Tuberculosis (TB) is a major global heath burden, with 1.6 million people succumbing to the disease every year. The search for new drugs to improve the current chemotherapeutic regimen is crucial to reducing this global health burden. The cell wall polymer peptidoglycan (PG) has emerged as a very successful drug target in bacterial pathogens, as many currently used antibiotics target the synthesis of this macromolecule. However, the multitude of genes encoding PG-synthesizing and PG-modifying enzymes with apparent redundant functions has hindered the identification of novel drug targets in PG synthesis in Mycobacterium tuberculosis. Here, we demonstrate that two PG-cleaving enzymes are important for virulence of M. tuberculosis. In particular, the d,l-endopeptidase RipA represents a potentially attractive drug target, as its depletion results in the clearance of M. tuberculosis from the host and renders the bacteria hypersusceptible to rifampin, a frontline TB drug, and to several cell wall-targeting antibiotics.
KEYWORDS: cell division, pathogenesis, tuberculosis
ABSTRACT
Synthesis and cleavage of the cell wall polymer peptidoglycan (PG) are carefully orchestrated processes and are essential for the growth and survival of bacteria. Yet, the function and importance of many enzymes that act on PG in Mycobacterium tuberculosis remain to be elucidated. We demonstrate that the activity of the N-acetylmuramyl-l-alanine amidase Ami1 is dispensable for cell division in M. tuberculosis in vitro yet contributes to the bacterium’s ability to persist during chronic infection in mice. Furthermore, the d,l-endopeptidase RipA, a predicted essential enzyme, is dispensable for the viability of M. tuberculosis but required for efficient cell division in vitro and in vivo. Depletion of RipA sensitizes M. tuberculosis to rifampin and to cell envelope-targeting antibiotics. Ami1 helps sustain residual cell division in cells lacking RipA, but the partial redundancy provided by Ami1 is not sufficient during infection, as depletion of RipA prevents M. tuberculosis from replicating in macrophages and leads to dramatic killing of the bacteria in mice. Notably, RipA is essential for persistence of M. tuberculosis in mice, suggesting that cell division is required during chronic mouse infection. Despite the multiplicity of enzymes acting on PG with redundant functions, we have identified two PG hydrolases that are important for M. tuberculosis to replicate and persist in the host.
INTRODUCTION
Tuberculosis (TB) is the current leading cause of mortality in the world due to a single infectious agent, causing 1.6 million deaths annually (1). Mycobacterium tuberculosis, the causative agent of tuberculosis, owes a lot of its success as a pathogen to its complex cell envelope. The thick waxy coat provides an impermeable barrier that is composed of lipids, including mycolic acids, a defining feature of mycobacteria that is capable of modulating the host immune response (2, 3). Many components of the cell envelope provide attractive targets for drug development. In fact, the current chemotherapy regimen for treating TB includes two drugs, isoniazid and ethambutol, that inhibit the synthesis of cell envelope mycolic acids. Improving our understanding of how the components of the cell envelope are generated, maintained, and modified may provide novel drug targets for antimycobacterial drug development.
Peptidoglycan (PG), the rigid polysaccharide layer that lies between the outer “mycomembrane” and the plasma membrane, is composed of multiple strands of a glycan backbone comprising repeating disaccharide units of N-acetylglucosamine (NAG) and N-acetyl-muramic acid (NAM) (Fig. 1A). Peptide stems project from the backbone, anchored at the NAM disaccharide, and are cross-linked to one another, providing the strength and rigor of this mesh surrounding the cell. PG is unusual in mycobacteria as it contains predominantly 3-3 cross-links between the third residues of the peptide stems (Fig. 1A) (4, 5). These 3-3 cross-links are formed by l,d-transpeptidases (LDT), PG-maturing enzymes, and are important for the survival of the bacterium in the chronic stage of infection (2, 3, 6, 7). Although PG is rigid, it is not a static structure; it must be expanded, broken apart, and modified during the cell cycle to allow the cell to grow and divide (8). Indeed, during cell elongation, the existing PG macromolecule must be cleaved for the incorporation of new PG material near the poles. Also, during the final steps of cell division when the septum is formed, PG needs to be cleaved to allow the physical separation of two daughter cells. Thus, the synthesis and cleavage of PG during cell growth and division is a carefully coordinated process that allows PG to remain intact and retain the cellular structure. Several enzymes (PG hydrolases) are capable of cleaving PG and do so at different positions on the macromolecule (Fig. 1A). Lytic transglycosylases, such as the resuscitation-promoting factors (Rpfs) are believed to cleave between the NAG and NAM disaccharides of the glycan backbone (9–11). d,l-Endopeptidases such as RipA and RipB cleave within the peptide stem (12), while NAM l-alanine amidases cleave the peptide stem from the NAM residue on the glycan backbone (Fig. 1A).
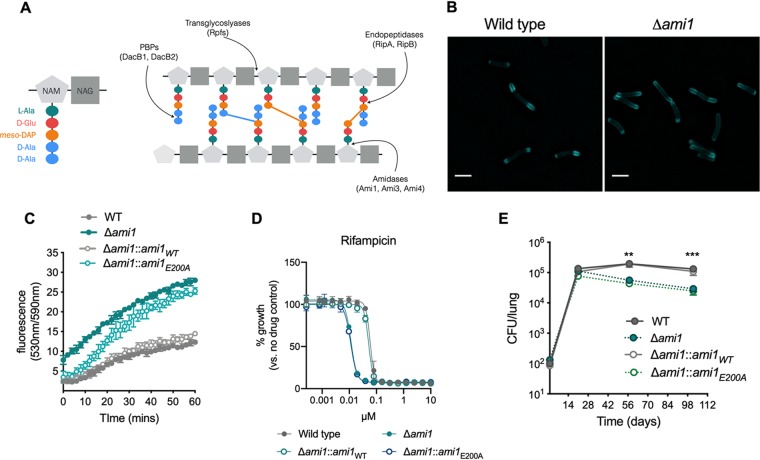
FIG 1.
The peptidoglycan amidase Ami1 is dispensable for M. tuberculosis cell division in vitro but is important for persistence in vivo. (A) Schematic diagram of peptidoglycan (PG). N-Acetyl muramic acid (NAM) and N-acetylglucosamine (NAG) residues make up the glycan backbone of PG. The pentapeptide stem constitutes of l-alanine, d-glutamate, meso-diaminopimelic acid (mDAP), and two d-alanine residues (left panel). Different classes of PG-hydrolyzing enzymes cleave the PG macromolecule at different sites (right panel). The colored bars between peptide stems represent cross-links (blue, 4-3; orange, 3-3). (B) Micrographs of M. tuberculosis cells labeled with the fluorescent d-alanine analogue HADA for 20 h. Bar, 2 μm. (C) Ethidium bromide (EtBr) uptake assay. Bacteria were incubated with EtBr, and the fluorescence (indicating uptake of EtBr and binding to DNA) was monitored over the course of 60 min. (D) MIC profiles of Δami1, Δami1::ami1WT, and Δami1::ami1E200A strains relative to that of the wild type for the frontline TB drug rifampin. (E) Bacterial burden in the lungs of CB57BL/6 mice infected by aerosol inhalation with the following strains: wild-type H37Rv (WT), Δami1, Δami1::ami1WT, and Δami1::ami1E200A variant. Significance was determined by one-way analysis of variance (ANOVA) and adjusted for multiple comparisons. **, adjusted P (adj-P) < 0.005; ***, adj-P < 0.0005. All data are representative of two independent experiments. Data in panel E are from 2 independent experiments, each with 4 mice per group and time point.
The M. tuberculosis genome encodes four predicted NAM l-alanine amidases, Ami1 (Rv3717), Ami2/CwlM (Rv3915), Ami3 (Rv3811), and Ami4 (Rv3594) (13). Recent studies have discovered that Ami2/CwlM, which is essential in M. tuberculosis and in Mycobacterium smegmatis (a nonpathogenic and fast-growing mycobacterium), lacks amidase activity but instead regulates cell growth within the cytoplasm by interacting with several PG biosynthesis enzymes, depending on its phosphorylation status (14, 15). While the activities of Ami3 and Ami4 remain unknown, the crystal structure of Ami1 has been solved by two independent groups, and Ami1 PG hydrolase activity has been demonstrated in vitro (16, 17). Interestingly, M. smegmatis cells lacking the Ami1 homologue (MSMEG_6281) form multiseptated chains with a high frequency of lateral branching, indicative of a cell division defect (18). However, the role of Ami1 in cell division in M. tuberculosis has not yet been investigated.
