SUMMARY
Although the vinegar fly Drosophila melanogaster is arguably the most studied organism on the planet, fundamental aspects of this species’ natural ecology have remained enigmatic [1]. We have here investigated a wild population of D. melanogaster from a mopane forest in Zimbabwe. We find that these flies are closely associated with marula fruit (Sclerocarya birrea), and propose that this seasonally abundant and predominantly Southern African fruit is a key ancestral host of D. melanogaster. Moreover, when fruiting, marula is nearly exclusively used, suggesting that these forest-dwelling D. melanogaster are seasonal specialists, in a similar manner to e.g. Drosophila erecta on screw pine cones [2]. We further demonstrate that the main chemicals released by marula activate odorant receptors that mediate species-specific host choice (Or22a)[3, 4] and oviposition site selection (Or19a)[5]. The Or22a expressing neurons -– ab3A – respond strongly to the marula ester ethyl isovalerate: a volatile rarely encountered in high amounts in other fruit. We also show that Or22a differs among African populations sampled from a wide range of habitats, in line with a function associated with host fruit usage. Flies from Southern Africa, of which most carry a distinct allele at the Or22a/ Or22b locus, have ab3A neurons that are more sensitive to ethyl isovalerate than e.g. European flies. Finally, we discuss the possibility that marula, which is also a culturally and nutritionally important resource to humans, may have helped the transition to commensalism in D. melanogaster.
Graphical Abstract
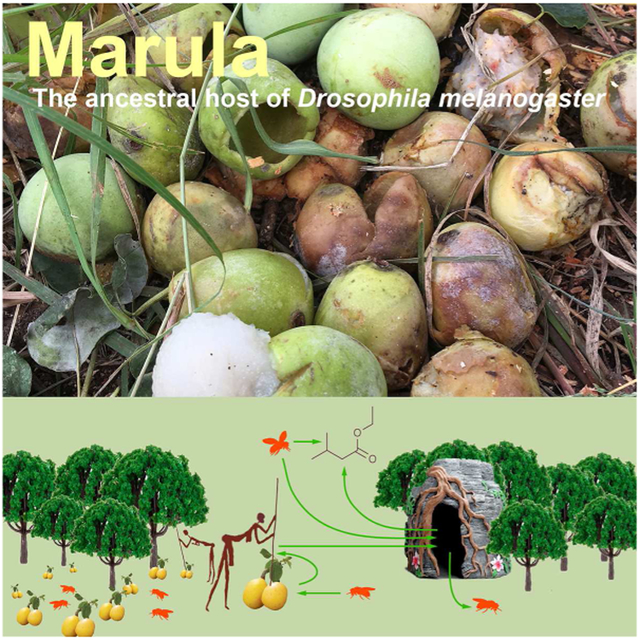
eTOC Blurb
The origin and biology of wild Drosophila melanogaster has remained enigmatic. Mansourian et al. report here wild flies from a mopane forest in Zimbabwe and show that these flies are seasonally associated with marula. Marula is an important human resource, and may have facilitated human commensalism in D. melanogaster.
RESULTS AND DISCUSSION
Marula – candidate ancestral host of Drosophila melanogaster
The vinegar fly Drosophila melanogaster displays preference towards certain fruit and strongly favors citrus for egg-laying [5]. The presence of a distinct host partiality is intriguing, and implies that D. melanogaster during its evolutionary history likely has had a close association with a specific fruit, or group of fruit, with characteristics akin to citrus. This ancestral host is, however, likely not found among members of the Asian genus Citrus, but rather among fruit found within the Miombo and Mopane forests of the fly’s predicted Urheimat in Southern Africa, more precisely present day Zimbabwe and Zambia [6](Figure 1A).
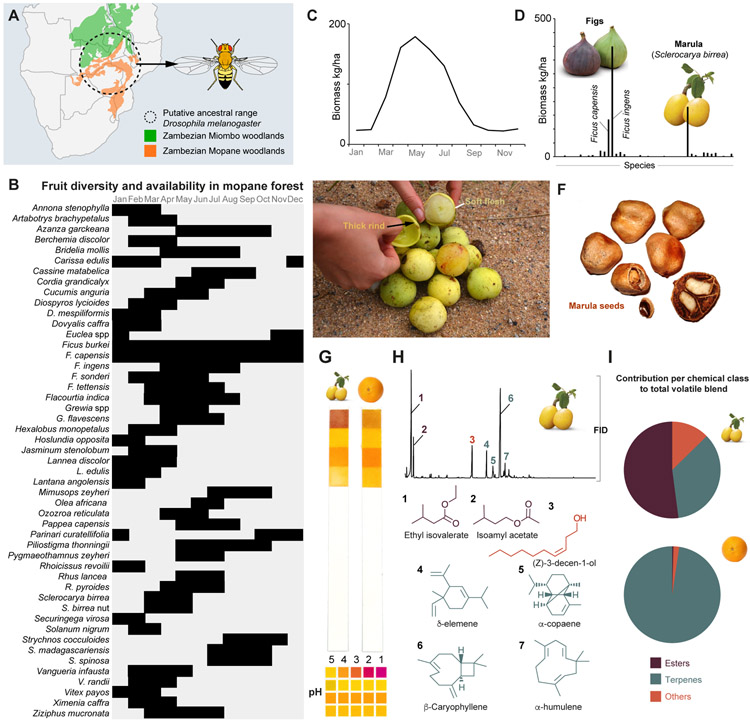
Figure 1. Fruit diversity in a Mopane forests and marula characteristics.
(A) Predicted ancestral range of D. melanogaster and the dominant vegetation zones.
(B) Diversity and availability of fruit in a Mopane forest, (C) Total fruit biomass per month, and (D) yearly biomass per fruit variety (as listed in (B)). Data from the Matopos national park and adapted from [7].
(E) Ripe marula (Sclerocarya birrea) fruit (photo: E. Jirle), and (F) marula seeds.
(G) pH test sticks exposed to marula (left) and orange (right) fruit pulp, with scale below (pH1-5).
(H) Flame Ionization Detection (FID) traces from a headspace collection of marula volatiles. Numbers refer to the primary volatile constituents, structures of which shown below. Color code as per panel (I).
(I) Contribution (%) per chemical class to the total volatile blend of marula (above) and orange (below).
