Summary
In response to elevated ambient temperature Arabidopsis thaliana seedlings display a thermomorphogenic response that includes elongation of hypocotyls and petioles. Phytochrome B and cryptochrome 1 are two photoreceptors also playing a role in thermomorphogenesis. Downstream of both environmental sensors PHYTOCHROME INTERACTING FACTOR 4 (PIF4) is essential to trigger this response at least in part through the production of the growth promoting hormone auxin.
Using a genetic approach, we identified PHYTOCHROME INTERACTING FACTOR 7 (PIF7) as a novel player for thermomorphogenesis and compared the phenotypes of pif7 and pif4 mutants. We investigated the role of PIF7 during temperature‐regulated gene expression and the regulation of PIF7 transcript and protein by temperature.
Furthermore, pif7 and pif4 loss‐of‐function mutants were similarly unresponsive to increased temperature. This included hypocotyl elongation and induction of genes encoding auxin biosynthetic or signalling proteins. PIF7 bound to the promoters of auxin biosynthesis and signalling genes. In response to temperature elevation PIF7 transcripts decreased while PIF7 protein levels increased rapidly.
Our results reveal the importance of PIF7 for thermomorphogenesis and indicate that PIF7 and PIF4 likely depend on each other possibly by forming heterodimers. Elevated temperature rapidly enhances PIF7 protein accumulation, which may contribute to the thermomorphogenic response.
Keywords: Arabidopsis thaliana, auxin, PIF4, PIF7, thermomorphogenesis
Introduction
Ambient temperature influences plants in numerous ways. Their distribution, phenology, defence capacity, growth and development are altered by modest changes in average temperature (Quint et al., 2016; Gangappa et al., 2017; Lau et al., 2018; Casal & Balasubramanian, 2019). In response to mild temperature elevation Arabidopsis displays a number of growth and developmental responses known as thermomorphogenesis, which include accelerated flowering, hypocotyl and petiole elongation, a reduction of the stomatal index and leaf hyponasty (Quint et al., 2016; Casal & Balasubramanian, 2019). Some of these responses improve the cooling capacity of Arabidopsis rosettes, which is likely important for plants to cope with increased temperature (Crawford et al., 2012).
Thermomorphogenesis and photomorphogenesis are similar at different levels. This is particularly obvious when comparing shade and elevated temperature responses (Legris et al., 2017). In both cases environmental sensing depends at least in part on the photoreceptors phytochrome B (phyB) and cryptochrome 1 (cry1) (Jung et al., 2016; Legris et al., 2016; Ma et al., 2016; Pedmale et al., 2016; Casal & Balasubramanian, 2019). Other signalling components including ELONGATED HYPOCOTYL 5 (HY5), CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) and DE‐ETIOLATED 1 (DET1), which were initially identified for their role in light responses, also play an important role in thermomorphogenesis (Delker et al., 2014; Gangappa & Kumar, 2017; Park et al., 2017).
PHYTOCHROME INTERACTING FACTOR 4 (PIF4) is an essential component for high temperature response under most tested conditions, while the role of PIF1, PIF3 and PIF5 is minor (Koini et al., 2009; Stavang et al., 2009; Nomoto et al., 2012; Zhu et al., 2016; Qiu et al., 2019). PIF4 is inhibited by phyB and cry1 (Ma et al., 2016; Qiu et al., 2019), while PIF4 function depends on HEMERA that regulates both PIF4 abundance and its trans‐activating potential (Qiu et al., 2019). A key role of PIF4 is to induce expression of auxin biosynthetic and signalling genes ultimately leading to hypocotyl elongation (Franklin et al., 2011; Sun et al., 2012; Raschke et al., 2015). Hypocotyl elongation in response to shade and temperature elevation also depends on other phytohormones including gibberellic acid (GA) and brassinosteroids (BR) (Quint et al., 2016; Legris et al., 2017; Casal & Balasubramanian, 2019). BR acts in the hypocotyl while auxin biosynthesis mainly occurs in cotyledons before being transported to the hypocotyl to promote elongation (Stavang et al., 2009; Oh et al., 2012; Kohnen et al., 2016; Procko et al., 2016; Ibanez et al., 2018; Martinez et al., 2018; Bellstaedt et al., 2019).
Given the overlap of signalling components regulating temperature and shade responses and the central role of PHYTOCHROME INTERACTING FACTOR 7 (PIF7) in the phyB‐mediated neighbour proximity response, we decided to test whether PIF7 is required for elevated temperature‐induced growth responses.
Materials and Methods
Plant material
Arabidopsis thaliana Columbia (Col‐0) ecotype was used. The mutants phyB‐9 (Neff et al., 1998), cry1‐304 (Mockler et al., 1999), yuc2yuc5yuc8yuc9 (Nozue et al., 2015), pif4‐101, phyBpif4 (Lorrain et al., 2008), phyBpif7 (Galvao et al., 2019), phyB‐9pif4‐101pif5-3‐pif7‐1 (Goyal et al., 2016), pif4‐101pif5‐3pif7‐1 (de Wit et al., 2015), pif7‐1 and pif7‐2 (Leivar et al., 2008), were previously characterized. The transgenic PIF7‐HA line (pif7‐2/pPIF7::PIF7‐3HA‐tPIF7) was previously described (Galvao et al., 2019). Furthermore, cry1‐304phyB‐9, pif4‐101pif7‐2, cry1‐304pif4‐101pif5‐3 and cry1‐304pif4‐101pif5‐3pif7‐1, yuc2yuc5yuc8, yuc2yuc5yuc9, yuc2yuc8yuc9 and yuc5yuc8yuc9 were generated by crosses and confirmed by genotyping using oligonucleotides listed in the Supporting Information Table S1. The yuc alleles are as in Nozue et al. (2015).
