Summary
Soils provide a heterogeneous environment varying in space and time; consequently, the biodiversity of soil microorganisms also differs spatially and temporally. For soil microbes tightly associated with plant roots, such as arbuscular mycorrhizal fungi (AMF), the diversity of plant partners and seasonal variability in trophic exchanges between the symbionts introduce additional heterogeneity. To clarify the impact of such heterogeneity, we investigated spatiotemporal variation in AMF diversity on a plot scale (10 × 10 m) in a grassland managed at low intensity in southwest Germany. AMF diversity was determined using 18S rDNA pyrosequencing analysis of 360 soil samples taken at six time points within a year. We observed high AMF alpha‐ and beta‐diversity across the plot and at all investigated time points. Relationships were detected between spatiotemporal variation in AMF OTU richness and plant species richness, root biomass, minimal changes in soil texture and pH. The plot was characterized by high AMF turnover rates with a positive spatiotemporal relationship for AMF beta‐diversity. However, environmental variables explained only ≈20% of the variation in AMF communities. This indicates that the observed spatiotemporal richness and community variability of AMF was largely independent of the abiotic environment, but related to plant properties and the cooccurring microbiome.
Introduction
Understanding spatial and temporal patterns in species diversity is one of the fundamental goals of biodiversity research (Gaston and Spicer, 2013). Soil microbial communities exhibit spatial patterns at scales from sub‐millimetre to hundreds of metres, determined by heterogeneous environmental conditions at respective scale‐dependencies (Grundmann et al., 2001; Ettema and Wardle, 2002; Nunan et al., 2003; Bahram et al., 2015). Simultaneously, dynamic variations in abiotic soil conditions lead to fluctuating soil microbial abundances and functions over time, documented in agricultural (Kandeler and Böhm, 1996; Kandeler et al., 1999), tundra (Björk et al., 2008) and forest ecosystems (Görres et al., 1998; Nacke et al., 2016). Moreover, plant growth and development or changes in vegetation within a year are able to shift soil microbial communities (Chaparro et al., 2014; Nacke et al., 2016). This is especially relevant for obligate biotrophic plant mutualists such as arbuscular mycorrhizal fungi (AMF; Smith and Read, 2008).
Recent studies have aimed to identify general patterns of and major influences on AMF community composition. Some findings have included, for instance, high impacts of land‐use intensity (Bouffaud et al., 2017), soil properties (Kivlin et al., 2011; Lekberg et al., 2012), plant community composition (van der Heijden et al., 1998; König et al., 2010; Neuenkamp et al., 2018) and/or host‐plant identity (Sanders, 2003). Since AMF are obligate root mutualists, most studies have focused on fungus‐plant‐relationships. Conflicting results have been observed, however, regarding the interactions between plant community composition and AMF communities, ranging from enhanced (Wu et al., 2007; Hiiesalu et al., 2014) to reduced plant diversity in the presence of AMF (Antoninka et al., 2011) to no relationship between plant and AMF diversity (Öpik et al., 2008). These contradictory findings may be related in part to study scales (Hempel, 2018), since different environmental forces work at different scales (Chase, 2014); this also applies to AMF (Vályi et al., 2016).
According to ecological theory, niche‐related (environmental/deterministic; MacArthur and Wilson, 1967) and neutral (stochastic; Hubbell, 2001) processes in particular shape community composition and habitat colonization. However, these processes appear to have different strengths at various scales: niche‐related effects are more common at larger (e.g., regional or global) scales, while neutral processes operate mainly at small spatial scales (Chase, 2014). Many studies have been conducted at broad geographical scales (Öpik et al., 2006; Hazard et al., 2013; Davison et al., 2015; Bouffaud et al., 2016, 2017), but to date little is known about richness and occurrence patterns of AMF at or within plot scales (≤ 50 m × 50 m) in grassland ecosystems (Lekberg et al., 2012; Horn et al., 2014). One advantage of such small‐scale study designs is the focus on environmental conditions and variations in plant communities within a specific habitat, thereby excluding overriding effects of large‐scale heterogeneity at the landscape level (Berner et al., 2011; Regan et al., 2017). Thus, fundamental influences on AMF communities can be studied at such plot or subplot scales, ranging from centimetre to metre. Repeating such sampling scales at one plot adds information on temporal autocorrelations (Tobler, 1970), providing an opportunity to investigate spatial hot spots and temporal hot moments simultaneously.
To understand temporal influences and to identify hot moments (Kuzyakov and Blagodatskaya, 2015) in changing AMF communities (Dumbrell et al., 2011), a study would need to cover the entire vegetation period by sampling soils at several time points. To date, few of the studies focused on temporal variation have sampled AMF communities more than twice during the growing season (Bainard et al., 2014; Liu et al., 2014). Repeated sampling is necessary, however, to account for seasonal variations in plant cover, which is likely coupled with changes in soil moisture, temperature and nutrient fluxes (e.g. phosphate and nitrate), and thus reflected in dynamic soil microbial communities (Bardgett et al., 2005). Even though a direct connection between AMF diversity, its abundance, and changes in plant diversity is not always apparent: Dumbrell et al. (2011) showed that during spring and summer, when plant growth is strong, environmental conditions and AMF distribution patterns are not constant. However, even fewer studies have investigated both spatial and temporal variations in AMF communities (Davison et al., 2012; Koorem et al., 2014; Barnes et al., 2016), and these studies have mainly been done on forest sites. Davison et al. (2012) found seasonal differences in AMF richness as well as distance decay in community similarity at three 10 m × 10 m forest plots sampled four times within one year, while Koorem et al. (2014) confirmed the seasonal variability in AMF by fatty acid analyses at small spatial scales (1.05 m × 1.05 m) sampled twice during one summer. Combining spatial and temporal sampling also makes it possible to quantify the beta‐diversity of AMF communities, which describes how species composition changes over spatial scales and over time. However, analyses of beta‐diversity have only rarely included microorganisms (e.g. Gossner et al., 2016).
Within the research platform ‘Biodiversity Exploratories’ (Fischer et al., 2010) the project SCALEMIC Experiment established a spatiotemporal sampling design in a low land‐use intensity grassland at the plot scale (10 m × 10 m) and assigned six sampling dates from spring to autumn in one vegetation season. Through an interdisciplinary approach, it was previously clarified that plant growth changes plot‐scale spatial heterogeneity of soil microorganisms during the vegetation period, and elucidated driving forces behind this observed microbial heterogeneity (Regan et al., 2014). We linked existing measures of seasonal and spatial changes in plant diversity, abiotic soil properties and general microbial community composition (Regan et al., 2014, 2015, 2017; Klaus et al., 2016) to AMF diversity and community patterns. Using high‐throughput sequencing technology, this study aimed to answer the following questions: (i) how much variability in AMF alpha‐ and beta‐diversity exists on a spatial scale of 10 m × 10 m and a temporal scale of one season?; (ii) are spatial and temporal AMF patterns coupled?; and (iii) which environmental drivers are responsible for the observed patterns? We expected a strong relationship between the AMF community and its changing environment, primarily vegetation and phosphate availability.
Results
Taxonomical distribution of AMF
We recovered 1 088 162 AMF SSU rDNA gene reads from all 360 soil samples. After a quality filtering step that included removal of 22 042 potential chimera and non‐AMF reads, we had a total of 562 320 AMF reads representing 1562 reads per sample, and which were clustered into 155 abundant operational taxonomical units (OTUs). As described in detail in the ‘experimental procedures’ section, the removal of rare OTUs (OTUs represented by ≤ 3 reads) had no significant effect on AMF beta‐diversity. Thus, the AMF matrix including only abundant OTUs was used for further analyses.
The 155 abundant AMF OTUs were assigned to seven genera: Acaulospora (2 OTUs), Ambispora (2), Archaeospora (3), Claroideoglomus (20), Diversispora (9), Glomus (117) and Paraglomus (2). Taxonomical distribution based on the number of observed AMF OTUs differed slightly between sampling dates. The genus Glomus was most abundant throughout the entire growing season, ranging from 69% in April to 77% in November (Fig. 1, Supporting Information Table S1) with the highest diversity (a total of 106 OTUs) detected in October. Besides Glomus, other AMF genera displayed temporal peaks; e.g. Claroideoglomus in June (16.4%) and Diversispora in April (8%).
Figure 1.
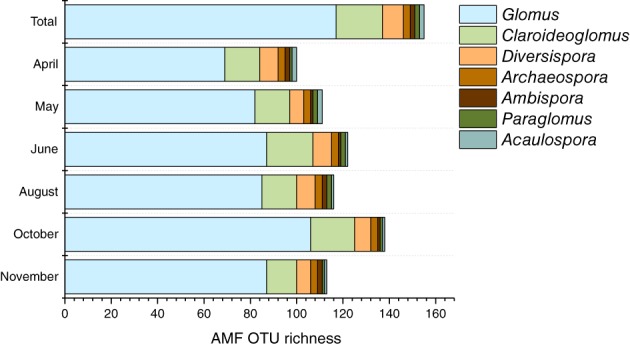
Bar graphs representing the temporal distribution of AMF OTUs of Glomeromycota genera detected across the entire plot.
