Abstract
BACKGROUND
Fungicide resistance is a growing problem affecting many crop pathogens owing to the low success rate in finding novel chemical classes of fungicides. Pyridachlometyl is a new fungicide that seems to belong to a new chemical class of tubulin polymerization promoters.
RESULTS
Pyridachlometyl exhibited potent antifungal activity against a broad range of fungal species belonging to the phyla Ascomycota and Basidiomycota. No cross‐resistance was observed with other fungicide classes, such as ergosterol biosynthesis inhibitors, respiratory inhibitors, or tubulin polymerization inhibitors in Zymoseptoria tritici. Pyridachlometyl‐resistant strains were obtainable by UV mutagenesis in Z. tritici and Penicillium digitatum. Mutations in tubulin‐coding genes were found in all laboratory mutants but the patterns of mutation were distinct from that of tubulin polymerization inhibitors, such as benzimidazole fungicides.
CONCLUSION
Pyridachlometyl is a promising new tool for disease control. However, strict resistance management strategies should be recommended for the practical use of pyridachlometyl. © 2019 The Authors. Pest Management Science published by John Wiley & Sons Ltd on behalf of Society of Chemical Industry.
Keywords: pyridachlometyl, fungicide resistance, α‐tubulin, β‐tubulin
Locations of substitutions leading to resistance against pyridachlometyl and carbendazim. The former is near the interface between the two tubulin dimers. The latter is β‐tubulin.

1. INTRODUCTION
Although social pressure to reduce chemical use in agriculture is strengthening, some scientists believe that intensive farming is the least damaging option for food supply and environmental conservation in terms of effects on wildlife populations and greenhouse gas emissions because attaining a higher yield from the same amount of land reduces the demand for farmland expansion.1 Agrochemicals are essential to intensive farming practices to protect crops from a number of pests, including fungal diseases,2, 3 but effective disease control has become more difficult because of increasing fungicide resistance.3, 4 Given this background, a shortage of new fungicides with a novel mechanism of action is a serious concern because in most cases the mechanism of action is related to cross‐resistance among fungicides. Fungicides with the same mechanism of action, which act via the same target protein, generally result in cross‐resistance because the mutations in target site proteins can impair the binding of fungicides. In recent years, only three classes of fungicides, DMI (sterol C14 demethylation inhibitor in ergosterol biosynthesis), QoI (Qo site of complex III inhibitor in the respiratory chain), and SDHI (succinate dehydrogenase inhibitor in the respiratory chain), have accounted for approximately 60% of the global fungicide market5 because of their high efficacy and the broad spectrum of these three chemical classes. Frequent usage of these three classes of fungicides has led to the development of resistance in crop pathogens, such as Zymoseptoria tritici,6, 7 Erysiphe necator,8, 9, 10 Podosphaera xanthii,11, 12 Microdochium spp.,13, 14 and Cercospora beticola.15, 16 As a result, agrochemical research and development seek new chemical classes of fungicides to respond to the needs of crop growers.5 Despite these efforts, few successful novel fungicides have been developed in the last decade.
Tubulin polymerization promoters stand out as a potential chemical class because they are highly active against various phytopathogenic fungi.17, 18, 19, 20, 21 This chemical class targets tubulins, which are protein dimers consisting of two closely related 55‐KD α and β subunits (α‐ and β‐tubulin, respectively). The polymerization of tubulin into microtubules and also the depolymerization of microtubules into tubulin play a central role in all eukaryotic cells (e.g. in the process of nuclear division).22, 23 Although some anti‐tubulin agents, such as carbendazim, thiabendazole, and diethofencarb (Fig. 1) and their analogs, have been used as agricultural fungicides for years, they are all classified as polymerization inhibitors, which target the colchicine site in β‐tubulin.22, 24 On the contrary, the depolymerization of tubulin associated with the hydrolysis of GTP from GDP is also essential for cells.25 Interestingly, triazolopyrimidine compounds, such as BAS600F (Fig. 1), have been reported as a new type of anti‐tubulin agent, which promotes polymerization of tubulin in vitro.21 Thus, these compounds are expected to inhibit depolymerization of tubulin in vivo; a similar action in cancer cells has been reported for paclitaxel, which is also known to promote polymerization of tubulin in vitro.23 It has also been shown that triazolopyrimidine compounds bind to the vinblastine site,26 which is distinct from the colchicine site and is known to be the binding site of Vinka alkaloids such as vincristine.21, 27
Figure 1.
