Summary
Background
The incidence of epidermal keratinocyte‐derived cutaneous squamous cell carcinoma (cSCC) is increasing worldwide.
Objectives
To study the role of the complement classical pathway components C1q, C1r and C1s in the progression of cSCC.
Methods
The mRNA levels of C1Q subunits and C1R and C1S in cSCC cell lines, normal human epidermal keratinocytes, cSCC tumours in vivo and normal skin were analysed with quantitative real‐time polymerase chain reaction. The production of C1r and C1s was determined with Western blotting. The expression of C1r and C1s in tissue samples in vivo was analysed with immunohistochemistry and further investigated in human cSCC xenografts by knocking down C1r and C1s.
Results
Significantly elevated C1R and C1S mRNA levels and production of C1r and C1s were detected in cSCC cells, compared with normal human epidermal keratinocytes. The mRNA levels of C1R and C1S were markedly elevated in cSCC tumours in vivo compared with normal skin. Abundant expression of C1r and C1s by tumour cells was detected in invasive sporadic cSCCs and recessive dystrophic epidermolysis bullosa‐associated cSCCs, whereas the expression of C1r and C1s was lower in cSCC in situ, actinic keratosis and normal skin. Knockdown of C1r and C1s expression in cSCC cells inhibited activation of extracellular signal‐related kinase 1/2 and Akt, promoted apoptosis of cSCC cells and significantly suppressed growth and vascularization of human cSCC xenograft tumours in vivo.
Conclusions
These results provide evidence for the role of tumour‐cell‐derived C1r and C1s in the progression of cSCC and identify them as biomarkers and putative therapeutic targets in cSCC.
What's already known about this topic?
The incidences of actinic keratosis, cutaneous squamous cell carcinoma (cSCC) in situ and invasive cSCC are increasing globally.
Few specific biomarkers for progression of cSCC have been identified, and no biological markers are in clinical use to predict the aggressiveness of actinic keratosis, cSCC in situ and invasive cSCC.
What does this study add?
Our results provide novel evidence for the role of complement classical pathway components C1r and C1s in the progression of cSCC.
What is the translational message?
Our results identify complement classical pathway components C1r and C1s as biomarkers and putative therapeutic targets in cSCC.
Short abstract
Linked Comment: https://doi.org/10.1111/bjd.18419.
https://doi.org/10.1111/bjd.18821 available online
Keratinocyte‐derived cutaneous squamous cell carcinoma (cSCC) causes 20% of all skin‐cancer‐related deaths, with an estimated 5‐year metastasis rate of 5%.1, 2, 3, 4 Currently, the incidence of cSCC is increasing globally.5 The progression of cSCC takes place from actinic keratosis (AK) to cSCC in situ (cSCCIS) and eventually to invasive and metastatic cSCC. The main risk factors for cSCC include long‐term exposure to ultraviolet (UV) radiation from sunlight, immunosuppression and chronic dermal ulcers.6 Moreover, chronic inflammation has been recognized as an important factor in the development of cSCC.7
The complement system connects innate and acquired immunity and initiates the inflammatory responses in host defence.8 The complement system is activated in a sequential manner via three distinct pathways (classical, lectin and alternative pathways), which converge in cleavage of the central component C3 to C3a and C3b fragments. Covalent binding of C3b to target cells promotes phagocytosis and initiates activation of the terminal lytic pathway and formation of the membrane attack complex.9 In addition, activation of the complement system generates an inflammatory response and stimulates macrophage and B‐cell activities. The small cleavage products C3a and C5a of the main complement components C3 and C5 function as anaphylatoxins by increasing vascular permeability and promoting contraction of smooth muscle cells.10
The classical pathway of complement is typically activated by binding of C1 complex to antibodies bound to their target antigens. The C1 complex consists of six subcomponents of C1q, each with a collagenous triple helix of subunits C1qA, C1qB and C1qC and two copies each of the C1r and C1s subunits.11 The binding of C1 complex to a target results in a conformational change in C1q, which initiates a stepwise proteolytic activation of serine proteinases C1r and C1s.12 The activities of C1r and C1s can be inhibited by the serine proteinase inhibitor C1INH. C1s then cleaves C4 to C4a and C4b fragments, and C4‐bound C2 to C2a and C2b. This leads to generation of the C3 convertase C4bC2a, which activates C3 and initiates the lytic pathway.13 Recent observations on the diversity of C1q ligands and C1s substrates suggest that C1 has functions outside the complement system.14, 15, 16, 17, 18, 19, 20
Our previous studies have shown that the expression of complement factor H and complement factor I, two regulators of the alternative pathway, and two activating components complement factor B and C3, are significantly upregulated in tumour cells in cSCCs, and that complement factor I, complement factor B and C3 promote growth of cSCC in vivo.21, 22, 23 However, the role of the classical pathway of complement in the progression of cSCC is not known. The results of the present study show marked upregulation of the expression of C1r and C1s by cSCC cells in culture and by cSCC tumour cells in vivo. Furthermore, our results show that knockdown of C1r and C1s inhibits activation of the extracellular signal‐related kinase (ERK)1/2 and phosphoinositide 3‐kinase (PI3K) signalling pathways, promotes apoptosis of cSCC cells and suppresses vascularization and growth of cSCC xenografts in vivo. These results provide novel evidence for the role of C1r and C1s in the progression of cSCC and identify them as biomarkers and potential therapeutic targets in cSCC.
Materials and methods
Detailed information on the materials and methods is provided in Appendix S1 (see Supporting Information).
Ethical issues
Collection of normal skin and cSCC tissues and the use of archival tissue specimens were approved by the ethics committee of the Hospital District of Southwest Finland (4/2006). The research was carried out according to the Declaration of Helsinki. All studied patients gave informed and written permission before surgery, and the study was carried out with the authorization of Turku University Hospital (Turku, Finland). All experiments with mice were carried out with the permission of the State Provincial Office of Southern Finland, according to institutional guidelines.