Redundancy is often observed for PG amidases in other bacteria, such as Escherichia coli, where deletion of several amidases is required before substantial defects in cell division are observed (19). While the genome of M. tuberculosis suggests that there is an abundance of PG-hydrolyzing enzymes, it is not known whether these enzymes are indeed functionally redundant. We sought to investigate the physiological role of the PG amidase Ami1 and the endopeptidases RipA and RipB in M. tuberculosis. We discovered that the PG-hydrolyzing activity of a single amidase, Ami1, contributes to normal persistence of M. tuberculosis in the host but does not appear to play an important role in cell division in vitro under regular growth conditions. Ami1 does, however, contribute to the residual cell division activity in cells lacking the dominant PG hydrolase, RipA. Deletion of the d,l-endopeptidase RipA significantly impaired cell division in culture, and its depletion attenuated M. tuberculosis replication within macrophages and led to substantial killing of M. tuberculosis in mice. Furthermore, we observed that depletion of RipA renders M. tuberculosis more susceptible to several cell wall-targeting drugs and to the frontline anti-TB drug rifampin. Deletion of RipB did not affect cell division, yet the enzyme is essential in the absence of RipA. Together, this work shows that although multiple PG hydrolases are encoded in the M. tuberculosis genome, these enzymes fulfill important distinct roles in different contexts. This work may provide new strategies for the development of PG-targeting drugs against M. tuberculosis.
RESULTS
Ami1 is dispensable for cell division in vitro but participates in the maintenance of cell wall integrity and antibiotic resistance.
Deletion of the Ami1 homologue in M. smegmatis (MSMEG_6281) resulted in multiseptated chains with a high frequency of branching indicative of a cell division defect (18). To assess the importance of Ami1 for cell division in M. tuberculosis, we generated an Ami1 deletion (Δami1) mutant (see Fig. S1A and B in the supplemental material). Using the fluorescent d-alanine analogue (FDAA) HCC-amino-d-alanine (HADA) (20–22) that incorporates into newly synthesized peptidoglycan (Fig. 1B), we observed that Ami1 deficiency affected neither cell length nor the frequency of septa-containing cells (Fig. S1C and D). Nevertheless, M. tuberculosis lacking Ami1 or expressing an amidase-inactive mutant (Ami1E200A) (17) accumulated ethidium bromide faster and to a larger extent than the wild type or the mutant strain complemented with the wild-type enzyme (Fig. 1C). The levels of Ami1E200A were confirmed to be similar to that of Ami1 expressed in the wild-type and complemented strains (Fig. S1E), demonstrating that the active site mutation did not affect protein stability. In line with the observed increased cell wall permeability, the M. tuberculosis Δami1 mutant showed increased susceptibility to the frontline TB drug rifampin (Fig. 1D) and slightly increased susceptibility to the PG-targeting antibiotic vancomycin (see Fig. S2A in the supplemental material). Together, these data suggest that while Ami1 is dispensable for cell division in M. tuberculosis, the absence of this hydrolase leads to cell wall alterations, resulting in increased permeability and increased antibiotic susceptibility.
Validation of the M. tuberculosis Δami1 mutant. (A) Schematic diagram of the ami1 locus in wild-type (WT) and Δami1 mutant bacteria. Red bars represent the labeled oligonucleotide probe. Labeling and detection of probe DNA were carried out using the ECL Direct nucleic acid labeling and detection system (GE Healthcare). (B) Southern blot of SacI-digested genomic DNA, probed with ami1 horseradish peroxidase (HRP)-labeled probe. (C) Quantification of cell length measurements of WT and Δami1 mutant populations. WT n = 90; Δami1 mutant n = 113. The solid line represents the median. The area between the dashed lines represents the interquartile range. Significance was determined by the Kruskal-Wallis test; ns, not significant. (D) The percentage of the population of cells with septa. WT n = 113; Δami1 mutant n = 131. Significance was determined by the Mann-Whitney test; ns, not significant. (E) Immunoblotting to confirm the expression of the amidase-inactive variant of Ami1 (Ami1E200A). Whole-cell lysates were prepared from wild-type (WT), and Δami1 (M), Δami1::ami1WT (CWT), and Δami1::ami1E200A (CE200A) variant mid-log-phase cultures. Total protein (40 μg) was run by SDS-PAGE, and Ami1 protein was detected using rabbit anti-Ami1 sera. DlaT was used as a loading control. Download FIG S1, TIF file, 0.7 MB (739.4KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenotypic analysis of Δami1. (A) MIC profiles of wild-type Δami1, Δami1::ami1WT, and Δami1::ami1E200A strains for several cell wall-targeting drugs. (B) Bacterial burden in the spleens of CB57BL/6 mice infected by aerosol inhalation with the following strains: wild-type H37Rv (WT) and Δami1, Δami1::ami1WT, and Δami1::ami1E200A variant. (C) CFU burden of the wild type (WT) and the Δami1 mutant in naive and IFN-γ-activated murine BMDMs. Macrophages were infected at a multiplicity of infection (MOI) of 0.1. (D) Survival in phosphate-buffered saline (PBS) nutrient starvation model. (E) Survival during nitric oxide (NO) exposure for 3 days using diethylenetriamine NONOate (DETA-NO), supplemented every 24 hrs. All data are representative of at least two independent experiments. Data in panel B are from 2 independent experiments, each with 4 mice per group and time point. Download FIG S2, TIF file, 0.5 MB (501.4KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ami1 amidase activity is required for persistence of M. tuberculosis in mice.
The lack of any obvious cell division phenotype of M. tuberculosis Δami1 suggests that other PG amidases or hydrolases are compensating for the loss of Ami1. The enzyme may, however, be important under specific conditions, such as during infection. To test this, we infected mice with the Δami1 mutant, the complemented mutant (Δami1::ami1) and an Δami1 mutant expressing the inactive amidase (Δami1::ami1E200A) (17). The Δami1 mutant replicated similarly to wild-type M. tuberculosis during the acute phase of infection (up to 28 days postinfection), but a decline in CFU burden was observed during the chronic phase of infection (Fig. 1E). This persistence defect was fully complemented by expressing Ami1 in the Δami1 mutant background (Δami1::ami1WT). However, expression of Ami1E200A did not complement the persistence defect, indicating that the amidase activity of Ami1 is important for persistence (Fig. 1E). Deletion of Ami1 did not have a significant impact on dissemination of M. tuberculosis to the spleen (Fig. S2B). This was surprising, as a transposon library screen predicted that Ami1 is important for persistence of M. tuberculosis in the spleen of mice (23); perhaps the persistence defect in the spleen is only revealed during competition with Ami1 expressing M. tuberculosis. Thus, the amidase activity of Ami1 is dispensable for replication during the acute phase of mouse infection but contributes to the ability of M. tuberculosis to persist in the lungs during the chronic phase of infection. We did not observe any attenuation of the M. tuberculosis Δami1 mutant during macrophage infection (Fig. S2C), and the mutant survived nutrient deprivation and nitric oxide (NO) exposure, two host-relevant stresses, similarly to wild-type M. tuberculosis (Fig. S2D and E).
Genome-wide fitness screening predicts synthetic lethality between Ami1 and RipA.
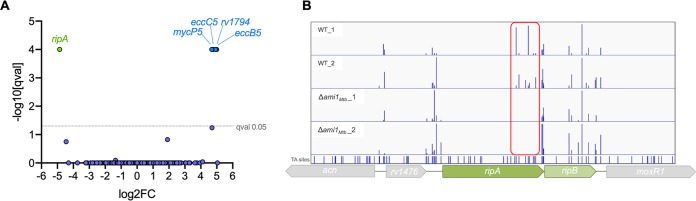
To identify enzymes that compensate for the activity of Ami1 during growth in culture, we utilized transposon mutagenesis coupled with next-generation sequencing (Tn-seq). Transposon mutant libraries were generated in wild-type M. tuberculosis and the Δami1 mutant, and the frequency of transposon mutants within the libraries was determined by sequencing the transposon-genome junctions to identify the insertion sites. We used the TRANSIT Tn-seq analysis tool (24) to identify mutants that were over- or underrepresented in the Δami1 mutant libraries compared to the wild-type libraries (log2 fold change, greater than 2 or less than −0.5) with statistical significance after correction for multiple comparisons (q < 0.05). The enrichment of mutants in the Δami1 mutant transposon library with disruption of genes encoding Esx5 type VII secretion system components (MycP5, EccB5, and EccC5) is in agreement with our finding that loss of Ami1 results in a leaky cell envelope, rendering the Esx5 system nonessential, as previously observed in Mycobacterium marinum (25). Surprisingly, only transposon insertions in a single gene, ripA, were identified as significantly underrepresented in the Δami1 mutant libraries (Fig. 2A). RipA is a peptidoglycan d,l-endopeptidase that cleaves peptidoglycan in the peptide stem (Fig. 1A); it is thus not surprising that this enzyme can compensate for Ami1 activity. Interestingly, RipA is predicted to be essential for growth of M. tuberculosis (26). Our results show that transposon insertions occurred only within the C-terminal hydrolase-encoding domain of ripA (Fig. 2B). The complete lack of insertions in the N-terminal domain is likely because this domain regulates the hydrolase activity of RipA (C-terminal domain), which, if not controlled correctly, could severely damage the cell wall and lead to mortality (27). In order to confirm the predicted synthetic lethal relationship between RipA and Ami1, it was necessary to first determine whether RipA is essential in M. tuberculosis.
