Abstract
The mononuclear phagocytic system consists of many cells, in particular macrophages, scattered throughout the body. However, there is increasing evidence for the heterogeneity of tissue‐resident macrophages, leading to a pressing need for new tools to discriminate mononuclear phagocytic system subsets from other hematopoietic lineages. Macrophage‐expressed gene (Mpeg)1.1 is an evolutionary conserved gene encoding perforin‐2, a pore‐forming protein associated with host defense against pathogens. Zebrafish mpeg1.1:GFP and mpeg1.1:mCherry reporters were originally established to specifically label macrophages. Since then more than 100 peer‐reviewed publications have made use of mpeg1.1‐driven transgenics for in vivo studies, providing new insights into key aspects of macrophage ontogeny, activation, and function. Whereas the macrophage‐specific expression pattern of the mpeg1.1 promoter has been firmly established in the zebrafish embryo, it is currently not known whether this specificity is maintained through adulthood. Here we report direct evidence that beside macrophages, a subpopulation of B‐lymphocytes is marked by mpeg1.1 reporters in most adult zebrafish organs. These mpeg1.1 + lymphoid cells endogenously express mpeg1.1 and can be separated from mpeg1.1 + macrophages by virtue of their light‐scatter characteristics using FACS. Remarkably, our analyses also revealed that B‐lymphocytes, rather than mononuclear phagocytes, constitute the main mpeg1.1‐positive population in irf8 null myeloid‐defective mutants, which were previously reported to recover tissue‐resident macrophages in adulthood. One notable exception is skin macrophages, whose development and maintenance appear to be independent from irf8, similar to mammals. Collectively, our findings demonstrate that irf8 functions in myelopoiesis are evolutionary conserved and highlight the need for alternative macrophage‐specific markers to study the mononuclear phagocytic system in adult zebrafish.
Keywords: B lymphocytes, Macrophages, mpeg1, Perforin‐2
Mpeg1 not only is a restricted macrophage marker, but also labels B cells in the adult zebrafish; thus previously identified irf8‐independent macrophages likely consist of B lymphocytes.
Abbreviations
- cldnh
claudin h
- csf1ra
colony stimulating factor 1 receptor a
- ef1α
elongation factor 1 alpha
- FSC
forward scatter
- Ighz
immunoglobulin heavy constant zeta
- Ighm
immunoglobulin heavy constant mu
- irf8
interferon regulatory factor 8
- kdrl
kinase insert domain receptor like
- mhc2dab
major histocompatibility complex class II DAB gene
- mpeg
macrophage-expressed gene
- marco
macrophage receptor with collagenous structure
- SSC
side scatter
- WKM
whole kidney marrow
1. INTRODUCTION
Over the last decades, the zebrafish has become a powerful model to study biologic processes conserved with mammals.1, 2 In the fields of hematopoiesis and immunology, zebrafish has provided a new level of precision in understanding key aspects of developmental immunity and disease,3, 4 some of which are currently being tested in the clinic.5 Consequently, many efforts have been engaged to thoroughly characterize the zebrafish immune system. Using proximal promoter elements of lineage‐specific genes, we and others have established fluorescent transgenic lines that specifically mark the major blood cell lineages in the zebrafish.6, 7 Combined with light scatter separation, these lines have allowed the prospective isolation of zebrafish immune cells by flow cytometry, and their subsequent morphologic, phenotypic, and functional characterization. Collectively, these studies have highlighted the high degree of conservation between zebrafish and mammalian immune cell types.
Macrophages, which represent an important branch of the innate immune system8 and also exhibit key developmental functions,9 are, together with neutrophils, one of the most studied leukocyte populations in the zebrafish model. This is mainly due to the availability of several reporter lines, among which the mpeg1.1 promoter‐driven fluorescent reporters are the most frequently used.10 The mpeg1.1 gene encodes a pore‐forming protein named Perforin‐2, which is highly related to Perforin‐1 and the C9 subunit of the complement.11, 12 Whereas the two latter proteins are only found in vertebrates and act by killing extracellular targets, Mpeg1/Perforin‐2 is already present in early multicellular organisms like sponges, and targets intracellular pathogens.13 Mpeg1.1:GFP and mpeg1.1:mCherry reporter fish have been instrumental in characterizing the behavior of macrophages through live imaging in transgenic embryos, and the mpeg1.1:Gal4 line for analyzing macrophage‐targeted gene function. Altogether, these studies have tremendously contributed to increasing our knowledge on the roles of macrophages in multiple processes of developmental physiology,14, 15, 16 as well as in pathologic mechanisms involved in human disease, such as inflammation, infection,17, 18 and cancer.19, 20, 21 Whereas most of the field has initially focused on early macrophages taking advantage of the optical transparency of the zebrafish embryo and larvae, a growing number of investigators are now using these lines to address multiple aspects of macrophage biology in adults. This raises important questions, as the specificity of the mpeg1.1 driver in the adult hematopoietic system still remains to be determined.22 Indeed, although mpeg1.1 was originally described as a macrophage‐specific gene in mammals,11 recent evidence demonstrates that its expression is not restricted to mononuclear phagocytes.13
In this study, we initially aimed at characterizing different subsets of macrophages in the adult zebrafish, by combining mpeg1.1 transgenics with available lines marking the blood compartment. These extended analyses revealed a previously unappreciated cellular heterogeneity of the mpeg1.1 + population, as we identified a subset of mpeg1.1‐positive cells that are distinct from macrophages. Molecular characterization showed that these cells consisted of B lymphocytes. A detailed examination of their tissue distribution further demonstrated that mpeg1.1‐expressing B cells were present in all major adult lymphoid organs in zebrafish. In addition, mpeg1.1 expression was also found outside the hematopoietic system, in a subset of epithelial cells located in the skin, as recently described.23 Finally, we show that adult zebrafish deficient for irf8 recover mpeg1.1 + cells, in agreement with previous observations.24 However, based on our findings, we propose that in most tissues recovering mpeg1.1 + cells consist of B lymphocytes and do not represent irf8‐independent macrophages. One exception is the epidermis, where the presence of mpeg1.1 + macrophages is mildly affected by the absence of irf8.
2. MATERIAL AND METHODS
2.1. Zebrafish husbandry
Zebrafish were maintained under standard conditions, in accordance with institutional (Université Libre de Bruxelles, Brussels, Belgium; ULB) and national ethical welfare guidelines and regulations. All experimental procedures were approved by the ULB ethical committee for animal welfare (CEBEA). The following lines were used: Tg(mhc2dab:GFP LT)sd6 7; Tg(mpeg1.1:eGFP)gl22 [10]; Tg(mpeg1.1:mCherry)gl23 [10]; Tg(kdrl:Cre)s898 [25]; Tg(actb2:loxP‐STOP‐loxP‐DsRedexpress)sd5 [25]; Tg(Cau.Ighv‐ighm:EGFP)sd19 [26] is referred to as ighm:GFP. Homozygous irf8std95/std95 mutants were derived from heterozygous incrosses of irf8std95/ + fish.24 Special care was taken to control reporter gene dosage through experiments (with all control and mutant animals used in this study known to carry similar hemizygous or homozygous doses of the GFP transgenes). Unless specified, the term “adult” fish refers to animals aged between 3 and 6 mo.
2.2. Flow cytometry and cell sorting
Single‐cell suspensions of dissected adult zebrafish organ were prepared as previously described.7 Flow cytometry and cell sorting were performed with a FACS ARIA II (Becton Dickinson, Franklin Lakes, NJ, USA). Analyses were performed using the FlowJo software (Flow‐Jo LLC, Ashland, OR, USA). Cytospins and May‐Grunwald/Giemsa stains were performed as previously described.27
2.3. Quantitative PCR
RNA extraction from sorted cells and cDNA synthesis were performed as described.28 Biologic triplicates were compared for each subset. Relative amount of each transcript was quantified via the ΔCt method, using elongation‐factor‐1‐alpha (ef1α) expression for normalization, or via the ΔΔCt method, using ef1α and whole kidney marrow (WKM) for normalization. Primers are listed in Table 1.
