Abstract
Endometrial carcinoma (EC) molecular classification based on four molecular subclasses identified in The Cancer Genome Atlas (TCGA) has gained relevance in recent years due to its prognostic utility and potential to predict benefit from adjuvant treatment. While most ECs can be classified based on a single classifier (POLE exonuclease domain mutations – POLEmut, MMR deficiency – MMRd, p53 abnormal – p53abn), a small but clinically relevant group of tumours harbour more than one molecular classifying feature and are referred to as ‘multiple‐classifier’ ECs. We aimed to describe the clinicopathological and molecular features of multiple‐classifier ECs with abnormal p53 (p53abn). Within a cohort of 3518 molecularly profiled ECs, 107 (3%) tumours displayed p53abn in addition to another classifier(s), including 64 with MMRd (MMRd–p53abn), 31 with POLEmut (POLEmut–p53abn), and 12 with all three aberrations (MMRd–POLEmut–p53abn). MMRd–p53abn ECs and POLEmut–p53abn ECs were mostly grade 3 endometrioid ECs, early stage, and frequently showed morphological features characteristic of MMRd or POLEmut ECs. 18/28 (60%) MMRd–p53abn ECs and 7/15 (46.7%) POLEmut–p53abn ECs showed subclonal p53 overexpression, suggesting that TP53 mutation was a secondary event acquired during tumour progression. Hierarchical clustering of TCGA ECs by single nucleotide variant (SNV) type and somatic copy number alterations (SCNAs) revealed that MMRd–p53abn tumours mostly clustered with single‐classifier MMRd tumours (20/23) rather than single‐classifier p53abn tumours (3/23), while POLEmut–p53abn tumours mostly clustered with single‐classifier POLEmut tumours (12/13) and seldom with single‐classifier p53abn tumours (1/13) (both p ≤ 0.001, chi‐squared test). Finally, the clinical outcome of patients with MMRd–p53abn and POLEmut–p53abn ECs [stage I 5‐year recurrence‐free survival (RFS) of 92.2% and 94.1%, respectively] was significantly different from single‐classifier p53abn EC (stage I RFS 70.8%, p = 0.024 and p = 0.050, respectively). Our results support the classification of MMRd–p53abn EC as MMRd and POLEmut–p53abn EC as POLEmut. © 2019 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: POLE, molecular classification, endometrial cancer
Introduction
Of the many advances in the field of endometrial cancer (EC) during the last decade, perhaps the one with most impact is the molecular classification proposed by The Cancer Genome Atlas (TCGA) 1, which has gained prominence in recent years 2. This classifies ECs into four molecular subtypes – POLE/ultramutated (POLE), microsatellite instability‐high/hypermutated (MSI), somatic copy‐number alteration high/serous‐like (SCNA‐high), and somatic copy‐number alteration low (SCNA‐low) – with significantly different prognoses 3, and thus of potential clinical relevance. For example, the favourable outcome of POLE (exonuclease domain) mutant EC, independent of adjuvant treatment 4, has led to proposals to de‐escalate post‐operative therapy in this subgroup 4. In contrast, the consistently poor clinical outcome of patients with SCNA‐high (serous‐like) ECs suggests that intensification of adjuvant treatment may be worthwhile. The reported response of MSI tumours to immune checkpoint inhibitors 5, 6, as well as the possibility of a better response to adjuvant radiotherapy 7, also opens new treatment opportunities for this group of patients. A randomised controlled clinical trial (PORTEC4a 8) is currently testing the added value of integrating this molecular approach into risk assessment, to individualise adjuvant treatment and reduce over‐ and under‐treatment in patients 9.
One important contributor to the impact and rapidly increasing adoption of the TCGA classifier is the relative ease in which subgroups analogous to those originally described can be identified by techniques in routine clinical practice 9, 10, 11, 12, 13, 14. ECs analogous to the SCNA‐high subclass can be identified by p53 immunohistochemistry (p53abn EC) 15, the MSI subclass by immunohistochemistry for MMR proteins (MMRd EC), and the POLE subclass by targeted sequencing of the POLE exonuclease domain (POLEmut EC) (the latter being the most difficult to implement in routine practice). Tumours lacking these three prior features are classified as p53 wild‐type (p53wt EC) 11 or no specific molecular profile subtype (NSMP EC) 10, analogous to the SCNA‐low subclass.
Using this surrogate marker approach, most ECs can be classified into a single molecular class (henceforward referred to as ‘single‐classifier’ ECs). However, 3–6% of tumours have more than one molecular classifying feature (hereafter referred to as ‘multiple‐classifier’ ECs) 9, 10, 11, 12, and include those with combined POLE exonuclease domain mutation (EDM) and abnormal p53 (POLEmut–p53abn), combined DNA mismatch repair deficiency (MMRd) and abnormal p53 (MMRd–p53abn), combined MMRd and POLE EDM (MMRd–POLEmut), and all three defects (MMRd–POLEmut–p53abn) 9, 10, 11, 12. There is currently no consensus on how these tumours should be classified or treated; some studies have excluded them from further analysis 10, while others have allocated them to one of the four subtypes 11 without detailing their clinicopathological or molecular features. Consequently, the biology and prognostic significance of multiple‐classifier ECs are unclear, and how they should be managed is unanswered. This creates an important problem for tumours that carry opposite features, as one may favour treatment de‐escalation (POLEmut) and the other intensified treatment (p53abn).
In this study, we aimed to perform a comprehensive analysis of the clinical, morphological, and molecular characteristics of EC with abnormal p53 immunostaining and/or TP53 mutation in combination with POLEmut and/or MMRd in order to inform clinical management of these multiple‐classifier ECs.
Materials and methods
Patient and tissue selection
Patient identity was protected by study‐specific patient numbers. Informed consent and ethical approval were obtained according to the local protocol in each participating centre.