FIG 2.
Transposon mutagenesis coupled with next-generation sequencing (Tn-seq) predicts synthetic lethality between ami1 and ripA. (A) Identification of genetic interactors of ami1. For each gene, the ratio of normalized sequence reads per insertion site (Δami1 pool/wild-type [WT] pool) is plotted on the x axis (log2 fold change). The y axis represents the significance of each of these changes in representation (q value). Data are from two biological replicate transposon mutant libraries per strain (B) Transposon insertion sites across the ripAB locus. The frequencies of TA dinucleotide sites with transposon insertions across the ripAB locus are represented by the height of the bars. The transposon insertions are shown for two independent replicate transposon mutant libraries generated in the wild type and the Δami1 mutant. The red box highlights the insertions within the C-terminal hydrolase domain that are detected only in WT libraries.
RipA and RipB are not essential in M. tuberculosis.
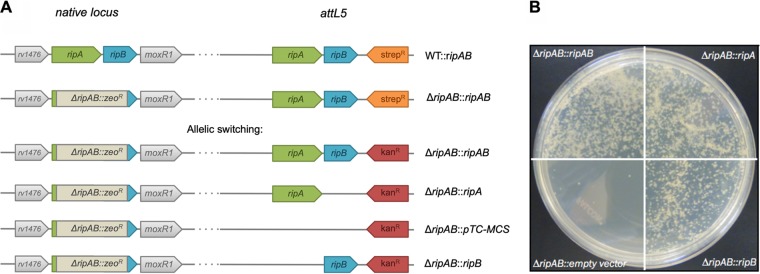
A recent study in M. smegmatis demonstrated that ripA is not essential, provided that ripB, coding for another d,l-endopeptidase and located downstream of ripA within the same operon, is expressed (28). To examine whether ripA and ripB are essential in M. tuberculosis, we first generated a merodiploid strain by expressing the ripA and ripB genes from an integrated streptomycin resistance-conferring plasmid in the attL5 locus (WT::ripAB) (Fig. 3A). Subsequently, the native ripAB locus was replaced by recombineering with a zeocin resistance cassette (ΔripAB::ripAB). The recombination was confirmed by PCR targeting the junctions between the zeocin cassette and the adjacent genes at the native ripAB locus (see Fig. S3A and B in the supplemental material). Allelic switching experiments were carried out to replace the integrated plasmid at the attL5 site containing the ripAB genes with plasmids expressing either ripA alone (ΔripAB::ripA), ripB alone (ΔripAB::ripB), or an empty plasmid (ΔripAB::pTC-MCS) (Fig. 3A). Allelic switching with replacement plasmids expressing only ripA or ripB was successful (Fig. 3B). Several colonies were selected, and we confirmed by PCR that the ripAB plasmid was replaced with one that expressed a single rip endopeptidase gene (Fig. S3D). Attempts to replace the ripAB genes at the attL5 site with an empty plasmid were unsuccessful (Fig. 3B; see also Fig. S3D, bottom). This confirmed that the PG endopeptidases RipA and RipB are individually dispensable in M. tuberculosis, but deletion of both ripA and ripB genes is either bactericidal or growth inhibitory for M. tuberculosis.
FIG 3.
Allelic switching demonstrates that ripA and ripB are individually dispensable but that at least one of the two enzymes is required for viability. (A) Schematic diagram of the native ripAB locus and the attL5 site for the various allelic switching strategies. (B) Photographs of agar plates of allelic switching transformations to replace the streptomycin resistance ripAB plasmid at the attL5 locus with kanamycin resistance plasmids expressing both ripA and ripB (ΔripAB::ripAB), ripA (ΔripAB::ripA) or ripB (ΔripAB::ripB) alone, or an empty plasmid (pTC-MCS).
Confirmation of the replacement of the native ripAB locus with the ΔripAB::zeoR cassette in an ripAB merodiploid background and subsequent allelic switching at the attL5 locus. (A) Schematic diagram displaying the ΔripAB::zeoR cassette at the native ripAB locus and the ripAB genes in the attL5 integrated plasmid. Arrows indicate the primers used to confirm the ΔripAB::zeoR cassette at the native ripAB locus. The line below indicates the DNA region used for recombination. (B). PCR results confirming two candidates using the primer pairs indicated below each gel image. (C) Kanamycin sensitivity check on cured ΔripAB::ripAB candidates 3 and 4 (confirmed in panel B). Colonies picked after curing on 5% sucrose to ensure loss of the pNIT-sacB plasmid were checked for kanamycin sensitivity by spotting onto 7H10 agar plates with or without kanamycin. Candidate 3A was chosen to perform subsequent allelic switching experiments. (D) PCR to confirm the presence of the kanamycin resistance replacement plasmids at the attL5 locus. The red asterisk indicates the confirmed candidate that was chosen for subsequent characterization. Download FIG S3, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RipA is the major PG septal hydrolase required for cell division in M. tuberculosis.
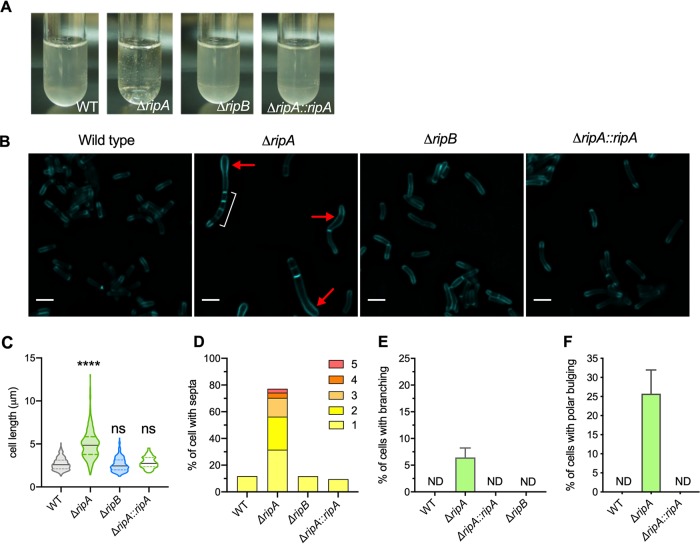
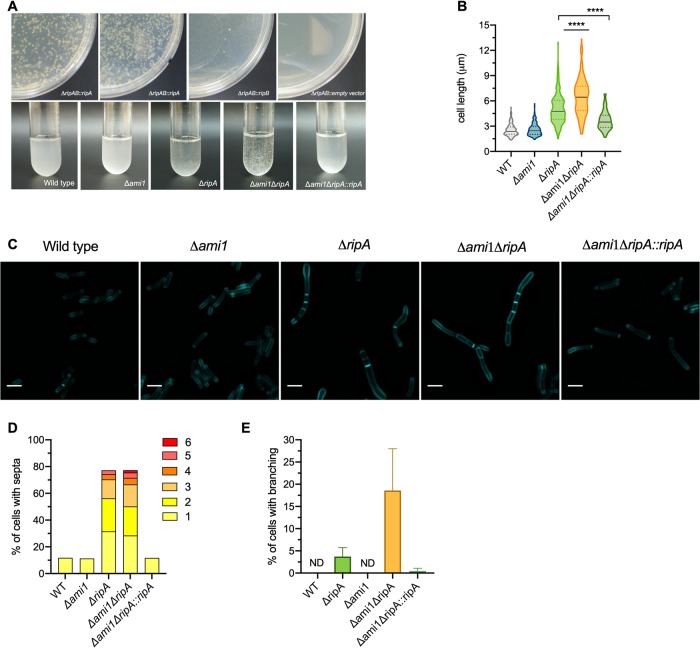
Upon culturing cells lacking RipA (ΔripAB::ripB, which we will subsequently refer to as the ΔripA mutant) in liquid medium, the cells were noticeably clumpy (Fig. 4A). We examined the cell division phenotype of cells lacking RipA by labeling the peptidoglycan with the fluorescent d-alanine analogue HADA. This revealed elongated multiseptated cells (Fig. 4B). Cells lacking RipA had an increased median cell length and a larger variation in cell length than the wild type and the complemented mutant (ΔripA::ripA) (Fig. 4C). Approximately 10% of wild-type and ΔripA::ripA mutant cells contained septa. In contrast, the majority of ΔripA mutant cells (>70%) contained at least one septum, with half of that population containing more than one septum, indicating that M. tuberculosis without RipA elongates and forms septa but cannot efficiently separate into two daughter cells (Fig. 4D). Branching was also observed in a small fraction of cells lacking RipA, indicating a dysregulation of the PG elongation complex machinery that causes it to localize to the lateral cell wall instead of at the poles (Fig. 4E). However, further investigation is needed to confirm this hypothesis. Some ΔripA cells contained bulges, most commonly at the poles, which might suggest that lack of RipA led to a weakened cell wall in the polar region, where the incorporation of new PG material occurs during cell elongation (Fig. 4F). In contrast, cells lacking RipB (ΔripB) did not have any increase in cell length or in the frequency of cells with septa, indicating that RipB is dispensable for cell division in M. tuberculosis under regular in vitro conditions (Fig. 4).
