Summary
Powdery mildew disease, elicited by the obligate fungal pathogen Blumeria graminis f.sp. tritici (Bgt), causes widespread yield losses in global wheat crop. However, the molecular mechanisms governing wheat defense to Bgt are still not well understood.
Here we found that TuACO3, encoding the 1‐aminocyclopropane‐1‐carboxylic acid (ACC) oxidase functioning in ethylene (ET) biosynthesis, was induced by Bgt infection of the einkorn wheat Triticum urartu, which was accompanied by increased ET content. Silencing TuACO3 decreased ET production and compromised wheat defense to Bgt, whereas both processes were enhanced in the transgenic wheat overexpressing TuACO3.
TuMYB46L, phylogenetically related to Arabidopsis MYB transcription factor AtMYB46, was found to bind to the TuACO3 promoter region in yeast‐one‐hybrid and EMSA experiments. TuMYB46L expression decreased rapidly following Bgt infection. Silencing TuMYB46L promoted ET content and Bgt defense, but the reverse was observed when TuMYB46L was overexpressed.
Hence, decreased expression of TuMYB46L permits elevated function of TuACO3 in ET biosynthesis in Bgt‐infected wheat. The TuMYB46L‐TuACO3 module regulates ET biosynthesis to promote einkorn wheat defense against Bgt. Furthermore, we found four chitinase genes acting downstream of the TuMYB46L‐TuACO3 module. Collectively, our data shed a new light on the molecular mechanisms underlying wheat defense to Bgt.
Keywords: ACC oxidase (ACO), Blumeria graminis, einkorn wheat, ethylene (ET), gene module, MYB transcription factor, plant defense
Introduction
Common wheat (Triticum aestivum, AABBDD) is the most widely cultivated staple crop in the world (IWGSC et al., 2018). Efficient and stable wheat production is essential for maintaining global food security. Powdery mildew disease, caused by the obligate biotrophic fungal pathogen Blumeria graminis f.sp. tritici (Bgt), leads to widespread yield reductions in many wheat‐producing regions (Singh et al., 2016). The infection cycles of cereal powdery mildew fungi (including Bgt) are similar to each other (Hückelhoven & Panstruga, 2011; Wicker et al., 2013). Under favorable conditions, the conidium germinates and produces a primary germ tube and an appressorial germ tube (AGT). The AGT then develops into an appressorium from which an infection peg emerges. In a compatible interaction, the infection peg enters host cells and forms a haustorium. This structure absorbs nutrients from host cells and promotes rapid secondary hyphae growth on the leaves, leading to the formation of powdery mildew colonies and the production of a new generation of conidia. To manage the damages inflicted by Bgt, substantial efforts are being devoted to study the genetic and molecular mechanisms of wheat defense to powdery mildew disease in order to develop effective controlling measures (Yahiaoui et al., 2004; Bhullar et al., 2010; He et al., 2018; McNally et al., 2018; Xing et al., 2018; Zou et al., 2018).
To date, two layers of plant immunity to pathogen attacks have been elucidated (Bigeard et al., 2015; Cui et al., 2015). Pattern triggered immunity (PTI), initiated by recognition of pathogen associated molecular patterns by host cell surface receptors, is a basal level of defense. However, effector triggered immunity (ETI), augmented after detecting the avirulent effector of pathogen by host intracellular immunoreceptor molecules, is an intensified form of defense often with high race specificity. Many defense proteins, such as various types of pathogen‐related (PR) proteins including chitinases, are induced upon the activation of PTI and ETI, and contribute to the limitation of pathogen growth. The mechanisms underlying the induction of defense genes are highly complex, which frequently involve phytohormone signaling processes and vary among different pathosystems (Pieterse et al., 2012; Bigeard et al., 2015; Cui et al., 2015). Detailed characterization of these mechanisms is essential for completely understanding the genetic and molecular basis of plant defense to different pathogen attacks.
Through molecular and genetic analysis, ethylene (ET) biosynthesis and signal transduction have been found to regulate plant defense to diverse pathogens (Broekaert et al., 2006; Tintor et al., 2013; Broekgaarden et al., 2015). In Arabidopsis thaliana, ET biosynthesis was activated in response to the infection by the necrotroph Botrytis cinerea (Gravino et al., 2015); ET induction also was shown to play a positive role in defense against the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Guan et al., 2015). In both studies, the infection‐induced ET biosynthesis was caused by increased activity of 1‐aminocyclopropane‐1‐carboxylic acid synthase (ACS), a key enzyme required for ET biosynthesis in plant cells (Booker & Delong, 2015). In line with the above findings, a recent work shows that rice defense against the hemi‐biotrophic fungal pathogen Magnaporthe oryzae also involved the upregulation of ET production through the function of ACS. Apart from ACS, several transcriptional factor (TF) genes acting in the ET signaling pathway also have been demonstrated to regulate plant defense against pathogens. TaPIE1, a pathogen‐induced ET responsive factor (ERF), was reported to positively regulate the resistance response to Rhizoctonia cerealis by activating the transcription of defense‐related genes downstream of the ET signaling pathway (Zhu et al., 2014). Overexpression of GmERF5 was shown to enhance the resistance of soybean to Phytophthora sojae via positively regulating the expression of the defense genes PR10, PR1‐1 and PR10‐1 (Dong et al., 2015a). Activation of two TFs functioning in ET signaling in rice increased the resistance to M. oryzae by elevating the production of reactive oxygen species (ROS) and phytoalexins (Yang et al., 2017). Finally, ectopic expression of ERF1‐V, an ET‐responsive element‐binding factor gene of the AP2/ERF transcription factor gene family, conferred high resistance to Bgt infection in wheat (Xing et al., 2017). Despite the progress outlined above, there is still little molecular evidence for the function of ET biosynthesis and signaling in plant defense to obligate biotrophic fungal pathogens, such as powdery mildew fungi.
Higher plants generally possess a large family of MYB TF genes, many of which have been found to regulate plant defense against pathogens. For example, the knockout of AtMYB46, an R2R3 type MYB TF originally found to regulate secondary cell wall biosynthesis in Arabidopsis (Zhong et al., 2007; Zhong & Ye, 2012), confers enhanced resistance to B. cinerea through increasing the expression of cell wall remodeling genes encoding type III peroxidases and two defense‐related genes coding for PR3 and PDF1.2a (Ramírez et al., 2011a,b). Another Arabidopsis R2R3 MYB TF, MYB15, positively regulates Arabidopsis basal immunity to the bacterial pathogen Pseudomonas syringae through promoting defense‐induced lignification (Chezem et al., 2017). MYBS1, a MYB TF gene in rice, contributes to rice defense against M. oryzae through suppressing bsr‐d1 gene expression, which resulted in elevated ROS accumulation and, thus, limited pathogen growth (W. Li et al., 2017). Finally, several MYB TF genes inducible by ET treatment, including MYB108 from cotton, and TaMYB4 and TaPIMP2 from wheat (Al‐Attala et al., 2014; Cheng et al., 2016; Wei et al., 2017), are found to contribute to plant defense against pathogens. Nevertheless, there is still no detailed report on the regulation of plant pathogen defense by MYB TFs through upregulating the transcription of ET biosynthesis gene(s) and the resultant elevation of ET production.
In order to efficiently analyze the defense mechanisms to Bgt, we used the diploid einkorn wheat Triticum urartu (AA), which has a much smaller genome than that of common wheat, as an experimental host (Zhang et al., 2016; Ling et al., 2018). Triticum urartu accessions from around the world differed extensively in their response to Bgt infection; T. urartu genes and networks associated with different types of resistance responses to Bgt have been revealed by transcriptome comparison and selection sweep mapping analyses (Zhang et al., 2016; Ling et al., 2018). Further to the studies above, we here report the function of TuACO3 and TuMYB46L, encoding an active 1‐aminocyclopropane‐1‐carboxylic acid (ACC) oxidase (ACO) enzyme and a R2R3 myeloblastosis (MYB) TF, respectively, in wheat defense against Bgt. We found that the expression of TuACO3 was increased by Bgt infection in diverse T. urartu accessions, and its protein promoted ET biosynthesis in Bgt‐infected plants. Silencing TuACO3 lowered Bgt infection‐induced ET production and weakened the defense against Bgt. Through yeast‐one‐hybrid (Y1H) assay and complementary molecular experiments, we identified TuMYB46L that could bind to the promoter region of TuACO3 and acted as a negative regulator of TuACO3 expression. TuMYB46L was rapidly downregulated by Bgt infection, thus permitting enhanced expression of TuACO3 in the infected plants. Therefore, TuMYB46L and TuACO3 comprise a functional gene module that regulates ET biosynthesis to promote einkorn wheat defense against Bgt. We further identified four chitinase genes that function downstream of the TuMYB46L‐TuACO3 module through transcriptome comparison. Together, our data provide new information on the function of ET biosynthesis in wheat defense against Bgt and the transcriptional control of plant ACO gene in powdery mildew‐infected plants.
Materials and Methods
Plant materials
The Triticum urartu accessions used in this work included PI428193, PI428202, PI428214, PI428220, PI428224 and G1812 (Zhang et al., 2016; Ling et al., 2018). They were all susceptible to the Blumeria graminis f.sp. tritici (Bgt) race E09. G1812 has previously been used for determining the genomic sequence of T. urartu (Ling et al., 2018). The common wheat cultivar Kenong 199 (KN199), highly susceptible to E09 (Wang et al., 2014; Zou et al., 2018), was used for propagating Bgt and as a recipient for developing the transgenic wheat lines overexpressing TuACO3 or TuMYB46L (ACO, 1‐aminocyclopropane‐1‐carboxylic acid (ACC) oxidase; MYB, myeloblastosis).
Bgt infection and phenotyping
Wheat seeds were germinated in the glasshouse at 20–22°C under a 16 h : 8 h, light : dark photoperiod for 1 wk to yield seedlings at the one‐leaf stage. They were inoculated with E09 spores as described by Wang et al. (2014). At 72 h post‐inoculation (hpi), the leaves were cut into 5‐cm segments, and subjected to microcolony staining using Coomassie blue (Wang et al., 2014). For each inoculation, 10–15 leaves from ≥ 10 infected seedlings were examined, with the results used to calculate the percentage of microcolonies developing from the total number of examined Bgt spores. For observing the development of Bgt colonies, the infected leaves were photographed at 8 d post‐inoculation (dpi), with Bgt colony area determined using imagej software (https://imagej.nih.gov/ij/).
