Abstract
Uveal melanoma is the most common primary malignancy of the eye, and a number of discoveries in the last decade have led to a more thorough molecular characterization of this cancer. However, the prognosis remains dismal for patients with metastases, and there is an urgent need to identify treatments that are effective for this stage of disease. Animal models are important tools for preclinical studies of uveal melanoma. A variety of models exist, and they have specific advantages, disadvantages, and applications. In this review article, these differences are explored in detail, and ideas for new models that might overcome current challenges are proposed.
Keywords: melanoma, mouse, transgenic mouse, uveal, xenograft
1. INTRODUCTION
Uveal melanoma is a rare (estimated incidence of 6 cases per million) and unique subtype of melanoma that arises in the uveal tract of the eye, most commonly in the choroid (Damato & Damato, 2012; McLaughlin et al., 2005). Local interventions, such as radiation therapy and enucleation, are effective at treating the primary tumor (Krantz, Dave, Komatsubara, Marr, & Carvajal, 2017). However, up to half of the patients will develop metastatic disease, predominantly to the liver (Rietschel et al., 2005). For these patients, liver‐directed therapy and participation in clinical trials are recommended, but most die from their disease, and median survival is only 10.2 months (Khoja et al., 2019; Kujala, Makitie, & Kivela, 2003; National Comprehensive Cancer Network).
Despite this, great strides have been made in understanding the molecular features of uveal melanoma. In the past decade, the collective work from several groups has led to the identification of important recurrent mutations and overactive signaling pathways in this cancer. Early oncogenic driver mutations occur in a nearly mutually exclusive pattern in the guanine nucleotide‐binding protein subunit alpha‐q/11 signaling pathway (Field et al., 2018; Moore et al., 2016; Robertson et al., 2017). This includes constitutively active variants of GNAQ and GNA11, which are found in over 90% of cases (Van Raamsdonk et al., 2009, 2010). A smaller subset of tumors harbor activating mutations in the G protein‐coupled receptor cysteinyl leukotriene receptor 2 (CYSLTR2) or phospholipase C beta 4 (PLCB4) (Johansson et al., 2016; Moore et al., 2016). There is a second node of nearly mutually exclusive mutations that classifies uveal melanomas and affects prognosis. Inactivating mutations are found in BRCA1‐associated protein 1 (BAP1), while recurrent point mutations are observed in the eukaryotic translation initiation factor 1A X‐linked (EIF1AX) or a splicing factor such as SF3B1 (Field et al., 2018; Harbour et al., 2010, 2013; Martin et al., 2013).
The molecular makeup of a particular uveal melanoma has significant implications for predicting metastasis. Most importantly, tumors with loss‐of‐function BAP1 mutations carry the worst prognosis, as approximately 84% of metastatic uveal melanomas are of this subtype (Harbour et al., 2010; Shain et al., 2019). Specific cytogenetic alterations have also been well described in this cancer (Aalto, Eriksson, Seregard, Larsson, & Knuutila, 2001; Anbunathan, Verstraten, Singh, Harbour, & Bowcock, 2019). Monosomy 3 co‐occurs with BAP1 mutation, thereby eliminating both functional alleles (Field et al., 2018; Robertson et al., 2017). 6q loss, 1q gain, and 8q gain are also significantly enriched in uveal melanoma metastases (Ehlers, Worley, Onken, & Harbour, 2005; Hammond et al., 2015; Shain et al., 2019).
These discoveries were largely enabled by the analysis of patient tumor specimens and have greatly advanced our understanding of the molecular underpinnings of uveal melanoma tumorigenesis and their prognostic significance. Various animal models have likewise been indispensable in elucidating the biology and potential therapeutic vulnerabilities of this cancer (Cao & Jager, 2015; Stei, Loeffler, Holz, & Herwig, 2016; Yang, Cao, & Grossniklaus, 2015). In the past several years, there have been many promising preclinical studies that have used these models to identify novel treatment strategies, several of which are now in the early stages of clinical trials (Vivet‐Noguer, Tarin, Roman‐Roman, & Alsafadi, 2019; Yang, Manson, Marr, & Carvajal, 2018).
In this review article, we discuss the strengths and weaknesses of existing animal models of uveal melanoma, with an emphasis on mouse models. We also identify unmet needs that will require future model development and refinement. The goal of any animal model of uveal melanoma should be to faithfully recapitulate the processes of tumor initiation, growth, metastasis, and response to therapy as observed in patients with this disease.
2. ANIMAL MODELS OF UVEAL MELANOMA
Though the focus of this review is mouse models of uveal melanoma, other species certainly have their advantages. Rabbits (Oryctolagus cuniculus), for example, have large eyes that facilitate the implantation of tumor cells and subsequent monitoring using techniques such as fundoscopy, ultrasound, and magnetic resonance imaging (Bontzos & Detorakis, 2017; Gao, Tang, Liu, Yang, & Liu, 2018). The zebrafish (Danio rerio) is a model organism that has been used more widely in many scientific fields in recent years (Meyers, 2018). Both xenograft (Fornabaio et al., 2018; van der Ent et al., 2014) and transgenic (Mouti, Dee, Coupland, & Hurlstone, 2016; Perez, Henle, Amsterdam, Hagen, & Lees, 2018) zebrafish models of uveal melanoma have been developed. These models are excellent for high‐throughput pharmacologic screening and in vivo microscopy. The genetic models have yielded valuable insights into uveal melanoma signaling, such as the establishment of the importance of YAP activation in the initiation of this cancer. However, tumorigenesis in these models required mutation of p53, and metastasis was difficult to assess because of the induction of multiple primary tumors (Mouti et al., 2016; Perez et al., 2018).
Mice (Mus musculus) are the most widely used laboratory animal in the study of uveal melanoma (Cao & Jager, 2015). Their fecundity, gestation time, and size make them the most cost‐effective mammalian model (Zuberi & Lutz, 2016). Furthermore, genetic manipulation of mice has produced various strains that are used in many uveal melanoma models. The primary goals of this review are to compare the different types of mouse models of uveal melanoma and propose directions for further development.
3. INOCULATION SITES
The majority of murine models of uveal melanoma require the inoculation of cells or tumors into mice. Some uveal melanoma cell lines can be grown subcutaneously, which is convenient for measuring growth and response to therapy. However, others grow poorly subcutaneously but flourish in the tissue from which they were derived (Ozaki et al., 2016). In these cases, orthotopic models are preferable and may better model the human disease. Models of primary uveal melanoma in which the route of inoculation results in growth in the iris, ciliary body, or choroid are considered orthotopic (Figure 1). Inoculation of cells into the anterior chamber of the eye was one of the first techniques developed and reliably produces tumors in the iris that are capable of metastasis (Niederkorn, 1984). In 2000, a suprachoroidal injection technique was described in which cells are deposited into the posterior compartment (not to be confused with posterior chamber) of the eye (Dithmar, Rusciano, & Grossniklaus, 2000). In this approach, the needle is inserted through the limbus and into the choroid. Injected cells occupy the suprachoroidal space and likely spill into the subretinal space and vitreous. This technique is advantageous because it rapidly produces tumors in the choroid and ciliary body, the sites at which uveal melanoma most commonly occurs in patients. Furthermore, it reduces extraocular growth as compared to transconjunctival inoculations and consistently produces distant metastases (Tables 1 and 2a,b). A third type of orthotopic model is intravitreal injection. Although uveal melanoma does not arise in the vitreous humor, this environment is supportive of tumor growth and injected cells mimic human disease by invading and involving the uveal tract (Kilian et al., 2016; Yoo et al., 2016). All three of the above inoculation methods are amenable to combination with enucleation, which allows for longer follow‐up and the study of metastatic outgrowth.
Figure 1.
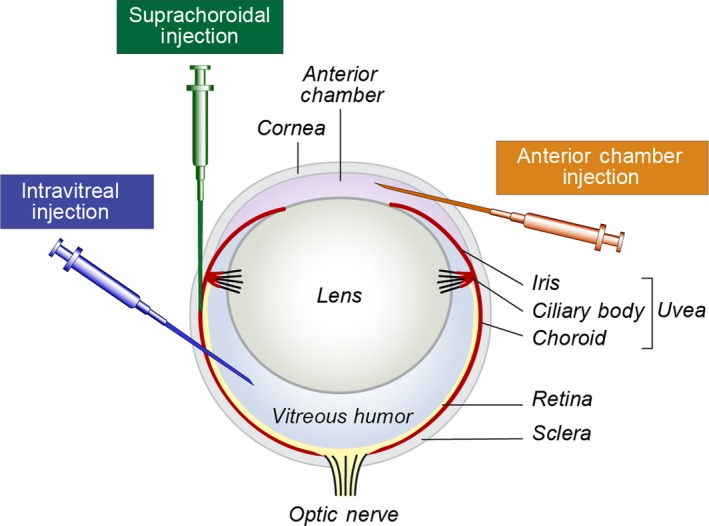
Routes of injection for orthotopic models of primary uveal melanoma. The needle trajectories for the three most commonly used types of injections are depicted (anterior chamber in orange, suprachoroidal in green, and intravitreal in blue). All three result in growth of cells in the uveal tract and therefore produce orthotopic models of uveal melanoma
Table 1.
Syngeneic mouse cutaneous melanoma models for simulating uveal melanoma
| Cell line | Source | Original publication (Laboratory of origin) | Inoculation method | Metastasis | References |
|---|---|---|---|---|---|
| B16LS9 | Mouse cutaneous melanoma | Rusciano et al. (1994) (Max Burger) | Suprachoroidal | Liver, lungs, and lymph nodes | Jones et al. (2019); Dong et al. (2019); Xue et al. (2015); Yang et al. (2016), Yang and Grossniklaus (2010), Yang, Jager, and Grossniklaus (2010), Yang, Xu, Iuvone, and Grossniklaus (2006); Lattier et al. (2013); Zhang et al. (2011); Alizadeh et al. (2003); Dithmar, Rusciano, and Grossniklaus (2000), Dithmar, Rusciano, Lynn, et al. (2000); Diaz, Rusciano, Dithmar, and Grossniklaus (1999) |
| Ant. chamber | Not reported | Han, Brown, and Niederkorn (2016) | |||
| Intravitreal | Liver | Han et al. (2016); Yang et al. (2011) | |||
| Intrasplenic | |||||
| Intrahepatic | Not reported | Xue et al. (2015) | |||
| B16F10 | Mouse cutaneous melanoma | Fidler, Gersten, and Budmen (1976) (Marilyn Budmen) | Suprachoroidal | None | Grossniklaus, Barron, and Wilson (1995) |
| Ant. chamber | Not reported | el Filali et al. (2012); de Lange et al. (2012); Ly et al. (2010); Grossniklaus et al. (1995); Knisely and Niederkorn (1990); | |||
| Lungs | Harning and Szalay (1987); Niederkorn, Sanborn, and Gamel (1987), Niederkorn (1984) | ||||
| Tail vein | Lungs | Sanborn, Niederkorn, and Gamel (1992); Niederkorn et al. (1987) | |||
| Queens | Mouse cutaneous melanoma | Harning et al. (1987) (Jeanne Szalay) | Suprachoroidal | Lungs | Rajaii et al. (2014); Grossniklaus et al. (1995) |
| Ant. chamber | None | Grossniklaus et al. (1995) | |||
| Lungs | Sanborn, Niederkorn, Kan‐Mitchell, and Albert (1992), Sanborn, Niederkorn, and Gamel (1992); Harning and Szalay (1987) | ||||
| Tail vein | Lungs | Sanborn, Niederkorn, and Gamel (1992) | |||
| HCmel12 | Mouse cutaneous melanoma | Kilian et al. (2016) (Thomas Tüting) | Intravitreal | Lungs and lymph nodes | Stei, Loeffler, Kurts, et al. (2016); Kilian et al. (2016) |
| Oncogene‐transduced melan‐A cells | Immortalized mouse melanocyte | Bennett, Cooper, and Hart (1987) (Ian Hart) | Subcutaneous | Not reported | Moore et al. (2016) |
| Lungs and liver | Van Raamsdonk et al. (2010) |
Abbreviation: Ant. chamber, anterior chamber.