Cell cultures
Human cSCC cell lines (n = 8) were initiated from surgically removed cSCCs.24 Five cSCC cell lines were derived from primary cSCCs: UT‐SCC‐12A, UT‐SCC‐91, UT‐SCC‐105, UT‐SCC‐111 and UT‐SCC‐118. Three cSCC cell lines were from metastatic cSCCs: UT‐SCC‐7, UT‐SCC‐59A and UT‐SCC‐115. These cell lines were authenticated by short tandem repeat DNA profiling.24 Primary normal human epidermal keratinocytes (NHEKs) were obtained from PromoCell (Heidelberg, Germany). NHEKs were cultured from normal skin of patients (n = 11) who had undergone mammoplastic surgery at Turku University Hospital, Turku, Finland. Cell cultures were performed as previously described.21, 22, 23
Tissue samples
Primary cSCC samples (n = 6) were obtained from surgically removed tumours in Turku University Hospital.25 Normal human skin samples (n = 10) were collected from the upper arm of healthy volunteers and during mammoplasty operation in Turku University Hospital. Human liver RNA was obtained from Human MTC Panel I (Clontech, Mountain View, CA, U.S.A.). Altogether 260 archival formalin‐fixed paraffin‐embedded tissue samples from sporadic, UV‐induced cSCC (n = 115; mean age 79 years, range 45–102), cSCCIS (Bowen disease; n = 63; mean age 79 years, range 59–95), AK (n = 61; mean age 78 years, range 58–95) and normal skin (n = 21) were obtained from the archives of the Department of Pathology, Turku University Hospital. In addition, recessive dystrophic epidermolysis bullosa‐associated cSCC (RDEBSCC) tissue samples (n = 16; mean age 33 years, range 12–56) were stained and analysed.26, 27
Statistical analysis
SPSS software (IBM, Armonk, NY, U.S.A.) was used for statistical analysis to determine the significance of differences between two sample groups. For quantitative real‐time polymerase chain reaction (qRT‐PCR) and staining analyses for Ki‐67, CD34 and active caspase‐3, a two‐tailed Mann–Whitney U‐test was used. Student's t‐test was used to analyse differences in xenograft tumour volume and assays of cell number, viability, apoptosis and migration, and the χ2‐test was used for comparison of intensities of immunohistochemistry (IHC) staining.
Results
Increased expression of C1r and C1s in cutaneous squamous cell carcinoma cells and tumours
The mRNA levels of C1R, C1S and C1Q subunits in cSCC cell lines and NHEKs were determined by qRT‐PCR (Table S1; see Supporting Information). The mean expression levels of C1R and C1S mRNAs were significantly higher in cSCC cell lines (n = 8) than in NHEKs (n = 10) (Fig. 1a). The mRNA levels of C1Q subunits C1QA and C1QB and C1QC variants 1 and 2 were very low in cSCC cell lines (n = 8) and NHEKs (n = 7) compared with those in liver (Fig. S1; see Supporting Information). Marked levels of C1r (80 kDa) and C1s (76 kDa) proteins were detected in conditioned media of cSCC cell lines by Western blotting, whereas the production of both C1r and C1s by NHEKs was very low (Fig. 1b, upper panel). No correlation between C1r and C1s protein levels produced by cSCC cells was detected (Fig. 1b, upper panel).
Figure 1.
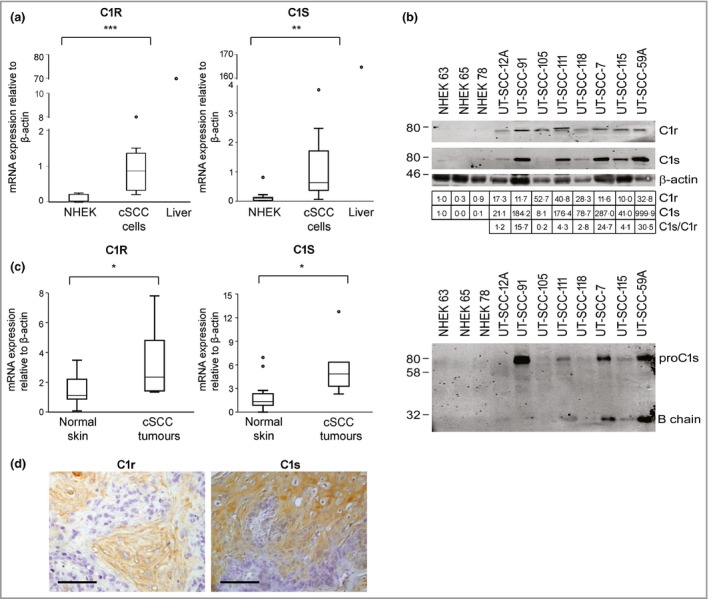
Expression of C1r and C1s is upregulated in cutaneous squamous cell carcinoma (cSCC) cells. (a) C1R and C1S mRNA levels in normal human epidermal keratinocytes (NHEKs; n = 10) and cSCC cell lines (n = 8) were determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). RNA from human liver was used as a positive control. (b) The levels of C1r and C1s in NHEKs and cSCC cell lines were determined by Western blotting of conditioned media under nonreducing conditions (upper panel), with β‐actin in cell lysates as a sample control. C1r and C1s protein levels were quantitated densitometrically and corrected for β‐actin levels, and the ratio of C1s to C1r was calculated. The levels of the cleaved form of C1s in conditioned media of NHEKs and cSCC cell lines were determined by Western blotting under reducing conditions (lower panel). (c) Levels of C1R and C1S mRNA in normal skin (n = 10) and cSCC (n = 6) samples were analysed by qRT‐PCR. (d) Xenografts established with human cSCC cells were stained for immunohistochemistry using C1r and C1s antibodies. Scale bar = 100 μm. Statistical analysis was performed with Mann–Whitney two‐way U‐test. *P < 0·05, **P < 0·01, ***P < 0·001.
The cleaved form of C1s (B chain, 28 kDa) was detected in conditioned media of cSCC cells under reducing conditions, indicating C1r activity in these cells (Fig. 1b, lower panel). Elevated C1R and C1S mRNA levels were also noted in cSCC tumour tissue (n = 6) compared with normal skin tissue samples (n = 10) (Fig. 1c). In addition, IHC showed tumour‐cell‐specific expression of C1r and C1s in xenograft tumours established with human metastatic cSCC cells (UT‐SCC‐7) in the skin of severe combined immunodeficient (SCID) mice (Fig. 1d).
Expression of C1r and C1s by tumour cells in cutaneous squamous cell carcinomas in vivo
The expression of C1r and C1s in cSCC tumour tissue in vivo was examined by IHC in tissue microarrays containing tissue samples from different stages of epidermal carcinogenesis, namely sporadic UV‐induced cSCC (n = 115), cSCCIS (n = 63), premalignant precursor lesion (AK, n = 61) and normal skin (n = 21). In addition, tissue samples from RDEBSCC, an aggressive form of cSCC (n = 16), were included in the IHC analysis.
Notable tumour‐cell‐specific expression of C1r was detected in cSCCs (Fig. 2a, b) and RDEBSCC samples (Fig. 2c). In general, the staining intensity for C1r was stronger in cSCC tissue sections than in cSCCIS, AK and normal skin tissue samples (Fig. 2d–f). The staining intensity was scored negative (−), weak (+), moderate (++) or strong (+++). Semiquantitative analysis revealed mainly strong or moderate staining in tissue sections in cSCC. Moreover, in RDEBSCC sections the cytoplasmic staining of C1r was strong or moderate. On the other hand, in the cSCCIS, AK and normal skin groups, the proportion of strong or moderate staining was significantly lower than in cSCC or RDEBSCC samples (Fig. 2g).
Figure 2.
