Environmental exposure has a significant impact on human health. While some airborne fungi can cause life-threatening infections, the impact of environment on fungal spore dispersal and transmission is poorly understood. The democratization of shotgun metagenomics allows us to explore important questions about fungal propagation. We focus on Pneumocystis, a genus of host-specific fungi that infect mammals via airborne particles. In humans, Pneumocystis jirovecii causes lethal infections in immunocompromised patients if untreated, although its environmental reservoir and transmission route remain unclear.
KEYWORDS: airborne microorganisms, epidemiology, host-parasite relationship, metagenomics, opportunistic fungi
ABSTRACT
Environmental exposure has a significant impact on human health. While some airborne fungi can cause life-threatening infections, the impact of environment on fungal spore dispersal and transmission is poorly understood. The democratization of shotgun metagenomics allows us to explore important questions about fungal propagation. We focus on Pneumocystis, a genus of host-specific fungi that infect mammals via airborne particles. In humans, Pneumocystis jirovecii causes lethal infections in immunocompromised patients if untreated, although its environmental reservoir and transmission route remain unclear. Here, we attempt to clarify, by analyzing human exposome metagenomic data sets, whether humans are exposed to different Pneumocystis species present in the air but only P. jirovecii cells are able to replicate or whether they are selectively exposed to P. jirovecii. Our analysis supports the latter hypothesis, which is consistent with a local transmission model. These data also suggest that healthy carriers are a major driver for the transmission.
OPINION/HYPOTHESIS
Exposure to environmental factors plays a key role in human health. However, the impact of biological and chemical particles in the environment on human health is poorly understood. High-throughput sequencing of DNA is a powerful approach to identify fungal pathogens in environmental sources (1). Fungi represent a significant fraction of microorganisms identified in environmental metagenomics data sets.
Pneumocystis species are opportunistic fungi that exclusively infect mammals, yet each species displays a high level of host specificity. As extracellular parasites, Pneumocystis organisms have been found almost exclusively in the alveolar space in the lungs of mammals (2). These organisms cause severe pneumonia in immunocompromised individuals and are also able to asymptomatically infect healthy individuals. In humans, Pneumocystis pneumonia (PCP) is caused by Pneumocystis jirovecii and is a serious public health issue with a high mortality rate if untreated (3). Renal transplant and untreated HIV-infected patients have an exceptionally high risk of developing PCP.
The Pneumocystis life cycle is thought to alternate primarily between vegetative active trophic forms and metabolically inert ascii (cysts) (4). Trophic forms are believed to replicate by asexual reproduction via binary fission, and ascii, by sexual reproduction (5). Pneumocystis organisms require a living host to survive and remain unculturable in vitro. The parasitic nature of their relationship with mammalian hosts is reflected in their genomes, which lack many genes involved in de novo biosynthesis of essential nutrients and, instead, encode mechanisms to scavenge nutrients from the host (we refer interested readers to references 2 and 6 and the references therein). Their metabolic machinery seems ill-equipped to survive outside their hosts; that is, viable trophic forms are unlikely to be found in the environment.
Pneumocystis infection is mediated by transmission of ascii (7) between an infected host and a susceptible host (8, 9). Although Pneumocystis DNA has been reported to be in the environment (see reference 2), it remains unclear if and for how long a Pneumocystis cell can remain viable outside its host.
The epidemiology of Pneumocystis infection is poorly understood and has been almost exclusively studied in humans in the context of drug resistance or outbreaks. Most of the studies utilize a multilocus sequencing approach targeting a few mitochondrial and nuclear genes. The structure of P. jirovecii populations is ambiguous. Some studies have reported a strong local population structure mediated by clusters of infected patients (10); however, this is not supported by other studies (11). These conflicting results could be explained by the presence of viable ascii in the environment over extended periods of time. If Pneumocystis ascii are able to survive in the environment, spores could disperse over large geographical distances. In such a scenario, individuals would be constantly infected with new strains, therefore creating highly heterogeneous populations, which would be consistent with the observed high prevalence of coinfections with multiple P. jirovecii strains (11, 12). Pneumocystis ascii share similarities with other long-distance fungal dispersers, for example, the presence of glucans in the cell wall, which likely promote survival in a harsh environment. The scenario of a long-distance dispersal of Pneumocystis ascii has not been explored to date. If Pneumocystis ascii are ubiquitous, then humans should simultaneously be exposed to different Pneumocystis species, but only P. jirovecii cells would be able to replicate. Under this model, we should expect a widespread coexistence of ascii from different Pneumocystis species in the environment.
Although patients with PCP have high organism burdens which likely facilitate transmission, healthy carriers also potentially play a role. The transmission of Pneumocystis through immunocompetent individuals has been demonstrated in animal models (13). In this situation, Pneumocystis organisms are able to actively infect new hosts and complete their life cycle with the ability to release newly formed ascii before the host immune system clears the infection. Assuming that recurrent infections are frequent, healthy individuals would be the primary source of ascii. In that case, we should expect a high density of P. jirovecii ascii in the vicinity of humans, irrespective of the presence of immunocompromised individuals.