Table 2.
Human uveal melanoma cell lines derived from (a) primary tumors used in mouse xenograft experiments (b) metastases used in mouse xenograft experiments
| (a) Cell line (mutations) | Source | Original publication (Laboratory of origin) | Inoculation method | Metastasis | References |
|---|---|---|---|---|---|
| Mel92.1 (GNAQQ209L; EIF1AXG6D) | Primary tumor | De Waard‐Siebinga et al. (1995) (Martine Jager) | Subcutaneous | Not reported | Faiao‐Flores et al. (2019); Forsberg et al. (2019); Kines et al. (2018); Chen et al. (2017, 2014); Ambrosini, Sawle, Musi, and Schwartz (2015); Ambrosini, Musi, Ho, Stanchina, and Schwartz, (2013); Musi, Ambrosini, Stanchina, and Schwartz, (2014); Surriga et al. (2013); Ho et al. (2012); Samadi et al. (2012); Landreville et al. (2012) |
| Suprachoroidal | Liver | Dong et al. (2019) | |||
| Not reported | Yu et al. (2014) | ||||
| Ant. chamber | Liver | Ma and Niederkorn (1998), Ma, Luyten, Luider, Jager, and Niederkorn (1996) | |||
| Tail vein | Liver and lungs | Matatall et al. (2013) | |||
| Intrasplenic | Liver |
Barisione et al. (2015); |
|||
| Mel202 (GNAQQ209L; SF3B1R625G) | Primary tumor | Ksander, Rubsamen, Olsen, Cousins, and Streilein (1991) (J. Wayne Streilein) | Subcutaneous | Not reported | Forsberg et al. (2019) |
| Ant. chamber | Liver | Ma and Niederkorn (1998), Ma, Luyten, Luider, Jager, and Niederkorn (1996) | |||
| Intravitreal | Not reported | Yoo et al. (2016) | |||
| Tail vein | Liver | Niederkorn, Mellon, Pidherney, Mayhew, and Anand (1993) | |||
| Mel270 (GNAQQ209P) | Primary tumor | Verbik, Murray, Tran, and Ksander (1997) (Bruce Ksander) | Suprachoroidal | Not reported | Yu et al. (2014) |
| Subcutaneous | Liver and lungs | Tafreshi et al. (2019) | |||
| Lot reported | Voropaev et al. (2019); Annala et al. (2019); Kaochar et al. (2018) | ||||
| Intrasplenic | Liver | Barisione et al. (2015); Gangemi et al. (2014, 2012) | |||
| MP41 (GNA11Q209L) | PDX from a primary tumor | Amirouchene‐Angelozzi et al. (2014) (Sergio Roman‐Roman) | Tail vein | Liver | Faiao‐Flores et al. (2019) |
| T105 and T142 (GNA11Q209L) | Primary tumors | Mouriaux et al. (2016) (Sylvain Guérin) | Subcutaneous | Not reported | Mouriaux et al. (2016) |
| UMT2 (GNA11Q209L) | Primary tumors | Suesskind et al. (2013) (Sigrid Henke‐Fahle) | Suprachoroidal | None | Süsskind, Hurst, Rohrbach, and Schnichels (2017) |
| (b) Cell line | Source | Original publication (Laboratory of origin) | Inoculation site | Metastasis | References |
|---|---|---|---|---|---|
| OMM1 (GNA11Q209L) | Subcutis metastasis | Luyten et al. (1996) (Theo Luider) | Subcutaneous | Not reported | Zhou, Jin, Jin, Liu, and Pan (2017); Wang, Liu, Jin, Jiang, and Pan (2017); Sutmuller et al. (2000) |
| Ant. chamber | Liver | Repp, Mayhew, Howard, Alizadeh, and Niederkorn (2001) | |||
| Tail vein | Liver | Ma and Niederkorn (1995) | |||
| OMM1.3a = OMM2.3 (GNAQQ209P) | Liver metastasis | Verbik et al. (1997) (Bruce Ksander) | Subcutaneous | Not reported | Ambrosini et al. (2019); Jin et al. (2018); Vaqué et al. (2013) |
| Suprachoroidal | Liver | Liang et al. (2012); Zhu et al. (2010) | |||
| Retro‐orbital | Liver and lungs | Surriga et al. (2013) | |||
| Intrasplenic | Liver | Jin et al. (2018) | |||
| TJU‐UM001 (GNAQQ209P) | Liver metastasis | Yoshida et al. (2014) (Takami Sato) | Intrahepatic | Peritoneum, lymph nodes | Kageyama et al. (2017); Ozaki et al. (2016); Cheng et al. (2015) |
| Intrasplenic | Liver | Piquet et al. (2019); Ozaki et al. (2016) | |||
| TJU‐UM004 (GNAQQ209P) | Orbital metastasis | Cheng et al. (2015) (Takami Sato) | Intrahepatic | None | Kageyama et al. (2017) |
Abbreviation: Ant. chamber: anterior chamber.
The Mel270 cell line was derived from this patient's primary tumor. The OMM2.5 cell line (also called OMM1.5) is derived from another liver metastasis in the same patient.
The eye is bypassed in some models in order to more quickly and reliably produce large tumors in visceral organs, especially the liver. Intravenous injection into either the retro‐orbital sinus or tail vein mimics the latter part of the metastatic cascade—hematogenous dissemination, arrest and extravasation in distant sites, and metastatic colony formation and growth. The liver and lungs are the most frequently reported sites of experimental metastasis with these routes of injection (Tables 1 and 2a,b). Others have developed the intrasplenic inoculation, which consistently produces tumors in the liver (Barisione et al., 2015; Gangemi et al., 2014, 2012; Jin et al., 2018). Finally, direct implantation of cells or tumors into the liver also results in florid growth in an orthotopic model of metastatic uveal melanoma (Kageyama et al., 2017; Ozaki et al., 2016).
Irrespective of the location of injection, disease progression (e.g., tumor growth and/or metastatic dissemination) can be studied in real time using non‐invasive imaging methods such as bioluminescence imaging (Barisione et al., 2015; Surriga et al., 2013). For this technique, the injected cells have been transduced to stably express a luciferase reporter. When the graft‐bearing mice are injected with luciferin, the tumor cells emit light that can be detected by an optical imaging instrument such as Perkin Elmer's In Vivo Imaging System (IVIS). The intensity of the signal has been demonstrated to be a suitable surrogate for tumor size and thus enables dynamic evaluation of the effects of different experimental conditions on tumor progression (Cosette et al., 2016; Poeschinger, Renner, Weber, & Scheuer, 2013).
4. SYNGENEIC CUTANEOUS MELANOMA MOUSE MODELS FOR SIMULATING UVEAL MELANOMA
The syngeneic cutaneous melanoma mouse model has been used for decades in uveal melanoma research. In this system, cutaneous melanoma cells are implanted in mice of the same genetic background as the mice from which the line was derived. Although the cell lines used are not uveal in origin, this system allows for the investigation of intraocular growth and metastasis of melanoma cells, as many of these lines metastasize to the liver (Table 1). This mimics the behavior of uveal melanoma in humans and allows for the study of the full metastatic process, including local invasion, intravasation, survival in the blood, extravasation, and growth in distant organs. The ability to examine the interaction between tumor and host cells as the cancer progresses in an immunocompetent animal is arguably the greatest strength of this model. Additionally, recipient mice may be genetically altered in order to study specific contributions of the host in melanoma progression (Lattier, Yang, Crawford, & Grossniklaus, 2013; Stei, Loeffler, Kurts, et al., 2016).
The most widely used syngeneic model is the inoculation of C57BL/6 mice with the B16LS9 cell line, a derivative of the B16 cutaneous melanoma line that was enriched for hepatic metastatic propensity through serial in vivo passaging (Rusciano, Lorenzoni, & Burger, 1994). This cell line metastasizes to the liver from the eye, and its use has led to valuable insights into the behavior of metastatic melanoma. For instance, this model was used to show that natural killer cells and pigment‐derived epithelial factor play distinct roles in counteracting intrahepatic growth of melanoma cells (Jones, Yang, Zhang, Morales‐Tirado, & Grossniklaus, 2019). Although B16LS9 cutaneous melanoma cells were used, the histological growth patterns of the hepatic metastases in the mouse model were similar to those observed in the livers of patients with metastatic uveal melanoma (Grossniklaus et al., 2016).
The primary disadvantage of the syngeneic model is that available mouse melanoma cell lines are of cutaneous origin, so the mutations and other molecular drivers of these cells differ from those found in human uveal melanoma. Therefore, their behavior, especially their response to therapy, may differ from what is observed in patients. Interestingly, there are a few syngeneic models that do carry canonical uveal melanoma mutations. Immortalized mouse melanocytes transduced with driver mutations found in patients undergo oncogenic transformation and are capable of producing tumors and even metastases (Moore et al., 2016; Van Raamsdonk et al., 2010). Additionally, the HCmel12 mouse cutaneous melanoma cell line has been reported to carry a GNA11Q209L variant (Schrage et al., 2015). Further details on other mutations in this cell line would allow for a more complete assessment of its suitability as a model for uveal melanoma. In the future, if mouse uveal melanoma cell lines could be derived from the genetically engineered mouse models discussed below, they would be powerful tools for syngeneic models. This strategy would allow for the controlled manipulation and study of bona fide uveal melanoma in an immunocompetent host.
5. XENOGRAFT MOUSE MODELS OF UVEAL MELANOMA
Xenograft models are another widely used approach. As the name implies, cells or tumors from a foreign source are grafted into mice. Most commonly, human uveal melanoma cell lines are used. The primary advantage of these models is that the cells are derived from patients. As such, they largely retain molecular features of the original tumor (Amirouchene‐Angelozzi et al., 2014; Griewank et al., 2012; Jager, Magner, Ksander, & Dubovy, 2016). This technique is therefore well‐suited for studying tumor signaling and response to treatment. Many recent publications detailing new potential treatments for uveal melanoma utilize xenograft models (Table 2a,b). Another advantage of xenografts is reproducibility from mouse to mouse (Gould, Junttila, & de Sauvage, 2015). Many human uveal melanoma cell lines have been described, although some are not commercially available. Frequently used cell lines with validated uveal melanoma mutations are included in Table 2a,b. Many of these xenograft models are useful for studying metastasis, as they produce tumors in organs such as the liver and lungs. It is also worth noting that some cell lines were derived from human uveal melanoma metastases. These are especially applicable for studying tumor growth in visceral organs such as the liver.
Authentication of uveal melanoma cell lines for use in xenograft models is critical. Some lines historically thought to be uveal melanoma have been found to harbor BRAFV600E mutations and are now recognized as being of cutaneous origin (Griewank et al., 2012; Yu et al., 2015). Furthermore, several of these were found by short tandem repeat (STR) analysis to be the same cell line (Folberg et al., 2008; Yu et al., 2015). Validation of uveal melanoma cell lines (including species confirmation, STR analysis, and pathogen detection) by individual laboratories is strongly encouraged. However, even after careful molecular characterization of any cancer cell line, the ability of the cells to faithfully recapitulate the behavior of their parental tumors has been questioned due to changes in molecular features that can result from culturing them in vitro (Ben‐David et al., 2018; Gillet, Varma, & Gottesman, 2013; Goodspeed, Heiser, Gray, & Costello, 2016). An example of this is that the karyotypes, including the status of chromosome 3, of several of the older cell lines differ from those of the patients’ original tumors (Jager et al., 2016). Additionally, it has been demonstrated that the gene expression profiles of uveal melanoma cell lines in culture diverge from their source tumors even after short‐term passaging (Mouriaux et al., 2016). One way to avoid these problems is to implant human tumor specimens directly into mice; this is the basis of patient‐derived xenografts.