(See also Figure S1)
The Miombo and Mopane forests carry an impressive diversity of fruit-bearing plants [7](Figure 1B, C). Based on what we know of D. melanogaster’s physiology and preference we can, however, deduce some of this hypothetical ancestral host’s characteristics (Figure S1), and thereby narrow down the list of likely candidates. In this context, marula stands out. This fruit is extremely abundant, only matched in terms of biomass by figs (Ficus spp.)[7](Figure 1D), and displays physical and chemical properties that fit with the known preference of D. melanogaster. Briefly, marula has a thick rind similar to citrus, which encloses a sugary (and highly fermentable) juicy pulp (Figure 1E), with a pH similar to orange (Figure 1G), features all favored by D. melanogaster (Figure S1). Marula emit terpenes and esters, which in terms of total emission contribution, as well as in numbers, are the primary chemical components, as determined via gas chromatography-mass spectroscopy analysis of headspace collections (Figure 1H, I). The two main chemicals, ethyl isovalerate (an ester) and ß-caryophyllene (a sesquiterpene), together make up ~55% of the headspace. Both terpenes and esters are known to be important and ecologically relevant olfactory cues for D. melanogaster [5, 8]. In short, marula fulfills the criteria on essentially all counts, and is accordingly a good candidate ancestral host.
Wild D. melanogaster in the ancestral habitat utilize marula
Do flies from the native habitats then use marula? To answer this question, we mounted an expedition to Southern Africa in search of forest-dwelling D. melanogaster and marula. Specifically, we searched mopane woodlands of the Matopos national park in Southwestern Zimbabwe (Figure 2A); a site situated within the predicted ancestral range [6]. The Matopos covers 424 km2, hosts no permanent human habitation and is covered in Mopane and kopje woodlands (Figure 2B).
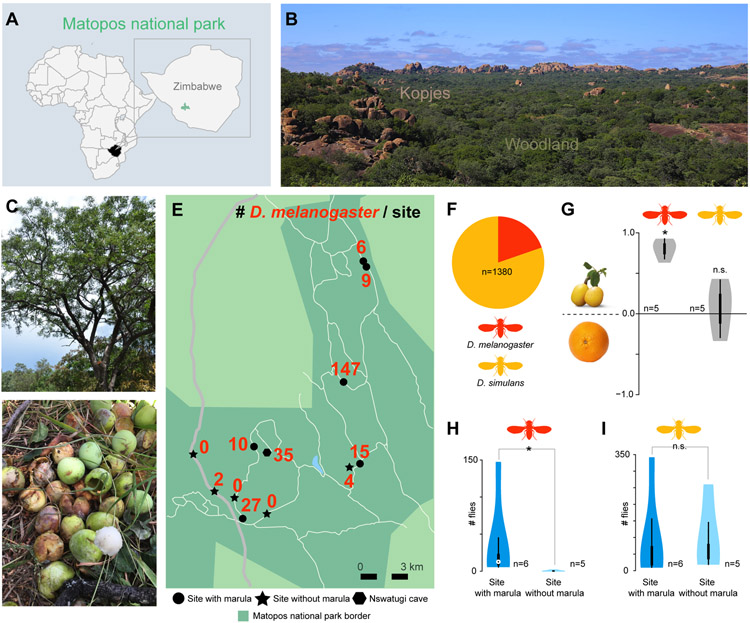
Figure 2. Wild D. melanogaster from mopane woodlands are closely associated with marula.
(A) Location of the Matopos national park, Zimbabwe.
(B) View of the park, showing extensive mopane woodland cover with interspersed kopje rock formations. Photo: M. Stensmyr
(C) A marula (Sclerocarya birrea) tree and (D) fermenting fruit. Photo: E. Jirle and M. Stensmyr
(E) The Matopos national park and the collection sites with total numbers of specimens of Drosophila melanogaster caught.
(F) Proportion of D. melanogaster to Drosophila simulans from all collection sites.
(G) Violin plots showing OI of wild D. melanogaster and D. simulans (color code as per (F)) provided a choice between traps baited with marula or orange. White circle show the median, box the 25th-75th percentiles, extended by whiskers indicating 1.5x the interquartile range from the 25th-75th percentiles; shape denotes density estimate and extend to extreme values. Deviation of the OI against zero was analyzed for significance (* p < 0.05) with a one-sample Wilcoxon test (p < 0.05).
(H) Violin plots showing the number of D. melanogaster and D. simulans (I) caught at sites with or without marula. Violin plots as per (G). Difference between the means was analyzed for significance (*) with a Mann-Whitney Utest (p < 0.05).
Once in the Matopos we localized marula trees (Figure 2C), as well as fruiting trees with fermenting fruit below (Figure 2D), among which we placed fly-traps – baited with marula. Over the next days, these traps caught numerous D. melanogaster (n=147 from this single site). Traps placed under an additional 5 marula trees yielded another 67 D. melanogaster specimens (Figure 2E). At all examined sites though, D. simulans outnumbered D. melanogaster (Figure 2F). We forthwith refer to these flies as “wild”, in line with their presence in undisturbed wilderness, with the caveat that their ultimate origin remains unknown. For more information about these flies, and other D. melanogaster specimens caught from wilderness areas in Southern Africa, we refer interested readers to an accompanying paper [9].
We next provided the forest flies with a choice of marula versus orange, the favorite breeding substrate of domestic D. melanogaster [5]. We placed paired traps, containing either marula or orange, under a fruiting marula tree. Similar to the laboratory strain, the wild D. melanogaster showed a strong preference for marula (Figure 2G). Interestingly though, D. simulans displayed no such preference (Figure 2G), indicating that the marula preference is exclusive to D. melanogaster and, moreover, that marula is not simply overall a more suitable fruit resource to Drosophila spp. We next dissected marula in search of fly eggs and larvae, and in all fruit examined we localized drosophilid larvae, from which D. melanogaster adults later emerged. In short, wild African D. melanogaster are drawn to the odor of marula, prefer marula to orange, and use marula as breeding substrate.
Wild D. melanogaster are seasonal specialists on marula
To investigate the general distribution of D. melanogaster in the Matopos, we next placed traps (baited with fermenting marula) at locations (n=5) with no fruiting marula trees nearby, but with otherwise similar vegetation (including other fruiting trees). Strikingly, D. melanogaster was absent, or very sparse in traps at these locations (Figure 2H). On the other hand, D. simulans was as abundant at sites with marula as without (Figure 2I). The distribution pattern of D. melanogaster in the Matopos hence indicates niche confinement, and in turn a specialized lifestyle. D. melanogaster as a seasonal fruit specialist would actually not be surprising given i) the scarcity of the species in Miombo and Mopane forests outside of marula season [9], ii) the observed presence of a distinct egg-laying preference [5], and iii) the fact that host specialization is a prevalent feature in the melanogaster subgroup. Drosophila sechellia exclusively breeds in noni fruit [10], whereas Drosophila erecta and Drosophila orena are seasonal specialists on Pandanus cones [2] and Syzygium waterberrys [11] respectively. Drosophila teissieri is closely associated with Parinari fruit, which limits its geographic range [2, 11, 12], whereas Drosophila santomea is found with figs from Ficus clamydocarpa trees [13]. Thus, seasonal host specialization in D. melanogaster would fall into the pattern displayed by most (if not all) of its close relatives. Out of marula season, these forest flies may go into diapause, much like they do in temperate regions [14], or switch to opportunism, utilizing alternate breeding substrates. One such alternative could be figs, which are present year round in the Matopos (Figure 1B) and in terms of biomass even more abundant than marula (Figure 1D). D. melanogaster has moreover been reared from figs in Africa [15], which are also an alternate host for the seasonal specialist D. erecta outside of Pandanus season [16].