Spatiotemporal variation in AMF richness
The OTU richness of total AMF was spatially modelled and checked for autocorrelation. Kriged maps were generated for all sampling dates except October, at which date the empirical variogram model was a pure nugget, indicating no spatial autocorrelation at the measured scale (Fig. 2A‐E). The observed patterns occurred and were distributed throughout the entire AMF community over the entire sampling season. In April and May (Fig. 2A and B), AMF diversity was homogeneous with low OTU richness across the plot. An increase in AMF OTU richness was detected in June (yellow areas in Fig. 2C). Moreover, first patches developed in June, and became more pronounced in August and November (Fig. 2D and E). In general, total AMF OTU richness decreased at the end of the growing season (increase of dark green in the kriged maps), but discrete hot spots and cold spots with high or low AMF OTU richness appeared.
Figure 2.
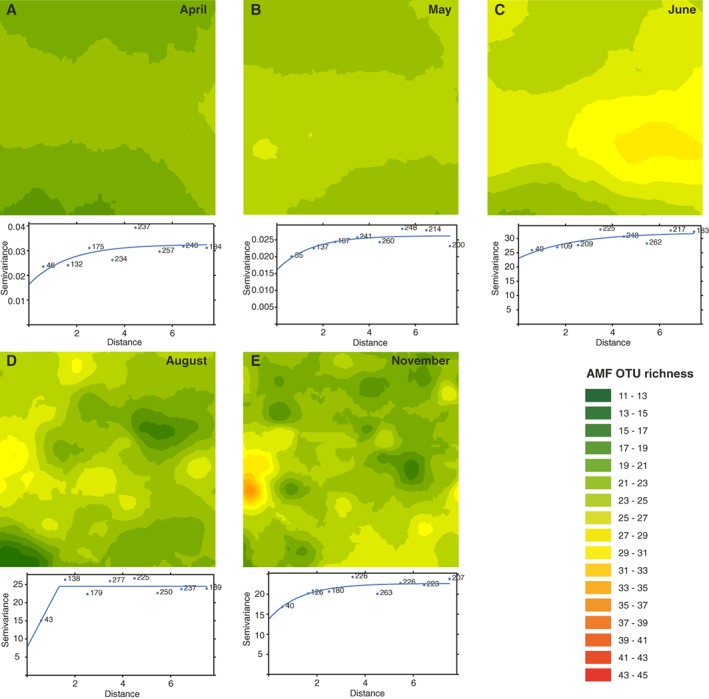
Geostatistical data analysis of AMF OTU richness with all AMF OTUs grouped together per sampling date: (A) April, (B) May, (C) June, (D) August and (E) November. Spatial patterns within the data were analysed and calculated as semivariogram models (lower panels in figure) and visualized as kriged maps using these models (corresponding upper panels in figure). Dimensions of all maps are 10 m × 10 m.
For the two most abundant AMF genera, Glomus and Claroideoglomus, OTU richness was also spatially modelled and could be visualized through kriged maps (Supporting Information Figs S1 and S2). Spatial distribution of Glomus could be modelled in May, June, August and November, while the spatial distribution of Claroideoglomus could only be modelled in August and November. As was the case for all AMF OTUs, Glomus OTU richness was low to medium in May and June (Supporting Information Figs S1a, b), tending towards spatial patches of low or high richness. Heterogeneity of distribution became more pronounced in August with two spots of high OTU richness (Supporting Information Fig. S1c). However, a shift in OTU richness occurred in November (Supporting Information Fig. S1d) with areas of low and high OTU richness of Glomus. Claroideoglomus OTUs exhibited similar spatial patterns in August with areas of low and intermediate richness (Supporting Information Fig. S2a) and in November generally lower richness but more heterogeneous distribution across the plot (Supporting Information Fig. S2b).
The effect of sampling date on AMF richness, assessed by linear mixed effect models (LMEM), was plotted for all OTUs and additionally for the OTUs of the two most abundant genera, Glomus and Claroideoglomus (Supporting Information Fig. S3). Sampling date significantly influenced richness of both total AMF and Glomus OTUs (P < 0.0001), but not Claroideoglomus OTUs. OTU richness significantly increased in all AMF OTUs from April to June, dropping in August. Total AMF OTU richness peaked in October and dropped significantly in November. The richness of Glomus OTUs was similar with significantly lower richness in April and a peak in October. The OTU richness of Claroideoglomus did not change over the sampling season.
Environmental impacts on AMF richness
Linear mixed effect models, taking into account the impact of all available environmental factors (n = 34) on total AMF OTU richness, revealed significant effects of soil‐ and plant‐related parameters in 21 cases, as well as 23 significant effects of environmental variables on Glomus OTU richness. Claroideoglomus OTU richness was significantly affected by soil carbon content, but this explained only 3% of its variance. To detect those environmental variables most closely associated with the observed temporal effect on total AMF and Glomus OTU richness, we combined environmental variables and sampling date as fixed effects in LMEMs. This combination indicated that the measured environmental variables explained less unique variance in total AMF and Glomus OTU richness than did sampling date. The final LMEM (Table 1) for total AMF OTU richness pointed to plant species richness, percent silt content and sampling date as the three main drivers at the investigated site, which, taken together, explained 36% of the variance. Silt content had a positive effect, while plant species richness was slightly negatively associated with total AMF OTU richness. Temporal variation was reflected by the significant effects of sampling time and indicated by the different intercepts of the single months (Table 1). Glomus richness was driven by plant species richness, root biomass, percent silt content, pH and sampling date, which together explained 38% of the variance in Glomus. Here, we found a slightly negative impact of plant species richness, while root biomass, silt content and pH positively affected the OTU richness of Glomus. For both total AMF and Glomus OTU richness, sampling date explained large proportions of the variance, 27% and 28%, respectively. As plant species richness was only assessed at three sampling dates, we additionally fitted models without this variable for total AMF and Glomus OTU richness to determine the best predictor variables across the whole season. When all six sampling dates were analysed, total AMF OTU richness was not influenced by plant variables; instead, there was a slight negative association with soil NH4 + content. Glomus OTU richness was best predicted by legume and root biomass, NH4 +, silt content, pH, the fungal to bacterial ratio and sampling date over the entire season (see Table 1).
Table 1.
LMEM results for richness of entire AMF, Glomus, and Claroideoglomus for three and six sampling dates.
| Sampling dates | Target | Model coefficients of fixed effects | n | Percentage explained variance | ||
|---|---|---|---|---|---|---|
| Random effects | Fixed effects | |||||
| Environmental variables | Sampling time | |||||
| 3 | all AMF OTU = | −0.57 * plant species no. + 1.7 * silt content + 23.39 (for sampling time May) + 25.47 (for sampling time June) + 30.95 (for sampling time October) | 180 | 48 | 9 | 27 |
| 6 | all AMF OTU = | −0.7 * NH4 + + 1.3 * silt content + 22.25 (for sampling time April) + 22.81 (for sampling time May) + 25.3 (for sampling time June) + 22.82 (for sampling time August) + 30.5 (for sampling time October) + 22.96 (for sampling time November) | 360 | 45 | 6 | 27 |
| 3 | Glomus OTU = | −0.41 * plant species no. + 0.5 * root biomass + 1.5 * silt content + 0.75 * pH + 15.78 (for sampling time May) + 16.73 (for sampling time June) + 22.9 (for sampling time October) | 180 | 60 | 10 | 28 |
| 6 | Glomus OTU = | 0.37 * legume biomass +0.34 * root biomass − 0.65 * NH4 + + 1.25 * silt content +0.35 * pH − 12.73 * fungi:bacteria ratio + 15.46 (for sampling time April) + 16.73 (for sampling time May) + 18.06 (for sampling time June) + 17.41 (for sampling time August) + 23.82 (for sampling time October) + 17.96 (for sampling time November) | 360 | 54 | 7 | 14 |
| 6 | Claroideoglomus OTU | 0.57 * Ctotal | 360 | 11 | 3 | — |
Given are significant environmental variables with their coefficients (data z‐transformed for comparison between coefficients), number of samples in the model as well as explained variances. Subplot number was used as random effect (intercepts not displayed). n = number of samples.
Spatiotemporal variation in AMF community composition
AMF beta‐diversity (βSOR) was lower within time points and within subplots than between these groups (ANOSIM P‐value = 0.001). When the data set was stratified by sampling date, silt content, pH, microbial biomass, soil C content and K2SO4‐extractable organic N significantly explained variation in βSOR. However, these variables together explained less than 10% of the variation in βSOR. No relationship between AMF βSOR and any plant variable could be detected. Additionally, a spatial gradient explaining 4% of the variability was observed (Supporting Information Tables S2 and S3).
The AMF βSOR of the subplots between one time point and the one immediately following varied slightly (Supporting Information Fig. S4). However, there was no stronger correlation between subplots near each other in comparison to those subplots further distant (Supporting Information Fig. S5; for data on Glomus and Claroideoglomus see Supporting Information Figs S6 and S7, respectively). No significant correlations between the temporal development of βSOR and environmental variables were observed. Turnover (βSIM), meaning OTU replacement between time points, and nestedness (βSNE), meaning OTU gain and loss from one time point to the next, are summarized in Fig. 3 (for data on Glomus and Claroideoglomus, see Supporting Information Fig. S8). The turnover in AMF community composition appeared to be constant between 0.3 and 0.4 during the sampling season. The highest AMF βSIM was detected between June and August, which is likely linked to the fact that this difference represented a duration of two months. βSNE peaked later in the season, particularly between August and October, but also between October and November. However, kriged maps revealed hot spots of turnover from April to May as well as from October to November (Supporting Information Fig. S9).