Quantification of ethylene content
Ethylene (ET) content was measured using a gas chromatography instrument (Yin et al., 2015). For the samples without Bgt inoculation, the seedlings (30 for T. urartu and 10 for common wheat) at the one‐leaf stage were transferred into a 50‐ml Falcon tube with their roots immersed in water. After 24 h at 22°C, the tube was tightly sealed with a rubber cap for 24 h, with the emitted ET subsequently measured. In the case of Bgt inoculation, the desired seedlings were placed into the Falcon tube, followed by Bgt inoculation as described above, with the tube sealed immediately post‐inoculation. The ET content was measured at 24 hpi, and repeated three times, with each measurement including three biological replicates.
Gene expression analysis by qRT‐PCR
Gene expression levels were determined using the samples collected from at least 10 seedlings (including untreated controls and those treated for 24 h) by quantitative reverse transcription (qRT)‐PCR with gene‐specific primers (Supporting Information Table S1; Methods S1).
Single‐cell functional assay
Single‐cell functional tests, based on transient overexpression (TOE) of cloned cDNA or transiently induced gene silencing (TIGS) in wheat epidermal cells, were accomplished essentially as described in previous publications (Douchkov et al., 2005; Shen et al., 2007). The reporter plasmid contained the expression cassette of β‐glucuronidase gene. The effector constructs were pUbi:TuACO3, pUbi:TuMYB46L, pUbi:TuACO3as or pUbi:TuMYB46Las. The former two constructs transiently expressed the full‐length cDNA of TuACO3 and TuMYB46L, respectively, whereas the latter two plasmids contained a short segment from TuACO3 or TuMYB46L coding sequence in an antisense orientation (Methods S1).
Immunoblotting assay
Immunoblotting was carried out using either the antibody to a recombinant TuACO3 protein or the FLAG tag (Sigma, F7425). The recombinant TuACO3 was prepared by expressing a histidine‐tagged protein in Escherichia coli, with the purified protein used to raise a mouse polyclonal antibody as described by Qin et al. (2008). More details are described in Methods S1.
Virus‐induced gene silencing
Virus‐induced gene silencing (VIGS) was carried out using the barley stripe mosaic virus (BSMV) vector (Yuan et al., 2011). Two constructs, pCaBS‐γ:ACO3as and pCaBS‐γ:MYB46Las harboring a short antisense fragment derived from the coding sequence of TuACO3 (122 bp) or TuMYB46L (130 bp), were prepared. Together with pCaBS‐α and pCaBS‐β, the two recombinant viruses, BSMV:ACO3as and BSMV:MYB46Las, were formed, and used to silence endogenous TuACO3 and TuMYB46L in G1812 leaf cells, respectively. The two viruses also were used to silence the orthologous genes of TuACO3 or TuMYB46L in the common wheat cultivar KN199 because the VIGS‐inducing fragments carried by BSMV:ACO3as and BSMV:MYB46Las were conserved in the respective orthologs (Methods S1).
Overexpression of TuACO3 and TuMYB46L in common wheat
The constructs pUbi:TuACO3‐FLAG and pUbi:TuMYB46L‐FLAG, expressing TuACO3‐FLAG and TuMYB46L‐FLAG fusion proteins, respectively, were prepared with the vector pHZ206 (carrying a maize ubiquitin gene promoter) and the primers listed in Table S1. They were introduced into the immature embryos of KN199 by biolistic transformation (Wang et al., 2014). Homozygous T2 lines were selected from selfed T1 individuals, with the expression of TuACO3‐FLAG or TuMYB46L‐FLAG identified by immunoblotting using the antibodies recognizing FLAG tag or TuACO3. In total, 12 and 17 independent lines overexpressing TuACO3‐FLAG or TuMYB46L‐FLAG were identified. TuACO3‐OX1 and ‐OX2 (overexpressing TuACO3‐FLAG) and TuMYB46L‐OX1 and ‐OX2 (overexpressing TuMYB46L‐FLAG) were used as representatives for more detailed analysis. These transgenic lines resembled wild‐type (WT) KN199 in their growth and development in the glasshouse (Fig. S1). The transgenic plants and the WT segregate controls (WTS7 and WTS9) were inoculated with Bgt, followed by examination of microcolonies and colonies as detailed above. At least 30 plants were inoculated with Bgt per line per experiment. Measurement of ET content, either before or after Bgt inoculation, was carried out as described above. We were unable to overexpress TuACO3 and TuMYB46L in T. urartu because of the difficulty to genetically transform this species.
Yeast‐one‐hybrid (Y1H) assay
A Y1H library was generated using the vector pGADT7 and the SMART cDNA Library Construction Kit (634901; Clontech, Dalian, China). The cDNAs used for library construction were reverse‐transcribed from total RNAs extracted from the G1812 leaves collected at 0, 4 and 24 hpi with Bgt. This library was screened using the 2‐kb promoter region of TuACO3 cloned in the bait vector pABAi (pABAi:TuACO3pro). The positive clones obtained from the initial screen were analyzed by DNA sequencing, with those carrying in frame inserts subjected to a second screen. To validate the binding of TuMYB46L to the TuACO3 promoter region, the cDNA coding sequence of TuMYB46L was cloned into pGADT7, generating pGADT7:TuMYB46L. This construct was co‐transformed with pABAi:TuACO3pro into the yeast strain Y1H Gold. Positive binding of TuMYB46L to TuACO3 promoter region was indicated by abundant growth of the transformants on the SD/‐Leu/‐Ura/+AbA medium. As controls, pABAi:TuACO3mpro was prepared by mutating each of the four predicted MYB46 binding sites (E1–E4) within the 2‐kb promoter region of TuACO3 into AAAAAAA; pGADT7:EVC (empty vector control) and pGADT7:GFP (expressing green fluorescent protein, GFP) also were used to test the binding specificity (Methods S1).
Chromatin immunoprecipitation (ChIP)‐quantitative (q)PCR analysis
Approximately 1 × 107 protoplasts, isolated according to Wang et al. (2014), were obtained from c. 150 G1812 seedlings. The pUbi:TuMYB46L‐FLAG plasmid (20 µg) and the control vector pHZ206 (20 µg) (see above) each were transformed into c. 1 × 106 protoplasts using polyethylene glycol. After 18 h of culture in the dark, the protoplasts were collected and treated with 1% formaldehyde at room temperature for 15 min to promote protein‐chromatin cross‐linking (Bao et al., 2014). The nuclei were then isolated, lysed and used for preparing chromatin complexes. The resulting complexes were ultrasonically treated to obtain a suitable input, in which the chromosomal DNA had been fragmented to a size of c. 500 bp, for ChIP. The anti‐FLAG antibody (F7425; Sigma, Shanghai, China) was used to precipitate the chromatins, which were subsequently analyzed by qPCR with 20 different primer sets (Table S1) covering the 2‐kb promoter region of TuACO3. As control, a mock precipitation with normal mouse IgG was executed. Two independent ChIP‐qPCR experiments were performed with similar results obtained.
Electrophoretic mobility shift assay (EMSA)
The cDNA coding sequence of TuMYB46L was cloned into the pET‐32a vector to express a recombinant TuMYB46L‐HIS tag protein in the E. coli Rosetta (DE3) strain, which was purified to homogeneity using Ni‐NTA Agarose (30210; Qiagen, Beijing, China). Three oligonucleotide probes (E1, E3 and E4, 39 bp each; Table S1) were synthesized and labeled with biotin at the 3′ end (Invitrogen, Beijing, China). An EMSA was performed using a LightShift Chemiluminescent EMSA kit (20148; Thermo Scientific, Beijing, China) according to the supplier's instructions. The biotin‐labeled probes each were incubated in a reaction mixture (1 × binding buffer, 2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA) with or without TuMYB46L protein at room temperature for 20 min. The following controls were included in the experiment to test the binding specificity: WT competitor (unlabeled E1, E3 and E4), mutant competitor (mutated and unlabeled E1, E3 and E4, with their MYB46 binding site changed to AAAAAAA, Table S1), and mutant probe (mutated and labeled E1, E3 and E4). The EMSA assay was carried out five times with identical findings made.
RNA sequencing and GO analysis of differentially regulated genes
Total RNA was extracted from the leaf samples collected from control G1812 seedlings and those treated with Bgt (at 24 hpi) or ET (at 24 h post‐treatment). They were then subjected to RNA sequencing as reported by Dong et al., (2015b). Three independent biological replicates were sequenced for each treatment at each time point. Two criteria were used to compute differentially expressed genes (DEGs), (1) a log2 ratio > 1.0 and (2) the expression difference was consistently observed in between different biological replicates. Gene ontology (GO) enrichment analysis of the DEGs shared by the two treatments was accomplished using the software agrigo (http://bioinfo.cau.edu.cn/agriGO/).
Analysis of chitinase genes and chitinase enzyme assay
The effects of ET treatment (10 ppm) or Bgt infection (for 24 h) on the expression of four chitinase genes were validated by qRT‐PCR with gene‐specific primers (Table S1). Transcript levels of the four genes in the G1812 leaf samples infected by BSMV:EVC, BSMV:ACO3as or BSMV:MYB46Las were evaluated by qRT‐PCR. Total chitinase activities were assayed with a commercial kit (Solarbio, BC0820) following the manufacturer's guidelines (Methods S1).
Sequence information and raw data
Sequence information for TuACO3, TuMYB46L, TaACO3, TaMYB46L, VIGS‐induced fragments and the DEGs identified by RNA sequencing, as well as the raw data used for calculating the means (± SE), are provided in Dataset S1.
Results
TuACO3 enhances wheat defense against Bgt via increasing ET biosynthesis
In our previous study (Zhang et al., 2016), a gene encoding a putative ACO was found induced by Bgt infection in multiple T. urartu accessions. This gene (TuG1812G0600003491), located on chromosome 6A of T. urartu (Ling et al., 2018), is orthologous to rice OsACO3 based on phylogenetic analysis (Fig. S2). Because of the poor understanding of ET function in plant defense against obligate biotrophic fungal pathogens and the fact that ACO catalyzes the last step of ET biosynthesis (Kende, 1993; Booker & Delong, 2015), we decided to analyze TuACO3 further to gain insights into the function of ET in wheat defense against Bgt.