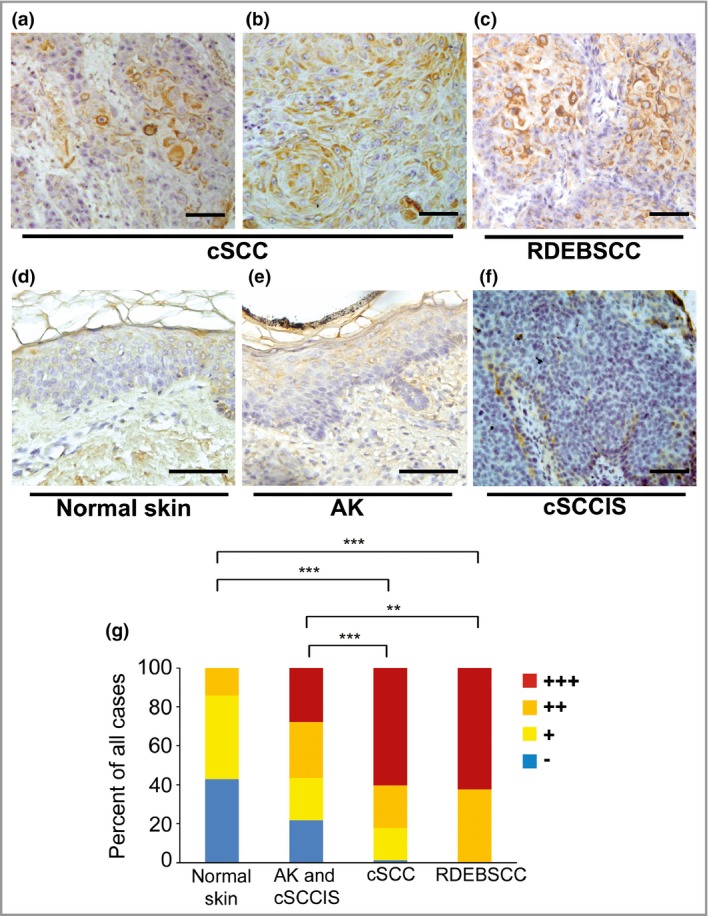
Expression of C1r by tumour cells in cutaneous squamous cell carcinoma (cSCC). (a–f) Sections of tissue microarray blocks containing samples from ultraviolet‐induced sporadic cSCC (n = 106), recessive dystrophic epidermolysis bullosa‐associated cSCCs (RDEBSCC, n = 15), cSCC in situ (cSCCIS, n = 61), premalignant lesions (actinic keratosis, AK, n = 61) and normal skin (n = 8) were stained with C1r antibody. Strong cytoplasmic staining was detected in tumour cells in cSCC (a, b) and RDEBSCC (c). Staining for C1r was absent or weak in normal skin (d). In AK (e) and cSCCIS (f) staining was weak. Scale bar = 100 μm. (g) C1r immunostaining was scored as negative (−), weak (+), moderate (++) or strong (+++) based on the specific staining intensity. **P < 0·01, ***P < 0·001 by χ2‐test.
Prominent expression of C1s was also detected in tumour cells in cSCC tissue samples (Fig. 3a, b) and in RDEBSCC tissue sections (Fig. 3c). Normal skin stained weakly for C1s (Fig. 3d). In premalignant lesions (AK) (Fig. 3e) and in cSCCIS (Fig. 3f) the proportion of strong or moderate staining for C1s was significantly lower than in cSCC samples. Semiquantitative analysis revealed strong or moderate staining for C1s in the majority of tissue sections in the cSCC and RDEBSCC groups (Fig. 3g). In comparison, the proportion of negative and weak staining for C1s was significantly higher in AK and cSCCIS than in cSCC tissues. In normal skin the expression of C1s was negative or weak in most tissue samples (Fig. 3g).
Figure 3.

Expression of C1s by tumour cells in cutaneous squamous cell carcinoma (cSCC). (a–f) Sections of tissue microarray blocks containing samples from ultraviolet‐induced sporadic cSCC (n = 115), recessive dystrophic epidermolysis bullosa‐associated cSCCs (RDEBSCC, n = 16), cSCC in situ (cSCCIS, n = 63), premalignant lesions (actinic keratosis, AK, n = 57) and normal skin (n = 21) were stained with C1s antibody. Cytoplasmic staining for C1s was moderate (a) or strong (b) in cSCC and strong in RDEBSCC (c). In normal skin the staining for C1s was mainly weak (d). In AK (e) and cSCCIS (f) staining was weak or moderate. Scale bar = 100 μm. (g) C1s immunostaining was scored as negative (−), weak (+), moderate (++) or strong (+++) based on the specific staining intensity. *P < 0·05, ***P < 0·001 by χ2‐test.
Knockdown of C1r and C1s promotes apoptosis of cutaneous squamous cell carcinoma cells
To elucidate the functional role of C1r and C1s in cSCC cells, their expression was silenced by specific small interfering (si)RNAs (Fig. 4a, b; and Figs S2–S4; see Supporting Information). Knockdown of C1r (Fig. 4c; and Fig. S2c–g) and C1s (Fig. 4d; and Fig S2a, c–g) resulted in a significant decrease in the growth of cSCC cells. Knockdown of C1r or C1s had no marked effect on the growth of NHEKs (Fig. S3; see Supporting Information). Additionally, significant reduction in the viability of cSCC cells was noted 72 h after transfection with C1r siRNAs (Fig. S4c) and C1s siRNAs (Fig. S4d).
Figure 4.
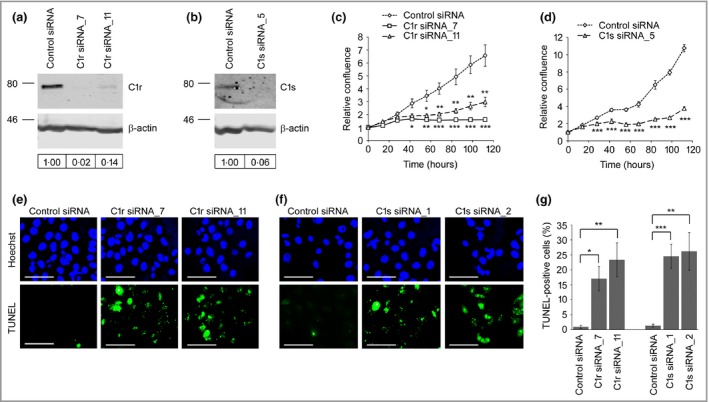
Knockdown of C1r and C1s promotes apoptosis of cutaneous squamous cell carcinoma (cSCC) cells. (a, b) cSCC cells (UT‐SCC‐12A) were transfected with C1R small interfering (si)RNA (C1r siRNA_7 or C1r siRNA_11; 75 nmol L−1) (a) or C1S siRNA (C1s siRNA_5; 120 nmol L−1) (b) and control siRNA. Cell lysates were analysed by Western blotting 8 days after transfections. The levels of C1R and C1S were quantitated densitometrically and corrected for β‐actin levels in the same samples. (c, d) cSCC cells (UT‐SCC‐12A) were transfected with control siRNA, C1R (75 nmol L−1) (c) or C1S siRNAs (120 nmol L−1) (d). The confluency of the cells was determined at the indicated time points using IncuCyte ZOOM (n = 6–8; mean ± SEM). (e–g) UT‐SCC‐12A cells were transfected with C1R (75 nmol L−1) or C1S (120 nmol L−1) siRNAs or control siRNA; 48 h after transfection apoptotic cells were detected with TUNEL staining, and the relative number of TUNEL‐positive cells was counted (mean ± SEM) (g). Representative images after C1r (e) and C1s (f) knockdown are shown. Scale bar = 1 μm. *P < 0.05, **P < 0.01, ***P < 0.001 by t‐test
An increased number of TUNEL‐positive apoptotic cells was detected 48 h after transfection with C1r (Fig. 4e, g; and Fig. S5; see Supporting Information) and C1s siRNA (Fig. 4f, g; and Fig. S5a, b, d) compared with cells transfected with control siRNA. Similar results on cSCC cell viability (UT‐SCC‐7, ‐12A, ‐59A, ‐91, ‐105, ‐111) and apoptosis (UT‐SCC‐7, ‐12A, ‐91, ‐105, ‐111, ‐115) were obtained with six cSCC cell lines. The migration rate of cSCC cells in a scratch wound model was significantly reduced after knockdown of C1r (Fig. 5a, b) and C1s (Fig. 5c, d). Similar results on the migration rate of cSCC cells were obtained with two cSCC cell lines (UT‐SCC‐7 and ‐12A). Knockdown of C1r and C1s upregulated the production of matrix metalloproteinase (MMP)‐9 in cSCC cells (UT‐SCC‐7) (Fig. S6; see Supporting Information).