We obtained patient data and tumour tissue from ECs with more than one classifying feature (a pathogenic POLE exonuclease domain variant, MMR protein loss of expression or p53 abnormal expression) from previously published datasets 1, 10, 12, 13, 16, 17. A total of 2988 ECs had been molecularly classified in previous studies 10, 12, 13, 16, 17, in which MMR and p53 status were determined by immunohistochemistry (IHC) and POLE variants by Sanger sequencing or NGS of the complete exonuclease domain (exons 9–14) or targeted sequencing of exons 9, 13, and 14. For this current study, all POLE variants were reviewed and tumours were excluded when the POLE variant was not considered pathogenic following the recommended approach presented by León‐Castillo et al 18. Additionally, following the criteria mentioned previously, four tumours reported as having all three molecular features (POLEmut–MMRd–p53abn) but with a non‐pathogenic POLE EDM based on review were classified as MMRd–p53abn ECs. ECs with MMR loss and a pathogenic POLE variant (MMRd–POLEmut ECs) were excluded from this study, as they are described by León‐Castillo et al 18. Clinical follow‐up and, when available, slides were centrally collected for further analyses. Follow‐up data were provided by each centre in an anonymised dataset and datasets were combined into a final password‐protected database.
Our study cohort was further extended by 530 TCGA ECs in which a combination of a pathogenic POLE EDM, a TP53 variant (excluding TP53 variants classified as benign by SIFT or neutral by VEP), and/or MSI‐H (based on Bethesda protocol classification 19 was obtained from the Genome Data Analysis Center (GDAC) (available at http://www.broadinstitute.org/cancer/cga). TP53 mutations were assessed using the public databases COSMIC 20, ClinVar 21, and IARC TP53 mutations database 22, as well as the in silico tools SIFT 23 and PolyPhen 24; only mutations classified as (likely) pathogenic or variants of unknown significance (VUS) were included in the study. This resulted in a final study cohort of 3518 molecularly profiled ECs.
Scoring MMR and p53 IHC
As multiple‐classifier ECs are identified by IHC, we re‐evaluated or restained tumours with available unstained slides to exclude potential misinterpretation in the original study. Unstained sections were stained for either two (PMS2 and MSH6) or four mismatch repair proteins (MLH1, PMS2, MSH2, and MSH6) dependent on slide availability [MLH1 (ES05, 1:100; Agilent DAKO, Amstelveen, North Holland, The Netherlands), PMS2 (EP51, 1:50; Agilent DAKO), MSH2 (FE11, 1:200; Agilent DAKO), and MSH6 (EPR 3945, 1:800; GeneTex, Irvine, CA, USA)] and p53 (DO‐7, 1:200; Agilent DAKO).
MMRd was defined as loss of MMR nuclear staining, for at least one MMR protein, with positive internal control. Subclonal loss of MMR expression 25 was defined as abrupt and complete regional loss of expression of an MMR protein with intervening stromal positivity serving as an internal control in the regions of absent tumour cell staining. Cases with heterogeneous or non‐abrupt patchy staining thought to be a result of sub‐optimal pre‐analytic handling or those with absent internal control staining were not considered MMRd. An abnormal/mutant p53 IHC stain was defined as strong nuclear expression in over 75% of the tumour cells (overexpression), complete loss of nuclear stain in the presence of internal control staining (complete absence), or cytoplasmic staining (cytoplasmic), following scoring described by Singh et al 15. Subclonal abnormal p53 IHC staining was defined as abrupt and complete regional abnormal p53 expression, in which the subclonal region was at least 10% of the total tumour volume. If the p53 IHC staining pattern could not be classified into one of the previously mentioned patterns (inconclusive p53 stain), TP53 mutational status was used for molecular profiling through Sanger sequencing of exons 5–8.
Histopathological review
Tumours with at least one haematoxylin–eosin (H&E) slide available were selected for histological review, blinded for molecular classification, by one pathologist (AL, TB, BG or RS). This review consisted of scoring for each case predefined histological features: hobnailing, slit‐like spaces, papillary growth, squamous metaplasia, tumour intraepithelial lymphocytes (TILs), peritumoural lymphocytes, solid growth greater than 50%, and the presence of tumour giant cells. For the purpose of this study, we did not reassess tumour histotype. Type of myometrial invasion and presence of lymphovascular space invasion (LVSI) were also annotated.
Evaluation of somatic nucleotide and copy number variation in TCGA ECs
Somatic mutations were classified into the 96 categories described by Alexandrov et al 26. Distance metrics for the difference between pairs of samples were generated using the mutational changes (1 – cosine similarity/2) or the proportion of genome that had a copy number change (both of these range between 0 and 1). These distances were combined into a single metric using Euclidean distance. Individual and combined metrics were used to cluster the samples and plot a heatmap using the function heatmap.2 from the package ‘gplots’ to visualise similarity between samples.
Statistical analysis
We assessed 5‐year recurrence‐free survival (RFS) and 5‐year overall survival (OS) comparing Kaplan–Meier curves with the log‐rank test. Follow‐up time was calculated with the reverse Kaplan–Meier method. We used Mann–Whitney tests to compare non‐parametric continuous variables; categorical variables were assessed with Fisher's exact test or the chi‐squared test. Two‐sided P value less than 0.05 was considered significant.
Results
Identification and clinicopathological characteristics of multiple‐classifier ECs
Our initial cohort comprised 3518 tumours, 3353 being classified to a single molecular subtype. Of the remaining 167 tumours, 30 were classified as MMRd–POLEmut ECs and will be reported separately (León‐Castillo et al 18), leaving 138 tumours (3.9%) that were assigned a provisional status of multiple‐classifier with abnormal p53 (mutant p53 expression by IHC or a mutation in TP53). Stringent quality control including central pathological review resulted in the exclusion of 35 of these ECs for the following reasons: (1) reassignment of p53 immunostaining from mutant to wild‐type pattern (n = 27, 21 being initially evaluated using tissue microarrays); (2) non‐pathogenic POLE variant (n = 3); and (3) lack of evidence of MMRd on IHC review (n = 1) (Figure 1). The remaining 107 tumours (3% of total) met the inclusion criteria and were used for subsequent analyses.
Figure 1.