Using qRT‐PCR, we validated the induction of TuACO3 by Bgt in G1812 and another five diverse T. urartu accessions (Figs 1a, S3a). Immunoblot analysis revealed increased accumulation of native TuACO3 in G1812 leaf tissues at 24 hpi (Fig. 1b). Concomitant to this increase, ET production was significantly upregulated (by 50.2%) at 24 hpi (Fig. 1c). Similar rises of ET production were found in the other five T. urartu accessions at 24 hpi (Fig. S3b). In single‐cell functional assays, Bgt infection level, as measured by the haustorium index, was decreased by overexpressing TuACO3, but uplifted by knocking down TuACO3 expression via TIGS (Fig. 1d). Consistent with these findings, lowering TuACO3 expression by BSMV‐mediated VIGS decreased Bgt infection‐induced ET production, and promoted susceptibility to Bgt in G1812 (Fig. 1e–h). Based on the reference genome sequence of Chinese Spring (CS) (International Wheat Genome Sequencing Consortium et al., 2018), the orthologous gene of TuACO3 in common wheat was TraesCS6A02G325600 (TaACO3) (Fig. S3c). Silencing of this gene in the common wheat cultivar KN199 also increased the susceptibility to Bgt (Fig. S3d,e).
Figure 1.
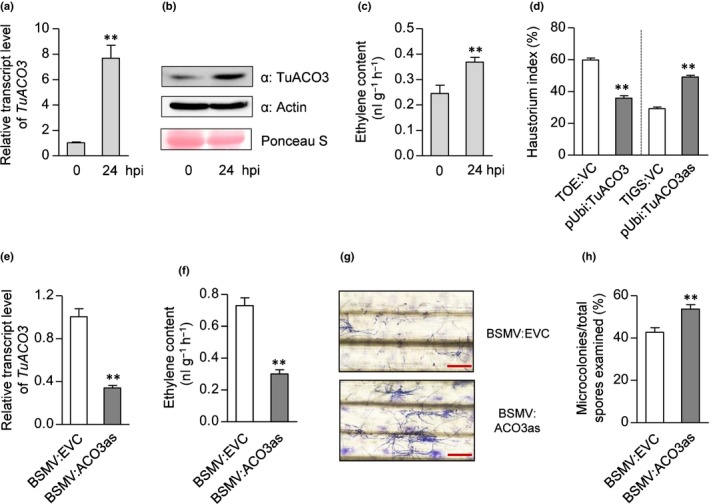
Functional analysis of TuACO3 in the Triticum urartu accession G1812. (a–c) Elevation of TuACO3 expression and ethylene (ET) production at 24 h post‐inoculation (hpi) of Blumeria graminis f.sp. tritici (Bgt). Transcript and protein increases were revealed by quantitative reverse transcription (qRT‐PCR) (a) and immunoblotting with a TuACO3‐specific antibody (b). Detection of actin served as an internal control. Upregulation of ET biosynthesis was determined by gas chromatography analysis (c). (d) Bgt haustorium growth in the leaf cells, suppressed by transiently expressing TuACO3 from the construct pUbi:TuACO3 but stimulated by transiently silencing TuACO3 with the construct pUbi:TuACO3as. TOE:VC and TIGS:VC, empty vector controls for the transient overexpression and silencing assays, respectively. Each mean (± SE) was calculated from the data of three technical repeats (with more than 200 transfected cells examined in each repeat). The results shown were typical of three independent experiments. **, P < 0.01 (Student's t‐test). (e–h) Effects of silencing TuACO3 on Bgt defense and ET production. Silencing was achieved using the recombinant virus BSMV:ACO3as. BSMV:EVC, an empty BSMV vector, served as a control. BSMV:ACO3as lowered the transcript level of TuACO3 (e), decreased ET production (f), and increased the development of Bgt microcolonies as shown by Coomassie blue staining of fungal structures (g) and quantitative comparison with the control (h). The datasets presented each were representative of three independent experiments. Each mean (± SE) was calculated from at least three biological replicates. **, P < 0.01 (Student's t‐test). Bars, 200 μm. ACO, 1‐aminocyclopropane‐1‐carboxylic acid oxidase.
In order to further examine the function of TuACO3 in ET biosynthesis and Bgt defense, we analyzed two independent common wheat transgenic lines (TuACO3‐OX1, and ‐OX2) designed to overexpress TuACO3 with a C‐terminal FLAG tag. In both lines, TuACO3‐FLAG was confirmed to be overexpressed using immunoblotting with the antibodies to FLAG or TuACO3 (Fig. 2a). Compared to WTS7, a WT segregate control line, ET production was elevated by c. 1.7‐fold in TuACO3‐OX1 and 1.4‐fold in TuACO3‐OX2 (Fig. 2b). When infected by Bgt, the two OX lines showed significantly reduced fungal colonies on the leaf surface (Fig. 2c–f). These results demonstrated that the transgenically expressed TuACO3 functioned in ET biosynthesis, with concurrent improvement in Bgt defense.
Figure 2.
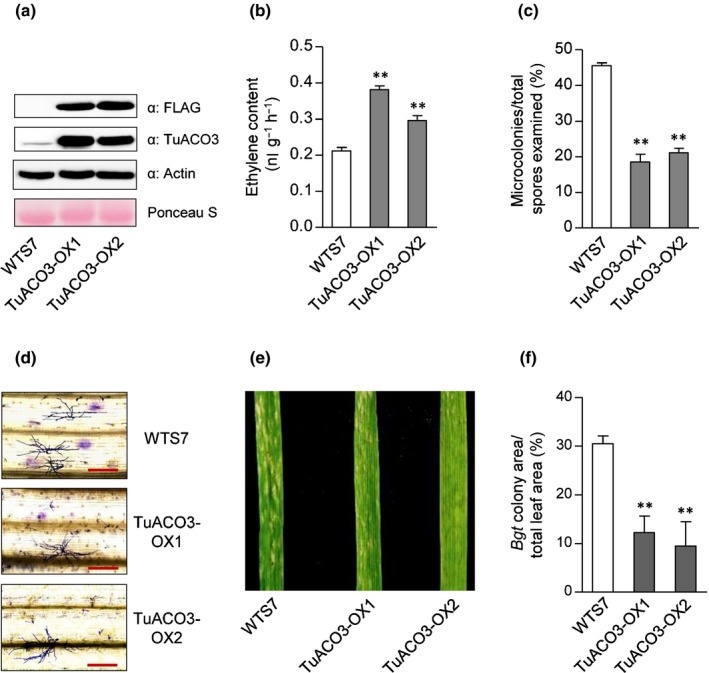
Validation of TuACO3 function using transgenic common wheat plants. TuACO3‐OX1 and ‐OX2 were two independent representative homozygous transgenic lines. WTS7 was a wild‐type (WT) segregate served as a control. (a) Overexpression of TuACO3‐FLAG protein in TuACO3‐OX1 and ‐OX2 revealed by immunoblotting with the antibodies specific for the FLAG tag or TuACO3. Equal loading was checked by detection of actin and Ponceau S staining. (b) Increased ethylene (ET) production in TuACO3‐OX1 and ‐OX2 relative to WTS7. (c, d) Reduced development of Blumeria graminis f.sp. tritici (Bgt) microcolonies in TuACO3‐OX1 and ‐OX2 recorded at 72 h post‐inoculation (hpi), as shown by quantitative comparison with WTS7 (c) and Coomassie blue staining of fungal structures (d). (e, f) Differences in Bgt colony growth among WTS7 and TuACO3‐OX1 and ‐OX2 at 8 d post‐inoculation (dpi), as indicated by the photographs of Bgt‐infected leaves (e) and quantitative comparison of the percentages of Bgt colony area. The datasets shown each were representative of at least three independent experiments. The numerical data shown were means ± SE, with those in (b) and (c) determined from three biological replicates. The means in (f) each were obtained by scanning nine Bgt‐infected leaves from three separate experiments. **, P < 0.01 (Student's t‐test). Bars, 200 μm. ACO, 1‐aminocyclopropane‐1‐carboxylic acid oxidase.
The above findings prompted us to test if applying ET, or the chemicals known to stimulate or inhibit ET biosynthesis, may alter wheat defense against Bgt (Methods S1). This test was conducted using both G1812 and the common wheat variety KN199. Exogenous application of ET or ACC (a precursor of ET biosynthesis) significantly increased the resistance to Bgt in both G1812 and KN199, whereas the reverse was found when aminoethoxyvinylglycine (AVG, an inhibitor of ET biosynthesis) was applied (Fig. 3). The increased resistance against Bgt by exogenous ET was additionally confirmed using five more T. urartu accessions (Fig. S4). Together, the analyses presented above corroborate with each other, and suggest that TuACO3, induced by Bgt infection, promotes wheat defense against Bgt through significantly upregulating the content of ET.
Figure 3.