Figure 5.
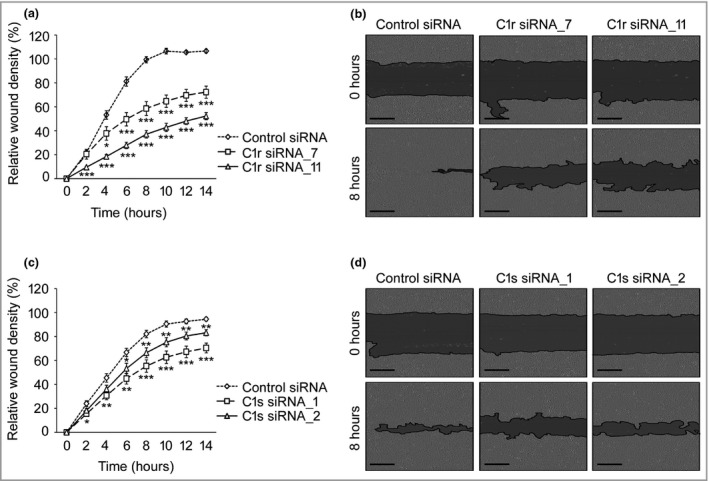
Knockdown of C1r and C1s inhibits migration of cutaneous squamous cell carcinoma (cSCC) cells. UT‐SCC‐12A cells were transfected with C1R (75 nmol L−1) (a) or C1S (120 nmol L−1) (c) small interfering (si)RNAs or control siRNA, and incubated for 48 h (n = 8) (mean ± SEM). A wound was made using 96‐well WoundMaker and incubation was continued in 0·5 mmol L−1 hydroxycarbamide. Representative images after C1r (b) and C1s (d) knockdown are shown. Scale bar = 300 μm. *P < 0·05, **P < 0·01, ***P < 0·001 by t‐test.
Knockdown of C1r and C1s suppresses growth of cutaneous squamous cell carcinoma in vivo
The role of C1r and C1s on cSCC growth in vivo was examined in a xenograft model. cSCC cells (UT‐SCC‐91) were transfected with C1R, C1S or control siRNA, incubated for 72 h and injected (7 × 106 cells) subcutaneously into the back of SCID mice. The growth of cSCC xenograft tumours with C1r or C1s knockdown was significantly reduced compared with control siRNA tumours (Fig. 6a). Histological analysis of xenografts harvested 16 days after implantation showed that the C1r and C1s knockdown tumours contained less tumour tissue than the control tumours (Fig. 6b). Furthermore, the relative number of proliferating Ki‐67‐positive cells (Fig. 6b, c) and the density of CD34‐positive blood vessels were significantly lower in C1r and C1s knockdown tumours (Fig. 6b, d). On the other hand, the percentage of active caspase‐3‐positive apoptotic cells was significantly higher in C1r and C1s knockdown tumours than in control tumours (Fig. 6b, e).
Figure 6.

Knockdown of C1r and C1s suppresses growth of cutaneous squamous cell carcinoma (cSCC) in vivo. (a) cSCC cells (UT‐SCC‐91) were transfected with C1R small interfering (si)RNA_5 (n = 7), C1S siRNA_5 (n = 8) (120 nmol L−1) or control siRNA (n = 8) and incubated for 72 h. Cells (7 × 106) were injected subcutaneously into the back of severe combined immunodeficient mice and the size of tumours was measured twice a week (mean ± SEM). (b) Xenografts were harvested after 16 days and stained with haematoxylin and eosin (HE), and for immunohistochemistry for the proliferation marker Ki‐67, the vascular endothelial marker CD34 and the apoptotic marker active caspase‐3, with Mayer's haematoxylin as counterstain. Representative stainings from each group are shown. Arrows indicate CD34‐positive blood vessels. Scale bar = 100 μm. (c–e) The percentage of Ki‐67‐positive cells (c), the number of CD34‐positive blood vessels (d) and the percentage of active caspase‐3‐positive cells (e) were counted. *P < 0·05, **P < 0·01, ***P < 0·001 by Mann–Whitney U‐test.
Alteration of the gene expression profile in cutaneous squamous cell carcinoma cells after C1s knockdown
To gain mechanistic insight into the functional role of C1r and C1s in cSCC progression, C1s expression was knocked down in three cSCC cell lines and gene expression profiling was performed by mRNA sequencing. Ingenuity Pathway Analysis revealed significant downregulation of metastasis‐related biofunctions after C1s knockdown (Fig. 7a). Furthermore, the genes significantly regulated following C1s knockdown were associated with gene ontology terms related to signalling pathways regulating cell proliferation [negative regulation of MAP kinase activity MAPK cascade and negative regulation of MAPK kinase activity] and cell viability (Phosphatidylinositol 3‐kinase activity and phosphatidylinositol‐4,5‐bisphosphate 3‐kinase activity) (Fig. 7). Expression of several dual‐specificity phosphatases was significantly upregulated in gene ontology term negative regulation of MAPK cascade (Fig. 8a). Additionally, gene ontology term Phosphatidylinositol 3‐kinase activity contained significantly downregulated genes including PIK3R3, which codes for PI3K regulatory subunit 3 and PIK3IP1, which codes for phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit β, which positively regulate the PI3K signalling pathway (Fig. 8b). Analysis of cell lysates by Western blotting after C1s knockdown showed that the levels of phosphorylated Akt and phosphorylated ERK1/2 were decreased, indicating inhibition in the activation of the PI3K and ERK1/2 signalling pathways, respectively (Fig. 9).
