Abstract
Purpose
In this study, we demonstrate the first combination of 3D FID proton MRSI and spatial encoding via concentric‐ring trajectories (CRTs) at 3T. FID‐MRSI has many benefits including high detection sensitivity, in particular for J‐coupled metabolites (e.g., glutamate/glutamine). This makes it highly attractive, not only for clinical, but also for, potentially, functional MRSI. However, this requires excellent reliability and temporal stability. We have, therefore, augmented this 3D‐FID‐MRSI sequence with single‐echo, imaging‐based volumetric navigators (se‐vNavs) for real‐time motion/shim‐correction (SHMOCO), which is 2× quicker than the original double‐echo navigators (de‐vNavs), hence allowing more efficient integration also in short‐TR sequences.
Methods
The tracking accuracy (position and B0‐field) of our proposed se‐vNavs was compared to the original de‐vNavs in phantoms (rest and translation) and in vivo (voluntary head rotation). Finally, the intra‐session stability of a 5:40 min 3D‐FID‐MRSI scan was evaluated with SHMOCO and no correction (NOCO) in 5 resting subjects. Intra/inter‐subject coefficients of variation (CV) and intra‐class correlations (ICC) over the whole 3D volume and in selected regions of interest ROI were assessed.
Results
Phantom and in vivo scans showed highly consistent tracking performance for se‐vNavs compared to the original de‐vNavs, but lower frequency drift. Up to ~30% better intra‐subject CVs were obtained for SHMOCO (P < 0.05), with values of 9.3/6.9/6.5/7.8% over the full VOI for Glx/tNAA/tCho/m‐Ins ratios to tCr. ICCs were good‐to‐high (91% for Glx/tCr in motor cortex), whereas the inter‐subject variability was ~11–19%.
Conclusion
Real‐time motion/shim corrected 3D‐FID‐MRSI with time‐efficient CRT‐sampling at 3T allows reliable, high‐resolution metabolic imaging that is fast enough for clinical use and even, potentially, for functional MRSI.
Keywords: concentric rings, dynamic functional magnetic resonance spectroscopic imaging, intra‐subject reproducibility, real time motion correction, reliability
1. INTRODUCTION
Proton MRSI (1H‐MRSI) allows the non‐invasive assessment of comprehensive neurochemical profiles in vivo1 and has been used to study many different neurologic, neuropsychiatric, and oncologic pathologies. 1 The simultaneous acquisition of multiple voxels allows for regional mapping of various neurochemicals at 3T, including NAA, Cr, Cho, myo‐inositol (m‐Ins), and glutamate + glutamine (Glx).2
At higher B0 of 7T or higher, FID‐based MRSI sequences with short acquisition delays have shown great potential for high‐resolution metabolic mapping.3, 4, 5 The benefits include negligible signal losses caused by T2‐relaxation or J‐modulation, low specific absorption rates, low chemical‐shift displacement errors, and reduced sensitivity to errors. At 7T, FID‐MRSI sequences have been accelerated by non‐Cartesian k‐space sampling based on concentric‐ring trajectories (CRTs).6 For high spectral bandwidths, the self‐rewinding and constant‐angular velocity properties of CRTs render them more SNR‐efficient, faster, and less susceptible to gradient imperfections than other spatial‐spectral encoding approaches. Although 3T MRI scanners are more widely available, so far only a few studies have used CRTs,7, 8 only 2 studies used FID‐MRSI,9, 10 and no application has yet been reported for a combination of both at 3T. Besides being highly beneficial for clinical application, preliminary results11 also raise the hope that the efficient combination of FID‐MRSI and rapid CRT encoding could provide 3D mapping of glutamate with temporal and spatial resolutions high enough to observe stimuli‐induced changes via functional MRSI. However, this would require also excellent intra‐session stability, which is challenging to achieve.
One of the most common sources of artifacts in MRSI is subject motion. Motion artifacts are less obvious to recognize in MRSI compared to MR imaging,12 but can nevertheless severely degrade localization accuracy and spectral quality (e.g., line broadening, lipid contamination, and spectral peak splitting).13, 14 Rapid gradient switching within a heavy duty cycle sequence (e.g., CRTs, spirals, echo‐planar spectroscopic imaging [EPSI]) causes temporal B0 changes because of heating of the gradient coils and passive shims.15 A prospective (real‐time) correction that updates the position of the imaging volume, carrier frequency, and B0‐shim using volumetric, dual‐echo EPI navigators (vNavs) has already been demonstrated.16, 17, 18, 19 These vNavs are well‐suited to use in long‐TR sequences, where they can be deployed in the dead times between signal readout and the subsequent excitation19, 20 but prolong scan times of short‐TR sequences.
The aims of this study were 3‐fold: (1) to demonstrate that 3D‐FID‐MRSI accelerated by CRTs can provide high‐resolution metabolic maps in sufficiently short scan times at 3T; (2) to evaluate real‐time motion/shim correction based on shorter, single‐echo vNavs compared to previously proposed double‐echo vNavs; and (3) to report the intra‐session stability of real‐time motion/shim corrected 3D‐FID‐MRSI for major neurochemicals.
2. METHODS
2.1. Volunteers and hardware
This study was performed on a 3T Prisma MR scanner with a 64‐channel receive‐only head coil (all Siemens Healthineers) in 6 healthy volunteers (male/female, 4/2; age, 28.8 ± 5.4 y). Institutional Review Board approval and written, informed consent as well as a questionnaire to exclude abnormal medical conditions were obtained before the MR examinations.