FIG 4.
RipA is the major septal PG hydrolase required for cell division in M. tuberculosis. (A) Photographs of liquid cultures of the indicated strains. (B) Micrographs of cells labeled with HCC-amino-d-alanine (HADA) for 20 h. White bracket indicates multiple septa. Red arrow indicates polar bulging. Bar, 2 μm. (C) Violin plots showing the cell lengths of the WT (n = 312) and the ΔripA (n = 268), ΔripB (n = 210), and ΔripA::ripA (n = 125) mutants. The black line represents the median; the area between the dashed lines is the interquartile range. Significance was determined by Kruskal-Wallis test; ****, P < 0.0001; ns, not significant. (D) Quantification of the proportion of cells with multiple septa in the WT (n = 361) and ΔripA (n = 347), ΔripB (n = 392), and ΔripA::ripA mutants (n = 378). The bar height represents the percentage of cells that contain 1 or more septa. The colored segments within the bars represent the proportions of cells with different numbers of septa. (E) Quantification of the proportion of cells with branching. (F) Quantification of the proportion of cells with bulging in the polar region. ND, none detected. Data are representative of at least two independent experiments.
Loss of Ami1 further exaggerates the cell division defects of the M. tuberculosis ΔripA mutant.
After we established that RipA can be deleted from M. tuberculosis, we next examined the predicted synthetic lethality between RipA and Ami1 by attempting to delete ripA in the Δami1 mutant background. We followed the same strategy as that described above, replacing the native ripAB with a zeocin resistance cassette in a ripAB merodiploid background of the Δami1 mutant. Allelic switching with plasmids expressing one of either rip hydrolase gene was next attempted. Replacement of the ripAB genes with ripB alone resulted in very small, slow-growing colonies, suggesting that M. tuberculosis cells lacking both Ami1 and RipA are viable but severely compromised in growth (Fig. 5A, top). When the ripAB genes were replaced by ripA alone, the resulting colonies grew similarly to the positive control (where the integrated streptomycin resistance ripAB-expressing plasmid is replaced with a kanamycin resistance ripAB-expressing plasmid). Attempts to replace the integrated ripAB plasmid with an empty vector (pTC-MCS) were unsuccessful, suggesting that, as is the case in wild-type M. tuberculosis, at least one of the endopeptidases (RipA or RipB) is required for growth or viability of the Δami1 mutant. Candidates resulting from each allelic replacement transformation were confirmed by PCR (see Fig. S4 in the supplemental material). Growth of the M. tuberculosis Δami1 ΔripA mutant in liquid medium was very slow, and the cells aggregated. The aggregates were noticeably bigger than those of the ΔripA mutant (Fig. 5A, lower). M. tuberculosis Δami1 ΔripA mutant cells varied drastically in length, with a median length of 6.47 μm, which is significantly longer than that of cells lacking only RipA (4.75 μm) (Fig. 5B). Cells labeled with HADA showed elongated cells with multiple septa and a noticeable proportion of cells with branching (Fig. 5C and D). We observed more branching in Δami1 ΔripA mutant cells than in the ΔripA mutant (Fig. 5E). Expression of RipA in Δami1 ΔripA (Δami1 ΔripA::ripA) rescued these phenotypes, further showing that RipA is required for efficient cell division. Deletion of ripB in the Δami1 mutant background did not have any impact on cell division or length (see Fig. S5 in the supplemental material).
FIG 5.
Deletion of ami1 amplifies cell division defects in the ΔripA mutant. (A) Agar plates of allelic switching transformations in the Δami1 ΔripAB::ripAB strain (top) and photographs of liquid cultures (bottom). (B) Violin plots of cell lengths for WT (n = 189) and Δami1 (n = 187), ΔripA (n = 196), ΔripA::ripA (n = 143), Δami1 ΔripA (n = 214), and Δami1 ΔripA::ripA (n = 172) mutants. The black line represents the median; the area between the dashed lines is the interquartile range. Significance was determined by a Kruskal-Wallis test; ****, P < 0.0001. (C) Micrographs of cells labeled with HADA for 20 h. Bar, 2 μm. (D) Quantification of the proportion of cells with at least one septum. The bar height represents the percentage of cells that contain 1 or more septa. The colored segments within the bars represent the proportions of cells with different numbers of septa. WT (n = 195); Δami1 (n = 380), ΔripA (n = 206), ΔripA::ripA (n = 314), Δami1 ΔripA (n = 225), and Δami1 ΔripA::ripA (n = 317) mutants. (E) Proportion of cells with branching. ND, none detected. Data in panels B to E are representative of two independent experiments.
Confirmation of the replacement of the native ripAB locus with the ΔripAB::zeoR cassette in the Δami1 ripAB merodiploid background and subsequent allelic switching at the attL5 locus (A) Confirmation of the replacement of the native ripAB locus with a zeocin resistance cassette (upper panel). Red box indicates the confirmed candidate that was chosen for subsequent curing of the pNIT-sacB plasmid. Kanamycin sensitivity phenotype confirmation of the cured candidate (lower panel). (B) PCR to confirm the presence of the kanamycin resistance replacement plasmids at the attL5 locus. The red asterisk indicates the confirmed Δami1 ΔripB mutant candidate that was chosen for subsequent characterization. Download FIG S4, TIF file, 0.6 MB (671.3KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cells lacking Ami1 and RipB do not have an increased cell length. Violin plots showing the cell lengths of the WT (n = 162) and the Δami1 ΔripA (n = 140) and Δami1 ΔripB (n = 89) mutants. The black line represents the median. The area between the dashed lines is the interquartile range. Significance was determined by the Kruskal-Wallis test; ****, P < 0.0001; ns, not significant. Data are representative of two independent experiments. Download FIG S5, TIF file, 0.1 MB (91.8KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RipA depletion increases the sensitivity of M. tuberculosis to multiple cell wall-targeting drugs and to the frontline TB drug rifampin.