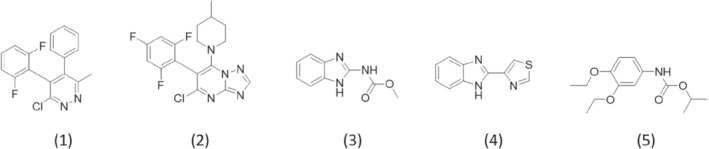
Structures of pyridachlometyl (1), BAS600F (2), carbendazim (3), thiabendazole (4), and diethofencarb (5).
Pyridachlometyl (Fig. 1) is a new fungicide characterized by a central pyridazine ring.28 Considering the partial similarity in chemical structure, pyridachlometyl appears to be a tubulin polymerization promoter, similar to BAS600F.19 Although the structure–activity relationships of some pyridazine derivatives have been reported,19 there are no reports specifically on pyridachlometyl, and the antifungal activity of pyridazine derivatives has been demonstrated for only a few fungal species. Therefore, we report details on the activity of pyridachlometyl, with an emphasis on the lack of cross‐resistance with existing fungicides. Additionally, this study focuses on the future risk of development of resistance to pyridachlometyl by pathogens.
2. MATERIALS AND METHODS
2.1. Chemical materials
Carbendazim, thiabendazole, diethofencarb, fluxapyroxad, and prothioconazole‐desthio were purchased from Sigma‐Aldrich Japan (Tokyo, Japan). Pyridachlometyl and BAS600F were synthesized by Sumitomo Chemical, as previously described.18, 28 For in vitro experiments, chemical compounds were dissolved in dimethyl sulfoxide (DMSO) as stock solutions.
2.2. Fungal materials
Z. tritici strains were isolated from the leaves of field crops collected from European countries (single pycnidia isolates). The sampling locations of the isolates are listed in Table S1. Samples of Z. tritici strains were stored before assaying in 25% glycerol at −80 °C. Other fungal materials used in Table 1 were stored as slant cultures at 12 °C in a collection at Sumitomo Chemical and precultured before assaying, as shown in Table S2.
Table 1.
In vitro antifungal spectrum of pyridachlometyl
| Division | Class | Order | Species | EC50 (mg L−1)a |
|---|---|---|---|---|
| Ascomycota | Dothideomycetes | Capnodiales | Zymoseptoria tritici | 0.056 |
| Passalora fulva | 0.034 | |||
| Pseudocercospora fijiensis | 0.016 | |||
| Cercospora beticola | 0.17 | |||
| Pleosporales | Alternaria solani | 0.22 | ||
| Cochliobolus miyabeanus | 0.21 | |||
| Corynespora cassiicola | 2.3 | |||
| Venturiales | Venturia inaequalis | 0.040 | ||
| Sordariomycetes | Magnaporthales | Pyricularia oryzae | 0.055 | |
| Glomerellales | Colletotrichum gloeosporioides | 0.023 | ||
| Colletotrichum acutatum | 1.2 | |||
| Xylariales | Microdochium majus | 0.075 | ||
| Hypocreales | Fusarium graminearum | 1.4 | ||
| Leotiomycetes | Helotiales | Botrytis cinerea | 0.059 | |
| Sclerotinia sclerotiorum | 1.7 | |||
| Clarireedia homoeocarpa | 0.038 | |||
| Eurotiomycetes | Eurotiales | Penicillium digitatum | 1.4 | |
| Penicillium expansum | 2.4 | |||
| Basidiomycota | Agaricomycetes | Cantharellales | Rhizoctonia solani AG4 | 0.66 |
| Atheliales | Athelia rolfsii | 0.40 | ||
| Ustilaginomycetes | Ustilaginales | Ustilago maydis | 0.12 | |
| (Stramenopiles; non‐fungi)b | Oomycetes | Peronosporales | Phytophthora infestans | >10 |
| Pythium ultimum | >10 | |||
| Pythium aphanidermatum | >10 |
Each value is based on the mean of two individual EC50 values; 95% confidence intervals of all EC50 values ranged between 50% (MIN) and 300% (MAX) of the respective values.
Division in classical taxonomic rank is not assigned for Oomycete.