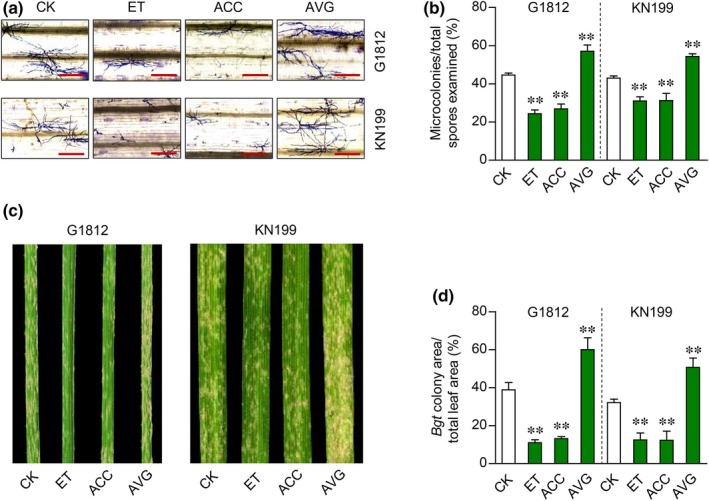
Effect of changing ethylene (ET) concentration on Blumeria graminis f.sp. tritici (Bgt) defense in the Triticum urartu accession G1812 and the common wheat cultivar Kenong 199 (KN199). Ethylene (10 ppm) was applied by placing plants in sealed containers. 1‐Aminocyclopropane‐1‐ carboxylic acid (ACC, 20 μM) and aminoethoxyvinylglycine (AVG, 10 μM) were applied by spraying. The plants were inoculated with Bgt at 24 h after the treatment. (a, b) Influence of Bgt microcolony development by ET, ACC or AVG as revealed by Coomassie blue staining of fungal structures (a) and quantitative comparison with the control (CK) (b). The samples were analyzed at 72 h post‐inoculation (hpi). (c, d) Differences in Bgt colony growth on the leaves treated by ET, ACC or AVG at 8 d post‐inoculation (dpi), as shown by the photographs of Bgt‐infected leaves (c) and quantitative comparison of the percentages of Bgt colony area (d). The data displayed were typical of four independent experiments. The means (± SE) in (b) each were calculated from eight seedlings; those in (d) each were obtained by scanning 12 Bgt‐infected leaves from four separate experiments. **, P < 0.01 (Student's t‐test). Bars, 200 μm.
TuMYB46L is a negative regulator of TuACO3 expression
In order to identify the TF(s) controlling Bgt infection‐induced expression of TuACO3, we screened a Y1H library using TuACO3 promoter region as bait. This led to the identification of six genes whose proteins could bind to the promoter of TuACO3 in two independent Y1H screens (Table S2). One of them, namely TuMYB46L (TuG1812G0500001166), was predicted to encode a MYB TF carrying a putative R2R3 MYB domain (Fig. S5a), whose full‐length protein showed 35.2% identity to AtMYB46, a previously characterized MYB TF regulating secondary cell wall biosynthesis and host defense against B. cinerea in Arabidopsis (Zhong et al., 2007; Ko et al., 2009; Ramírez et al., 2011a,b; Zhong & Ye, 2012; Kim et al., 2014). Consistent with the identification of TuMYB46L by Y1H, the 2‐kb promoter region of TuACO3 was found to carry four cis‐elements highly similar to the consensus binding site of AtMYB46, ACC(A/T)A(A/C)(T/C) (Zhong & Ye, 2012) (Fig. S5b). We therefore focused on analyzing the potential regulation of TuACO3 transcription by TuMYB46L.
The specific binding of TuMYB46L to TuACO3 promoter was verified by including additional controls (Fig. 4a). The binding was not observed when the four putative MYB46 recognition sites in the promoter region of TuACO3 were all mutated to AAAAAAA; neither was it observed when free GFP was used as prey and WT TuACO3 promoter region as bait. In the effector–promoter reporter assay conducted in Nicotiana benthamiana leaves, expression of TuMYB46L, but not free GFP, suppressed the promoter activity of TuACO3 (Fig. 4b,c). TuMYB46L did not suppress the activity of the mutant promoter of TuACO3 (with all four MYB46 recognition sites changed to AAAAAAA) as effectively as it did for the WT promoter of TuACO3 (Fig. 4b,c). Phylogenetic analysis validated the relatedness of TuMYB46L to its homologs (including AtMYB46) from monocot and dicot plants (Fig. S5c). However, the predicted proteins of TuMYB46L and its close cereal homologs (ZmMYB46 and OsMYB46) were considerably larger than those of AtMYB46 and the homologs from dicot plants (Fig. S5c). As anticipated, TuMYB46L was targeted to the nucleus when transiently expressed as a YFP fusion protein in N. benthamiana leaf cells (Methods S1; Fig. S5d).
Figure 4.
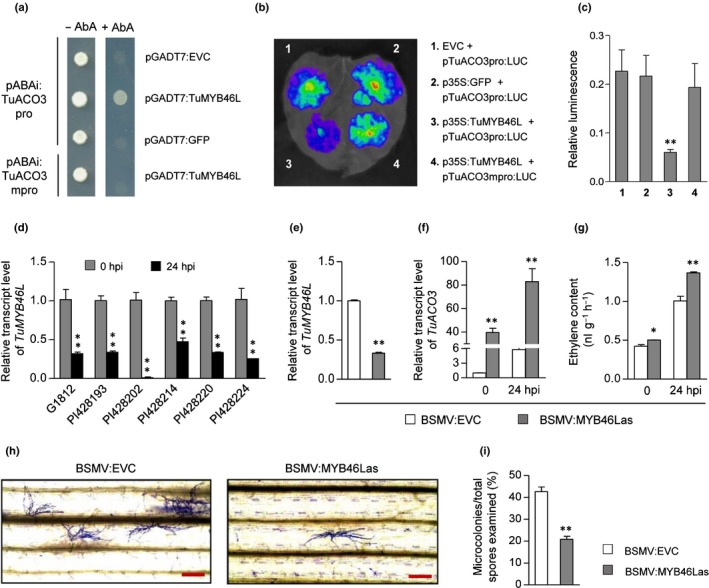
Functional analysis of TuMYB46L. (a) Binding of TuMYB46L to TuACO3 promoter of in yeast‐one‐hybrid (Y1H) assay. The constructs pABAi:TuACO3pro and pABAi:TuACO3mpro carried wild‐type (WT) or mutated versions of TuACO3 promoter (2 kb); the mutant was created by changing each of the four MYB46 recognition sites into AAAAAAA. The plasmids pGADT7:TuMYB46L and pGADT7:GFP expressed TuMYB46L or GFP. The plasmid pGADT7:EVC was used as empty vector control. Positive binding of TuMYB46L to the WT promoter of TuACO3 was indicated by abundant growth of the yeast carrying both pABAi:TuACO3pro and pGADT7:TuMYB46L in the presence of aureobasidin A (AbA). (b, c) Suppression of TuACO3 promoter (carried by pTuACO3pro:LUC) activity by TuMYB46L in Nicotiana benthamiana leaf cells, as evidenced by luminescence imaging (b) and quantitative comparison of luciferase signals (c). The luciferase signals specified by TuACO3 promoter were strongly suppressed by TuMYB46L (expressed from p35S:TuMYB46L) but not by green fluorescent protein (GFP) (expressed from the control construct p35S:GFP). No suppression was obtained with the empty vector control (EVC). TuMYB46L did not suppress the activity of a mutant TuACO3 promoter (carried in pTuACO3mpro:LUC, with all four MYB46 recognition sites changed to AAAAAAA) as effectively as it did for the WT promoter of TuACO3. (d) Decreased expression of TuMYB46L in six different Triticum urartu accessions at 24 h post inoculation (hpi) as detected by qRT‐PCR. (e–i) BSMV mediated silencing of TuMYB46L in the T. urartu accession G1812. Compared to the empty viral vector control (BSMV:EVC), the recombinant virus BSMV:MYB46Las carrying the silencing‐inducing fragment lowered TuMYB46L transcript level (e), and promoted TuACO3 expression (f) and ethylene production (g) in both the control and Blumeria graminis f.sp. tritici (Bgt)‐inoculated plants. Bgt microcolony development was strongly inhibited in the plants in which TuMYB46L was silenced as indicated by Coomassie blue staining of fungal structures (h) and quantitative comparison (i). The datasets shown each were representative of three independent experiments. Each mean (± SE) was determined from three biological replicates. *, P < 0.05; **, P < 0.01 (Student's t‐test). Bars, 200 μm. ACO, 1‐aminocyclopropane‐1‐carboxylic acid oxidase; MYB, myeloblastosis.
The expression of TuMYB46L was strongly and rapidly decreased by Bgt infection in all six T. urartu accessions examined (Fig. 4d). Silencing TuMYB46L expression by BSMV‐mediated VIGS resulted in a significantly greater induction of TuACO3 at 24 hpi of Bgt, and substantially more ET was produced in TuMYB46L silenced plants relative to their controls (Fig. 4e–g). The Bgt microcolonies recorded for TuMYB46L silenced plants were substantially fewer than those found for the controls (Fig. 4h,i). In agreement with these results, the defense against Bgt was compromised by overexpressing TuMYB46L, but strengthened after silencing TuMYB46L by TIGS, in the single‐cell transfection assays conducted with T. urartu leaves (Fig. S6a,b). BSMV‐mediated VIGS also was conducted for the orthologous gene of TuMYB46L in common wheat (TraesCS5A02G101000, TaMYB46L), which led to substantial increase of TaACO3 transcript level and considerable decrease of the susceptibility to Bgt (Fig. S6d–f).
We further analyzed two independent transgenic lines (TuMYB46L‐OX1 and TuMYB46L‐OX2) overexpressing a TuMYB46L‐FLAG tag protein in the common wheat cultivar KN199. Expression of TuMYB46L‐FLAG in the transgenic lines was confirmed by immunoblotting (Fig. 5a). Infection with Bgt significantly upregulated ET production in WTS9, a corresponding WT segregate control line, but this upregulation was diminished in TuMYB46L‐OX1 and TuMYB46L‐OX2 (Fig. 5b). Consistently, substantially more Bgt microcolonies and stronger Bgt colony growth were observed on TuMYB46L‐OX1 and TuMYB46L‐OX2 leaves than on those of WTS9 (Fig. 5c–f). These findings, together with the foregoing results (Fig. 4), indicate that TuMYB46L is a negative regulator of TuACO3 expression, and that its downregulation is required for the enhancement of TuACO3 expression, ET content and Bgt defense in wheat.
Figure 5.