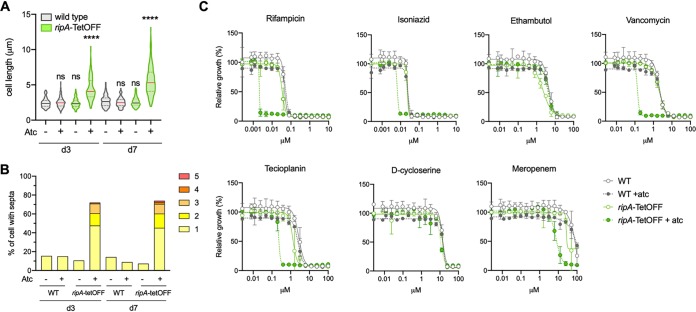
The poor growth and formation of aggregates of the M. tuberculosis ΔripA mutant made it difficult to characterize the consequences of ripA deletion. We therefore generated a strain in which ripA could be transcriptionally silenced by the addition of anhydrotetracycline (atc) (ripA-TetOFF) (29). Silencing ripA slowed growth (see Fig. S6 in the supplemental material), and the bacteria formed elongated multiseptated chains similar to those of the ΔripA mutant (Fig. 6A and B). We used ripA-TetOFF to determine whether the activities of frontline TB drugs and cell wall-targeting drugs that are active against mycobacteria are altered when RipA is depleted. Notably, the MICs of rifampin, isoniazid, teicoplanin, vancomycin, and meropenem were between 4-fold and 33-fold decreased when ripA was silenced (ripA-TetOFF + atc) (Fig. 6C). No shift in MIC was observed for ethambutol and d-cycloserine. Addition of atc did not alter the MICs against wild-type M. tuberculosis, showing that atc itself was not affecting the sensitivity of M. tuberculosis to the drugs tested.
FIG 6.
RipA depletion sensitizes M. tuberculosis to multiple cell wall-targeting drugs and to the frontline TB drug rifampin. (A) Violin plots of cell lengths upon ripA silencing with the addition of anhydrotetracycline (atc). Significance was determined by a Kruskal-Wallis test; ****, P < 0.0001; ns, not significant. (B) Quantification of the proportion of cells with at least one septum. The bar height represents the percentage of cells that contain 1 or more septa. The colored segments within the bars represent the proportions of cells with different numbers of septa. (C) MIC profiles of wild-type and ripA-tetOFF strains. Anhydrotetracycline (500 ng/ml) was added to cultures at the time of drug exposure. Growth was measured 14 days after addition of drug and atc. Data are representative of two independent experiments.
Transcriptional silencing of ripA attenuates growth of M. tuberculosis. Growth curve of WT and ripA-TetOFF strains with or without 500 ng/ml anhydrotetracycline (atc). Data are representative of two independent experiments. Download FIG S6, TIF file, 0.1 MB (96.7KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RipA depletion leads to reduced replication in macrophages and to clearance of M. tuberculosis from infected mice.
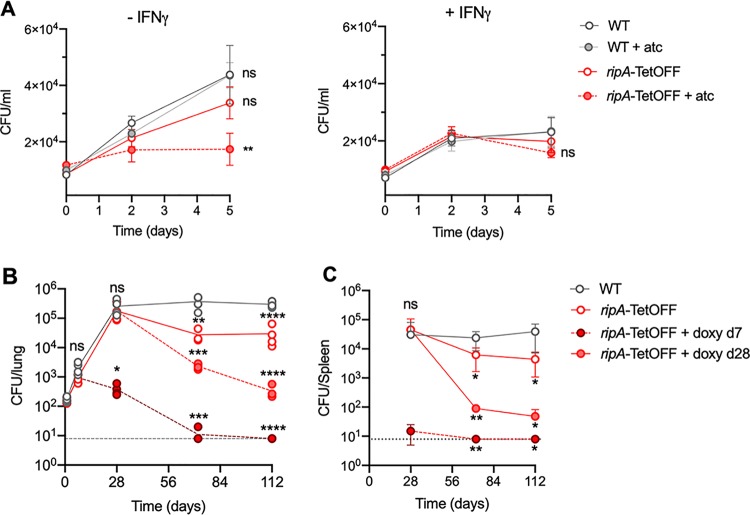
Next, we determined the effects of RipA depletion on M. tuberculosis during infection of bone marrow-derived macrophages (BMDMs) ex vivo and during infection of mice. During macrophage infections, ripA silencing was initiated (by addition of atc to cell culture medium) after the bacteria had been phagocytosed by the macrophages (4 h postinfection [p.i.]). In resting BMDMs, ripA silencing led to growth inhibition, as the number of CFU remained unchanged during the 5-day infection period (Fig. 7A). Replication of ripA-TetOFF in the absence of atc was slightly lower than that of the wild type (plus or minus atc) but not significantly so. In interferon gamma (IFN-γ)-activated BMDMs, there was no significant difference in CFU between any of the strains, but a slight reduction in CFU of the RipA-depleted strain (ripA-TetOFF + atc) on day 5 p.i. was observed (Fig. 7A). RipA depletion thus halted replication of M. tuberculosis in macrophages but did not render the bacteria more prone to being killed upon macrophage activation. We next determined the effects of RipA depletion during infection in an animal model. We infected mice with wild-type and ripA-TetOFF strains by aerosol inhalation and induced depletion of RipA during the early acute phase of infection (day 7 p.i.) and also once the chronic phase of infection was established (day 28 p.i.) by feeding mice with chow containing doxycycline (doxy). Depleting RipA both at day 7 and day 28 lead to a reduction in CFU in the lungs and spleen (Fig. 7 B and C). Depleting RipA beginning on day 7 caused CFU to decline below the limit of detection by day 72. At day 112 p.i., the lungs from mice infected with the ripA-TetOFF mutant silenced at day 7 had no visible lesions, while those silenced at day 28 had very few lesions compared to the wild-type- and ripA-TetOFF mutant-infected lungs from mice that did not receive doxy (see Fig. S7 in the supplemental material). These data demonstrate that RipA is required for cell division in vivo, as silencing ripA expression halts M. tuberculosis replication within macrophages and prevents growth during mouse infection. Moreover, ripA silencing led to substantial killing of M. tuberculosis in chronically infected mice, suggesting that RipA is required for cell division during persistence and renders the bacteria susceptible to host-derived antibacterial stresses.
FIG 7.
RipA depletion halts replication within macrophages and leads to clearance of M. tuberculosis from infected mice. (A) CFU quantification of the WT and the ripA-tetOFF (plus or minus atc) mutant during infection of naive and IFN-γ-activated murine BMDMs. Bacteria were incubated for 4 h at a multiplicity of infection (MOI) of 0.1 before washing to remove any nonphagocytosed bacteria. Macrophages were lysed in 0.01% Triton X-100, and the lysates were serially diluted and plated on 7H10 agar to determine the number of intracellular CFU. Anhydrotetracycline (atc) was added to wells at 4 h postinfection to silence ripA expression in intracellular M. tuberculosis. CFU burden in the lungs (B) and spleen (C) of mice infected by aerosol inhalation. The dashed lines indicate the limit of detection. At each time point, CFU of each strain was analyzed in groups of 4 mice. Doxycycline (doxy) was provided in chow to silence ripA expression in infected mice. The data are representative of two independent experiments. Significance was determined by one-way ANOVA and adjusted for multiple comparisons. ns, not significant; *, adj-P < 0.05; **, adj-P < 0.005; ***, adj-P < 0.0005; ****, adj-P < 0.0001.
Gross pathology of mouse lungs infected with the ripA-TetOFF strain. The left upper lobes of infected mice at 112 days post infection were fixed in 4% formaldehyde. CFU data from these animals are shown in Fig. 7. Download FIG S7, TIF file, 0.9 MB (970.5KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The importance of PG for the survival and proliferation of bacteria is evident from the number of broad-spectrum antibiotics currently in use that target the enzymes that build and modify it. However, the multiplicity of enzymes synthesizing and/or modifying PG in M. tuberculosis and in other bacteria suggests redundancy between these enzymes and makes investigating the roles of individual enzymes difficult (13). Nevertheless, despite their similar enzymatic functions, these enzymes can be individually important for different processes of PG synthesis, degradation, and re-modeling across diverse environmental conditions. For example, two class A penicillin-binding proteins (PBPs), MrcB and MrcA, are specifically important for the growth of E. coli at acidic and alkaline pH, respectively (30). Salmonella enterica serovar Typhimurium expresses a PG synthase that is selectively active when Salmonella resides in acidic environments, including those inside macrophages (31). Our data reveal that Ami1 is specifically required in M. tuberculosis during chronic mouse infection yet completely dispensable for growth in culture. In contrast, M. smegmatis depends on Ami1 for normal cell division in vitro (18). M. smegmatis lacking RipA elongated and formed chains in acidic medium but was indistinguishable from wild-type M. smegmatis under regular culture conditions, indicating that RipA is dispensable for cell division in M. smegmatis unless the bacteria are exposed to acid stress (28, 32). However, in M. tuberculosis, we discovered that RipA is important for cell division during regular growth in vitro and during infection. There are notable differences in PG synthesis dynamics between M. smegmatis and M. tuberculosis (22), and our work reveals differences between the enzymes that functionally dominate septal separation during cell division in M. smegmatis and M. tuberculosis. Ami1 is the dominant septal PG hydrolase mediating cell division in M. smegmatis (18), while RipA fulfills this role in M. tuberculosis. Notwithstanding, M. tuberculosis and M. smegmatis both depend on either RipA or RipB for viability and replication. Their activities might not only be required for septal PG degradation that allows cell division but may also facilitate cell growth by cleaving PG at the polar region, allowing for the incorporation of new PG. It has been demonstrated that PG hydrolases are necessary for cell growth in other bacteria (33–35). The branching and polar bulging evident in ΔripA and Δami1 ΔripA mutant cells suggest that RipA may also play a role in the incorporation or modification of new PG material during elongation that occurs at the poles in mycobacteria. Consistent with this hypothesis, RipA was shown to localize to the poles, as well as to the septa, in Mycobacterium bovis BCG (9). RipA is also known to interact with other cell wall enzymes. Its interaction with the lytic transglycosylase RpfB leads to a synergistic increase in their capacity to cleave PG (10). RipA also interacts with and is regulated by PonA1 (PBP1), a PG synthase that localizes to both the poles and septa and contributes to cell growth and virulence in mice (36, 37). The absence of RipA might thus lead to defects in cell growth and division not only by lack of its own enzymatic activity but also by perturbing its interacting partners, such as PonA1 and RpfB. This is supported by the branching observed in RipA-deficient cells, which suggests a disturbance in the localization of enzymes that normally congregate to the poles and elongate the sidewall.