Patient‐derived xenograft (PDX) models are relatively new in the uveal melanoma field but have demonstrated considerable translational potential. The research group led by Didier Decaudin has been the most successful and prolific in generating PDX models of uveal melanoma (Table 3). They implant fresh primary and metastatic tumor specimens in the interscapular fat pad of severe combined immunodeficient (SCID) mice and achieve an engraftment rate of 28% (Némati et al., 2010). Importantly, the tumors that grow in these mice maintain mutations, chromosomal imbalances, and histopathological features of the tumors from which they were derived (Carita, Nemati, & Decaudin, 2015). These PDX models have also been used for the derivation of new cell lines with clinically relevant features such as loss of BAP1 expression (Amirouchene‐Angelozzi et al., 2014). They have also been effective for assessing the efficacy of novel combination therapies to treat uveal melanoma (Amirouchene‐Angelozzi et al., 2016; Carita et al., 2016).
Table 3.
Patient‐derived mouse xenograft models of uveal melanoma
| PDX model | Source | Original Publication (Laboratory of origin) | Inoculation site | References |
|---|---|---|---|---|
| 6 cases successfully grafted 3 times | Liver metastases | Kageyama et al. (2017) (Takami Sato) | Liver | Kageyama et al. (2017) |
| MP34, MP38, MP41, MP42, MP46, MP47, MP55, MP71, MP77, and MP80 | Primary tumors | Némati et al. (2010) (Didier Decaudin) | Interscapular fat pad | Carita et al. (2016); Amirouchene‐Angelozzi et al. (2016, 2014); Némati et al. (2014, 2010); Madic et al. (2012) |
| MM33 | Subcutis metastasis | |||
| MM26, MM28, MM52, MM66, and MM74 | Liver metastases | |||
| ØPI‐204 | Primary tumor | Heegaard, Spang‐Thomsen, and Prause (2003) (Jan Ulrik Prause) | Subcutaneous | Heegaard et al. (2003) |
Another exciting recent development has been the generation of PDX models from hepatic uveal melanoma metastases (Kageyama et al., 2017). In these models, tumor specimens obtained after surgery or biopsy were surgically implanted into the livers of NOD SCID gamma mice. The authors achieved an 83% engraftment rate and found that the histology, genetics, and proteomics of the implanted tumors resembled corresponding features of patient metastases. Tumors could also be monitored by CT imaging. PDX models such as these hold promise for preclinical evaluation of experimental therapeutic compounds and the realization of personalized medicine.
Like all models, xenografts have disadvantages. The chief among these is the necessity of using immunocompromised mice. This can partially be avoided by taking advantage of the immune‐privileged nature of the anterior chamber of the eye (Niederkorn, 2012). However, this approach can only be used to study the primary tumor, and the majority of grafts spontaneously regress (Sutmuller et al., 2000). In this new era of immunotherapy, the inability to study the interplay between the tumor and host immune system, especially in sites of metastasis, is a major limitation. In uveal melanoma, this is somewhat tempered by the low response rate of patients to PD‐1 and/or CTLA4 inhibition (Algazi et al., 2016; Carvajal et al., 2017). However, other immunomodulatory pathways and cell types have been implicated in this cancer and are being actively investigated (Dougall, Kurtulus, Smyth, & Anderson, 2017; Robertson et al., 2017; Yang et al., 2016). Mice with humanized immune systems would be ideal recipients for xenograft models of all tumor types. Efforts to create such mice are ongoing but are complicated by, among other issues, graft‐versus‐host disease and interspecies differences in cytokine specificity (Allen et al., 2019; Wege, 2018). Other criticisms of xenografts, particularly PDX models, include their high cost, low engraftment rate, and low throughput (Siolas & Hannon, 2013). These are valid concerns, and the actual utility of these models in informing the treatment of patients with uveal melanoma will become more apparent in coming years.
Another approach to avoiding artifacts induced by two‐dimensional cell culturing is the use of three‐dimensional (3D) culture systems. Such “tumor organoid” models now exist for several cancers, including those arising in the colon, breast, and pancreas (Drost & Clevers, 2018; Yang, Sun, Liu, & Mao, 2018). 3D cultures derived from patient tumor specimens can be grafted into mice (patient‐derived organoid xenografts) and faithfully match the molecular phenotypes and even treatment responses of the source tumors (Sachs et al., 2018; Vlachogiannis et al., 2018). Some even allow for the study of the tumor microenvironment, as they incorporate stromal cells such as cancer‐associated fibroblasts and lymphocytes (Neal et al., 2018). In the uveal melanoma literature, there have been a few reports of 3D cultures in which cells form tumorspheres (Angi, Versluis, & Kalirai, 2015; Lapadula et al., 2019; Valyi‐Nagy et al., 2018). Further work is needed to determine the feasibility of generating such cultures from patient tumors and whether these 3D cell models better reflect the biology of their parental tumors. If so, they may serve as superior tools for both in vitro assays and xenograft models.
6. GENETICALLY ENGINEERED MOUSE MODELS (GEMMS) OF UVEAL MELANOMA
The third class of mouse models of uveal melanoma encompasses mice that have been genetically engineered to produce tumors. The primary advantage of these models is that they make it possible to study autochthonous tumorigenesis in an immunocompetent host. In particular, the contribution of specific genetic alterations to oncogenic signaling and disease progression can be assessed (Kersten, de Visser, van Miltenburg, & Jonkers, 2017; Zitvogel, Pitt, Daillere, Smyth, & Kroemer, 2016).
Older models include transgenic mice in which pigment cell‐specific promoters of genes such as Tyrosinase drive expression of the SV40 large T antigen or HRAS, although some of these tumors originate from the retinal pigment epithelium rather than the uvea (Kramer, Powell, Wilson, Salvatore, & Grossniklaus, 1998; Syed et al., 1998; Tolleson et al., 2005). In the Tg(Grm1) model, the Dopachrome tautomerase (Dct) promoter controls expression of the metabotropic glutamate receptor to produce both uveal melanoma and cutaneous melanoma (Schiffner et al., 2014). RET‐driven GEMMs develop melanocytic neoplasms throughout the body, including in the uveal tract (Eyles et al., 2010; Kato et al., 1998). The major weakness of all of these models is that they are driven by molecular changes not observed in patients with uveal melanoma; this limits their clinical applicability.
In the years since the discovery of GNAQ and GNA11 as the main oncogenic drivers of uveal melanoma, three genetically engineered mouse models using these genes have been published (Table 4). In the first, a Tet‐on system was used to induce GNAQQ209L expression in mice deficient for p16Ink4a and p19Ink4b (Feng et al., 2014). Although over half of the mice developed melanocytic cutaneous lesions by 9 months, there was no report of uveal melanoma. Despite this, cutaneous tumors in this model demonstrated YAP activation downstream of oncogenic GNAQ. Another seminal paper published simultaneously reached the same conclusion and demonstrated in vivo efficacy of a YAP inhibitor using a xenograft model of uveal melanoma (Yu et al., 2014).
Table 4.
Genetically engineered mouse models of uveal melanoma
| Model genotype | Induction | Phenotype | Original Publication (Laboratory of origin) |
|---|---|---|---|
| Dct‐rtTA/+; tet‐HA‐GNAQ Q209L/+; p16p19KO | 5‐ to 6‐week‐old mice; doxycycline in food | >50% of mice developed cutaneous melanoma; no report of lesions in the uveal tract | Feng et al. (2014) (J. Silvio Gutkind) |
| Rosa26‐floxed stop‐GNAQ Q209L/+; Mitf‐cre/+ | Embryonic (E15.5) activation by constitutive Cre driver | Skin hyperpigmentation overt uveal melanoma and occasional dermal melanoma at 3 months in 15/15 mice; melanocytic neoplasia of the leptomeninges, harderian gland, cochlea, and vestibular system; putative metastases in the lungs at 3 months in 18/19 mice | Huang et al. (2015) (Catherine Van Raamsdonk) |
| Rosa26‐floxed stop‐GNAQ Q209L/+; Tyrosinase‐creER/+ | 8‐week‐old mice; daily IP injection of tamoxifen and tail dip in 4‐HT for 5 days | Skin hyperpigmentation; melanocytic hyperplasia of the uveal tract (but not overt melanoma) in 3/3 mice | |
| R26‐LSL‐GNA11 Q209L/+; Tyrosinase‐creERT2/+ | 4‐week‐old mice; single IP injection of tamoxifen | Skin hyperpigmentation; overt uveal and dermal melanoma at 6 months in 50% of mice; melanocytic neoplasia of the leptomeninges, third ventricle, harderian gland, and heart; putative metastases in axillary lymph nodes and lungs at 3‐6 months in 100% of mice | Moore et al. (2018) (Yu Chen) |
| R26‐LSL‐GNA11 Q209L/+; BAP1lox/lox; Tyrosinase‐creERT2/+ | Compared to above: increased dermal melanoma burden and proliferative index, no change in number or size of uveal melanoma tumors or lung lesions | ||
| AAV5‐CMV‐Cre or AAV5‐Trp2‐GFPCre; Lats1f/f; Lats2f/f | 2‐ to 4‐month‐old mice; suprachoroidal injection of AAV | Eye bulging at 2 months and uveal melanoma formation at 6 months in 12/14 and 8/10 mice, respectively | Li et al. (2019) (Junhao Mao) |
| AAV5‐Trp2‐GFPCre; Lats1f/f; Lats2f/f; LSL‐KrasG12D | Compared to above: larger uveal melanoma tumors in 7/7 mice and reduced survival (<4 months) |
In a different model, the expression of GNAQQ209L in a lox–stop–lox conditional knock‐in allele inserted at the Rosa26 locus produced uveal melanoma in 3 months with 100% penetrance (Huang, Urtatiz, & Van Raamsdonk, 2015). Furthermore, it appears that cells from these tumors intravasate into blood vessels and metastasize to the lungs. Mice also developed dermal melanomas and melanocytic neoplasms at other sites, including the leptomeninges and inner ear. This model uses Mitf‐cre to initiate oncogene expression. Lastly, another model in which a similar conditional knock‐in allele encoding GNA11Q209L is activated by the inducible Tyrosinase‐creERT2 produced a comparable phenotype, albeit at a later timepoint (Moore et al., 2018). When Bap1 deletion was combined with GNA11Q209L expression, uveal melanomas were unexpectedly smaller. However, skin melanoma burden increased, as did cellular proliferation of these tumors. Comparative genomics from this model identified RasGRP3 as a critical signaling node upstream of MAPK pathway activation, a finding that had been independently reported by another group that used orthogonal methods (Chen et al., 2017).
These models have shed light on key features of uveal melanomagenesis. First, they demonstrate that GNAQ and GNA11 are potent oncogenes. The deletion of tumor suppressors was not required to form uveal melanoma; indeed, in the second model, the expression of the human GNAQ transgene was only 3.3% of that of the murine wild‐type allele as measured by RT‐PCR of primary melanocyte cultures from the affected mice (Huang et al., 2015). Second, they illuminate differences between the pigment cell‐specific promoters used in induction. The constitutive expression of Mitf‐cre beginning at E15.5 likely explains the earlier onset of tumor formation in the GNAQQ209L model as compared to the GNA11Q209L model in which Tyr‐creERT2 is induced in 4‐week‐old mice (Huang et al., 2015; Moore et al., 2018). Interestingly, induction of Tyr‐creER in 8‐week‐old mice in the GNAQQ209L model did not produce overt uveal melanoma. Whether this is simply due to the differences in induction (mouse age and type of inducible Cre recombinase) or the result of differing potencies of the oncogenic drivers remains to be explored. Finally, these GEMMs illustrate which populations of melanocytes are susceptible to oncogenic transformation by these mutations and downstream activated pathways.