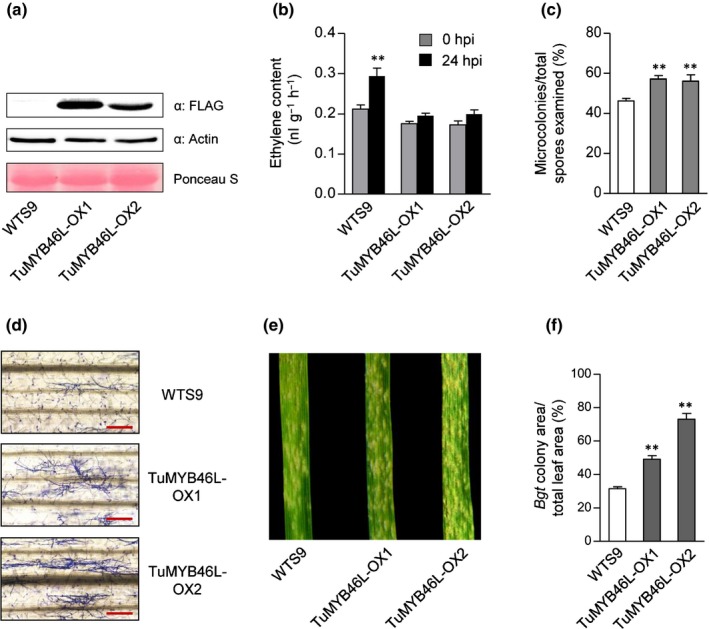
Verification of TuMYB46L function using transgenic plants developed with the common wheat cultivar Kenong 199. TuMYB46L‐OX1 and ‐OX2 were two independent representative homozygous transgenic lines. WTS9 was a wild‐type (WT) segregate used as a control. (a) Overexpression of TuMYB46L‐FLAG protein in TuMYB46L‐OX1 and ‐OX2 revealed by immunoblotting with the antibody specific for FLAG. Detection of actin and staining by Ponceau S indicated equal loading. (b) Upregulation of ethylene biosynthesis by Blumeria graminis f.sp. tritici (Bgt) infection was depressed in TuMYB46L‐OX1 and ‐OX2 but not in WTS9. (c, d) Increased development of Bgt microcolonies in TuMYB46L‐OX1 and ‐OX2 at 72 h post‐inoculation (hpi), as shown by quantitative comparison with WTS9 (c) and Coomassie blue staining of fungal structures (d). (e, f) Stronger Bgt colony growth observed in TuMYB46L‐OX1 and ‐OX2 relative to that in WTS9 at 8 dpi, as indicated by the photographs of Bgt‐infected leaves (e) and quantitative comparison of the percentages of Bgt colony area. The data displayed each were typical of three independent experiments. The means (± SE) in (b) and (c) each were calculated from three biological replicates; those in (f) each were obtained by scanning nine Bgt‐infected leaves from three separate experiments. **, P < 0.01 (Student's t‐test). Bars, 200 μm. MYB, myeloblastosis.
TuMYB46L binds to cis‐elements in the promoter region of TuACO3
Among the four putative MYB46 binding sites in the 2‐kb sequence upstream of the initiation codon of TuACO3, the first one (E1, ACCAAAG) was located near the ATG codon, and the third (E3, ACCTAAA) and fourth (E4, ATTTGGT) were more distant (Figs 6a, S5b). We first performed ChIP‐qPCR assays to test the binding of TuMYB46L to different regions of TuACO3 promoter. A TuMYB46L‐FLAG tag protein was transiently expressed in T. urartu leaf protoplasts, followed by ChIP with the antibody against FLAG. Co‐precipitating DNAs were quantified by qPCR using primer sets covering the 2 kb promoter. Compared to the control in which the FLAG antibody was replaced by normal mouse IgG, the inclusion of FLAG antibody led to the precipitation of substantially more DNA fragments carrying E1 or E3 + E4, but not that harboring E2, with the average enrichment obtained for the two regions being c. 3.5‐fold and 2.3‐fold, respectively (Fig. 6b).
Figure 6.
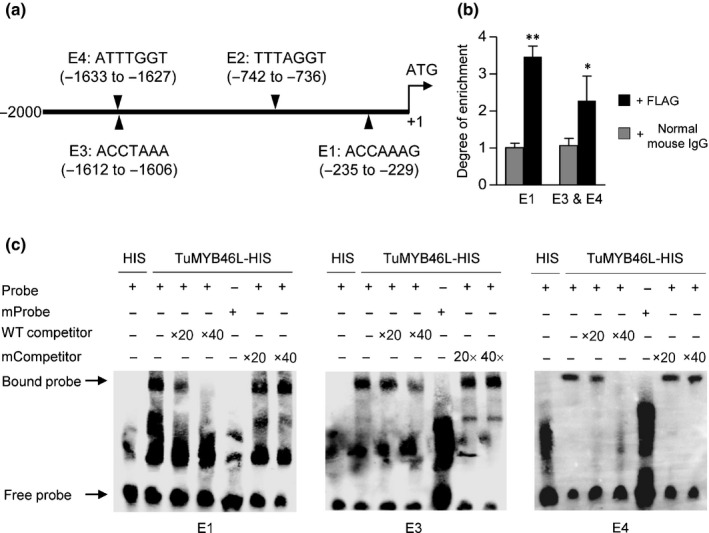
Binding of TuMYB46L to TuACO3 promoter region. (a) A diagram showing the positions of four putative MYB46 binding sites (E1–E4) in the 2‐kb promoter region of TuACO3. (b) Chromatin immunoprecipitation‐quantitative (ChIP‐q)PCR assay. The assay was conducted by expressing a TuMYB46L‐FLAG protein in G1812 leaf protoplasts, with the chromatins bound to TuMYB46L‐FLAG precipitated using the FLAG antibody and subsequently quantified by qPCR. The chromatin fragments carrying the E1 element or harboring the closely spaced E3 and E4 elements were strongly enriched in the precipitation with FLAG antibody compared to the mock precipitation with normal mouse IgG. The means (± SE) each were calculated from three biological replicates, and statistically compared using Student's t‐test. *, P < 0.05; **, P < 0.01. (c) Validation of the binding of TuMYB46L to E1, E3 and E4 by EMSA. A TuMYB46L‐HIS protein expressed in Escherichia coli was purified; its binding to biotin‐labeled E1, E3 or E4 probes was tested in the absence or presence of unlabeled wild‐type (WT) probes. No specific binding was obtained with the labeled mutant probes in which the putative binding sites were all changed to AAAAAAA. Moreover, the specific binding of TuMYB46L‐HIS to E1, E3 and E4 was not affected by the presence of the unlabeled mutant probes. Three sets of independent assays were performed, which all validated the binding of TuMYB46L‐HIS to E1, E3 and E4. ACO, 1‐aminocyclopropane‐1‐carboxylic acid oxidase; MYB, myeloblastosis.
We thus conducted EMSA to validate the binding of E1, E3 and E4 by a bacterially expressed, histidine‐tagged TuMYB46L protein. As anticipated, the TuMYB46L‐HIS tag protein was able to bind all three elements in repeated EMSA experiments (Fig. 6c). The binding was specific because it was not found with the mutant probes, it decreased by the WT competitor probes, but it was not affected by the mutated competitor probes (Fig. 6c).
Multiple processes and genes function in ET‐mediated defense against Bgt
In order to gain insight into the biological processes and genes functioning in ET‐mediated wheat defense against Bgt, we carried out two types of transcriptome assays. The first type used the G1812 leaf samples without or with Bgt inoculation for 24 h, and the second employed the G1812 leaves without or with ET treatment for 24 h. For Bgt treatment, the up‐ and down‐regulated genes were 2307 and 1516, respectively; for ethylene treatment the differentially expressed genes were 1406 (up‐regulated) and 1222 (down‐regulated), respectively (Fig. 7a). The up‐regulated and down‐regulated genes shared by the two treatments amounted to 691 and 559, respectively (Fig. 7b). For the shared upregulated genes, the significantly enriched biological processes were related to chitin catabolism, cellulose biosynthesis and isoprenoid biosynthesis; for the shared downregulated genes, the highly enriched biological processes included oxidation reduction and metal ion transport (Table S3).
Figure 7.
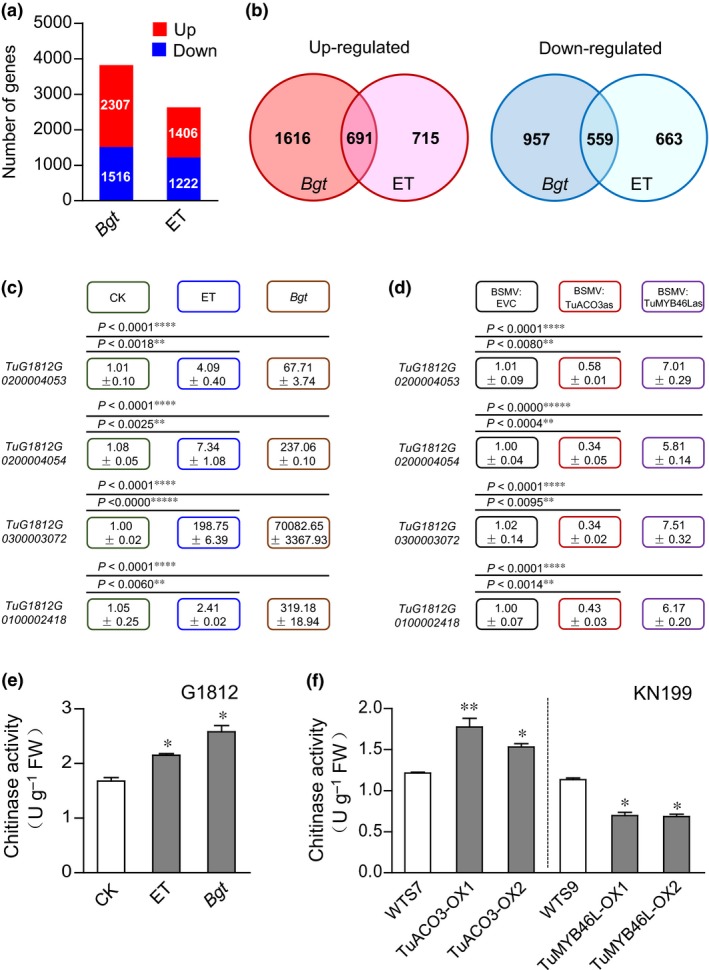
Analysis of the genes regulated by Blumeria graminis f.sp. tritici (Bgt) inoculation and ethylene (ET) treatment. (a) The numbers of up‐ and downregulated genes triggered by Bgt inoculation or ET treatment in the Triticum urartu accession G1812. (b) The numbers of up‐ and downregulated genes shared by Bgt inoculation and ET treatment. (c) Validation by quantitative reverse transcription (qRT)‐PCR of the four chitinase genes upregulated by Bgt inoculation and ET treatment, with the obtained relative expression values shown in the color‐coded boxes. The actual significance levels as compared to the control (CK) also were displayed. (d) Effects of silencing TuACO3 or TuMYB46L on the expression of four chitinase genes. Transcript levels of the four genes in the G1812 plants inoculated with BSMV‐EVC (empty vector control), BSMV:ACO3as (silencing TuACO3), or BSMV:MYB46Las (silencing TuMYB46L) were evaluated by qRT‐PCR with gene‐specific primers. The relative expression values scored were indicated in the color‐coded boxes, and the actual significance levels as compared to the control (BSMV:EVC) also were provided. (e) Elevation of total chitinase activities in the G1812 plants treated by ET or Bgt inoculation compared to the untreated controls (CK). The samples used for the assay were collected at 24 h after ET (10 ppm) treatment or at 24 h post‐inoculation (hpi) of Bgt. (f) Total chitinase activities were increased in the transgenic lines overexpressing TuACO3 (TuACO3‐OX1 and ‐OX2), but decreased in the other two transgenic lines overexpressing TuMYB46L (TuMYB46L‐OX1 and ‐OX2) relative to their corresponding WTS controls. In (c) to (f), the data presented were representative of three independent experiments, with each mean (± SE) calculated from three biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; *****, P < 0.00001 (Student's t‐test). ACO, 1‐aminocyclopropane‐1‐carboxylic acid oxidase; MYB, myeloblastosis.