A recent study published in Cell (14) exploring the dynamics of the human environmental exposome provided an opportunity to investigate our hypotheses. The authors collected and analyzed metagenomic sequences from air filter devices attached to 15 self-declared healthy adults. These individuals were followed for up to 890 days over 66 different geographical locations. The data set is available as 594 SRA files in the NCBI SRA database (∼5 TB of raw data). We also utilized data from two additional metagenomics studies from surface swabs across New York City (NYC) subway systems, the Gowanus Canal, and public parks (1,573 SRA files) (15) and the collection of 63 uncultured hospital air samples collected over a 6-month period (6,888 SRA files) (16). Here, we show clear evidence of P. jirovecii, but no other Pneumocystis species, in air filters from 4 out of 15 individuals in the human exposome study (14), whereas no evidence of Pneumocystis was identified in the other two studies, possibly because the sampling methods were not suitable for Pneumocystis detection. The significance of this finding is discussed in the context of epidemiology and host species specificity.
SUMMARY OF PERSONAL ENVIRONMENTAL EXPOSURE SAMPLE COLLECTION
We reproduce here a summary of the sampling methodology of the original exposome study (14). All 15 participants recruited lived in the northern California Greater Bay Area, and some traveled to other locations. All participants were age 18 or older and self-reported as healthy. A MicroPEM v3.2 personal exposure monitor (RTI International, Research Triangle Park, NC, USA) was modified and used to simultaneously collect biotic and abiotic samples of personal aerosol exposure for the period of August 2014 to January 2017. Before sampling and data collection, a 3.0-mm pore-size polytetrafluoroethylene (PTFE) 25-mm Teflon filter (Pall Corporation, Port Washington, NY, USA) or a 0.8-mm pore-size polyethersulfone (PES) 25-mm filter (Sterlitech, Kent, WA, USA) was placed in MicroPEM filter cassettes to collect aerosol particulates for biotics extraction. Participants were instructed to either carry the monitor on their arm or place the monitor near them within a 2-m radius at all times during the sampling period. At the end of the sampling and monitoring period, filters and cartridges were removed from the monitor and stored at –80°C until analysis. None of the individuals were health care workers or involved in animal models of PCP.
Detection of Pneumocystis in air metagenomes.
Raw data were download from NCBI (last accessed June 2019) and mapped to the following genomes using Bowtie 2 (17): nuclear genomes of three P. jirovecii strains (RU7 [18], SE8 [19], and SE2178 [20]); single strains of Pneumocystis sp. macacae that infect macaques (O. H. Cissé, L. Ma, J. Brenchley, and J. A. Kovacs, unpublished), Pneumocystis carinii that infect rats (18), and Pneumocystis murina that infect mice (18); and mitochondrial genomes of P. jirovecii, P. carinii, and P. murina (21).
Mapped reads were assembled using metaSPades (22). Small-size contigs (<500 nucleotides) and/or those with a high GC content (>50%; because Pneumocystis genomes have a GC content of ∼30%) were separated. Analysis of small-size and/or high-GC content contigs indicated that they were heavily enriched in ambiguous ribosomal DNA (rDNA) sequences and low complexity repeats, the vast majority of which could not be reliably categorized. The remaining contigs were aligned to the reference genome sequences using BLAT (23) with a minimum nucleotide alignment score of 200 as the threshold. Ribosomal DNA and low-complexity sequences were excluded because of their high rates of false positivity. A total of 1,711 contigs were retained for further investigation. Contigs were annotated with AUGUSTUS (24) using Pneumocystis built-in models (25). Orthology was inferred using the reciprocal best BLASTn hit (26) with an E value of 10−10 as the cutoff. For each orthologous group, sequence orientations were assessed using Revseq from the EMBOSS package (27), aligned using MUSCLE (28), and visually inspected for alignment inconsistencies using AliView (29). Maximum likelihood phylogenies were inferred using RAxML-NG (30) with 100 replicates as support values. Pairwise nucleotide identities were calculated using Water from EMBOSS.
Variant calling was performed by aligning retrieved reads from the NCBI SRA to P. jirovecii genome assembly strain RU7 (NCBI accession number GCF_001477535.1) using the Burrows-Wheeler Aligner MEM algorithm (BWA-MEM) (31). To assess the levels of different mammal DNAs, Illumina raw reads were aligned using BWA-MEM to the following genomes: NCBI accession numbers GCF_000001405.39 (human), GCF_000002285.3 (dog), GCF_000181335.3 (cat), GCF_000151735.1 (guinea pig), GCF_000001895.5 (rat), and GCF_000001635.25 (mouse). Alignment files (BAM) were filtered and sorted with SAMtools (32) using “-f 3 -F 4 -F 8 -F 256 -F 2048 -q 30” to keep high-quality reads (mapping quality ≥ 30 and properly paired). Duplicates were marked using Picard MarkDuplicates v2.1.1 (http://broadinstitute.github.io/picard). Per base genome, sequencing depths (coverage breaths) were computed from BAM files using BEDTools (33) with the “genomeCoverageBed” option. Variants were identified using FreeBayes v1.3.1-dirty (34) with the parameters “–ploidy 1” and vcffilter “-f QUAL > 30.” Variant positions were annotated using BEDTools with the “annotate” option. Analyses were conducted with Perl v5.22.0 scripts (http://www.perl.org/) using Bioperl v1.7.6 (35). Statistical analyses were conducted in R v3.3.2 (36). Hierarchical clustering was done using pheatmap v1.0.12 (37). Phylogenetic trees were visualized using FigTree v1.4.4 (38). Pipelines were written using Snakemake (39). All analyses, including alignments, accessions, and custom scripts are available at https://github.com/ocisse/Exposome.