Like other models, these GEMMs are not without their disadvantages. Disease progression is considerably slower than in syngeneic or xenograft models due to the time required for tumor initiation. A problem specific to the GNAQQ209L model is the microphthalmia caused by the Mitf‐cre allele (Alizadeh, Fitch, Niswender, McKnight, & Barsh, 2008). Additionally, inserting the oncogenes in the Rosa26 locus is somewhat artificial. A model in which an activatable allele is targeted to the endogenous mouse Gnaq or Gna11 locus might better model physiologic expression of these genes. This approach has been successful in generating valuable Braf V600E GEMMs of cutaneous melanoma (Dankort et al., 2007; Mercer et al., 2005).
A serious obstacle encountered in the above uveal melanoma GEMMs is the induction of transgene expression in melanocytes throughout the entire body. This complicates the models in numerous ways. First, melanocytic neoplasms in other organs may cause pathology, such as the ataxic phenotype caused by melanocytosis of the vestibular system. Second, mice sometimes have to be euthanized before the ocular tumor can be fully studied because of rapid growth of melanomas arising from the dermis. Third, the study of metastasis is difficult because of the number of primary tumors, including some that develop in vital organs such as the heart (Huang et al., 2015; Moore et al., 2018).
An exciting recent publication describes a new method to overcome these issues by utilizing adeno‐associated viral delivery of Cre recombinase to the uveal tract (Li et al., 2019). In this model, the suprachoroidal injection of an AAV5‐CMV‐Cre vector produced ocular melanocytic tumors in adult mice carrying conditional null alleles of the Hippo kinases Lats1 and Lats2, which normally function to suppress YAP/TAZ signaling. Furthermore, a similar vector in which Cre expression is under the control of the pigment cell‐specific tyrosinase‐related protein 2 (Trp2) promoter produced a comparable phenotype. Importantly, cells of these tumors were positive for melanoma markers Melan‐A/Mart1 and HMB45 but negative for RPE65. This indicates that they arose from uveal melanocytes and not cells of the retinal pigment epithelium. Remarkably, the authors found that activation of the YAP pathway alone was both necessary and sufficient for initiation of uveal melanoma. Activation of the MAPK pathway using an inducible Kras G12D allele was not sufficient for tumor formation but did accelerate tumor growth and mortality in the Lats double knockout mice. They explored this intriguing synergy between MAPK and Hippo signaling and discovered an interactive transcriptional network in which AP1 factors amplify the oncogenic output of YAP/TEAD in uveal melanoma. The use of this AAV‐Cre system represents a significant improvement upon the aforementioned Cre driver mouse strains in that it limits oncogenic transformation to melanocytes within the uveal tract of adult mice. This is a powerful new tool that could be used in conjunction with both existing and new alleles to generate genetic mouse models that would enable the study of the entire disease process of uveal melanoma in vivo.
In addition to this AAV approach, the RCAS‐TVA system might achieve similar results. This method has been used to generate numerous GEMMs of cutaneous melanoma (Cho et al., 2015; Kircher et al., 2019; VanBrocklin, Robinson, Lastwika, Khoury, & Holmen, 2010). RCAS subgroup A is an avian retrovirus capable of infecting cells that express the TVA receptor. Dopachrome tautomerase‐TVA transgenic mice express this receptor in pigment‐producing cells, and this strain could be crossed with one of the conditional knock‐in alleles described above. Intraocular injection of RCAS virus that encodes for Cre would then activate oncogene expression in melanocytes of the eye. An advantage of this model over the AAV approach is targeted delivery to cells of interest such that there is no requirement for inclusion of a pigment cell‐specific promoter within the virus. This provides more room for genes of interest, which can be linked with Cre within the same viral vector to enable delivery to the same cells. Additionally, high titers of RCAS are easily produced in vitro using the chicken fibroblast DF‐1 cell line (Fisher et al., 1999), and there is no need for helper virus in these cells. Retroviruses also permit long‐term expression of genes due to genome integration, though this requires that the cells are dividing.
Despite these advancements in genetic models of uveal melanoma, the inherent differences in tumor biology between mice and humans cannot be ignored. Putative metastases that have been observed in published genetic models occur in the lungs, not the liver (Huang et al., 2015; Moore et al., 2018). Additionally, the loss of Bap1 did not enhance the aggressiveness of uveal melanomas; in fact, the ocular phenotype was weaker, and there was no increase in size or incidence of lung lesions compared with mice expressing GNA11Q209L alone (Moore et al., 2018). The basis for these differences is not understood and merits further investigation. It must also be acknowledged that the chromosomal abnormalities and epigenetic modifications observed in patients with uveal melanoma are nearly impossible to model in a mouse. Thus, while these GEMMs, as well as improved models, will continue to provide valuable insights into the progression of uveal melanoma in vivo, it is unlikely that any one model will fully recapitulate the human disease in all of its intricacies.
7. CONCLUSIONS
In summary, although there is no perfect mouse model of uveal melanoma, currently available models have been instrumental in elucidating critical signaling pathways and testing new therapeutic strategies for this cancer. Each type of model has distinctive strengths and weaknesses. Syngeneic models are excellent for the investigation of tumor progression in an immunocompetent host but use cutaneous melanoma cell lines. Xenograft models allow for the study of human uveal melanoma cells and tumors in a living organism, but this does not include the immune response because recipient mice must be severely immunocompromised. Genetically engineered models allow for studies of autochthonous uveal melanoma formation and dissemination, yet tumors in these mice differ from those in patients in terms of molecular complexity and metastatic behavior. Investigators should leverage the models best suited to address their specific scientific questions. Future model development should aim to overcome current limitations and further enable efforts to investigate uveal melanoma biology and develop therapies most likely to succeed in patients afflicted with this cancer.
CONFLICT OF INTERESTS
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Diana Lim for her assistance with graphic design and Sheri Holmen and Amanda Truong for critically reviewing the manuscript.
Richards JR, Yoo JH, Shin D, Odelberg SJ. Mouse models of uveal melanoma: Strengths, weaknesses, and future directions. Pigment Cell Melanoma Res. 2020;33:264–278. 10.1111/pcmr.12853
Funding information
This work was funded by grants to Jackson R. Richards, Jae Hyuk Yoo, and Shannon J. Odelberg from the National Cancer Institute of the National Institutes of Health (F30CA217184, K99CA230312, and R01CA202778) and the Melanoma Research Foundation (Cure OM Junior Fellowship Award). The content is solely the responsibility of the authors and does not necessarily represent the official views of funding agencies.
REFERENCES
- Aalto, Y. , Eriksson, L. , Seregard, S. , Larsson, O. , & Knuutila, S. (2001). Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Investigative Ophthalmology & Visual Science, 42(2), 313–317. [PubMed] [Google Scholar]
- Algazi, A. P. , Tsai, K. K. , Shoushtari, A. N. , Munhoz, R. R. , Eroglu, Z. , Piulats, J. M. , … Sullivan, R. J. (2016). Clinical outcomes in metastatic uveal melanoma treated with PD‐1 and PD‐L1 antibodies. Cancer, 122(21), 3344–3353. 10.1002/cncr.30258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh, A. , Fitch, K. R. , Niswender, C. M. , McKnight, G. S. , & Barsh, G. S. (2008). Melanocyte‐lineage expression of Cre recombinase using Mitf regulatory elements. Pigment Cell Melanoma Res, 21(1), 63–69. [DOI] [PubMed] [Google Scholar]
- Alizadeh, H. , Howard, K. , Mellon, J. , Mayhew, E. , Rusciano, D. , & Niederkorn, J. Y. (2003). Reduction of liver metastasis of intraocular melanoma by interferon‐beta gene transfer. Investigative Ophthalmology & Visual Science, 44(7), 3042–3051. [DOI] [PubMed] [Google Scholar]
- Allen, T. M. , Brehm, M. A. , Bridges, S. , Ferguson, S. , Kumar, P. , Mirochnitchenko, O. , … PrabhuDas, M. (2019). Humanized immune system mouse models: Progress, challenges and opportunities. Nature Immunology, 20(7), 770–774. 10.1038/s41590-019-0416-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini, G. , Do, C. , Tycko, B. , Realubit, R. B. , Karan, C. , Musi, E. , … Schwartz, G. K. (2019). Inhibition of NF‐κB‐dependent signaling enhances sensitivity and overcomes resistance to BET inhibition in uveal melanoma. Cancer Research, 79(9), 2415–2425. 10.1158/0008-5472.CAN-18-3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini, G. , Musi, E. , Ho, A. L. , de Stanchina, E. , & Schwartz, G. K. (2013). Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK‐dependent autophagic cell death. Molecular Cancer Therapeutics, 12(5), 768–776. 10.1158/1535-7163.MCT-12-1020 [DOI] [PubMed] [Google Scholar]
- Ambrosini, G. , Sawle, A. D. , Musi, E. , & Schwartz, G. K. (2015). BRD4‐targeted therapy induces Myc‐independent cytotoxicity in Gnaq/11‐mutatant uveal melanoma cells. Oncotarget, 6(32), 33397–33409. 10.18632/oncotarget.5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirouchene‐Angelozzi, N. , Frisch‐Dit‐Leitz, E. , Carita, G. , Dahmani, A. , Raymondie, C. , Liot, G. , … Schoumacher, M. (2016). The mTOR inhibitor Everolimus synergizes with the PI3K inhibitor GDC0941 to enhance anti‐tumor efficacy in uveal melanoma. Oncotarget, 7(17), 23633–23646. 10.18632/oncotarget.8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirouchene‐Angelozzi, N. , Nemati, F. , Gentien, D. , Nicolas, A. , Dumont, A. , Carita, G. , … Roman‐Roman, S. (2014). Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Molecular Oncology, 8(8), 1508–1520. 10.1016/j.molonc.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbunathan, H. , Verstraten, R. , Singh, A. D. , Harbour, J. W. , & Bowcock, A. M. (2019). Integrative copy number analysis of uveal melanoma reveals novel candidate genes involved in tumorigenesis including a tumor suppressor role for PHF10/BAF45a. Clinical Cancer Research, 25(16), 5156–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angi, M. , Versluis, M. , & Kalirai, H. (2015). Culturing uveal melanoma cells. Ocular Oncology and Pathology, 1(3), 126–132. 10.1159/000370150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annala, S. , Feng, X. , Shridhar, N. , Eryilmaz, F. , Patt, J. , Yang, J. , … Kostenis, E. (2019). Direct targeting of Gαq and Gα11 oncoproteins in cancer cells. Science Signalling, 12(573), eaau5948. [DOI] [PubMed] [Google Scholar]