Because previous studies have shown the increase of host chitinase gene expression in fungal‐infected plant tissues and the involvement of ET in the induction process (Jacobs et al., 1999; Akagi et al., 2011), we further analyzed the four chitinase genes (i.e., TuG1812G0100002418, TuG1812G0200004053, TuG1812G0200004054 and TuG1812G0300003072, Ling et al., 2018) that were upregulated by both Bgt and ET treatments (Table S4). The fold‐induction of the four genes varied approximately from three to nine (after Bgt infection) or two to four (post ET application) (Table S4). Their upregulated expression after Bgt or ET treatment was confirmed by qRT‐PCR (Fig. 7c). Importantly, the expression of the four chitinase genes all were repressed in the T. urartu leaf tissues in which TuACO3 was silenced by BSMV‐mediated VIGS, but promoted in those with downregulated TuMYB46L expression (Fig. 7d).
Consistent with the foregoing results, total chitinase activities were substantially elevated by Bgt infection (1.5‐fold) or ET treatment (1.3‐fold) in T. urartu (Fig. 7e). Moreover, total chitinase activity levels in the two TuACO3 overexpression transgenic lines were significantly higher than that of the control (WTS7) (Fig. 7f). By contrast, total chitinase activities in the two TuMYB46L overexpression lines were substantially lower relative to those of the control WTS9 (Fig. 7f).
Discussion
In this work, we studied the functions of TuACO3 and TuMYB46L, and discovered a previously uncharacterized gene module (i.e. TuMYB46L‐TuACO3; ACO, 1‐aminocyclopropane‐1‐carboxylic acid (ACC) oxidase; MYB, myeloblastosis) that regulates ethylene (ET) biosynthesis and content in einkorn wheat defense against Blumeria graminis f.sp. tritici (Bgt). The findings were made primarily in the einkorn wheat Triticum urartu, and subsequently confirmed in common wheat by barley stripe mosaic virus (BSMV)‐mediated virus‐induced gene silencing (VIGS) and using the KN199 transgenic lines overexpressing TuACO3 or TuMYB46L.
The expression and function of TuACO3 are enhanced in Bgt‐infected wheat
Although ACO has long been known to catalyze the last step of ET biosynthesis (Spanu & Boller, 1989; Kende, 1993; Booker & DeLong, 2015), it is only recently that several studies have uncovered the importance of transcriptional control in the function of ACO during cotton fiber development, apple fruit ripening and Arabidopsis root development (Li et al., 2014; T. Li et al., 2017; Hu et al., 2019; Park et al., 2018) . A number of studies have recorded changes in the expression of various ACO gene members accompanying the infections by different pathogens (Shan & Goodwin, 2006; Yu et al., 2011; Vilanova et al., 2017), but the function of the concerned ACO and the underlying regulatory mechanism were generally not analyzed using molecular genetic means.
Based on the data gathered here, we suggest that the function of TuACO3 is enhanced in Bgt‐infected wheat, which contributes to host defense against Bgt via significantly increasing ET biosynthesis and content. Importantly, we validated the promotion of wheat defense against Bgt by TuACO3 via elevating ET biosynthesis through analyzing two independent common wheat transgenic lines overexpressing TuACO3 (Fig. 2). Thus, our work has generated molecular and functional evidence for the positive contribution of an ACO gene to plant defense against pathogen infection through elevating ET concentration.
Before this work, there has been some evidence for the involvement of ET in plant defense against powdery mildew fungi. The Arabidopsis mutant cev1 displays constitutively active jasmonic acid/ET induced defense responses and shows increased resistance to three different powdery mildew pathogens (Ellis & Turner, 2001). In grapevine, powdery mildew infection and artificial treatment with ethephon induce an overlapping set of defense‐related proteins (Jacobs et al., 1999). The present work demonstrates that ET biosynthesis evidently is upregulated in Bgt‐infected wheat and contributes to host defense against Bgt. Although this contribution did not offer a complete protection to the disease, it probably reduced disease severity because silencing TuACO3 not only decreased ET production, but also increased the development of powdery mildew colonies in Bgt‐infected wheat (Fig. 1e–h). Furthermore, we showed that exogenous application of ET enhanced wheat resistance to Bgt and that inhibition of ET biosynthesis by aminoethoxyvinylglycine (AVG) aggravated susceptibility to Bgt (Fig. 3). Consequently, our work suggests that ET‐mediated defense can be activated by an adapted powdery mildew pathogen in a compatible interaction in wheat. This provides a new molecular insight into the role of ET in plant defense to biotrophic fungal pathogens.
Consistent with the increased TuACO3 expression and ET production discussed above, the transcript level of one 1‐aminocyclopropane‐1‐carboxylic acid synthase (ACS) gene (i.e. TuACS2), which functions immediately upstream of ACO (Booker & DeLong, 2015), was elevated in Bgt‐infected T. urartu plants (Fig. S7). The activity of TuACS2 may enhance the generation of the precursor ACC for ET biosynthesis in Bgt‐invaded wheat tissues. However, further work is needed to validate this possibility.
Because the ACO gene family is well conserved in higher plants (Booker & DeLong, 2015; Sun et al., 2017; Park et al., 2018), and powdery mildew diseases occur in diverse plant species (Glawe 2008), it is worth investigating whether the enhancement of plant defense by ACO, as revealed here, also may happen in other powdery mildew pathosystems. Considering the limited understanding on most of the powdery mildew diseases recorded to date (Glawe 2008), molecular and functional analysis of ACO may provide a generally useful entry point for studying host defense mechanisms to different powdery mildew fungi.
Downregulation of TuMYB46L is required for elevated expression of TuACO3 after Bgt infection
Judging from the molecular genetic data obtained (Figs 4, 5), we suggest that downregulation of TuMYB46L is a prerequisite for the elevated expression of TuACO3 in Bgt‐infected wheat and its subsequent function in increasing ET biosynthesis. Central to the action of TuMYB46L is its ability to bind to multiple cis‐elements in the promoter region of TuACO3 (Fig. 6). The rapid decline of TuMYB46L expression following Bgt infection diminishes the binding of its protein to the promoter of TuACO3, which permits transcriptional upregulation of TuACO3 in Bgt‐infected wheat. Thus, TuMYB46L is likely a negative regulator of TuACO3 expression and function under normal growth conditions.
As a vital phytohormone, the biosynthesis of ET must be tightly regulated according the environmental conditions to which the plants are exposed (Booker & DeLong, 2015; Broekgaarden et al., 2015). Failure to adjust ET biosynthesis and signaling lessens plant adaptation to harmful environments (Guo & Ecker, 2003; Booker & DeLong, 2015; Broekgaarden et al., 2015). Therefore, TuMYB46L regulates ET biosynthesis through controlling the expression of TuACO3 according to the presence or absence of Bgt infection in wheat. The significance of the TuMYB46L‐TuACO3 module in wheat defense against Bgt via regulating ET production is clearly demonstrated in this work. Further study is needed to examine if the negative regulation of TuACO3 by TuMYB46L also may be required for efficient plant growth under normal environmental conditions.
Among the homologs of TuMYB46L (Fig. S5c), only AtMYB46 from Arabidopsis has so far been shown to affect plant response to pathogen challenge (Ramírez et al., 2011a,b). The knockout plants of AtMYB46 showed increased disease resistance to Botrytis cinerea because of changed cell wall integrity and more efficient induction of the genes involved in cell wall remodeling or pathogen defense than WT plants (Ramírez et al., 2011a,b). In view of the relatedness of TuMYB46L to AtMYB46, on the one hand, it will be interesting to investigate if the contribution of the TuMYB46L‐TuACO3 module to wheat defense against Bgt also may involve changed cell wall integrity and enhanced expression of cell wall remodeling genes, in addition to the upregulation of TuACO3 expression and ET production observed in this work. On the other hand, it also may be worthwhile to investigate the potential existence of an AtMYB46‐AtACO module and its possible contribution to Arabidopsis defense against pathogens. In a preliminary analysis, we found that AtMYB46 could suppress the promoter activity of AtACO4, which is the closest Arabidopsis homolog of TuACO3 (65.8% protein identity), and that the promoter region of AtACO4 also carries five predicted AtMYB46 binding sites (Fig. S8). Whether the suppression of AtACO4 promoter activity by AtMYB46 may take part in the regulation of AtACO4 transcription during Arabidopsis pathogen defense is an important question for future research. Also important for future study is to examine if TuMYB46L may regulate secondary cell wall development as this is a shared function among MYB46 homologs (Zhong et al., 2011; Xie et al., 2018); TuMYB46L may resemble AtMYB46 and the MYB46 homolog from birch in being able to regulate multiple plant processes (Ramírez et al., 2011a,b; Guo et al., 2017). Nevertheless, certain functional difference between TuMYB46L and AtMYB46 is to be expected because the identity level of their full‐length proteins (35.2%) is not high and their proteins differ substantially in size (Fig. S5a,c).