2.2. Volumetric echo‐planar‐imaging navigators
Originally, head pose and B0‐field changes were obtained in real‐time from dual‐echo, volumetric EPI navigators (de‐vNavs).16, 19 For this, each TR starts with a navigator acquisition followed by an immediate online EPI reconstruction to create B0‐maps and magnitude images (Figure 1A,C). Position changes are computed using PACE21 by co‐registering the magnitude images (TE1) of different time points.16, 18, 19 B0‐maps are generated per TR from phase images of both TEs. The total navigator block, including acquisition, reconstruction, update calculations, and feedback transmission of the current updates back to the MRSI sequence required ~760 ms.19
Figure 1.
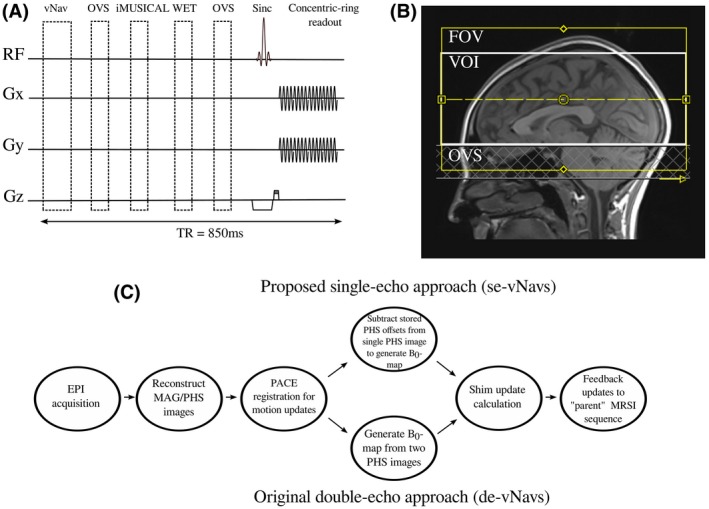
(A) Schematic diagram of the navigated 3D‐FID‐MRSI sequence: volumetric navigators (vNavs), iMUSICAL coil combination pre‐scan, water suppression enhanced through T1 effects (WET), outer‐volume‐suppression (OVS), and the 3D‐FID‐MRSI sequence with concentric‐ring readout (acquisition delay = 0.8 ms, TR = 850 ms). (B) Positioning of the MRSI volume including the placement of OVS slabs below the VOI. (C) Online reconstruction pipeline for our proposed single‐echo navigators and the original double‐echo navigators. The pipelines differ only by how the B0‐maps are created: for the single‐echo approach, the pre‐calculated phase offsets are subtracted from the single phase images, whereas for the double‐echo approach, B0‐maps are generated by subtracting the phase images from the 2 TEs
In contrast, our proposed single‐echo, volumetric EPI navigators (se‐vNavs) acquire only a single echo, resulting in only 1 magnitude/phase image per TR (Figure 1C). Calculating motion updates remains unchanged, because only the magnitude image from the first echo is used for calculating updates. Although B0‐maps are originally generated from the 2 TE phase images, we can reconstruct B0‐maps from the single phase images after subtracting pre‐stored, coil‐dependent phase offsets.22, 23 These are calculated using ASPIRE24 from a dual‐echo EPI reference pre‐scan whose parameters matched those of the se‐vNavs listed below, except for the dual‐echo TE1/TE2 of 7/14 ms (ASPIRE requires TE2 = 2 × TE1). The remaining processing pipeline (update calculation, feedback sending) is identical to the original pipeline (Figure 1C). Using our se‐vNavs, the total navigator block requires only ~360 ms.
The parameters for se‐vNavs were: TR = 17 ms; TE = 7 ms (de‐vNavs = TE1/TE2, 7/9.4 ms); matrix = 32 × 32; slices = 18; FOV = 256 × 256 × 144 mm3; bandwidth = 4734 Hz/pixel; flip angle = 4°; echo train length = 32; water excitation only; slice partial Fourier = 6/8.
2.3. Data acquisition
All sessions started with a 3D, T1‐weighted, MPRAGE sequence to position the MRSI volume (Figure 1B). B0‐map‐based, 1st and 2nd order B0‐shimming using standard Siemens routines was performed over the MRSI volume‐of‐interest (VOI).
The position of the navigator was identical to that of the phase offset reference scan covering the subject’s brain and was set using a rapid “setter” sequence19 before the MRSI sequence.
The 3D‐FID‐MRSI sequence (Figure 1A) used the following settings: TR= 850 ms; acquisition delay = 0.8 ms; flip angle = 70°; 600 µs sinc excitation pulse; B1 = 13.2 µT; VOI = 220 × 220 × 76 mm3; FOV = 220 × 220 × 126 mm3; in vivo matrix size = 50 × 50 × 21 (in phantoms = 32 × 32 × 21); complex spectral data points = 360; acquisition bandwidth = 1030 Hz; no temporal interleaving; acquisition window = 350 ms; averages = 1; maximum slew rate = 200 mT/m/ms; maximum gradient strength = 80 mT/m; spherical k‐space coverage; water suppression enhanced through T1‐effects (WET); outer‐volume saturation (OVS) band (30 mm thick) below VOI covering nasal cavity and skull base; TA = 5:40 min.
2.4. Data processing
All measured data were processed with an in‐house developed pipeline25 based on Bash (Free Software Foundation, Boston, MA) and MATLAB (The MathWorks, Natick, MA). The post‐processing pipeline included a modified Pipe‐Menon pre‐gridding density compensation,26 an off‐resonance correction,27 convolution gridding28 using a Kaiser‐Bessel kernel (width 3), coil‐wise L2‐lipid regularization29 and iMUSICAL coil combination.30 Detailed steps have been described previously.6, 25 LCModel 6.331 was used to fit in vivo spectra in the spectral range of 1.8–4.2 ppm.