2.3. Antifungal tests
The antifungal activity of each test compound against each pathogen species was evaluated by either microtiter plate or agar plate fungitoxicity assays under the incubation conditions detailed in Table S2. The resistance factor (RF) was calculated using the following formula:
2.3.1. 96‐well microtiter plate method
Growth of Z. tritici, Passalora fulva, and Ustilago maydis was evaluated on 96‐well microtiter plates. Inoculum of each fungus was harvested at more than 100× density in distilled water and suspended in the appropriate medium at the density shown for each species (Table S2). A series of dilutions of test compound in DMSO was prepared at 150 times the desired test concentrations of active ingredients for each test (Table S2). For example, to obtain the final concentrations of 1, 0.3, and 0.1 mg L−1, the test compound concentrations 150, 45, and 15 mg L−1 were diluted 150‐fold, respectively. One microliter aliquots of each fungicide were mixed with 149 μL of prepared inoculum in media with two replicates for each concentration. After the incubation periods (Table S2), growth was measured by determining optical density at a wavelength of 600 nm using a microplate reader SH‐9000 Lab (Corona Electric, Ibaraki, Japan) with a 3 × 3 matrix of scanning points. Optical density values were corrected against the value of the blank well that had no inoculum. The 50% effective concentrations (EC50) were calculated by probit analysis.
2.3.2. Agar plate method
Except for Z. tritici, P. fulva, and U. maydis, each fungal strain was cultured on agar media amended with a dilution series of fungicides at designated concentrations (Table S2). Mean mycelium radial length from two different inoculums in each plate medium was measured at designated periods after inoculation and EC50 values were calculated by probit analysis.
2.4. Generation of fungicide‐resistant UV mutants
To generate the fungicide‐resistant UV mutants of Z. tritici, the Set1 strain isolated in the 1980s in Japan was used. The yeast‐like cells of the parental strain (Set1) on the Malt Yeast Agar (MYA) medium were exposed to UV light at 30 mJ cm−2, leading to approximately 50% lethality after UV treatment. The yeast‐like cells were distributed onto potato dextrose agar (PDA) plates amended with pyridachlometyl at 1.0 mg L−1 or carbendazim at 1.0 mg L−1. The plates were incubated in the dark at 18 °C for 8 days. Visible colonies were chosen from the primary selection plates and re‐grown on another selection plate under the same conditions to harvest for storage. To generate the fungicide‐resistant UV mutants of Penicillium digitatum, the PD5 strain isolated in the 1980s in Japan was used. A conidial suspension was prepared at a concentration of 1.0 × 108 conidia mL−1, followed by the addition of 1‐methyl‐3‐nitro‐1‐nitrosoguanidine (MNNG) at a concentration of 50 mg L−1. After incubating the suspension at 20 °C for 30 min, it was centrifuged and washed four times with sterilized water. The resulting conidia were spread onto the PDA medium amended with 1 mg L−1 of BAS600F. After 4 days of incubation at 23 °C, visible colonies were chosen from the primary selection plates and re‐grown on another selection plate under the same conditions to harvest for storage. The strains of Z. tritici and P. digitatum were stored in 25% glycerol at −80 °C for conducting further assays.
2.5. DNA sequencing of tubulin genes
The DNA of each fungus was extracted using PrepMan Ultra Reagent (Applied Biosystems, Waltham, USA) according to the manufacturer's instructions. A trace amount of the yeast‐like Z. tritici cells or P. digitatum conidia were picked from a colony on the plate and suspended in 50 μL PrepMan Ultra reagent and incubated at 96 °C for 10 min. After centrifugation at 8500 × g for 2 min, the supernatant was removed and used immediately or stored at −20 °C until use. The amplification of tubulin‐subunit encoding genes by PCR reaction was conducted using the Ex Taq Kit (Takara, Kyoto, Japan) with the primer pairs listed in Table S3. After DNA amplification, PCR products were purified using a Wizard DNA Clean‐Up System (Promega Corporation, WI, USA) and then sequenced using an ABI3100 DNA system (Applied Biosystems, CA, USA). Sequences were aligned using GENETYX Ver. 12 software (Genetyx Corporation, Tokyo, Japan).