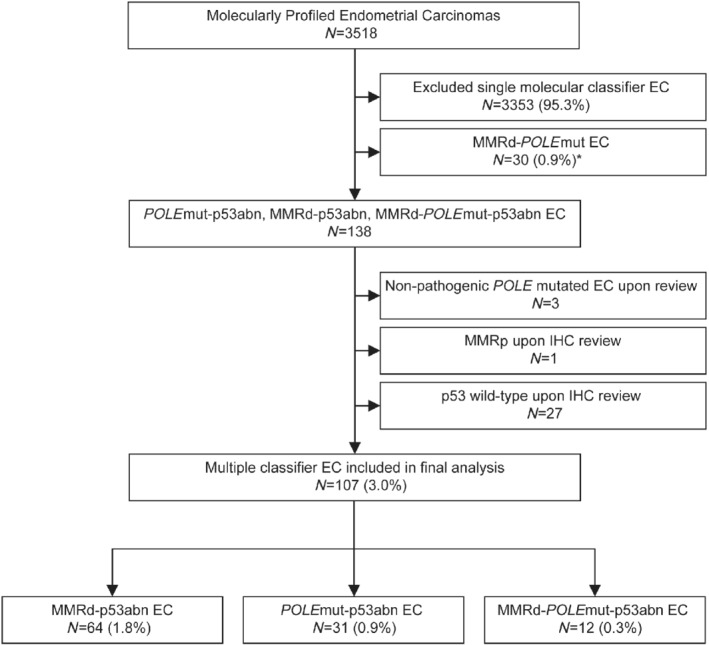
Flow chart of sample analysis. POLEmut–MMRd ECs are reported separately in León‐Castillo et al 18.
The clinicopathological characteristics of the patients with these 104 ECs are shown in Table 1 27, 28. The mean age was 62 (range 35–87) years. The FIGO 2009 stage was as follows: 80 (76.9%) stage I and 24 (23.1%) stage II–IV. The most common combination of classifiers was MMRd–p53abn (64; 1.8% of 3518), followed by POLEmut–p53abn (31; 0.9% of 3518) and MMRd–POLEmut–p53abn (12; 0.3% of 3518).
Table 1.
Clinicopathological characteristics of multiple‐classifier EC with abnormal p53
| Total n = 107 (%) | MMRd–p53abn EC n = 64 (%) | POLEmut–p53abn EC n = 31 (%) | MMRd–POLEmut–p53abn EC | |
|---|---|---|---|---|
| n = 12 (%) | ||||
| Age, years | ||||
| Mean [range] | 61.6 [35–87] | 61.7 [35–87] | 62.1 [50–83] | 59.9 [47–74] |
| < 60 | 51 (47.7) | 28 (43.8) | 16 (51.6) | 7 (58.3) |
| 60–70 | 33 (30.8) | 22 (34.4) | 8 (25.8) | 3 (25.0) |
| > 70 | 22 (20.6) | 14 (21.9) | 6 (19.4) | 2 (16.7) |
| Missing | 1 (0.9) | 0 (0) | 1 (3.2) | 0 (0) |
| Stage | ||||
| IA | 41 (38.3) | 25 (39.1) | 9 (29) | 7 (58.3) |
| IB | 41 (38.3) | 22 (34.4) | 15 (48.4) | 4 (33.3) |
| II | 3 (2.8) | 2 (3.1) | 1 (3.2) | 0 (0) |
| III | 16 (15) | 11 (17.2) | 4 (12.9) | 1 (8.3) |
| IV | 6 (5.6) | 4 (6.3) | 2 (6.5) | 0 (0) |
| Histology | ||||
| Endometrioid | 77 (72) | 46 (71.9) | 22 (71) | 9 (75.0) |
| Serous | 9 (8.4) | 6 (9.4) | 2 (6.5) | 1 (8.3) |
| Mixed | 16 (15) | 8 (12.5) | 6 (19.4) | 2 (16.7) |
| Clear cell | 3 (2.8) | 2 (3.1) | 1 (3.2) | 0 (0) |
| Undifferentiated | 2 (1.9) | 2 (3.1) | 0 (0) | 0 (0) |
| Grade | ||||
| 1–2 | 25 (23.4) | 16 (25) | 7 (22.6) | 2 (16.7) |
| 3 | 82 (76.6) | 48 (75) | 24 (77.4) | 10 (83.3) |
| Myometrium invasion | ||||
| Intramucosal | 4 (3.7) | 2 (3.1) | 0 (0) | 2 (16.7) |
| < 50% | 45 (42.1) | 28 (43.8) | 11 (35.5) | 6 (50.0) |
| > 50% | 53 (49.5) | 31 (48.4) | 19 (61.3) | 4 (33.3) |
| Missing | 5 (4.7) | 3 (4.7) | 1 (3.2) | 0 (0) |
| LVSI | ||||
| Absent | 44 (41.1) | 22 (34.4) | 15 (48.4) | 7 (58.3) |
| Present | 32 (29.9) | 23 (35.9) | 5 (16.1) | 4 (33.3) |
| Missing | 31 (29.0) | 19 (29.7) | 11 (35.5) | 1 (8.3) |
| Treatment | ||||
| Radiotherapy | 18 (16.8) | 12 (18.8) | 5 (16.1) | 1 (8.3) |
| Chemotherapy | 9 (8.4) | 4 (6.3) | 4 (12.9) | 1 (8.3) |
| Radiochemotherapy | 10 (9.3) | 7 (10.9) | 3 (9.7) | 0 (0) |
| None | 15 (14.0) | 9 (14.1) | 4 (12.9) | 2 (16.7) |
| Missing | 55 (51.4) | 32 (50) | 15 (48.4) | 8 (66.7) |
| Risk classification (ESMO clinical practice guidelines, 2013 27) | ||||
| Low risk | 9 (8.4) | 6 (9.4) | 3 (9.7) | 0 (0) |
| Intermediate risk | 31 (29.0) | 16 (25) | 8 (25.8) | 7 (58.3) |
| High risk | 51 (47.7) | 30 (46.9) | 17 (54.8) | 4 (33.3) |
| Advanced stage I | 13 (12.1) | 10 (15.6) | 2 (6.5) | 1 (8.3) |
| Metastatic | 3 (2.8) | 2 (3.1) | 1 (3.2) | 0 (0) |
| Risk classification (ESMO–ESTRO–ESGO clinical practice guidelines, 2016 28) | ||||
| Low risk | 4 (3.7) | 2 (3.1) | 2 (6.5) | 1 (8.3) |
| Intermediate | 11 (10.3) | 7 (10.9) | 3 (9.7) | 0 (0) |
| High–intermediate | 22 (20.6) | 11 (17.2) | 5 (16.1) | 6 (50) |
| High | 59 (55.1) | 37 (57.8) | 17 (54.8) | 5 (41.7) |
| Advanced or metastatic | 6 (5.6) | 4 (6.3) | 2 (6.5) | 0 (0) |
| Not assessable | 5 (4.7) | 3 (4.7) | 2 (6.5) | 0 (0) |
DNA mismatch repair‐deficient, p53 abnormal (MMRd–p53abn) ECs
From the total study population, 64 tumours had both MMRd/MSI and abnormal p53 (MMRd–p53abn) (Table 1). Of these, 40 (62.5%) were classified based on MMR protein expression, with an unusually high number of ECs (n = 25, 62.5%) presenting with loss of MSH6 +/− MSH2, or single PMS2 loss 25. The mean patient age in this subgroup was 62 years. Most tumours were endometrioid (46/64, 71.9%), with 65.2% being grade 3. 76.7% (49/64) of these patients had early‐stage ECs (stage I–II). Central histological review was possible in 54 ECs (median of two slides per case, range 1–17) (supplementary material, Table S1) and typically revealed a dense lymphoid infiltrate (peritumoural lymphocytes in 59.3% and tumour‐infiltrating lymphocytes in 42.6% of tumours). LVSI was observed in 33.3% of tumours. Solid growth (≥ 50% of the tumour) was present in 24 (44.4%) ECs, and squamous metaplasia was identified in 13 (24.1%). Hobnailing, slit‐like spaces or papillary growth was present in eight (14.8%), 12 (22.2%), and 14 (25.9%) tumours, respectively.