Pneumocystis organisms are present in the human exposome.
We identified Pneumocystis reads in 24 of the 594 SRA files provided by the human exposome study (14). These 24 SRA files represent air filters from 4 of 15 individuals (Table 1). We found no conclusive evidence of Pneumocystis presence in the metagenomic data sets from the surface swabs of the NYC subway system (15) or air samples from hospitals (16). The latter study used filters to capture particles between 0.1 and 10 μm in size, which should capture Pneumocystis ascii if they are present. However, the depth of sequencing appears lower than for those of other data sets. Only data from the human exposome study (14) were used for subsequent analyses.
TABLE 1.
Proportion of Pneumocystis and selected mammal DNA in air filter metagenomes
| SRA accession no. | Sourcea | Dateb (M/D/Y) | No. of paired-end reads | Proportion of genome bases covered by at least two sequencing reads (2× coverage) (%)c |
Geographic location | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P. jirovecii | Human | Dog | Cat | Guinea pig | Rat | Mouse | |||||
| SRR6399753 | P1 | 10/29/14 | 46,823,961 | 0.16 | 0.05 | 0.02 | 0.02 | 0.01 | 0.02 | 0.01 | Cambridge, MA, USA |
| SRR6399562 | P1 | 11/6/14 | 81,370,159 | 0.77 | 0.09 | 0.05 | 0.06 | 0.04 | 0.05 | 0.01 | Bethesda, MD, USA |
| SRR6399671 | P1 | 2/1/15 | 57,429,806 | 0.51 | 0.04 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | San Francisco, CA, USA |
| SRR6399870 | P1 | 8/2/15 | 49,422,187 | 0.37 | 0.06 | 0.04 | 0.03 | 0.02 | 0.01 | 0.01 | Palo Alto, CA, USA |
| SRR6399877 | P1 | 8/11/15 | 64,558,662 | 0.67 | 0.10 | 0.08 | 0.08 | 0.07 | 0.06 | 0.05 | Palo Alto, CA, USA |
| SRR6399596 | P1 | 10/3/15 | 66,441,164 | 0.09 | 0.06 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | Lexington, KY, USA |
| SRR6399808 | P1 | 12/3/15 | 45,397,952 | 0.18 | 0.03 | 0.01 | 0.01 | 0.007 | <0.01 | <0.01 | Bethesda, MD, USA |
| SRR6399953 | P1 | 1/2/16 | 94,139,119 | 2.53 | 1.25 | 0.40 | 0.37 | 0.18 | 0.19 | 0.16 | West Hartford, CT, USA |
| SRR6399569 | P1 | 1/7/16 | 87,515,965 | 1.04 | 0.16 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | San Diego, CA, USA |
| SRR6399527 | P1 | 1/10/16 | 68,493,408 | 0.28 | 0.04 | 0.02 | 0.02 | 0.01 | 0.01 | <0.01 | Los Angeles, CA, USA |
| SRR6399888 | P1 | 4/6/16 | 71,048,240 | 0.58 | 0.61 | 0.24 | 0.24 | 0.07 | 0.04 | 0.04 | Pottstown, PA, USA |
| SRR6399614 | P1 | 5/12/16 | 56,634,040 | 0.42 | 0.07 | 0.01 | 0.01 | 0.03 | 0.04 | 0.02 | Cold Spring Harbor, NY, USA |
| SRR6399464 | P1 | 6/17/16 | 75,614,307 | 0.51 | 0.08 | 0.06 | 0.06 | 0.04 | 0.05 | 0.04 | San Diego, CA, USA |
| SRR6399789 | P1 | 11/28/16 | 61,447,835 | 1.59 | 0.37 | 0.11 | 0.11 | 0.09 | 0.07 | 0.08 | Palo Alto, CA, USA |
| SRR6399684 | P1 | 12/5/16 | 69,122,523 | 1.68 | 0.23 | 0.30 | 0.23 | 0.09 | 0.11 | 0.12 | Palo Alto, CA, USA |
| SRR6399520 | P1 | 1/2/17 | 66,800,060 | 0.65 | 0.19 | 0.14 | 0.10 | 0.04 | 0.05 | 0.04 | West Hartford, CT, USA |
| SRR6399939 | P3 | 3/7/16 | 51,995,194 | 2.00 | 0.61 | 0.08 | 0.08 | 0.05 | 0.04 | 0.04 | Redwood City, CA, USA |
| SRR6399879 | P3 | 3/29/16 | 82,285,688 | 1.13 | 1.45 | 0.23 | 0.24 | 0.30 | 0.13 | 0.12 | San Mateo, CA, USA |
| SRR6399803 | P3 | 11/28/16 | 67,067,766 | 1.07 | 0.42 | 0.12 | 0.11 | 0.20 | 0.12 | 0.09 | San Mateo, CA, USA |
| SRR6399681 | P3 | 12/5/16 | 56,853,387 | 0.56 | 0.18 | 0.06 | 0.11 | 0.09 | 0.12 | 0.04 | San Mateo, CA, USA |
| SRR6399685 | P3 | 12/11/16 | 62,867,255 | 0.61 | 0.11 | 0.09 | <0.01 | 0.18 | 0.08 | 0.07 | New York, NY, USA |
| SRR6399802 | P5 | 11/27/16 | 54,602,220 | 0.31 | 0.13 | 0.04 | 0.04 | 0.03 | 0.03 | 0.03 | Cupertino, CA, USA |
| SRR6399682 | P5 | 12/6/16 | 62,121,563 | 0.53 | 0.09 | 0.06 | 0.05 | 0.12 | 0.04 | 0.04 | Cupertino, CA, USA |
| SRR6399512 | P8 | 2/23/16 | 59,583,085 | 0.35 | 0.20 | 0.04 | 0.04 | 0.02 | 0.02 | 0.01 | Palo Alto, CA, USA |
Identification codes of the tracked individuals.
