Abstract
Significant efforts are necessary to introduce new dietary protein sources to feed a growing world population while maintaining food supply chain sustainability. Such a sustainable protein transition includes the use of highly modified proteins from side streams or the introduction of new protein sources that may lead to increased clinically relevant allergic sensitization. With food allergy being a major health problem of increasing concern, understanding the potential allergenicity of new or modified proteins is crucial to ensure public health protection. The best predictive risk assessment methods currently relied on are in vivo models, making the choice of endpoint parameters a key element in evaluating the sensitizing capacity of novel proteins. Here, we provide a comprehensive overview of the most frequently used in vivo and ex vivo endpoints in murine food allergy models, addressing their strengths and limitations for assessing sensitization risks. For optimal laboratory‐to‐laboratory reproducibility and reliable use of predictive tests for protein risk assessment, it is important that researchers maintain and apply the same relevant parameters and procedures. Thus, there is an urgent need for a consensus on key food allergy parameters to be applied in future food allergy research in synergy between both knowledge institutes and clinicians.
Keywords: animal models, biomarkers, food allergy, prevention
Abbreviations
- Ag
allergen
- ALA
alpha‐lactalbumin
- BAT
basophil activation test
- BLG
beta‐lactoglobulin
- BN
Brown Norway
- DC
dendritic cell
- e.c.
epicutaneously
- ELISA
enzyme‐linked immunosorbent assay
- GM‐CSF
granulocyte‐macrophage colony‐stimulating factor
- i.c.
intracutaneous
- i.d.
intradermally
- i.g.
intragastric
- i.n.
intranasal
- i.p.
intraperitoneally
- Ig
immunoglobulin
- ILC
innate lymphoid cell
- i.v.
intravenous
- KSCN
potassium thiocyanate
- NA
not analysed
- NKT
natural killer T cell
- OVA
ovalbumin
- PCR
polymerase chain reaction
- RBL
rat basophil leukemia
- s.c.
subcutaneously
- SPT
skin prick test
- Treg
regulatory T cell
- TSLP
thymic stromal lymphopoietin
- WPC
whey protein concentrate
- WPH
whey protein hydrolysate
1. INTRODUCTION
A variety of in vitro and in vivo models have been developed that address the factors and mechanisms involved in the sensitization to food proteins.1, 2, 3, 4 Currently, approaches are being developed using protein chemistry and in vitro and in silico methods to characterize food proteins and derivatives that arise during product processing and reformulation, which may explain why certain food proteins induce sensitization of the immune system, while others are tolerated.5, 6 However, elucidating the mechanisms underlying allergen sensitization is a complex, multidimensional problem that often requires a wide range of additional in vivo and ex vivo experimentation,5 as a wide range of molecules, tissues, and cells play a role in the mechanisms underlying food allergen sensitization.1 For instance, epithelial release of thymic stromal lymphopoietin (TSLP), granulocyte‐macrophage colony‐stimulating factor (GM‐CSF), IL‐25, and IL‐33 upon local epithelial stress support type 2 helper T (Th2) cell pathology by attracting IL‐4 secreting lymphoid cells, basophils, and invariant natural killer T (iNKT) cells.7 Il‐4 promotes surface expression of Th2‐costimulatory molecule OX40 ligand on dendritic cells (DCs) 8 and cytokine secretion by Th2 lymphoid cells (ILC2s), which further augments DC activity and suppresses allergen‐specific regulatory T (Treg) cells.9, 10 This complexity, as depicted in Figure 1, illustrates the need for experimental food allergy models that integrate such complex cell‐tissue communication to assess the sensitization potential of new protein sources. Murine food allergy models, even though they have their limitations, are currently the best predictive models available to evaluate the food‐sensitizing capacity of new food proteins before introducing them into the human diet. Although researchers aim to reduce the use of experimental animals to address the 3R principle that guides animal experimentation to replace (alternative model), reduce (minimize number of animals), and refine (minimize animal pain and enhance animal welfare), there is a lack of replacement models such as in silico prediction models, in vitro primary cell assays, or tissue explants assays that are able to characterize and predict the human responses to food proteins. In the past, numerous experimental food allergy models have been developed to assess food allergenicity. However, interlaboratory differences in the models used with respect to sensitization and elicitation route, choice of adjuvant, clinical signs, genetic background of the animals, housing conditions, and microbiome composition and metabolic activity in the different vivaria often make it difficult to draw generalized conclusions.5 It is important to note that almost all models (except genetic models) require adjuvants to trigger sensitization. Therefore, the choice of the adjuvants together with the exposure route are crucial points to consider. In addition, there are numerous in vivo, ex vivo, and in vitro parameters evaluated for the assessment of food allergy. Figure 2 illustrates the types of in vivo (inside a living organism) or ex vivo (outside an organism) methodology and endpoints used in experimental murine models of food allergy. However, there is a need to establish a list of reliable, validated, and effective endpoint parameters to guide researchers working with animal models of food allergy. In this review, we describe a selective list of the most commonly used experimental applied endpoints in food allergies with a focus on milk, egg, and peanut allergens and critically evaluate their applicability for evaluating sensitization potency. Each endpoint was selected and critically described with strengths and limitation based on consortium experience and occurrence in literature.
Figure 1.
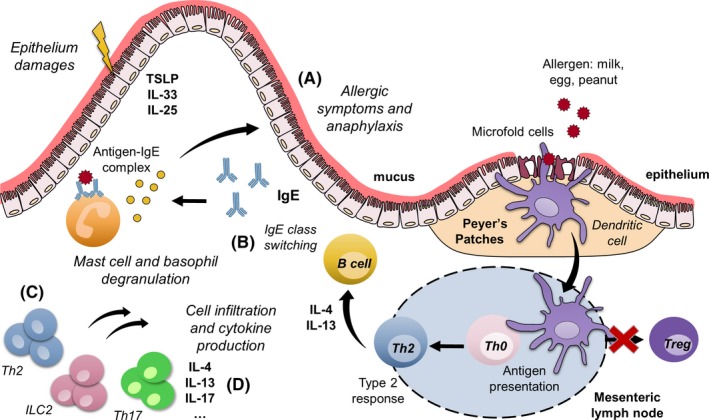
Immune mechanisms of food allergy and its associated principal measured endpoints. A, Assessment of allergic symptoms (body temperature) after allergen challenge. B, Evaluation of immunoglobulin (IgE) in serum. C, Phenotyping of T‐cell population. D, Cytokine production in response to allergen restimulation (ex vivo assay)
Figure 2.