2.6. Model construction
The homology model of Z. tritici tubulin tetramer (two α and two β subunits) was constructed using the standard settings of the modelling tool in MOE (Molecular Operating Environment, Version 2018.01, Chemical Computing Group Inc., Montreal, Canada). The X‐ray structure of the tubulin tetramer from Bos taurus stabilized by triazolopyrimidine (PDB 5NJH) with a resolution of 2.4 Å was chosen as a structural template for the pyridachlometyl‐bound model. The overall sequence identity to α‐tubulin of Z. tritici was 78.4%, whereas that to β‐tubulin (S221P mutant) was 81.6%. Alignments of amino acid sequences were performed using the BLOSUM62 substitution matrix. The structure was refined and prepared for docking using ‘Structure Preparation’ and ‘Protonate 3D’ in MOE. Then, the template docking procedure of MOE was utilized to dock pyridachlometyl to the triazolopyrimidine binding site of this structure. However, the carbendazim‐bound model of Z. tritici tubulin tetramer was constructed using the X‐ray structure of two α‐tubulins from Sus barbatus and two β‐tubulins from Gallus gallus stabilized by nocodazole (PDB 5CA1; resolution 2.4 Å). Carbendazim was docked to the nocodazole binding site in the same way.
2.7. Growth speed (calculation of doubling time)
Mutants of Z. tritici were inoculated in Yeast Bacto Glycerol medium (YBG) liquid medium at a density of 104 mL−1. Growth was measured by optical density after 0, 24, 48, 72, and 96 h of incubation on the rotary shaker at 120 rpm, at a wavelength of 600 nm with a SH‐9000 Lab microplate reader (Colona Electric) using a 3 × 3 matrix of scanning points with 16 replicated wells for each mutant. Mean growth (ΔOD) of each well was used for the calculation of the specific growth rate [μ(h−1) and doubling time, t d(h)]. Specific growth rate μ was calculated by linear approximation of ΔOD at the log phase (0–96 h for 12 °C, 0–72 h for 18 °C, and 0–72 h for 27 °C) with the least squares method. Under these conditions, the R‐squared values of the calculated formula were more than 0.9 for each mutant. Doubling time [t d(h)] was calculated using the formula t d = loge 2/μ.
3. RESULTS AND DISCUSSION
3.1. Antifungal spectrum of pyridachlometyl
As shown in Table 1, all fungal species tested were sensitive to pyridachlometyl (EC50 < 5 mg L−1). A correlation between pyridachlometyl's effectiveness and fungal classification was expected because of the similarity in the primary structure of tubulin proteins, but a relationship was not observed. For example, sensitivity was significantly different between two closely related species, such as Colletotrichum gloeosporioides and C. acutatum, although the primary structures of their tubulin proteins, including the key amino acids at putative binding sites, are almost identical.29 This difference might be ascribed to the differential expression level of the gene encoding β‐tubulin rather than the structural difference of tubulin proteins.29 In C. acutatum, the gene encoding β‐tubulin was overexpressed in response to benomyl, a profungicide of carbendazim, and was mediated by a transcription factor‐like protein containing a leucine‐zipper motif.29 Furthermore, the tubulin gene copy numbers were different across species of fungi.30 For example, Fusarium graminearum, which was less sensitive to pyridachlometyl, is known to possess two divergent β‐tubulin‐encoding genes exhibiting 76.6% similarity in their putative amino acid sequences.30 Such redundancy of tubulin genes might also be related to the varying sensitivity of each species to pyridachlometyl.
3.2. Activity of pyridachlometyl to Zymoseptoria tritici field strains
The Z. tritici strains isolated in 2017 from Germany, UK, and Ireland were subjected to the pyridachlometyl sensitivity test. In this test, two commercial fungicides, SDHI fungicide fluxapyroxad and DMI fungicide prothioconazole (desthio form), were included as reference fungicides because they are frequently used in these countries. Table 2 shows the EC50 values of some strains with varying degrees of sensitivity to fluxapyroxad or prothioconazole‐desthio. However, they did not exhibit cross‐resistance to pyridachlometyl. Additionally, the majority of these strains were highly resistant to carbendazim (EC50 > 10 mg L−1; data not shown). The high frequency of carbendazim‐resistant strains could be explained by the historic use of carbendazim to control Z. tritici as well as by the current use of thiophanate‐methyl (a profungicide of carbendazim) to control wheat head blight.31, 32 We confirmed that such carbendazim‐insensitive field strains are sensitive to pyridachlometyl.
Table 2.