Figure 3.
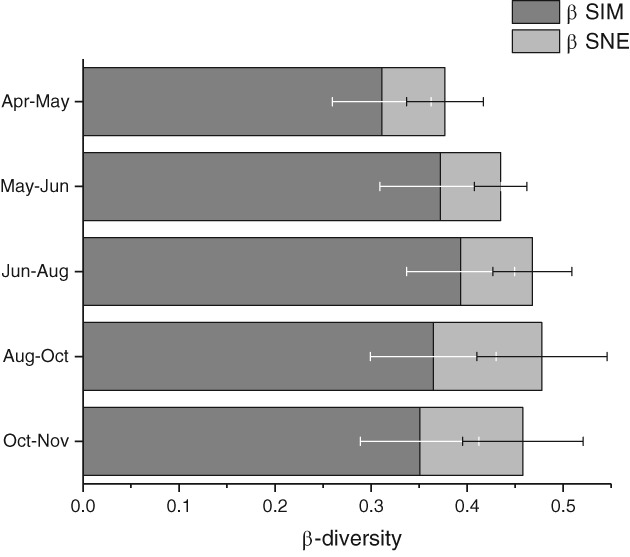
Patterns of variability within AMF assemblages across the studied plot from one time point to the next. Stacked bars represent overall beta‐diversity (βSOR) observed in the partial data sets, computed using the R package betapart (Baselga & Orme, 2012); dark grey sections of the bars represent the contribution of the turnover of AMF (βSIM), light grey sections account for the nestedness of AMF (βSNE); error bars represent variability between SCALEMIC subplots.
Analysis of spatial AMF βSOR demonstrated some continuity within subplots early in the growing season (see supplemental material for more details; Supporting Information Figs S10 and S11). The relationship between temporal βSOR (the average βSOR over time) and spatial βSOR (the average AMF βSOR with the neighbouring subplots) of each subplot displayed a positive trend (Fig. 4), indicating that subplots with AMF communities that differed strongly from neighbouring subplots also changed more over time. The positive relationship was significantly stronger than the relationship observed in null‐models of βSOR, which were based on random community permutations that maintained each sample's richness and the overall or sampling date point‐specific probability of OTU occurrence (Supporting Information Fig. S12). Concurrently, no significant relationships between OTU richness or changes in alpha‐diversity with spatial or temporal βSOR were observed. The same pattern was observed for the βSOR of Glomus OTUs, while Claroideoglomus βSOR did not differ significantly from the null‐models (Supporting Information Fig. S12). No explanatory power was gained by adding environmental variables to the linear model explaining temporal βSOR with spatial βSOR. Among the environmental variables, the mean grass biomass best explained spatial βSOR of all OTUs (P‐value 0.01, adjusted R 2 = 0.43).
Figure 4.
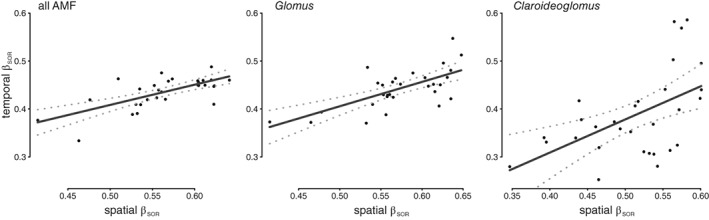
Relationship between spatial and temporal βSOR of total AMF, Glomus or Claroideoglomus, respectively. Spatial indices of AMF turnover (x‐axis) represent the AMF turnover between each subplot and its neighbours averaged over all sampling dates. Temporal indices (y‐axis) represent the mean delta (turnover from one time point to the subsequent time point). Regression lines (black) are based on linear models and 95% confidence intervals (grey dotted lines).
Discussion
General characterization of AMF
AMF form a multispecies mutualism with over 80% of terrestrial plants, i.e. with more than one fungus per host plant (Smith and Read, 2008). With a total of 155 abundant AMF OTUs on a 10 m × 10 m plot over a vegetation period that extended from April to November, our study found relatively high AMF richness compared to recent studies (Dumbrell et al., 2010; Davison et al., 2012; Horn et al., 2014).
All four orders of the phylum Glomeromycota and seven genera were represented in the observed OTUs. Dominant at all time points was the AMF genus Glomus. This dominance is in agreement with previously published studies in grasslands, forests and agricultural ecosystems (Daniell et al., 2001; Gai et al., 2009; Öpik et al., 2009). Although AMF OTU richness was high, genera such as Rhizophagus or Funneliformis, which have often been observed in a comparable German grassland of low land‐use intensity (Horn et al., 2014), were not detected in our dataset.
Spatiotemporal variation in AMF alpha‐diversity
Our current study determined whether AMF richness exhibits spatiotemporal variation at a small spatial scale in a grassland soil, and which environmental variables shape differences in AMF alpha‐diversity. Studies in which other results of the SCALEMIC Experiment have been published observed an increase in biomass of grasses and forbs until June and additionally a gain in biomass of legumes in October as well as significant shifts in mineral nitrogen content of soils over time (Regan et al., 2014). Moreover, temporal shifts in plant biomass and nutrient availability were detected (Klaus et al., 2016). Results of our spatial analyses illustrate clearly how AMF OTU richness varied across the sampled plot and also over the season. The appearance of hot or cold spots of AMF richness showed a dynamic process that developed during the vegetation period. The detected spatial autocorrelation of AMF OTUs with ranges below 10 m across our plot for five of six time points is in accordance with previous studies, e.g., by Bahram et al. (2015), who reported autocorrelation ranges around 9 m. Richness of AMF OTUs therefore shows distance‐decay relationships at the investigated plot scale.
Interestingly, although more than 100 OTUs were detected on the entire 10 m × 10 m plot at each sampling date, many of these AMF appeared in patches of 20‐30 OTUs per sampling point. This discrepancy between total observed OTU richness across the plot and OTU richness per individual sampling point may be related to carrying capacity (Allen, 1989). We define carrying capacity as the maximum number of AMF OTUs the studied SCALEMIC grassland plot can sustainably support. Accordingly, the local AMF carrying capacity appears to have been patchy but potentially dynamic over time. The observed temporal dynamic could be connected to changes in resource availability. This could be related to asynchronous growth of plants across the site (Yachi and Loreau, 1999). For instance, a changing supply of photo‐assimilates could be accompanied by dense AMF population sizes at one sampling point (hot spot), but reduced AMF richness at another point (cold spot) on the plot within or across sampled time points. This is in line with findings of the linear mixed effect models, which found a connection between AMF richness and changing environmental variables such as plant species richness, root biomass, pH and NH4 +. To the best of our knowledge, this has not been shown previously and underscores the need for more temporal investigations.
Restricted impact of environmental variables on AMF richness
Linear mixed effect model analyses revealed a significant effect of soil texture on the total richness of both AMF and the genus Glomus across the plot. The heterogeneous distribution of silt in our soil modified important habitat conditions. Hot spots of high silt content are characterized by larger volumes of medium‐sized pore space and improved aeration in the surrounding microenvironment (Horn et al., 2010), resulting in favourable habitat conditions for AMF, which lead to increased AMF OTU richness. Although the dependence of AMF on soil texture in grassland soils was shown in a large scale study (Oehl et al., 2017), no studies have yet demonstrated that this effect occurs with small textural changes (changes in silt content < 10%) at the plot scale. Soil texture significantly affected temporal variations in AMF OTU richness even though it was temporally stable. We suggest that this is because soil texture influences a number of habitat conditions such as nutrient availability, pore space distribution and thereby also the hydrological budget and oxygen supply (Horn et al., 2010), which themselves vary over time. That soil texture was a better measure than single effects, e.g. soil water content, indicates its value as a measurement that captures a range of temporal variations in texture‐dependent habitat conditions. This result emphasizes the importance of microhabitat conditions for AMF.
In addition to soil texture, plant species richness was a significant driver of OTU richness for both total AMF and Glomus in the months of May, June and October. Interestingly, a reduction in plant species richness led to an increase in total AMF and Glomus OTU richness. Argüello et al. (2016) described a positive feedback mechanism for AMF‐plant mutualism, leading to stronger cooperation between AMF and plants under the condition of high AMF diversity per plant. In addition, newly emerging plant species over the season may have had different root architectures such as less root biomass, leading to a reduction in AMF OTU richness, as higher root biomass significantly increased the OTU richness of the genus Glomus. It has also been suggested that both AMF and plants actively control their associated symbiotic partners (van der Heijden et al., 2015), in which case changes that resulted in a more cooperative plant community could influence AMF OTU richness. Similarly, the newly emerging plant species may have had more acidic root exudates, which would have reduced the OTU richness of Glomus, as Glomus OTU richness decreased with decreasing soil pH. A similar effect of pH on AMF OTU richness in grassland ecosystems was also observed by Heyburn et al. (2017). Even when they were evaluated over all six sampling dates throughout the season, the effects of soil texture, pH and sampling date remained significant. Over the longer time, the impact of plants on total AMF OTU richness was not significant; instead, a negative effect of NH4 + was detected. The same was true for OTU richness of Glomus. As NH4 + reduces soil pH, this was likely a combined effect of the two soil properties. In addition, an increase in legume biomass led to an increase in Glomus OTU richness, which may have been due to increased coverage of the mycorrhizal plant partners such as Trifolium pratense (L.) (van der Heijden et al., 1998) and Vicia sepium (L.) (Closa and Goicoechea, 2011) over time. The negative relationship between the fungal:bacterial ratio of phospholipid fatty acids and Glomus richness was related to an overall increase in saprotrophic fungi at the site (Regan et al., 2014), which was accompanied by a reduction in Glomus richness, indicating both competition for resources (Hodge et al., 2001) and interactions due to fungal community composition (Tiunov and Scheu, 2005) at the study site. However, not all AMF genera reacted in the same way. Claroideoglomus was affected neither by soil texture nor by any of the above‐mentioned environmental properties; instead, a small but significant effect of soil carbon content on this genus was detected.