p53 IHC was available for central review in 28 of the 64 MMRd–p53abn tumours. Nine (30%) showed p53 overexpression, while one tumour had an inconclusive staining pattern in combination with a confirmed pathogenic TP53 mutation. Interestingly, the remaining 18 tumours (60%) showed abrupt strong p53 nuclear overexpression (> 75% of all tumour nuclei) in a well‐defined area (at least 10% of tumour volume), a pattern defined as subclonal p53abn staining (Figure 2).
Figure 2.

MMRd–p53abn EC IHC and molecular features. Subclonal p53 staining and loss of PMS2 are shown in A (×5 original magnification). Percentage of cases with subclonal and diffuse abnormal p53 staining patterns is depicted in B. (C) Heatmap showing hierarchical clustering of MMRd–p53abn, single‐classifier MMRd, and single‐classifier p53abn ECs in TCGA, based on mutational changes and copy number changes. Individual similarity metrics for copy number and mutational changes were combined using Euclidean distance. Patients were classified in groups based on MMRd–p53abn (brown), single‐classifier MMRd (green), and single‐classifier p53abn (red). Samples were ordered based on hierarchical clustering.
To further characterise the group of MMRd–p53abn ECs, we used sequencing data from the TCGA study, comparing analogous tumours with combined microsatellite instability and TP53 mutation (MMRd–p53abn EC, n = 23) with those with one of these defects in isolation (henceforth referred to as single‐classifier MMRd EC if microsatellite‐unstable and single‐classifier p53abn EC if TP53‐mutant). Fifteen (65%) MMRd–p53abn ECs had a (likely) pathogenic TP53 mutation and eight (35%) had a TP53 VUS. Preliminary analysis revealed a significantly higher frequency of multiple TP53 mutations in MMRd–p53abn tumours when compared with single‐classifier p53abn (i.e. lacking POLE mutation or MSI‐H) ECs (36.4% versus 2.7%, p < 0.001, Fisher's exact test). To further define the molecular characteristics of MMRd–p53abn ECs, we performed hierarchical clustering of TCGA ECs according to the proportion of SNVs of each trinucleotide context and SCNA burden (see the Materials and methods section), revealing that MMRd–p53abn ECs clustered mostly with single‐classifier MMRd tumours (which are typically SCNA‐low) rather than with single‐classifier p53abn ECs (20/23 versus 3/23, respectively; p ≤ 0.001) (Figure 2). Similar results were obtained when analysing individually SNVs and SCNA (supplementary material, Figure S1).
POLE‐mutant, p53‐abnormal (POLEmut–p53abn) ECs
A total of 31 ECs (0.9%) had both a POLE mutation and abnormal p53 (POLEmut–p53abn) (Table 1). In all of these POLEmut–p53abn ECs, the POLE variant was a known pathogenic mutation including p.Pro286Arg (16), p.Val411Leu (10), Ala456Pro (3), p.Ser459Phe (1), and p.Pro436Arg (1), as described by León‐Castillo et al 18. Patients with POLEmut–p53abn ECs had a mean age of 62 years. Histologically, these tumours were most frequently grade 3 endometrioid EC (n = 15, 48.4%). Twenty‐four patients had (77.4%) stage I tumours, 1 (3.2%) had a stage II, 4 (12.9%) had stage III, and 2 had (6,5%) stage IV tumours. Thirty cases were available for morphological review (median two slides available per case, range 1–15) (supplementary material, Table S1). This revealed a prominent lymphoid infiltrate in the majority of ECs (90.9% with peritumoural lymphocytes and 59.1% with TILs). Tumour giant cells, previously reported in 33–40% of POLE‐mutant single‐classifier ECs 29, 30, were observed in 12 (40%) tumours and substantial LVSI was present in 10%. Solid growth ≥ 50% was identified in 16 cancers (53.3%) and squamous metaplasia in 4 (13.3%). Hobnailing was observed in four (13.3%), slit‐like spaces in six (20%), and papillary growth in four (13.3%) tumours.
Central review of p53 immunostaining was possible in 15 tumours. Six (37.5%) had diffuse overexpression of p53; one showed complete loss of p53 expression; and one had inconclusive p53 staining with a confirmed TP53 mutation. Intriguingly, similar to our results in MMRd–p53 multiple‐classifier ECs, subclonal abnormal p53 staining (subclonal overexpression) was frequent (n = 7, 46.7%) (Figure 3).