The amidase activity of Ami1 contributes to the ability of M. tuberculosis to persist within the host, most likely by supporting cell division in a bacterial subpopulation, as the stable bacterial burden during the chronic phase of infection has been demonstrated to be a result of balanced replication and death (38). Previous work in our lab identified an integral membrane protein involved in cell division, PerM, that is also specifically required for persistence in mice (39, 40). RipA is essential for replication in the acute phase of infection and also during persistence in the chronic phase. Together, these findings indicate that M. tuberculosis depends on replication and/or PG turnover to survive during chronic infection. We speculate that during persistence in vivo, a specific cell division or cell envelope modification complex forms that, in addition to the dominant PG hydrolase RipA, also includes Ami1, PerM, and other enzymes that strengthen PG against immune-driven host stresses.
Our work demonstrates that RipA represents an attractive drug target, as its inhibition results in clearance of M. tuberculosis during infection and renders M. tuberculosis significantly more vulnerable to drugs currently used to treat TB, as well as to PG-targeting glycopeptides. Furthermore, its localization in the periplasm facilitates access to inhibiting compounds. Despite the multitude of d,l-endopeptidases and amidases in M. tuberculosis, we found that RipA and Ami1 are individually important. We hope that a greater understanding of the specific roles of individual enzymes that synthesize and modify PG will motivate the development of novel cell wall-targeting drugs against M. tuberculosis.
MATERIALS AND METHODS
Bacterial cultures and manipulation.
Mycobacteria were grown in liquid Middlebrook 7H9 medium supplemented with 0.2% glycerol, 0.05% tyloxapol, and ADN (0.5% bovine serum albumin, 0.2% dextrose, 0.085% NaCl) and on solid agar plates with Middlebrook 7H10 agar supplemented with 0.2% glycerol and Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment (Becton, Dickinson). The following antibiotics were added for selection of genetically modified strains at the following concentrations: hygromycin (50 μg/ml), kanamycin (25 μg/ml), streptomycin (25 μg/ml), and zeocin (25 μg/ml); anhydrotetracycline (500 ng/ml) was added to induce gene silencing. The strains used in this study are listed in Table S1 in the supplemental material.
List of strains used in this study. Download Table S1, PDF file, 0.03 MB (28.6KB, pdf) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutant generation and complementation.
Knockout mutants were generated in M. tuberculosis H37Rv by recombineering as previously described (41, 42). All plasmids were generated using Gateway cloning technology (Life Technologies). The knockout cassettes for ami1 (rv3717) and ripAB (rv1477 and rv1478) consisted of 500 bp upstream and 500 bp downstream of the genes to be replaced. ami1 was replaced, apart from the first and last 20 bp of the gene, by a hygromycin resistance cassette (see Fig. S1 in the supplemental material). The ripA and ripB genes were replaced with a zeocin resistance cassette. The knockout cassettes were cut from their vector backbones by restriction digest and transformed into the recombineering strain (H37Rv::pNIT-RecET) (42, 43). Recombinants were selected on 7H10 agar plates containing the appropriate antibiotics. Recombinant candidates were confirmed by PCR and then cured of the pNIT-RecET plasmid by counterselection on 5% sucrose 7H10 agar plates and confirmed by kanamycin sensitivity (see Fig. S3C and S4A in the supplemental material). Complementation of Δami1 was achieved by expressing ami1 under the control of its native promoter (200 bp upstream of ami1). Complementation with ripA and ripB was achieved by expressing either both genes or a single rip gene under the control of a constitutive promoter (Pmyc1-tetO [44]). The plasmids used in this study are listed in Table S2 in the supplemental material.
Plasmids used in this study. Download Table S2, PDF file, 0.02 MB (26.5KB, pdf) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of Ami1 antiserum and immunoblotting.
An N-terminal hexahistidine-tagged rv3717 (ami1) gene fusion was generated by cloning the rv3717 gene (without the secretion signal peptide) into the pET-300/NT-DEST plasmid. Expression of the His-tagged Ami1 protein in the BL21 (DE3) strain of E. coli was induced at 25°C for 6 h before purification using a His Trap high-performance (HP) column. The purified protein was checked by SDS-PAGE and then concentrated and dialyzed against phosphate-buffered saline (PBS). Rabbit polyclonal antiserum against recombinant Ami1 was generated by Thermo Fisher Scientific.
Transposon mutant library sequencing.
Transposon mutant libraries were constructed in wild-type H37Rv and the Δami1 mutant by himar1 mutagenesis as described previously (43). Genomic DNA was extracted from the transposon libraries, and the library mutant composition was determined by sequencing amplicons of the transposon-genome junctions as described previously (26, 45). Mapping and quantification of transposon insertions was done as described previously (26, 43). Genes affecting the fitness/viability in the Δami1 mutant background were identified using the resampling test module in the TRANSIT analysis platform (24). P values were defined as the proportion of values within 106 permutations that had a value more extreme than the observed experimental result, and these P values were adjusted for multiple comparisons (q value) using the Benjamini-Hochberg procedure. Tn-seq fold changes (TnSeq-FC) were computed as log2-transformed ratios of the normalized read counts between the wild type and Δami1 mutant libraries. We defined genes having a q value of <0.05 according to the permutation test to be significant determinants of fitness in the absence of ami1.
Peptidoglycan labeling with fluorescent d-alanine analogues.
Fluorescent d-alanine analog (FDAA) HADA (HCC-amino-d-alanine) was synthesized by the Memorial Sloan Kettering Cancer Center (MSKCC) Organic Synthesis Core Facility following the procedure described by Kuru and colleagues (21). For incorporation of fluorescent d-alanine analogues into the peptidoglycan of M. tuberculosis, cultures with an optical density at 580 nm (OD580) of 0.1 to 0.3 were incubated in 7H9 medium containing 1 mM HADA at 37°C for the indicated times. Cells were washed three times in PBS-0.05% tyloxapol and fixed with 4% formaldehyde.
High-resolution microscopy.
Microscopy imaging was performed using equipment and methods described previously (22). Fixed bacterial suspensions were spread onto soft agar pads (7H9 medium plus 1% agarose) and visualized with the appropriate filter set. Training and advice were provided at the Bio-Imaging Resource Center at Rockefeller University. Images were analyzed using ImageJ software.
Mouse infection.
The animal experiments were performed in accordance with National Institutes of Health guidelines for housing and care of laboratory animals and according to institutional regulations after protocol review and approval by the Institutional Animal Care and Use Committee of Weill Cornell Medicine (protocol number 0601441A). Female 7- to 8-week-old C57BL/6 mice (Jackson Laboratory) were infected with ∼100 CFU using an inhalation exposure system (Glas-Col). CFU burden of lungs and spleens at each time point was determined by plating dilutions of organ homogenates on 7H10 agar. Four mice were euthanized at each time point for each group. For ripA silencing in vivo, mice were provided with chow containing doxycycline (2,000 ppm; Research Diets).
Preparation and infection of murine bone marrow-derived macrophages.