Phenotypic characterization and growth conditions
Seed sterilization and stratification, plant growth and light conditions were described previously (de Wit et al., 2015; Kohnen et al., 2016). Long‐day (LD) or short‐day (SD) photoperiods correspond to 16 h light : 8 h dark or 8 h light : 16 h dark, respectively, with c. 120 µmoles m−2 s−1 of photosynthetically active radiation (PAR) in LD and SD. For hypocotyl elongation measurements, seeds were sown on sterile nylon meshes on the growth media. Seedlings were grown on vertical plates in an incubator (Model AR‐22L; CLF Plant Climatics, Wertingen, Germany) for 4 d at 21°C. High temperature treatment (28°C) started on day 5 at ZT2. For picloram (Sigma‐Aldrich, Steinheim, Germany, P5575) treatment, nylon meshes were transferred on day 5 before the temperature shift to half strength MS medium with the indicated picloram concentration (0.1% dimethyl sulphoxide (DMSO) for Mock). Seedlings imaging and measurements were described previously (de Wit et al., 2018). Petiole measurements were performed as described (de Wit et al., 2015). Following 14 d in a LD growth room, plants were transferred to AR‐22L incubators and acclimated for 1 d to constant 21°C (LD). The next morning (ZT3), temperature in one incubator was shifted to constant 28°C. Petiole length of leaf 3 was measured after 3 d of treatment.
RNA isolation and quantitative RT‐PCR
RNA isolation and reverse transcription quantitative polymerase chain reaction (RT‐qPCR) reactions were performed as previously described (Kohnen et al., 2016). Oligonucleotides are listed in Table S1.
ChIP‐qPCR
Briefly, 6‐d‐old PIF7‐HA (Galvao et al., 2019) seedlings grown in LD at 21°C were either kept at 21°C or shifted at ZT2 to 28°C for 2 h before harvesting in liquid nitrogen. Chromatin extraction was performed as described previously (Bourbousse et al., 2018) except that samples were crosslinked only with formaldehyde. Immunoprecipitation was performed as described previously (Gendrel et al., 2005) using an anti‐HA antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA; sc‐7392 X). The qPCR was done in triplicates on input and immunoprecipitated DNA. Oligonucleotides are listed in Table S1.
Western‐blot analysis
Total protein extracts from PIF7‐HA seedlings were obtained as previously described (Galvao et al., 2019). For PIF4 Western‐blot 20–25 seedlings were collected in liquid nitrogen and proteins extracted in 90 µl extraction buffer (100 mM Tris‐HCl pH 6.8, 5% SDS, 20% glycerol, 80 µM MG132, 20 mM DTT, 1× protease inhibitor cocktail (P9599; Sigma‐Aldrich), 1 mM bromophenolblue), boiled at 95°C for 5 min and centrifuged for 2 min. Protein samples were separated on 4–20% Mini‐Protean TGX gels (Bio‐Rad, Hercules, CA, USA) and blotted on nitrocellulose membrane (Bio‐Rad) using Turbo transfer system (Bio‐Rad). Membranes were blocked with 5% milk overnight at 4°C for αPIF4, and 1 h at room temperature for αHA, before probing with anti‐HA coupled with horseradish peroxidase (HRP) (Roche, Mannheim, Germany; Cat. 12013819001), polyclonal H3 (1 : 2000; Abcam, Cambridge, UK; Cat. no. 1791), polyclonal PIF4 (1 : 3000, Abiocode R2534‐4) or DET3 (1 : 20 000) antibodies. HRP‐conjugated anti‐rabbit was used as secondary antibody. Chemiluminescence signal were obtained with Immobilon Western Chemiluminescent HRP Substrate (Millipore, Merck KGaA, Darmstadt, Germany) on an ImageQuant LAS 4000 mini (GE Healthcare, Buckinghamshire, UK). Relative intensities correspond to the average of HA/H3 of six biological replicates obtained with imagej (https://imagej.nih.gov/ij/).
Yeast two‐hybrid assay
PIF7 and PIF4 full length coding sequences were cloned into the pGBKT7 and pGADT7 vectors (Clontech, Mountain View, CA, USA). After co‐transformation of yeast strain TATA (Hybrigenics, Paris, France) and selection of transformants, serial cell suspensions were spotted on synthetic drop‐out medium lacking leucine and tryptophan (SD‐LW) and plates were put at 30°C for 2 d. A β‐galactosidase assay was performed directly on yeast spots as previously described (Duttweiler, 1996).
Statistical analysis
We performed two‐way analysis of variance (ANOVA) (aov) and computed Tukey's Honest Significance Differences (HSD) test (agricolae package) with default parameters using R software (https://www.r-project.org/).
Results
The thermomorphogenic response depends on PIF7
We analysed the thermomorphogenic response in 4‐d‐old seedlings grown under LDs that were either kept at 21°C or transferred to 28°C for three additional days. We used this shift protocol to allow us to investigate the early response to increasing temperature. Consistent with previous reports (Koini et al., 2009; Stavang et al., 2009), wild‐type Col‐0 (WT) hypocotyl elongated robustly at 28°C while pif4 was largely unresponsive (Fig. 1a). The phenotype of both tested pif7 alleles was slightly less severe than pif4 while pif4pif7 was similar to pif4 (Fig. 1a). We also analysed the thermomorphogenic hypocotyl elongation response in SDs and found that pif7 like pif4 was largely unresponsive to temperature elevation (Fig. S1). We conclude that PIF7 is required for elevated ambient temperature‐induced hypocotyl elongation irrespective of day length and conducted all subsequent experiments in LDs because in nature higher temperatures are more common when days get long.
Figure 1.