Figure 3.

POLEmut–p53abn EC IHC and molecular features. Subclonal p53 staining and H&E stain of a POLEmut–p53abn EC are shown in A (×5 original magnification). Percentage of cases with subclonal and diffuse abnormal p53 staining patterns is depicted in B. (C) Heatmap showing hierarchical clustering of POLEmut–p53abn, single‐classifier POLEmut, and single‐classifier p53abn ECs in TCGA, based on mutational changes and copy number changes. Individual similarity metrics for copy number and mutational changes were combined using Euclidean distance. Patients were classified in groups based on POLEmut–p53abn (pink), single‐classifier POLEmut (blue), and single‐classifier p53abn (red). Samples were ordered based on hierarchical clustering.
We used the TCGA EC cohort to compare the 13 ECs with combined POLE mutation and TP53 mutation (POLEmut–p53abn) with tumours with either defect alone (single‐classifier POLEmut and single‐classifier p53abn EC). Eight (62%) POLEmut–p53abn ECs had at least one TP53 (likely) pathogenic mutation, the remaining five (38%) having at least one VUS. POLEmut–p53abn ECs more frequently had multiple TP53 mutations when compared with single‐classifier p53abn tumours (75% versus 2.7%, p < 0.001, Fisher's exact test). Additionally, known TP53 mutational hot‐spot codons Arg 175, Gly 245, Arg 248, Arg 249, Arg 273, and Arg 282 31 were seldom mutated in POLEmut–p53abn ECs, in contrast to single‐classifier TP53mut ECs where these alterations were common, although this difference was not statistically significant [1/12 (8%) versus 57/184 (31.5%); p = 0.11].
Further analysis of TP53 mutations by trinucleotide context demonstrated that TCT>TAT alterations (characteristic of POLEmut carcinomas 26, 32) were uncommon in POLEmut–p53abn ECs (4.2%) and absent in single‐classifier p53abn ECs (0/116) (p = 0.31). However, TCG>TTG substitutions (also substantially enriched in POLEmut carcinomas 32) accounted for 6/24 (25%) of the TP53 variants in POLEmut–p53abn in contrast to single‐classifier p53abn ECs in which these alterations did not occur (p ≤ 0.001).
Hierarchical clustering of ECs by SNV and SCNA proportions revealed that 12 of 13 POLEmut–p53abn ECs clustered with single‐classifier POLE‐mutant ECs, while a single case clustered with the single‐classifier p53abn tumours (p ≤ 0.001, chi‐squared test) (Figure 3). Separate analysis of SNV and SCNAs rendered similar results (supplementary material, Figure S2).
POLE‐mutant, DNA mismatch repair‐deficient p53‐abnormal (MMRd–POLEmut–p53abn) ECs
From our pooled cohort, 12 ECs (0.3%) demonstrated concomitant pathogenic POLE EDM, mismatch repair deficiency, and abnormal p53 (MMRd–POLEmut–p53abn ECs) (Table 1). All cancers had a known pathogenic POLE mutation, as described by León‐Castillo et al 18, including p.Pro286Arg (1), p.Val411Leu (4), p.Ser297Phe (2), p.Ser459Phe (1), p.Phe367Ser (2), p.Leu424Ile (1), and p.Met295Arg (1). These patients had a mean age of 60 years and presented with early‐stage tumours (58.3% stage IA and 33.3% stage IB). Most MMRd–POLEmut–p53abn ECs were endometrioid (n = 9, 75%), whereas two were classified as mixed ECs (serous and endometrioid) and one as serous EC. Eleven tumours were available for morphological review (supplementary material, Table S1). An abundant lymphoid infiltrate was present in these tumours [8 (72.7%) had prominent TILs and 10 (90.9%) peritumoural lymphocytes]. Hobnailing was observed in one (9.1%) tumour, slit‐like spaces were present in two (18.2%), and three (27.3%) had papillary growth. In 4/11 of these MMRd–POLEmut–p53abn ECs, we were able to re‐evaluate p53 and MMR IHC. All four displayed subclonal p53abn expression and two of them additionally showed subclonal loss of MLH1 with concomitant loss of PMS2 expression.
We further analysed the seven MMRd–POLEmut–p53abn ECs available in the TCGA. Five tumours had a TP53 (likely) pathogenic mutation and two had a VUS. Hierarchical clustering based on SNV and SCNA proportions (supplementary material, Figure S3) showed five tumours clustering with single‐classifier POLEmut ECs, two with single‐classifier MMRd ECs, and none with single‐classifier p53abn ECs. Of note, the two ECs that clustered with single‐classifier MMRd cancers had a V411L and L424I POLE variant, respectively, with low C>A substitutions and high indels 18 and pathogenic TP53 mutations.
The limited number of MMRd–POLEmut–p53abn ECs with available clinical data (n = 9) did not allow for survival analysis.
Clinical outcome of multiple‐classifier ECs versus single‐classifier ECs
Finally, we investigated the outcome of MMRd–p53abn ECs and POLEmut–p53abn ECs in the pooled study population (supplementary material, Table S2). Fourty‐four MMRd–p53abn ECs were available for survival analysis. Patients had a 5‐year RFS and OS of 83.4% and 82%, respectively (supplementary material, Figure S4). RFS and OS could be analysed for 23 patients with POLEmut–p53abn ECs. The survival analysis revealed that these ECs were associated with a favourable outcome with a 90.9% 5‐year RFS and 5‐year OS of 95.2% (supplementary material, Figure S4).
We further analysed the clinical outcome of multiple‐classifier ECs by comparing their 5‐year RFS with single‐classifier p53abn ECs. For this purpose, we used a cohort of 187 single‐classifier p53abn ECs described previously 12, 13. These patients had a lower proportion of stage I disease (93, 49.7%) compared with MMRd–p53abn ECs (31, 70.5%) and POLEmut–p53abn ECs (19, 82.6%). Due to the differences in stage distribution, only stage I ECs were analysed. Thus, patients with stage I single‐classifier p53abn ECs had a 5‐year RFS of 70.8%, while those with stage I MMRd–p53abn ECs (n = 31) had an RFS of 92.2% (p = 0.024) and patients with stage I POLEmut–p53abn EC (n = 19) showed an RFS of 94.1% (p = 0.050) (Figure 4).