Table 1.
qPCR primers used throughout the paper
| Gene | Forward primer | Reverse primer |
|---|---|---|
| ef1α | GAGAAGTTCGAGAAGGAAGC | CGTAGTATTTGCTGGTCTCG |
| mpeg1.1 | CCCACCAAGTGAAAGAGG | GTGTTTGATTGTTTTCAATGG |
| csf1ra | ATGACCATACCCAACTTTCC | AGTTTGTTGGTCTGGATGTG |
| marco | ACGACAGCTTCGATAATTTG | AAAATACTGCTCTCGGTTCC |
| cd45 | AGTTCCTGAAATGGAAAAGC | GCACAGAAAAGTCCAGTACG |
| cldnh1 | TTACAACCCGTTACTGCCCG | TGCCGGCTTGTACTTCTTCT |
2.4. Immunostaining and confocal microscopy analyses
Guts were dissected from adult fish, fixed in 4% paraformaldehyde (PFA) overnight (O/N) and incubated in 30% sucrose:PBS O/N before snap‐freezing in OCT. Scales were manually detached from euthanized fish and pretreated with 100 mM DTT before O/N fixation in 4% PFA. Immunostaining on gut cryosections or floating scales was performed as described,28 using the following primary and secondary antibodies: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/chicken anti‐GFP https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/polyclonal-antibodies (1:500; Abcam, Cambridge, UK), https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/leporidae anti‐DsRed https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/polyclonal-antibodies (1:500; Clontech, Mountain view, CA, USA), https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/alexa-fluor 488‐conjugated anti‐chicken https://www.sciencedirect.com/topics/neuroscience/immunoglobulin-g-antibody (1:500; Invitrogen, Carlsbad, CA, USA), https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/alexa-fluor 594‐conjugated anti‐rabbit IgG (1:500; Abcam). Images were taken with a Zeiss LSM 780 inverted https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/microscopes (Zeiss, Oberkochen, Germany), using a Plan Apochromat 20× objective. Image postprocessing (contrast and gamma adjust) were performed with the Zeiss Zen Software.
2.5. IPEX collection and phagocytosis assay
Intraperitoneal exudate (IPEX) of adult zebrafish was collected as previously described.29 For phagocytosis assay, 2.5 µl of FluoSpheres 1 µm, blue fluorescent (Termofisher scientific, Waltham, MA, USA) were injected in the peritoneal cavity, resulting in a cell:beads ratio ranging between 1:50 and 1:100. IPEX was collected at different time points, resuspended in 1% inactivated fetal bovine serum and analyzed by flow cytometry with an LSRFortessa machine (Becton Dickinson, Franklin Lakes, NJ, USA).
2.6. Single‐cell transcriptome analysis
Whole‐kidney marrows were isolated from three separate adult wild‐type fish (3‐9 mo old) and subjected to inDrop single‐cell RNA sequencing.30 These data are publicly available at the NCBI GEO database under accession ID GSE100913 and can be accessed directly through the website https://molpath.shinyapps.io/zebrafishblood/
2.7. Statistical analyses
For Figure 3D, t‐test (with multiple‐testing correction) was used to derive P‐values. All plots and statistical analysis were performed using R.31
Figure 3.
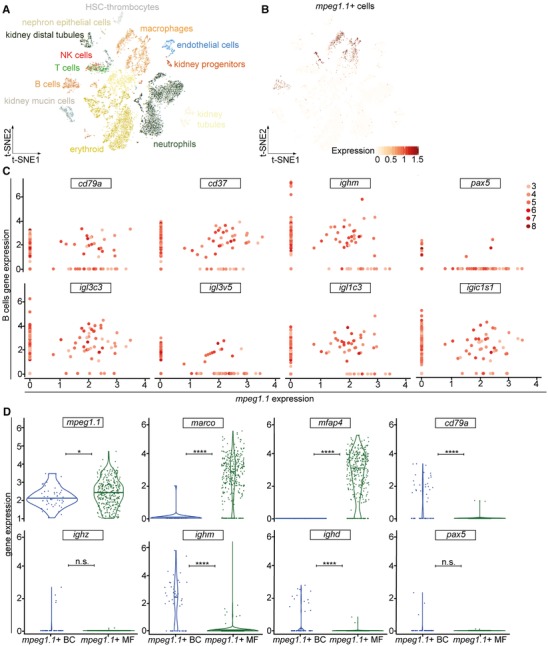
Characterization of mpeg1.1 expression in adult hematopoiesis by single‐cell RNAseq analysis. (A) 2D projection of the tSNE analysis showing the hematopoietic and nonhematopoietic cell types of the adult WKM, identified by single‐cell InDrops RNA sequencing. (B) Analysis of mpeg1.1 expression (red) across the clusters in the tSNE plot. Intensity of the color is proportional to the expression level. (C) Log of normalized and scaled expression of B‐cell genes (y‐axis) and mpeg1.1 (x‐axis) in single cells from the WKM. Each scatterplot represents the expression of a B‐cell gene, indicated on top. Dots correspond to individual cells and the color code on the right describes the overall number of B‐cell markers expressed by each cell. (D) Violin plot analysis comparing gene expression (y‐axis) between mpeg1.1 + macrophages and B cells
3. RESULTS
3.1. The mpeg1 transgene marks distinct populations of leukocytes in the zebrafish WKM
In mpeg1.1:GFP or mpeg1.1:mCherry transgenic adult zebrafish, parenchymal microglia can be isolated from other CNS‐associated macrophages by flow cytometry based on fluorophore expression levels.28 We therefore sought to investigate whether mpeg1.1 reporters could similarly discriminate distinct macrophage subsets in other tissues. To facilitate our study, we used mpeg1.1:mCherry; mhc2dab:GFP LT animals, where GFP marks antigen‐presenting cells within the adult hematopoietic tissue.7 This strategy was expected to specifically colabel mononuclear phagocytes, allowing tracking of these cells according to multiple parameters. Accordingly, we detected double positive cells by flow cytometry in the WKM (Fig. 1). This population could be fractionated into two subsets, based on levels of GFP and mCherry: the double positivelo population (mpeg1.1 lo mhc2dab +; Dplo) and the double positivehi population (mpeg1.1 hi mhc2dab +/hi; Dphi), accounting for approximately 70% and 30% of total double positive cells, respectively. Because mature mononuclear phagocytes were previously identified as a distinct forward scatter (FSC)hi side scatter (SSC)int population, overlapping the conventional myeloid and precursor fractions,7 we next examined the scatter profile of these two populations. Surprisingly, we found that only the Dphi cells localized in the myeloid/progenitor scatter fraction, whereas nearly all Dplo cells belonged to the FSCloSSClo lymphoid fraction. Because this latter fraction primarily contains lymphocytes,7, 32 these observations suggested that fluorophore expression in the mpeg1.1:mCherry line was not restricted to macrophages only.
Figure 1.