Even though environmental soil properties explained a portion of the variance in AMF alpha‐diversity, sampling date was the most important driver of total AMF and Glomus OTU richness. Previous studies have reported an increase in AMF OTU richness during the growing season with a decrease in autumn, which could be explained by changing weather conditions within a sampling year (i.e. temperature and precipitation; Kabir et al., 1998; Staddon et al., 2003). In our study, there were significantly fewer AMF OTUs in November than in June or October. This phenomenon may have been related to cold temperatures and less precipitation at this sampling date [see supplemental Fig. A1 of Regan et al. (2014)]. Also, the observed decline in AMF OTU richness in August may have been associated with a suppression of plant growth and reduced carbon supply from plant to fungus after the mowing event (Gehring and Whitham, 2002). This mowing event and the subsequent regrowth of plants could have led to the high number of AMF OTUs detected in October. It was shown recently that more AMF propagules are present in mown than in unmown soils (Binet et al., 2013), benefiting new AMF infections after mowing. In addition, this is likely connected to increasing root exudation following aboveground plant biomass removal (Waters and Borowicz, 1994). More diverse exudates are likely to recruit a greater AMF diversity (Hugoni et al., 2018). Consequently, mowing leads to emerging micro‐niches, which favour a higher variability in AMF. Thereby, AMF can be considered as stress tolerant since they can cope with partition and destruction of their hyphae (Buscot, 2015). Since up to only 10% of the explained variance was directly attributable to measured environmental effects (see Table 1), our results could indicate that neutral processes, stochasticity or randomness due to natural variability may play a role in the formation of unpredictable AMF patchiness in addition to the contribution from deterministic processes. This applied to both total AMF OTUs and OTUs of the genera Glomus and Claroideoglomus.
Pronounced spatiotemporal relationships in AMF beta‐diversity
In addition to alpha‐diversity, this study sought to understand whether or not beta‐diversity in AMF exhibited similar spatiotemporal patterns. With respect to OTU richness, AMF beta‐diversity expressed as Sørenson index indicated spatiotemporal relationships. Theoretically, local AMF communities should be of a common and predictable composition since the species pool at the plot scale is limited; thus, beta‐diversity in both spatial and temporal senses should be low according to Powell and Bennett (2016). However, in our study, AMF beta‐diversity appeared high, with a particularly high turnover rate from one observed time point to the next. Although the turnover rates were high (consistently ≈40% of the AMF community changed from one time point to the subsequent one), a certain spatiotemporal stability of AMF communities was observed. Our results suggest that AMF community composition at the first three time points (April‐June) was determined by prior communities. It is possible that DNA measures either detected defective and dead cells (Carini et al., 2016), or dormant AMF stages, such as spores from the previous year, which impacted the observed AMF community at the beginning of the vegetation period. This effect was lost during the summer, which suggests that either legacy effects due to the cyclic character of seasons in temperate regions (Bahram et al., 2015) or the appearance of priority effects (Viana et al., 2016) shaped AMF community composition, as has been shown for soil bacteria (Francioli et al., 2016, 2018). The plant‐AMF interaction may have been set back to zero during winter, resulting in a random start of plant growth and fungal infections during spring. These priority effects could have resulted in high heterogeneity within subplots, which decreases over the season. Coupled with this is the fact that competition among AMF emerges only over time (Maherali and Klironomos, 2012). Alternatively, AMF detected from spores in our analyses transformed from spores while colonizing growing plant roots during the second half of the vegetation period. In the second half of the year, these AMF may have dropped below the molecular detection limit. Also, the mowing event before sampling in August shuffled AMF community composition, since mowing is known to multiply AMF propagule numbers (Binet et al., 2013). However, we could not identify a direct link between either the plant community taken together or between single plant species and the AMF community. This missing link between these two communities (Hart et al., 2001) supports the ‘independence hypothesis’ which suggests that neither plants nor AMF express any covariation at all in this mutualism (Zobel and Öpik, 2014).
Recent studies mention dispersal limitation of AMF (Davison et al., 2015) as one reason for patchiness in community composition at small scales. This results in ‘unpredictable assembly’ (Powell and Bennett, 2016) of AMF, which corresponds well with our findings that approximately 80% of variation in βSOR could not be explained by environmental variables. This could indicate that stochastic rather than niche‐related processes shape AMF βSOR. However, one general pattern could be identified: over time, AMF community composition differed less within than between subplots. More similar environmental conditions found within a subplot appeared to result in significantly lower beta‐diversity over time. This indicates that even small environmental differences between two subplots affected AMF community composition. Indeed, it has previously been shown that pH, C, N, P and soil water content shape AMF grassland communities (Horn et al., 2014). In our study, around 20% of the observed variation in AMF beta‐diversity could be explained by measured environmental factors. Nevertheless, it is possible that the low variance explained by environmental variables in our study indicates the influence of important but unmeasured variables, and not stochastic processes as such. To confirm that neutral processes shape spatiotemporal AMF beta‐diversity, future studies should consider microscale effects such as root exudates and pore space to identify currently unidentifiable drivers.
Conclusions
Our study of AMF alpha‐ and beta‐diversity found spatiotemporal distribution patterns at the observed plot scale of 10 m × 10 m. We were able to illustrate well both the dynamism of AMF OTU richness, and community development across one vegetation season. Thereby, we demonstrated, albeit indirectly, that stochastic recruitment processes largely shaped our observed patterns of AMF OTU richness and community composition. If seasonal variations in carrying capacity are considered, then shifts in plant growth, diversity and dominance are likely to favour AMF species already engaged in the symbioses. However, our results revealed high AMF turnover over time, suggesting ongoing recruitment of AMF from formerly dormant propagules. We acknowledge that the detection of niche‐based processes could have been limited by the choice of our measured environmental parameters, which were either unable to detect them or wrongly selected for this purpose. Nonetheless, both the scale and spatiotemporal approach of the SCALEMIC Experiment have expanded our understanding of biotic and abiotic interactions at scales that had heretofore not been examined in such detail. Further research, ideally on more than one site, is needed for a deeper and more comprehensive understanding of the spatiotemporal assembly of soil microbes at small scales by assessing and linking functions of bacteria and fungi with plant traits. Likewise, and within the frame of an emerging discussion as to whether AM fungal communities are more structured by the abiotic or biotic environment (Hempel, 2018), future studies should incorporate balanced consideration of environmental variables.
Experimental procedures
Study site and soil sampling
The studied grassland plot (48°27′31.37″N, 9°27′36.26″E) is one of 300 experimental plots in the large and long‐term interdisciplinary research project ‘Biodiversity Exploratories’, which aims to understand relationships between land‐use, multitrophic biodiversity and ecosystem functioning across Germany (Fischer et al., 2010). The grassland plot is located in the Schwäbische Alb in southwest Germany. The plot has never received mineral fertilizers and has never been ploughed. Characterized by a rather nutrient‐poor substrate, this plot's soil type is a Rendzic Leptosol (FAO classification). The plot is dominated by Plantago lanceolata L., Festuca rubra L and Helictotrichon pubescens (Huds.) Pilg. and belongs to the phytosociological class of Festuco‐Brometea (Oberdorfer et al., 2001; Klaus et al., 2016). Furthermore, the grassland is usually mown once per year, and grazed briefly by sheep for 1‐2 weeks in late summer or early autumn. In 2011, the year of investigation, mowing took place on 30th July and sheep herds grazed on this site in May for five days, in September for seven days and in October for one day.
The SCALEMIC Experiment (Regan et al., 2014) encompasses a 10 m × 10 m plot divided into 30 subplots, each 2 m × 1.67 m (Supporting Information Fig. S13). Within each subplot six pairs of sampling locations (each 20 cm × 20 cm) were randomly assigned, with one pair sampled at each of six dates over one growing season. This provided a randomized complete block design for temporal data analysis with sampling date as ‘treatment’ factor, subplots as complete blocks and pair of sampling locations as randomization unit. Sample pairs were separated by 50 cm to provide appropriate lag distances for geostatistical analyses. Sampling dates were chosen along a seasonal gradient with the following characterization: (i) beginning of vegetation (5th April), (ii) stage of main plant growth (17th May), (iii) peak of plant biomass (27th June), (iv) two weeks after mowing (16th August), (v) nine weeks after mowing (5th October) and (vi) after the first frost (21st November). Accordingly, a total of 360 soil samples were collected (60 per date × 6 dates) in the year 2011.
Soil samples were collected with core augers (diameter 58 mm). The upper 10 cm layer was taken at each sampling point (Supporting Information Fig. S13) after vegetation was removed and the top one cm, consisting of litter, was discarded from the sample. Subsequently, the soil was immediately stored at 4 °C and sieved (< 5 mm) within 24 h after sampling to remove stones, roots and macrofauna. An aliquot for molecular analyses was stored at −20°C before processing in the laboratory. A detailed description of the sampling design and procedure can also be found in Regan et al. (2014).