- Barisione, G. , Fabbi, M. , Gino, A. , Queirolo, P. , Orgiano, L. , Spano, L. , … Gangemi, R. (2015). Potential role of soluble c‐Met as a new candidate biomarker of metastatic uveal melanoma. JAMA Ophthalmology, 133(9), 1013–1021. 10.1001/jamaophthalmol.2015.1766 [DOI] [PubMed] [Google Scholar]
- Ben‐David, U. , Siranosian, B. , Ha, G. , Tang, H. , Oren, Y. , Hinohara, K. , … Golub, T. R. (2018). Genetic and transcriptional evolution alters cancer cell line drug response. Nature, 560(7718), 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, D. C. , Cooper, P. J. , & Hart, I. R. (1987). A line of non‐tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. International Journal of Cancer, 39(3), 414–418. 10.1002/ijc.2910390324 [DOI] [PubMed] [Google Scholar]
- Bontzos, G. , & Detorakis, E. T. (2017). Animal models of uveal melanoma for localized interventions. Critical Reviews in Oncogenesis, 22(3–4), 187–194. 10.1615/CritRevOncog.2018024510 [DOI] [PubMed] [Google Scholar]
- Cao, J. , & Jager, M. J. (2015). Animal eye models for uveal melanoma. Ocular Oncology and Pathology, 1(3), 141–150. 10.1159/000370152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carita, G. , Frisch‐Dit‐Leitz, E. , Dahmani, A. , Raymondie, C. , Cassoux, N. , Piperno‐Neumann, S. , … Decaudin, D. (2016). Dual inhibition of protein kinase C and p53‐MDM2 or PKC and mTORC1 are novel efficient therapeutic approaches for uveal melanoma. Oncotarget, 7(23), 33542–33556. 10.18632/oncotarget.9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carita, G. , Nemati, F. , & Decaudin, D. (2015). Uveal melanoma patient‐derived xenografts. Ocular Oncology and Pathology, 1(3), 161–169. 10.1159/000370154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal, R. D. , Schwartz, G. K. , Tezel, T. , Marr, B. , Francis, J. H. , & Nathan, P. D. (2017). Metastatic disease from uveal melanoma: Treatment options and future prospects. British Journal of Ophthalmology, 101(1), 38–44. 10.1136/bjophthalmol-2016-309034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. U. , Wu, Q. , Depeille, P. , Chen, P. , Thornton, S. , Kalirai, H. , … Bastian, B. C. (2017). RasGRP3 mediates MAPK pathway activation in GNAQ mutant uveal melanoma. Cancer Cell, 31(5), 685–696 e686. 10.1016/j.ccell.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Wu, Q. , Tan, L. , Porter, D. , Jager, M. J. , Emery, C. , & Bastian, B. C. (2014). Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene, 33(39), 4724–4734. 10.1038/onc.2013.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H. , Terai, M. , Kageyama, K. , Ozaki, S. , McCue, P. A. , Sato, T. , & Aplin, A. E. (2015). Paracrine effect of NRG1 and HGF drives resistance to MEK inhibitors in metastatic uveal melanoma. Cancer Research, 75(13), 2737–2748. 10.1158/0008-5472.CAN-15-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J. H. , Robinson, J. P. , Arave, R. A. , Burnett, W. J. , Kircher, D. A. , Chen, G. , … Holmen, S. L. (2015). AKT1 activation promotes development of melanoma metastases. Cell Reports, 13(5), 898–905. 10.1016/j.celrep.2015.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosette, J. , Ben Abdelwahed, R. , Donnou‐Triffault, S. , Sautes‐Fridman, C. , Flaud, P. , & Fisson, S. (2016). Bioluminescence‐based tumor quantification method for monitoring tumor progression and treatment effects in mouse lymphoma models. Journal of Visualized Experiments, (113), e53609 10.3791/53609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damato, E. M. , & Damato, B. E. (2012). Detection and time to treatment of uveal melanoma in the United Kingdom: An evaluation of 2,384 patients. Ophthalmology, 119(8), 1582–1589. 10.1016/j.ophtha.2012.01.048 [DOI] [PubMed] [Google Scholar]
- Dankort, D. , Filenova, E. , Collado, M. , Serrano, M. , Jones, K. , & McMahon, M. (2007). A new mouse model to explore the initiation, progression, and therapy of BRAFV600E‐induced lung tumors. Genes & Development, 21(4), 379–384. 10.1101/gad.1516407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange, J. , Ly, L. V. , Lodder, K. , Verlaan‐de Vries, M. , Teunisse, A. F. , Jager, M. J. , & Jochemsen, A. G. (2012). Synergistic growth inhibition based on small‐molecule p53 activation as treatment for intraocular melanoma. Oncogene, 31(9), 1105–1116. 10.1038/onc.2011.309 [DOI] [PubMed] [Google Scholar]
- De Waard‐Siebinga, I. , Blom, D.‐J. , Griffioen, M. , Schrier, P. I. , Hoogendoorn, E. D. , Beverstock, G. , … Jager, M. J. (1995). Establishment and characterization of an uveal‐melanoma cell line. International Journal of Cancer, 62(2), 155–161. 10.1002/ijc.2910620208 [DOI] [PubMed] [Google Scholar]
- Diaz, C. E. , Rusciano, D. , Dithmar, S. , & Grossniklaus, H. E. (1999). B16LS9 melanoma cells spread to the liver from the murine ocular posterior compartment (PC). Current Eye Research, 18(2), 125–129. 10.1076/ceyr.18.2.125.5380 [DOI] [PubMed] [Google Scholar]
- Dithmar, S. , Rusciano, D. , & Grossniklaus, H. E. (2000). A new technique for implantation of tissue culture melanoma cells in a murine model of metastatic ocular melanoma. Melanoma Research, 10(1), 2–8. 10.1097/00008390-200002000-00001 [DOI] [PubMed] [Google Scholar]
- Dithmar, S. , Rusciano, D. , Lynn, M. J. , Lawson, D. H. , Armstrong, C. A. , & Grossniklaus, H. E. (2000). Neoadjuvant interferon alfa‐2b treatment in a murine model for metastatic ocular melanoma: A preliminary study. Archives of Ophthalmology, 118(8), 1085–1089. [DOI] [PubMed] [Google Scholar]
- Dong, L. , You, S. , Zhang, Q. , Osuka, S. , Devi, N. S. , Kaluz, S. , … Van Meir, E. G. (2019). Arylsulfonamide 64B inhibits hypoxia/HIF‐induced expression of c‐Met and CXCR4 and reduces primary tumor growth and metastasis of uveal melanoma. Clinical Cancer Research, 25(7), 2206–2218. 10.1158/1078-0432.CCR-18-1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougall, W. C. , Kurtulus, S. , Smyth, M. J. , & Anderson, A. C. (2017). TIGIT and CD96: New checkpoint receptor targets for cancer immunotherapy. Immunological Reviews, 276(1), 112–120. 10.1111/imr.12518 [DOI] [PubMed] [Google Scholar]
- Drost, J. , & Clevers, H. (2018). Organoids in cancer research. Nature Reviews Cancer, 18(7), 407–418. 10.1038/s41568-018-0007-6 [DOI] [PubMed] [Google Scholar]
- Ehlers, J. P. , Worley, L. , Onken, M. D. , & Harbour, J. W. (2005). DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clinical Cancer Research, 11(10), 3609–3613. 10.1158/1078-0432.CCR-04-1941 [DOI] [PubMed] [Google Scholar]
- el Filali, M. , Ly, L. V. , Luyten, G. P. , Versluis, M. , Grossniklaus, H. E. , van der Velden, P. A. , & Jager, M. J. (2012). Bevacizumab and intraocular tumors: An intriguing paradox. Molecular Vision, 18, 2454–2467. [PMC free article] [PubMed] [Google Scholar]
- Eyles, J. O. , Puaux, A.‐L. , Wang, X. , Toh, B. , Prakash, C. , Hong, M. , … Abastado, J.‐P. (2010). Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. Journal of Clinical Investigation, 120(6), 2030–2039. 10.1172/JCI42002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiao‐Flores, F. , Emmons, M. F. , Durante, M. A. , Kinose, F. , Saha, B. , Fang, B. , … Smalley, K. S. M. (2019). HDAC inhibition enhances the in vivo efficacy of MEK inhibitor therapy in uveal melanoma. Clinical Cancer Research, 25(18), 5686–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, X. , Degese, M. S. , Iglesias‐Bartolome, R. , Vaque, J. P. , Molinolo, A. A. , Rodrigues, M. , … Gutkind, J. S. (2014). Hippo‐independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio‐regulated rho GTPase signaling circuitry. Cancer Cell, 25(6), 831–845. 10.1016/j.ccr.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler, I. J. , Gersten, D. M. , & Budmen, M. B. (1976). Characterization in vivo and in vitro of tumor cells selected for resistance to syngeneic lymphocyte‐mediated cytotoxicity. Cancer Research, 36(9 pt.1), 3160–3165. [PubMed] [Google Scholar]
- Field, M. G. , Durante, M. A. , Anbunathan, H. , Cai, L. Z. , Decatur, C. L. , Bowcock, A. M. , … Harbour, J. W. (2018). Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nature Communications, 9(1), 116 10.1038/s41467-017-02428-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, G. H. , Orsulic, S. , Holland, E. , Hively, W. P. , Li, Y. I. , Lewis, B. C. , … Varmus, H. E. (1999). Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene, 18(38), 5253–5260. 10.1038/sj.onc.1203087 [DOI] [PubMed] [Google Scholar]
- Folberg, R. , Kadkol, S. S. , Frenkel, S. , Valyi‐Nagy, K. , Jager, M. J. , Pe'er, J. , & Maniotis, A. J. (2008). Authenticating cell lines in ophthalmic research laboratories. Investigative Ophthalmology & Visual Science, 49(11), 4697–4701. 10.1167/iovs.08-2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornabaio, G. , Barnhill, R. L. , Lugassy, C. , Bentolila, L. A. , Cassoux, N. , Roman‐Roman, S. , … Del Bene, F. (2018). Angiotropism and extravascular migratory metastasis in cutaneous and uveal melanoma progression in a zebrafish model. Scientific Reports, 8(1), 10448 10.1038/s41598-018-28515-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, E. M. V. , Lindberg, M. F. , Jespersen, H. , Alsén, S. , Bagge, R. O. , Donia, M. , … Nilsson, J. A. (2019). HER2 CAR‐T cells eradicate uveal melanoma and T‐cell therapy‐resistant human melanoma in IL2 Transgenic NOD/SCID IL2 receptor knockout mice. Cancer Research, 79(5), 899–904. 10.1158/0008-5472.CAN-18-3158 [DOI] [PubMed] [Google Scholar]
- Gangemi, R. , Amaro, A. , Gino, A. , Barisione, G. , Fabbi, M. , Pfeffer, U. , … Ferrini, S. (2014). ADAM10 correlates with uveal melanoma metastasis and promotes in vitro invasion. Pigment Cell & Melanoma Research, 27(6), 1138–1148. 10.1111/pcmr.12306 [DOI] [PubMed] [Google Scholar]
- Gangemi, R. , Mirisola, V. , Barisione, G. , Fabbi, M. , Brizzolara, A. , Lanza, F. , … Ferrini, S. (2012). Mda‐9/syntenin is expressed in uveal melanoma and correlates with metastatic progression. PLoS ONE, 7(1), e29989 10.1371/journal.pone.0029989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M. , Tang, J. , Liu, K. , Yang, M. , & Liu, H. (2018). Quantitative evaluation of vascular microcirculation using contrast‐enhanced ultrasound imaging in rabbit models of choroidal melanoma. Investigative Ophthalmology & Visual Science, 59(3), 1251–1262. 10.1167/iovs.17-22197 [DOI] [PubMed] [Google Scholar]