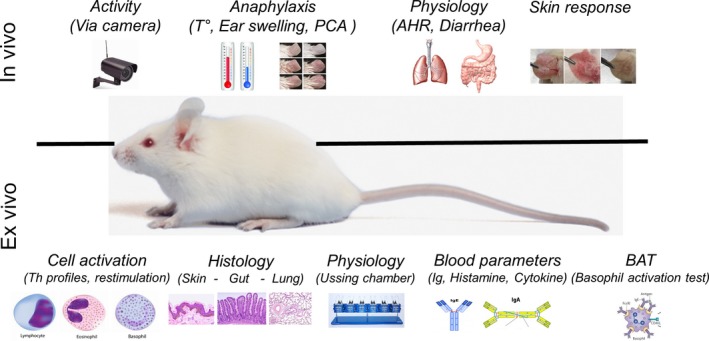
In vivo and ex vivo methodological endpoints used in murine food allergy models
2. MEASUREMENT OF BODY TEMPERATURE
In murine‐type models of food allergy to milk, eggs, and peanuts, a drop in the core body temperature is often observed after repetitive allergen challenge. This change in body temperature is an indicator of anaphylaxis (Table 1). Temperature is measured before and 30 minutes to 1 hour after allergen challenge, but this parameter can also be monitored over time.21, 22 Animals sensitized to a given food matrix or protein may display a significant reduction in body temperature (0.5‐10°C)3, 4 compared with that of naive animals. For an adequate level of sensitivity, 5‐16 animals per group should have their temperatures measured using a rectally inserted thermal probe,29 but it is also possible to measure changes over time for individual animals using an electronic ID transponder implanted subcutaneously.11, 12 To refine, improve, and objectify the currently applied manual monitoring methods, an automatic imaging method has been developed.14 It involves a noninvasive measurement of the whole‐body surface temperature paired with assessment of activity (see also Data S1 about activity/behavior via camera). Anaphylaxis imaging has been used in three in vivo allergy mouse models for (a) milk allergy, (b) egg allergy, and (c) peanut allergy in proof‐of‐principle experiments and suggests that imaging technology represents a reliable noninvasive method for objective monitoring of small animals during anaphylaxis over time. This method can be useful for monitoring diseases associated with changes in both body temperature and physical behavior.
Table 1.
Table of studies measuring body temperature in allergic reactions
| Mouse/rat model | Allergen | Number of animals | δT° | Therapeutic or preventive strategy | Conditions of measure | System of measurement | Ref. |
|---|---|---|---|---|---|---|---|
| C3H/HeOuJ | Whey | N = 6‐12 | 4°C | Prevention with omega‐3 long chain polyunsaturated fatty acids | 1 h after challenge | Implantable electronic ID transponder | 11 |
| C3H/HeOuJ | OVA | N = 4‐6 | 5°C | Prevention with prebiotics: scGOS/lcFOS/pAOS | 30 min after challenge | Implantable electronic ID transponder | 12 |
| BALB/c | Beta‐lact | N = 10 | 1.7°C | Prevention with ratios of omega‐6 and omega‐3 fatty acids | Before, 30 and 45 min after challenge | Rectal probe | 13 |
| BALB/c | Peanut, egg, milk | N = 3‐5 | 3°C | Anaphylaxis imaging | Monitoring after challenge | Imaging method for whole‐body surface temperature | 14 |
| C3H/HeN | Whey | N = 12‐15 | 4°C | Microbiota composition and allergy protection | Before and 45 min after challenge | Rectal probe | 15 |
| BN | OVA | N = 8 | 3°C | To develop an effective and rapid model of FA in Brown Norway rats | 60, 90 and 120 min after challenge | Rectal probe | 16 |
| BALB/c | BLG | N = 5 | 5°C | Nitration process of allergen | Before and 15 and 30 min after iv | Rectal probe | 17 |
| BALB/c | OVA | N = 10 | 0.5°C | Microbiota composition and allergy protection | Before and 5 and 10 min after iv OVA challenge | Rectal probe | 18 |
| BALB/c | OVA | N = 5 | 1.5°C | Anti‐acid medication for risk of food allergy | Before and 15 min after challenge | Rectal probe | 19 |
| BALB/c | OVA | N = 10 | None | Heat process on allergen | 30 min after challenge | Rectal probe | 20 |
| C3H/HeOuJ | Whey | N = 10 | 2‐10°C | Hydrolyze process on allergen | Measurement of temperature over time: 0, 15, 30, 60, 120 min after challenge | A programmable temperature transponder implanted subcutaneously | 21 |
| C3H/HeOuJ | Caseinate | N = 6 | 5°C | Transglutaminase cross‐linked caseinate process | Measurement of temperature over time: 0, 15, 30, 60, after challenge | A programmable temperature transponder implanted subcutaneously | 22 |
| BALB/cTac | OVA | N = 5‐10 | 5°C | Microbiota signature in allergy | Measurement every 5 min: 5‐60 min | Rectal probe coupled to a Physitemp Thermalert Model TH‐5 | 23 |
| BALB/c | OVA | N = 10 | 1°C | To develop models of food allergy and oral tolerance | 30 min after challenge | Rectal probe | 24 |
| C57BL/6 | Peanut | N = 4‐10 | 2°C | Commensal bacteria for allergy protection | After challenge | Rectal probe | 25 |
| C3H/HeJ and BALB/c | Peanut | N = 5 | 3‐5°C | Skin sensitization study | 30 min after challenge | Rectal thermometer (WPI Instruments) | 26 |
| C3H/HeJ and BALB/c | OVA or peanut | N = 5 | 4°C | Epicutaneous immunotherapy | 30 min after challenge | Rectal thermometer (WPI Instruments) | 27 |
| C3H/HeJ | BLG | N = 10 | 3°C | Pasteurization process on milk allergen | After challenge | Rectal probe | 28 |
2.1. Strengths
The measurement of core body temperature is a cost‐effective, reliable assessment of the allergic reaction.
Therapeutic or preventative strategies for the reduction of allergic reactions can be easily evaluated.
Can be used to evaluate the severity of allergic shock and differences between allergens subjected to physical transformations (ie, native versus processed).
2.2. Limitations
The occurrence of anaphylaxis is dependent on the mouse strain used: Balbc or C3H mice are prone to develop anaphylaxis, whereas C57BL/6 or A/J mice necessitate stringent exposure protocols to achieve sensitization.
The clinical score may be biased as a consequence of the laboratory environment, stress level, animal strain, and technical experimenter.
A decrease in temperature is only observed after a food/allergen challenge after a previous sensitization event; this endpoint therefore contains no predictive value for the sensitization potential of a food protein.
2.3. Technical recommendations
Using a rectal probe, mice or rats must be acclimated to the experimental room at least 1 hour before starting the temperature measurements to obtain stable values.
The rectal temperature must be evaluated 10 minutes to 1.5 hours after the challenge.
The animal temperature can be registered over time using a programmable temperature transponder implanted subcutaneously.