2.5. Part 1: tracking accuracy—se‐vNavs versus de‐vNavs
The accuracy and correlation of the position and B0‐field tracking (i.e., translation, rotation, frequency, and 1st‐order shims) were compared between se‐vNavs and de‐vNavs in phantom and in vivo measurements. To ensure comparable conditions for the se‐vNavs and de‐vNavs, the MRSI TR was set to 1120 ms to accommodate the more time‐consuming de‐vNavs. A head‐shaped gel phantom (6.3 L 1% agarose, 2 µmol/kg gadolinium, 0.1% NaCl, and 0.05% NaN3) was used.32 “Rest” and “push” phantom measurements were performed, where in the latter, the phantom was pushed once (4 mm into the scanner at 1:45 min). For the in vivo measurements, an MR‐experienced volunteer was trained and acoustically instructed to perform a head rotation of ~3° left–right (at 1:30 min), return to the initial position (at 2:00 min), perform a head rotation of ~3° left–right (at 3:00 min), and finally return to the initial position (at 3:30 min). A Styrofoam insert inside the coil providing the volunteer with a reference for the current head position ensured the reproducibility of the motion pattern.
2.6. Part 2: variability of 3D‐FID‐MRSI—no correction versus shim/motion‐correction
Five volunteers were scanned each within 1 session and without repositioning with a total of 8 3D‐FID‐MRSI scans: 4 no correction (NOCO) and 4 shim/motion‐correction (SHMOCO) in an interleaved fashion.
Metabolic concentration ratios (Glx/tCr, tNAA/tCr, tCho/tCr, and m‐Ins/tCr) were obtained on a voxel‐by‐voxel basis and voxels with poor spectral quality (i.e., CRLBs of tCr, tCh, tNAA, Glx, and m‐Ins >20%) were excluded from analysis. Mean SNR, FWHM, and CRLBs of Glx, tNAA, tCr, tCho, and m‐Ins were compared between SHMOCO and NOCO using paired t‐tests (P < 0.05 was considered statistically significant). Further, the intra‐subject (within volunteers) reproducibility and inter‐subject (between volunteers) variability were assessed by linear mixed effects models and are reported as coefficients of variation (CV). As a measure of method reliability, intra‐class correlation coefficients (ICC) were calculated by an absolute‐agreement, 2‐way, mixed‐effects model among the 4 SHMOCO and NOCO scans, respectively. Besides the entire VOI, specific brain regions were investigated in a region‐of‐interest (ROI)‐based analysis. ROIs with a volume of ~1 cm3 were manually placed in the upper part of the visual cortex (occipital lobe), in the motor cortex (frontal lobe), in the dorsolateral prefrontal cortex (DLPFC), and in the auditory cortex (temporal lobe). High‐resolution T1‐weighted images guided the consistent placing of ROIs in the same anatomic locations.
3. RESULTS
3.1. Part 1: tracking accuracy—se‐vNavs versus de‐vNavs
Figure 2 and Supporting Information Figure S1 show motion and B0‐field logs from the phantom and in vivo (volunteer 1) measurements. High similarities were found between the tracking of se‐vNavs and de‐vNavs. The mean differences of the total translation were 0.00 ± 0.01 mm, 0.01 ± 0.15 mm, and 0.01 ± 0.16 mm for the phantom “rest” and “push” as well as in vivo measurements, respectively (all R > 0.99, P < 0.001). The mean differences in 1st‐order shims (cor/sag/tra) were small and resulted in 0.01 ± 0.02/0.01 ± 0.03/0.05 ± 0.10 Hz/mm (all R > 0.97, P < 0.001) for the phantom in rest and 0.10 ± 0.06/0.06 ± 0.05/0.08 ± 0.05 Hz/mm (all R > 0.95, P < 0.001) for the in vivo scan. The frequency drift curves were similar in shape, but the total drifts for de‐vNavs were ~2.5‐fold (phantom) and ~1.7‐fold (in vivo) higher than for the se‐vNavs.
Figure 2.
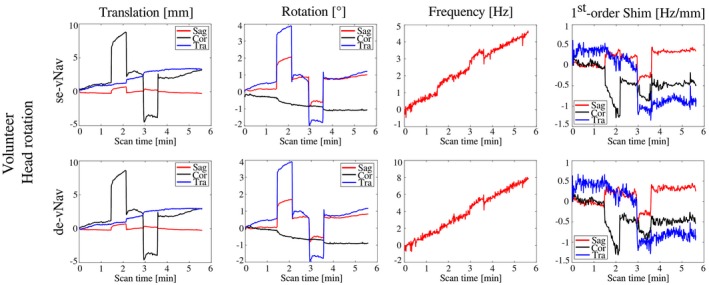
Comparison of the tracking performance of our proposed single‐echo navigators (se‐vNavs) and the original double‐echo navigators (de‐vNavs). Translation, rotation, frequency, and 1st‐order shim logs are shown for both navigator approaches. For the in vivo measurement, a MR‐trained volunteer was acoustically instructed to perform a predefined head rotation pattern
3.2. Part 2: variability of 3D‐FID‐MRSI—NOCO versus SHMOCO
Sample spectra for volunteer 2 from the above mentioned ROIs are shown in Figure 3. Figure 4 depicts the 4 Glx/tCr metabolic ratio maps with SHMOCO for volunteer 3 in 3 adjacent slices. Supporting Information Table S1 shows mean SNR, FWHM, and CRLBs of Glx, tNAA, tCr, tCho, and m‐Ins averaged over all 5 volunteers for the NOCO and SHMOCO measurements. Slightly improved results (higher SNR, lower FWHM, and lower CRLBs) were obtained for SHMOCO compared to NOCO, with only few values reaching statistical significance (e.g., SNR in DLPFC). Supporting Information Figure S2 shows scatter plots of the longitudinal measurements per subject for Glx/tCr and tNAA/tCr in the visual and motor cortex ROI.
Figure 3.