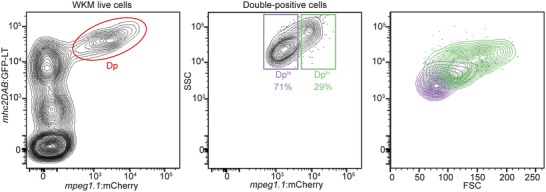
Two populations of mpeg1.1:mcherry‐positive cells in the adult WKM. The WKM of Tg(mpeg1.1:mcherry; mhc2dab:GFP LT) adult reporter fish was dissociated and analyzed by flow cytometry. The whole mpeg1.1 +; mhc2dab:GFP + double‐positive (Dp) population is circled in red (left panel) and is further subdivided into a mpeg1.1 lo; mhc2dab lo (Dplo) and a mpeg1.1 hi; mhc2dab hi (Dphi) fraction based on fluorescence intensity (middle panel). Right panel: separation of the Dphi and Dplo populations by light‐scatter characteristics. SSC = side scatter; FSC = forward scatter. Percentages of each population refer to a single individual and are relative to the total Dp population (mean ± sd of 3 fish: see text)
To evaluate whether expression within the WKM lymphoid gate was a common feature of all mpeg1.1 transgenics, we next turned to mpeg1.1:GFP adult fish. Flow cytometry analysis revealed that, similar to their mCherry counterparts, mpeg1.1:GFP‐labeled cells represented approximately 13% of the WKM (data not shown). This prompted us to examine in more details GFP expression among the different hematopoietic subsets (Fig. 2Ai). Within the combined myeloid and progenitor fractions, mpeg1.1:GFP + cells accounted for 4.0 ± 0.3% of total cells (Fig. 2Aii, red gate), in accordance with our previous estimations of mononuclear phagocytes abundance in this tissue.7 As expected, GFP was not detected within the eosinophil fraction (FSChiSSChi; data not shown). However, and validating our initial observations in mpeg1:mCherry animals, the analysis of the lymphoid fraction revealed the presence of a prominent mpeg1:GFP lo population, accounting for 53.2 ± 8.3% of cells within the lymphoid gate (n = 3) (Fig. 2Aiii, green gate). We conclude that the mpeg1 reporters show a previously unappreciated expression pattern within the lymphoid lineage in the WKM.
Figure 2.
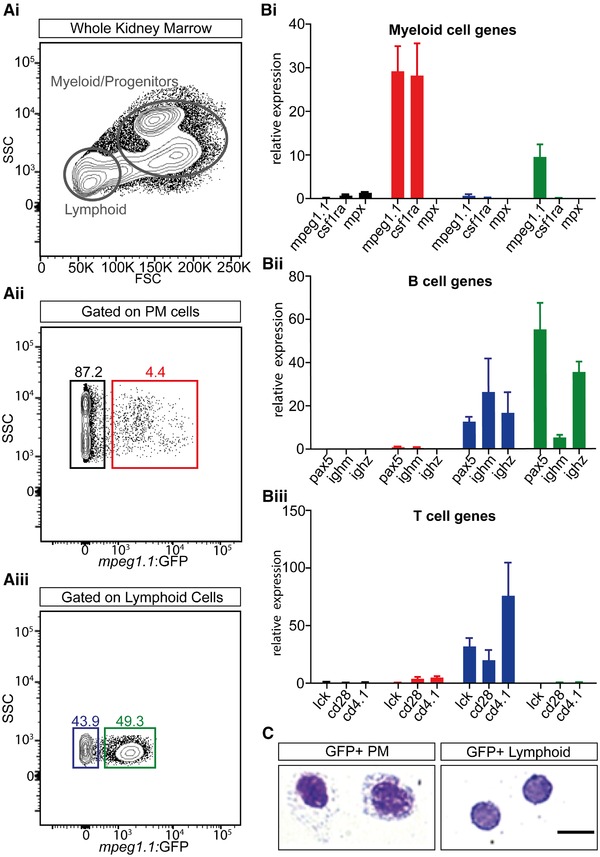
Mpeg1.1 expression identifies a subset of B cells in the adult WKM. (A) Gating strategy to isolate lymphoid and myeloid‐progenitors (MP) cells from the WKM using light‐scattering characteristics (Ai). Expression of mpeg1.1:GFP in the MP (Aii) and lymphoid (Aiii) fractions. Throughout the figure, the GFP− MP fraction is denoted by a black gate and bars, GFP+ MP by a red gate and bars, GFP− lymphoid by a blue gate and bars and GFP+ lymphoid by a green gate and bars. Percentages represent a single individual and are relative to the total live cells (mean ± sd of 3 fish: see text) (B) Q‐PCR expression for genes specific to the myeloid (Bi), B‐ (Bii), and T‐ (Biii) cell lineages in sorted mpeg1.1:GFP+ and mpeg1.1:GFP− cells. Units on the y‐axis represent changes (fold) above WKM, which is set at 1.0. Error bars indicate sem (n = 3). (C) Morphology of mpeg1.1:GFP+ cells isolated from the lymphoid and MP fractions in WKM. Cells were cytospun and stained with May‐Grunwald‐Giemsa. Myeloid GFP+ cells show the characteristics of macrophages, with kidney‐shaped nuclei and vacuoles, whereas lymphoid GFP+ cells revealed a typical lymphocytic morphology, with a nonlobed nucleus and a high nuclear:cytoplasmic ratio. Images were taken with a Zeiss AxioImager Z1 micorscope, using a 100× oil‐immersion objective. Scale bar: 20 µm
3.2. The lymphoid mpeg1‐positive cells are B cells
Because we initially observed that lymphoid mpeg1:GFP + cells co‐expressed the antigen‐presenting cell mhc2dab transgene, we hypothesized they may be B lymphocytes.7 We sorted GFP+ and GFP− subsets from the WKM lymphoid fraction of mpeg1.1:GFP fish and scored them for hematopoietic markers by qPCR, comparing them to GFP subsets isolated from the combined myeloid and precursor fractions (hereafter referred to as the PM fraction) (Fig. 2Ai, 2B). Consistent with their expected monocyte/macrophage identity, GFP+ PM cells showed strong expression of mpeg1.1 and csf1ra, but not mpx, a neutrophil marker (Fig. 2Bi). Within the lymphoid gate, GFP+ cells also expressed mpeg1.1, although at a lower level. Expression of mpeg1.1 mRNA was undetectable in the lymphoid GFP− population, thus indicating that fluorophore expression in the mpeg1 reporter line reflects endogenous gene expression. Interestingly, lymphoid GFP+ cells did not express csf1ra (Fig. 2Bi), supporting the hypothesis that this population represents a leukocyte subset distinct from mononuclear phagocytes. Rather, examination of canonic B‐cell‐associated genes revealed strong expression of pax5, ighz, and ighm (Fig. 2Bii). The expression of B‐cell genes was not restricted to mpeg1:GFP + cells within the lymphoid fraction, as similar levels of transcripts were also found in the GFP− subset. Additionally, the lymphoid GFP+ cells also lacked expression of T‐cell markers such as lck, cd28, and cd4.1, which were found to be highly enriched in the GFP− subset (Fig. 2Biii). Altogether, these results indicate that mpeg1:GFP marks a subpopulation of B lymphocytes that contains a mixture of IgM‐ and IgZ‐expressing cells. We concluded that, in addition to mononuclear phagocytes, mpeg1.1 also marks cells from the B‐cell lineage. These findings were further supported by imaging using May‐Grünwald‐Giemsa staining, showing that the majority of lymphoid mpeg1:GFP + cells exhibit the expected morphology of lymphocytes, whereas PM GFP+ cells harbored features of macrophages (Fig. 2C), as previously described.7, 32
3.3. Single‐cell analyses of mpeg1‐expressing WKM cells
Our results revealed the existence of mpeg1.1 + B lymphocytes in the adult WKM. However, B‐cell transcripts were also found in the lymphoid mpeg1.1 − cell population, suggesting that not all B lymphocytes express mpeg1.1. To further explore the identity of lymphoid mpeg1.1 + cells, we turned to single‐cell analysis and examined transcript expression among individual hematopoietic cells using our previously reported single‐cell transcriptional profiling of zebrafish WKM.30 By querying this dataset, we first confirmed the existence of a mpeg1.1+ cluster associated with the B‐cell lineage (Fig. 3A, B). Extending our initial findings, we further found that these cells co‐expressed a large array of B‐specific genes including cd79a, cd37, ighm, pax5, as well as several transcripts encoding different light chain isotypes (Fig. 3C). As expected, B‐cell gene expression was negligible in cell clusters positive for the macrophage transcripts marco or mfap4 (Fig. S1). From these analyses, we estimated that mpeg1.1+ cells accounted for approximately 28.5% of total B lymphocytes (N = 47 out of 165 B cells found in the WKM), in line with our cytometry analysis. Next, we performed a comparative expression analysis between WKM‐derived mpeg1.1+ B cells and macrophages. Violin plots show the distribution of cells expressing each transcript (Fig. 3D). In agreement with the levels of fluorophores observed by flow cytometry, mpeg1.1 was expressed slightly higher in macrophages than in B cells. As expected, marco and mfap4 belonged to genes that were uniquely expressed in mpeg1.1+ macrophages, whereas cd79a, ighm, and ighd were expressed significantly higher in mpeg1.1+ B cells. Ighz and pax5 were also more expressed in B cells than in macrophages, but the very low cell number did not allow reaching significance. Collectively, these unbiased analyses of single‐cell gene expression showed that lymphoid mpeg1.1 + cells display a robust B lymphocyte signature.