Figure 7.
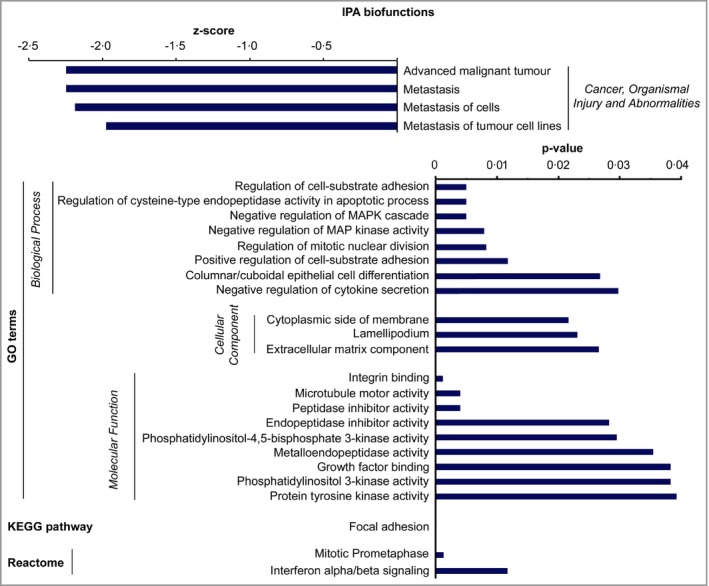
Alteration of the gene expression profile in cutaneous squamous cell carcinoma (cSCC) cells after C1s knockdown. cSCC cell lines (UT‐SCC‐12A, ‐59A and ‐91) were transfected with C1S small interfering (si)RNA_5 or control siRNA (120 nmol L−1) and mRNA sequencing was performed 72 h after transfection. Summary of Ingenuity Pathway Analysis biofunctions, gene ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Reactome related to C1s knockdown (P < 0·05, fold change log2 1·0).
Figure 8.
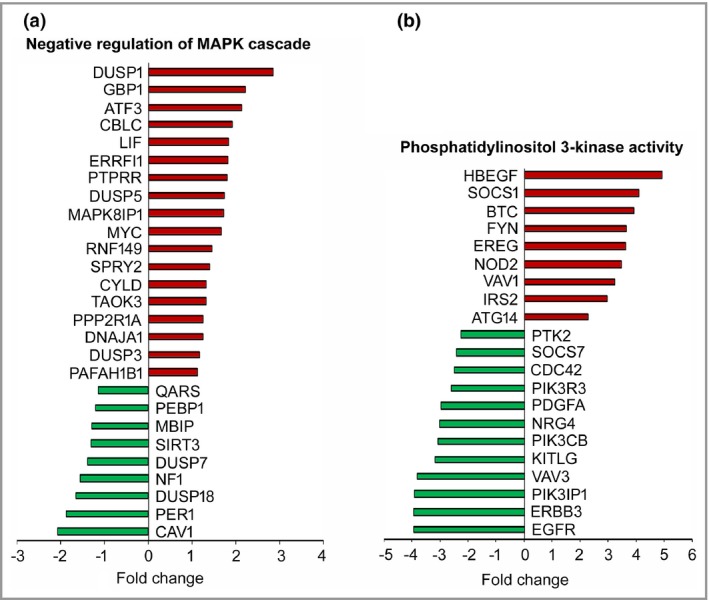
Alteration of the gene expression profile in cutaneous squamous cell carcinoma (cSCC) cells after C1s knockdown. (a) Significantly regulated genes belonging to the gene ontology term ‘negative regulation of MAPK cascade’ are shown in gene blot. (b) Significantly regulated genes belonging to the gene ontology term ‘phosphatidylinositol 3‐kinase activity’ are shown in gene blot.
Figure 9.
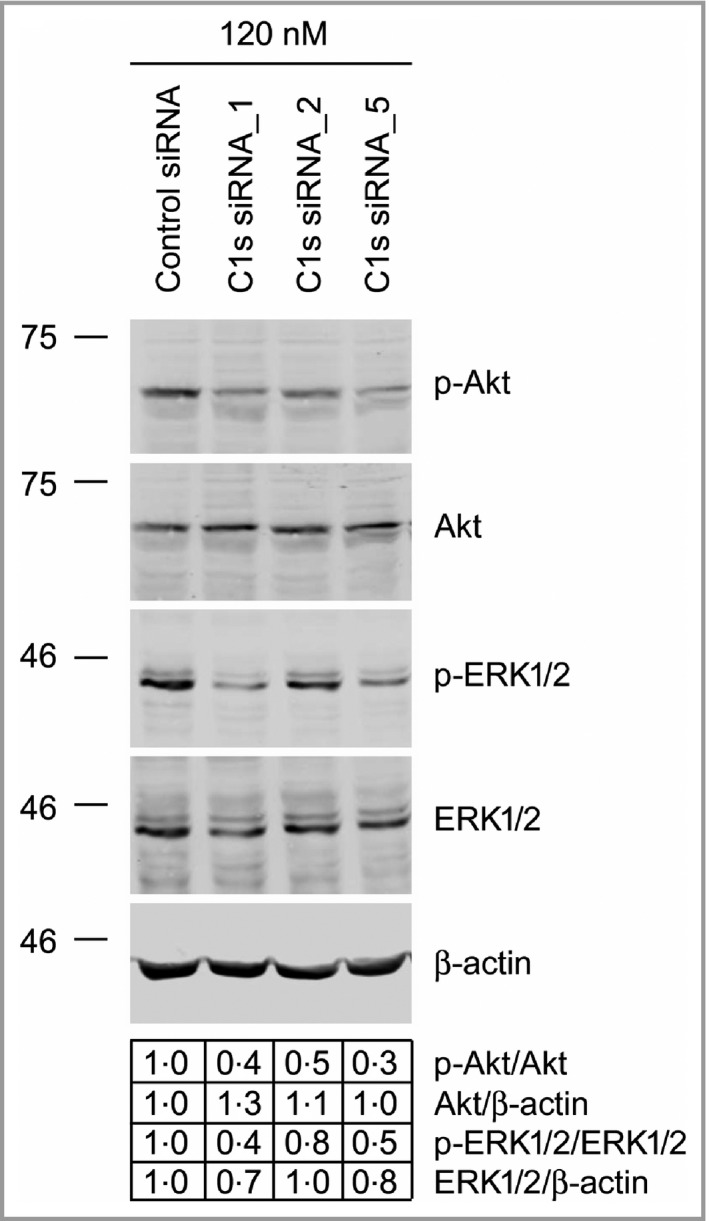
Decreased activation of Akt and ERK1/2 in cutaneous squamous cell carcinoma (cSCC) cells after C1s knockdown. cSCC cells (UT‐SCC7) were transfected with C1S small interfering (si)RNAs and control siRNA (120 nmol L−1) and the levels of phosphorylated Akt (p‐Akt), phosphorylated extracellular signal‐related kinase 1/2 (p‐ERK1/2), total Akt and total ERK1/2 were analysed by Western blotting 72 h after transfection. The levels of p‐Akt and p‐ERK1/2 in blots were determined densitometrically and corrected for the levels of total Akt and ERK1/2 in the same samples, respectively (values below the blots). β‐actin was used as loading control.