3. EVALUATION OF IMMUNOGLOBULINS IN SERUM
While in vivo measurements are essential to assess the elicitation of an allergic response, they do not provide insight into de novo allergen sensitization. Therefore, blood, tissue, or organs must be collected and further analyzed by ex vivo methods. Serum immunoglobulin (Ig) content is the most common parameter measured when evaluating sensitization to food allergens in animal models, followed by fecal IgA (see Data S1), as antibody responses are considered a direct indicator of allergen sensitization together with mast cell and basophil degranulation. IgE is the most common Ig isotype measured when evaluating the allergenicity of food proteins and is regularly quantified in parallel with IgG1 (Table 2). Total and antigen‐specific Ig levels can be analyzed, where the latter is a measure of how dosing with a given food or protein influences the overall level of IgE or IgG. Serum‐specific IgE and IgG can be quantified by a series of different ex vivo methods, where ELISAs are the most commonly applied, followed by immunoblotting methods and mediator release assays (Figure 3). Whereas specific IgG in general is measured by means of an indirect ELISA,43 specific IgE is most often measured by antibody‐capture ELISA.44 In fact, IgE is the least abundant Ig isotype in serum (with an approximate amount of only one IgE for every 50 000 IgGs45), making it difficult for IgE to compete for binding to proteins coated on ELISA plates. Other methods of measuring specific IgE include enzyme allergosorbent test (EAST) immunoblotting.34 When measuring specific IgEs by means of in‐house‐developed antibody‐capture ELISAs, there is a need for coupling the protein of interest to a molecule against which labeled secondary Igs are commercially available, as secondary Igs for direct binding to the proteins of interest can rarely be purchased. Molecules coupled to the protein of interest are most often digoxigenin (DIG)43 or biotin,30 with the additional advantage that they serve as signal amplifiers (Figure 3). Not only is the total level of specific Igs of interest in evaluating the sensitization response in animal models, the increase in affinity between Igs and the allergen is also important. Studies have shown that the binding strength between specific IgEs and the corresponding allergens is of great importance for the induction of a degranulation response and thereby the severity of the allergic disease.46, 47 The avidity can be measured by means of simple potassium thiocyanate (KSCN) ELISAs which have shown that no general relationship exists between the level and avidity of specific Igs,48, 49 though a correlation may be observed during a multiple antigen exposure immune responses. This method, although not very sensitive, is based on the ratio of the areas derived from the curves obtained by plotting the OD and log of the sera dilution in the ELISA experiment with and without thiocyanate treatment. Where measures of specific IgE only allow for evaluation of sensitization, they provide no indication of the biological relevance of the IgEs present in the serum and thereby the clinical relevance of the food allergy model. To provide insights into the biological relevance of secreted IgEs, functional tests should be performed, such as the in vivo temperature drop, a skin prick test (SPT), or evaluation of challenge‐derived symptoms. Further, ex vivo mediator release tests such as the rat basophilic leukemia (RBL) assay and basophil activation test (BAT) enable an evaluation of the biological relevance of the IgE raised in food allergy animal models (see Data S1 for description and opinion about mediator release assays and additional passive cutaneous anaphylaxis (PCA) and active cutaneous anaphylaxis (ACA) models).
Table 2.
Studies using measurements of Igs from serum
| Mouse/rat model | Allergen | Sensitization and challenge | Ig measured in serum | Aim of the study | Ref. |
|---|---|---|---|---|---|
| BALB/c | OVA | I.g. + CT followed by i.g. challenge |
IgG1, IgG2a: indirect ELISA IgE: Ab‐capture ELISA |
To elucidate the class of bioactive polyphenols that exhibit a beneficial anti‐allergic effect and to assess whether the protective effect matches the in vivo bioavailable metabolite concentrations | 30 |
| BALB/c | OVA, | I.p. followed by i.g. | IgG1, IgG2a, IgA: indirect ELISA | To investigate how thermal processing influences the ability of ovalbumin (OVA) to induce allergic symptoms and immune responses in a mouse model of food allergy | 20 |
| C57BL/6J | OVA | Oral (by feeding) + s.c. + alum followed by i.d. (ear test) challenge |
IgG1: indirect ELISA IgE: Ab‐capture ELISA |
To investigate the potential of hydrolyzed egg and whole egg to induce tolerance by the oral route (ie, by feeding) | 31 |
| BN | OVA | I.p. + alum + tBp followed by i.g. challenge |
IgG1, IgG2a, IgG2b, IgA: indirect ELISA IgE: Ab‐capture ELISA |
To develop an effective and rapid model of food allergy with only one i.p. injection of the allergen with alum together with toxin from Bordetella pertussis (tBp) to promote IgE production and 2 wk later the i.g. administration of allergens | 16 |
| BALB/c | WPC, WPH, or BLG | I.p. with BLG + alum followed by oral (solution in drinking bottle) challenge with WPC and WPH | IgE: Ab‐capture ELISA | To develop an experimental murine model of food allergy to the cow's protein ß‐lactoglobulin (BLG) that mimics the main clinical characteristics of human disease as well as to examine the allergenic and immunological properties of extensively hydrolyzed whey proteins | 32 |
| BALB/c | BLG | I.p. + alum or FCA or FIA | IgG1, IgG2a, IgE: indirect ELISA | To study the sensitizing capacity of BLG and the influence of the use of adjuvant. | 33 |
| BALB/c | Whole milk | I.g. +/− CT followed by i.g. challenge |
IgG1, IgG2a: indirect ELISA IgE: EAST (enzyme allergosorbent test) |
To map epitopes of the major soybean allergen Gly m 5 that are corecognized by casein‐specific antibodies and to identify a peptide responsible for the cross‐reactivity | 34 |
|
C3H HeOuJ |
WPC, ALA, BLG | I.g. + CT | IgG1, IgE: indirect ELISA | To test a panel of high and low allergenic proteins | 35 |
|
C3H HeOuJ PF |
WPC or WPH | I.g. + CT followed by i.g. + i.d. challenge | IgG1, IgE: indirect ELISA | To validate a mouse model for cow's milk allergy to assess the potential allergenicity of hydrolyzed cow's milk‐based infant formulas | 21 |
| BN | WPH, BLG | I.p. + alum |
IgG2a, IgE: indirect ELISA IgG1: inhibitory ELISA |
To provide a thorough analysis of the immunogenicity and allergenicity of hydrolyzed cow's milk proteins for use in infant formulas | 36 |
| BALB/c | Extract | I.c. + CpG + CT or + non/CpG + CT followed by i.g. + CT challenge | IgG1, IgG2a, IgE, IgA: indirect ELISA | To evaluate the effect of the application of peanut extract (PE) alone or mixed with CT and unmethylated sequences (CpG) as adjuvant on the intact skin | 37 |
| BALB/cJ | Roasted extract or Ara h 1 | I.n. or e.c. followed by i.g. + CT challenge |
IgG1: indirect ELISA IgE: Ab‐capture ELISA |
To assess the impact of repeated short‐term epicutaneous (e.c.) applications on intact skin or after repeated intranasal (i.n). administration of food allergens from roasted peanut | 38 |
| BALB/c | Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 6 with no S‐S bridges | I.p. + alum | IgG1, IgE: indirect ELISA (protein G for IgG removal) | To investigate the impact of heat processing of peanut seed on the sensitization to native Ara h 6 | 39 |
| C3H/HeJ | Extract | I.g. + CT followed by i.p. challenge |
IgG1, IgG2a: indirect ELISA IgE, IgA: Ab‐capture ELISA |
To reveal the immune responses that are induced against peanuts allergens during sensitization, including the very early responses | 40 |
| C3H/HeJ | Whole peanut | I.g. + CT followed by i.g. challenge | IgE: indirect ELISA | To develop a murine model of IgE‐mediated peanut allergy that closely mimics human peanut allergy | 41 |
| BN | Ara h 1 | I.p. |
IgG1, IgG2a: indirect ELISA and inhibitory ELISA for IgG1 IgE: Ab‐capture ELISA Total IgE: sandwich ELISA |
To study the sensitizing capacity of four different 7S proteins and to determine whether related proteins would induce similar sensitization when removed from their “normal” matrix | 42 |
| BN | Ara h 1 | I.p. |
IgG1, IgG2a: indirect ELISA IgE: Ab‐capture ELISA |
To investigate the ability of digested protein—Ara h 1 to sensitize | 43 |
Figure 3.