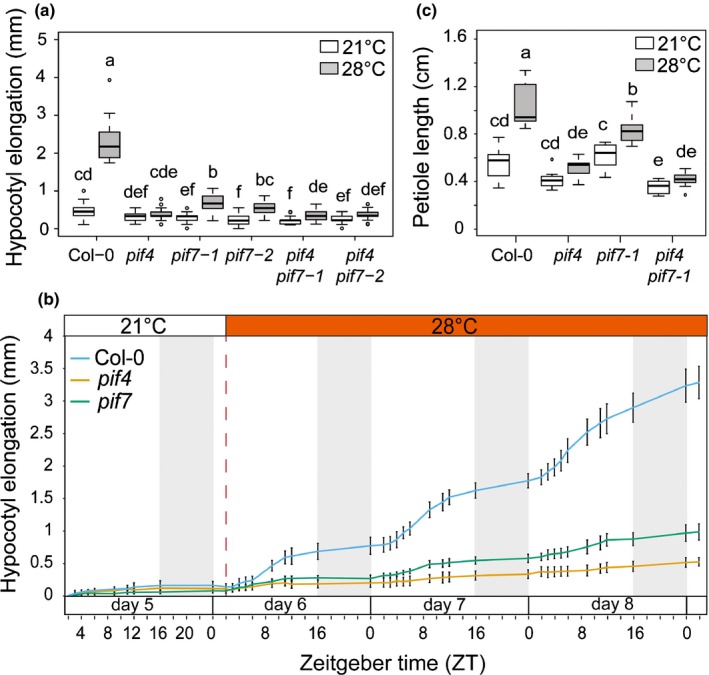
Thermomorphogenic response requires both PIF4 and PIF7 for hypocotyl and petiole elongation in Arabidopsis. (a) Hypocotyl elongation of wild‐type (Col‐0) and pif mutants grown in long days (LDs) at 21°C for 4 d then either kept at 21°C or transferred to 28°C (at ZT2 on day 5) for three additional days. Elongation during the last 3 d is indicated. Different letters indicate significant difference (two‐way ANOVA with Tukey's HSD test, P < 0.05, n > 25). (b) High‐resolution growth analysis of Col‐0, pif4‐101, and pif7‐1 seedlings. Hypocotyl elongation from LD‐grown seedlings (21°C) was measured from time‐lapse images with indicated intervals starting from ZT0 on day 5. The red dashed line indicates start of 28°C treatment at ZT2 on day 6. The grey zone represents the dark period. Data represent means ± 2 SE; n > 8. (c) Petiole lengths (leaf 3) of Col‐0 and pif mutants grown in LD at 21°C for 15 d then either kept at 21°C or transferred to 28°C for three additional days. Different letters indicate significant difference (two‐way ANOVA with Tukey's HSD test, P < 0.05, n = 10). For (a) and (c) the horizontal bar represents the median, boxes extend from the 25th to the 75th percentile, while whiskers extend to 1.5 times the interquartile range of the lower and upper quartiles, respectively, outliers are indicated with circles.
To determine whether the phenotype observed after 3 d reflects a similar defect in the growth pattern we followed growth kinetics of the WT, pif4 and pif7. Elevated temperature enhanced growth during the day while growth at night was limited in both conditions (Fig. 1b) (Park et al., 2017). Enhanced elongation triggered at 28°C during the first day of treatment depended on PIF4 and PIF7 (Fig. 1b). Consistent with the phenotype observed after 3 d, pif7 grew slightly more than pif4 during the next 2 d (Fig. 1b). We conclude that in response to temperature elevation growth during the day depends on PIF4 and PIF7. In constant light and LD, PIF4 controls day growth downstream of phyB and cry1 (Ma et al., 2016; Qiu et al., 2019). The importance of PIF7 in warm LD (Fig. 1a,b) prompted us to measure hypocotyl growth of phyBpif and cry1pif mutant combinations. Both phyB and cry1 mutants showed robust temperature‐induced elongation while the phyBcry1 double mutant was unresponsive suggesting that both photoreceptors are crucial for temperature‐controlled hypocotyl elongation (Fig. S2a). However, we note that cry1phyB double mutant had very long hypocotyls at 21°C possibly limiting further elongation at 28°C. As observed previously (Qiu et al., 2019), pif4 partially suppressed phyB (Fig. S2a,b). However, phyBpif7 had shorter hypocotyls than phyBpif4, both at 21°C and 28°C, highlighting the importance of PIF7 for phyB repressed hypocotyl elongation (Fig. S2a,b). The phyB phenotype was almost totally suppressed in phyBpif4pif5pif7 (Fig. S2b). Consistent with the dominant function of PIF4 downstream of cry1 (Ma et al., 2016), pif4pif5 was epistatic over cry1 with no further suppression observed in cry1pif4pif5pif7 (Fig. S2c). We conclude that PIF4 and PIF7 both promote hypocotyl elongation in response to increased temperature in LD, while their regulation by photoreceptors differs at least partially.
Later in development high temperature leads to petiole elongation (Koini et al., 2009), which we analysed in young rosettes that were either maintained at 21°C or transferred for 3 d to 28°C. Elevated temperature‐induced petiole elongation was most affected in pif4, but also impaired in pif7 and the response of pif4pif7 was very similar to pif4 (Fig. 1c). Taken together our results indicate that PIF7 is almost as important as PIF4 for thermomorphogenic growth responses.
PIF7 controls temperature‐induced expression of ‘auxin’ genes
PIF4 is essential to induce expression of YUC genes leading to higher auxin levels and growth (Sun et al., 2012). Transfer to 28°C led to significantly increased expression of YUC8 and YUC9 (after 90 and 180 min) while YUC2 induction was modest and not significant (Figs 2a, S3). PIF4 and PIF7 were required for temperature‐induced expression of YUC8 and the auxin signalling genes IAA29 and SAUR22 (Fig. 2a), demonstrating the requirement of both phytochrome‐interacting factors (PIFs) for enhanced ‘auxin gene’ expression and growth (Figs 1, 2). To test whether PIF7 may directly control the expression of YUC8 and IAA29 we performed ChIP experiments using a full genomic PIF7‐HA line (Galvao et al., 2019). This experiment showed that after 2 h at 28°C PIF7 was bound to the promoter of YUC8 and IAA29 at a position where PIF4 binding was reported previously (Fig. 2b) (Hornitschek et al., 2012; Sun et al., 2012). To assess the functional importance of temperature‐induced YUC expression we analysed hypocotyl elongation in a yuc2yuc5yuc8yuc9 quadruple mutant and all possible triple mutants. This experiment confirmed the importance of YUC8 and revealed a role for YUC2 in thermomorphogenesis (Fig. 2c) (Sun et al., 2012). In response to a lower red to far‐red (R : FR) ratio indicative of neighbouring plants PIF7 plays a particularly important role to enhance auxin production while PIF4 also regulates the response to auxin (Nozue et al., 2011; Hornitschek et al., 2012; Li et al., 2012; Pucciariello et al., 2018). We therefore compared the sensitivity of pif4, pif7 and pif4pif7 mutants to the synthetic auxin picloram in seedlings grown at 28°C (Fig. 2d). This experiment showed that although pif7 had a very small response to 28°C (mock) it responded like the WT to 2 µM picloram, while pif4 had a reduced response. At higher picloram concentrations pif7 also responded less than the WT. We note that the lower picloram response of pif4 compared to pif7 correlates with the growth phenotypes of the mutants after prolonged elevated temperature treatments (Figs 1, 2d).