3.4. Tissue distribution of mpeg1.1 + B cells
Because we demonstrated that mpeg1.1+ lymphocytes and myeloid cells could be discriminated by their expression of ighm, we next crossed mpeg1.1:mCherry fish to ighm:GFP transgenic animals, which specifically mark IgM‐expressing B lymphocytes.26 As expected, flow cytometry analysis of adult double transgenic fish identified a prominent double positive cell subset in the WKM (Fig. 4Ai). This population, defined as mpeg1.1 lo ighm+, accounted for 11.5 ± 7.4% of total WKM blood cell numbers (n = 6). Two other subsets with differential levels of mCherry and GFP expression (mpeg1.1 hi ighm − and mpeg1.1 − ighm hi) were also present, although less abundant (4.2 ± 0.7% and 4.2 ± 1.7% of total WKM cell numbers, respectively; n = 6). Based on our previous gene expression analyses, the mpeg1.1+ighm− population likely contains a mixture of mononuclear phagocytes and mpeg1.1+ IgZ‐expressing B lymphocytes, which are not labeled in the ighm:GFP line.26 The fractionation of ighm:GFP+ cells into two subpopulations according to mpeg1.1 expression was also consistent with our initial identification of ighm transcripts in both mpeg1.1:GFP+ and mpeg1.1:GFP− lymphoid subsets, and reflects the heterogeneity of the ighm:GFP+ population. Importantly, mpeg1.1+ ighm+ B cells (mpeg1 lo ighm +) appeared to constitute the prominent cell subset among IgM‐expressing cells, as approximately 70% of GFP+ cells co‐expressed mpeg1.1:mCherry.
Figure 4.
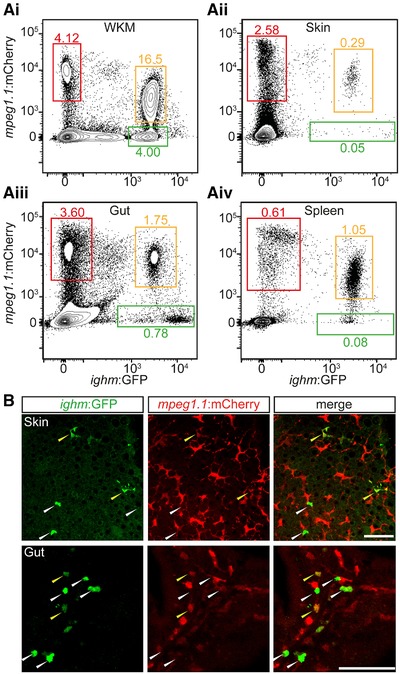
Distribution of mpeg1.1‐positive B cells in adult tissues. (A) Distribution of fluorescence in cell suspensions prepared from WKM (Ai), skin (Aii), gut (Aiii), and spleen (Aiv) of an adult Tg(ighm:GFP; mpeg1.1:mCherry) fish. In each organ, three main populations can be discriminated: mpeg1.1 hi; ighm− cells (red gate); mpeg1.1−; ighm hi B cells (green gate); and mpeg lo; ighm+ cells (orange gate). Percentages represent the single individual (mean ± sd of 5 fish: see text). (B) Immunofluorescence on skin scales (upper panels) and gut sections (lower panels) from an Tg(ighm:GFP; mpeg1.1:mCherry) reporter fish. The GFP (left), mCherry (middle), and merged (right) images are shown. White arrowheads indicate ighm+; mpeg1.1− cells; yellow arrowheads indicate ighm+; mpeg+ cells. Images were taken with a Zeiss LSM 780 inverted confocal microscope, using a 20× plan apochromat objective. Scale bar: 50 µm
We next sought to determine whether the two subsets of B cells existed in peripheral tissues. Using the same mpeg1.1:mCherry; ighm:GFP double transgenic fish, we analyzed the spleen, gut, and skin, known to host IgM‐expressing B lymphocytes.26, 33 Whereas the proportion of IgM + cells greatly varied among tissues, the majority of IgM+ cells showed co‐expression with mpeg1.1 in all samples examined (Fig. 4A). Confocal imaging and immunohistochemistry analyses confirmed the presence of double positive cells scattered in the skin or located along the basal lamina propria of the gut in double transgenic animals (Fig. 4B). Interestingly, the skin double positive cells displayed a dendritic morphology that was barely distinguishable from single mpeg1.1 + ighm − cells, probably due to the fact that all these cells are located between epithelial cells in the epidermis. Collectively, these results indicate that mpeg1.1 expression is persistent in mature B lymphocytes and labels nearly all IgM+ B cells in the periphery.
3.5. Mpeg1 expression does not discriminate a subset of phagocytic B cells in zebrafish
Previous studies have established the central role of mpeg1 in killing intracellular bacteria,13 as well as the phagocytic capabilities of teleost B cells.34 Given the clear distribution of ighm+ B cells into mpeg1.1+ and mpeg1.1− populations, we thus next wondered whether expression of the microbicidal mpeg1.1 gene could discriminate a subset of phagocytic B cells. We injected 1 µm blue‐fluorescent FluoSpheres in the peritoneal cavity of mpeg1.1:mCherry; ighm:GFP adult fish, where mpeg1.1 + B cells are present in steady‐state conditions (Fig. 5A). We then collected the IPEX at different time points and quantified the percentage of phagocytic cells within each fraction by flow cytometry. As expected, both the mpeg1.1+ighm− and mpeg1.1−ighm− cell populations, which contain professional phagocytes (macrophages and polynuclear cells, respectively) actively engulfed blue beads between 2 and 8 hr postinjection (hpi) (Fig. 5B). By contrast, the phagocytic capacity of ighm+ B cells was negligible, as we did not observe any particle internalization for either mpeg1.1+ or mpeg1.1− ighm+ B cells (Fig. 5B). Importantly, all phagocytic ighm− cells clustered within the myeloid gate (data not shown), therefore ruling out the presence of ighm− B cells in the phagocytic population. Together, these results indicate that zebrafish ighm+ B cells lack phagocytic activity, in line with our previous observations in this model.26
Figure 5.