Laboratory D. melanogaster shows oviposition preference for marula and marula volatiles
Wild African D. melanogaster hence not only utilize marula – for parts of the year, marula appears to be exclusively utilized. We next wondered how domestic flies react to this fruit. To this end, we used a two-choice assay [17] to examine egg-laying preference in Canton-S wild type flies. The Canton-S(pecial) strain was established sometime before 1916 from a population in Canton, Ohio [18], well outside the sub-Saharan range of marula. We first verified the citrus preference of these flies in the oviposition assay. Given a choice between oranges and banana, the flies clearly preferred citrus as oviposition substrate (Figure 3A). Having confirmed the assay, we subsequently tested orange versus marula, and indeed, flies provided this choice strongly preferred marula, similar to Wild African D. melanogaster (Figure 3B). The ancestral marula preference is accordingly conserved in non-African flies.
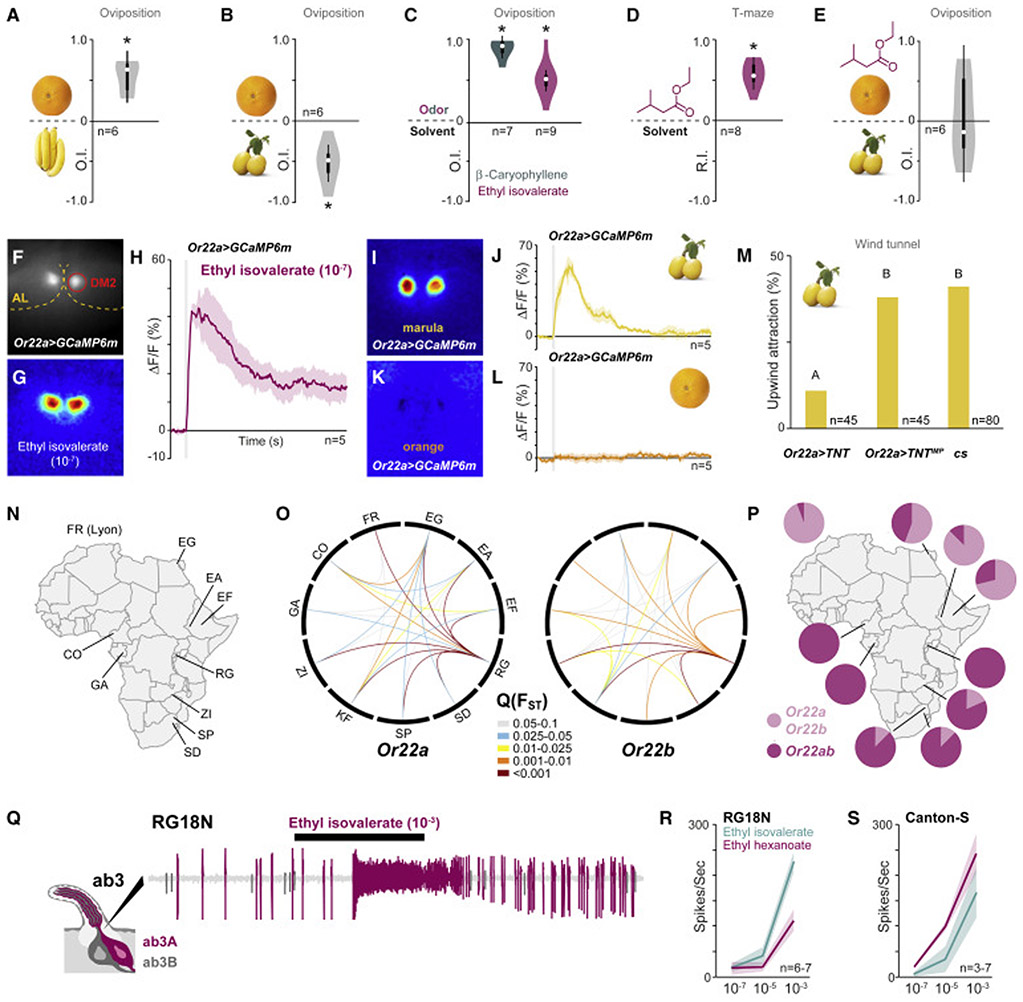
Figure 3. Wild D. melanogaster from mopane woodlands are closely associated with marula.
(A) Violin plots showing oviposition indices (OI) of Canton-S flies from a binary-choice test between standard cornmeal fly food mixed with orange or banana pulp. Violin plots as per Figure 2G. Deviation of the OI against zero was analyzed for significance (*) with a one-sample Wilcoxon test (p < 0.05).
(B) Violin plots showing OI of Canton-S flies from a binary-choice test between standard cornmeal fly food mixed with orange or marula pulp, or (C) fly food mixed with ß-caryophyllene or ethyl isovalerate against fly food alone. Violin plots as per Figure 2G. Deviation of the OI against zero was analyzed for significance (*) with a one-sample Wilcoxon test (p < 0.05).
(D) Violin plots showing the Response Index (RI) of canton-S flies towards ethyl isovalerate (10−4) in a mini T-maze (depicted left). Violin plots as per Figure 2G. Deviation of the RI against zero was analyzed for significance (*) with a one-sample Wilcoxon test (p < 0.05).
(E) Violin plots showing OI of canton-S flies from a binary-choice test between standard cornmeal fly food mixed with orange and ethyl isovalerate against fly food with marula. Violin plots as per Figure 2G. Deviation of the OI against zero was analyzed for significance (*) with a one-sample Wilcoxon test (p < 0.05).
(F) Prestimulation view of Or22a-Gal4>UAS-GCaMP6m showing intrinsic fluorescence from the DM2 glomerulus.
(G) Pseudocolored image showing ethyl isovalerate-induced fluorescence changes in the antennal lobe (AL) of a Or22a-Gal4>UAS-GCaMP6m fly.