Month/day/year.
Genome sizes: P. jirovecii, 8.3 Mb; human, 3.2 Gb; dog, 2.4 Gb; cat, 2.5 Gb; guinea pig, 2.7 Gb; rat, 2.8 Gb; mouse, 2.8 Gb.
Humans seem to be selectively exposed to Pneumocystis jirovecii.
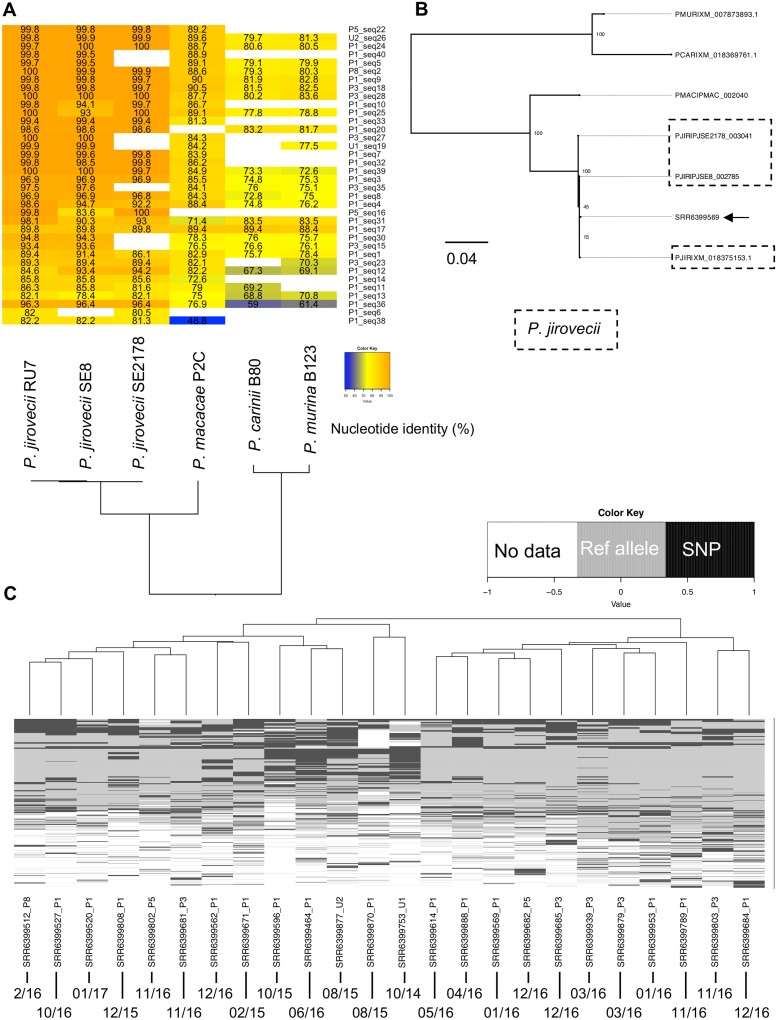
The de novo assembly of Pneumocystis reads retrieved from 24 SRA files yielded 45 contigs larger than 500 nucleotides (Table 1). Maximum likelihood phylogeny and pairwise alignment identity scores for 37 of 45 contigs with their one-to-one ortholog unambiguously identified all contigs as being from P. jirovecii and clearly distinct from all other Pneumocystis species analyzed (Fig. 1 A and B). The remaining eight contigs, which lack homologs that are present in other Pneumocystis species, were clearly identified as P. jirovecii based on pairwise alignments. This was a somewhat surprising finding given that multiple Pneumocystis species are distributed in the ambient air surrounding mammals. While Pneumocystis species show a strict host species specificity for productive infection, air sampling does not depend on replication of the organism but should randomly sample the proximal ambient air. Given that Pneumocystis species appear unable to replicate outside the mammalian host, including in air filters, the only source of aerosolized organisms appears to be an infected host of the same species. While Pneumocystis can cause pneumonia in immunosuppressed hosts, it can also lead to subclinical infection or colonization in immunocompetent hosts, and sampling of, e.g., wild rodents demonstrates the presence of Pneumocystis DNA in ∼30 to 90% of animals (40, 41). None of the individuals involved in the study were reported to have an immunodeficiency, although we cannot rule out undisclosed medical conditions or contact with an infected immunocompromised patient. We have information that at least some individuals included in the study had close contact with animals such as pets, laboratory animals, or zoo animals (see https://www.technologynetworks.com/proteomics/news/exposing-the-exposome-309789).