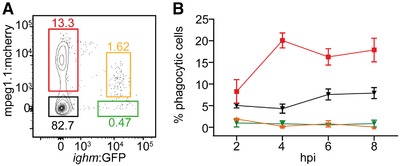
Mpeg1.1 expression does not discriminate a subset of phagocytic B cells in zebrafish. (A) Distribution of fluorescence in cell suspension from the IPEX of adult Tg(ighm:GFP; mpeg1.1:mCherry) fish. Percentages of cells in each gate refer to the total live cell fraction and represent an average of 3 individuals. (B) Percentage of phagocytic cells within each of the fractions described in (A), at different hours postinjection (hpi) of blue fluorescent beads. Each time point represents the average of 3 individuals
3.6. Adult tissue macrophages, except in the skin, develop in an irf8‐dependent manner
The unexpected observation that B lymphocytes represent a significant proportion of the total mpeg1.1:GFP+ population in adults prompted us to examine in more details the phenotype of macrophage‐deficient mutants previously characterized using mpeg1.1 reporters. Notably, we focused on mutants for the macrophage‐specific transcription factor irf8, as they are devoid of mature macrophages during embryonic life, but seem to recover later in life, based on the presence of mpeg1.1:GFP+ cells in juvenile and adult fish.24 To rigorously assess the lineage identity of irf8‐independent mpeg1.1+ cells, we analyzed hemato‐lymphoid organs isolated from irf8 null; mpeg1.1:GFP adult animals by flow cytometry. In the WKM, GFP lo B lymphocytes were present in proportion similar to that of their wild‐type siblings (Fig. 6A, red gate). However, the mutants exhibited a profound deficit in macrophages, as demonstrated by the complete loss of the minor GFP hi population (Fig. 6A, blue gate). In the periphery, examination of spleen, gut, and skin cell suspensions showed no reproducible alterations in the frequency of mpeg1.1 + cells. However, flow cytometry profiling revealed important differences between controls and irf8 null fish (Fig. 6B,C). When back‐gated to the lymphoid or myeloid lineages, we found, as expected, that the GFP+ populations in control animals scattered through both the lymphoid and myeloid gates (Fig. 6B). In contrast, the majority of the GFP+ cells in the spleen and gut of irf8 null mutants segregated in the lymphoid gate only, thus indicating a deficit in macrophages. However, irf8 null mpeg1.1 + cells in the skin segregated in both the lymphoid and myeloid fractions with frequencies that mirrored the wild‐type controls, thus suggesting a normal mononuclear phagocyte compartment.
Figure 6.
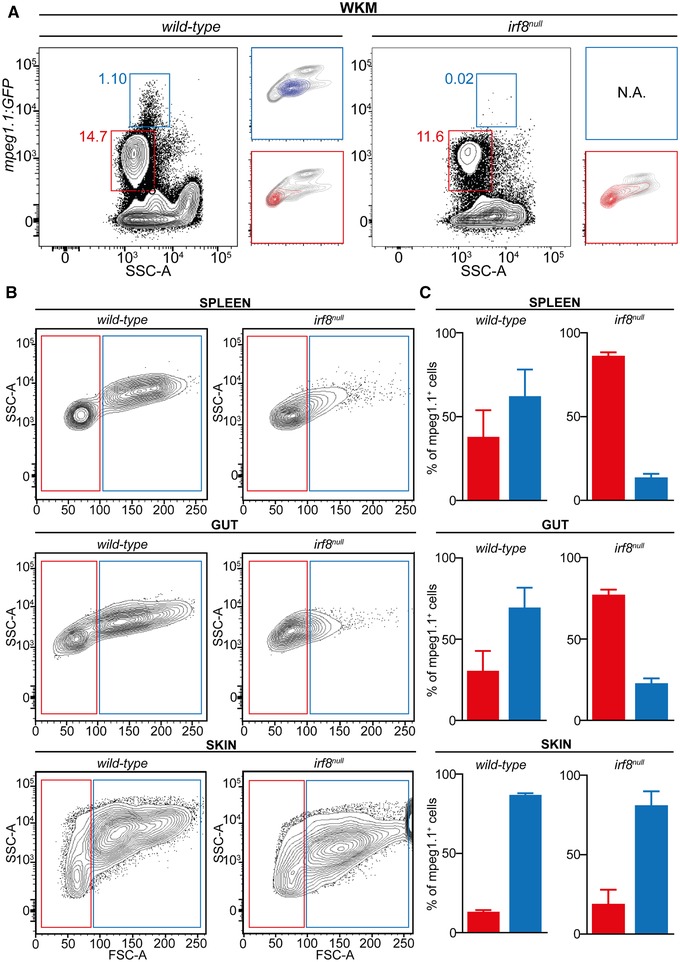
Adult zebrafish irf8 mutants display myeloid deficiencies. (A, B) Flow‐cytometry analysis of organ cell suspensions from adult wild‐type and irf8 null fish carrying the mpeg1.1:GFP reporter. (A) The whole mpeg1.1:GFP+ cell cluster of the WKM is split into an mpeg1.1 lo (red frame) and a mpeg1.1 hi (blue frame) subsets. Lateral panels show the projection of the mpeg1.1 lo and mpeg1.1 hi clusters into the FSC/SSC plot. Percentages relative to the total mpeg1.1:GFP+ cells are indicated aside of each gate and refer to a single individual (mean ± sd of 3 fish: see text). (B) FSC/SSC plots showing the distribution of mpeg1.1:GFP+ cells from the spleen, gut, and skin in the lymphoid (red) and MP (blue) gates. (C) Quantification of mpeg1.1:GFP+ lymphoid (red) and MP (blue) cells depicted in (B). Columns represent the percentage relative to the total mpeg1.1:GFP+ population of each organ. Error bars represent sd (n = 3)
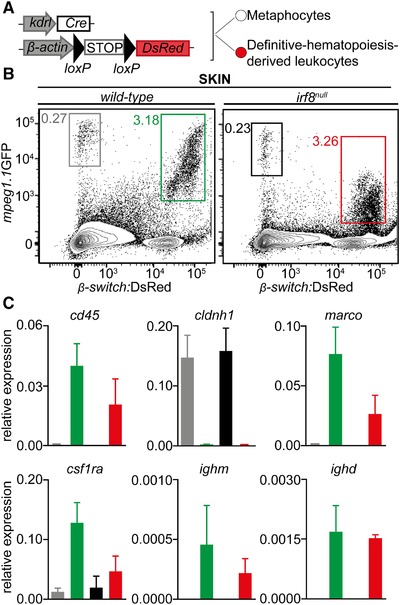
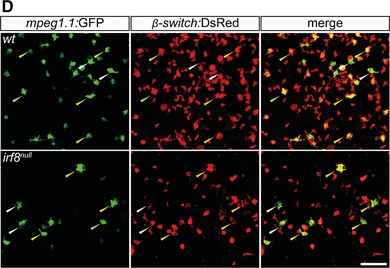
Because the identification of a distinct mpeg1.1 + cell population with a nonhematopoietic origin has been recently reported in the zebrafish epidermis, we next sought to evaluate to which extend these cells, referred to as metaphocytes,23 contributed to the mpeg1.1 + population found in the skin of irf8 null mutants. To do so, we took advantage of Tg(kdrl:Cre; ßactin2:loxP‐STOP‐loxP‐DsRed), a fate‐mapping model where constitutive Cre expression is targeted to endothelial cells,25 resulting in permanent labeling of hemogenic endothelium‐derived adult hematopoietic cells (Fig. 7A). Due to their distinct ectodermal origin,23 mpeg1.1 + metaphocytes were not expected to be labeled in this system. We thus crossed the irf8 mutant line to Tg(kdrl:Cre; ßactin2:loxP‐STOP‐loxP‐DsRed; mpeg1.1:eGFP), and analyzed the adult skin for GFP and DsRed using flow cytometry. In line with our hypothesis, in WT controls, two major mpeg1.1:GFP+ populations could be observed: GFP+DsRed− and GFP+DsRed+ (Fig. 7B). To further characterize these fractions, we analyzed them for hematopoietic and epithelial markers (Fig. 7C). Using these criteria, we concluded that the GFP+ DsRed− cells were nonhematopoietic, as demonstrated by the complete lack of expression of the pan‐leukocytic cd45 and other myeloid and lymphoid genes. Rather, GFP+ DsRed− cells expressed high level of cldnh1 transcripts and likely qualified as the so‐called metaphocytes (Fig. 7C). As expected, mpeg1.1 + mononuclear phagocytes and B cells belong to the GFP+DsRed+ positive population, which was negative for cldnh1 expression. As shown in Figure 6B, the relative representation of both populations was not significantly changed in irf8 null fish skin. This indicated that neither the ontogeny nor maintenance of metaphocytes rely on irf8. In addition, these results also validated our previous observations that the irf8 deficiency does not affect skin macrophage development in zebrafish. These data were corroborated by fluorescent confocal analyses performed on fish scales (Fig. 7D). Collectively, these findings demonstrated that irf8 mutant fish have a profound deficit of mononuclear phagocytes in the WKM, spleen, and gut but subnormal numbers of mononuclear phagocytes in the skin. We conclude that, with the exception of the skin, mpeg1.1+ hematopoietic cells in most organs mainly consist of B lymphocytes, which development is largely independent on irf8.
