMed] [Google Scholar]
- 24.Staal J, Abràmoff MD, Niemeijer M, Viergever MA, Van Ginneken B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging. 2004;23(4):501–509. doi: 10.1109/TMI.2004.825627. [DOI] [PubMed] [Google Scholar]
- 25.Tao X, Gao H, Wang Y, Shen X, Wang J, Jia J: Scale-recurrent network for deep image deblurring, 2018. arXiv:180201770
- 26.Villalobos-Castaldi FM, Felipe-Riverȯn EM, Sȧnchez-Fernȧndez LP. A fast, efficient and automated method to extract vessels from fundus images. J Vis. 2010;13(3):263–270. doi: 10.1007/s12650-010-0037-y. [DOI] [Google Scholar]
- 27.Wu X: An efficient antialiasing technique.. In: Acm Siggraph Computer Graphics, ACM, vol 25, 1991, pp 143–152
- 28.Xingjian S, Chen Z, Wang H, Yeung DY, Wong WK, Woo WC : Convolutional lstm network: A machine learning approach for precipitation nowcasting.. In: Advances in neural information processing systems, 2015, pp 802–810
- 29.Yi X, Babyn P: Sharpness-aware low-dose ct denoising using conditional generative adversarial network. Journal of Digital Imaging, 2018. 10.1007/s10278-018-0056-0 [DOI] [PMC free article] [PubMed]
- 30.Yin B, Li H, Sheng B, Hou X, Chen Y, Wu W, Li P, Shen R, Bao Y, Jia W. Vessel extraction from non-fluorescein fundus images using orientation-aware detector. Med Image Anal. 2015;26(1):232–242. doi: 10.1016/j.media.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Zuiderveld K: Contrast limited adaptive histogram equalization. Graphics Gems, 474–485, 1994