Femurs and tibias of female C57BL/6 mice were extracted, and bone marrow cells were aseptically flushed using PBS. Cells were resuspended in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS), 10 mM HEPES, and 20% (vol/vol) L929 culture filtrate (LCM) and incubated for 6 days to allow differentiation into macrophages. Cells were harvested and seeded at 6 × 104 cells per well in 96-well plates in 10% LCM complete DMEM overnight before infection. Macrophages were activated with IFN-γ (20 ng/ml) overnight before infection and throughout the infection. Mycobacteria were washed in PBS + 0.05% tyloxapol, and a single cell suspension was generated by low-speed centrifugation to pellet clumped cells. The bacteria were diluted into 10% LCM and added to macrophages at a multiplicity of infection (MOI) of 0.1. After 4 h, extracellular bacteria were removed by washing the macrophages three times with warm PBS. The number of intracellular bacteria was determined by lysing macrophages with 0.01% Triton X-100 and culturing serial dilutions of macrophage lysates on 7H10 agar plates.
Determination of the MIC.
Cultures were grown to the mid-log phase and washed once in 7H9 medium, and a single cell suspension was prepared by low-speed centrifugation to pellet clumps. Single cell suspensions were used to inoculate 384-well black plates with clear flat bottoms that contained a range of drug concentrations. Drugs were dispensed using an HP D300e Digital Dispenser (Hewlett Packard). The drug dispensing was randomized using the HP Digital Dispenser software (version 3.2.2), and the dimethyl sulfoxide (DMSO) concentration in each well was normalized to 1%. After incubation at 37°C (5% CO2) for 12 days, the optical density (OD580) in each well was read using a SoftMax M2 plate reader. The data were derandomized using HP Digital Dispenser Data Merge software. The percentage of growth relative to that of the no-drug controls was calculated for each drug.
ACKNOWLEDGMENTS
We thank Katarzyna Cialowicz at the Bio-Imaging Resource Center (Rockefeller University) and Hélène Botella for teaching and advice with microscopy.
This work was supported by NIH grant R01 AI063446 and by the Tri-Institutional TB Research Unit (NIH grant U19AI111143).
Footnotes
Citation Healy C, Gouzy A, Ehrt S. 2020. Peptidoglycan hydrolases RipA and Ami1 are critical for replication and persistence of Mycobacterium tuberculosis in the host. mBio 11:e03315-19. https://doi.org/10.1128/mBio.03315-19.
REFERENCES
- 1.World Health Organization. 2019. Global tuberculosis report 2018. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Jarlier V, Nikaido H. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett 123:11–18. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- 3.Karakousis PC, Bishai WR, Dorman SE. 2004. Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell Microbiol 6:105–116. doi: 10.1046/j.1462-5822.2003.00351.x. [DOI] [PubMed] [Google Scholar]
- 4.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi J-L. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J Bacteriol 190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, Boshoff HIM, Barry CII. 2012. Meropenem inhibits d,d-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol 86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta R, Lavollay M, Mainardi J-L, Arthur M, Bishai WR, Lamichhane G. 2010. The Mycobacterium tuberculosis protein Ldt. Nat Med 16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baranowski C, Welsh MA, Sham L-T, Eskandarian HA, Lim HC, Kieser KJ, Wagner JC, Mckinney JD, Fantner GE, Ioerger TR, Walker S, Bernhardt TG, Rubin EJ, Rego EH. 2018. Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. Elife 7:100. doi: 10.7554/eLife.37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieser KJ, Rubin EJ. 2014. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol 12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. 2007. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol 66:658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 10.Hett EC, Chao MC, Deng LL, Rubin EJ. 2008. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog 4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, Kaplan G, Mizrahi V. 2008. The resuscitation‐promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol 67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Böth D, Schneider G, Schnell R. 2011. Peptidoglycan remodeling in Mycobacterium tuberculosis: comparison of structures and catalytic activities of RipA and RipB. J Mol Biol 413:247–260. doi: 10.1016/j.jmb.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Machowski EE, Senzani S, Ealand C, Kana BD. 2014. Comparative genomics for mycobacterial peptidoglycan remodelling enzymes reveals extensive genetic multiplicity. BMC Microbiol 14:75. doi: 10.1186/1471-2180-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutte CC, Baer CE, Papavinasasundaram K, Liu W, Chase MR, Meniche X, Fortune SM, Sassetti CM, Ioerger TR, Rubin EJ. 2016. A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. Elife 5:a021113. doi: 10.7554/eLife.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turapov O, Forti F, Kadhim B, Ghisotti D, Sassine J, Straatman-Iwanowska A, Bottrill AR, Moynihan PJ, Wallis R, Barthe P, Cohen-Gonsaud M, Ajuh P, Vollmer W, Mukamolova GV. 2018. Two faces of CwlM, an essential PknB substrate, in Mycobacterium tuberculosis. Cell Rep 25:57–67.e5. doi: 10.1016/j.celrep.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A, Kumar S, Kumar D, Mishra A, Dewangan RP, Shrivastava P, Ramachandran S, Taneja B. 2013. The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis. Acta Cryst 69:2543–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prigozhin DM, Mavrici D, Huizar JP, Vansell HJ, Alber T. 2013. Structural and biochemical analyses of Mycobacterium tuberculosis N-acetylmuramyl-l-alanine amidase Rv3717 point to a role in peptidoglycan fragment recycling. J Biol Chem 288:31549–31555. doi: 10.1074/jbc.M113.510792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senzani S, Li D, Bhaskar A, Ealand C, Chang J, Rimal B, Liu C, Kim SJ, Dhar N, Kana B. 2017. An amidase_3 domain-containing N-acetylmuramyl-l-alanine amidase is required for mycobacterial cell division. Sci Rep 7:1–16. doi: 10.1038/s41598-017-01184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priyadarshini R, Popham DL, Young KD. 2006. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol 188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, De Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. 2015. Synthesis of fluorescent d-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc 10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Botella H, Yang G, Ouerfelli O, Ehrt S, Nathan CF, Vaubourgeix J. 2017. Distinct spatiotemporal dynamics of peptidoglycan synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. mBio 8:e01183-17. doi: 10.1128/mBio.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dejesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ates LS, Ummels R, Commandeur S, van de Weerd R, van der Weerd R, Sparrius M, Weerdenburg E, Alber M, Kalscheuer R, Piersma SR, Abdallah AM, Abd El Ghany M, Abdel-Haleem AM, Pain A, Jiménez CR, Bitter W, Houben ENG. 2015. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet 11:e1005190. doi: 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dejesus MA, Gerrick ER, Gerrick ER, Xu W, Xu W, Park SW, Long JE, Boutte CC, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao MC, Kieser KJ, Minami S, Mavrici D, Aldridge BB, Fortune SM, Alber T, Rubin EJ. 2013. Protein complexes and proteolytic activation of the cell wall hydrolase RipA regulate septal resolution in mycobacteria. PLoS Pathog 9:e1003197. doi: 10.1371/journal.ppat.1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinelli DJ, Martinelli DJ, Pavelka MS Jr, Pavelka MS. 2016. The RipA and RipB peptidoglycan endopeptidases are individually nonessential to Mycobacterium smegmatis. J Bacteriol 198:1464–1475. doi: 10.1128/JB.00059-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehrt S, Schnappinger D. 2014. Regulated expression systems for mycobacteria and their applications. Microbiol Spectr 2:MGM2-0018-2013. doi: 10.1128/microbiolspec.MGM2-0018-2013. [DOI] [PubMed] [Google Scholar]