Discussion
The complement system plays an important role in innate immunity and is activated via three distinct pathways (classical, lectin and alternative), all of which lead to lytic pathway activation and target cell lysis. In the classical pathway, the first component (the C1qr2s2 complex) is activated by at least two surface‐bound Fc regions of antigen‐bound IgMs or IgGs. The tertiary structure of C1q is first altered resulting in autoactivation of serine proteinase C1r, which then activates C1s, also a serine proteinase.12 The activation of the classical pathway can also be initiated in an antibody‐independent manner by binding of C1q to other ligands, including C‐reactive protein,28 serum amyloid P component29 and membrane blebs of apoptotic cells.30 Recently, the role of complement in cancer progression has gained attention.31, 32, 33, 34, 35 It has been shown that tumour‐cell‐derived complement components can promote cancer growth in an autocrine manner.36 In addition, a high expression level of complement component C3 in primary tumour was recently shown to correlate with the rate of metastasis across the blood–brain barrier.37
Recent observations show that locally produced C1q can act as a cancer‐promoting factor in the tumour microenvironment independently of complement activation.38 Our results show that the mRNAs for C1q subunits are not expressed by cSCC cells. In addition, our previous studies showed that cSCC cells do not express mRNAs for the C4 or C2 components of the classical pathway.21, 22 However, the results of the present study show that cSCC cell‐derived C1s is specifically cleaved and activated in culture in the absence of C1q. Therefore, the data presented in this study suggest that C1r and C1s contribute to cSCC tumour progression independently of complement classical pathway activation and can act without C1q, C4 or C2.
It is possible that C1r is activated autocatalytically in the absence of C1q and subsequently activates C1s in the tumour microenvironment of cSCC. However, several types of cells produce C1q subunits, such as macrophages,38 bone‐marrow‐derived cells39 and dermal microvascular endothelial cells in wounds.40 It is therefore also possible that C1q derived from stromal cells or circulation is present in the cSCC tumour microenvironment, allowing assembly of the C1 complex. In our experiments, C1r and C1s were secreted to the medium of cSCC cells in culture, indicating the extracellular function of C1r and C1s in the tumour microenvironment. Interestingly, knockdown of C1s resulted in significant downregulation of biofunctions related to metastasis in cSCC cells. As a serine proteinase, C1s has been shown to activate latent MMP‐9,41 suggesting a putative mechanism for C1s in promoting cancer growth, angiogenesis and metastasis. Recently, knockdown of MMP‐9 has been shown to increase migration and invasion of oral SCC cells.42 Accordingly, increased expression of MMP‐9 was noted in cSCC cells after knockdown of C1r and C1s, suggesting that the stimulatory effect of C1r and C1s on cSCC cell migration may involve MMP‐9.
Our results also show that silencing the expression of C1r or C1s in cSCC cells in culture resulted in significant reduction in cell viability and promoted apoptosis of cSCC cells. These findings are in accordance with the results of our studies in a xenograft model in SCID mice showing suppression of tumour growth and vascularization and an increased number of apoptotic cells in vivo following knockdown of C1r or C1s in cSCC cells. Together these observations provide evidence for the important roles of C1r and C1s in cSCC tumour growth in vivo.
As C1s is proteolytically activated by C1r and serves as the effector of C1r we studied the mechanistic role of C1r and C1s in cSCC progression by RNA‐sequencing‐based gene expression profiling after knockdown of C1s expression in three distinct cSCC cell lines. Interestingly, several gene ontology terms related to cell viability and proliferation were significantly regulated in cSCC cells following knockdown of C1s, and this was associated with inhibition of the activity of the PI3K and ERK1/2 signalling pathways. In accordance with our results, complement activation along the vasculature of the tumours has been shown to be associated with the growth and increased angiogenesis of tumours.43, 44, 45 Activation of the classical pathway has previously been reported in papillary thyroid carcinoma,46 in lung cancer44 and in a mouse model of cervical cancer.47 Moreover, deposition of classical pathway components has been detected in oral and oropharyngeal SCCs,48 follicular and mucosa‐associated lymphoid tissue lymphomas49 and astrocytomas.50 These results are in accordance with those of the present study and provide additional evidence for the role of the classical pathway of complement in cancer progression.
Solar UV radiation, chronic inflammation and immunosuppression are important risk factors for development of cSCC. In recent studies, the role of the complement system in tumour growth by altering the host immune response and promoting chronic inflammation has been emphasized.51, 52, 53 Complement activation leads to the formation of anaphylatoxins C3a, C4a and C5a, which may alter the tumour microenvironment by attracting macrophages, increasing histamine release from mast cells and increasing vascular permeability.54 It has also been shown that C1s can cleave and inactivate the alarmin high‐mobility group box 1, and this may regulate inflammation by suppressing lipopolysaccharide‐induced production of proinflammatory cytokines by monocytes, macrophages and dendritic cells.20
The complement system is a complex network of effectors, receptors and regulators, some of which exert functions beyond complement activation. Complement components serve as important effectors of monoclonal antibodies used in cancer therapy, and therefore it is important to identify the expression and function of distinct complement components in different types of cancers. Recently, the efficacy of immune checkpoint inhibitor therapy in advanced and metastatic cSCC has been demonstrated.55 In this respect it is interesting that anaphylatoxin C5a has been shown to promote tumour progression, and that blocking complement receptor C5a results in decreased expression of the immune checkpoint molecule programmed death ligand 1 in a murine lung cancer model.52 In addition to markers for personalized cancer therapy, complement components can serve as diagnostic and prognostic biomarkers in cancer. Combinations of complement components could serve as biomarkers for predicting the aggressiveness of cSCC for different patients, as demonstrated by detection of different complement components by a multiplex detection array to predict progression of oral SCC.56
In summary, the results of the present study show tumour‐cell‐specific overexpression of complement system components C1r and C1s in cSCC cells in vivo. Furthermore, the results demonstrate that C1r and C1s promote the growth of cSCC xenografts in vivo by enhancing tumour vascularization and tumour cell viability. These results provide evidence for important roles for C1r and C1s in cSCC progression and suggest them as tumour‐cell‐derived molecular markers in cSCC. The feasibility of this approach in assessing disease progression has recently been demonstrated in SCC.56 At present, a number of novel pharmacological inhibitors targeted against specific complement components are under development.57 It is therefore of importance to elucidate the role of individual complement components in the growth and development of cSCC in order to identify targets for these new drugs in the therapy of metastatic and unresectable advanced cSCC.
Supporting information
Appendix S1 Supplementary materials and methods.
Appendix S2 Supplementary references.