(H) Averaged traces from DM2 glomerulus of Or22a-Gal4>UAS-GCaMP6m flies stimulated with ethyl isovalerate. Shaded areas represent SEM. Gray bar represents stimulus duration (1 s).
(I) Pseudocolored image showing marula- and orange- (J) induced fluorescence changes in the AL of Or22a- Gal4>UAS-GCaMP6m flies.
(K) Averaged traces from DM2 glomerulus of Or22a-Gal4>UAS-GCaMP6m flies stimulated with marula and orange (L). Shaded areas represent SEM. Gray bar represents stimulus duration (0.5 s).
(M) Marula odor-mediated upwind flight attraction of Or22a-Gal4>UAS-TNT flies in comparison to Or22a-Gal4>UAS-TNTMP and Canton-S (cs). Bars labeled with different letters indicate significant difference as analyzed by a binomial generalized linear model (GLM) followed by Tukey’s test (p < 0.05).
(N) Geographic origin of examined D. melanogaster populations. Abbreviations as per [5].
(O) Genetic differentiation among populations at Or22a and Or22b is depicted via Circos plots [39] based on FST quantiles (Q(FST)). Only connections between populations with unusually high FST values (elevated genetic differentiation) are shown. The red color, for example, indicates that between this pair of populations, less than 0.1% of windows on the same chromosome arm have an FST value this high.
(P) Frequency of the Or22ab allele across the examined D. melanogaster populations.
(Q) Representative single sensillum recording trace from an ab3 sensillum. The larger-amplitude spiking neuron, i.e. ab3A, responds to ethyl isovalerate. The duration of stimulus delivery (0.5 s) is marked by the black bar.
(R) Dose-response curve of ab3A neurons from the RG18N strain and Canton-S (S) toward ethyl isovalerate and ethyl hexanoate. Shaded area shows standard deviation.
Which chemicals then mediate the marula preference? We used the same two-choice assay and next tested the major chemical components of the headspace individually. We have previously shown that fly food spiked with terpenes confer positive egg-laying site selection [5], thus we only re-tested the main terpene (ß-caryophyllene), which as expected generated preferential oviposition (Figure 3C). The main ester component, ethyl isovalerate, also conferred oviposition preference (Figure 3C), as well as attraction in a T-maze assay (Figure 3D). The preference of marula over orange may hence be mediated by the high presence of esters in the former. In line with this reasoning, flies provided a choice of orange spiked with ethyl isovalerate against marula failed to make a choice (Figure 3E).
The marula volatile ethyl isovalerate activates Or22a expressing neurons
In D. sechellia and D. erecta, host specialization is linked to the Or22a circuit, which in both species is activated by distinct esters from the respective hosts [3, 4]. We thus wondered if the primary marula ester ethyl isovalerate also activates Or22a-expressing olfactory sensory neurons (OSNs) in D. melanogaster. To investigate this issue, we performed functional imaging of the antennal lobe from flies expressing the calcium reporter GCaMP6m [19] under the control of Or22a-Gal4 [20](Figure 3F). Stimulation with ethyl isovalerate yielded strong calcium signals in the DM2 glomerulus (the target of the Or22a-expressing OSNs [20, 21] already at 10−7 dilution (Figure 3G, H). In line with the chemistry, marula odor also triggered strong Ca2+ signals from DM2 (Figure 3I, J), whereas orange odor triggered weak to no activity from the same glomerulus (Figure 3K, L). Thus, similar to its specialized siblings, the main ester from the preferred host activates Or22a. Silencing of the Or22a pathway via Or22a-Gal4>UAS-TNT did not, however, abolish the marula oviposition preference (data not shown), suggesting that additional pathways are involved in this behavior. Rather than mediating egg-laying preference, the primary function of Or22a may instead be to locate the host over distance. Hence, we next examined up-wind flight navigation towards marula of flies with Or22a silenced (via Or22a-Gal4>UAS-TNT) in a wind tunnel assay [22]. Flies with non-functional Or22a input showed a reduced ability to localize marula compared to control flies (Figure 3M), suggesting that these neurons predominant function is to assist the fly in locating its host over distance. The importance of these neurons in this context is also evident from D. sechellia, which has a numerical increase of Or22a-expressing OSNs which likely affords an improved ability to find noni over distance [3].
Or22a shows signs of local adaptation
Since marula is restricted to sub Saharan Africa, most D. melanogaster have to make do on alternative hosts. If Or22a indeed is linked to the specific chemistry of the host, we would accordingly expect to see local adaptation of the Or22a locus between D. melanogaster populations from diverse environments that may utilize disparate hosts. Thus, we next estimated local genetic differentiation (as indexed by FST [23, 24]) within the OR family between genomes from 10 African populations, plus one European (Figure 3N). For each window centered on an olfactory receptor gene, we then evaluated the FST quantile for each pairwise population comparison (the proportion of all windows on the same chromosome arm that showed stronger allele frequency differences (higher FST) between these same two populations (Figure 3O). The Or22a locus, and the adjacent tandem paralog Or22b, shows striking genetic differentiation between almost all population pairs (Figure 3O), in stark contrast to most of the other ORs, for which little or no sign of local adaptation can be discerned (Figure S2).
In cases where other ORs did show strong FST outliers (quantiles <0.0001), often differentiation in one or a few populations was most apparent. These genes included Or33a, Or65b, and Or67a (Figure S2). Interestingly, these receptors also appear to have important functions. Or33a has unknown function [25], but like Or22a, it shows variable expression across species [26]. Or67a detects aromatic esters (e.g. methyl benzoate)[27] and has undergone serial duplication in Drosophila suzukii and Drosophila biarmipes [28]. Or65b is expressed in pheromone sensing neurons [29] but its function has not been established. In short, unlike most members of the OR family in D. melanogaster, Or22a (and its closely linked paralogs Or22b) shows strong signs of local adaptation, in line with a function associated with host specific chemistry.
At the molecular level, Or22a (and Or22b) thus differs between populations, but does this local differentiation also translate into functional changes in the ab3A neurons where these genes are expressed [20, 21]? The most conspicuous alteration among the investigated populations in the Or22a/ Or22b locus is a deletion allele, whereby a segment stretching from the second exon of Or22a to the start of the second exon of Or22b has been deleted, generating a chimeric receptor, Or22ab (Figure S3A)[30]. In light of the chimeric appearance of Or22ab, this variant appears to be a derived deletion (following a more ancient duplication to create these paralogs), rather than representing the ancestral state of the Or22locus [30].