DNA extraction and pyrosequencing of AMF amplicons
DNA was extracted from two replicates of each homogenized soil subsample (300 mg each) according to the manufacturer's protocol using the FastDNA® SPIN Kit for Soil (MP Biomedicals, Solon, OH) as described in Stempfhuber et al. (2016). Independent measurements of DNA concentration from both sample replicates were made on a NanoDrop® ND‐1000 spectrophotometer (Thermo Scientific, Wilmington, DE). The replicates were subsequently pooled and re‐measured, confirming the final DNA concentration of each sample, which was diluted to a PCR template concentration of 5 ng DNA ml−1 with ultra‐pure water. A semi‐nested PCR protocol was used to amplify the ≈630 bp‐long small subunit (SSU) region of the AMF 18S rDNA via pyrosequencing analysis (454 GS‐FLX, Roche). In the first PCR run (PCRI) a Glomeromycota‐specific region was amplified with the primer set GLOMERWT0/GLOMER1536 (Wubet et al., 2006), followed by the semi‐nested second PCR reaction with the forward general fungal primer NS31 (Simon et al., 1992) including the A adaptor and a 10 bp multiplex identifier (1 of 60 different MIDs), and the B adaptor including the reverse modified AMF primer AM1a and AM1b (Morris et al., 2013). The first PCR was carried out at a 25 μl reaction volume with 0.5 μl of diluted DNA template (5‐20 ng μl−1), 12.5 μl GoTaq Green Mastermix 2× (Promega, Mannheim, Germany), 1 μl of each primer (25 μM) on an Eppendorf Mastercycler DNA Engine Thermal Cycler PCR (Eppendorf, Hamburg, Germany) with the following PCR conditions: 98°C for 30 s, 5 cycles of 94°C for 30 s, 60°C (−1 °C/cycle, 4 cycles) for 30 s, 72°C for 1 min and 25 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and for extension 72°C for 5 min. For the semi‐nested PCR, two separate amplifications were performed using 1 μl of the diluted amplified product of PCRI (1:10), 25 μl GoTaq Green Mastermix 2x, and 1 μl of each primer (25 μM); these 50 μl reactions were run under the following conditions: 98°C for 30 s, followed by 30 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 1 min and 72°C for 5 min.
Each sample in both PCR amplification steps was amplified in triplicate and accompanied by a negative control. The semi‐nested PCR amplified products were pooled per sample, taking into account the amplicon concentration (checked by a 1.5% agarose gel). Pooled samples (30 μl each) were purified with the QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's recommended protocol. The purified products were quantified by fluorometry using Quant‐iT™ PicoGreen® dsDNA Assay Kit (Life Technologies GmbH, Darmstadt, Germany) as suggested by Roche Diagnostics GmbH (Mannheim, Germany) for amplicon library preparation. Equimolar concentrations of 60 MID tagged amplicons were loaded into individual lanes on a GS‐FLX LUMITRAC 600 plate (Titanium Series) separated with a four‐lane gasket and sequenced at the Department of Soil Ecology, UFZ – Helmholtz‐Centre for Environmental Research (Halle/Saale, Germany).
Bioinformatic analysis of sequence data
Sequence read quality filtering and splitting of the dataset into individual samples was performed using mainly MOTHUR (Schloss et al., 2009). Sequences were trimmed using the ‘keepfirst’ command in order to discard sequences with less than 300 bp and chopping at least 50 bp from potential noisy sequence ends. Simultaneously, all sequences with average quality scores of below 20 as well as MID‐ and primer sequences were removed. Sequences were then downsampled to the smallest read number per sample (1562 sequences per sample) and potential chimeric sequences were identified and removed by UCHIME (Edgar et al., 2011) as implemented in MOTHUR. These quality‐filtered sequences were clustered into OTUs based on the algorithm implemented in CD‐HIT‐EST (Huang et al., 2010) with a sequence similarity threshold of 97%. The representative sequence for each resulting OTU was compared to a Global Alignment for Sequence Taxonomy (GAST)‐based taxonomic assignment of an NCBI based fungal reference data set (Huse et al., 2008) at the 97% similarity level. All non‐Glomeromycota OTUs were removed from the dataset (≈11% of sequences). Representative sequences (most abundant sequence per OTU) of the Glomeromycota OTUs were further taxonomically assigned by using the MaarjAM virtual taxa reference database (web‐based database for studies of the diversity of arbuscular mycorrhizal fungi, version 0.8.1 beta; Öpik et al., 2010).
The raw SSU DNA sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under study accession number SRP137677. In addition to all measured values, the analysed and processed data used in this study can be found in the BExIS database (https://www.bexis.uni-jena.de/).
Environmental properties
The interdisciplinary approach of the SCALEMIC Experiment permits data from previous works on various environmental properties of the site to be used in the present study. We collected information on plant diversity (richness, Shannon index) on three sampling dates (May, June and October) and plant biomass (aboveground biomass of grasses, herbs and legumes; root and litter biomass; Regan et al. (2014), Regan et al. (2015), Klaus et al. (2016)). We also measured soil texture, bulk density, water content, mineral nitrogen (Nmin = NH4 + + NO3 −), total carbon and nitrogen, extractable organic carbon and extractable nitrogen (EOC, EN), bioavailable phosphate (PO4 3−), pH, bacterial and fungal biomass (phospholipid fatty acid (PLFA) content; Regan et al. (2014)); as well as bacterial and archaeal abundances (qPCR on 16S rRNA; Regan et al. (2017)). These variables were used in statistical analyses to determine their explanatory power with respect to AMF OTU richness and community composition (see detailed list of environmental properties in Supporting Information Table S4).
Statistical analyses
Statistical analyses were performed using the software R (version 3.4.0; R Development Core Team, 2017) unless stated otherwise. First, to test whether rare AMF taxa (OTUs represented by ≤ 3 sequence reads per sample) affected estimates of beta‐diversity, we performed Procrustes correlation analysis based on Bray Curtis dissimilarity using the protest function (Peres‐Neto and Jackson, 2001) of the ‘vegan’ R package (Oksanen et al., 2018). This approach provides information about congruence between two non‐metric multidimensional scaling (NMDS) ordinations; in our case AMF data matrices comprising all OTUs and only abundant OTUs (OTUs represented by > 3 sequence reads per sample) with 999 permutations. Results indicated nearly identical ordinations in the presence or absence of rare AM fungal OTUs on AMF beta‐diversity (Procrustes correlation coefficient = 0.9915, P = 0.001). Hence, all subsequent analyses were performed using the relative abundance AMF community matrix excluding singletons, doubletons and tripletons.
To assess the spatial distribution of the richness of all AMF OTUs, and of the OTUs belonging to the genera Glomus and Claroideoglomus, semivariogram analyses were performed using the R package ‘gstat’ (Pebesma, 2004). Data were checked for normality of distribution and were log or square root transformed if necessary according to McBratney and Webster (1986). As environmental data did not show general distribution trends across the study site in preliminary analyses, isotropy was assumed for semivariogram analysis. Subsequently, empirical semivariograms for the three AMF groups at each sampling date were computed separately. In cases where empirical semivariograms indicated spatial autocorrelation, semivariogram models were fitted using the ‘fit.variogram’ function. Bin sizes were restricted to minimum 35 points per bin; spherical, exponential and linear models were fitted using the default method of the ‘fit.variogram’ function. The model with the lowest sum of squared error (SSErr) was selected. To estimate the amount of variance that was spatially correlated, the percent spatial structure was calculated by subtracting the nugget effect from the sill, and dividing the remaining, or partial sill, variance by the sill variance. Kriged maps for semivariogram models were generated with ArcGIS (ESRI, 2010, Environmental Research Institute, Redlands, CA).
The effects of sampling date and environmental variables on the OTU richness of all AMF and of the genera Glomus and Claroideoglomus were assessed using LME models accounting for the spatial sampling design. First, different model structures were tested with SAS 9.4 (SAS Institute Inc., Cary, NC) using subplot number as random block effect, and models were evaluated and chosen based on Akaike's information criterion (AIC; see Supporting Information Table S5). The addition of spatial autocorrelation structures as well as addition of a random effect for pairs of sampling locations did not substantially improve model fit, and this was also the case for an addition of temporal autocorrelation structure. As the model was to be used repeatedly for selection of important covariates, we chose a model that included a subplot effect and residual error as random effects. The resulting model structure was as follows:
where y ijk is the value of the response variable for the i‐th sampling date on the j‐th subplot at the k‐th sampling location, b j is the random effect for the j‐th subplot, β 1−n are the slopes of the regression on the predictor variables (= fixed effects) x ijk(1−n), and e ijk is the independently normally distributed error term with constant variance. This model structure was subsequently used for all LMEMs, which were computed in R using the package ‘nlme’ (Pinheiro et al., 2017). We separately assessed the effects of sampling date and environmental variables in univariate models for each independent variable for total AMF OTUs, Glomus and Claroideoglomus. To detect those environmental variables which were most strongly related to the observed temporal effects, additional LMEMs were computed on the combination of individual environmental variables together with sampling date. The best predictors among plants as well as abiotic and biotic soil properties were then included in the final LMEMs. These contained all significant drivers, and were set up separately for total AMF OTUs, Glomus and Claroideoglomus. Models with the best predictor variables were selected based on lowest AIC (based on full maximum likelihood) using the ‘stepAIC’ function with forward and backward selection, and checked for homoscedasticity and normal distribution of residuals. Spearman correlations of environmental variables in the final models revealed no considerable multicollinearity. Explained unique variance of dependent variables by independent variables and random effects was assessed following the approach of Nakagawa and Schielzeth (2013) using the function ‘sem.model.fits’ of the ‘piecewiseSEM’ package (Lefcheck, 2016). Separate models were calculated for models that contained plant diversity data because these were only available at three sampling dates.