- Gillet, J. P. , Varma, S. , & Gottesman, M. M. (2013). The clinical relevance of cancer cell lines. Journal of the National Cancer Institute, 105(7), 452–458. 10.1093/jnci/djt007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed, A. , Heiser, L. M. , Gray, J. W. , & Costello, J. C. (2016). Tumor‐derived cell lines as molecular models of cancer pharmacogenomics. Molecular Cancer Research, 14(1), 3–13. 10.1158/1541-7786.MCR-15-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S. E. , Junttila, M. R. , & de Sauvage, F. J. (2015). Translational value of mouse models in oncology drug development. Nature Medicine, 21(5), 431–439. 10.1038/nm.3853 [DOI] [PubMed] [Google Scholar]
- Griewank, K. G. , Yu, X. , Khalili, J. , Sozen, M. M. , Stempke‐Hale, K. , Bernatchez, C. , … Woodman, S. E. (2012). Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res, 25(2), 182–187. 10.1111/j.1755-148X.2012.00971.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, H. E. , Barron, B. C. , & Wilson, M. W. (1995). Murine model of anterior and posterior ocular melanoma. Current Eye Research, 14(5), 399–404. 10.3109/02713689508999938 [DOI] [PubMed] [Google Scholar]
- Grossniklaus, H. E. , Zhang, Q. , You, S. , McCarthy, C. , Heegaard, S. , & Coupland, S. E. (2016). Metastatic ocular melanoma to the liver exhibits infiltrative and nodular growth patterns. Human Pathology, 57, 165–175. 10.1016/j.humpath.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, D. W. , Al‐Shammari, N. S. , Danson, S. , Jacques, R. , Rennie, I. G. , & Sisley, K. (2015). High‐resolution array CGH analysis identifies regional deletions and amplifications of chromosome 8 in uveal melanoma. Investigative Ophthalmology & Visual Science, 56(6), 3460–3466. 10.1167/iovs.14-16215 [DOI] [PubMed] [Google Scholar]
- Han, Z. , Brown, J. R. , & Niederkorn, J. Y. (2016). Growth and metastasis of intraocular tumors in aged mice. Investigative Ophthalmology & Visual Science, 57(6), 2366–2376. 10.1167/iovs.16-19156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour, J. W. , Onken, M. D. , Roberson, E. D. , Duan, S. , Cao, L. , Worley, L. A. , … Bowcock, A. M. (2010). Frequent mutation of BAP1 in metastasizing uveal melanomas. Science, 330(6009), 1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour, J. W. , Roberson, E. D. , Anbunathan, H. , Onken, M. D. , Worley, L. A. , & Bowcock, A. M. (2013). Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nature Genetics, 45(2), 133–135. 10.1038/ng.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harning, R. , & Szalay, J. (1987). Ocular metastasis of in vivo and in vitro derived syngeneic murine melanoma. Investigative Ophthalmology & Visual Science, 28(9), 1599–1604. [PubMed] [Google Scholar]
- Heegaard, S. , Spang‐Thomsen, M. , & Prause, J. U. (2003). Establishment and characterization of human uveal malignant melanoma xenografts in nude mice. Melanoma Research, 13(3), 247–251. 10.1097/00008390-200306000-00004 [DOI] [PubMed] [Google Scholar]
- Ho, A. L. , Musi, E. , Ambrosini, G. , Nair, J. S. , Deraje Vasudeva, S. , de Stanchina, E. , & Schwartz, G. K. (2012). Impact of combined mTOR and MEK inhibition in uveal melanoma is driven by tumor genotype. PLoS ONE, 7(7), e40439 10.1371/journal.pone.0040439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. L. , Urtatiz, O. , & Van Raamsdonk, C. D. (2015). Oncogenic G protein GNAQ induces uveal melanoma and intravasation in mice. Cancer Research, 75(16), 3384–3397. 10.1158/0008-5472.CAN-14-3229 [DOI] [PubMed] [Google Scholar]
- Jager, M. J. , Magner, J. A. , Ksander, B. R. , & Dubovy, S. R. (2016). Uveal melanoma cell lines: Where do they come from? (An American Ophthalmological Society Thesis). Transactions of the American Ophthalmological Society, 114, T5. [PMC free article] [PubMed] [Google Scholar]
- Jin, Y. , Zhang, P. , Wang, Y. , Jin, B. , Zhou, J. , Zhang, J. , & Pan, J. (2018). Neddylation blockade diminishes hepatic metastasis by dampening cancer stem‐like cells and angiogenesis in uveal melanoma. Clinical Cancer Research, 24(15), 3741–3754. 10.1158/1078-0432.CCR-17-1703 [DOI] [PubMed] [Google Scholar]
- Johansson, P. , Aoude, L. G. , Wadt, K. , Glasson, W. J. , Warrier, S. K. , Hewitt, A. W. , … Hayward, N. K. (2016). Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget, 7(4), 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, N. M. , Yang, H. , Zhang, Q. , Morales‐Tirado, V. M. , & Grossniklaus, H. E. (2019). Natural killer cells and pigment epithelial‐derived factor control the infiltrative and nodular growth of hepatic metastases in an Orthotopic murine model of ocular melanoma. BMC Cancer, 19(1), 484 10.1186/s12885-019-5712-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, K. , Ohara, M. , Saito, K. , Ozaki, S. , Terai, M. , Mastrangelo, M. J. , … Sato, T. (2017). Establishment of an orthotopic patient‐derived xenograft mouse model using uveal melanoma hepatic metastasis. Journal of Translational Medicine, 15(1), 145 10.1186/s12967-017-1247-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaochar, S. , Dong, J. , Torres, M. , Rajapakshe, K. , Nikolos, F. , Davis, C. M. , … Poulaki, V. (2018). ICG‐001 exerts potent anticancer activity against uveal melanoma cells. Investigative Opthalmology & Visual Science, 59(1), 132–143. 10.1167/iovs.17-22454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , Takahashi, M. , Akhand, A. A. , Liu, W. , Dai, Y. , Shimizu, S. , … Nakashima, I. (1998). Transgenic mouse model for skin malignant melanoma. Oncogene, 17(14), 1885–1888. 10.1038/sj.onc.1202077 [DOI] [PubMed] [Google Scholar]
- Kersten, K. , de Visser, K. E. , van Miltenburg, M. H. , & Jonkers, J. (2017). Genetically engineered mouse models in oncology research and cancer medicine. EMBO Molecular Medicine, 9(2), 137–153. 10.15252/emmm.201606857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoja, L. , Atenafu, E. G. , Suciu, S. , Leyvraz, S. , Sato, T. , Marshall, E. , … Joshua, A. M. (2019). Meta‐analysis in metastatic uveal melanoma to determine progression‐free and overall survival benchmarks: An International Rare Cancers Initiative (IRCI) ocular melanoma study. Annals of Oncology, 30(8), 1370–1380. 10.1093/annonc/mdz176 [DOI] [PubMed] [Google Scholar]
- Kilian, M. M. , Loeffler, K. U. , Pfarrer, C. , Holz, F. G. , Kurts, C. , & Herwig, M. C. (2016). Intravitreally injected HCmel12 melanoma cells serve as a mouse model of tumor biology of intraocular melanoma. Current Eye Research, 41(1), 121–128. 10.3109/02713683.2015.1004721 [DOI] [PubMed] [Google Scholar]
- Kines, R. C. , Varsavsky, I. , Choudhary, S. , Bhattacharya, D. , Spring, S. , McLaughlin, R. , … Schiller, J. T. (2018). An infrared dye‐conjugated virus‐like particle for the treatment of primary uveal melanoma. Molecular Cancer Therapeutics, 17(2), 565–574. 10.1158/1535-7163.MCT-17-0953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, D. A. , Trombetti, K. A. , Silvis, M. R. , Parkman, G. L. , Fischer, G. M. , Angel, S. N. , … Holmen, S. L. (2019). AKT1(E17K) activates focal adhesion kinase and promotes melanoma brain metastasis. Molecular Cancer Research, 17(9), 1787–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knisely, T. L. , & Niederkorn, J. Y. (1990). Immunologic evaluation of spontaneous regression of an intraocular murine melanoma. Investigative Ophthalmology & Visual Science, 31(2), 247–257. [PubMed] [Google Scholar]
- Kramer, T. R. , Powell, M. B. , Wilson, M. M. , Salvatore, J. , & Grossniklaus, H. E. (1998). Pigmented uveal tumours in a transgenic mouse model. British Journal of Ophthalmology, 82(8), 953–960. 10.1136/bjo.82.8.953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz, B. A. , Dave, N. , Komatsubara, K. M. , Marr, B. P. , & Carvajal, R. D. (2017). Uveal melanoma: Epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol, 11, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksander, B. R. , Rubsamen, P. E. , Olsen, K. R. , Cousins, S. W. , & Streilein, J. W. (1991). Studies of tumor‐infiltrating lymphocytes from a human choroidal melanoma. Investigative Ophthalmology & Visual Science, 32(13), 3198–3208. [PubMed] [Google Scholar]
- Kujala, E. , Makitie, T. , & Kivela, T. (2003). Very long‐term prognosis of patients with malignant uveal melanoma. Investigative Ophthalmology & Visual Science, 44(11), 4651–4659. 10.1167/iovs.03-0538 [DOI] [PubMed] [Google Scholar]
- Landreville, S. , Agapova, O. A. , Matatall, K. A. , Kneass, Z. T. , Onken, M. D. , Lee, R. S. , … Harbour, J. W. (2012). Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clinical Cancer Research, 18(2), 408–416. 10.1158/1078-0432.CCR-11-0946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapadula, D. , Farias, E. , Randolph, C. E. , Purwin, T. J. , McGrath, D. , Charpentier, T. H. , … Benovic, J. L. (2019). Effects of oncogenic Gαq and Gα11 inhibition by FR900359 in uveal melanoma. Molecular Cancer Research, 17(4), 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattier, J. M. , Yang, H. , Crawford, S. , & Grossniklaus, H. E. (2013). Host pigment epithelium‐derived factor (PEDF) prevents progression of liver metastasis in a mouse model of uveal melanoma. Clinical & Experimental Metastasis, 30(8), 969–976. 10.1007/s10585-013-9596-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Li, Q. I. , Dang, K. , Ma, S. , Cotton, J. L. , Yang, S. , … Mao, J. (2019). YAP/TAZ activation drives uveal melanoma initiation and progression. Cell Reports, 29(10), 3200–3211 e3204. 10.1016/j.celrep.2019.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Zhan, W. , Zhu, A. , Yoon, Y. , Lin, S. , Sasaki, M. , … Shim, H. (2012). Development of a unique small molecule modulator of CXCR4. PLoS ONE, 7(4), e34038 10.1371/journal.pone.0034038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyten, G. P. M. , Naus, N. C. , Mooy, C. M. , Hagemeijer, A. , Kan‐Mitchell, J. , Van Drunen, E. , … Luider, T. M. (1996). Establishment and characterization of primary and metastatic uveal melanoma cell lines. International Journal of Cancer, 66(3), 380–387. [DOI] [PubMed] [Google Scholar]
- Ly, L. V. , Baghat, A. , Versluis, M. , Jordanova, E. S. , Luyten, G. P. M. , van Rooijen, N. , … Jager, M. J. (2010). In aged mice, outgrowth of intraocular melanoma depends on proangiogenic M2‐type macrophages. The Journal of Immunology, 185(6), 3481–3488. 10.4049/jimmunol.0903479 [DOI] [PubMed] [Google Scholar]
- Ma, D. , Luyten, G. P. , Luider, T. M. , Jager, M. J. , & Niederkorn, J. Y. (1996). Association between NM23‐H1 gene expression and metastasis of human uveal melanoma in an animal model. Investigative Ophthalmology & Visual Science, 37(11), 2293–2301. [PubMed] [Google Scholar]