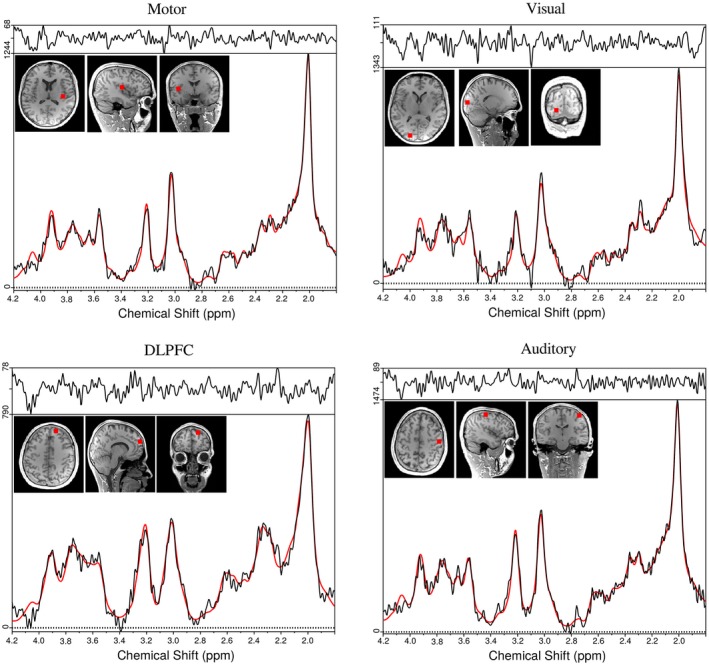
Representative spectra for volunteer 2 from 4 different ROIs (motor cortex, visual cortex, dorsolateral prefrontal cortex [DLPFC], and auditory cortex). For every ROI, the voxel position is marked on T1‐weighted images. The spectra are 1st‐order phase corrected for display
Figure 4.
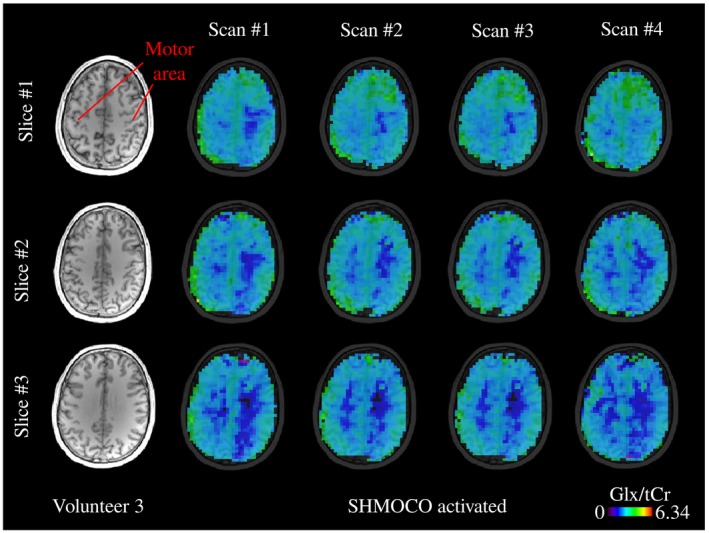
Metabolic ratio maps (Glx/tCr) for volunteer 3 depicted in 3 adjacent slices for the 4 scans with real‐time motion/shim correction turned on (SHMOCO); TR = 850 ms, acquisition delay = 0.8 ms, 50 × 50 × 21 matrix, nominal voxel size = 0.12 mL; TA = 5:40 min
The average maximum translations from the NOCO (0.9 ± 0.4 mm) and SHMOCO (1.2 ± 0.4 mm) scans were statistically non‐significant (P = 0.1).
Table 1 summarizes the means and SDs of the intra‐subject CVs obtained for the different metabolites ratios and brain regions averaged over all 5 volunteers. Intra‐subject CVs obtained with SHMOCO were all lower (i.e., better) compared to NOCO, with improvements of up to ~30%. Intra‐subject CVs of 9.3%, 6.9%, 6.5%, and 7.8% were obtained across the full VOI for Glx/tCr, tNAA/tCr, tCho/tCr, and m‐Ins/tCr with SHMOCO. For Glx/tCr, values of 9.8% and 8.2% were obtained in visual and motor cortex. Significant differences were found between NOCO and SHMOCO over the whole VOI (P < 0.001), but no moderation effect (P = 0.67) with the metabolites (i.e., no evidence that the difference between NOCO and SHMOCO) was more pronounced for certain metabolites. No moderation effect was also found with the ROIs and metabolites (i.e., no significant evidence that SHMOCO performed better for certain metabolites in certain ROIs) (P = 0.92). Paired t‐tests (NOCO vs. SHMOCO) for the individual ROIs and metabolites revealed significant differences in intra‐subject CVs for all metabolites in the full VOI, but only in 12 of 28 combinations of ROI and metabolite (e.g., Glx/tCr in the visual cortex) (Table 1). As measure of method reliability, ICCs are summarized in Supporting Information Table S2. Consistently better ICCs were found for SHMOCO compared to NOCO. Overall good (ICC >75%) to high (ICC >85%) measurement reliability was observed for SHMOCO with values of 85.1% and 91.1% for Glx/tCr in the visual and motor cortex, respectively. The inter‐subject variability for both SHMOCO and NOCO was generally (up to 2‐fold) higher than the intra‐subject variability and is summarized in Supporting Information Table S3.
Table 1.