The relationships among ET biosynthesis and signaling, cell wall development, organ growth and pathogen defense are very complex, and may involve growth–defense trade‐offs (Ramírez et al., 2011a,b; Souza et al., 2017; Xie et al., 2018). The finding of the participation of an Arabidopsis ACS protein (ACS5) in cell wall biosynthesis independent of ET concentration adds further complexity to the interactions (Xu et al., 2008). However, we demonstrated here that the TuMYB46L‐TuACO3 module increased ET production in Bgt‐infected wheat, and that exogenous application of ET or ACC enhanced wheat defense against Bgt. Thus, further research on the TuMYB46L‐TuACO3 module may help to improve understanding of the interplays among ET biosynthesis and signaling, cell wall development and function, plant growth and pathogen defense.
Chitinase genes participate in ET‐mediated defense against Bgt
Through transcriptome comparison and validation by quantitative reverse‐trancription (qRT)‐PCR, the expression of four different chitinase genes was confirmed to be upregulated by both Bgt infection and ET treatment (Fig. 7a–c; Table S4). Furthermore, the four genes were depressed by silencing TuACO3 but elevated after suppressing TuMYB46L (Fig. 7d), suggesting that they act downstream of the TuACO3‐TuMYB46L module. This proposition is additionally supported by (1) increased total chitinase activities in the wheat plants infected by Bgt or treated with ET (Fig. 7e) and in those overexpressing TuACO3 (Fig. 7f), and (2) reduced total chitinase activities in the wheat lines overexpressing TuMYB46L (Fig. 7f).
Plant chitinase isozymes are potent players in host defense against fungal pathogens (Pusztahelyi, 2018), and past studies have reported the involvement of ET in the induction of host chitinase gene expression in fungal‐infected plants (Jacobs et al., 1999; Akagi et al., 2011). This work improves the understanding of chitinase gene induction by a pathogenic fungus (Bgt) through identifying a specific host gene module (TuMYB46L‐TuACO3) that produces the endogenous ET signal needed for the induction. The next challenge is to reveal how ET orchestrates chitinase gene expression. In this context, it is interesting to note that overexpression of certain ET responsive factors (ERFs) can enhance the expression of pathogenesis‐related proteins including chitinase (Berrocal‐Lobo et al., 2002; Akagi et al., 2011; Zhu et al., 2014). It also will be interesting to test if reactive oxygen species (ROS) and additional pathogenesis‐related genes may be involved in the defense processes regulated by TuMYB46L‐TuACO3 in further research.
Apart from chitinase genes, our transcriptome analysis also detected upregulation of the genes involved in the biosynthesis of cellulose or isoprenoids in Bgt‐infected or ET‐treated wheat plants (Table S3). On the one hand, there is now increasing evidence that alterations in cellulose biosynthesis and cell wall integrity modulate plant defense against pathogens (Caño‐Delgado et al., 2003; Hernández‐Blanco et al., 2007; Souza et al., 2017). On the other, the phytoalexins derived from phenylpropanoid metabolism are active defense compounds against plant pathogens (Ahuja et al., 2012; Liu et al., 2015). Therefore, elevated expression of the genes functioning in cellulose or isoprenoid biosynthesis may help to strengthen the defense against Bgt in wheat. We noted that the 2‐kb promoter region of the three upregulated cellulose synthase genes, as well as the two upregulated phenylalanine ammonia lyase genes, each carried two or three putative MYB46 binding sites (Table S5). We are now in the process of investigating if these genes also may be regulated by the TuMYB46L‐TuACO3 module.
In summary, this study establishes that the TuMYB46L‐TuACO3 module regulates ET biosynthesis to promote einkorn wheat defense against Bgt. The insights obtained improve our knowledge on wheat defense mechanisms to Bgt and on the ACO enzyme that is critical in generating the ET signal for plant adaptation to changing environment (Booker & DeLong, 2015). Our work points to the possibility of controlling wheat powdery mildew disease through manipulating ET concentration, and may stimulate further research into the regulation and function of ET biosynthesis in plant defense against diverse powdery mildew fungi.
Author contributions
HZ, KZ and DW designed the project; HZ, LD and XH performed the experiments; HZ, LD, HJ and CY performed the analyses; KZ, YH, BL, HQ, QS and JZ contributed to data collection; and HZ and DW wrote the manuscript. HZ and LD contributed equally to this work.
Supporting information
Dataset S1 Sequence information and raw data.
Fig. S1 Morphology of transgenic wheat lines overexpressing TuACO3 or TuMYB46L.
Fig. S2 Phylogenetic analysis of ACO genes from different plant species.
Fig. S3 Elevation of TuACO3 expression and ET production by Bgt infection in five different T. urartu accessions and analysis of TaACO3 function in common wheat using BSMV‐mediated VIGS.
Fig. S4 Inhibition of Bgt colony development and growth by ET treatment in five different T. urartu accessions.
Fig. S5 Primary structure, phylogeny and nuclear localization of TuMYB46L and predicted MYB46 binding sites in the 2‐kb promoter region of TuACO3.
Fig. S6 Effects of transiently overexpressing or silencing TuMYB46L on Bgt haustorium growth and analysis of TaMYB46L function by VIGS in common wheat.
Fig. S7 Elevation of TuACS2 transcript level by Bgt infection at 24 hpi.
Fig. S8 Suppression of AtACO4 promoter activity by AtMYB46.
Methods S1 Additional description of methods.
Table S1 PCR primers used in this study.
Table S2 List of six T. urartu genes found in two independent Y1H screens.
Table S3 Significantly enriched biological processes identified by GO analysis.
Table S4 Expression changes of the four chitinase genes upregulated by both ET treatment and Bgt infection.
Table S5 Presence of putative MYB46 recognition sites in the 2‐kb promoter region of the five genes related to cellulose or isoprenoid synthesis.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This study was supported by the key research and development program of the Ministry of Science and Technology of the People's Republic of China (2016YFD0101004, 2017YFD0100600). We thank Mingyue Gou for constructive suggestions on manuscript preparation. The RNA‐Seq data generated in this study were deposited in the Genome Sequence Archive of Beijing Institute of Genomics (accession no. CRA001335).
Contributor Information
Kunpu Zhang, Email: zkp66@126.com.
Daowen Wang, Email: dwwang@genetics.ac.cn.
References
- Ahuja I, Kissen R, Bones AM. 2012. Phytoalexins in defense against pathogens. Trends in Plant Science 17: 73–90. [DOI] [PubMed] [Google Scholar]
- Akagi A, Dandekar AM, U Stotz H. 2011. Resistance of Malus domestica fruit to Botrytis cinerea depends on endogenous ethylene biosynthesis. Phytopathology 101: 1311–1321. [DOI] [PubMed] [Google Scholar]
- Al‐Attala MN, Wang X, Abou‐Attia MA, Duan X, Kang Z. 2014. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Molecular Biology 84: 589–603. [DOI] [PubMed] [Google Scholar]
- Bao Z, Zhang N, Hua J. 2014. Endopolyploidization and flowering time are antagonistically regulated by checkpoint component MAD1 and immunity modulator MOS1. Nature Communications 5: 5628. [DOI] [PubMed] [Google Scholar]
- Berrocal‐Lobo M, Molina A, Solano R. 2002. Constitutive expression of ETHYLENE‐RESPONSE‐FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal 29: 23–32. [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. 2015. Signaling mechanisms in pattern‐triggered immunity (PTI). Molecular Plant 8: 521–539. [DOI] [PubMed] [Google Scholar]
- Booker MA, DeLong A. 2015. Producing the ethylene signal: regulation and diversification of ethylene biosynthetic enzymes. Plant Physiology 169: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Delaure SL, De Bolle MF, Cammue BP. 2006. The role of ethylene in host‐pathogen interactions. Annual Review of Phytopathology 44: 393–416. [DOI] [PubMed] [Google Scholar]
- Broekgaarden C, Caarls L, Vos IA, Pieterse C, van Wees S. 2015. Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiology 169: 2371–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar NK, Zhang Z, Wicker T, Keller B. 2010. Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biology 10: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño‐Delgado A, Penfield S, Smith C, Catley M, Bevan M. 2003. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana . The Plant Journal 34: 351–362. [DOI] [PubMed] [Google Scholar]
- Cheng HQ, Han LB, Yang CL, Wu XM, Zhong NQ, Wu JH, Wang FX, Wang HY, Xia GX. 2016. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. Journal of Experimental Botany 67: 1935–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chezem WR, Memon A, Li FS, Weng JK, Clay NK. 2017. SG2‐Type R2R3‐MYB transcription factor MYB15 controls defense‐induced lignification and basal immunity in Arabidopsis . Plant Cell 29: 1907–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. 2015. Effector‐triggered immunity: from pathogen perception to robust defense. Annual Review Plant Biology 66: 487–511. [DOI] [PubMed] [Google Scholar]
- Dong L, Cheng Y, Wu J, Cheng Q, Li W, Fan S, Jiang L, Xu Z, Kong F, Zhang D et al 2015a. Overexpression of GmERF5, a new member of the soybean EAR motif‐containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. Journal of Experimental Botany 66: 2635–2647. [DOI] [PubMed] [Google Scholar]
- Dong L, Liu H, Zhang J, Yang S, Kong G, Chu JS, Chen N, Wang D. 2015b. Single‐molecule real‐time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genomics 16: 1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P. 2005. A high‐throughput gene‐silencing system for the functional assessment of defense‐related genes in barley epidermal cells. Molecular Plant–Microbe Interactions 18: 755–761. [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG. 2001. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawe DA. 2008. The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Annual Review of Phytopathology 46: 27–51. [DOI] [PubMed] [Google Scholar]
- Gravino M, Savatin DV, Macone A, De Lorenzo G. 2015. Ethylene production in Botrytis cinerea‐ and oligogalacturonide‐induced immunity requires calcium‐dependent protein kinases. The Plant Journal 84: 1073–1086. [DOI] [PubMed] [Google Scholar]
- Guan R, Su J, Meng X, Li S, Liu Y, Xu J, Zhang S. 2015. Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae . Plant Physiology 169: 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. 2003. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)‐dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677. [DOI] [PubMed] [Google Scholar]