Figure 2.
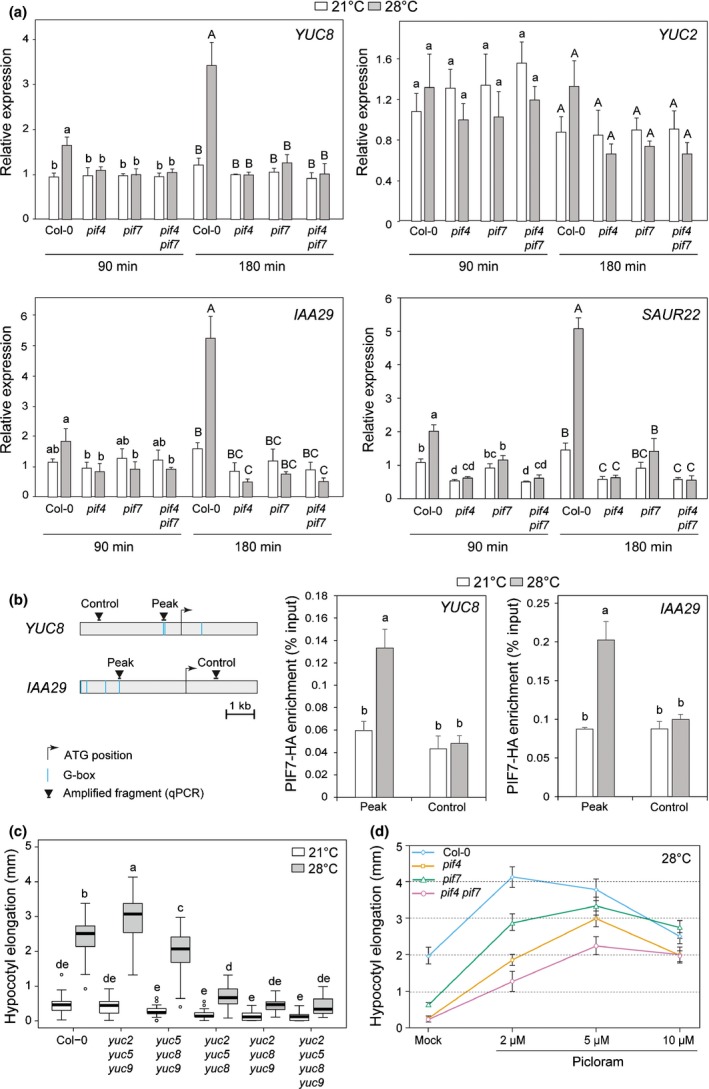
PIF4 and PIF7 regulate the auxin pathway during thermomorphogenesis in Arabidopsis. (a) Relative expression of auxin biosynthesis (YUC2 and YUC8) and auxin response (IAA29 and SAUR22) genes in 5‐d‐old Col‐0 and pif mutants either kept at 21°C or transferred to 28°C at ZT2; samples at 21 and 28°C were harvested at the same ZT. Gene expression values were calculated as fold induction relative to a Col‐0 sample at 21°C, t = 90 min. n = 3 (biological) with three technical replicas for each RNA sample. Data are means, ± 2 SE. Different letters indicate significant differences within timepoints (P < 0.05). (b) PIF7‐HA binding to the promoter of YUC8 and IAA29 evaluated by ChIP‐qPCR in 6‐d‐old seedlings either kept at 21°C or transferred for 2 h to 28°C at ZT2. Input and immunoprecipitated DNA were quantified by qPCR using primers shown on the schematic representation of the genes with ‘Peak’ indicating where PIF4 binding was identified before (left). PIF7‐HA enrichment is presented as IP/Input and error bars show standard deviation from three technical replicas. Different letters indicate significant differences (P < 0.05). Data from one representative experiment are shown. (c) Hypocotyl length of wild‐type (Col‐0) and yuc mutants grown in long day (LD) at 21°C for 4 d then kept at 21°C or transferred to 28°C for three additional days. Growth during the last 3 d is indicated. The horizontal bar represents the median, boxes extend from the 25th to the 75th percentile, while whiskers extend to 1.5 times the interquartile range of the lower and upper quartiles, respectively, outliers are indicated with circles. Different letters indicate significant difference (two‐way ANOVA with Tukey's HSD test, P < 0.05 n > 25). (d) Hypocotyl elongation at 28°C of Col‐0 and pif mutants in response to indicated concentrations of exogenously applied synthetic auxin, picloram. Seedlings were grown and measured as indicated in (a) picloram was applied at the time of transfer to 28°C. Data represent means ± 2 SE; n > 25.
To investigate whether PIF4 and PIF7 regulate the same process required for temperature‐induced hypocotyl elongation rather than different independently required steps, we analysed expression of hormone biosynthetic genes that were previously implicated in thermomorphogenesis (Stavang et al., 2009). In our conditions expression of the BR biosynthetic gene BRASSINOSTEROID‐6‐OXIDASE 2 (BR6ox2) was induced by higher temperature in WT plants but not in pif7 and pif4 (Figs S3, S4). Temperature‐induced expression of CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD) depended more on PIF4 than PIF7 while higher expression of the gibberellic acid biosynthesis gene GIBBERELLIN‐3‐OXIDASE 1 (GA3OX1) was largely independent of PIF4 or PIF7 (Figs S3, S4). Similarly, strong induction of a small heat‐shock gene (HSP17.6B) was unaffected in the tested pif mutants (Figs S3, S4). We therefore conclude that pif4 and pif7 show a similar temperature‐regulated gene expression pattern with a particularly obvious effect on auxin biosynthesis and response genes (Figs 2, S3, S4).