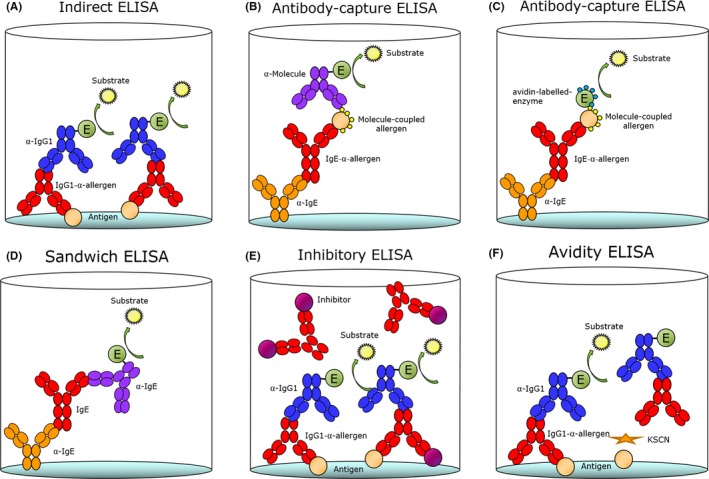
ELISA methods. Antibodies (Abs) can be evaluated by means of different ELISA methods for assessment of their amount, specificity, and avidity. Specific IgG1 Abs are most often analyzed by means of an indirect ELISA (A), while specific IgE is most often analyzed by means of an Ab‐capture ELISA (B, C). Total IgG1 and total IgE are analyzed by a sandwich ELISA (D). Furthermore, the specific Ab responses can also be evaluated for specificity with an inhibitory ELISA (E) or for binding strength with an avidity ELISA (F)
3.1. Strengths
Specific IgE antibody analysis is the most trustworthy measure of sensitization.
Measures of specific IgE antibodies are often used to evaluate not only sensitization but also the potential severity of the allergic reaction after a second encounter.
Measurements of antibodies can be performed without the use of advanced equipment such as a cytometer or robotics.
3.2. Limitations
Assays often need to be developed in‐house, restricting the possibilities for comparison between laboratories.
IgE only accounts for a fraction of all serum antibodies, requiring more advanced ELISAs for analysis of specific IgE.
IgE levels do not predict the clinical severity of a food allergy model, and other ex vitro experiments are needed to further address this parameter.
Measures with optical density (OD) as the unit only allow for one serum dilution.
3.3. Technical recommendations
Antibody‐capture ELISAs should be used for the measurement of specific IgE.
Other antibody parameters in addition to the amount of total and specific antibodies are relevant and should be measured, such as clonality and avidity.
Measures of total and specific antibodies should always be expressed as titer values or as concentrations with no upper or lower limit for dilutions.
Serum depleted of IgG using protein G columns before use in indirect ELISAs needs to be considered.
4. PHENOTYPING OF T‐CELL POPULATIONS
Assessment of serum Ig levels provides important information about the sensitization phase but does not allow for quantification of immune cell responses, including cellular infiltration to sites of allergic inflammation. The phenotyping of innate (eg, macrophages, eosinophils, basophils, neutrophils, dendritic cells) and adaptive (B and T cells) responses is indispensable for assessing the mechanisms of allergic sensitization (Table 3). Immune cells are generally isolated from organs, including the mesenteric lymph nodes, spleen, lung, skin, or intestine, and analyzed by flow cytometry. Typically, allergic inflammation is characterized by a predominantly type 2 immune response and secretion of the canonical type 2 cytokines IL‐4, IL‐5, IL‐9, and IL‐13 by innate immune cells (eg, eosinophils, basophils, mast cells (MCs), type 2 innate lymphoid cells, and polarized Th2 cells).44, 45 Indeed, in mice specifically expressing the ovalbumin T‐cell receptor, sensitization to ovalbumin in their diet induced the expansion of IL‐4‐producing CD4+ T cells in mesenteric lymph nodes, the spleen, and Peyer's patches.60 Importantly, adoptive transfer of antigen‐specific CD4+ T cells derived from mesenteric lymph nodes of OVA‐sensitized mice is sufficient to transfer allergen‐induced diarrhea to naïve recipients. The recipient mice also display an upregulation of the Th2‐related chemokines CCL17 and CCL22 in the small intestine.61 In addition to polarized Th2 responses, the proportion of other common T‐cell subtypes, such as Th1 and Th17 that are characterized by the production of IFN‐γ and IL‐17, respectively, can also be elevated in lymphoid organs of allergic mice. In contrast, expansion and/or the regulatory capacity of CD25+ Foxp3+ T cells associated with tolerance are often compromised in many food allergy models.62 Additionally, other T‐cell subtypes can be involved in food allergy pathogenesis. The recently discovered Th9 subset and associated IL‐9 secretion were found to be involved in food allergy and especially in peanut allergies.63 IL‐9 is mainly responsible for the production of IL‐4 by Th2 cells to promote mucosal mast cell accumulation and secretion of mucus and chemokines by epithelial cells to sustain allergic inflammation.64 To a lesser extent, γδT cells found in the intestinal epithelium and in the lamina propria were also shown to be involved in food allergy. These cells are involved in blocking the induction of tolerance and modulating inflammatory responses.65,66
Table 3.