- Guo H, Wang Y, Wang L, Hu P, Wang Y, Jia Y, Zhang C, Zhang Y, Zhang Y, Wang C et al 2017. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla . Plant Biotechnology Journal 15: 107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Zhu S, Zhao R, Jiang Z, Ji Y, Ji J, Qiu D, Li H, Bie T. 2018. Pm21, encoding a typical CC‐NBS‐LRR protein, confers broad‐spectrum resistance to wheat powdery mildew disease. Molecular Plant 11: 879–882. [DOI] [PubMed] [Google Scholar]
- Hernández‐Blanco C, Feng DX, Hu J, Sanchez‐Vallet A, Deslandes L, Llorente F, Berrocal‐Lobo M, Keller H, Barlet X, Sanchez‐Rodriguez C et al 2007. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19: 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R, Panstruga R. 2011. Cell biology of the plant‐powdery mildew interaction. Current Opinion in Plant Biology 14: 738–746. [DOI] [PubMed] [Google Scholar]
- Hu DG, Yu JQ, Han PL, Xie XB, Sun CH, Zhang QY, Wang JH, Hao YJ. 2019. The regulatory module MdPUB29‐MdbHLH3 connects ethylene biosynthesis with fruit quality in apple. New Phytologist 221: 1966–1982. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) , Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A et al 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: pii: eaar7191. [DOI] [PubMed] [Google Scholar]
- Jacobs AK, Dry IB, Robinson SP. 1999. Induction of different pathogenesis‐related cDNAs in grapevine infected with powdery mildew and treated with ethephon. Plant Pathology 48: 325–336. [Google Scholar]
- Kende H. 1993. Ethylene biosynthesis. Annual Review of Plant Physiology and Plant Molecular Biology 44: 283–307. [Google Scholar]
- Kim WC, Kim JY, Ko JH, Kang H, Han KH. 2014. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Molecular Biology 85: 589–599. [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim WC, Han KH. 2009. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis . The Plant Journal 60: 649–665. [DOI] [PubMed] [Google Scholar]
- Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C et al 2014. Genome sequence of the cultivated cotton Gossypium arboreum . Nature Genetics 46: 567–572. [DOI] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A. 2017a. The jasmonate‐activated transcription factor MdMYC2 regulates ethylene responsive factor and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29: 1316–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J et al 2017b. A natural allele of a transcription factor in rice confers broad‐spectrum blast resistance. Cell 170: 114–126. [DOI] [PubMed] [Google Scholar]
- Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y et al 2018. Genome sequence of the progenitor of wheat A subgenome Triticum urartu . Nature 557: 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P. 2015. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Molecular Plant 8: 689–708. [DOI] [PubMed] [Google Scholar]
- McNally KE, Menardo F, Lüthi L, Praz CR, Müller MC, Kunz L, Ben‐David R, Chandrasekhar K, Dinoor A, Cowger C et al 2018. Distinct domains of the AVRPM3A2/F2 avirulence protein from wheat powdery mildew are involved in immune receptor recognition and putative effector function. New Phytologist 218: 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Roh J, Youn JH, Son SH, Park JH, Kim SY, Kim TW, Kim SK. 2018. Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development. Molecules and Cells 41: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon‐Reyes A, Van Wees SC. 2012. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Pusztahelyi T. 2018. Chitin and chitin‐related compounds in plant‐fungal interactions. Mycology 9: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Qian W, Wang W, Wu Y, Yu C, Jiang X, Wang D, Wu P. 2008. GDP‐mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 105: 18308–18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez V, Agorio A, Coego A, Garcia‐Andrade J, Hernandez MJ, Balaguer B, Ouwerkerk PB, Zarra I, Vera P. 2011a. MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis . Plant Physiology 155: 1920–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez V, García‐Andrade J, Vera P. 2011b. Enhanced disease resistance to Botrytis cinerea in myb46 Arabidopsis plants is associated to an early down‐regulation of CesA genes. Plant Signaling & Behavior 6: 911–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan XC, Goodwin PH. 2006. Silencing an ACC oxidase gene affects the susceptible host response of Nicotiana benthamiana to infection by Colletotrichum orbiculare . Plant Cell Reports 25: 241–247. [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ülker B, Somssich IE, Schulze‐Lefert P. 2007. Nuclear activity of MLA immune receptors links isolate‐specific and basal disease‐resistance responses. Science 315: 1098–1103. [DOI] [PubMed] [Google Scholar]
- Singh RP, Singh PK, Rutkoski J, Hodson DP, He X, Jorgensen LN, Hovmoller MS, Huerta‐Espino J. 2016. Disease impact on wheat yield potential and prospects of genetic control. Annual Review of Phytopathology 54: 303–322. [DOI] [PubMed] [Google Scholar]
- Spanu P, Boller T. 1989. Ethylene biosynthesis in tomato plants infected by Phytophthora infestans . Journal of Plant Physiology 134: 533–537. [Google Scholar]
- Souza CA, Li S, Lin AZ, Boutrot F, Grossmann G, Zipfel C, Somerville SC. 2017. Cellulose‐derived oligomers act as damage‐associated molecular patterns and trigger defense‐like responses. Plant Physiology 173: 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Li Y, He W, Ji C, Xia P, Wang Y, Du S, Li H, Raikhel N, Xiao J et al 2017. Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase. Nature Communications 8: 15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nürnberger T, Tsuda K, Saijo Y. 2013. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proceedings of the National Academy of Sciences, USA 110: 6211–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilanova L, Vall‐llaura N, Torres R, Usall J, Teixidó N, Larrigaudière C, Giné‐Bordonaba J. 2017. Penicillium expansum (compatible) and Penicillium digitatum (non‐host) pathogen infection differentially alter ethylene biosynthesis in apple fruit. Plant Physiology and Biochemistry 120: 132–143. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. 2014. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology 32: 947–951. [DOI] [PubMed] [Google Scholar]
- Wei X, Shan T, Hong Y, Xu H, Liu X, Zhang Z. 2017. TaPIMP2, a pathogen‐induced MYB protein in wheat, contributes to host resistance to common root rot caused by Bipolaris sorokiniana . Scientific Reports 7: 1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Oberhaensli S, Parlange F, Buchmann JP, Shatalina M, Roffler S, Ben‐David R, Dolezel J, Simkova H, Schulze‐Lefert P et al 2013. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nature Genetics 45: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen JG, Muchero W. 2018. Regulation of lignin biosynthesis and its role in growth‐defense tradeoffs. Frontier in Plant Science 9: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Di Z, Yang W, Liu J, Li M, Wang X, Cui C, Wang X, Wang X, Zhang R et al 2017. Overexpression of ERF1‐V from Haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Frontiers in Plant Science 8: 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Hu P, Liu J, Witek K, Zhou S, Xu J, Zhou W, Gao L, Huang Z, Zhang R et al. 2018. Pm21 from Haynaldia villosa encodes a CC‐NBS‐LRR protein conferring powdery mildew resistance in wheat. Molecular Plant 11: 874–878. [DOI] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ. 2008. Two leucine‐rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis . Plant Cell 20: 3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiaoui N, Srichumpa P, Dudler R, Keller B. 2004. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. The Plant Journal 37: 528–538. [DOI] [PubMed] [Google Scholar]
- Yang C, Li W, Cao J, Meng F, Yu Y, Huang J, Jiang L, Liu M, Zhang Z, Chen X et al. 2017. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. The Plant Journal 89: 338–353. [DOI] [PubMed] [Google Scholar]
- Yin CC, Ma B, Collinge DP, Pogson BJ, He SJ, Xiong Q, Duan KX, Chen H, Yang C, Lu X et al 2015. Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase‐mediated abscisic acid pathway. Plant Cell 27: 1061–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Shen L, Zhang A, Sheng J. 2011. Methyl jasmonate‐induced defense responses are associated with elevation of 1‐aminocyclopropane‐1‐carboxylate oxidase in Lycopersicon esculentum fruit. Journal of Plant Physiology 168: 1820–1827. [DOI] [PubMed] [Google Scholar]
- Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D. 2011. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zheng H, Li Y, Li H, Liu X, Qin H, Dong L, Wang D. 2016. Coexpression network analysis of the genes regulated by two types of resistance responses to powdery mildew in wheat. Scientific Reports 6: 23805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH. 2011. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiology 52: 1856–1871. [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. 2007. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis . Plant Cell 19: 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. 2012. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiology 53: 368–380. [DOI] [PubMed] [Google Scholar]
- Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z. 2014. The wheat ethylene response factor transcription factor pathogen‐induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiology 164: 1499–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Wang H, Li Y, Kong Z, Tang D. 2018. The NB‐LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytologist 218: 298–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 Sequence information and raw data.
Fig. S1 Morphology of transgenic wheat lines overexpressing TuACO3 or TuMYB46L.
Fig. S2 Phylogenetic analysis of ACO genes from different plant species.
Fig. S3 Elevation of TuACO3 expression and ET production by Bgt infection in five different T. urartu accessions and analysis of TaACO3 function in common wheat using BSMV‐mediated VIGS.
Fig. S4 Inhibition of Bgt colony development and growth by ET treatment in five different T. urartu accessions.
Fig. S5 Primary structure, phylogeny and nuclear localization of TuMYB46L and predicted MYB46 binding sites in the 2‐kb promoter region of TuACO3.
Fig. S6 Effects of transiently overexpressing or silencing TuMYB46L on Bgt haustorium growth and analysis of TaMYB46L function by VIGS in common wheat.
Fig. S7 Elevation of TuACS2 transcript level by Bgt infection at 24 hpi.
Fig. S8 Suppression of AtACO4 promoter activity by AtMYB46.
Methods S1 Additional description of methods.
Table S1 PCR primers used in this study.
Table S2 List of six T. urartu genes found in two independent Y1H screens.
Table S3 Significantly enriched biological processes identified by GO analysis.
Table S4 Expression changes of the four chitinase genes upregulated by both ET treatment and Bgt infection.
Table S5 Presence of putative MYB46 recognition sites in the 2‐kb promoter region of the five genes related to cellulose or isoprenoid synthesis.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