Figure 4.
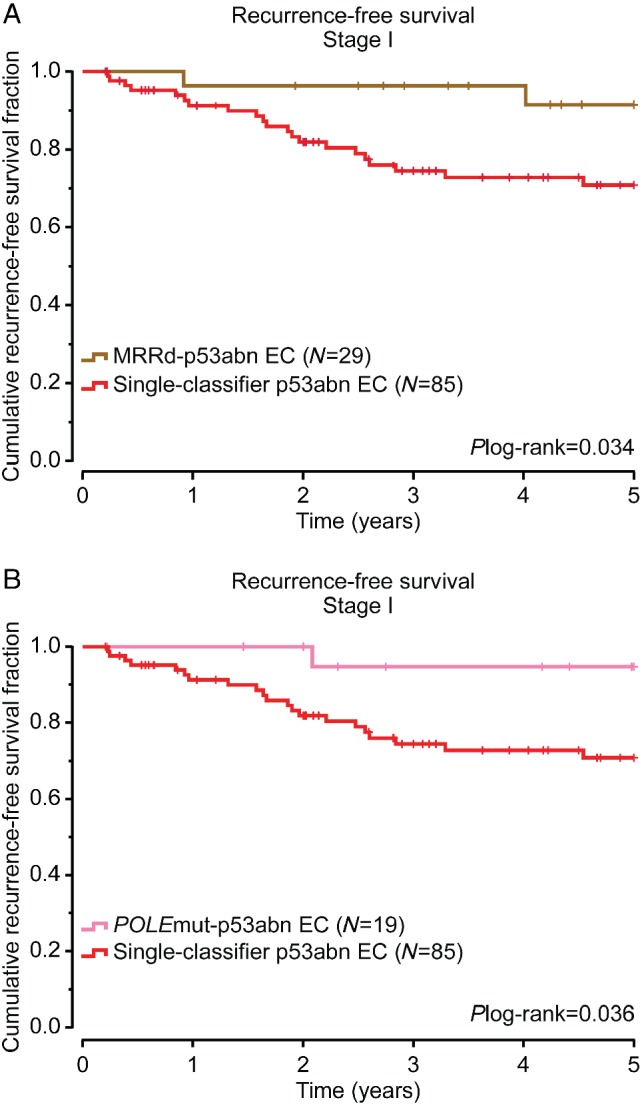
Survival analysis of multiple‐classifier EC compared with single‐classifier p53abn EC. Five‐year recurrence‐free survival analysis of stage I MMRd–p53abn EC (A) and POLEmut–p53abn EC (B) compared with single‐classifier p53abn EC. Two‐sided P values were obtained from log‐rank testing. Patients were classified based on POLEmut–p53abn (pink), MMRd–p53abn (brown), and single‐subtype p53abn (red).
Discussion
The molecular EC classification developed by TCGA has the potential to become the new standard in diagnostic pathology. Together with stage, the molecular subtype of EC provides powerful prognostic information; unlike stage, this information can be very accurately determined at the time of initial biopsy 33. When using the surrogate approach to determine the four molecular subtypes (POLEmut, MMRd, p53abn, and NSMP EC), a small but clinically relevant number of ECs (3–5%) have been unclassifiable due to the presence of more than one molecular classifying feature. These so‐called ‘multiple‐classifier’ ECs create a hurdle in the application of the molecular EC classification in practice, as it is not immediately apparent how these should be classified/treated. This issue is particularly relevant for the co‐occurrence of abnormal p53 IHC in the context of MMRd or POLEmut, as these features, when present singly, are associated with opposing clinical outcomes. The present study is the first to provide an extensive characterisation of these uncommon multiple‐classifier ECs and by doing so, provides guidance on how these should be interpreted.
The molecular landscape of single‐classifier POLEmut EC, p53abn EC or MMRd EC cases is considered to be shaped by their driver alterations, being POLE EDM, TP53 mutation or MMR deficiency. Interestingly, we observed similar mutational changes and SCNA between MMRd–p53abn EC and POLEmut–p53abn EC with single‐classifier MMRd and POLEmut EC, respectively. This strongly suggests that TP53 variants occurring in the context of an MMRd or POLEmut EC are likely passenger events, not affecting the molecular landscape of the tumour. This is further supported by the phenotype of the multiple‐classifier ECs, as pathology review revealed an enrichment for features associated with MMRd or POLEmut EC (TILs, peritumoural lymphocytes, squamous metaplasia) and not serous‐like (p53abn) features. Another important observation in support of this interpretation is the unusual high frequency (47–60%) of subclonal abnormal p53 staining in the multiple‐classifier EC. Of note, cases designated as single‐classifier p53 EC from the previously published PORTEC‐1/‐2 cohorts (n = 74) did not show subclonal expression 10. This same observation was made independently by Singh et al 15. Together, these data strongly support the interpretation that the TP53 mutation is a later event during tumour progression in multiple‐classifier EC, without affecting the molecular landscape or the phenotype.
Multiple‐classifier EC can be identified on the basis of sequencing approaches (e.g. presence of POLE and TP53 mutations by sequencing) or by the use of surrogate markers (e.g. abnormal p53 IHC). By sequencing, more TP53 mutations will be identified, resulting in the higher number of multiple‐classifiers present in TCGA (TP53 mutational status‐based) compared with the cases identified using the surrogate marker approach (p53 IHC‐based) 34. This is likely the result of the high number of ‘non‐hotspot’ TP53 mutations in POLEmut–p53abn EC compared with single‐classifier TP53mut EC, as well as the higher sensitivity of sequencing approaches to identify subclonal events (low allele frequency). These non‐hotspot TP53 mutations appear to not always impact the expression and function of p53. This is in line with Singh et al 15, where a high proportion of cases with such a TP53 variant and p53 wild‐type staining pattern were identified in multiple‐classifier ECs.