FIG 1.
Evidence of Pneumocystis DNA in air metagenomes. (A) Pairwise nucleotide identity heatmap of 37 of 45 contigs. Only contigs for which a 1-to-1 ortholog could be established were used. Blank spaces in the heatmap represent a lack of an identifiable ortholog. These missing orthologs likely represent an incomplete genome or annotation rather than real biological gene loss. (B) Maximum likelihood phylogeny of a representative contig encoding the 3-oxoacyl-[acyl-carrier-protein] synthase (arrow). The contig was recovered from the NCBI SRA accession number SRR6399569. Bootstrap values of 100 replicates are presented on the tree nodes. Sequences are labeled with the following species identifiers: PJIR, Pneumocystis jirovecii; PMAC, Pneumocystis sp. macacae; PCAR, P. carinii; PMUR, P. murina. P. jirovecii sequences are boxed. (C) Single nucleotide polymorphisms (SNP) were identified from the alignments of raw reads from each SRA file to P. jirovecii reference genome assembly strain RU7. The heatmap shows the variant calling results for 1,852 genomic positions encoded as follows: black, presence of a SNP; gray, region covered by at least two sequencing reads with the allele as the reference genome; white, absence of coverage. Each row represents a unique SNP, and each column represents the 15 samples with detectable Pneumocystis sequences. Collection dates (month/year) are presented at the bottom of the figure.
Dog and cats cannot be infected by P. jirovecii, but they can be infected by genetically distinct Pneumocystis species. Pneumocystis pneumonia is rare in dogs (42), although severe cases with development of spontaneous pneumomediastinum have been reported (43, 44). The agent Pneumocystis sp. canis is phylogenetically related but clearly distinct from P. jirovecii and other Pneumocystis species (45). The Pneumocystis species that infects cats is referred as Pneumocystis sp. catus and constitutes another distinct species (46). While spontaneous PCP has not been observed in cats, PCP is frequently found in feline immunodeficiency virus-infected (47) or corticoid-treated cats (48). The low occurrence of PCP in healthy dogs and cats suggests that their immune system clears or maintains Pneumocystis organism loads at a low level. Therefore, healthy animals are expected to be colonized or infected subclinically, similar to healthy humans. Given that the phylogenetic placement of both Pneumocystis sp. canis and Pneumocystis sp. catus is known and that the nucleotide divergence of P. jirovecii with these two species exceeds 16% (46, 49), our unbiased approach would have recovered sequences from these species if present, even in the absence of complete genome sequences. If present, Pneumocystis sp. canis and Pneumocystis sp. catus sequences would be easy to identify in a phylogenetic tree because they will not cluster with any of the species used here and, instead, have their own branches located between primate Pneumocystis species (P. jirovecii and Pneumocystis sp. macacae) and rodent Pneumocystis species (P. carinii and P. murina).
Unlike other fungi that can replicate in the environment (e.g., in soil) and for which even a single organism has the potential to replicate wherever it lands, a Pneumocystis cyst, the infective form of the organism, must be inhaled and presumably deposited into the alveoli to initiate productive infection. We found high levels of mammal DNA, including that of humans and animals, in the air metagenomes where P. jirovecii DNA was detected (Table 1), which suggest that other Pneumocystis species would have been detected if present. The absence of non-P. jirovecii sequences in the human exposome suggests that Pneumocystis cysts can travel for only a limited range from the infected host, or if they travel long distances, the likelihood of being inhaled (or sampled in the current study) diminishes very rapidly. Thus, efficient transmission likely requires close proximity to an infected host (50). The fact that none of the filters contained other species of Pneumocystis strengthens the likelihood of a selective exposure of humans to P. jirovecii, though the limited number of species included in our database represents a potential caveat.
Participants were instructed to either carry the monitor on their arm or place the monitor near them within a 2-m radius at all times during the sampling period. Assuming that the filter was worn on the upper arm at all times, it is unlikely that this would create a significant barrier to the deposition of Pneumocystis from four-legged mammals, particularly from smaller animals. The reason is that experimental data from animal models suggest that Pneumocystis ascii remain in suspension in the ambient air. Noninfected rats acquired P. carinii and developed PCP when exposed to the exhaled air from infected rats (51, 52). In addition, Pneumocystis ascii from different species are of similar size, 3.5 to 8 μm (reviewed in reference 53), which suggests similar ascii dispersion capabilities, although interspecies differences cannot be ruled out. At the time of this writing, we are not aware of any study that has compared ascii diffusion properties in multiple Pneumocystis species. Another confounding factor is that P. jirovecii populations would be expected to outnumber those of other Pneumocystis populations in human-dense areas. This would allow a higher recovery yield for P. jirovecii versus other Pneumocystis species but should not influence the recovery of other species.