Figure 7.
Skin adult macrophages develop independently of irf8. (A) Scheme of the transgenic lines used to mark endothelial‐derived definitive hematopoiesis (B) Flow cytometry analysis on a skin cell suspension from an adult Tg(kdrl:Cre; β‐actin:switch‐DsRed; mpeg1.1:GFP) triple transgenic (left panel) discriminates two mpeg1.1+ populations based on DsRed fluorescence levels: mpeg1.1:GFP+; β‐actin:switch‐DsRed− (grey gate) and mpeg1.1:GFP+; β‐actin:switch‐DsRed+ (green gate) cells. The frequency of each population is unchanged in irf8 null fish (right panel, grey and red gates). (C) For each genotype, gene expression in mpeg1.1:GFP+; β‐actin:switch‐DsRed− and mpeg1.1:GFP+; β‐actin:switch‐DsRed+ cells examined for the presence of hematopoietic (cd45), epithelial (cldnh1), macrophage (marco, csf1ra), and B‐cell‐ (ighm, ighd) specific transcripts. The color code matches the FACS plots. Units on the y‐axis represent transcript expression normalized to ef1α expression levels. Error bars represent sem (n = 3). (D) Fluorescence of GFP (left panels) and DsRed (middle panels) in the skin of adult Tg(mpeg1.1:GFP; β‐actin:switch‐DsRed+) wild‐type (upper panels), and irf8 null (lower panels) fish. The right panels show a merge of both fluorescent channels (n = 3). Yellow arrowheads indicate mpeg1.1:GFP+; β‐actin:switch‐DsRed+ cells; white arrowheads indicate mpeg1.1:GFP+; β‐actin:switch‐DsRed− cells. Images were taken with a Zeiss LSM 780 inverted confocal microscope, using a 20× plan apochromat objective. Scale bar: 50 µm
4. DISCUSSION
The adult zebrafish has recently emerged as a promising model system to perform immunologic investigations. However, as a relatively latecomer to the field of immunology, only a limited number of antibodies are available against hematopoietic cells, thus precluding their prospective isolation using techniques traditionally performed in the mouse model. To overcome these limitations, most studies in adult zebrafish take advantage of transgenic lines marking leukocytes. Among these, many have employed mpeg1.1‐driven transgenes to be a specific reporter for adult macrophages. However, these macrophage‐reporter lines were originally created to perform live‐imaging studies in transparent embryos and larvae, and their reliability in adults has never been evaluated.
In this work, we disprove the wide assumption that mpeg1.1 serves as a specific pan‐macrophage marker in the adult zebrafish, as we found that B cells also express mpeg1.1. Importantly, we showed that fluorophore expression driven by the 1.2 kb‐promoter recapitulates the endogenous expression pattern of mpeg1.1 in B cells, thus resulting in labeling of mpeg1.1+ B lymphocytes in vivo, alongside mononuclear phagocytes. Interestingly, our findings correlate with current knowledge of the Mpeg1 expression in mammals. Indeed, although Mpeg1 was originally described in human and mouse as a macrophage‐specific gene (hence its name, macrophage‐expressed gene 1),11 recent evidence indicated that it is more broadly expressed that initially thought. For example, Mpeg1 was shown to be constitutively expressed in splenic marginal zone (MZ) B cells, where it stands as a specific marker to differentiate them from follicular (FO) B cells.35 Similarly, we identified two subsets of B cells based on mpeg1.1 expression in zebrafish. However, histologic examination of secondary lymphoid tissues did not allow identifying any specific anatomic areas where these two populations would segregate. As teleosts lack well‐organized lymphoid structures and do not form germinal centers (a feature that arose in birds across the vertebrate phylum36), whether mpeg1.1 can discriminate different zebrafish B cell subsets based on their locations thus remains an open question. In addition, Mpeg1 expression in mammals was also reported in a large array of innate immune cells, including dendritic cells, neutrophils, and NK cells.13 Based on our analyses in zebrafish, we conclude that mpeg1.1 is not expressed in neutrophils, or in other lymphocytes. Interestingly, zebrafish possess three mpeg1 paralogues37 and it remains possible that either mpeg1.2 or mpeg1.3, whose expression pattern is poorly characterized, will mark other hematopoietic populations. Finally, it was shown that in inflammatory conditions, Mpeg1 can also be induced in nonhematopoietic cells, such as barrier cells (keratinocytes and mucosal epithelia).13 Whereas our study did not address whether expression of zebrafish mpeg1.1 was similarly induced upon inflammation, we did find both endogenous and transgenic expression of mpeg1.1 outside of the hematopoietic tissue in steady‐state conditions, in a subset of cells present in the adult skin. Based on their nonhematopoietic identity and their expression of cldnh, these cells likely represent metaphocytes, ectodermal‐derived myeloid‐like cells with antigen‐presenting properties that were recently described in the zebrafish epidermis.23 However, in this study, the authors reported metaphocytes to represent 30% of the whole mpeg1.1+ population present in the skin, which is not concordant with our findings where only 8% of total mpeg1.1+ cells are nonhematopoietic. This underestimation is likely due to the nature of the tissue sample being analyzed, as our quantification refers to the whole skin (consisting of the three layers: epidermis, dermis, and hypodermis) whereas Lin et al. analyzed the epidermis only.23
Our work also provides strong evidence that zebrafish irf8 is vital for the proper development of macrophages throughout life and identified most mpeg1.1+ hematopoietic cells found in adult irf8 mutants as B lymphocytes. These results challenge a previous report that concluded, based on the progressive, albeit incomplete, recovery of mpeg1.1+ cells in irf8 mutant fish, that irf8 was required for the ontogeny of embryonic macrophages but dispensable for the formation of adult macrophages.24 In this study, however, the assignment to the macrophage identity exclusively relied on the detection of mpeg1.1:GFP+ cells by fluorescent imaging and was not supported by expression data. Here, by performing a detailed phenotypic characterization of these cells, we unambiguously demonstrated their B‐cell identity. Such conclusions are further supported by the fact that the timing of appearance of irf8‐independent mpeg1.1+ cells (starting at the juvenile stage) coincides with that of the ontogeny of B cells, which occurs at around 3 wk of development.26
Collectively, our data demonstrated that adult zebrafish irf8 mutants are devoid of most tissue macrophages, a phenotype that is consistent with the well‐established role of Irf8 as a critical regulator of myelopoiesis in mammals.38 One exception is the skin, where we found mpeg1.1‐positive cells harboring a myeloid gene signature. We postulate that these cells may constitute the equivalent of mammalian Langerhans cells, whose development and maintenance appears to be independent of Irf8 in both humans and mice.39, 40 However, further characterization of their cytochemical properties, ultrastructure, and additional functions will be required to test whether these cells are the true counterparts of mammalian Langerhans cells. In addition, by excluding a role for irf8 in the regulatory network that controls the ontogeny and maintenance of metaphocytes, this study also provides valuable insights into the biology of this newly discovered cell population.