Mean and SDs of the intra‐subject CV obtained with SHMOCO and NOCO for different metabolic concentration ratios
| Intra‐subject CV (%) | VOI | Visual | Motor_L | Motor_R | DLPFC_L | DLPFC_R | Auditory_L | Auditory_R |
|---|---|---|---|---|---|---|---|---|
| SHMOCO | ||||||||
| Glx/tCr | 9.3 ± 1.0* | 9.8 ± 1.1* | 8.3 ± 0.7 | 8.2 ± 0.3 | 10.9 ± 1.9 | 10.3 ± 0.5 | 9.3 ± 0.3 | 10.1 ± 0.5 |
| tNAA/tCr | 6.9 ± 0.7* | 8.1 ± 1.4 | 6.3 ± 0.9* | 6.1 ± 0.4* | 7.7 ± 1.1* | 8.5 ± 1.0* | 8.1 ± 1.5 | 8.8 ± 0.3 |
| tCho/tCr | 6.5 ± 1.0* | 7.2 ± 1.0 | 6.0 ± 1.4 | 6.0 ± 0.8 | 7.0 ± 0.8* | 7.2 ± 0.6* | 6.9 ± 0.5* | 7.6 ± 0.6* |
| m‐Ins/tCr | 7.8 ± 1.4* | 8.0 ± 1.3 | 6.7 ± 1.3 | 7.0 ± 1.5 | 8.7 ± 1.1* | 9.0 ± 0.7* | 8.2 ± 0.5* | 9.1 ± 0.8 |
| NOCO | ||||||||
| Glx/tCr | 12.0 ± 1.6* | 12.8 ± 1.2* | 11.0 ± 1.8 | 11.3 ± 1.8 | 11.6 ± 1.8 | 12.2 ± 1.8 | 11.7 ± 1.3 | 12.5 ± 1.9 |
| tNAA/tCr | 9.6 ± 0.7* | 10.3 ± 0.5 | 8.3 ± 0.6* | 9.0 ± 0.7* | 10.2 ± 0.8* | 11.5 ± 0.8* | 10.6 ± 0.4 | 10.9 ± 1.8 |
| tCho/tCr | 8.6 ± 0.7* | 8.6 ± 1.3 | 7.6 ± 0.7 | 8.1 ± 1.1 | 9.3 ± 0.4* | 10.0 ± 0.4* | 9.8 ± 0.6* | 9.5 ± 0.7* |
| m‐Ins/tCr | 9.5 ± 1.4* | 8.3 ± 0.9 | 8.3 ± 1.3 | 8.8 ± 1.2 | 10.7 ± 1.7* | 11.1 ± 1.2* | 10.8 ± 1.0* | 10.8 ± 1.7 |
Abbreviations: CV, coefficients of variation; NOCO, no correction; SHMOCO, motion/shim‐correction.
P‐value of < 0.05 was considered statistically significant.
4. DISCUSSION
We presented the first use of CRTs in a 3D‐FID‐MRSI sequence at 3T to generate high‐resolution metabolic maps in 5:40 min. The sequence was further equipped with a shorter, single‐echo volumetric EPI navigator for real‐time motion/shim correction. The performance of this novel navigator approach was compared to the previously published double‐echo navigator implementation, which had twice the duration. Finally, the temporal stability of the 3D‐FID‐MRSI sequence was assessed and the benefits of real‐time motion/shim correction were investigated in healthy volunteers.
Distinct anatomic contrasts could be observed (e.g., gray/white matter contrast in Glx or tCr)9 in the high‐resolution metabolic maps. The MRSI sequence was initially developed for 7T,6 but was adapted for 3T in this study and optimized by using a sub‐millisecond acquisition delay of 0.8 ms to maximize the signal and by shortening the water suppression module to reduce the TR.33 We acquired nominal voxel volumes of 0.12 mL in clinically highly feasible 5:40 min, which is significantly faster than most 3D‐MRSI reports from 3T. EPSI‐based studies report 0.31 mL voxels in 16.4 min34 or in 18 min with additional lipid inversion nulling,35 whereas non‐water‐suppressed multi‐band (3 slices) MRSI using density‐weighted CRTs has featured 0.25 mL voxels in 19.2 min.36 Overall, spectral quality in this study was good and allowed for reliable fitting of high‐concentrated metabolites (tNAA, tCr, tCho, m‐Ins, and Glx). However, the available SNR and spectral resolution at 3T limited the quantification of lower concentrated metabolites (e.g., glutathione, γ‐aminobutyric acid). Using a short acquisition delay of 0.8 ms made an improved optimization of lipid contamination via L2‐lipid regularization and macromolecular signals37 necessary because of their fast T2‐relaxation.
The potential for correction of motion and scanner‐instability related artifacts using vNavs has been shown in a broad range of MR techniques including anatomic imaging,38 diffusion,39 chemical exchange saturation transfer,40 and edited/non‐edited MRS/MRSI.16, 20, 41 All of these methods provide sufficient sequence dead time to incorporate vNavs without prolonging scan times. Because of their shortened acquisition time, our proposed se‐vNavs can also be readily integrated in MR sequences with little dead time, such as FID‐MRSI. Similar to Dymerska et al.,22 who generated B0‐maps for the dynamic correction of single‐echo EPI at 7T, we also used a dual‐echo reference scan for the calculation of phase offsets, which have been shown to be stable during long measurements and for large head motions. However, instead of a gradient echo pre‐scan, we used the same EPI readout as for the vNavs to minimize geometrical misalignments. Phantom and in vivo tests showed a high agreement in the position and B0‐field tracking between our se‐vNavs and the original de‐vNavs. An additional advantage of se‐vNavs can be observed in the frequency drift curves, where the drift is reduced by a factor of ~2 compared to de‐vNavs because of less gradient‐intensive EPI readout. Using SHMOCO led to moderately higher SNR and lower CRLBs compared to NOCO, whereas the FWHMs were little affected. However, the intra‐session stability of metabolic ratio maps was significantly improved, which could potentially have important implications for functional MRSI (fMRSI).