- Ma, D. , & Niederkorn, J. Y. (1995). Transforming growth factor‐beta down‐regulates major histocompatibility complex class I antigen expression and increases the susceptibility of uveal melanoma cells to natural killer cell‐mediated cytolysis. Immunology, 86(2), 263–269. [PMC free article] [PubMed] [Google Scholar]
- Ma, D. , & Niederkorn, J. Y. (1998). Role of epidermal growth factor receptor in the metastasis of intraocular melanomas. Investigative Ophthalmology & Visual Science, 39(7), 1067–1075. [PubMed] [Google Scholar]
- Madic, J. , Piperno‐Neumann, S. , Servois, V. , Rampanou, A. , Milder, M. , Trouiller, B. , … Stern, M.‐H. (2012). Pyrophosphorolysis‐activated polymerization detects circulating tumor DNA in metastatic uveal melanoma. Clinical Cancer Research, 18(14), 3934–3941. 10.1158/1078-0432.CCR-12-0309 [DOI] [PubMed] [Google Scholar]
- Martin, M. , Maßhöfer, L. , Temming, P. , Rahmann, S. , Metz, C. , Bornfeld, N. , … Zeschnigk, M. (2013). Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nature Genetics, 45(8), 933–936. 10.1038/ng.2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K. A. , Agapova, O. A. , Onken, M. D. , Worley, L. A. , Bowcock, A. M. , & Harbour, J. W. (2013). BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer, 13, 371 10.1186/1471-2407-13-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, C. C. , Wu, X. C. , Jemal, A. , Martin, H. J. , Roche, L. M. , & Chen, V. W. (2005). Incidence of noncutaneous melanomas in the U.S. Cancer, 103(5), 1000–1007. [DOI] [PubMed] [Google Scholar]
- Mercer, K. , Giblett, S. , Green, S. , Lloyd, D. , DaRocha Dias, S. , Plumb, M. , … Pritchard, C. (2005). Expression of endogenous oncogenic V600EB‐raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Research, 65(24), 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, J. R. (2018). Zebrafish: Development of a vertebrate model organism. Current Protocols Essential Laboratory Techniques, 16, e19. [Google Scholar]
- Moore, A. R. , Ceraudo, E. , Sher, J. J. , Guan, Y. , Shoushtari, A. N. , Chang, M. T. , … Chen, Y. U. (2016). Recurrent activating mutations of G‐protein‐coupled receptor CYSLTR2 in uveal melanoma. Nature Genetics, 48(6), 675–680. 10.1038/ng.3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A. R. , Ran, L. , Guan, Y. , Sher, J. J. , Hitchman, T. D. , Zhang, J. Q. , … Chen, Y. U. (2018). GNA11 Q209L mouse model reveals RasGRP3 as an essential signaling node in uveal melanoma. Cell Reports, 22(9), 2455–2468. 10.1016/j.celrep.2018.01.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouriaux, F. , Zaniolo, K. , Bergeron, M.‐A. , Weidmann, C. , De La Fouchardière, A. , Fournier, F. , … Guérin, S. L. (2016). Effects of long‐term serial passaging on the characteristics and properties of cell lines derived from uveal melanoma primary tumors. Investigative Ophthalmology & Visual Science, 57(13), 5288–5301. 10.1167/iovs.16-19317 [DOI] [PubMed] [Google Scholar]
- Mouti, M. A. , Dee, C. , Coupland, S. E. , & Hurlstone, A. F. (2016). Minimal contribution of ERK1/2‐MAPK signalling towards the maintenance of oncogenic GNAQQ209P‐driven uveal melanomas in zebrafish. Oncotarget, 7(26), 39654–39670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musi, E. , Ambrosini, G. , de Stanchina, E. , & Schwartz, G. K. (2014). The phosphoinositide 3‐kinase α selective inhibitor BYL719 enhances the effect of the protein kinase C inhibitor AEB071 in GNAQ/GNA11‐mutant uveal melanoma cells. Molecular Cancer Therapeutics, 13(5), 1044–1053. 10.1158/1535-7163.MCT-13-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Comprehensive Cancer Network . Uveal Melanoma (Version 1.2019). Retrieved from https://www.nccn.org/professionals/physician_gls/pdf/uveal.pdf [DOI] [PubMed] [Google Scholar]
- Neal, J. T. , Li, X. , Zhu, J. , Giangarra, V. , Grzeskowiak, C. L. , Ju, J. , … Kuo, C. J. (2018). Organoid modeling of the tumor immune microenvironment. Cell, 175(7), 1972–1988 e1916. 10.1016/j.cell.2018.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Némati, F. , de Montrion, C. , Lang, G. , Kraus‐Berthier, L. , Carita, G. , Sastre‐Garau, X. , … Decaudin, D. (2014). Targeting Bcl‐2/Bcl‐XL induces antitumor activity in uveal melanoma patient‐derived xenografts. PLoS ONE, 9(1), e80836 10.1371/journal.pone.0080836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemati, F. , Sastre‐Garau, X. , Laurent, C. , Couturier, J. , Mariani, P. , Desjardins, L. , … Decaudin, D. (2010). Establishment and characterization of a panel of human uveal melanoma xenografts derived from primary and/or metastatic tumors. Clinical Cancer Research, 16(8), 2352–2362. 10.1158/1078-0432.CCR-09-3066 [DOI] [PubMed] [Google Scholar]
- Niederkorn, J. Y. (1984). Enucleation in consort with immunologic impairment promotes metastasis of intraocular melanomas in mice. Investigative Ophthalmology & Visual Science, 25(9), 1080–1086. [PubMed] [Google Scholar]
- Niederkorn, J. Y. (2012). Ocular immune privilege and ocular melanoma: Parallel universes or immunological plagiarism? Frontiers in Immunology, 3, 148 10.3389/fimmu.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederkorn, J. Y. , Mellon, J. , Pidherney, M. , Mayhew, E. , & Anand, R. (1993). Effect of anti‐ganglioside antibodies on the metastatic spread of intraocular melanomas in a nude mouse model of human uveal melanoma. Current Eye Research, 12(4), 347–358. 10.3109/02713689308999459 [DOI] [PubMed] [Google Scholar]
- Niederkorn, J. Y. , Sanborn, G. E. , & Gamel, J. W. (1987). Suicide enzyme inhibition as a chemotherapeutic strategy for controlling metastases derived from intraocular melanomas. Investigative Ophthalmology & Visual Science, 28(11), 1844–1850. [PubMed] [Google Scholar]
- Ozaki, S. , Vuyyuru, R. , Kageyama, K. , Terai, M. , Ohara, M. , Cheng, H. , … Sato, T. (2016). Establishment and characterization of orthotopic mouse models for human uveal melanoma hepatic colonization. American Journal of Pathology, 186(1), 43–56. 10.1016/j.ajpath.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, D. E. , Henle, A. M. , Amsterdam, A. , Hagen, H. R. , & Lees, J. A. (2018). Uveal melanoma driver mutations in GNAQ/11 yield numerous changes in melanocyte biology. Pigment Cell Melanoma Res, 31(5), 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquet, L. , Dewit, L. , Schoonjans, N. , Millet, M. , Bérubé, J. , Gerges, P. R. A. , … Landreville, S. (2019). Synergic interactions between hepatic stellate cells and uveal melanoma in metastatic growth. Cancers (Basel), 11(8), 1043 10.3390/cancers11081043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeschinger, T. , Renner, A. , Weber, T. , & Scheuer, W. (2013). Bioluminescence imaging correlates with tumor serum marker, organ weights, histology, and human DNA levels during treatment of orthotopic tumor xenografts with antibodies. Molecular Imaging and Biology, 15(1), 28–39. 10.1007/s11307-012-0559-x [DOI] [PubMed] [Google Scholar]
- Rajaii, F. , Asnaghi, L. , Enke, R. , Merbs, S. L. , Handa, J. T. , & Eberhart, C. G. (2014). The demethylating agent 5-Aza reduces the growth, invasiveness, and clonogenicity of uveal and cutaneous melanoma. Invest Ophthalmol Vis Sci, 55(10), 6178–6186. [DOI] [PubMed] [Google Scholar]
- Repp, A. C. , Mayhew, E. S. , Howard, K. , Alizadeh, H. , & Niederkorn, J. Y. (2001). Role of fas ligand in uveal melanoma‐induced liver damage. Graefes Archive for Clinical and Experimental Ophthalmology, 239(10), 752–758. 10.1007/s004170100363 [DOI] [PubMed] [Google Scholar]
- Rietschel, P. , Panageas, K. S. , Hanlon, C. , Patel, A. , Abramson, D. H. , & Chapman, P. B. (2005). Variates of survival in metastatic uveal melanoma. Journal of Clinical Oncology, 23(31), 8076–8080. 10.1200/JCO.2005.02.6534 [DOI] [PubMed] [Google Scholar]
- Robertson, A. G. , Shih, J. , Yau, C. , Gibb, E. A. , Oba, J. , Mungall, K. L. , … Zmuda, E. (2017). Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell, 32(2), 204–220 e15. 10.1016/j.ccell.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusciano, D. , Lorenzoni, P. , & Burger, M. (1994). Murine models of liver metastasis. Invasion and Metastasis, 14(1–6), 349–361. [PubMed] [Google Scholar]
- Sachs, N. , de Ligt, J. , Kopper, O. , Gogola, E. , Bounova, G. , Weeber, F. , … Clevers, H. (2018). A living biobank of breast cancer organoids captures disease heterogeneity. Cell, 172(1–2), 373–386 e10. 10.1016/j.cell.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Samadi, A. K. , Cohen, S. M. , Mukerji, R. , Chaguturu, V. , Zhang, X. , Timmermann, B. N. , … Person, E. A. (2012). Natural withanolide withaferin A induces apoptosis in uveal melanoma cells by suppression of Akt and c‐MET activation. Tumour Biology, 33(4), 1179–1189. 10.1007/s13277-012-0363-x [DOI] [PubMed] [Google Scholar]
- Sanborn, G. E. , Niederkorn, J. Y. , & Gamel, J. W. (1992). Efficacy of dacarbazine (DTIC) in preventing metastases arising from intraocular melanomas in mice. Graefes Archive for Clinical and Experimental Ophthalmology, 230(2), 192–196. 10.1007/BF00164663 [DOI] [PubMed] [Google Scholar]
- Sanborn, G. , Niederkorn, J. , Kan‐Mitchell, J. , & Albert, D. (1992). Prevention of metastasis of intraocular melanoma in mice treated with difluoromethylornithine. Graefes Archive for Clinical and Experimental Ophthalmology, 230(1), 72–77. 10.1007/BF00166766 [DOI] [PubMed] [Google Scholar]
- Schiffner, S. , Braunger, B. M. , de Jel, M. M. , Coupland, S. E. , Tamm, E. R. , & Bosserhoff, A. K. (2014). Tg(Grm1) transgenic mice: A murine model that mimics spontaneous uveal melanoma in humans? Experimental Eye Research, 127, 59–68. 10.1016/j.exer.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Schrage, R. , Schmitz, A.‐L. , Gaffal, E. , Annala, S. , Kehraus, S. , Wenzel, D. , … Kostenis, E. (2015). The experimental power of FR900359 to study Gq‐regulated biological processes. Nature Communications, 6, 10156 10.1038/ncomms10156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain, A. H. , Bagger, M. M. , Yu, R. , Chang, D. , Liu, S. , Vemula, S. , … Kiilgaard, J. F. (2019). The genetic evolution of metastatic uveal melanoma. Nature Genetics, 51(7), 1123–1130. 10.1038/s41588-019-0440-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas, D. , & Hannon, G. J. (2013). Patient‐derived tumor xenografts: Transforming clinical samples into mouse models. Cancer Research, 73(17), 5315–5319. 10.1158/0008-5472.CAN-13-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stei, M. M. , Loeffler, K. U. , Holz, F. G. , & Herwig, M. C. (2016). Animal models of uveal melanoma: Methods, applicability, and limitations. BioMed Research International, 2016, 4521807 10.1155/2016/4521807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stei, M. M. , Loeffler, K. U. , Kurts, C. , Hoeller, T. , Pfarrer, C. , Holz, F. G. , & Herwig‐Carl, M. C. (2016). Impact of macrophages on tumor growth characteristics in a murine ocular tumor model. Experimental Eye Research, 151, 9–18. 10.1016/j.exer.2016.07.008 [DOI] [PubMed] [Google Scholar]