Our data support the prior suggestion [30] that the Or22ab fusion variant is quite ancient. This variant is at very high frequency within the ancestral range (e.g. 88% in Zambia). Nucleotide diversity of flanking sequences, which should accrue on the order of 4Ne ≈ 10 million generations in this species, is at or above typical levels among Zambia haplotypes carrying this deletion (Figure S3B). Hence, it is likely that the fusion variant existed well before the species expanded beyond its ancestral range on the order of 150,000 generations ago, or ~10,000 years ago [9, 31]. In contrast, putatively ancestral full-length Or22a/ Or22b haplotypes from Zambia show strongly reduced diversity across the deletion region (Figure S3B). This pattern could reflect a low long-term population size of the full-length allele, in accordance with its current rarity in the ancestral range. In some populations, such as in Europe or the Ethiopian highlands, the full-length allele has become predominant (Figure 3P). Many of these haplotypes show identical or nearly identical sequences (Figure S3C, D), in line with prior evidence for positive selection linked to the Or22a/ Or22b haplotype in Europe [30]. We note that some populations with similarly high frequencies of the fusion variant are strongly differentiated from each other at the Or22a/b locus (Figure 3O), which could imply either parallel increases of the fusion variant on distinct haplotypes, or additional variants under spatially varying selection at this locus.
Consequently, most D. melanogaster in Southern Africa will likely carry the Or22ab allele, which prompts the question: do their ab3A neurons respond to the marula ester? We selected a strain in which Or22ab is fixed (RG18N) and subsequently performed single sensillum recordings (SSR). Measurements from ab3A neurons revealed strong responses to stimulation with ethyl isovalerate (Figure 3Q). The ab3A neurons in RG18N actually responded more strongly to ethyl isovalerate than to ethyl hexanoate – the primary ligand of Or22a [27](Figure 3R), in contrast to ab3A neurons from Canton-S flies (which carry both Or22a and Or22b [32]) where ethyl hexanoate yielded a stronger response than ethyl isovalerate (Figure 3S). In short, African D. melanogaster not only detect ethyl isovalerate, they are even more sensitive to this marula compound than flies from outside Africa. We note that the distribution of populations with a high frequency of Or22ab overlaps with the distribution of marula. However, whether or not the Or22ab allele is an adaptation towards marula remains to be shown. Heterologous expression and detailed functional characterization of this interesting receptor variant will be a topic for future studies.
Marula as a vehicle for the domestication of D. melanogaster
The Matopos is best known for its elaborately painted caves (Figure 4A) – made by now-vanished San tribes during Late Pleistocene to Early Holocene [7]. For these tribes, marula played a pivotal role and archeological excavations of their cave homes have uncovered enormous quantities of marula stones [7, 33](Figure 1F). From the Pomongwe cave alone, remains of at least 24 million marula stones were recovered, which only represents the carbonized remains, and hence but a fraction of the marula that must have once been brought into this cave [33]. The San evidently spent considerable time collecting and processing marula, which would have been the staple food item during many months of the year. Thus, just like D. melanogaster, these San tribes appear to have been seasonal specialists on marula as well.
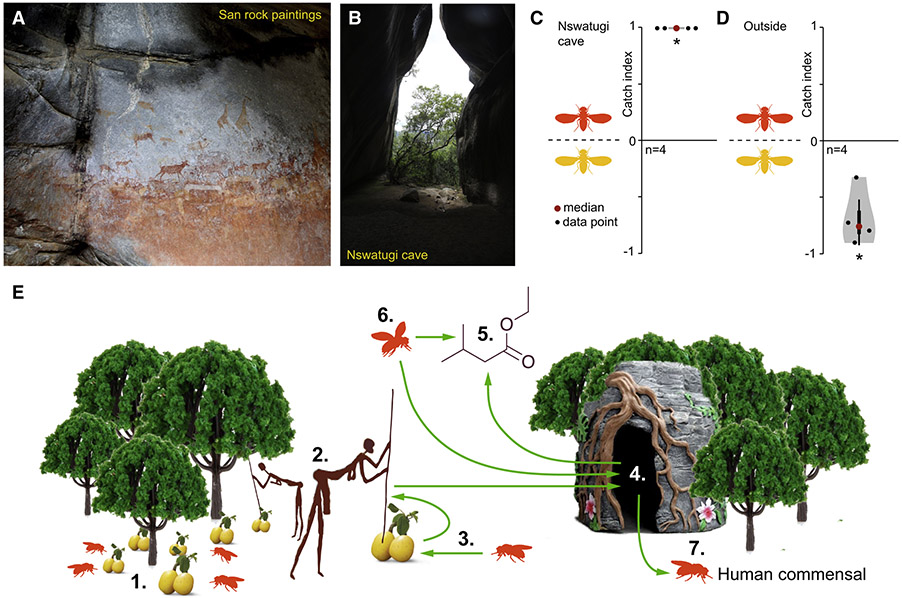
Figure 4. Marula and the domestication of D. melanogaster.
(A) San rock paintings from the Nswatugi cave depicting the local wildlife. Photo: M. Stensmyr.
(B) View from inside the Nswatugi cave. Photo: M. Stensmyr.
(C) Violin plots showing proportion of wild D. melanogaster and D. simulans caught in traps placed inside Nswatugi cave, and outside (D). Violin plots as per Figure 2B. Deviation of the OI against zero was analyzed for significance (*) with a one-sample Wilcoxon test (p < 0.05).
(E) Schematic model of the process leading up to commensalism. (1) Wild D. melanogaster on fallen marula; a resource of equal importance to now extinct San hunter-gatherers (2)(adapted from [33]) that co-inhabited the same habitat. The San brought large numbers of marula into their cave homes, and with the fruit likely also flies (3). The massive amounts of marula that evidently were stored in these caves (4) would have generated a potent scent trail, dominated by ethyl isovalerate (5), attracting flies (6). Inside the caves, flies would have adapted to their new environment and preference of their close neighbors — ultimately leaving as human commensals (7).
The marula-San link offers a plausible scenario by which D. melanogaster became a human commensal. The smell of the marula stores emanating from the caves would have attracted flies from far and wide. Flies would inside the caves have found a steady supply of marula, and fermenting leftovers – long after the fruit’s presence in the surrounding woodlands. In other words, the time frame for using the optimal breeding substrate would have been increased considerably. Inside the caves, the flies would also have benefitted from a reduced risk of predation as well as protection from adverse weather conditions. Over time, the cave flies would have accumulated adaptations helpful for human commensalism. Relevant traits may have included a willingness to enter darker enclosures [34] and an increased tolerance of ethanol, both of which differentiate D. melanogaster from its closest relatives [35]. Thus, we next wondered if D. melanogaster actually enter these caves. To this end, we placed traps (n=4), baited with fermenting marula along the far wall of the Nswatugi cave [see 7 for a description](Figure 4B). Over three days these traps caught a number of D. melanogaster specimens (n=35), but no D. simulans (Figure 4C), in contrast to the closest traps (n=3) placed under fruiting marula trees outside the cave, where D. simulans greatly outnumbered D. melanogaster (Figure 4D).