Functional MRS studies have reported changes in glutamate42 range from only subtle increases of 2–4% after visual or motor stimuli to more pronounced changes (up to 22%) after pain stimuli. Although fMRSI has not yet been conducted so far, this report contributes to paving the way to fMRSI by assessing the stability of 3D‐FID‐MRSI that makes it possible to judge if small metabolite changes can actually be resolved on a single‐subject basis or on a group level. To date, most reproducibility MRSI studies have focused on longitudinal measurements (repeated scans across 1 or several days with repositioning). Zhang et al35 have reported mean intra‐subject CVs in a longitudinal study (3 separate sessions) of 7.6% for metabolites relative to Cr, while Maudsley et al43 obtained median intra‐subject CVs of 6.2%, 7.2%, and 9.7%, for NAA, Cr, and Cho, respectively, from 5 separate sessions. Only a small number of studies have investigated the intra‐session stability of 3D‐MRSI to date. Comparisons between these inter‐ and intra‐session CVs are not straight‐forward, but generally lower CVs are expected without repositioning. Ding et al44 have assessed the intra‐session reproducibility and found metabolite CVs of 12.8%, 19.3%, 14.5%, 30.6%, and 30.6% for NAA, tCho, tCr, Glx, and m‐Ins, respectively, whereas Bian et al45 have reported an intra‐session CV of the metabolite peak height of 12.4%. The intra‐subject CVs obtained in this study (Glx/tCr: 9.3%, tNAA/tCr: 6.9%, tCho/tCr: 6.5%, m‐Ins/tCr: 7.8%) were significantly lower than those obtained in the above‐mentioned intra‐session reports. Further improvements in intra‐subject CVs compared to 8–10% in Glx/tCr as observed here can be expected with higher SNR and spectral resolution at ≥7T,9, 46 potentially enabling functional MRSI studies. Our results also confirm that the use of SHMOCO gives overall significantly better intra‐subject CVs than NOCO, which mainly reflects SHMOCO’s ability to correct for temporal frequency drifts and involuntary subject movements. Method reliabilities measured as ICCs were good to high and improved for SHMOCO compared to NOCO. The inter‐subject variability driven by differences in positioning, shim settings, and subject condition was generally higher than the intra‐session reproducibility, which is in accordance with literature.47
4.1. Limitations
As an explorative proof‐of‐principle study, the number of volunteers was limited, thereby limiting the statistical power of the analysis. Nevertheless, a clear trend toward improved results when using SHMOCO compared to NOCO has been observed. Concerning motion correction, comparisons of methods without a ground truth (i.e., an ideally non‐moving subject) always need to be interpreted with caution.
For a real dynamic/functional MRSI experiment, instead of performing repeated MRSI scans, it would be more suitable to run a single scan with multiple measurements, which has not yet been implemented nor possible because of the amount of raw data generated.
The duration of se‐vNavs is still fairly long for integration into some MR sequences. Further acceleration could be achieved by k‐space undersampling such as shown for FatNavs.48
5. CONCLUSIONS
3D‐FID‐MRSI at 3T allows for reliable, high‐resolution metabolic imaging in clinically attractive scan times. Implementing accelerated, single‐echo, volumetric, imaging‐based navigators reduced the susceptibility to motion and B0‐instabilities and increased the spectral quality, while providing similar tracking accuracy as previously published double‐echo navigators. Using real‐time motion/shim correction improved the intra‐session stability of 3D‐FID‐MRSI and makes it a potential basis for 3D‐fMRSI.
Supporting information
FIGURE S1 Comparison of the tracking performance of our proposed single‐echo navigators (se‐vNavs) and the original double‐echo navigators (de‐vNavs). Translation, rotation, frequency, and 1st‐order shim logs are shown for both navigator approaches. Phantom measurements were performed for “rest” and “push” conditions, where in the latter the phantom was manually pushed 4 mm into the scanner after 1:45 min
FIGURE S2 Scatter plots of the longitudinal measurements for all 5 subjects in the visual and motor cortex ROI. The 4 time points from the SHMOCO scans are depicted for Glx/tCr and tNAA/tCr (i.e., the mean concentrations within the aforementioned ROIs obtained from the 4 SHMOCO scans)
TABLE S1 Mean and SDs of SNR, FWHM, and metabolic CRLB values obtained with (SHMOCO) and without (NOCO) shim/motion correction for different metabolic concentration ratios. *P‐value of <0.05 was considered statistically significant
TABLE S2 Method reliability measured as intra‐class correlation coefficients (ICCs). All values are given in percent and include mean ICC and lower and upper bounds (in brackets) with alpha level of significance of 0.5. A clear trend toward higher method reliability was observed for SHMOCO compared to NOCO
TABLE S3 Means and SDs of the inter‐subject variability expressed as coefficients of variation (CV) for SHMOCO and NOCO. Slightly better (i.e., lower) values were obtained for SHMOCO
ACKNOWLEDGMENTS
This study was supported by the Austrian Science Fund (FWF) (KLI718, P30701, P31452, and J4124) and the Christian Doppler Laboratory for Clinical Molecular MR Imaging.