To investigate whether the air filters were seeded with a single strain or multiple strains of P. jirovecii, we aligned raw reads to P. jirovecii reference genome assembly strain RU7 (19) and identified single nucleotide polymorphisms (SNP). We filtered out singletons and analyzed 1,852 SNP positions. SNP profiles from different air filters appear to be heterogeneous, which indicates a wide dispersion of samples from individuals (Fig. 1C) and could reflect the differences in sampling times or exposure to different infected hosts. In addition, 115 of 1,852 positions overlap genomic regions encoding highly polymorphic major surface glycoproteins (Msg), which would presumably further enhance the genetic heterogenicity among isolates. In fact, the hypervariability of msg genes has been extensively used to study strain variation of P. jirovecii (54, 55). Overall, the source of the Pneumocystis that was sampled cannot be determined, but it presumably was either the volunteer or someone else in close proximity, which would be consistent with the dynamics of Pneumocystis air shedding (8, 56). Whatever the source, the frequent detection of Pneumocystis DNA suggests frequent shedding of organisms, which is consistent with serologic data suggesting that most humans have been infected by Pneumocystis at an early age (57, 58).
Concluding remarks.
Our observations highlight the potential impact of metagenomics to improve our understanding of fungal pathogen transmission and epidemiology. Our results support a short-range transmission model and suggest that close-range exposure to infected hosts may play a significant role in transmission of Pneumocystis. Analyses of publicly available metagenomic data sets provide new opportunities for modeling and exploring human exposure to airborne infectious pathogens.
ACKNOWLEDGMENTS
We thank Lisa Bishop, Shelly Samet, and Yueqin Liu for productive discussions.
This study used the Office of Cyber Infrastructure and Computational Biology (OCICB) high-performance computing (HPC) cluster at the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD.
This project was funded in whole or in part with federal funds from the Intramural Research Program of the United States National Institutes of Health Clinical Center. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Citation Cissé OH, Ma L, Jiang C, Snyder M, Kovacs JA. 2020. Humans are selectively exposed to Pneumocystis jirovecii. mBio 11:e03138-19. https://doi.org/10.1128/mBio.03138-19.
REFERENCES
- 1.Tong X, Xu H, Zou L, Cai M, Xu X, Zhao Z, Xiao F, Li Y. 2017. High diversity of airborne fungi in the hospital environment as revealed by meta-sequencing-based microbiome analysis. Sci Rep 7:39606. doi: 10.1038/srep39606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma L, Cisse OH, Kovacs JA. 2018. A molecular window into the biology and epidemiology of Pneumocystis spp. Clin Microbiol Rev 31:e00009-18. doi: 10.1128/CMR.00009-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Aliouat-Denis CM, Martinez A, Aliouat el M, Pottier M, Gantois N, Dei-Cas E. 2009. The Pneumocystis life cycle. Mem Inst Oswaldo Cruz 104:419–426. doi: 10.1590/s0074-02762009000300004. [DOI] [PubMed] [Google Scholar]
- 5.Hauser PM, Cushion MT. 2018. Is sex necessary for the proliferation and transmission of Pneumocystis? PLoS Pathog 14:e1007409. doi: 10.1371/journal.ppat.1007409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cisse OH, Hauser PM. 2018. Genomics and evolution of Pneumocystis species. Infect Genet Evol 65:308–320. doi: 10.1016/j.meegid.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Cushion MT, Linke MJ, Ashbaugh A, Sesterhenn T, Collins MS, Lynch K, Brubaker R, Walzer PD. 2010. Echinocandin treatment of pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLoS One 5:e8524. doi: 10.1371/journal.pone.0008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choukri F, Aliouat el M. M, Menotti J, Totet A, Gantois N, Garin YJ, Bergeron V, Dei-Cas E, Derouin F. 2011. Dynamics of Pneumocystis carinii air shedding during experimental pneumocystosis. J Infect Dis 203:1333–1336. doi: 10.1093/infdis/jir018. [DOI] [PubMed] [Google Scholar]
- 9.Le Gal S, Damiani C, Rouille A, Grall A, Treguer L, Virmaux M, Moalic E, Quinio D, Moal MC, Berthou C, Saliou P, Le Meur Y, Totet A, Nevez G. 2012. A cluster of Pneumocystis infections among renal transplant recipients: molecular evidence of colonized patients as potential infectious sources of Pneumocystis jirovecii. Clin Infect Dis 54:e62–e71. doi: 10.1093/cid/cir996. [DOI] [PubMed] [Google Scholar]