The archeological record indicates that systematic and intensive marula use begun ~12,000 years ago. At ~9,500 years ago, marula harvesting reached massive proportions, to finally ebb out at ~8,000 years ago [7]. These dates coincides with demographic data from D. melanogaster, which points to a within Africa expansion starting at ~10,000 years ago [9, 31] – an expansion presumably representing the dispersal of the commensal population throughout its new niche. In short, archeological and demographic data would support the notion that marula use by the San may have been a factor in turning the woodland species D. melanogaster into the cosmopolitan species of today (Figure 4E).
Conclusion
We have here demonstrated that D. melanogaster from a mopane forest within the predicted ancestral range are seasonal specialists on marula fruit. The odor of this seasonally abundant, and widely distributed fruit activates select key odorant receptors, previously implicated as having particular importance to D. melanogaster, and we argue that marula is the ancestral primary host of the fly. We moreover show that flies from sub Saharan Africa carry a specific allele of one of these odorant receptors, and are also more responsive to a key marula chemical. Finally we speculate that the marula specialization might have been important in driving commensalism.
The finding of a woodland population of D. melanogaster within the ancestral habitat opens up for a range of interesting questions to be addressed. For example, how do these flies differ from commensal relatives, i.e. which genetic factors underlie this shift in lifestyle? The finding that D. melanogaster appears to have a close association with a single host fruit will furthermore facilitate studies relating to host specific chemosensory adaptations, which so far have had to be conducted in other insects in which the wealth of tools available in D. melanogaster are unavailable.
STAR★ Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marcus Stensmyr (marcus.stensmyr@bio.lu.se)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly collections and husbandry
Field traps were made from standard 0.51 PET water bottles (purchased at a supermarket in Bulawayo, Zimbabwe) with a horizontal slit cut to allow flies to enter. The traps were baited with marula (or with oranges for certain experiments). The traps were placed at ground level in the vegetation. Flies were aspirated from the bottles, frozen and then transferred to 90% ethanol for later identification (using morphological characters). Laboratory strains of D. melanogaster were reared on standard yeast corn meal medium and kept at 23 °C under a 12 h/12 h light cycle. The following strains were used; Canton-S (gift from Dr Stefan Baumgartner), RG18N (Pool lab), Or22a-Gal4 (Sachse lab), UAS-TeTxLC.tnt.E2 (BDSC 28837), UAS-TeTxLC.(−)V.A2 (BDSC 28840), and 20XUAS-IVS-GCaMP6m (BDSC 42748 and 42750).
METHOD DETAILS
Odor analysis and GC-MS
Fruit collected in the field were enclosed in cooking bags (Matlagningspåse M, Toppits) and volatiles evacuated through custom made Tenax (GR 60/80, Grace Davison Discovery Science) filters for 2-3 h via modified aquarium pumps (of unknown original make) drawing air at 0.5 l min−1. The filters were subsequently flushed with heptane (Sigma). Eluates were then injected into an Agilent 7890A gas chromatograph equipped with a 5975C Network Mass Selective Detector (Agilent Technologies), fitted with a HP-5MS column (30 m, 0.25 mm, 0.25 μm). Helium was used as carrier gas at a constant flow of 1 ml min−1. The oven temperature was set at 60 °C for 1 min, which was followed by a heating gradient of 10 °C min−1 to 230 °C, and then held for 10 min. Chromatograms were analyzed using ChemStation (Agilent Technologies), with compounds tentatively identified by comparison to reference spectra in the NIST library and finally verified using synthetic standards of highest purity available (Sigma).
Electrophysiology
For single sensillum recordings, flies were first aspirated, then inserted and immobilized in pipette tips (200 μl, VWR). Recordings were performed using electrolytically sharpened (KNO2) tungsten microelectrodes (TW5-3, Harvard Apparatus). The recording electrode was positioned through a DC- 3K/PM10 piezo driven micromanipulator (Märzhäuser), whereas the reference electrode was inserted into the eye using a manually controlled micromanipulator (MM-3, Narishige). Odors were delivered via a Syntech CS-55 stimulus controller into a humidified air stream (1 l min-1) via cartridges made from Pasteur pipettes (VWR) containing a small piece of filter paper (0.5 cm x 0.5 cm, Grade: 1002, Munktell) soaked with 10 μl of the stimulus solution, or solvent only. Stimulus duration was set to 0.5 sec. Recordings were digitally converted via a IDAC 4 acquisition controller (Syntech) and stored on a PC (Custom configured) and analyzed (i.e. spikes sorted and counted) using the AutoSpike software (Syntech).
In vivo calcium imaging
For imaging, flies were cold anesthetized and immobilized with paraffin wax on a custom made stage, the dorsal side of the head was then covered with artificial haemolymph solution and a small window opened in the cuticle to expose the brain. A pE-300 CoolLED (Nikon instruments) was used as light source (488 nm excitation, dichroic 500 nm, long-pass filter 515 nm) and emitted light captured with a Andor Zyla sCMOS camera (Andor Technology) fitted onto a Nikon Eclipse FN1 microscope (Nikon Instruments) equipped with a NIR Apo 40x 0.8 NA objective (Nikon Instruments). Data was acquired at 256 x 256 pixels at a rate of 4 Hz using the NIS elements software (Nikon Instruments). To deliver odor stimulus, a Syntech CS-55 stimulus controller (Syntech) was used to switch a charcoal-filtered airstream (1 l min−1) between a 4 ml vial (VWR) containing the solvent and a 4 ml vial containing the stimulus. In control experiment the air stream was switched between two vials. Analysis of fluorescence intensity dynamics was performed in Fiji [36, 37] using the measure stack function.