- Suesskind, D. , Gauss, S. , Faust, U. E. , Bauer, P. , Schrader, M. , Bartz‐Schmidt, K. U. , & Henke‐Fahle, S. (2013). Characterisation of novel uveal melanoma cell lines under serum‐free conditions. Graefes Archive for Clinical and Experimental Ophthalmology, 251(8), 2063–2070. 10.1007/s00417-013-2292-9 [DOI] [PubMed] [Google Scholar]
- Surriga, O. , Rajasekhar, V. K. , Ambrosini, G. , Dogan, Y. , Huang, R. , & Schwartz, G. K. (2013). Crizotinib, a c‐Met inhibitor, prevents metastasis in a metastatic uveal melanoma model. Molecular Cancer Therapeutics, 12(12), 2817–2826. 10.1158/1535-7163.MCT-13-0499 [DOI] [PubMed] [Google Scholar]
- Süsskind, D. , Hurst, J. , Rohrbach, J. M. , & Schnichels, S. (2017). Novel mouse model for primary uveal melanoma: A pilot study. Clinical & Experimental Ophthalmology, 45(2), 192–200. 10.1111/ceo.12814 [DOI] [PubMed] [Google Scholar]
- Sutmuller, R. P. M. , Schurmans, L. R. H. M. , van Duivenvoorde, L. M. , Tine, J. A. , van der Voort, E. I. H. , Toes, R. E. M. , … Offringa, R. (2000). Adoptive T cell immunotherapy of human uveal melanoma targeting gp100. The Journal of Immunology, 165(12), 7308–7315. 10.4049/jimmunol.165.12.7308 [DOI] [PubMed] [Google Scholar]
- Syed, N. A. , Windle, J. J. , Darjatmoko, S. R. , Lokken, J. M. , Steeves, R. A. , Chappell, R. , … Albert, D. M. (1998). Transgenic mice with pigmented intraocular tumors: Tissue of origin and treatment. Investigative Ophthalmology & Visual Science, 39(13), 2800–2805. [PubMed] [Google Scholar]
- Tafreshi, N. K. , Tichacek, C. J. , Pandya, D. N. , Doligalski, M. L. , Budzevich, M. M. , Kil, H. J. , … Morse, D. L. (2019). Melanocortin 1 receptor‐targeted α‐particle therapy for metastatic uveal melanoma. Journal of Nuclear Medicine, 60(8), 1124–1133. 10.2967/jnumed.118.217240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleson, W. H. , Doss, J. C. , Latendresse, J. , Warbritton, A. R. , Melchior, W. B. Jr , Chin, L. , … Albert, D. M. (2005). Spontaneous uveal amelanotic melanoma in transgenic Tyr‐RAS+ Ink4a/Arf‐/‐ mice. Archives of Ophthalmology, 123(8), 1088–1094. 10.1001/archopht.123.8.1088 [DOI] [PubMed] [Google Scholar]
- Valyi‐Nagy, T. , Fredericks, B. , Ravindra, A. , Hopkins, J. , Shukla, D. , & Valyi‐Nagy, K. (2018). Herpes simplex virus 1 infection promotes the growth of a subpopulation of tumor cells in three‐dimensional uveal melanoma cultures. Journal of Virology, 92(19), e00700–e718. 10.1128/JVI.00700-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ent, W. , Burrello, C. , Teunisse, A. F. A. S. , Ksander, B. R. , van der Velden, P. A. , Jager, M. J. , … Snaar‐Jagalska, B. E. (2014). Modeling of human uveal melanoma in zebrafish xenograft embryos. Investigative Ophthalmology & Visual Science, 55(10), 6612–6622. 10.1167/iovs.14-15202 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk, C. D. , Bezrookove, V. , Green, G. , Bauer, J. , Gaugler, L. , O'Brien, J. M. , … Bastian, B. C. (2009). Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature, 457(7229), 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk, C. D. , Griewank, K. G. , Crosby, M. B. , Garrido, M. C. , Vemula, S. , Wiesner, T. , … Bastian, B. C. (2010). Mutations in GNA11 in uveal melanoma. New England Journal of Medicine, 363(23), 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBrocklin, M. W. , Robinson, J. P. , Lastwika, K. J. , Khoury, J. D. , & Holmen, S. L. (2010). Targeted delivery of NRASQ61R and Cre‐recombinase to post‐natal melanocytes induces melanoma in Ink4a/Arflox/lox mice. Pigment Cell & Melanoma Research, 23(4), 531–541. 10.1111/j.1755-148X.2010.00717.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaqué, J. P. , Dorsam, R. T. , Feng, X. , Iglesias‐Bartolome, R. , Forsthoefel, D. J. , Chen, Q. , … Gutkind, J. S. (2013). A genome‐wide RNAi screen reveals a Trio‐regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein‐coupled receptors. Molecular Cell, 49(1), 94–108. 10.1016/j.molcel.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbik, D. J. , Murray, T. G. , Tran, J. M. , & Ksander, B. R. (1997). Melanomas that develop within the eye inhibit lymphocyte proliferation. International Journal of Cancer, 73(4), 470–478. [DOI] [PubMed] [Google Scholar]
- Vivet‐Noguer, R. , Tarin, M. , Roman‐Roman, S. , & Alsafadi, S. (2019). Emerging therapeutic opportunities based on current knowledge of uveal melanoma biology. Cancers (Basel), 11(7), 1019 10.3390/cancers11071019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis, G. , Hedayat, S. , Vatsiou, A. , Jamin, Y. , Fernandez‐Mateos, J. , Khan, K. , … Valeri, N. (2018). Patient‐derived organoids model treatment response of metastatic gastrointestinal cancers. Science, 359(6378), 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voropaev, H. , Gimmelshein Vatkin, M. , Shneor, D. , Luski, S. , Honigman, A. , & Frenkel, S. (2019). Infectious knockdown of CREB and HIF‐1 for the treatment of metastatic uveal melanoma. Cancers (Basel), 11(8), 1056 10.3390/cancers11081056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Liu, M. , Jin, Y. , Jiang, S. , & Pan, J. (2017). In vitro and in vivo anti‐uveal melanoma activity of JSL‐1, a novel HDAC inhibitor. Cancer Letters, 400, 47–60. [DOI] [PubMed] [Google Scholar]
- Wege, A. K. (2018). Humanized mouse models for the preclinical assessment of cancer immunotherapy. BioDrugs: Clinical Immunotherapeutics, Biopharmaceuticals and Gene Therapy, 32(3), 245–266. 10.1007/s40259-018-0275-4 [DOI] [PubMed] [Google Scholar]
- Xue, S. , Yang, H. , Qiao, J. , Pu, F. , Jiang, J. , Hubbard, K. , … Yang, J. J. (2015). Protein MRI contrast agent with unprecedented metal selectivity and sensitivity for liver cancer imaging. Proceedings of the National Academy of Sciences of the USA, 112(21), 6607–6612. 10.1073/pnas.1423021112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Brackett, C. M. , Morales‐Tirado, V. M. , Li, Z. , Zhang, Q. , Wilson, M. W. , … Grossniklaus, H. E. (2016). The Toll‐like receptor 5 agonist entolimod suppresses hepatic metastases in a murine model of ocular melanoma via an NK cell‐dependent mechanism. Oncotarget, 7(3), 2936–2950. 10.18632/oncotarget.6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Cao, J. , & Grossniklaus, H. E. (2015). Uveal melanoma metastasis models. Ocular Oncology and Pathology, 1(3), 151–160. 10.1159/000370153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , & Grossniklaus, H. E. (2010). Constitutive overexpression of pigment epithelium‐derived factor inhibition of ocular melanoma growth and metastasis. Investigative Ophthalmology & Visual Science, 51(1), 28–34. 10.1167/iovs.09-4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Jager, M. J. , & Grossniklaus, H. E. (2010). Bevacizumab suppression of establishment of micrometastases in experimental ocular melanoma. Investigative Ophthalmology & Visual Science, 51(6), 2835–2842. 10.1167/iovs.09-4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Sun, L. , Liu, M. , & Mao, Y. (2018). Patient‐derived organoids: A promising model for personalized cancer treatment. Gastroenterology Report (Oxf), 6(4), 243–245. 10.1093/gastro/goy040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Xu, Z. , Iuvone, P. M. , & Grossniklaus, H. E. (2006). Angiostatin decreases cell migration and vascular endothelium growth factor (VEGF) to pigment epithelium derived factor (PEDF) RNA ratio in vitro and in a murine ocular melanoma model. Molecular Vision, 12, 511–517. [PubMed] [Google Scholar]
- Yang, J. , Manson, D. K. , Marr, B. P. , & Carvajal, R. D. (2018). Treatment of uveal melanoma: Where are we now? Therapeutic Advances in Medical Oncology, 10, 1758834018757175 10.1177/1758834018757175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , Li, H. , Mayhew, E. , Mellon, J. , Chen, P. W. , & Niederkorn, J. Y. (2011). NKT cell exacerbation of liver metastases arising from melanomas transplanted into either the eyes or spleens of mice. Investigative Ophthalmology & Visual Science, 52(6), 3094–3102. 10.1167/iovs.10-7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, J. H. , Shi, D. S. , Grossmann, A. H. , Sorensen, L. K. , Tong, Z. Z. , Mleynek, T. M. , … Li, D. Y. (2016). ARF6 is an actionable node that orchestrates oncogenic GNAQ signaling in uveal melanoma. Cancer Cell, 29(6), 889–904. 10.1016/j.ccell.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, M. , Selvan, S. , McCue, P. A. , DeAngelis, T. , Baserga, R. , Fujii, A. , … Sato, T. (2014). Expression of insulin‐like growth factor‐1 receptor in metastatic uveal melanoma and implications for potential autocrine and paracrine tumor cell growth. Pigment Cell & Melanoma Research, 27(2), 297–308. 10.1111/pcmr.12206 [DOI] [PubMed] [Google Scholar]
- Yu, F.‐X. , Luo, J. , Mo, J.‐S. , Liu, G. , Kim, Y. C. , Meng, Z. , … Guan, K.‐L. (2014). Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell, 25(6), 822–830. 10.1016/j.ccr.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Ambrosini, G. , Roszik, J. , Eterovic, A. K. , Stempke‐Hale, K. , Seftor, E. A. , … Woodman, S. E. (2015). Genetic analysis of the 'uveal melanoma' C918 cell line reveals atypical BRAF and common KRAS mutations and single tandem repeat profile identical to the cutaneous melanoma C8161 cell line. Pigment Cell & Melanoma Research, 28(3), 357–359. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Yang, H. , Kang, S. J. , Wang, Y. , Wang, G. D. , Coulthard, T. , & Grossniklaus, H. E. (2011). In vivo high‐frequency, contrast‐enhanced ultrasonography of uveal melanoma in mice: Imaging features and histopathologic correlations. Investigative Ophthalmology & Visual Science, 52(5), 2662–2668. 10.1167/iovs.10-6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Jin, B. , Jin, Y. , Liu, Y. , & Pan, J. (2017). The antihelminthic drug niclosamide effectively inhibits the malignant phenotypes of uveal melanoma in vitro and in vivo. Theranostics, 7(6), 1447–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, A. , Zhan, W. , Liang, Z. , Yoon, Y. , Yang, H. , Grossniklaus, H. E. , … Shim, H. (2010). Dipyrimidine amines: A novel class of chemokine receptor type 4 antagonists with high specificity. Journal of Medicinal Chemistry, 53(24), 8556–8568. 10.1021/jm100786g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel, L. , Pitt, J. M. , Daillere, R. , Smyth, M. J. , & Kroemer, G. (2016). Mouse models in oncoimmunology. Nature Reviews Cancer, 16(12), 759–773. 10.1038/nrc.2016.91 [DOI] [PubMed] [Google Scholar]
- Zuberi, A. , & Lutz, C. (2016). Mouse models for drug discovery. Can new tools and technology improve translational power? ILAR Journal, 57(2), 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]