PIF7 does not regulate PIF4 accumulation but both PIFs can interact with each other
The central importance of PIF4 for thermomorphogenesis prompted us to determine whether PIF7 is required for PIF4 accumulation. We compared PIF4 mRNA levels in the WT and pif7 at 21°C and 28°C and did not detect a major effect of PIF7 on PIF4 expression (Fig. 3a,b). PIF4 protein levels at 21°C and the slight increase observed after 3 h at 28°C were similar in the WT and pif7 (Fig. 3c). Consistent with phyB promoting PIF4 degradation (de Lucas et al., 2008), we detected high PIF4 levels in phyB and phyBpif7 at 21°C and PIF4 levels increased at 28°C independently of PIF7 (Fig. 3c). Alternatively, PIF4 and PIF7 might be both required for thermomorphogenesis because they work as a heterodimer to regulate gene expression (Fig. 2). We used the yeast two‐hybrid assay to determine whether both proteins can interact and found that PIF4 and PIF7 form homodimers and heterodimers in yeast (Fig. S5). We conclude that the strong thermomorphogenic phenotype of pif7 cannot be explained by lower PIF4 protein levels but may be due to PIF4/PIF7 heterodimer‐mediated gene expression (Figs 2, S5).
Figure 3.
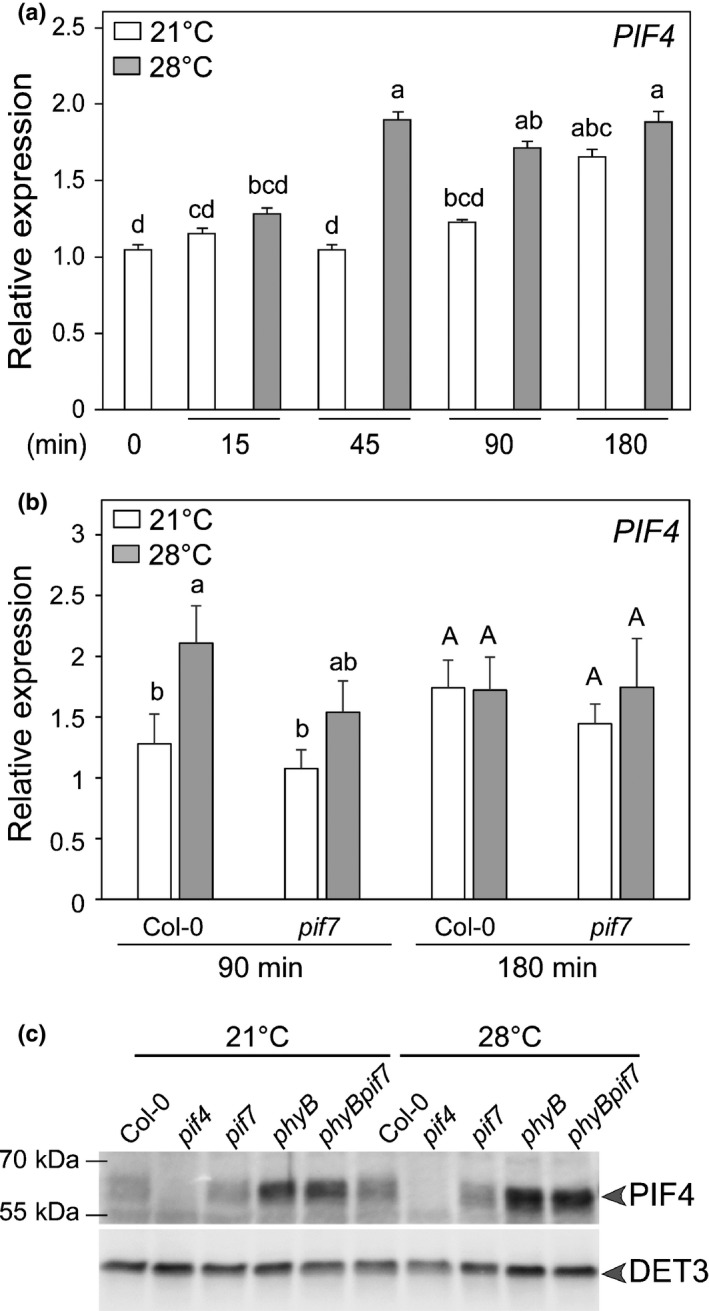
Regulation of PIF4 mRNA and protein levels in thermomorphogenesis in Arabidopsis. (a) Relative expression of PIF4 in 5‐d‐old Col‐0 and (b) in Col‐0 and pif7‐1 mutants. Seedlings were grown as in Fig. 2(a). Gene expression values were calculated as fold induction relative to a Col‐0 sample at 21°C, t = 0 (a) and t = 90 min (b). n = 3 (biological) with three technical replicas for each RNA sample. Data are means, ± 2 SE. Different letters indicate significant differences (P < 0.05). (c) PIF4 protein levels in the indicated genotypes detected with anti‐PIF4 antibody from total protein extracts after 3 h of 21°C and 28°C treatment in 5‐d‐old long day‐grown seedlings treated at ZT2. DET3 was used as a loading control.
PIF7 protein levels increase rapidly upon transfer to 28°C
The requirement of PIF7 for rapid temperature‐induced changes in gene expression and hypocotyl elongation (Figs 1, 2) suggested that PIF7 function and/or accumulation might be temperature‐regulated. Upon transfer to 28°C PIF7 transcript levels declined in the WT while in pif4 we observed a similar but not significant reduction (Fig. 4a,b). To analyse PIF7 protein we used a PIF7‐HA line (Galvao et al., 2019) and found that in contrast to PIF7 RNA, PIF7 protein levels increased significantly 90 min after the transfer to 28°C (Fig. 4c). As observed previously PIF7 was present as two major isoforms (Li et al., 2012) (Fig. 4c). Transfer to 28°C specifically led to increased abundance of the faster migrating isoform (Figs 4c, S6). We propose that temperature‐induced PIF7 levels may contribute to enhanced PIF7 activity required for rapid thermomorphogenic responses.