To assess AMF beta‐diversity, Sørensen distances (βSOR), as well as their turnover (βSIM) and nestedness (βSNE) components, were calculated using a function generalizing the ‘beta.sample’ algorithm from the R package ‘betapart’ (Baselga and Orme, 2012; see supplement material for further details). In accordance with the nested study design, the means of the distances between all combinations of within‐subplot repetitions were calculated. Significance of groupings of community composition by sampling date and subplot were assessed by analysis of similarities (ANOSIM), as implemented in the R package ‘vegan’ (Oksanen et al., 2018). To compare community structures at different sampling dates within and between subplots, the complete Sørensen distances between all samples were calculated and the median values of all pairwise distances matching each comparison were extracted and visualized using the R package ‘beanplot’ (Kampstra, 2008). Significance of comparisons was established using the non‐parametric Mann‐Whitney test.
To detect which environmental variables were sources of variation in βSOR, permutational multivariate analysis of variance based on the Sørensen distance matrix was performed using the function ‘adonis’ from the R package ‘vegan’ (Oksanen et al., 2018). First, all z‐transformed environmental variables were applied in separate univariate models, with stratification by sampling date. In a second model, all significant variables were combined in descending order of their significance in the first run, again with stratification by sampling date. Spearman correlations of the significant variables revealed no considerable multicollinearity.
Spearman correlations between the temporal developments of βSOR between consecutive sampling dates (delta) at each subplot were calculated and visualized by hierarchical clustering of the inverse correlation by Ward's criterion using the R packages ‘dendextend’ (Galili, 2015) and ‘vegan’ (Oksanen et al., 2018). Correlation between patterns in temporal development of mean turnover to environmental parameters at each subplot was assessed using the ‘vegan’ (Oksanen et al., 2018) implementation of Mantel's test.
In order to relate spatial and temporal patterns in βSOR, the approach of Mellin et al. (2014) was adapted to the present dataset. Briefly, for spatial βSOR, the average of the βSOR values for each sampling date of AMF communities between each subplot and its neighbours was calculated. For temporal βSOR, the mean delta at each sampling plot was calculated. Linear models between both βSOR terms were fitted for 1000 different rarefactions and compared to null‐models based on 1000 draws of species identities (based on their relative probability of occurrence among samples), while holding constant the total number of species in each sample. To detect further variables contributing to the spatiotemporal patterns in βSOR, OTU richness at each subplot and environmental variables and their changes over time were included in the models.
Author contributions
KG, RSB and SK contributed equally to this work. EK, SM and HPP designed the SCALEMIC Experiment. KG, RSB, SK, KMR, EK, FB and TW developed the concept of the current study and KG, RSB and SK drafted the manuscript. KMR conducted DNA extractions and SK conducted AMF DNA amplification and preparation of sequencing libraries. SK and TW performed bioinformatic processing of sequences. RSB, SK, KMR, MF, DP, DB and SM provided data. RSB performed geostatistics and together with HPP fitted linear mixed effect models. KG and AHB performed multivariate statistics. KMR edited and corrected the manuscript's English. All authors read and revised the manuscript.
Supporting information
Appendix S1: Supporting Information
Acknowledgements
We thank the managers of the three Exploratories, Kirsten Reichel‐Jung, Swen Renner, Katrin Hartwich, Sonja Gockel, Kerstin Wiesner, and Martin Gorke for their work in maintaining the plot and project infrastructure; Christiane Fischer and Simone Pfeiffer for giving support through the central office, Michael Owonibi and Andreas Ostrowski for managing the central data base, and Eduard Linsenmair, Dominik Hessenmöller, Jens Nieschulze, Ernst‐Detlef Schulze, Wolfgang W. Weisser and the late Elisabeth Kalko for their role in setting up the Biodiversity Exploratories project. The work has been funded by the DFG Priority Program 1374 ‘Infrastructure‐Biodiversity‐Exploratories’ (BU 941/22‐1, BU 941/22‐3, KA 1590/8‐2, KA 1590/8‐3). Field work permits were issued by the responsible state environmental office of Baden‐Württemberg (according to § 72 BbgNatSchG). Likewise, we kindly thank Beatrix Schnabel, Melanie Günther and Sigrid Härtling for 454 sequencing in Halle. AHB gratefully acknowledges the support of the German Centre for Integrative Biodiversity Research (iDiv) Halle‐Jena‐Leipzig funded by the German Research Foundation (FZT 118). Authors declare no conflict of interests.
References
- Allen, E.B. (1989) The restoration of disturbed arid landscapes with special reference to mycorrhizal fungi. J Arid Environ 17: 279–286. [Google Scholar]
- Antoninka, A. , Reich, P.B. , and Johnson, N.C. (2011) Seven years of carbon dioxide enrichment, nitrogen fertilization and plant diversity influence arbuscular mycorrhizal fungi in a grassland ecosystem. New Phytol 192: 200–214. [DOI] [PubMed] [Google Scholar]
- Argüello, A. , O'Brien, M.J. , van der Heijden, M.G.A. , Wiemken, A. , Schmid, B. , and Niklaus, P.A. (2016) Options of partners improve carbon for phosphorus trade in the arbuscular mycorrhizal mutualism. Ecol Lett 19: 648–656. [DOI] [PubMed] [Google Scholar]
- Bahram, M. , Peay, K.G. , and Tedersoo, L. (2015) Local‐scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytol 205: 1454–1463. [DOI] [PubMed] [Google Scholar]
- Bainard, L.D. , Bainard, J.D. , Hamel, C. , and Gan, Y. (2014) Spatial and temporal structuring of arbuscular mycorrhizal communities is differentially influenced by abiotic factors and host crop in a semi‐arid prairie agroecosystem. FEMS Microbiol Ecol 88: 333–344. [DOI] [PubMed] [Google Scholar]
- Bardgett, R.D. , Bowman, W.D. , Kaufmann, R. , and Schmidt, S.K. (2005) A temporal approach to linking aboveground and belowground ecology. Trends Ecol Evol 20: 634–641. [DOI] [PubMed] [Google Scholar]
- Barnes, C.J. , Burns, C.A. , van der Gast, C.J. , McNamara, N.P. , and Bending, G.D. (2016) Spatio‐temporal variation of core and satellite arbuscular mycorrhizal fungus communities in Miscanthus giganteus . Front Microbiol 7: 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga, A. , and Orme, C.D.L. (2012) Betapart: an R package for the study of beta diversity. Methods Ecol Evol 3: 808–812. [Google Scholar]
- Berner, D. , Marhan, S. , Keil, D. , Poll, C. , Schützenmeister, A. , Piepho, H.‐P. , and Kandeler, E. (2011) Land‐use intensity modifies spatial distribution and function of soil microorganisms in grasslands. Pedobiologia 54: 341–351. [DOI] [PubMed] [Google Scholar]
- Binet, M.N. , Sage, L. , Malan, C. , Clément, J.C. , Redecker, D. , Wipf, D. , et al (2013) Effects of mowing on fungal endophytes and arbuscular mycorrhizal fungi in subalpine grasslands. Fungal Ecol 6: 248–255. [Google Scholar]
- Björk, R.G. , Björkman, M.P. , Andersson, M.X. , and Klemedtsson, L. (2008) Temporal variation in soil microbial communities in alpine tundra. Soil Biol Biochem 40: 266–268. [Google Scholar]
- Bouffaud, M.‐L. , Creamer, R.E. , Stone, D. , Plassart, P. , van Tuinen, D. , Lemanceau, P. , et al (2016) Indicator species and co‐occurrence in communities of arbuscular mycorrhizal fungi at the European scale. Soil Biol Biochem 103: 464–470. [Google Scholar]
- Bouffaud, M.L. , Bragalini, C. , Berruti, A. , Peyret‐Guzzon, M. , Voyron, S. , Stockinger, H. , et al (2017) Arbuscular mycorrhizal fungal community differences among European long‐term observatories. Mycorrhiza 27: 331–343. [DOI] [PubMed] [Google Scholar]
- Buscot, F. (2015) Implication of evolution and diversity in arbuscular and ectomycorrhizal symbioses. J Plant Physiol 172: 55–61. [DOI] [PubMed] [Google Scholar]
- Carini, P. , Marsden, P.J. , Leff, J.W. , Morgan, E.E. , Strickland, M.S. , and Fierer, N. (2016) Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol 2: 16242. [DOI] [PubMed] [Google Scholar]
- Chaparro, J.M. , Badri, D.V. , and Vivanco, J.M. (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8: 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, J.M. (2014) Spatial scale resolves the niche versus neutral theory debate. J Veg Sci 25: 319–322. [Google Scholar]