Fig S1. Expression of C1QA, C1QB, C1QC variant 1 and C1QC variant 2 in cutaneous squamous cell carcinoma cells and normal human epidermal keratinocytes.
Fig S2. Knockdown of C1r and C1s inhibits growth of cutaneous squamous cell carcinoma cells.
Fig S3. Knockdown of C1r or C1s had no marked effect on the growth of normal human epidermal keratinocytes.
Fig S4. Knockdown of C1r and C1s inhibits viability of cutaneous squamous cell carcinoma cells.
Fig S5. Knockdown of C1r and C1s promotes apoptosis of cutaneous squamous cell carcinoma cells.
Fig S6. Knockdown of C1r and C1s upregulates the expression of matrix metalloproteinase‐9 in UT‐SCC‐7 cells.
Table S1 Primer and probe sequences used for quantitation of the mRNAs of human C1R, C1S, C1QA, C1QB, C1QC transcript variant 1, C1QC transcript variant 2 and β‐actin, with quantitative real‐time polymerase chain reaction.
Acknowledgments
We are grateful to Johanna Markola and Sinikka Kollanus for technical assistance. We thank the Bioinformatics Unit of Turku Centre for Biotechnology for the RNA‐sequencing data analysis. The unit is supported by University of Turku, Åbo Akademi University and Biocenter Finland.
Funding sources This study was supported by the Jane and Aatos Erkko Foundation, Finnish Cancer Research Foundation, Sigrid Jusélius Foundation and Turku University Hospital VTR grant (project 13336) and by personal grants to P.R. from the Finnish Medical Foundation, the Cancer Foundation of Southwest Finland and the Finnish Cultural Foundation Southwest Finland Regional Fund. K.V. is a doctoral candidate in the Turku Doctoral Program for Clinical Investigation.
Conflicts of interest None to declare.
https://doi.org/10.1111/bjd.18821 available online
References
- 1. Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol 2013; 68:957–66. [DOI] [PubMed] [Google Scholar]
- 2. Nehal KS, Bichakjian CK. Update on keratinocyte carcinomas. N Engl J Med 2018; 379:363–74. [DOI] [PubMed] [Google Scholar]
- 3. Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol 2018; 78:237–47. [DOI] [PubMed] [Google Scholar]
- 4. Schmults CD, Karia PS, Carter JB et al Factors predictive of recurrence and death from cutaneous squamous cell carcinoma: a 10‐year, single‐institution cohort study. JAMA Dermatol 2013; 149:541–7. [DOI] [PubMed] [Google Scholar]
- 5. Green AC, Olsen CM. Cutaneous squamous cell carcinoma: an epidemiological review. Br J Dermatol 2017; 177:373–81. [DOI] [PubMed] [Google Scholar]
- 6. Kivisaari A, Kähäri VM. Squamous cell carcinoma of the skin: emerging need for novel biomarkers. World J Clin Oncol 2013; 4:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ratushny V, Gober MD, Hick R et al From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest 2012; 122:464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 2010; 11:785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serna M, Giles JL, Morgan BP, Bubeck D. Structural basis of complement membrane attack complex formation. Nanomedicine 2014; 10:1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schraufstatter IU, Trieu K, Sikora L et al Complement c3a and c5a induce different signal transduction cascades in endothelial cells. J Immunol 2002; 169:2102–10. [DOI] [PubMed] [Google Scholar]
- 11. Almitairi JOM, Venkatraman Girija U, Furze CM et al Structure of the C1r–C1s interaction of the C1 complex of complement activation. Proc Natl Acad Sci U S A 2018; 23:768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Venkatraman Girija U, Gingras AR, Marshall JE et al Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc Natl Acad Sci U S A 2013; 110:13916–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lintner KE, Wu YL, Yang Y et al Early components of the complement classical activation pathway in human systemic autoimmune diseases. Front Immunol 2016; 15:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eriksson H, Nissen MH. Proteolysis of the heavy chain of major histocompatibility complex class I antigens by complement component C1s. Biochim Biophys Acta 1990; 1037:209–15. [DOI] [PubMed] [Google Scholar]
- 15. Nissen MH, Roepstorff P, Thim L et al Limited proteolysis of beta 2‐microglobulin at Lys‐58 by complement component C1s. Eur J Biochem 1990; 189:423–9. [DOI] [PubMed] [Google Scholar]
- 16. Busby WH Jr, Nam TJ, Moralez A et al The complement component C1s is the protease that accounts for cleavage of insulin‐like growth factor‐binding protein‐5 in fibroblast medium. J Biol Chem 2000; 275:37638–44. [DOI] [PubMed] [Google Scholar]
- 17. Kerr FK, O'Brien G, Quinsey NS et al Elucidation of the substrate specificity of the C1s protease of the classical complement pathway. J Biol Chem 2005; 280:395. [DOI] [PubMed] [Google Scholar]
- 18. Naito AT, Sumida T, Nomura S et al Complement C1q activates canonical Wnt signaling and promotes aging‐related phenotypes. Cell 2012; 149:1298–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cai Y, Teo BH, Yeo JG, Lu J. C1q protein binds to the apoptotic nucleolus and causes C1 protease degradation of nucleolar proteins. J Biol Chem 2015; 290:22570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeo JG, Leong J, Arkachaisri T et al Proteolytic inactivation of nuclear alarmin high‐mobility group box 1 by complement protease C1s during apoptosis. Cell Death Discov 2016; 2:16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Riihilä PM, Nissinen LM, Ala‐aho R et al Complement factor H: a biomarker for progression of cutaneous squamous cell carcinoma. J Invest Dermatol 2014; 134:498–506. [DOI] [PubMed] [Google Scholar]
- 22. Riihilä P, Nissinen L, Farshchian M et al Complement factor I promotes progression of cutaneous squamous cell carcinoma. J Invest Dermatol 2015; 135:579–88. [DOI] [PubMed] [Google Scholar]
- 23. Riihilä P, Nissinen L, Farshchian M et al Complement component C3 and complement factor B promote growth of cutaneous squamous cell carcinoma. Am J Pathol 2017; 187:1186–97. [DOI] [PubMed] [Google Scholar]
- 24. Farshchian M, Nissinen L, Grénman R, Kähäri VM. Dasatinib promotes apoptosis of cutaneous squamous carcinoma cells by regulating activation of ERK1/2. Exp Dermatol 2017; 26:89–92. [DOI] [PubMed] [Google Scholar]
- 25. Farshchian M, Kivisaari A, Ala‐aho R et al Serpin peptidase inhibitor clade A member 1 (SerpinA1) is a novel biomarker for progression of cutaneous squamous cell carcinoma. Am J Pathol 2011; 179:1110–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kivisaari AK, Kallajoki M, Mirtti T et al Transformation‐specific matrix metalloproteinases (MMP)‐7 and MMP‐13 are expressed by tumour cells in epidermolysis bullosa‐associated squamous cell carcinomas. Br J Dermatol 2008; 158:778–85. [DOI] [PubMed] [Google Scholar]
- 27. Kivisaari AK, Kallajoki M, Ala‐aho R et al Matrix metalloproteinase‐7 activates heparin‐binding epidermal growth factor‐like growth factor in cutaneous squamous cell carcinoma. Br J Dermatol 2010; 163:726–35. [DOI] [PubMed] [Google Scholar]