Moser P, Eckstein K, Hingerl L, et al. Intra‐session and inter‐subject variability of 3D‐FID‐MRSI using single‐echo volumetric EPI navigators at 3T. Magn Reson Med. 2020;83:1920–1929. 10.1002/mrm.28076
REFERENCES
- 1. Öz G, Alger JR, Barker PB, et al. Clinical proton MR spectroscopy in central nervous system disorders. Radiology. 2014;270:658–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maghsudi H, Schmitz B, Maudsley AA, et al. Regional metabolite concentrations in aging human brain: comparison of short‐TE whole brain MR spectroscopic imaging and single voxel spectroscopy at 3T. Clin Neuroradiol. 2019. 10.1007/s00062-018-00757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bogner W, Gruber S, Trattnig S, Chmelik M. High‐resolution mapping of human brain metabolites by free induction decay 1H MRSI at 7 T. NMR Biomed. 2012;25:873–882. [DOI] [PubMed] [Google Scholar]
- 4. Henning A, Fuchs A, Murdoch JB, Boesiger P. Slice‐selective FID acquisition, localized by outer volume suppression (FIDLOVS) for 1H‐MRSI of the human brain at 7 T with minimal signal loss. NMR Biomed. 2009;22:683–696. [DOI] [PubMed] [Google Scholar]
- 5. Nassirpour S, Chang P, Henning A. High and ultra‐high resolution metabolite mapping of the human brain using 1 H FID MRSI at 9.4T. NeuroImage. 2018;168:211–221. [DOI] [PubMed] [Google Scholar]
- 6. Hingerl L, Bogner W, Moser P, et al. Density‐weighted concentric circle trajectories for high resolution brain magnetic resonance spectroscopic imaging at 7T. Magn Reson Med. 2018;79:2874–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuyama JK, Wilson NE, Thomas MA. Spectroscopic imaging using concentrically circular echo‐planar trajectories in vivo. Magn Reson Med. 2012;67:1515–1522. [DOI] [PubMed] [Google Scholar]
- 8. Steel A, Chiew M, Jezzard P, et al. Metabolite‐cycled density‐weighted concentric rings k‐space trajectory (DW‐CRT) enables high‐resolution 1H magnetic resonance spectroscopic imaging at 3‐Tesla. Sci Rep. 2018;8:7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gruber S, Heckova E, Strasser B, et al. Mapping an extended neurochemical profile at 3 and 7 T using accelerated high‐resolution proton magnetic resonance spectroscopic imaging. Invest Radiol. 2017;52:631–639. [DOI] [PubMed] [Google Scholar]
- 10. Klauser A, Courvoisier S, Kasten J, et al. Fast high‐resolution brain metabolite mapping on a clinical 3T MRI by accelerated 1H‐FID‐MRSI and low‐rank constrained reconstruction. Magn Reson Med. 2019;81:2841–2857. [DOI] [PubMed] [Google Scholar]
- 11. Moser P, Hingerl L, Strasser B, et al. Full brain high resolution 3D‐MRSI at 7T via 3D‐CONCEPT. In Proceedings of the MRS Workshop, Metabolic Imaging, Utrecht, The Netherlands, 2018. Abstract 10. [Google Scholar]
- 12. Kreis R. Issues of spectral quality in clinical 1H‐magnetic resonance spectroscopy and a gallery of artifacts. NMR Biomed. 2004;17:361–381. [DOI] [PubMed] [Google Scholar]
- 13. Heckova E, Považan M, Strasser B, et al. Real‐time correction of motion and imager instability artifacts during 3D γ‐aminobutyric acid‐edited MR spectroscopic imaging. Radiology. 2018;286:666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lange T, Maclaren J, Buechert M, Zaitsev M. Spectroscopic imaging with prospective motion correction and retrospective phase correction. Magn Reson Med. 2012;67:1506–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lange T, Zaitsev M, Buechert M. Correction of frequency drifts induced by gradient heating in 1H spectra using interleaved reference spectroscopy. J Magn Reson Imaging. 2011;33:748–754. [DOI] [PubMed] [Google Scholar]
- 16. Hess AT, Dylan Tisdall M, Andronesi OC, Meintjes EM, van der Kouwe AJW. Real‐time motion and B0 corrected single voxel spectroscopy using volumetric navigators. Magn Reson Med. 2011;66:314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hess AT, Andronesi OC, Dylan Tisdall M, Gregory Sorensen A, van der Kouwe AJW, Meintjes EM. Real‐time motion and B0 correction for localized adiabatic selective refocusing (LASER) MRSI using echo planar imaging volumetric navigators. NMR Biomed. 2012;25:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tisdall MD, Hess AT, Reuter M, Meintjes EM, Fischl B, van der Kouwe AJW. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magn Reson Med. 2012;68:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bogner W, Hess AT, Gagoski B, et al. Real‐time motion‐ and B0‐correction for LASER‐localized spiral‐accelerated 3D‐MRSI of the brain at 3T. NeuroImage. 2014;88:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bogner W, Gagoski B, Hess AT, et al. 3D GABA imaging with real‐time motion correction, shim update and reacquisition of adiabatic spiral MRSI. NeuroImage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thesen S, Heid O, Mueller E, Schad LR. Prospective acquisition correction for head motion with image‐based tracking for real‐time fMRI. Magn Reson Med. 2000;44:457–465. [DOI] [PubMed] [Google Scholar]
- 22. Dymerska B, Poser BA, Barth M, Trattnig S, Robinson SD. A method for the dynamic correction of B0‐related distortions in single‐echo EPI at 7 T. NeuroImage. 2018;168:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poblador Rodriguez E, Moser P, Dymerska B, et al. A comparison of static and dynamic ΔB0 mapping methods for correction of CEST MRI in the presence of temporal B0 field variations. Magn Reson Med. 2019;82:633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eckstein K, Dymerska B, Bachrata B, et al. Computationally efficient combination of multi‐channel phase data from multi‐echo acquisitions (ASPIRE). Magn Reson Med. 2018;79:2996–3006. [DOI] [PubMed] [Google Scholar]
- 25. Považan M, Strasser B, Hangel G, et al. Automated routine for MRSI data processing. In Proceedings of the 2nd TRANSACT meeting – Quality Issues in Clinical MR Spectroscopy, University and Inselspital, Bern, Switzerland, 2014. p. S2. [Google Scholar]
- 26. Pipe JG, Menon P. Sampling density compensation in MRI: rationale and an iterative numerical solution. Magn Reson Med. 1999;41:179–186. [DOI] [PubMed] [Google Scholar]
- 27. Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized13C imaging. Magn Reson Med. 2006;56:932–937. [DOI] [PubMed] [Google Scholar]