Behavioral experiments
For the egg-laying assay, 15-20 newly mated females were introduced to two-choice petri dishes. Two- choice dishes were made by dividing a 47 mm Petri dish (VWR) into two halves. One half as a treatment; odorant (150 μl of a 10−2 dilution) mixed with standard cornmeal fly food and the other half as control; fly food mixed with solvent. Experiments with fruit were performed in a similar manner. About 5g of ripe fruit, either marula or orange (chopped and briefly run through a kitchen blender), was mixed with fly food. Two-choice dishes were covered by a 6-oz fly stock bottle (VWR). After 24 hours, the number of eggs in each side was counted and an oviposition index (OI) calculated (OI = (Number of eggs in treatment – Number eggs in control)/ Total number of eggs). T- maze experiments were carried out in an assay constructed from two transparent plastic 4 ml screw-cap vials (VWR) connected by a T-shaped tubing connector (VWR). Stimulus, as well as solvent control was pipetted (10 μl onto filter papers (0.5 cm x 0.5 cm, Grade: 1002, Munktell) and placed in respective chambers. 10 lightly cold anesthetized female flies were inserted into the assay through the T-connection. After allowing one minute for the flies to recover, the number of flies in respective chamber was recorded after three minutes. For the wind tunnel experiments, female flies (4-7 days old and mated) were starved 24 hours prior testing. Individual flies were released at the down-wind end of a wind tunnel (30x30x100 cm, made out of glass, and diffusely lit from above)[22] and exposed to a charcoal filtered air stream (0.15 m s−1) carrying a plume of marula odor released upwind. For odor delivery, marula fruit was kept inside a 250-ml glass jar (VWR) with a 38 mm wide opening that was covered with a metal mesh (mesh size 2 mm). The side of the jar was covered with aluminum foil to prevent visual fruit signals stimulating the flies. Charcoal filtered air (0.5 l min−1) was injected into the jar through a Pasteur pipette placed vertically above the mesh-covered opening. The air stream containing the fruit volatiles emanated as a wide plume from the opening of the jar into the center at the upwind end of the tunnel. Flies were recorded for upwind flight and landing at the odor source (i.e. on top of the metal mesh or the tip of the pipette) during 4 minutes.
Population genetic analysis
Genetic variation at olfactory receptor genes was analyzed based on previously-sequenced, town-collected population samples [38] and newly-sequenced genomes from Kafue National Park [9]. Sub-Saharan population samples with at least 10 sequenced genomes were included, in addition to population samples from Egypt and France. FST [23; 24] between each pair of analyzed populations was calculated for all genomic windows on euchromatic chromosome arms, where windows were scaled by their genetic diversity content to contain 250 non-singleton SNPs in the Zambia-Siavonga population sample. FST was also evaluated for a similarly-defined window centered on the transcription start site of each olfactory receptor gene. For each population pair, an olfactory receptor gene’s FST quantile was evaluated as the proportion of windows on the same chromosome arm for which this population pair showed a greater FST value than the focal gene’s window. Circos [39] was then applied to visualize population pairs showing low FST quantiles for each gene. Genomes carrying fusion (deletion) variants at the Or22 locus were readily detected based on a bimodal distribution of the number of sites with missing data within the known deletion region.
QUANTIFICATION AND STATISTICAL ANALYSIS
Values are shown as violin plots; white circle show the median, box the 25th-75th percentiles, extended by whiskers indicating 1.5x the interquartile range from the 25th-75th percentiles; shape denotes density estimate and extend to extreme values, as stated for each graph in the figure legends. All statistics were performed using R https://cran.r-project.org/). Statistical details related to sample size and p values are reported in the figure legends, with a star denoting p < 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Marula (Sclerocarya birrea) | Matopos forest | N/A |
| Orange (Citrus X sinensis) | Ica Supermarket, Lund | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ethyl isovalerate (CAS# 108-64-5) | Sigma-Aldrich | Cat#112283 |
| ß-caryophyllene (CAS# 87-44-5) | Sigma-Aldrich | Cat#22075 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster Or22a-Gal4 | Bloomington Drosophila Stock Center | BDSC:9951 and BDSC:9952 |
| D. melanogaster 20XUAS-IVS- GCaMP6m | Bloomington Drosophila Stock Center | BDSC: 42748 and BDSC:42750 |
| D. melanogaster UAS-TeTxLC.tnt.E2 | Bloomington Drosophila Stock Center | BDSC: 28837 |
| D. melanogaster UAS-TeTxLC.(−)V.A2 | Bloomington Drosophila Stock Center | BDSC 28840 |
| D. melanogaster Canton-S(pecial) | Baumgartner lab, Lund university | N/A |
| D. melanogaster RG18N | Pool lab, University of Wisconsin-Madison | N/A |
| D. melanogaster Matopos wt | Matopos forest | N/A |
| D. simulans Matopos wt | Matopos forest | N/A |
| Software and Algorithms | ||
| Fiji | [36, 37] | Fiji.sc |
| AutoSpike | Syntech | www.ockenfels-syntech.com/download-2/ |
| Circos | [39] | www.circos.ca |
| Illustrator CC 21.02 | Adobe | www.adobe.com |
| Photoshop CC | Adobe | www.adobe.com |
| R | R core team 2013 | cran.r-project.org |
| GC/MSD ChemStation | Agilent | www.agilent.com/en/products/software-informatics/massspec-workstations/gc-msd-chemstation-software |
| NIS elements | Nikon | www.nikoninstruments.com/Products/Software/ |
Highlights.
Wild African D. melanogaster are seasonally associated with marula fruit
Marula is the likely ancestral host of D. melanogaster
Marula odor activates a key odorant receptor that shows signs of regional adaptation
Marula use may have driven the switch to human commensalism
ACKNOWLEDGEMENT
We thank Drs Matthew Cobb, Lucia Prieto-Godino, and Daniel R. Matute for valuable comments, and Dr Nicholas J. Walker for kindly providing information regarding the San, marula, and the Matopos. We also wish to thank Dr Helena Fritz, Latifa Mrisho, and Lovisa Pettersson for assistance. Work in the Stensmyr lab is funded by the Crafoord Foundation, Carl-Tryggers foundation, and the Swedish Research council. The Pool lab acknowledges funding from the NIH (R01 GM111797).