- 30.Mueller EA, Egan AJ, Breukink E, Vollmer W, Levin PA. 2019. Plasticity of Escherichia coli cell wall metabolism promotes fitness and antibiotic resistance across environmental conditions. Elife 8:e4075. doi: 10.7554/eLife.40754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castanheira S, Cestero JJ, Rico-Pérez G, García P, Cava F, Ayala JA, Pucciarelli MG, García-Del Portillo F. 2017. A specialized peptidoglycan synthase promotes Salmonella cell division inside host cells. mBio 8:642. doi: 10.1128/mBio.01685-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botella H, Vaubourgeix J, Lee MH, Song N, Xu W, Makinoshima H, Glickman MS, Ehrt S. 2017. Mycobacterium tuberculosis protease MarP activates a peptidoglycan hydrolase during acid stress. EMBO J 36:536–548. doi: 10.15252/embj.201695028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vollmer W. 2012. Bacterial growth does require peptidoglycan hydrolases. Mol Microbiol 86:1031–1035. doi: 10.1111/mmi.12059. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto M, Ooiwa S, Sekiguchi J. 2012. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of d,l-endopeptidase activity at the lateral cell wall. J Bacteriol 194:796–803. doi: 10.1128/JB.05569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh SK, SaiSree L, Amrutha RN, Reddy M. 2012. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 36.Hett EC, Chao MC, Rubin EJ. 2010. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog 6:e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kieser KJ, Boutte CC, Kester JC, Baer CE, Barczak AK, Meniche X, Chao MC, Rego EH, Sassetti CM, Fortune SM, Rubin EJ. 2015. Phosphorylation of the peptidoglycan synthase PonA1 governs the rate of polar elongation in mycobacteria. PLoS Pathog 11:e1005010. doi: 10.1371/journal.ppat.1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill WP, Harik NS, Whiddon MR, Liao RP, Mittler JE, Sherman DR. 2009. A replication clock for Mycobacterium tuberculosis. Nat Med 15:211–214. doi: 10.1038/nm.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodsmith N, Guo XV, Vandal OH, Vaubourgeix J, Wang R, Botella H, Song S, Bhatt K, Liba A, Salgame P, Schnappinger D, Ehrt S. 2015. Disruption of an M. tuberculosis membrane protein causes a magnesium-dependent cell division defect and failure to persist in mice. PLoS Pathog 11:e1004645. doi: 10.1371/journal.ppat.1004645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang R, Kreutzfeldt K, Botella H, Vaubourgeix J, Schnappinger D, Ehrt S. 2019. Persistent Mycobacterium tuberculosis infection in mice requires PerM for successful cell division. Elife 8:e49570. doi: 10.7554/eLife.49570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gee CL, Gee CL, Papavinasasundaram KG, Papavinasasundaram KG, Blair SR, Blair SR, Baer CE, Falick AM, Falick AM, King DS, King DS, Griffin JE, Venghatakrishnan H, Venghatakrishnan H, Zukauskas A, Zukauskas A, Wei J-R, Dhiman RK, Dhiman RK, Crick DC, Rubin EJ, Sassetti CM, Alber T. 2012. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal 5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy KC, Papavinasasundaram K, Sassetti CM. 2015. Mycobacterial recombineering. Methods Mol Biol 1285:177–199. doi: 10.1007/978-1-4939-2450-9_10. [DOI] [PubMed] [Google Scholar]
- 43.Xu W, Dejesus MA, Rücker N, Engelhart CA, Wright MG, Healy C, Lin K, Wang R, Park SW, Ioerger TR, Schnappinger D, Ehrt S. 2017. Chemical genetic interaction profiling reveals determinants of intrinsic antibiotic resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e01334-17. doi: 10.1128/AAC.01334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res 33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long JE, DeJesus M, Ward D, Baker RE, Ioerger T, Sassetti CM. 2015. Identifying essential genes in Mycobacterium tuberculosis by global phenotypic profiling. Methods Mol Biol 1279:79–95. doi: 10.1007/978-1-4939-2398-4_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of the M. tuberculosis Δami1 mutant. (A) Schematic diagram of the ami1 locus in wild-type (WT) and Δami1 mutant bacteria. Red bars represent the labeled oligonucleotide probe. Labeling and detection of probe DNA were carried out using the ECL Direct nucleic acid labeling and detection system (GE Healthcare). (B) Southern blot of SacI-digested genomic DNA, probed with ami1 horseradish peroxidase (HRP)-labeled probe. (C) Quantification of cell length measurements of WT and Δami1 mutant populations. WT n = 90; Δami1 mutant n = 113. The solid line represents the median. The area between the dashed lines represents the interquartile range. Significance was determined by the Kruskal-Wallis test; ns, not significant. (D) The percentage of the population of cells with septa. WT n = 113; Δami1 mutant n = 131. Significance was determined by the Mann-Whitney test; ns, not significant. (E) Immunoblotting to confirm the expression of the amidase-inactive variant of Ami1 (Ami1E200A). Whole-cell lysates were prepared from wild-type (WT), and Δami1 (M), Δami1::ami1WT (CWT), and Δami1::ami1E200A (CE200A) variant mid-log-phase cultures. Total protein (40 μg) was run by SDS-PAGE, and Ami1 protein was detected using rabbit anti-Ami1 sera. DlaT was used as a loading control. Download FIG S1, TIF file, 0.7 MB (739.4KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenotypic analysis of Δami1. (A) MIC profiles of wild-type Δami1, Δami1::ami1WT, and Δami1::ami1E200A strains for several cell wall-targeting drugs. (B) Bacterial burden in the spleens of CB57BL/6 mice infected by aerosol inhalation with the following strains: wild-type H37Rv (WT) and Δami1, Δami1::ami1WT, and Δami1::ami1E200A variant. (C) CFU burden of the wild type (WT) and the Δami1 mutant in naive and IFN-γ-activated murine BMDMs. Macrophages were infected at a multiplicity of infection (MOI) of 0.1. (D) Survival in phosphate-buffered saline (PBS) nutrient starvation model. (E) Survival during nitric oxide (NO) exposure for 3 days using diethylenetriamine NONOate (DETA-NO), supplemented every 24 hrs. All data are representative of at least two independent experiments. Data in panel B are from 2 independent experiments, each with 4 mice per group and time point. Download FIG S2, TIF file, 0.5 MB (501.4KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmation of the replacement of the native ripAB locus with the ΔripAB::zeoR cassette in an ripAB merodiploid background and subsequent allelic switching at the attL5 locus. (A) Schematic diagram displaying the ΔripAB::zeoR cassette at the native ripAB locus and the ripAB genes in the attL5 integrated plasmid. Arrows indicate the primers used to confirm the ΔripAB::zeoR cassette at the native ripAB locus. The line below indicates the DNA region used for recombination. (B). PCR results confirming two candidates using the primer pairs indicated below each gel image. (C) Kanamycin sensitivity check on cured ΔripAB::ripAB candidates 3 and 4 (confirmed in panel B). Colonies picked after curing on 5% sucrose to ensure loss of the pNIT-sacB plasmid were checked for kanamycin sensitivity by spotting onto 7H10 agar plates with or without kanamycin. Candidate 3A was chosen to perform subsequent allelic switching experiments. (D) PCR to confirm the presence of the kanamycin resistance replacement plasmids at the attL5 locus. The red asterisk indicates the confirmed candidate that was chosen for subsequent characterization. Download FIG S3, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmation of the replacement of the native ripAB locus with the ΔripAB::zeoR cassette in the Δami1 ripAB merodiploid background and subsequent allelic switching at the attL5 locus (A) Confirmation of the replacement of the native ripAB locus with a zeocin resistance cassette (upper panel). Red box indicates the confirmed candidate that was chosen for subsequent curing of the pNIT-sacB plasmid. Kanamycin sensitivity phenotype confirmation of the cured candidate (lower panel). (B) PCR to confirm the presence of the kanamycin resistance replacement plasmids at the attL5 locus. The red asterisk indicates the confirmed Δami1 ΔripB mutant candidate that was chosen for subsequent characterization. Download FIG S4, TIF file, 0.6 MB (671.3KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cells lacking Ami1 and RipB do not have an increased cell length. Violin plots showing the cell lengths of the WT (n = 162) and the Δami1 ΔripA (n = 140) and Δami1 ΔripB (n = 89) mutants. The black line represents the median. The area between the dashed lines is the interquartile range. Significance was determined by the Kruskal-Wallis test; ****, P < 0.0001; ns, not significant. Data are representative of two independent experiments. Download FIG S5, TIF file, 0.1 MB (91.8KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptional silencing of ripA attenuates growth of M. tuberculosis. Growth curve of WT and ripA-TetOFF strains with or without 500 ng/ml anhydrotetracycline (atc). Data are representative of two independent experiments. Download FIG S6, TIF file, 0.1 MB (96.7KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gross pathology of mouse lungs infected with the ripA-TetOFF strain. The left upper lobes of infected mice at 112 days post infection were fixed in 4% formaldehyde. CFU data from these animals are shown in Fig. 7. Download FIG S7, TIF file, 0.9 MB (970.5KB, tif) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of strains used in this study. Download Table S1, PDF file, 0.03 MB (28.6KB, pdf) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, PDF file, 0.02 MB (26.5KB, pdf) .
Copyright © 2020 Healy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.