Studies using immune infiltrate as readout of allergic inflammation to egg, milk, and peanut proteins
| Mouse/rat Model | Allergen | Allergen sensitization | Allergen challenge | Immune infiltrate | Method | Therapeutic/preventive strategy | Ref. |
|---|---|---|---|---|---|---|---|
| C57BL/6 or BALB/c | OVA, crude peanut extract | Skin | Intragastric | Mast cells, eosinophils | Flow cytometry for intestinal eosinophils/mast cells, chloroacetate esterase staining for mast cells in jejunum, H&E staining | Anti‐TSLP treatment or basophil depletion limits food allergen sensitization and the development of intestinal food allergy | 50 |
| C57BL/6 | OVA | Skin | Intragastric | Mast cells, eosinophils | Flow cytometry for intestinal eosinophils/mast cells, chloroacetate esterase staining for mast cells in jejunum, H&E staining | Targeting basophil‐derived IL‐4 reduces food allergen sensitization and limits intestinal food allergy | 7 |
| BALB/c | Raw or roasted peanut extracts | Skin | Intragastric | Eosinophils | Flow cytometry for eosinophils in the small intestinal lamina propria; H&E staining jejunum | NA | 51 |
| BALB/c | OVA | Skin | Intragastric | Eosinophils | Flow cytometry for peripheral eosinophils; H&E staining jejunum | Basophil depletion attenuates intestinal allergy; CD4 T‐cell depletion limits TSLP‐mediated intestinal food allergy | 52 |
| BALB/c | OVA | Systemic | Intragastric | Mast cells, CD4 T cells | H&E staining jejunum, chloroacetate esterase staining of mast cells in the jejunum; flow cytometry of CD4 + T cells in the small intestinal lamina propria | Treatment with mast cell stabilizing cromolyn sodium protects against food allergen sensitization | 53 |
| BALB/c | OVA | Skin | Intragastric | Mast cells | Chloroacetate esterase staining of connective tissue mast cells in the jejunum | Targeting of IgE responses prevented intestinal mast cell expansion and anaphylaxis | 54 |
| Various | OVA | Intestine | Intragastric | Allergen‐specific Tregs that acquire Th2 mast cells | Flow cytometry of small intestinal Foxp3 + Tregs; chloroacetate esterase staining of connective tissue mast cells in the jejunum | NA | 55 |
| BALB/c | OVA; whole peanut extract | Intestine | Intragastric | Mast cells; eosinophils | H&E staining jejunum; pinacyanol erythrosine staining to determine mast cell numbers and granulation status in the jejunum | NA | 56 |
| BALB/c | OVA | Skin | Intragastric | Mast cells | Chloroacetate esterase staining of mast cells in the jejunum | ST2 blockade attenuates food‐induced anaphylaxis | 57 |
| C3H/HeJ; BALB/c | OVA; ground peanut | Skin; systemic | Intragastric | Lap + Tregs | Flow cytometry for Lap + Tregs in the lamina propria | Clinical protection induced by epicutaneous immunotherapy (EPIT) | 58 |
| BALB/c | BLG | Intestine | Intragastric | Lamina propria lymphocytes | ELISPOT IL‐12, IL‐17 producing lymphocytes from the intestinal lamina propria | Blocking TSLP signaling prevents food allergy | 59 |
4.1. Strengths
Precise mechanistic insights into the cellular response in isolated organs and tissues support the sensitizing potential of food proteins when combined with additional readouts.
Precise determination of the T‐cell profile by using specific markers of the T‐cell population.
Quantitative evaluation of the infiltrating cell population by flow cytometry.
4.2. Limitations
Analysis of cell populations without the contribution of neighboring cell tissue (loss of microenvironment).
Isolation of immune cells from tissues relies on enzymatic digestion protocols and may thus alter phenotypical and functional properties of the cells of interest.
Difficulty with the separation of minor subpopulations.
Sacrifice of the animal is required for organ and tissue sampling.
Need for sophisticated equipment such as FACS.
Type 2 immune response‐associated mucus production in tissues makes cell isolation difficult and can create bias in cell phenotyping and frequencies.
4.3. Technical recommendations
Remove fat and store organs, tissues and cells at 4°C to avoid uncontrolled cell death or degradation of surface markers.
Perform flow cytometry and culturing the same day as the animal kill.
Phenotyping of T cells can be achieved by intracellular cytokine/transcription factor staining using flow cytometry.
5. CYTOKINE PRODUCTION IN RESPONSE TO ALLERGEN RESTIMULATION
The logical follow‐up to the analysis of infiltration/expansion of innate and adaptive immune cells in the tissues and organs is the evaluation of cytokine secretion. This evaluation comes directly from serum or from lymphatic tissue cells restimulated ex vivo. Food allergen stimulation of only lymphatic tissue cells, or in coculture with dendritic cells, allows for the immunophenotyping of the immune cell populations specific for the exposed food antigen or matrix. To confirm allergen specificity, splenocytes, mesenteric lymph node cells, or lamina propria cells isolated from sensitized and/or challenged mice are restimulated with corresponding allergenic proteins or peptides. After culture for up to 5 days, cytokines associated with the inflammatory response (IL‐4, IL‐5, IL‐13, IL‐17, and IFN‐γ) and the regulatory response (IL‐10 and TGF‐β) are analyzed in the supernatants by ELISA60, 61, 62, 63, 64 or a multiplex system. The cytokine production indicates whether T cells were primed toward the challenged food proteins and distinguishes Th1 or Th2 cell type responses. The prototypical type 2 cytokines include IL‐4, IL‐5, and IL‐13. While IL‐4 is critical for the polarization of Th2 cells and IgE class‐switching in B cells,64 IL‐5 promotes the activation, proliferation, and survival of eosinophils, and IL‐13 induces mucus production from goblet cells. Additional assays may be used including proteomics and gene expression profiling by PCR or microarray technology, that provide mechanistic insights and potential drug targets.
5.1. Strengths
Precise assessment of the allergen specificity by restimulating cells with the same allergen used in the animal model.
Class determination of the T‐cell response by evaluation of cytokine production in the supernatant of sorted T cells.
Higher production of cytokines can be obtained after proliferation and restimulation with the antigen than by direct measurement in serum.
5.2. Limitations
Restimulation with allergens can activate nonspecific T cells due to certain cross‐reactivity.
Difficult to obtain a level above the sensitivity threshold with cells isolated from naïve mice.
Some mechanistic endpoints are not equally important in animals and humans.
5.3. Technical recommendations
For allergen presentation, presorted T cells need to be co‐cultured with dendritic cells.
MHC peptide—tetramers can be used to sort specific T cells and have better assessment of allergen specificity.
Need for positive (polyclonal anti‐CD3/anti‐CD28) and negative control (nonallergen) stimuli to ensure proper T‐cell responsiveness.
Endotoxin levels within the allergen extract need to be controlled to prevent bias in restimulation responses.
Ideally, when using gene expression sequencing data, this method should be confirmed with at least one other technology (eg, flow cytometry).
As cells and mediators associated with immune responses change rapidly, longitudinal assessments of mechanistic endpoints will be more informative than single time point assessments. The timing of the measurements will depend on the research question, for example, sensitization mechanisms vs mechanisms of acute allergic responses following (re)challenge.
6. FUTURE ANALYSIS OF FOOD ALLERGY MODELS
To date, the methods to study intestinal pathophysiology are in vitro culture systems with cell lines or explanted mucosa grown in monolayers,67, 68 intestinal organoid cultures,69, 70 and “gut‐on‐a‐chip” devices.71, 72 These technologies have offered many insights into gut physiology, but they lack cellular complexity, architecture, and immune and inflammatory responses that are crucial for a comprehensive understanding of underlying disease mechanisms and pathways. Alternatively, in vivo animal models provide the intact organ in the context of the vascular supply, systemic mediators, and circulating cells. However, in vivo experiments may be hampered by technical difficulties, including interindividual variability and maintenance of constant and reproducible experimental conditions.5 To address the limitations of in vitro and in vivo models of gut disease, Yissachar et al73 developed a chamber unit for culturing 12‐ to 14‐day‐old mouse colon or small intestine segments under highly controlled conditions. Of particular interest is that the chamber unit has two paired inputs and outputs that allow for controlled introduction of molecules or microbes into the lumen while simultaneously introducing continuous replenishment of medium to support tissue viability. The tissue remains intact, and the overall structure with epithelial cell layers is preserved for at least 24 hours, making this method suitable for studying epithelial transport of food allergens and their effect on epithelial integrity. However, other measurements are currently difficult due to the very short time that such tissue explants can be maintained. Furthermore, the enteric nervous system structure is maintained, and immune cells are detected as they found in healthy intestinal biopsies. It is possible to envisage the use of this type of ex vivo chamber unit in food allergy research by using intestinal fragments from naive, sensitized, and allergic animals to introduce a variety of food proteins. It is thus possible to further elucidate pathways involved in luminal physiology and antigen uptake and presentation and make comparisons between known allergenic and nonallergenic proteins. This approach may lead to novel insights into new proteins and cross‐reactive proteins and to the development of a predictive model for food allergy. Additional studies related to the survival and growth of anaerobic and aerobic microbiota revealed that the ex vivo colonization of cultured tissue with selected microbes may be possible. Indeed, changes in the composition and metabolic activity of gut microbes can influence all aspects of innate and adaptive immune processes within the mucosa (see also Data S1 for stool consistency as a readout in food allergy assessment). Thus, focusing on the effect of diverse microbiota profiles and specific bacteria on immunological responses upon the introduction of allergenic proteins may lead to novel mechanisms, therapeutic targets, or predictive models. However, intra‐ and interlaboratory variability in microbiome composition and metabolic activity after birth as a result of the breeding environment is also a major underlying cause for conflicting results between experiments. This variability must be taken into account beforehand in the experimental design of an animal trial.5 It is also noteworthy to consider the possible development of highly controlled chamber units for food allergy research used in combination with in vivo models to provide a new powerful strategy for studying mechanisms in the intestine.