Footnotes
DECLARATION OF INTERESTS
All authors declare no financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lachaise D, and Silvain JF (2004). How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster – D. simulans palaeogeographic riddle. Genetica 120, 17 – 39. [DOI] [PubMed] [Google Scholar]
- 2.Rio B, Couturier G, Lemeunier F, and Lachaise D (1983). Evolution d’une specialisation saisonniere chez Drosophila erecta (Dipt., Drosophilidae). Ann. Soc. Entomol. France 19, 235 – 248. [Google Scholar]
- 3.Dekker T, Ibba I, Siju KP, Stensmyr MC, and Hansson BS (2006) Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr. Biol 16, 101–109. [DOI] [PubMed] [Google Scholar]
- 4.Linz J, Baschwitz A, Strutz A, Dweck HKM, Sachse S, Hansson BS and Stensmyr MC (2013) Host plant-driven sensory specialization in Drosophila erecta. Proc. Royal Soc. London B Biol. Sci 280, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dweck HKM, Ebrahim SAM, Kromann S, Bown D, Hillbur Y, Sachse S, Hansson BS, and Stensmyr MC (2013). Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol 23, 2472–2480. [DOI] [PubMed] [Google Scholar]
- 6.Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, Duchen P, Emerson JJ, Saelao P, Begun DJ, and Langley CH (2012). Population genomics of sub-saharan Drosophila melanogaster. African diversity and non-African admixture. PLoS Genet. 8, e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker NJ (1995). Late Pleistocene and Holocene Hunter-Gatherers of the Matopos. An Archaeological Study of Change and Continuity in Zimbabwe. Studies in African archaeology, 0284-5040 ; 10 Societas Archaeologica Upsaliensis, Studies in African Archaeology, Uppsala: 284p. [Google Scholar]
- 8.Mansourian S, and Stensmyr MC (2015). The chemical ecology of the fly. Curr opinion neurobiol, 34, 95–102. [DOI] [PubMed] [Google Scholar]
- 9.Sprengelmeyer QD, Mansourian S, Lange JD, Matute DR, Cooper BS Jirle EV, Stensmyr MC and Pool JP (2018). Discovery of Drosophila melanogaster from Wild African Environments and Genomic Insights into Species History. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsacas L, and Bächli G (1981). Drosophila sechellia. n. sp., huitième espèce du sous-groupe melanogaster des iles Seychelles (Diptera, Drosophilidae). Rev. Fr. Entomol 3, 146–150. [Google Scholar]
- 11.Comeault AA, Serrato-Capuchina A, Turissini DA, McLaughlin PJ, David JR, and Matute DR (2017). A nonrandom subset of olfactory genes is associated with host preference in the fruit fly Drosophila orena. Evol Letters. 1, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David JR, Lemeunier F, Tsacas L, and Yassin A (2007). The historical discovery of the nine species in the Drosophila melanogaster species subgroup. Genetics 177, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llopart A, Lachaise D, and Coyne JA (2005). An anomalous hybrid zone in Drosophila. Evolution, 59, 2602–2607. [PubMed] [Google Scholar]
- 14.Schmidt PS, and Conde DR (2006). Environmental heterogeneity and the maintenance of genetic variation for reproductive diapause in Drosophila melanogaster. Evolution 60, 1602–1611. [PubMed] [Google Scholar]
- 15.Lachaise D (1974). Les drosophilidae des savanes préforestières de la région tropicale de Lamto (Côte-d’Ivoire). IV. b. – Synecologie fonctionelle du peuplement de Ficus capensis. Bull. Ecol 7, 79–104. [Google Scholar]
- 16.Lachaise D, and Tsacas L (1974). Les drosophilidae des savanes préforestières de la région tropicale de Lamto (ôte-d’Ivoire). 2. Le peuplement des fruits de Pandanus candelabrum (Pandanacées). Ann. Univ. d’Abidjan 7, 153–192. [Google Scholar]
- 17.Mansourian S, Corcoran J, Enjin A, Löfstedt C, Dacke M, and Stensmyr MC (2016). Fecal-derived phenol induces egg-laying aversion in Drosophila. Curr. Biol 26, 2762–2769. [DOI] [PubMed] [Google Scholar]
- 18.Bridges CB (1916). Non-disjunction as proof of the chromosome theory of heredity (concluded). Genetics, 1, 107–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, and Kim DS (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fishilevich E, and Vosshall LB (2005). Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol 15, 1548–1553. [DOI] [PubMed] [Google Scholar]
- 21.Couto A, Alenius M, and Dickson BJ (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol 15, 1535–1547. [DOI] [PubMed] [Google Scholar]
- 22.Becher PG, Bengtsson M, Hansson BS, and Witzgall P (2010). Flying the fly: long-range flight behavior of Drosophila melanogaster to attractive odors. J. Chem. Ecol 36, 599–607. [DOI] [PubMed] [Google Scholar]
- 23.Wright S (1931). Evolution in Mendelian populations. Genetics 16, 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson RR, Slatkin M, and Maddison WP (1992). Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stensmyr MC, Dweck HKM, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, et al. (2012). A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357. [DOI] [PubMed] [Google Scholar]
- 26.Pan JW, Li Q, Barish S, Okuwa S, Zhao S, Soeder C, Kanke M, Jones CD and Volkan PC (2017). Patterns of transcriptional parallelism and variation in the developing olfactory system of Drosophila species. Sci. Reports, 7, 8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Münch D, and Galizia CG (2016). DoOR 2.0-comprehensive mapping of Drosophila melanogaster odorant responses. Sci. Reports 6, 21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramasamy S, Ometto L, Crava CM, Revadi S, Kaur R, Horner DS, … & Rota-Stabelli O (2016). The evolution of olfactory gene families in Drosophila and the genomic basis of chemical-ecological adaptation in Drosophila suzukii. Genome Biol. Evol 8, 2297–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Naters WVDG, and Carlson JR (2007). Receptors and neurons for fly odors in Drosophila. Curr. Biol 17, 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguadé M (2008). Nucleotide and copy-number polymorphism at the odorant receptor genes Or22a and Or22b in Drosophila melanogaster. Mol. Biol. Evol 26, 61–70. [DOI] [PubMed] [Google Scholar]
- 31.Kern AD, & Hey J (2017). Exact calculation of the joint allele frequency spectrum for isolation with migration models. Genetics, 207, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elya C, Quan AS, Schiabor KM, and Eisen M (2017). Or22 allelic variation alone does not explain differences in discrimination of yeast-produced volatiles by D. melanogaster. bioRxiv, 186064. [Google Scholar]
- 33.Walker NJ (1989). King of foods: marula economics in the Matobos. Afr. Wildl 43, 281–285. [Google Scholar]
- 34.David JR, Allemand R, Capy P, Chakir M, Gibert P, Pétavy G, and Moreteau B (2004). Comparative life histories and ecophysiology of Drosophila melanogaster and D. simulans In Drosophila melanogaster, Drosophila simulans: So Similar, So Different (pp. 151–163). Springer, Dordrecht. [DOI] [PubMed] [Google Scholar]
- 35.McKenzie JA, and Parsons PA (1972). Alcohol tolerance: an ecological parameter in the relative success of Drosophila melanogaster and Drosophila simulans. Oecologia, 10, 373–388. [DOI] [PubMed] [Google Scholar]
- 36.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC bioinform. 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S Rueden C, Saalfeld S Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P and Cardona A (2012). Fiji: an open-source platform for biological-image analysis. Nature methods. 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lack JB, Lange JD, Tang AD, Corbett-Detig RB, and Pool JE (2016). A thousand fly genomes: an expanded drosophila genome nexus. Mol. Biol. Evol 33, 3308–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, and Marra MA (2009). Circos: an information aesthetic for comparative genomics. Genome res. 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.