Finally, our work sheds new lights on B cell immunity in zebrafish and may open new perspectives in the field of comparative immunology. As expression of perforin‐2 in professional phagocytes is tied to microbicidal activity,12, 13, 37, 41 it was tempting to speculate that mpeg1 could discriminate functionally distinct B cell populations in the zebrafish. This hypothesis was further supported by evidence from the literature that B cells in teleosts are capable to phagocytose particles and microorganisms and also display microbicidal activity.42, 43 However, the phagocytic ability of zebrafish B cells appears to be negligible, as we show here that they failed to uptake intraperitoneally injected latex beads, in contrast to professional phagocytes such as macrophages and neutrophils. Notably, whereas phagocytosis has been proposed as a common feature of teleost B cells,34 only a few studies so far have explored the phagocytic properties of zebrafish B cells, with contradicting results. Over the course of characterizing B cell ontogeny through the use of new fluorescent transgenic reporter lines, our group initially reported no significant phagocytosis of pHrodo‐labeled E.coli or latex beads by zebrafish IgM‐expressing B cells, either in vivo or in vitro.26 In another study by Zhu et al., internalization in vitro of soluble KLH by zebrafish B lymphocytes led the authors to consider these cells as phagocytic.44 However, endocytosis, rather than phagocytosis, is likely to be the main pathway for B cell acquisition of such small antigens.45 In support of this, the same study found that B cells poorly engulfed large bacteria,44 whose internalization more likely relies on active phagocytosis.45 As the phagocytic potential in teleost fish varies between different species and greatly depends on the anatomic source of B cells as well as the nature of the ingested particles,42, 46, 47, 48, 49 it could be hypothesized that zebrafish B cell populations from different compartments have distinct phagocytic capabilities. On the other hand, it is also possible that this feature was lost in zebrafish in the course of evolution, in line with the specific characteristics that separate many bony fish. Further investigation, out of the scope of the present study, will be required to address these important questions. Importantly, the poor phagocytic activity observed in B lymphocytes does not exclude a possible conservation of mpeg1 microbicidal properties within the zebrafish B cell lineage, as B lymphocytes can also be infected by pathogenic bacteria that can enter the cell through phagocytosis‐independent mechanisms.50, 51 It will be interesting to test this hypothesis using live invasive bacterial pathogens.
To conclude, our study unambiguously reveals that mpeg1.1 does not constitute an ideal marker to track macrophages in the adult zebrafish. Whereas the new observations made here do not complicate studies undertaken at ages before B‐lymphocytes appears (at around 3 wk of age), it is possible that due to misinterpretation, some of the conclusions drawn in previous reports relying on the specificity of mpeg1.1 expression in adults may need reassessment. For example, a recent study reported the existence of irf8‐independent macrophages in the gut.52 Although we cannot exclude that a subpopulation of intestinal macrophages may still develop in the absence of a functional irf8, based on our analyses it is more likely that these cells are in fact B cells. This hypothesis, which remains to be fully explored, is also supported by the severe reduction in macrophage‐specific C1q genes reported in the gut of irf8 mutants.52
Although the two hematopoietic mpeg1.1+ cell populations can be theoretically discriminated in adult tissues by flow cytometry based on the combination of their light‐scatter characteristics and different levels of mpeg1.1 expression (high for macrophages and low for B lymphocytes), it would be interesting to assess other markers such as csf1ra and mfap4, for which fluorescent reporters are available.53, 54 Whereas their specificity in adults remains to be determined, these lines may represent possible alternatives to the use of mpeg1.1‐driven transgenic lines for the study of macrophages in adult fish. Nevertheless, because none of these tools are ever specific, we would caution investigators working on adult immune cells not to rely exclusively on transgene expression for cell characterization. Rather, we would advise to always complement these analyses with cell isolation and gene expression approaches that will faithfully identify the cells of interest.
AUTHORSHIP
J.Y.B. and V.W. designed the research and directed the study. G.F., E.G., M.R., and M.M performed zebrafish experiments. S.I. and D.M.L. performed single‐cell RNA sequencing analysis. J.Y.B. and V.W. wrote the manuscript with comments from all authors. J.Y.B. and V.W. contributed equally to this work.
DISCLOSURES
The authors declare no conflicts of interest.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
We thank J.‐M. Vanderwinden and the LiMiF for technical support with confocal imaging, Christine Dubois for help with flow cytometry and Marianne Caron for technical assistance, and members of the Wittamer and Bertrand labs for critical comments and suggestions. V.W. is an investigator of WELBIO and is also supported by grants from the Fonds National de la Recherche Scientifique (FNRS) and The Minerve Foundation. J.Y.B. is funded by the Swiss National Fund (31003_166515). D.M.L. is supported by R24 OD016761 and R01 CA211734 from the National Institutes of Health. G.F. received funding from a FNRS fellowship (Research Fellow).
Ferrero G, Gomez E, lyer S, et al. The macrophage‐expressed gene (mpeg) 1 identifies a subpopulation of B cells in the adult zebrafish. J Leukoc Biol. 2020;107:431–443. 10.1002/JLB.1A1119-223R
Contributor Information
Julien Y. Bertrand, Email: julien.bertrand@unige.ch.
Valérie Wittamer, Email: vwittame@ulb.ac.be.
REFERENCES
- 1. Gut P, Baeza‐Raja B, Andersson O, et al. Whole‐organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol. 2013;9(2):97‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Rooijen E, Fazio M, Zon LI. From fish bowl to bedside: the power of zebrafish to unravel melanoma pathogenesis and discover new therapeutics. Pigment Cell Melanoma Res. 2017;30(4):402‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gore AV, Pillay LM, Venero Galanternik M, Weinstein BM. The zebrafish: a fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2018;7(3):e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perlin JR, Robertson AL, Zon LI. Efforts to enhance blood stem cell engraftment: recent insights from zebrafish hematopoiesis. J Exp Med. 2017;214(10):2817‐2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rennekamp AJ, Peterson RT. 15 years of zebrafish chemical screening. Curr Opin Chem Biol. 2015;24:58‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Svoboda O, Stachura DL, Machonova O, Zon LI, Traver D, Bartunek P. Ex vivo tools for the clonal analysis of zebrafish hematopoiesis. Nat Protoc. 2016;11(5):1007‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wittamer V, Bertrand JY, Gutschow PW, Traver D. Characterization of the mononuclear phagocyte system in zebrafish. Blood. 2011;117(26):7126‐7135. [DOI] [PubMed] [Google Scholar]
- 8. Watanabe S, Alexander M, Misharin AV, Budinger GRS. The role of macrophages in the resolution of inflammation. J Clin Invest. 2019;130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Theret M, Mounier R, Rossi F. The origins and non‐canonical functions of macrophages in development and regeneration. Development. 2019;146(9). [DOI] [PubMed] [Google Scholar]
- 10. Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage‐lineage expression in zebrafish. Blood. 2010;117(4):e49‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katrina Spilsbury M‐AOM, Wu WM, Peter B, Rowe GS, Yoichi T. Isolation of a novel macrophage‐specific gene by differential cDNA analysis. Blood. 1995;85(6):1620‐1629. [PubMed] [Google Scholar]
- 12. McCormack R, de Armas L, Shiratsuchi M, Podack ER. Killing machines: three pore‐forming proteins of the immune system. Immunol Res. 2013;57(1‐3):268‐278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCormack RM, de Armas LR, Shiratsuchi M, et al. Perforin‐2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Travnickova J, Tran Chau V, Julien E, et al. Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat Commun. 2015;6:6227. [DOI] [PubMed] [Google Scholar]