- 10.Alanio A, Gits-Muselli M, Guigue N, Desnos-Ollivier M, Calderon EJ, Di Cave D, Dupont D, Hamprecht A, Hauser PM, Helweg-Larsen J, Kicia M, Lagrou K, Lengerova M, Matos O, Melchers WJG, Morio F, Nevez G, Totet A, White LP, Bretagne S. 2017. Diversity of Pneumocystis jirovecii across Europe: a multicentre observational study. EBioMedicine 22:155–163. doi: 10.1016/j.ebiom.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parobek CM, Jiang LY, Patel JC, Alvarez-Martinez MJ, Miro JM, Worodria W, Andama A, Fong S, Huang L, Meshnick SR, Taylor SM, Juliano JJ. 2014. Multilocus microsatellite genotyping array for investigation of genetic epidemiology of Pneumocystis jirovecii. J Clin Microbiol 52:1391–1399. doi: 10.1128/JCM.02531-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alanio A, Gits-Muselli M, Mercier-Delarue S, Dromer F, Bretagne S. 2016. Diversity of Pneumocystis jirovecii during infection revealed by ultra-deep pyrosequencing. Front Microbiol 7:733. doi: 10.3389/fmicb.2016.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gigliotti F, Harmsen AG, Wright TW. 2003. Characterization of transmission of Pneumocystis carinii f. sp. muris through immunocompetent BALB/c mice. Infect Immun 71:3852–3856. doi: 10.1128/iai.71.7.3852-3856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Wang X, Li X, Inlora J, Wang T, Liu Q, Snyder M. 2018. Dynamic human environmental exposome revealed by longitudinal personal monitoring. Cell 175:277–291.e31. doi: 10.1016/j.cell.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afshinnekoo E, Meydan C, Chowdhury S, Jaroudi D, Boyer C, Bernstein N, Maritz JM, Reeves D, Gandara J, Chhangawala S, Ahsanuddin S, Simmons A, Nessel T, Sundaresh B, Pereira E, Jorgensen E, Kolokotronis S-O, Kirchberger N, Garcia I, Gandara D, Dhanraj S, Nawrin T, Saletore Y, Alexander N, Vijay P, Hénaff EM, Zumbo P, Walsh M, O’Mullan GD, Tighe S, Dudley JT, Dunaif A, Ennis S, O’Halloran E, Magalhaes TR, Boone B, Jones AL, Muth TR, Paolantonio KS, Alter E, Schadt EE, Garbarino J, Prill RJ, Carlton JM, Levy S, Mason CE. 2015. Geospatial resolution of human and bacterial diversity with city-scale metagenomics. Cell Syst 1:72–87. doi: 10.1016/j.cels.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King P, Pham LK, Waltz S, Sphar D, Yamamoto RT, Conrad D, Taplitz R, Torriani F, Forsyth RA. 2016. Longitudinal metagenomic analysis of hospital air identifies clinically relevant microbes. PLoS One 11:e0160124. doi: 10.1371/journal.pone.0160124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, Chen Z, Huang da W, Kutty G, Ishihara M, Wang H, Abouelleil A, Bishop L, Davey E, Deng R, Deng X, Fan L, Fantoni G, Fitzgerald M, Gogineni E, Goldberg JM, Handley G, Hu X, Huber C, Jiao X, Jones K, Levin JZ, Liu Y, Macdonald P, Melnikov A, Raley C, Sassi M, Sherman BT, Song X, Sykes S, Tran B, Walsh L, Xia Y, Yang J, Young S, Zeng Q, Zheng X, Stephens R, Nusbaum C, Birren BW, Azadi P, Lempicki RA, Cuomo CA, Kovacs JA. 2016. Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat Commun 7:10740. doi: 10.1038/ncomms10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cisse OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4:e00428-12. doi: 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid-Siegert E, Richard S, Luraschi A, Muhlethaler K, Pagni M, Hauser PM. 2017. Mechanisms of surface antigenic variation in the human pathogenic fungus Pneumocystis jirovecii. mBio 8. doi: 10.1128/mBio.01470-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Huang DW, Cuomo CA, Sykes S, Fantoni G, Das B, Sherman BT, Yang J, Huber C, Xia Y, Davey E, Kutty G, Bishop L, Sassi M, Lempicki RA, Kovacs JA. 2013. Sequencing and characterization of the complete mitochondrial genomes of three Pneumocystis species provide new insights into divergence between human and rodent Pneumocystis. FASEB J 27:1962–1972. doi: 10.1096/fj.12-224444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent WJ. 2002. BLAT: the BLAST-like alignment tool. Genome Res 12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanke M, Schoffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser PM, Burdet FX, Cisse OH, Keller L, Taffe P, Sanglard D, Pagni M. 2010. Comparative genomics suggests that the fungal pathogen Pneumocystis is an obligate parasite scavenging amino acids from its host’s lungs. PLoS One 5:e15152. doi: 10.1371/journal.pone.0015152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 28.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997.
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrison E. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907.
- 35.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehvaslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Core Team. 2018. R: a language and environment for statistical computing, Vienna, Austria. https://www.R-project.org/.
- 37.Kolde R. 2012. pheatmap: pretty heatmaps. R. package. https://cran.r-project.org/web/packages/pheatmap/index.html.