We also describe a limited number of MMRd–POLEmut–p53abn ECs (‘triple‐classifier’). The data available are insufficient to suggest how to treat patients with these cancers. The presence of p53abn subclonal staining in these cases suggests that, as in MMRd–p53abn and POLEmut–p53abn ECs, the TP53 mutations occur as a secondary event. However, it is difficult to assess, with the present data, whether POLEmut or MMRd are the driving events in these cancers. A larger number of these triple‐classifier cases will be required to study the biological behaviour of these ECs. Until then, considering the results reported by León‐Castillo et al 18 on MMRd–POLEmut ECs and the present study, we suggest that these triple‐classifiers be classified as POLEmut ECs if they have a pathogenic POLE EDM based on whole‐exome sequencing (WES) data or, in the case of absence of WES data, if the POLE EDM corresponds to one of the 11 pathogenic mutations described by León‐Castillo et al 18. In tumours in which the triple‐classifier ECs carry a POLE EDM that does not comply with the previous criteria, we recommend that they be classified as MMRd ECs, as discussed in depth by León‐Castillo et al 18.
Finally, we addressed the question of whether the biological behaviour of POLEmut and MMRd ECs is impacted by the presence of abnormal (subclonal) p53 expression. Although the number of cases is limited, the clinical outcomes of MMRd–p53abn ECs and POLEmut–p53abn ECs are strikingly different from what would be expected in single‐classifier p53abn (SCNA‐high/serous‐like) ECs 1, 9, 10, 11, 12, 13, 14. This supports the concept that passenger events do not affect biological behaviour. For clinical management, this means that the presence of TP53 mutations in the context of MMRd EC or POLEmut EC should not prompt intensified treatment.
In addition to multiple‐classifier ECs with mutant p53 expression/TP53 mutation, a fourth group of ECs with multiple classifying features can be encountered: MMRd–POLEmut ECs. The genomic architecture and clinical outcome of MMRd–POLEmut ECs differ depending on the pathogenicity of the POLE exonuclease domain variant and the ultramutated phenotype it confers to the tumour 18. Thus, the base change and indel proportion of ECs with pathogenic POLE EDM (as defined by the POLE score) and MSI are similar to MSS ECs with pathogenic POLE EDM 18. Additionally, when examining a cohort of MMRd ECs with pathogenic POLE EDM, although the number of patients was limited (n = 14), a good clinical outcome was observed (5‐year RFS 92.3%), in line with the prognosis described previously for single‐classifier POLEmut ECs (supplementary material, Figure S5) 9, 10, 11, 12, 13, 14, 18, 35, 36. These findings support the classification of tumours with a pathogenic POLE EDM and MMRd as single‐classifier POLEmut ECs.
In conclusion, this study is the first to describe evidence in support of categorising multiple‐classifier POLEmut–p53abn EC as single‐classifier POLEmut, and MMRd–p53abn EC as single‐classifier MMRd. Although rare (∼3%), correct designation of multiple‐classifier ECs facilitates the implementation of the molecular EC classification, enabling them to be included in future studies and, more importantly, providing valuable information for clinicians and patients to guide management.
Author contributions statement
AL and EG carried out experiments and analysed data. TB, CBG, DNC, JNM, and RN conceived experiments and analysed data. VTHBMS, MM, SK, SYB, JWC, EE, TTR, RAS, RG, XM, EO, and BTH analysed data. All the authors were involved in writing the paper and had final approval of the submitted and published versions.
Supporting information
Supplementary figure legends
Figure S1. Heatmap showing hierarchical clustering of MMRd–p53abn, single‐classifier MMRd, and single‐classifier p53abn ECs in TCGA
Figure S2. Heatmap showing hierarchical clustering of POLEmut–p53abn, single‐classifier POLEmut, and single‐classifier p53abn ECs in TCGA
Figure S3. Heatmap showing hierarchical clustering of MMRd–POLEmut–p53abn, single‐classifier MMRd, single‐classifier POLEmut, and single‐classifier p53abn ECs in TCGA
Figure S4. Overall survival and recurrence‐free survival of MMRd–p53abn and POLEmut–p53abn ECs
Figure S5. Clinical outcome of MMRd–POLEmut ECs
Table S1. Morphological features of MMRd–p53abn, POLEmut–p53abn, and MMRd–POLEmut–p53abn ECs
Table S2. Clinicopathological features of MMRd–p53abn and POLEmut–p53abn ECs with available survival data
Acknowledgements
This work is supported by the Dutch Cancer Society (KWF‐YIG 8232‐31648; AL and TB). This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748. We would like to thank Tissue Bank Bern for providing tissues for study purposes.
No conflicts of interest were declared.