6.1. Strengths
The tissue structure, cellular components, and neural system are highly preserved.
The model provides the possibility to study immediate responses generated after the introduction of different molecules and microbes.
6.2. Limitations
Only short‐term responses can be evaluated due to changes that can occur in the tissue over time.
Currently, only intestinal segments from 12‐ to 14‐day‐old mice have been tested.
Tissue preparation and assembly require specific skills.
7. CONCLUSION
The recent broadening of our knowledge of food allergy pathogenesis and development of murine food allergy models has enabled us to model the allergic elicitation reaction as well as the preceding sensitization events and observe relevant symptoms with different food proteins (milk, egg, and peanut). The principal endpoint parameters described in this review are critical parameters that should be evaluated in a correct manner so that they may be powerful in the different rodent models.
Characterizing a food allergy model using temperature, level of Igs, phenotyping of the cell infiltrate, and cytokine production gives an overview of the reaction while providing us insight into the degree of sensitizing capacity of the allergen used. Nevertheless, even though the in vivo measurements and the ex vivo experiments provide us with many answers about the immune response and the sensitization phase, we still do not have a complete overview of the immune mechanisms behind each reaction. There is still a strong need to better define the allergic reaction to predict the clinical outcomes of sensitization to novel food proteins. Although the current available models are suitable for studying the pathophysiology of food allergy, they still cannot predict the magnitude of the allergic potential of a particular allergen. Discovering and highlighting the molecules and cells involved in both sensitization and elicitation are necessary to improve risk assessment models and to facilitate the introduction of novel protein sources into our diet with a low risk of allergic sensitization.
CONFLICTS OF INTEREST
Dr Blanchard reports other from Nestec, outside the submitted work; Dr O'Mahony reports personal fees from AHL and grants from GSK, outside the submitted work. The other authors declare no conflict of interest.
Supporting information
ACKNOWLEDGMENT
The authors are all part of the COST Action FA1402 entitled: Improving Allergy Risk Assessment Strategy for New Food Proteins (ImpARAS).
Castan L, Bøgh KL, Maryniak NZ, et al. Overview of in vivo and ex vivo endpoints in murine food allergy models: Suitable for evaluation of the sensitizing capacity of novel proteins? Allergy. 2020;75:289–301. 10.1111/all.13943
Bouchaud and Bastiaan‐Net share senior authorship.
REFERENCES
- 1. Smit JJ, Noti M, O’Mahony L. The use of animal models to discover immunological mechanisms underpinning sensitization to food allergens. Drug Discov Today Dis Model. 2015;17‐18:63‐69. [Google Scholar]
- 2. Hussain M, Epstein MM, Noti M. Experimental food allergy models to study the role of innate immune cells as initiators of allergen‐specific Th2 immune responses. Drug Discov Today Dis Model. 2015;17‐18:55‐62. [Google Scholar]
- 3. Gavrovic‐Jankulovic M, Willemsen L. Epithelial models to study food allergen‐induced barrier disruption and immune activation. Drug Discov Today Dis Model. 2015;17‐18:29‐36. [Google Scholar]
- 4. Cubells‐Baeza N, Verhoeckx K, Larre C, Denery‐Papini S, Gavrovic‐Jankulovic M, Diaz PA. Applicability of epithelial models in protein permeability/transport studies and food allergy. Drug Discov Today Dis Model. 2015;17‐18:13‐21. [Google Scholar]
- 5. Bøgh KL, van Bilsen J, Głogowski R, et al. Current challenges facing the assessment of the allergenic capacity of food allergens in animal models. Clin Transl Allergy. 2016;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mazzucchelli G, Holzhauser T, Cirkovic Velickovic T, et al. Current (Food) allergenic risk assessment: is it fit for novel foods? Status quo and identification of gaps. Mol Nutr Food Res. 2018;62:1700278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hussain M, Borcard L, Walsh KP, et al. Basophil‐derived IL‐4 promotes epicutaneous antigen sensitization concomitant with the development of food allergy. J Allergy Clin Immunol. 2018;141(1):223‐234. [DOI] [PubMed] [Google Scholar]
- 8. Blazquez AB, Berin MC. Gastrointestinal dendritic cells promote Th2 Skewing via OX40L. J Immunol. 2008;180:4441‐4450. [DOI] [PubMed] [Google Scholar]
- 9. Noval Rivas M, Burton OT, Oettgen HC, Chatila T. IL‐4 production by group 2 innate lymphoid cells promotes food allergy by blocking regulatory T‐cell function. J Allergy Clin Immunol. 2016;138(3):801‐811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khodoun MV, Tomar S, Tocker JE, Wang YH, Finkelman FD. Prevention of food allergy development and suppression of established food allergy by neutralization of thymic stromal lymphopoietin, IL‐25, and IL‐33. J Allergy Clin Immunol. 2018;141(1):171‐179. [DOI] [PubMed] [Google Scholar]
- 11. van den Elsen LW, Meulenbroek LA, van Esch BC, et al. CD25+ regulatory T cells transfer n‐3 long chain polyunsaturated fatty acids‐induced tolerance in mice allergic to cow’s milk protein. Allergy. 2013;68:1562‐1570. [DOI] [PubMed] [Google Scholar]
- 12. Hogenkamp A, Knippels L, Garssen J, Van EB. Supplementation of mice with specific nondigestible oligosaccharides during pregnancy or lactation leads to diminished sensitization and allergy in the female. J Nutr. 2015;145:996‐1002. [DOI] [PubMed] [Google Scholar]
- 13. Thang CL, Boye JI, Shi HN, Zhao X. Effects of supplementing different ratios of omega‐3 and omega‐6 fatty acids in western‐style diets on cow’s milk protein allergy in a mouse model. Mol Nutr Food Res. 2013;57:2029‐2038. [DOI] [PubMed] [Google Scholar]
- 14. Manzano‐Szalai K, Pali‐Schöll I, Krishnamurthy D, Stremnitzer C, Flaschberger I, Jensen‐Jarolim E. Anaphylaxis imaging: non‐invasive measurement of surface body temperature and physical activity in small animals. PLoS ONE. 2016;11:e0150819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodriguez B, Prioult G, Hacini‐Rachinel F, et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T‐cell response. FEMS Microbiol Ecol. 2012;79:192‐202. [DOI] [PubMed] [Google Scholar]