Figure 4.
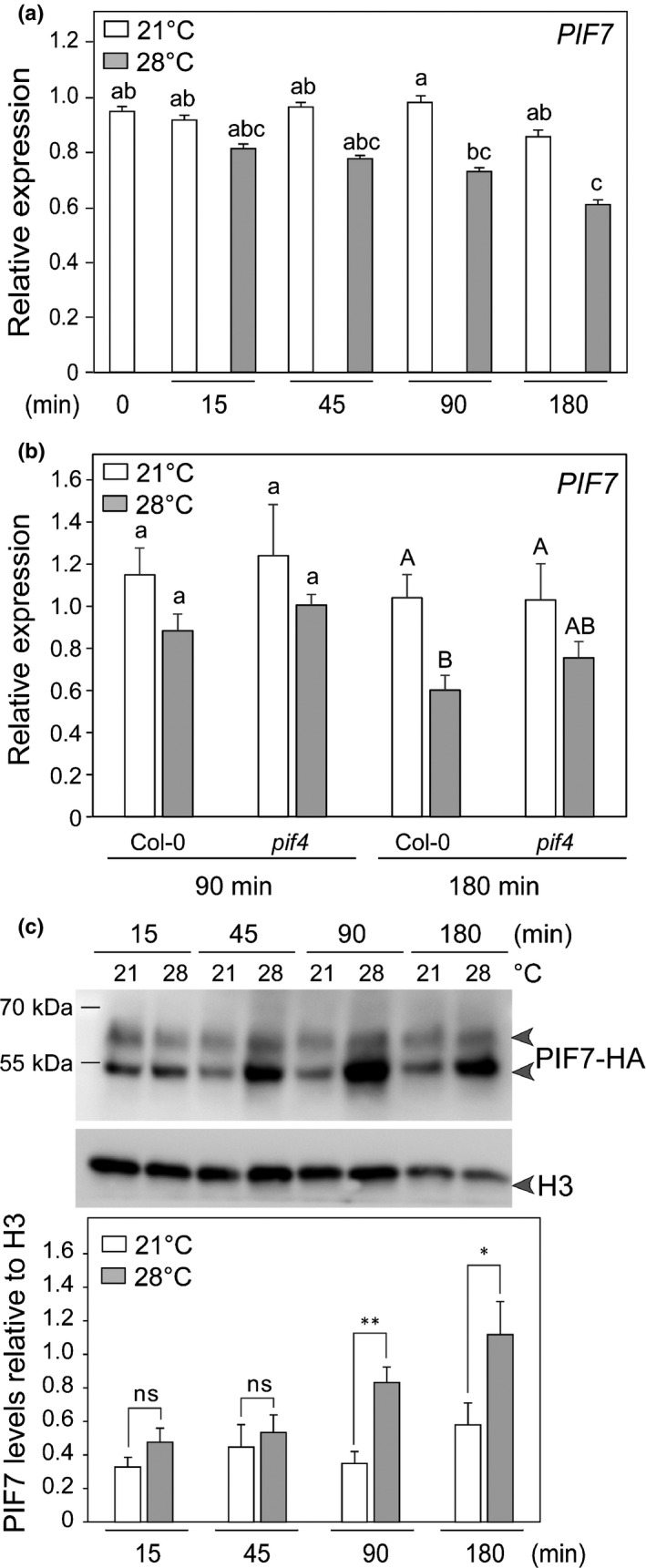
Regulation of PIF7 mRNA and protein levels in thermomorphogenesis in Arabidopsis. (a) Relative expression of PIF7 in 5‐d‐old Col‐0 and (b) in Col‐0 and pif4 mutant. Seedlings were grown as in Fig. 2(a). Gene expression values were calculated as fold induction relative to a Col‐0 sample at 21°C, t = 0 (a) and t = 90 min (b). n = 3 (biological) with three technical replicas for each RNA sample. Data are means, ± 2 SE. Different letters indicate significant differences (P < 0.05). (c) PIF7‐HA protein levels detected with an anti‐HA antibody from total protein extract after indicated time points at 21°C and 28°C in 5‐d‐old long day‐grown seedlings treated at ZT2. The HA signal was quantified and normalized to H3 signal (n = 6). Data are means, ±SE. Asterisks indicate significant (P values) between 28°C and 21°C samples at a given timepoint (Student's t‐test: * < 0.05; ** < 0.01), ns, nonsignificant.
Discussion
PIF4 was believed to have a uniquely important function in thermomorphogenesis (Koini et al., 2009; Sun et al., 2012; Ma et al., 2016; Casal & Balasubramanian, 2019; Qiu et al., 2019). Our work shows that in seedlings the role of PIF7 is almost as important as PIF4 (Figs 1, 2, S1, S2). However, upon prolonged growth at 28°C we observed a slightly greater growth response in pif7 than pif4 (Fig. 1b,c). In addition, we found that at 28°C pif4 responds less to picloram than pif7 (Fig. 2c). A greater role of PIF4 than PIF7 in controlling auxin responsiveness may explain the small phenotypic difference between both pif mutants. Our data on thermomorphogenesis reveals interesting similarities and differences with the shade avoidance response. A reduction of the R : FR ratio indicative of neighbour proximity leads to auxin synthesis that primarily depends on PIF7 (Li et al., 2012). Our data suggests that during thermomorphogenesis PIF4 and PIF7 are similarly important to promote auxin biosynthesis (Fig. 2a). PIF4 and PIF5 rather than PIF7 have been implicated in the control of auxin sensitivity (Nozue et al., 2011; Hornitschek et al., 2012; Li et al., 2012; Hersch et al., 2014; Pucciariello et al., 2018). We find that during thermomorphogenesis PIF4 also plays a more important function than PIF7 to promote auxin responsiveness (Fig. 2d). Finally, our data on temperature‐induced growth (Figs 1, 2) is consistent with a model emerging from the study of the low R : FR response with an early phase depending on auxin production (Tao et al., 2008; Li et al., 2012) and a prolonged response requiring PIF4‐controlled auxin sensitivity (Pucciariello et al., 2018).