- Closa, I. , and Goicoechea, N. (2011) Infectivity of arbuscular mycorrhizal fungi in naturally regenerating, unmanaged and clear‐cut beech forests. Pedosphere 21: 65–74. [Google Scholar]
- Daniell, T.J. , Husband, R. , Fitter, A.H. , and Young, J.P.W. (2001) Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microbiol Ecol 36: 203–209. [DOI] [PubMed] [Google Scholar]
- Davison, J. , Öpik, M. , Zobel, M. , Vasar, M. , Metsis, M. , and Moora, M. (2012) Communities of arbuscular mycorrhizal fungi detected in forest soil are spatially heterogeneous but do not vary throughout the growing season. PLoS One 7: e41938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison, J. , Moora, M. , Öpik, M. , Adholeya, A. , Ainsaar, L. , Bâ, A. , et al (2015) Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349: 970–973. [DOI] [PubMed] [Google Scholar]
- Dumbrell, A.J. , Nelson, M. , Helgason, T. , Dytham, C. , and Fitter, A.H. (2010) Idiosyncrasy and overdominance in the structure of natural communities of arbuscular mycorrhizal fungi: is there a role for stochastic processes? J Ecol 98: 419–428. [Google Scholar]
- Dumbrell, A.J. , Ashton, P.D. , Aziz, N. , Feng, G. , Nelson, M. , Dytham, C. , et al (2011) Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol 190: 794–804. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. , Haas, B.J. , Clemente, J.C. , Quince, C. , and Knight, R. (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema, C.H. , and Wardle, D.A. (2002) Spatial soil ecology. Trends Ecol Evol 17: 177–183. [Google Scholar]
- Fischer, M. , Bossdorf, O. , Gockel, S. , Hänsel, F. , Hemp, A. , Hessenmöller, D. , et al (2010) Implementing large‐scale and long‐term functional biodiversity research: the biodiversity Exploratories. Basic Appl Ecol 11: 473–485. [Google Scholar]
- Francioli, D. , Schulz, E. , Purahong, W. , Buscot, F. , and Reitz, T. (2016) Reinoculation elucidates mechanisms of bacterial community assembly in soil and reveals undetected microbes. Biol Fertil Soils 52: 1073–1083. [Google Scholar]
- Francioli, D. , Schulz, E. , Buscot, F. , and Reitz, T. (2018) Dynamics of soil bacterial communities over a vegetation season relate to both soil nutrient status and plant growth phenology. Microb Ecol 75: 216–227. [DOI] [PubMed] [Google Scholar]
- Gai, J.P. , Christie, P. , Cai, X.B. , Fan, J.Q. , Zhang, J.L. , Feng, G. , and Li, X.L. (2009) Occurrence and distribution of arbuscular mycorrhizal fungal species in three types of grassland community of the Tibetan plateau. Ecol Res 24: 1345–1350. [Google Scholar]
- Galili, T. (2015) Dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31: 3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston, K.J. , and Spicer, J.I. (2013) Biodiversity: An Introduction. Oxford: Blackwell Publishing. [Google Scholar]
- Gehring, C.A. , and Whitham, T.G. (2002) Mycorrhizae‐herbivore interactions: population and community consequences In Mycorrhizal Ecology Ecological Studies (Analysis and Synthesis), van der Heijden M.G.A., and Sanders I.R. (eds). Berlin, Heidelberg: Springer, pp. 295–320. [Google Scholar]
- Görres, J.H. , Dichiaro, M.J. , Lyons, J.B. , and Amador, J.A. (1998) Spatial and temporal patterns of soil biological activity in a forest and an old field. Soil Biol Biochem 30: 219–230. [Google Scholar]
- Gossner, M.M. , Lewinsohn, T.M. , Kahl, T. , Grassein, F. , Boch, S. , Prati, D. , et al (2016) Land‐use intensification causes multitrophic homogenization of grassland communities. Nature 540: 266–269. [DOI] [PubMed] [Google Scholar]
- Grundmann, G. , Dechesne, A. , Bartoli, F. , Flandrois, J. , Chasse, J. , and Kizungu, R. (2001) Spatial modeling of nitrifier microhabitats in soil. Soil Sci Soc Am J 65: 1709–1716. [Google Scholar]
- Hart, M.M. , Reader, R.J. , and Klironomos, J.N. (2001) Life‐history strategies of arbuscular mycorrhizal fungi in relation to their successional dynamics. Mycologia 93: 1186–1194. [Google Scholar]
- Hazard, C. , Gosling, P. , Van Der Gast, C.J. , Mitchell, D.T. , Doohan, F.M. , and Bending, G.D. (2013) The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. ISME J 7: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden, M.G.A. , Klironomos, J.N. , Ursic, M. , and Moutoglis, P. (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72. [Google Scholar]
- van der Heijden, M.G.A. , Martin, F.M. , Selosse, M. , and Sanders, I.R. (2015) Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205: 1406–1423. [DOI] [PubMed] [Google Scholar]
- Hempel, S. (2018) Passengers and drivers of arbuscular mycorrhizal fungal communities at different scales. New Phytol 220: 952–953. [DOI] [PubMed] [Google Scholar]
- Heyburn, J. , McKenzie, P. , Crawley, M.J. , and Fornara, D.A. (2017) Long‐term belowground effects of grassland management: the key role of liming. Ecol Appl 27: 2001–2012. [DOI] [PubMed] [Google Scholar]
- Hiiesalu, I. , Pärtel, M. , Davison, J. , Gerhold, P. , Metsis, M. , Moora, M. , et al (2014) Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytol 203: 233–244. [DOI] [PubMed] [Google Scholar]
- Hodge, A. , Campbell, C.D. , and Fitter, A.H. (2001) An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413: 297–299. [DOI] [PubMed] [Google Scholar]
- Horn, R. , Brümmer, G. , Kandeler, E. , Kögel‐Knabner, I. , Kretzschmar, R. , Stahr, K. , and Wilke, B. (2010) Scheffer/Schachtschabel: Lehrbuch der Bodenkunde. Berlin: Springer‐Verlag. [Google Scholar]
- Horn, S. , Caruso, T. , Verbruggen, E. , Rillig, M.C. , and Hempel, S. (2014) Arbuscular mycorrhizal fungal communities are phylogenetically clustered at small scales. ISME J 8: 2231–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Niu, B. , Gao, Y. , Fu, L. , and Li, W. (2010) CD‐HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26: 680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell, S.P. (2001) The unified neutral theory of biodiversity and biogeography. Princeton, Oxford: Princeton University Press. [Google Scholar]
- Hugoni, M. , Luis, P. , Guyonnet, J. , and Haichar, F.E.Z. (2018) Plant host habitat and root exudates shape fungal diversity. Mycorrhiza 28: 451–463. [DOI] [PubMed] [Google Scholar]
- Huse, S.M. , Dethlefsen, L. , Huber, J.A. , Mark Welch, D. , Relman, D.A. , and Sogin, M.L. (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4: e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir, Z. , O'Halloran, I. , Fyles, J. , and Hamel, C. (1998) Dynamics of the mycorrhizal symbiosis of corn (Zea mays L.): effects of host physiology, tillage practice and fertilization on spatial distribution of extra‐radical mycorrhizal hyphae in the field. Agric Ecosyst Environ 68: 151–163. [Google Scholar]
- Kampstra, P. (2008) Beanplot: A boxplot alternative for visual comparison of distributions. J Stat Softw 28: 1–9.27774042 [Google Scholar]
- Kandeler, E. , and Böhm, K.E. (1996) Temporal dynamics of microbial biomass, xylanase activity, N‐mineralisation and potential nitrification in different tillage systems. Appl Soil Ecol 4: 181–191. [Google Scholar]
- Kandeler, E. , Tscherko, D. , and Spiegel, H. (1999) Long‐term monitoring of microbial biomass, N mineralisation and enzyme activities of a Chernozem under different tillage management. Biol Fertil Soils 28: 343–351. [Google Scholar]
- Kivlin, S.N. , Hawkes, C.V. , and Treseder, K.K. (2011) Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biol Biochem 43: 2294–2303. [Google Scholar]
- Klaus, V.H. , Boch, S. , Boeddinghaus, R.S. , Hölzel, N. , Kandeler, E. , Marhan, S. , et al (2016) Temporal and small‐scale spatial variation in grassland productivity, biomass quality, and nutrient limitation. Plant Ecol 217: 843–856. [Google Scholar]
- König, S. , Wubet, T. , Dormann, C.F. , Hempel, S. , Renker, C. , and Buscot, F. (2010) TaqMan real‐time PCR assays to assess arbuscular mycorrhizal responses to field manipulation of grassland biodiversity: effects of soil characteristics, plant species richness, and functional traits. Appl Environ Microbiol 76: 3765–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koorem, K. , Gazol, A. , Öpik, M. , Moora, M. , Saks, Ü. , Uibopuu, A. , et al (2014) Soil nutrient content influences the abundance of soil microbes but not plant biomass at the small‐scale. PLoS One 9: e91998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyakov, Y. , and Blagodatskaya, E. (2015) Microbial hotspots and hot moments in soil: concept & review. Soil Biol Biochem 83: 184–199. [Google Scholar]
- Lefcheck, J.S. (2016) piecewiseSEM: piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol Evol 7: 573–579. [Google Scholar]