- 16. Abril‐Gil M, Garcia‐Just A, Pérez‐Cano FJ, Franch À, Castell M. Development and characterization of an effective food allergy model in Brown Norway rats. PLoS ONE. 2015;10:e0125314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diesner SC, Schultz C, Ackaert C, et al. Nitration of β‐lactoglobulin but not of ovomucoid enhances anaphylactic responses in food allergic mice. PLoS ONE. 2015;10:e0126279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diesner SC, Bergmayr C, Pfitzner B, et al. A distinct microbiota composition is associated with protection from food allergy in an oral mouse immunization model. Clin Immunol. 2016;173:10‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Diesner SC, Knittelfelder R, Krishnamurthy D, et al. Dose‐dependent food allergy induction against ovalbumin under acid‐suppression: a murine food allergy model. Immunol Lett. 2008;121:45‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Golias J, Schwarzer M, Wallner M, et al. Heat‐induced structural changes affect OVA‐antigen processing and reduce allergic response in mouse model of food allergy. PLoS ONE. 2012;7:e37156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Esch B, van Bilsen J, Jeurink PV, et al. Interlaboratory evaluation of a cow's milk allergy mouse model to assess the allergenicity of hydrolysed cow's milk based infant formulas. Toxicol Lett. 2013;220:95‐102. [DOI] [PubMed] [Google Scholar]
- 22. van Esch B, Gros‐van Hest M, Westerbeek H, Garssen J. Sensitizing capacity and allergenicity of enzymatically cross‐linked sodium caseinate in comparison to sodium caseinate in a mouse model for cow’s milk allergy. Toxicol Lett. 2013;218:50‐55. [DOI] [PubMed] [Google Scholar]
- 23. Noval Rivas M, Burton OT, Wise P, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perrier C, Corthésy B. Gut permeability and food allergies. Clin Exp Allergy. 2011;41:20‐28. [DOI] [PubMed] [Google Scholar]
- 25. Stefka AT, Feehley T, Tripathi P, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA. 2014;111:13145‐13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tordesillas L, Rahman AH, Hartmann BM, Sampson HA, Berin MC. Mass cytometry profiling the response of basophils and the complete peripheral blood compartment to peanut. J Allergy Clin Immunol. 2016;138(6):1741‐1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tordesillas L, Goswami R, Benedé S, et al. Skin exposure promotes a Th2‐dependent sensitization to peanut allergens. J Clin Invest. 2014;124:4965‐4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roth‐Walter F, Berin MC, Arnaboldi P, et al. Pasteurization of milk proteins promotes allergic sensitization by enhancing uptake through Peyer’s patches. Allergy. 2008;63:882‐890. [DOI] [PubMed] [Google Scholar]
- 29. Li X‐M, Srivastava K, Huleatt JW, Bottomly K, Burks AW, Sampson HA. Engineered recombinant peanut protein and heat‐killed Listeria monocytogenes coadministration protects against peanut‐induced anaphylaxis in a murine model. J Immunol. 2003;170:3289‐3295. [DOI] [PubMed] [Google Scholar]
- 30. Singh A, Demont A, Actis‐Goretta L, et al. Identification of epicatechin as one of the key bioactive constituents of polyphenol‐enriched extracts that demonstrate an anti‐allergic effect in a murine model of food allergy. Br J Nutr. 2014;112:358‐368. [DOI] [PubMed] [Google Scholar]
- 31. Hacini‐Rachinel F, Vissers YM, Doucet‐Ladevéze R, et al. Low‐allergenic hydrolyzed egg induces oral tolerance in mice. Int Arch Allergy Immunol. 2014;164:64‐73. [DOI] [PubMed] [Google Scholar]
- 32. Gomes‐Santos AC, Fonseca RC, Lemos L, et al. Hydrolyzed whey protein prevents the development of food allergy to β‐lactoglobulin in sensitized mice. Cell Immunol. 2015;298:47‐53. [DOI] [PubMed] [Google Scholar]
- 33. Adel‐Patient K, Nahori M‐A, Proust B, et al. Elicitation of the allergic reaction in beta‐lactoglobulin‐sensitized Balb/c mice: biochemical and clinical manifestations differ according to the structure of the allergen used for challenge. Clin Exp Allergy. 2003;33:376‐385. [DOI] [PubMed] [Google Scholar]
- 34. Curciarello R, Smaldini PL, Candreva AM, et al. Targeting a cross‐reactive Gly m 5 soy peptide as responsible for hypersensitivity reactions in a milk allergy mouse model. PLoS ONE. 2014;9:e82341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smit J, de Zeeuw‐Brouwer M, van Roest M, de Jong G, van Bilsen J. Evaluation of the sensitizing potential of food proteins using two mouse models. Toxicol Lett. 2016;262:62‐69. [DOI] [PubMed] [Google Scholar]
- 36. Bøgh KL, Barkholt V, Madsen CB. Characterization of the immunogenicity and allergenicity of two cow’s milk hydrolysates – a study in Brown Norway rats. Scand J Immunol. 2015;81:274‐283. [DOI] [PubMed] [Google Scholar]
- 37. Adel‐Patient K, Ah‐Leung S, Bernard H, Durieux‐Alexandrenne C, Créminon C, Wal J‐M. Oral sensitization to peanut is highly enhanced by application of peanut extracts to intact skin, but is prevented when CpG and cholera toxin are added. Int Arch Allergy Immunol. 2007;143:10‐20. [DOI] [PubMed] [Google Scholar]
- 38. Wavrin S, Bernard H, Wal JM, Adel‐Patient K. Cutaneous or respiratory exposures to peanut allergens in mice and their impacts on subsequent oral exposure. Int Arch Allergy Immunol. 2014;164:189‐199. [DOI] [PubMed] [Google Scholar]
- 39. Guillon B, Bernard H, Drumare MF, Hazebrouck S, Adel‐Patient K. Heat processing of peanut seed enhances the sensitization potential of the major peanut allergen Ara h 6. Mol Nutr Food Res. 2016;60:2722‐2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Wijk F, Hartgring S, Koppelman SJ, Pieters R, Knippels L. Mixed antibody and T cell responses to peanut and the peanut allergens Ara h 1, Ara h 2, Ara h 3 and Ara h 6 in an oral sensitization model. Clin Exp Allergy. 2004;34:1422‐1428. [DOI] [PubMed] [Google Scholar]
- 41. Li X‐M, Serebrisky D, Lee S‐Y, et al. A murine model of peanut anaphylaxis: T‐ and B‐cell responses to a major peanut allergen mimic human responses. J Allergy Clin Immunol. 2000;106:150‐158. [DOI] [PubMed] [Google Scholar]
- 42. Kroghsbo S, Bøgh KL, Rigby NM, Mills E, Rogers A, Madsen CB. Sensitization with 7S globulins from peanut, hazelnut, soy or pea induces IgE with different biological activities which are modified by soy tolerance. Int Arch Allergy Immunol. 2011;155:212‐224. [DOI] [PubMed] [Google Scholar]
- 43. Bøgh KL, Kroghsbo S, Dahl L, et al. Digested Ara h 1 has sensitizing capacity in Brown Norway rats. Clin Exp Allergy. 2009;39:1611‐1621. [DOI] [PubMed] [Google Scholar]