Several possibilities can explain the requirement of PIF4 and PIF7 for thermomorphogenesis. Each of them might control different essential steps for elevated temperature‐induced growth. Given the similar gene expression profile of pif4 and pif7, this is an unlikely explanation (Figs 2, S4). However, more research is required to test this hypothesis on a larger scale and with better spatial resolution (e.g. hypocotyls vs cotyledons). Given that both single mutants and the pif4pif7 double mutant have similar phenotypes (Figs 1, 2, S1), the function of these PIFs might depend on each other. We showed that PIF7 is not required for normal accumulation of PIF4 transcript or PIF4 protein and PIF7 mRNA expression is largely unaffected in pif4 (Figs 3, 4). As bHLH transcription factors bind DNA as dimers, an attractive hypothesis is that a PIF4/PIF7 heterodimer regulates expression of target genes such as YUC8 or IAA29 (Fig. 2) (Hornitschek et al., 2012; Li et al., 2012; Sun et al., 2012). Consistent with this hypothesis, PIF4 and PIF7 interact with each other when co‐expressed in mesophyll protoplasts (Kidokoro et al., 2009) and in the yeast two‐hybrid assay (Fig. S5). Collectively, these findings support the PIF4/PIF7 heterodimer hypothesis during thermomorphogenesis. However, in the phyB mutant background pif mutants act additively with almost full phyB suppression in phyBpif4pif5pif7 indicating that the different PIFs can act independently (Fig. S2b). Additive effects of PIF4 and PIF7 have also been observed during de‐etiolation and shade avoidance (Leivar et al., 2008; de Wit et al., 2016). Similarly, PIF4 and PIF7 act independently of each other to suppress cold tolerance during long days (Lee & Thomashow, 2012). We conclude that additional research is required to understand to what extent PIF4 and PIF7 activity depend on each other and how this dependency may be regulated by development or the environment.
Temperature elevation regulates PIF7 transcript and protein levels in opposite ways with a reduction of transcript but more PIF7 protein (Fig. 4). Reducing the R : FR ratio also leads to lower PIF7 transcripts, while PIF7 phosphorylation changes, which regulates PIF7 accumulation in the nucleus (Li et al., 2012; Huang et al., 2018). Photoperiod also regulates PIF7 protein and PIF7 mRNA with higher levels of PIF7 transcript and PIF7 protein in LD (Lee & Thomashow, 2012). In response to increasing ambient temperature PIF7 protein levels, particularly its faster isoform increased rapidly (Figs 4c, S6). We propose that the temperature‐enhanced PIF7 protein levels may contribute to the thermomorphogenic response. It will be interesting to decipher the mechanisms underlying this change in PIF7 accumulation and if/how this regulation contributes to enhanced PIF7 function at elevated temperature.
Author contributions
Conceptualization, ASF, VCG, YCI and CF; investigation, ASF, VCG, YCI and AB; resources, AG, MT and LAP; funding acquisition, CF; writing, CF; supervision, CF. ASF, VCG and YCI contributed equally to this work.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Thermomorphogenic response requires both PIF4 and PIF7 for hypocotyl elongation in short day (SD).
Fig. S2 PIF4 and PIF7 regulate thermomorphogenic hypocotyl elongation downstream of phyB and cry1.
Fig. S3 Relative expression of genes that were previously implicated in thermomorphogenesis.
Fig. S4 Relative expression of temperature‐induced genes in Col‐0 and pif mutants.
Fig. S5 PIF7 and PIF4 form homodimers and heterodimers in yeast.
Fig. S6 Regulation of the levels of both PIF7 isoforms in thermomorphogenesis.
Table S1 List of oligonucleotides used in this study.
Acknowledgements
The authors thank Mieke de Wit for the initial observation that pif7 mutants are unresponsive to elevated temperature, Martina Legris for comments on the manuscript, Philip A. Wigge (University of Potsdam) for sharing unpublished results and the Genomic Technologies Facility (GTF) for assistance with RT‐qPCR assays. Work in the Fankhauser laboratory is funded by the University of Lausanne and grants from the Swiss National Science Foundation (no. 310030B_179558 and CRSII3_154438). VCG was supported by an EMBO Long Term fellowship (ALTF 293‐2013).