- 28. Jackson JI, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding (computerised tomography application). IEEE Trans Med Imaging. 1991;10:473–478. [DOI] [PubMed] [Google Scholar]
- 29. Bilgic B, Gagoski B, Kok T, Adalsteinsson E. Lipid suppression in CSI with spatial priors and highly undersampled peripheral k‐space. Magn Reson Med. 2013;69:1501–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moser P, Bogner W, Hingerl L, et al. Non‐Cartesian GRAPPA and coil combination using interleaved calibration data ‐ application to concentric‐ring MRSI of the human brain at 7T. Magn Reson Med. 2019;82:1587–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. [DOI] [PubMed] [Google Scholar]
- 32. Lara, Frass‐Kriegl, Renner, et al. implementation, and evaluation of a head and neck MRI RF array integrated with a 511 keV transmission source for attenuation correction in PET/MR. Sensors (Basel). 2019;19:3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hangel G, Strasser B, Považan M, et al. Ultra‐high resolution brain metabolite mapping at 7 T by short‐TR Hadamard‐encoded FID‐MRSI. NeuroImage. 2018;168:199–210. [DOI] [PubMed] [Google Scholar]
- 34. Sabati M, Zhan J, Govind V, Arheart KL, Maudsley AA. Impact of reduced k‐space acquisition on pathologic detectability for volumetric MR spectroscopic imaging. J Magn Reson Imaging. 2014;39:224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y, Taub E, Salibi N, et al. Comparison of reproducibility of single voxel spectroscopy and whole‐brain magnetic resonance spectroscopy imaging at 3T. NMR Biomed. 2018;31:e3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emir U, Xia P, Zhou X, Chiew M, Thomas M, Dydak U. Density‐weighted concentric ring trajectory using simultaneous multi‐slice (SMS) acceleration: 3D metabolite‐cycled magnetic resonance spectroscopy imaging at 3 T. In Proceedings of the MRS Workshop: Metabolic Imaging, Utrecht, The Netherlands, 2018. Abstract 12. [Google Scholar]
- 37. Heckova E, Považan M, Strasser B, et al. Effects of different macromolecular models on reproducibility of FID‐MRSI at 7T. Magn Reson Med. 2020;83:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tisdall MD, Reuter M, Qureshi A, Buckner RL, Fischl B, van der Kouwe AJW. Prospective motion correction with volumetric navigators (vNavs) reduces the bias and variance in brain morphometry induced by subject motion. NeuroImage. 2016;127:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alhamud A, Taylor PA, van der Kouwe AJW, Meintjes EM. Real‐time measurement and correction of both B0 changes and subject motion in diffusion tensor imaging using a double volumetric navigated (DvNav) sequence. NeuroImage. 2016;126:60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simegn GL, van der Kouwe AJW, Robertson FC, Meintjes EM, Alhamud A. Real‐time simultaneous shim and motion measurement and correction in glycoCEST MRI using double volumetric navigators (DvNavs). Magn Reson Med. 2019;81:2600–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moser P, Hingerl L, Strasser B, et al. Whole‐slice mapping of GABA and GABA+ at 7T via adiabatic MEGA‐editing, real‐time instability correction, and concentric circle readout. NeuroImage. 2019;184:475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stanley JA, Raz N. Functional magnetic resonance spectroscopy: the “New” MRS for cognitive neuroscience and psychiatry research. Front Psychiatry. 2018;9:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maudsley AA, Domenig C, Sheriff S. Reproducibility of serial whole‐brain MR spectroscopic imaging. NMR Biomed. 2010;23:251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ding XQ, Maudsley AA, Sabati M, Sheriff S, Dellani PR, Lanfermann H. Reproducibility and reliability of short‐TE whole‐brain MR spectroscopic imaging of human brain at 3T. Magn Reson Med. 2015;73:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bian W, Li Y, Crane JC, Nelson SJ. Fully automated atlas‐based method for prescribing 3D PRESS MR spectroscopic imaging: toward robust and reproducible metabolite measurements in human brain. Magn Reson Med. 2018;79:636–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang P, Nassirpour S, Avdievitch N, Henning A. Non‐water‐suppressed 1H FID‐MRSI at 3T and 9.4T. Magn Reson Med. 2018;80:442–451. [DOI] [PubMed] [Google Scholar]
- 47. Veenith TV, Mada M, Carter E, et al. Comparison of inter subject variability and reproducibility of whole brain proton spectroscopy. PLoS ONE. 2014;9:e115304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gallichan D, Marques JP, Gruetter R. Retrospective correction of involuntary microscopic head movement using highly accelerated fat image navigators (3D FatNavs) at 7T. Magn Reson Med. 2016;75:1030–1039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Comparison of the tracking performance of our proposed single‐echo navigators (se‐vNavs) and the original double‐echo navigators (de‐vNavs). Translation, rotation, frequency, and 1st‐order shim logs are shown for both navigator approaches. Phantom measurements were performed for “rest” and “push” conditions, where in the latter the phantom was manually pushed 4 mm into the scanner after 1:45 min
FIGURE S2 Scatter plots of the longitudinal measurements for all 5 subjects in the visual and motor cortex ROI. The 4 time points from the SHMOCO scans are depicted for Glx/tCr and tNAA/tCr (i.e., the mean concentrations within the aforementioned ROIs obtained from the 4 SHMOCO scans)
TABLE S1 Mean and SDs of SNR, FWHM, and metabolic CRLB values obtained with (SHMOCO) and without (NOCO) shim/motion correction for different metabolic concentration ratios. *P‐value of <0.05 was considered statistically significant
TABLE S2 Method reliability measured as intra‐class correlation coefficients (ICCs). All values are given in percent and include mean ICC and lower and upper bounds (in brackets) with alpha level of significance of 0.5. A clear trend toward higher method reliability was observed for SHMOCO compared to NOCO
TABLE S3 Means and SDs of the inter‐subject variability expressed as coefficients of variation (CV) for SHMOCO and NOCO. Slightly better (i.e., lower) values were obtained for SHMOCO