References
- 1. Cancer Genome Atlas Research Network , Kandoth C, Schultz N, et al Integrated genomic characterization of endometrial carcinoma. Nature 2013; 497: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McAlpine J, Leon‐Castillo A, Bosse T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J Pathol 2018; 244: 538–549. [DOI] [PubMed] [Google Scholar]
- 3. Colombo N, Creutzberg C, Querleu D, et al Appendix 5: endometrial cancer: eUpdate published online 8 June 2017 (http://www.esmo.org/Guidelines/Gynaecological-Cancers). Ann Oncol 2017; 28(suppl_4): iv153–iv156. [DOI] [PubMed] [Google Scholar]
- 4. Van Gool IC, Rayner E, Osse EM, et al Adjuvant treatment for POLE proofreading domain‐mutant cancers: sensitivity to radiotherapy, chemotherapy, and nucleoside analogues. Clin Cancer Res 2018; 24: 3197–3203. [DOI] [PubMed] [Google Scholar]
- 5. Le DT, Durham JN, Smith KN, et al Mismatch repair deficiency predicts response of solid tumors to PD‐1 blockade. Science 2017; 357: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ott PA, Bang Y‐J, Berton‐Rigaud D, et al Pembrolizumab in advanced endometrial cancer: preliminary results from the phase Ib KEYNOTE‐028 study. J Clin Oncol 2016; 34(suppl_15): 5581. [Google Scholar]
- 7. Reijnen C, Küsters‐Vandevelde HVN, Prinsen CF, et al Mismatch repair deficiency as a predictive marker for response to adjuvant radiotherapy in endometrial cancer. Gynecol Oncol 2019; 154: 124–130. [DOI] [PubMed] [Google Scholar]
- 8.PORTEC‐4a: Randomised trial of standard or molecular profile‐based recommendation for radiotherapy after surgery for women with early stage endometrial cancer. 1 June 2016. [Accessed 2 December 2019]. Available from: https://www.isrctn.com/ISRCTN11659025
- 9. Stelloo E, Bosse T, Nout RA, et al Refining prognosis and identifying targetable pathways for high‐risk endometrial cancer; a TransPORTEC initiative. Mod Pathol 2015; 28: 836–844. [DOI] [PubMed] [Google Scholar]
- 10. Stelloo E, Nout RA, Osse EM, et al Improved risk assessment by integrating molecular and clinicopathological factors in early‐stage endometrial cancer – combined analysis of the PORTEC cohorts. Clin Cancer Res 2016; 22: 4215–4224. [DOI] [PubMed] [Google Scholar]
- 11. Talhouk A, McConechy MK, Leung S, et al A clinically applicable molecular‐based classification for endometrial cancers. Br J Cancer 2015; 113: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Talhouk A, McConechy MK, Leung S, et al Confirmation of ProMisE: a simple, genomics‐based clinical classifier for endometrial cancer. Cancer 2017; 123: 802–813. [DOI] [PubMed] [Google Scholar]
- 13. Kommoss S, McConechy MK, Kommoss F, et al Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population‐based case series. Ann Oncol 2018; 29: 1180–1188. [DOI] [PubMed] [Google Scholar]
- 14. Cosgrove CM, Tritchler DL, Cohn DE, et al An NRG oncology/GOG study of molecular classification for risk prediction in endometrioid endometrial cancer. Gynecol Oncol 2018; 148: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singh N, Piskorz A, Bosse T, et al p53 immunohistochemistry is an accurate surrogate for TP53 mutational analysis in endometrial carcinoma biopsies. J Pathol 2020; 250: 336–345. [DOI] [PubMed] [Google Scholar]
- 16. Bosse T, Nout RA, McAlpine JN, et al Molecular classification of grade 3 endometrioid endometrial cancers identifies distinct prognostic subgroups. Am J Surg Pathol 2018; 42: 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imboden S, Nastic D, Ghaderi M, et al Phenotype of POLE‐mutated endometrial cancer. PLoS One 2019; 14: e0214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. León‐Castillo A, Briton H, McConechy M, et al Interpretation of somatic POLE mutations in endometrial carcinoma. J Pathol 2020; 250: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Umar A, Boland CR, Terdiman JP, et al Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004; 96: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forbes SA, Beare D, Gunasekaran P, et al COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2014; 43(Database issue): D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Landrum MJ, Lee JM, Riley GR, et al ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2013; 42(Database issue): D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petitjean A, Mathe E, Kato S, et al Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007; 28: 622–629. [DOI] [PubMed] [Google Scholar]
- 23. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non‐synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 24. Adzhubei IA, Schmidt S, Peshkin L, et al A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stelloo E, Jansen AML, Osse EM, et al Practical guidance for mismatch repair‐deficiency testing in endometrial cancer. Ann Oncol 2017; 28: 96–102. [DOI] [PubMed] [Google Scholar]
- 26. Alexandrov LB, Nik‐Zainal S, Wedge DC, et al Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Colombo N, Preti E, Landoni F, et al Endometrial cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2013; 24(suppl_6): vi33–vi38. [DOI] [PubMed] [Google Scholar]
- 28. Colombo N, Creutzberg C, Amant F, et al ESMO–ESGO–ESTRO consensus conference on endometrial cancer: diagnosis, treatment and follow‐up. Ann Oncol 2016; 27: 16–41. [DOI] [PubMed] [Google Scholar]
- 29. Hussein YR, Weigelt B, Levine DA, et al Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod Pathol 2015; 28: 505–514. [DOI] [PubMed] [Google Scholar]
- 30. Van Gool IC, Ubachs JEH, Stelloo E, et al Blinded histopathological characterisation of POLE exonuclease domain‐mutant endometrial cancers: sheep in wolf's clothing. Histopathology 2018; 72: 248–258. [DOI] [PubMed] [Google Scholar]
- 31. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014; 25: 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alexandrov LB, Kim J, Haradhvala NJ, et al The repertoire of mutational signatures in human cancer. bioRxiv 2018. 10.1101/322859. [DOI] [Google Scholar]
- 33. Talhouk A, Hoang LN, McConechy MK, et al Molecular classification of endometrial carcinoma on diagnostic specimens is highly concordant with final hysterectomy: earlier prognostic information to guide treatment. Gynecol Oncol 2016; 143: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schultheis AM, Martelotto LG, De Filippo MR, et al TP53 mutational spectrum in endometrioid and serous endometrial cancers. Int J Gynecol Pathol 2016; 35: 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Church DN, Stelloo E, Nout RA, et al Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst 2015; 107: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McConechy MK, Talhouk A, Leung S, et al Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clin Cancer Res 2016; 22: 2865–2873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure legends
Figure S1. Heatmap showing hierarchical clustering of MMRd–p53abn, single‐classifier MMRd, and single‐classifier p53abn ECs in TCGA
Figure S2. Heatmap showing hierarchical clustering of POLEmut–p53abn, single‐classifier POLEmut, and single‐classifier p53abn ECs in TCGA
Figure S3. Heatmap showing hierarchical clustering of MMRd–POLEmut–p53abn, single‐classifier MMRd, single‐classifier POLEmut, and single‐classifier p53abn ECs in TCGA
Figure S4. Overall survival and recurrence‐free survival of MMRd–p53abn and POLEmut–p53abn ECs
Figure S5. Clinical outcome of MMRd–POLEmut ECs
Table S1. Morphological features of MMRd–p53abn, POLEmut–p53abn, and MMRd–POLEmut–p53abn ECs
Table S2. Clinicopathological features of MMRd–p53abn and POLEmut–p53abn ECs with available survival data