- 15. Peri F, Nusslein‐Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0‐ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133(5):916‐927. [DOI] [PubMed] [Google Scholar]
- 16. Gerri C, Marin‐Juez R, Marass M, Marks A, Maischein HM, Stainier DYR. Hif‐1alpha regulates macrophage‐endothelial interactions during blood vessel development in zebrafish. Nat Commun. 2017;8:15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tobin DM, Vary JC, Jr , Ray JP, et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140(5):717‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Madigan CA, Cambier CJ, Kelly‐Scumpia KM, et al. A macrophage response to mycobacterium leprae phenolic glycolipid initiates nerve damage in leprosy. Cell. 2017;170(5):973‐985. e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roh‐Johnson M, Shah AN, Stonick JA, et al. Macrophage‐dependent cytoplasmic transfer during melanoma invasion in vivo. Dev Cell. 2017;43(5):549‐562. e546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van den Berg MCW, MacCarthy‐Morrogh L, Carter D, et al. Proteolytic and opportunistic breaching of the basement membrane zone by immune cells during tumor initiation. Cell Rep. 2019;27(10):2837‐2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chia K, Mazzolini J, Mione M, Sieger D. Tumor initiating cells induce Cxcr4‐mediated infiltration of pro‐tumoral macrophages into the brain. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keightley MC, Wang CH, Pazhakh V, Lieschke GJ. Delineating the roles of neutrophils and macrophages in zebrafish regeneration models. Int J Biochem Cell Biol. 2014;56:92‐106. [DOI] [PubMed] [Google Scholar]
- 23. Lin X, Zhou Q, Zhao C, Lin G, Xu J, Wen Z. An ectoderm‐derived myeloid‐like cell population functions as antigen transporters for langerhans cells in zebrafish epidermis. Dev Cell. 2019;49(4):605‐617. e605. [DOI] [PubMed] [Google Scholar]
- 24. Shiau CE, Kaufman Z, Meireles AM, Talbot WS. Differential requirement for irf8 in formation of embryonic and adult macrophages in zebrafish. PLoS One. 2015;10(1):e0117513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464(7285):108‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Page DM, Wittamer V, Bertrand JY, et al. An evolutionarily conserved program of B‐cell development and activation in zebrafish. Blood. 2013;122(8):e1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stachura DL, Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2011;101:75‐110. [DOI] [PubMed] [Google Scholar]
- 28. Ferrero G, Mahony CB, Dupuis E, et al. Embryonic microglia derive from primitive macrophages and are replaced by cmyb‐dependent definitive microglia in zebrafish. Cell Rep. 2018;24(1):130‐141. [DOI] [PubMed] [Google Scholar]
- 29. Balla KM, Lugo‐Villarino G, Spitsbergen JM, et al. Eosinophils in the zebrafish: prospective isolation, characterization, and eosinophilia induction by helminth determinants. Blood. 2010;116(19):3944‐3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang Q, Iyer S, Lobbardi R, et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single‐cell resolution using RNA sequencing. J Exp Med. 2017;214(10):2875‐2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R. R: A language and environment for statistical computing.R Foundation for Statistical Computing, Vienna, Austria (retrieved 08/2018): URL = https://www.R-project.org/.
- 32. Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4(12):1238‐1246. [DOI] [PubMed] [Google Scholar]
- 33. Liu X, Li YS, Shinton SA, et al. Zebrafish B cell development without a pre‐B cell stage, revealed by CD79 fluorescence reporter transgenes. J Immunol. 2017;199(5):1706‐1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sunyer JO. Fishing for mammalian paradigms in the teleost immune system. Nat Immunol. 2013;14(4):320‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kleiman E, Salyakina D, De Heusch M, et al. Distinct transcriptomic features are associated with transitional and mature B‐cell populations in the mouse spleen. Front Immunol. 2015;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neely HR, Flajnik MF. Emergence and evolution of secondary lymphoid organs. Annu Rev Cell Dev Biol. 2016;32:693‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Benard EL, Racz PI, Rougeot J, et al. Macrophage‐expressed perforins Mpeg1 and Mpeg1.2 have an anti‐bacterial function in zebrafish. Journal of Innate Immunity. 2015;7(2):136‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yanez A, Goodridge HS. Interferon regulatory factor 8 and the regulation of neutrophil, monocyte, and dendritic cell production. Curr Opin Hematol. 2016;23(1):11‐17. [DOI] [PubMed] [Google Scholar]
- 39. Hambleton S, Salem S, Bustamante J, et al. IRF8 mutations and human dendritic‐cell immunodeficiency. N Engl J Med;365(2):127‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chopin M, Seillet C, Chevrier S, et al. Langerhans cells are generated by two distinct PU.1‐dependent transcriptional networks. J Exp Med. 2013;210(13):2967‐2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCormack RM, Szymanski EP, Hsu AP, et al. MPEG1/perforin‐2 mutations in human pulmonary nontuberculous mycobacterial infections. JCI Insight. 2017;2(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J, Barreda DR, Zhang YA, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol. 2006;7(10):1116‐1124. [DOI] [PubMed] [Google Scholar]
- 43. Zhang YA, Salinas I, Li J, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11(9):827‐835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu LY, Lin AF, Shao T, et al. B cells in teleost fish act as pivotal initiating APCs in priming adaptive immunity: an evolutionary perspective on the origin of the B‐1 cell subset and B7 molecules. J Immunol. 2014;192(6):2699‐2714. [DOI] [PubMed] [Google Scholar]
- 45. Flannagan RS, Jaumouille V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol. 2012;7:61‐98. [DOI] [PubMed] [Google Scholar]
- 46. Edholm ES, Bengten E, Stafford JL, et al. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J Immunol. 2010;185(7):4082‐4094. [DOI] [PubMed] [Google Scholar]
- 47. Ronneseth A, Ghebretnsae DB, Wergeland HI, Haugland GT. Functional characterization of IgM+ B cells and adaptive immunity in lumpfish (Cyclopterus lumpus L.). Dev Comp Immunol. 2015;52(2):132‐143. [DOI] [PubMed] [Google Scholar]
- 48. Yang S, Tang X, Sheng X, Xing J, Zhan W. Development of monoclonal antibodies against IgM of half‐smooth tongue sole (Cynoglossus semilaevis) and analysis of phagocytosis of fluorescence microspheres by mIgM+ lymphocytes. Fish Shellfish Immunol. 2017;66:280‐288. [DOI] [PubMed] [Google Scholar]
- 49. Yang S, Tang X, Sheng X, Xing J, Zhan W. Development of monoclonal antibodies against IgM of sea bass (Lateolabrax japonicus) and analysis of phagocytosis by mIgM+ lymphocytes. Fish Shellfish Immunol. 2018;78:372‐382. [DOI] [PubMed] [Google Scholar]
- 50. García‐Pérez BEDlC‐Lp JJ, Castañeda‐Sánchez JI, Muñóz‐Duarte AR, et al. Macropinocytosis is responsible for the uptake of pathogenic and non‐pathogenic mycobacteria by B lymphocytes (Raji cells). BMC Microbiology. 2012(12):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bloomfield G, Kay RR. Uses and abuses of macropinocytosis. J Cell Sci. 2016;129(14):2697‐2705. [DOI] [PubMed] [Google Scholar]
- 52. Earley AM, Graves CL, Shiau CE. Critical role for a subset of intestinal macrophages in shaping gut microbiota in adult zebrafish. Cell Rep. 2018;25(2):424‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walton EM, Cronan MR, Beerman RW, Tobin DM. The macrophage‐specific promoter mfap4 allows live, long‐term analysis of macrophage behavior during mycobacterial infection in zebrafish. PLoS One. 2015;10(10):e0138949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gray C, Loynes CA, Whyte MK, Crossman DC, Renshaw SA, Chico TJ. Simultaneous intravital imaging of macrophage and neutrophil behaviour during inflammation using a novel transgenic zebrafish. Thromb Haemost. 2011;105(5):811‐819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