- 44. Madsen JL, Kroghsbo S, Madsen CB, Pozdnyakova I, Barkholt V, Bøgh KL. The impact of structural integrity and route of administration on the antibody specificity against three cow’s milk allergens – a study in Brown Norway rats. Clin Transl Allergy. 2014;4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205‐217. [DOI] [PubMed] [Google Scholar]
- 46. El‐Khouly F, Lewis SA, Pons L, Burks AW, Hourihane JO. IgG and IgE avidity characteristics of peanut allergic individuals. Pediatr Allergy Immunol. 2007;18:607‐613. [DOI] [PubMed] [Google Scholar]
- 47. Wang J, Lin J, Bardina L, et al. Correlation of IgE/IgG4 milk epitopes and affinity of milk‐specific IgE antibodies with different phenotypes of clinical milk allergy. J Allergy Clin Immunol. 2010;125(3):695‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pullen GR, Fitzgerald MG, Hosking CS. Antibody avidity determination by ELISA using thiocyanate elution. J Immunol Methods. 1986;86(1):83‐90. [DOI] [PubMed] [Google Scholar]
- 49. Lew AM, Anders RF, Edwards SJ, Langford CJ. Comparison of antibody avidity and titre elicited by peptide as a protein conjugate or as expressed in vaccinia. Immunology. 1988;65:311. [PMC free article] [PubMed] [Google Scholar]
- 50. Noti M, Kim BS, Siracusa MC, et al. Exposure to food allergens through inflamed skin promotes intestinal food allergy through the thymic stromal lymphopoietin‐basophil axis. J Allergy Clin Immunol. 2014;133(5):1390‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moghaddam AE, Hillson WR, Noti M, et al. Dry roasting enhances peanut‐induced allergic sensitization across mucosal and cutaneous routes in mice. J Allergy Clin Immunol. 2014;134:1453‐1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han H, Thelen TD, Comeau MR, Ziegler SF. Thymic stromal lymphopoietin‐mediated epicutaneous inflammation promotes acute diarrhea and anaphylaxis. J Clin Invest. 2014;124:5442‐5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Forbes EE, Groschwitz K, Abonia JP, et al. IL‐9– and mast cell–mediated intestinal permeability predisposes to oral antigen hypersensitivity. J Exp Med. 2008;205:897‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bartnikas LM, Gurish MF, Burton OT, et al. Epicutaneous sensitization results in IgE‐dependent intestinal mast cell expansion and food‐induced anaphylaxis. J Allergy Clin Immunol. 2013;131(2):451‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Noval Rivas M, Burton O, Wise P, et al. Regulatory T cell reprogramming toward a Th2‐cell‐like lineage impairs oral tolerance and promotes food allergy. Immunity. 2015;42:512‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ganeshan K, Neilsen CV, Hadsaitong A, Schleimer RP, Luo X, Bryce PJ. Impairing oral tolerance promotes allergy and anaphylaxis: a new murine food allergy model. J Allergy Clin Immunol. 2009;123(1):231‐238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Galand C, Leyva‐Castillo JM, Yoon J, et al. IL‐33 promotes food anaphylaxis in epicutaneously sensitized mice by targeting mast cells. J Allergy Clin Immunol. 2016;138:1356‐1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tordesillas L, Mondoulet L, Blazquez AB, Benhamou P‐H, Sampson HA, Berin MC. Epicutaneous immunotherapy induces gastrointestinal LAP + regulatory T cells and prevents food‐induced anaphylaxis. J Allergy Clin Immunol. 2017;139(1):189‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Frossard CP, Zimmerli SC, Rincon Garriz JM, Eigenmann PA. Food allergy in mice is modulated through the thymic stromal lymphopoietin pathway. Clin Transl Allergy. 2015;6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nakajima‐Adachi H, Kikuchi A, Fujimura Y, et al. Peyer’s patches and mesenteric lymph nodes cooperatively promote enteropathy in a mouse model of food allergy. PLoS ONE. 2014;9:e107492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Knight AK, Bele A, Zhang S, Mayer L, Sampson HA, Berin MC. CD4 T cells activated in the mesenteric lymph node mediate gastrointestinal food allergy in mice. Am J Physiol Gastrointest Liver Physiol. 2007;293(6):G1234‐G1243. [DOI] [PubMed] [Google Scholar]
- 62. DeLong JH, Hetherington KA, Wambre E, James EA, Robinson D, Ara W. 1‐Reactive T cells in peanut allergic individuals. J Allergy Clin Immunol. 2011;127:1211‐1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brough HA, Cousins DJ, Munteanu A, et al. IL‐9 is a key component of memory THcell peanut‐specific responses from children with peanut allergy. J Allergy Clin Immunol. 2014;134(6):1329‐1338. [DOI] [PubMed] [Google Scholar]
- 64. Akdis M, Burgler S, Crameri R, et al. to 37, and interferon‐γ: receptors, functions, and roles in diseases. J Allergy Clin Immunol. 2011;127:701‐721. [DOI] [PubMed] [Google Scholar]
- 65. Frossard CP, Asigbetse KE, Burger D, Eigenmann PA. Gut T cell receptor‐γδ + intraepithelial lymphocytes are activated selectively by cholera toxin to break oral tolerance in mice. Clin Exp Immunol. 2015;180:118‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Saidova A, Hershkop AM, Ponce M, Eiwegger T. Allergen‐specific T cells in IgE‐mediated food allergy. Arch Immunol Ther Exp (Warsz). 2018;66:161‐170. [DOI] [PubMed] [Google Scholar]
- 67. Haller D, Bode C, Hammes WP, Pfeifer AM, Schiffrin EJ, Blum S. Non‐pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co‐cultures. Gut. 2000;47:79‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tsilingiri K, Barbosa T, Penna G, et al. Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex‐vivo organ culture model. Gut. 2012;61:1007‐1015. [DOI] [PubMed] [Google Scholar]
- 69. Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262‐265. [DOI] [PubMed] [Google Scholar]
- 70. Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586‐1597. [DOI] [PubMed] [Google Scholar]
- 71. Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut‐on‐a‐chip. Proc Natl Acad Sci. 2016;113:E7‐E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut‐on‐a‐chip inhabited by microbial flora that experiences intestinal peristalsis‐like motions and flow. Lab Chip. 2012;12:2165. [DOI] [PubMed] [Google Scholar]
- 73. Yissachar N, Zhou Y, Ung L, et al. An intestinal organ culture system uncovers a role for the nervous system in microbe‐immune crosstalk. Cell. 2017;168(6):1135‐1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