- 38.Rambaut A. 2009. FigTree, a graphical viewer of phylogenetic trees. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, United Kingdom: https://github.com/rambaut/figtree/releases. [Google Scholar]
- 39.Koster J, Rahmann S. 2012. Snakemake: a scalable bioinformatics workflow engine. Bioinformatics 28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 40.Petruzela J, Bryja J, Bryjova A, Katakweba A, Sabuni C, Baird SJE, de Bellocq JG. 2019. Evolutionary history of Pneumocystis fungi in their African rodent hosts. Infect Genet Evol 75:103934. doi: 10.1016/j.meegid.2019.103934. [DOI] [PubMed] [Google Scholar]
- 41.Palmer RJ, Settnes OP, Lodal J, Wakefield AE. 2000. Population structure of rat-derived Pneumocystis carinii in Danish wild rats. Appl Environ Microbiol 66:4954–4961. doi: 10.1128/aem.66.11.4954-4961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sukura A, Saari S, Jarvinen AK, Olsson M, Karkkainen M, Ilvesniemi T. 1996. Pneumocystis carinii pneumonia in dogs: a diagnostic challenge. J Vet Diagn Invest 8:124–130. doi: 10.1177/104063879600800124. [DOI] [PubMed] [Google Scholar]
- 43.Ramsey IK, Foster A, McKay J, Herrtage ME. 1997. Pneumocystis carinii pneumonia in two Cavalier King Charles spaniels. Vet Rec 140:372–373. doi: 10.1136/vr.140.14.372. [DOI] [PubMed] [Google Scholar]
- 44.Weissenbacher-Lang C, Fuchs-Baumgartinger A, Klang A, Kneissl S, Pirker A, Shibly S, von Ritgen S, Weissenböck H, Künzel F. 2017. Pneumocystis carinii infection with severe pneumomediastinum and lymph node involvement in a Whippet mixed-breed dog. J Vet Diagn Invest 29:757–762. doi: 10.1177/1040638717710237. [DOI] [PubMed] [Google Scholar]
- 45.English K, Peters SE, Maskell DJ, Collins ME. 2001. DNA analysis of Pneumocystis infecting a Cavalier King Charles spaniel. J Eukaryot Microbiol Suppl:106S. doi: 10.1111/j.1550-7408.2001.tb00471.x. [DOI] [PubMed] [Google Scholar]
- 46.Danesi P, Corro M, Falcaro C, Carminato A, Furlanello T, Cocchi M, Krockenberger MB, Meyer W, Capelli G, Malik R. 2019. Molecular detection of Pneumocystis in the lungs of cats. Med Mycol 57:813–824. doi: 10.1093/mmy/myy139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller C, Abdo Z, Ericsson A, Elder J, VandeWoude S. 2018. Applications of the FIV model to study HIV pathogenesis. Viruses 10:206. doi: 10.3390/v10040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiota T, Shimada Y, Kurimoto H, Oikawa H. 1990. Pneumocystis carinii infection in corticosteroid-treated cats. J Parasitol 76:441–445. doi: 10.2307/3282686. [DOI] [PubMed] [Google Scholar]
- 49.Ma L, Imamichi H, Sukura A, Kovacs JA. 2001. Genetic divergence of the dihydrofolate reductase and dihydropteroate synthase genes in Pneumocystis carinii from 7 different host species. J Infect Dis 184:1358–1362. doi: 10.1086/324208. [DOI] [PubMed] [Google Scholar]
- 50.Choukri F, Menotti J, Sarfati C, Lucet JC, Nevez G, Garin YJ, Derouin F, Totet A. 2010. Quantification and spread of Pneumocystis jirovecii in the surrounding air of patients with Pneumocystis pneumonia. Clin Infect Dis 51:259–265. doi: 10.1086/653933. [DOI] [PubMed] [Google Scholar]
- 51.Hughes WT. 1982. Natural mode of acquisition for de novo infection with Pneumocystis carinii. J Infect Dis 145:842–848. doi: 10.1093/infdis/145.6.842. [DOI] [PubMed] [Google Scholar]
- 52.Hughes WT, Bartley DL, Smith BM. 1983. A natural source of infection due to Pneumocystis carinii. J Infect Dis 147:595–595. doi: 10.1093/infdis/147.3.595. [DOI] [PubMed] [Google Scholar]
- 53.Chabe M, Aliouat-Denis CM, Delhaes L, Aliouat el M, Viscogliosi E, Dei-Cas E. 2011. Pneumocystis: from a doubtful unique entity to a group of highly diversified fungal species. FEMS Yeast Res 11:2–17. doi: 10.1111/j.1567-1364.2010.00698.x. [DOI] [PubMed] [Google Scholar]
- 54.Ripamonti C, Orenstein A, Kutty G, Huang L, Schuhegger R, Sing A, Fantoni G, Atzori C, Vinton C, Huber C, Conville PS, Kovacs JA. 2009. Restriction fragment length polymorphism typing demonstrates substantial diversity among Pneumocystis jirovecii isolates. J Infect Dis 200:1616–1622. doi: 10.1086/644643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rostved AA, Sassi M, Kurtzhals JAL, Sorensen SS, Rasmussen A, Ross C, Gogineni E, Huber C, Kutty G, Kovacs JA, Helweg-Larsen J. 2013. Outbreak of Pneumocystis Pneumonia in renal and liver transplant patients caused by genotypically distinct strains of Pneumocystis jirovecii. Transplantation 96:834–842. doi: 10.1097/TP.0b013e3182a1618c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desoubeaux G, Dominique M, Morio F, Thepault RA, Franck-Martel C, Tellier AC, Ferrandiere M, Hennequin C, Bernard L, Salame E, Bailly E, Chandenier J. 2016. Epidemiological outbreaks of Pneumocystis jirovecii pneumonia are not limited to kidney transplant recipients: genotyping confirms common source of transmission in a liver transplantation unit. J Clin Microbiol 54:1314–1320. doi: 10.1128/JCM.00133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vargas SL, Hughes WT, Santolaya ME, Ulloa AV, Ponce CA, Cabrera CE, Cumsille F, Gigliotti F. 2001. Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin Infect Dis 32:855–861. doi: 10.1086/319340. [DOI] [PubMed] [Google Scholar]
- 58.Pifer LL, Hughes WT, Stagno S, Woods D. 1978. Pneumocystis carinii infection: evidence for high prevalence in normal and immunosuppressed children. Pediatrics 61:35–41. [PubMed] [Google Scholar]