- 28. Agrawal A, Shrive AK, Greenhough TJ, Volanakis JE. Topology and structure of the C1q‐binding site on C‐reactive protein. J Immunol 2001; 166:3998–4004. [DOI] [PubMed] [Google Scholar]
- 29. Hicks PS, Saunero‐Nava L, Du Clos TW, Mold C. Serum amyloid P component binds to histones and activates the classical complement pathway. J Immunol 1992; 149:3689–94. [PubMed] [Google Scholar]
- 30. Navratil JS, Watkins SC, Wisnieski JJ, Ahearn JM. The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. J Immunol 2001; 166:3231–9. [DOI] [PubMed] [Google Scholar]
- 31. Afshar‐Kharghan V. The role of the complement system in cancer. J Clin Invest 2017; 127:780–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reis ES, Mastellos DC, Ricklin D et al Complement in cancer: untangling an intricate relationship. Nat Rev Immunol 2018; 18:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hajishengallis G, Reis ES, Mastellos DC et al Novel mechanisms and functions of complement. Nat Immunol 2017; 18:1288–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kourtzelis I, Stavros R. The dual role of complement in cancer and its implication in anti‐tumor therapy. Ann Transl Med 2016; 4:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nissinen L, Farshchian M, Riihilä P, Kähäri VM. New perspectives on role of tumor microenvironment in progression of cutaneous squamous cell carcinoma. Cell Tissue Res 2016; 365:691–702. [DOI] [PubMed] [Google Scholar]
- 36. Cho MS, Vasquez HG, Rupaimoole R et al Autocrine effects of tumor‐derived complement. Cell Rep 2014; 6:1085–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boire A, Zou Y, Shieh J et al Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Cell 2017; 168:1101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bulla R, Tripodo C, Rami D et al C1q acts in the tumour microenvironment as a cancer‐promoting factor independently of complement activation. Nat Commun 2016; 7:10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petry F, Botto M, Holtappels R et al Reconstitution of the complement function in C1q‐deficient (C1qa/) mice with wild‐type bone marrow cells. J Immunol 2001; 167:4033–7. [DOI] [PubMed] [Google Scholar]
- 40. Bossi F, Tripodo C, Rizzi L et al C1q as a unique player in angiogenesis with therapeutic implication in wound healing. Proc Natl Acad Sci U S A 2014; 111:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakiyama H, Inaba N, Toyoguchi T et al Immunolocalization of complement C1s and matrix metalloproteinase 9 (92kDa gelatinase/type IV collagenase) in the primary ossification center of the human femur. Cell Tissue Res 1994; 277:239–45. [PubMed] [Google Scholar]
- 42. Väyrynen O, Åström P, Nyberg P et al Matrix metalloproteinase 9 inhibits the motility of highly aggressive HSC‐3 oral squamous cell carcinoma cells. Exp Cell Res 2019; 376:18–26. [DOI] [PubMed] [Google Scholar]
- 43. Khan MA, Assiri AM, Broering DC. Complement and macrophage crosstalk during process of angiogenesis in tumor progression. J Biomed Sci 2015; 22:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ajona D, Pajares MJ, Corrales L et al Investigation of complement activation product C4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst 2013; 105:1385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nunez‐Cruz S, Gimotty PA, Guerra MW et al Genetic and pharmacologic inhibition of complement impairs endothelial cell function and ablates ovarian cancer neovascularization. Neoplasia 2012; 11:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lucas SD, Karlsson‐Parra A, Nilsson B et al Tumor‐specific deposition of immunoglobulin G and complement in papillary thyroid carcinoma. Hum Pathol 1996; 27:1329–35. [DOI] [PubMed] [Google Scholar]
- 47. Markiewski MM, DeAngelis RA, Benencia F et al Modulation of the antitumor immune response by complement. Nat Immunol 2008; 9:1225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ajona D, Pajares MJ, Chiara MD et al Complement activation product C4d in oral and oropharyngeal squamous cell carcinoma. Oral Dis 2015; 21:899–904. [DOI] [PubMed] [Google Scholar]
- 49. Bu X, Zheng Z, Wang C, Yu Y. Significance of C4d deposition in the follicular lymphoma and MALT lymphoma and their relationship with follicular dendritic cells. Pathol Res Pract 2007; 203:163–7. [DOI] [PubMed] [Google Scholar]
- 50. Mäkelä K, Helén P, Haapasalo H, Paavonen T. Complement activation in astrocytomas: deposition of C4d and patient outcome. BMC Cancer 2012; 12:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rutkowski MJ, Sughrue ME, Kane AJ et al The complement cascade as a mediator of tissue growth and regeneration. Inflamm Res 2010; 59:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Corrales L, Ajona D, Rafail S et al Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J Immunol 2012; 189:4674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gunn L, Ding C, Liu M et al Opposing roles for complement component C5a in tumor progression and the tumor microenvironment. J Immunol 2012; 189:2985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mamidi S, Höne S, Kirschfink M. The complement system in cancer: ambivalence between tumour destruction and promotion. Immunobiology 2017; 222:45–54. [DOI] [PubMed] [Google Scholar]
- 55. Migden MR, Rischin D, Schmults CD et al PD‐1 blockade with cemiplimab in advanced cutaneous squamous cell carcinoma. N Engl J Med 2018; 379:341–51. [DOI] [PubMed] [Google Scholar]
- 56. Gallenkamp J, Spanier G, Wörle E et al Novel multiplex detection array revealed systemic complement activation in oral squamous cell carcinoma. Oncotarget 2017; 9:3001–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harris CL. Expanding horizons in complement drug discovery: challenges and emerging strategies. Semin Immunopathol 2018; 40:125–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary materials and methods.
Appendix S2 Supplementary references.
Fig S1. Expression of C1QA, C1QB, C1QC variant 1 and C1QC variant 2 in cutaneous squamous cell carcinoma cells and normal human epidermal keratinocytes.
Fig S2. Knockdown of C1r and C1s inhibits growth of cutaneous squamous cell carcinoma cells.
Fig S3. Knockdown of C1r or C1s had no marked effect on the growth of normal human epidermal keratinocytes.
Fig S4. Knockdown of C1r and C1s inhibits viability of cutaneous squamous cell carcinoma cells.
Fig S5. Knockdown of C1r and C1s promotes apoptosis of cutaneous squamous cell carcinoma cells.
Fig S6. Knockdown of C1r and C1s upregulates the expression of matrix metalloproteinase‐9 in UT‐SCC‐7 cells.
Table S1 Primer and probe sequences used for quantitation of the mRNAs of human C1R, C1S, C1QA, C1QB, C1QC transcript variant 1, C1QC transcript variant 2 and β‐actin, with quantitative real‐time polymerase chain reaction.