- Lekberg, Y. , Schnoor, T. , Kjøller, R. , Gibbons, S.M. , Hansen, L.H. , Al‐Soud, W.A. , et al (2012) 454‐sequencing reveals stochastic local reassembly and high disturbance tolerance within arbuscular mycorrhizal fungal communities. J Ecol 100: 151–160. [Google Scholar]
- Liu, W. , Jiang, S. , Zhang, Y. , Yue, S. , Christie, P. , Murray, P.J. , et al (2014) Spatiotemporal changes in arbuscular mycorrhizal fungal communities under different nitrogen inputs over a 5‐year period in intensive agricultural ecosystems on the North China plain. FEMS Microbiol Ecol 90: 436–453. [DOI] [PubMed] [Google Scholar]
- MacArthur, R.H. , and Wilson, E.O. (1967) Theory of Island Biogeography. Princeton, Oxford: Princeton University Press. [Google Scholar]
- Maherali, H. , and Klironomos, J.N. (2012) Phylogenetic and trait‐based assembly of arbuscular mycorrhizal fungal communities. PLoS One 7: e36695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBratney, A. , and Webster, R. (1986) Choosing functions for semi‐variograms of soil properties and fitting them to sampling estimates. Eur J Soil Sci 37: 617–639. [Google Scholar]
- Mellin, C. , Bradshaw, C.J. , Fordham, D.A. , and Caley, M.J. (2014) Strong but opposing beta‐diversity‐stability relationships in coral reef fish communities. Proc Biol Sci 281: 20131993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, E.K. , Buscot, F. , Herbst, C. , Meiners, T. , Obermaier, E. , Wäschke, N.W. , et al (2013) Land use and host neighbor identity effects on arbuscular mycorrhizal fungal community composition in focal plant rhizosphere. Biodivers Conserv 22: 2193–2205. [Google Scholar]
- Nacke, H. , Goldmann, K. , Schöning, I. , Pfeiffer, B. , Kaiser, K. , Castillo‐Villamizar, G.A. , et al (2016) Fine spatial scale variation of soil microbial communities under European beech and Norway spruce. Front Microbiol 7: 2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, S. , and Schielzeth, H. (2013) A general and simple method for obtaining R2 from generalized linear mixed‐effects models. Methods Ecol Evol 4: 133–142. [Google Scholar]
- Neuenkamp, L. , Moora, M. , Öpik, M. , Davison, J. , Gerz, M. , Mannisto, M. , et al (2018) The role of plant mycorrhizal type and status in modulating the relationship between plant and arbuscular mycorrhizal fungal communities. New Phytol 220: 1236–1247. [DOI] [PubMed] [Google Scholar]
- Nunan, N. , Wu, K. , Young, I.M. , Crawford, J.W. , and Ritz, K. (2003) Spatial distribution of bacterial communities and their relationships with the micro‐architecture of soil. FEMS Microbiol Ecol 44: 203–215. [DOI] [PubMed] [Google Scholar]
- Oberdorfer, E. , Schwabe, A. , and Müller, T. (2001) Pflanzensoziologische Exkursionsflora für Deutschland und angrenzende Gebiete. Stuttgart (Hohenheim): Eugen Ulmer. [Google Scholar]
- Oehl, F. , Laczko, E. , Oberholzer, H.‐R. , Jansa, J. , and Egli, S. (2017) Diversity and biogeography of arbuscular mycorrhizal fungi in agricultural soils. Biol Fertil Soils 53: 777–797. [Google Scholar]
- Oksanen, J. , Blanchet, F. , Friendly, M. , Kindt, R. , Legendre, P. , McGlinn, D. , et al (2018) vegan: Community Ecology Package. R Package Version 2.2‐0. Vienna, Austria: R Foundation for Statistical Computing; Available at: http://cran.rproject.org/package=vegan. [Google Scholar]
- Öpik, M. , Moora, M. , Liira, J. , and Zobel, M. (2006) Composition of root‐colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J Ecol 94: 778–790. [Google Scholar]
- Öpik, M. , Moora, M. , Zobel, M. , Saks, Ü. , Wheatley, R. , Wright, F. , and Daniell, T. (2008) High diversity of arbuscular mycorrhizal fungi in a boreal herb‐rich coniferous forest. New Phytol 179: 867–876. [DOI] [PubMed] [Google Scholar]
- Öpik, M. , Metsis, M. , Daniell, T.J. , Zobel, M. , and Moora, M. (2009) Large‐scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol 184: 424–437. [DOI] [PubMed] [Google Scholar]
- Öpik, M. , Vanatoa, A. , Vanatoa, E. , Moora, M. , Davison, J. , Kalwij, J. , et al (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188: 223–241. [DOI] [PubMed] [Google Scholar]
- Pebesma, E.J. (2004) Multivariable geostatistics in S: the gstat package. Comput Geosci 30: 683–691. [Google Scholar]
- Peres‐Neto, P.R. , and Jackson, D.A. (2001) How well do multivariate data sets match? The advantages of a procrustean superimposition approach over the mantel test. Oecologia 129: 169–178. [DOI] [PubMed] [Google Scholar]
- Pinheiro, J. , Bates, D. , DebRoy, S. , Sarkar, D. , and R Development Core Team . (2017) nlme: Linear and nonlinear mixed effects models. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Powell, J.R. , and Bennett, A.E. (2016) Unpredictable assembly of arbuscular mycorrhizal fungal communities. Pedobiologia 59: 11–15. [Google Scholar]
- R Development Core Team . (2017) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Regan, K.M. , Nunan, N. , Boeddinghaus, R.S. , Baumgartner, V. , Berner, D. , Boch, S. , et al (2014) Seasonal controls on grassland microbial biogeography: are they governed by plants, abiotic properties or both? Soil Biol Biochem 71: 21–30. [Google Scholar]
- Regan, K.M. , Nunan, N. , Boeddinghaus, R.S. , Baumgartner, V. , Berner, D. , Boch, S. , et al (2015) Corrigendum to “seasonal controls on grassland microbial biogeography: are they governed by plants, abiotic properties or both?”[soil biology and biochemistry 71 (April 2014)]. Soil Biol Biochem 86: 212–214. [Google Scholar]
- Regan, K. , Stempfhuber, B. , Schloter, M. , Rasche, F. , Prati, D. , Philippot, L. , et al (2017) Spatial and temporal dynamics of nitrogen fixing, nitrifying and denitrifying microbes in an unfertilized grassland soil. Soil Biol Biochem 109: 214–226. [Google Scholar]
- Sanders, I.R. (2003) Preference, specificity and cheating in the arbuscular mycorrhizal symbiosis. Trends Plant Sci 8: 143–145. [DOI] [PubMed] [Google Scholar]
- Schloss, P.D. , Westcott, S.L. , Ryabin, T. , Hall, J.R. , Hartmann, M. , Hollister, E.B. , et al (2009) Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, L. , Lalonde, M. , and Bruns, T.D. (1992) Specific amplification of 18S fungal ribosomal genes from vesicular‐arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.E. , and Read, D.J. (2008) Mycorrhizal symbiosis. London: Academic Press. [Google Scholar]
- Staddon, P.L. , Thompson, K. , Jakobsen, I. , Grime, J.P. , Askew, A.P. , and Fitter, A.H. (2003) Mycorrhizal fungal abundance is affected by long‐term climatic manipulations in the field. Glob Chang Biol 9: 186–194. [Google Scholar]
- Stempfhuber, B. , Richter‐Heitmann, T. , Regan, K.M. , Kölbl, A. , Wüst, P.K. , Marhan, S. , et al (2016) Spatial interaction of archaeal ammonia‐oxidizers and nitrite‐oxidizing bacteria in an unfertilized grassland soil. Front Microbiol 6: 1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiunov, A.V. , and Scheu, S. (2005) Arbuscular mycorrhiza and Collembola interact in affecting community composition of saprotrophic microfungi. Oecologia 142: 636–642. [DOI] [PubMed] [Google Scholar]
- Tobler, W.R. (1970) A computer movie simulating urban growth in the Detroit region. Econ Geogr 46: 234–240. [Google Scholar]
- Vályi, K. , Mardhiah, U. , Rillig, M.C. , and Hempel, S. (2016) Community assembly and coexistence in communities of arbuscular mycorrhizal fungi. ISME J 10: 2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana, D.S. , Cid, B. , Figuerola, J. , and Santamaría, L. (2016) Disentangling the roles of diversity resistance and priority effects in community assembly. Oecologia 182: 865–875. [DOI] [PubMed] [Google Scholar]
- Waters, J.R. , and Borowicz, V.A. (1994) Effect of clipping, benomyl, and genet on 14 C transfer between mycorrhizal plants. Oikos 71: 246–252. [Google Scholar]
- Wu, B. , Hogetsu, T. , Isobe, K. , and Ishii, R. (2007) Community structure of arbuscular mycorrhizal fungi in a primary successional volcanic desert on the southeast slope of Mount Fuji. Mycorrhiza 17: 495–506. [DOI] [PubMed] [Google Scholar]
- Wubet, T. , Weiß, M. , Kottke, I. , and Oberwinkler, F. (2006) Two threatened coexisting indigenous conifer species in the dry Afromontane forests of Ethiopia are associated with distinct arbuscular mycorrhizal fungal communities. Botany 84: 1617–1627. [Google Scholar]
- Yachi, S. , and Loreau, M. (1999) Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci USA 96: 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel, M. , and Öpik, M. (2014) Plant and arbuscular mycorrhizal fungal (AMF) communities–which drives which? J Veg Sci 25: 1133–1140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information