References
- Bellstaedt J, Trenner J, Lippmann R, Poeschl Y, Zhang X, Friml J, Quint M, Delker C. 2019. A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiology 180: 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbousse C, Vegesna N, Law JA. 2018. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proceedings of the National Academy of Sciences, USA 115: E12453–E12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S. 2019. Thermomorphogenesis. Annual Review of Plant Biology 70: 321–346. [DOI] [PubMed] [Google Scholar]
- Crawford AJ, McLachlan DH, Hetherington AM, Franklin KA. 2012. High temperature exposure increases plant cooling capacity. Current Biology 22: R396–397. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez‐Falcon M, Pontin M, Iglesias‐Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- de Wit M, George GM, Ince YC, Dankwa‐Egli B, Hersch M, Zeeman SC, Fankhauser C. 2018. Changes in resource partitioning between and within organs support growth adjustment to neighbor proximity in Brassicaceae seedlings. Proceedings of the National Academy of Sciences, USA 115: E9953–E9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Keuskamp DH, Bongers FJ, Hornitschek P, Gommers CMM, Reinen E, Martinez‐Ceron C, Fankhauser C, Pierik R. 2016. Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Current Biology 26: 3320–3326. [DOI] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C. 2015. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytologist 208: 198–209. [DOI] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibanez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J et al 2014. The DET1‐COP1‐HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Reports 9: 1983–1989. [DOI] [PubMed] [Google Scholar]
- Duttweiler HM. 1996. A highly sensitive and non‐lethal beta‐galactosidase plate assay for yeast. Trends in Genetics 12: 340–341. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD et al 2011. Phytochrome‐interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences, USA 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao VC, Fiorucci AS, Trevisan M, Franco‐Zorilla JM, Goyal A, Schmid‐Siegert E, Solano R, Fankhauser C. 2019. PIF transcription factors link a neighbor threat cue to accelerated reproduction in Arabidopsis. Nature Communications 10: 4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Berriri S, Kumar SV. 2017. PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Current Biology 27: 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Kumar SV. 2017. DET1 and HY5 control PIF4‐mediated thermosensory elongation growth through distinct mechanisms. Cell Reports 18: 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. 2005. Profiling histone modification patterns in plants using genomic tiling microarrays. Nature Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Goyal A, Karayekov E, Galvao VC, Ren H, Casal JJ, Fankhauser C. 2016. Shade promotes phototropism through phytochrome B‐controlled auxin production. Current Biology 26: 3280–3287. [DOI] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C. 2014. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111: 6515–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, Lopez‐Vidriero I, Franco‐Zorrilla JM, Solano R, Trevisan M, Pradervand S et al 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. The Plant Journal 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Huang X, Zhang Q, Jiang Y, Yang C, Wang Q, Li L. 2018. Shade‐induced nuclear localization of PIF7 is regulated by phosphorylation and 14‐3‐3 proteins in Arabidopsis. eLife 7: pii: e31636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez C, Delker C, Martinez C, Burstenbinder K, Janitza P, Lippmann R, Ludwig W, Sun H, James GV, Klecker M et al 2018. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Current Biology 28: 303–310.e3. [DOI] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S et al 2016. Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889. [DOI] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K et al 2009. The phytochrome‐interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiology 151: 2046–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen MV, Schmid‐Siegert E, Trevisan M, Petrolati LA, Senechal F, Muller‐Moule P, Maloof J, Xenarios I, Fankhauser C. 2016. Neighbor detection induces organ‐specific transcriptomes, revealing patterns underlying hypocotyl‐specific growth. Plant Cell 28: 2889–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. 2009. High temperature‐mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Current Biology 19: 408–413. [DOI] [PubMed] [Google Scholar]
- Lau OS, Song Z, Zhou Z, Davies KA, Chang J, Yang X, Wang S, Lucyshyn D, Tay IHZ, Wigge PA et al 2018. Direct control of SPEECHLESS by PIF4 in the high‐temperature response of stomatal development. Current Biology 28: 1273–1280.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF. 2012. Photoperiodic regulation of the C‐repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 109: 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schafer E, Vierstra RD, Casal JJ. 2016. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900. [DOI] [PubMed] [Google Scholar]
- Legris M, Nieto C, Sellaro R, Prat S, Casal JJ. 2017. Perception and signalling of light and temperature cues in plants. The Plant Journal 90: 683–697. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al‐Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. 2008. The Arabidopsis phytochrome‐interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda‐Paz J, Cowing‐Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS et al 2012. Linking photoreceptor excitation to changes in plant architecture. Genes & Development 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. 2008. Phytochrome‐mediated inhibition of shade avoidance involves degradation of growth‐promoting bHLH transcription factors. The Plant Journal 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H. 2016. Cryptochrome 1 interacts with PIF4 to regulate high temperature‐mediated hypocotyl elongation in response to blue light. Proceedings of the National Academy of Sciences, USA 113: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez C, Espinosa‐Ruiz A, de Lucas M, Bernardo‐Garcia S, Franco‐Zorrilla JM, Prat S. 2018. PIF4‐induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO Journal 37: pii: e99552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. 1999. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082. [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. The Plant Journal 14: 387–392. [DOI] [PubMed] [Google Scholar]
- Nomoto Y, Kubozono S, Miyachi M, Yamashino T, Nakamichi N, Mizuno T. 2012. A circadian clock‐ and PIF4‐mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana . Plant and Cell Physiology 53: 1965–1973. [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN. 2011. Genomic analysis of circadian clock‐, light‐, and growth‐correlated genes reveals PHYTOCHROME‐INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiology 156: 357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Tat AV, Kumar Devisetty U, Robinson M, Mumbach MR, Ichihashi Y, Lekkala S, Maloof JN. 2015. Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genetics 11: e1004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. 2012. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Ha JH, Kim JY, Park CM. 2017. COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytologist 215: 269–280. [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR et al 2016. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J. 2016. The epidermis coordinates auxin‐induced stem growth in response to shade. Genes & Development 30: 1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciariello O, Legris M, Costigliolo Rojas C, Iglesias MJ, Hernando CE, Dezar C, Vazquez M, Yanovsky MJ, Finlayson SA, Prat S et al 2018. Rewiring of auxin signaling under persistent shade. Proceedings of the National Academy of Sciences, USA 115: 5612–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Li M, Kim RJ, Moore CM, Chen M. 2019. Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nature Communications 10: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. 2016. Molecular and genetic control of plant thermomorphogenesis. Nature Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Raschke A, Ibanez C, Ullrich KK, Anwer MU, Becker S, Glockner A, Trenner J, Denk K, Saal B, Sun X et al 2015. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4‐dependent auxin response genes. BMC Plant Biology 15: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego‐Bartolome J, Gomez MD, Yoshida S, Asami T, Olsen JE, Garcia‐Martinez JL, Alabadi D, Blazquez MA. 2009. Hormonal regulation of temperature‐induced growth in Arabidopsis. The Plant Journal 60: 589–601. [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. 2012. PIF4‐mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genetics 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ et al 2008. Rapid synthesis of auxin via a new tryptophan‐dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY. 2016. TOC1‐PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nature Communications 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Thermomorphogenic response requires both PIF4 and PIF7 for hypocotyl elongation in short day (SD).
Fig. S2 PIF4 and PIF7 regulate thermomorphogenic hypocotyl elongation downstream of phyB and cry1.
Fig. S3 Relative expression of genes that were previously implicated in thermomorphogenesis.
Fig. S4 Relative expression of temperature‐induced genes in Col‐0 and pif mutants.
Fig. S5 PIF7 and PIF4 form homodimers and heterodimers in yeast.
Fig. S6 Regulation of the levels of both PIF7 isoforms in thermomorphogenesis.
Table S1 List of oligonucleotides used in this study.