In vitro antifungal activity of pyridachlometyl for the field strains of Zymoseptoria tritici
| Pyridachlometyl | Fluxapyroxad | Prothioconazole‐desthio | ||||
|---|---|---|---|---|---|---|
| Strains | EC50 (mg L−1)a | 95% CIb | EC50 (mg L−1) | 95% CI | EC50 (mg L−1) | 95% CI |
| DE 17‐Septo 3.1 | 0.040 | 0.023–0.058 | 0.036 | 0.026–0.049 | 0.048 | 0.036–0.064 |
| DE 17‐Septo 3.3 | 0.068 | 0.050–0.088 | 0.180 | 0.147–0.224 | 0.412 | 0.334–0.520 |
| DE 17‐Septo 4.8 | 0.095 | 0.068–0.137 | 3.058 | 2.284–4.301 | 0.747 | 0.562–0.907 |
| UK 17‐Septo 8.10 | 0.020 | 0.014–0.029 | 5.300 | 2.638–14.466 | 0.020 | 0.014–0.028 |
| UK 17‐Septo 9.10 | 0.048 | 0.036–0.063 | 0.024 | 0.017–0.033 | 0.371 | 0.294–0.516 |
| UK 17‐Septo 10.8 | 0.027 | 0.018–0.039 | 0.049 | 0.034–0.066 | 0.556 | 0.418–0.724 |
| UK 17‐Septo 10.10 | 0.055 | 0.049–0.076 | 1.569 | 1.151–2.212 | 0.150 | 0.148–0.152 |
| IE 17‐Septo 6.4 | 0.084 | 0.066–0.110 | 0.335 | 0.243–0.472 | 0.265 | 0.185–0.405 |
Each value is based on two biological replicates.
95% CI: 95% confidence interval of EC50 values (mg L−1).
The ranges of EC50 distribution were 0.0034–0.18 mg L−1 for pyridachlometyl, 0.0043 to >10 mg L−1 for fluxapyroxad, and 0.0039–1.0 mg L−1 for the prothioconazole‐desthio (Fig. 2). A wide range of distributions was observed for fluxapyroxad (CV = 2.2) and prothioconazole‐desthio (CV = 1.1) because of the sensitivity shift primarily conferred by mutations in the target gene.6, 7 In contrast, the range for pyridachlometyl was considerably smaller (CV = 0.60).
Figure 2.
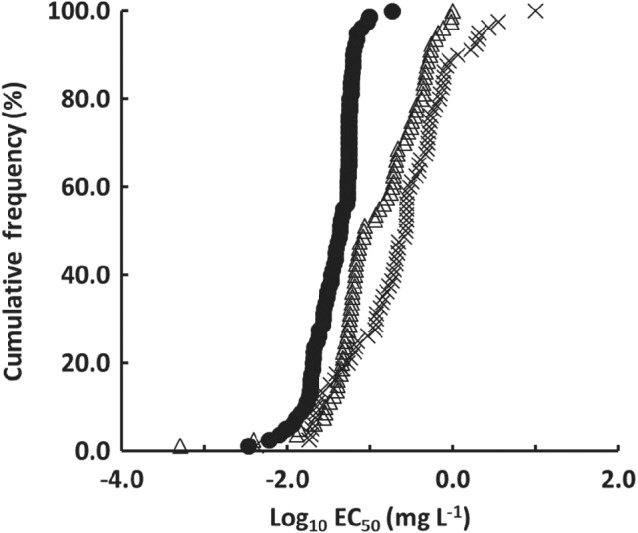
Sensitivity distribution of Z. tritici in European countries. Strains collected in 2017 (N = 80) are ranked according to increasing EC50 values (cumulative). Pyridachlometyl, fluxapyroxad, and prothioconazole‐desthio are represented by a black circle, cross, and white triangle, respectively. The cut‐off value for EC50 is 10 mg L−1.
3.3. Laboratory mutants selected by carbendazim and pyridachlometyl
Selection pressure was applied with 1 mg L−1 carbendazim or 1 mg L−1 pyridachlometyl, which produced seven and 17 resistant mutants, respectively (Table 3). All seven mutants selected by carbendazim had a point mutation in β‐tubulin (E198G for two mutants, compound mutations of Q134K with F265 L for two mutants, and E198D, E198A, and F200Y for one mutant, respectively). All mutants showed cross‐resistance to thiabendazole (Table S4). Among them, only mutants harboring E198G or E198A were sensitive to diethofencarb, as previously reported,22 whereas all mutants selected by carbendazim were fully sensitive to pyridachlometyl. Among the mutations that may confer resistance to carbendazim, the amino acids E198, F200Y, and Q134 are likely to be involved in the interaction between carbendazim and β‐tubulin because their three‐dimensional positions are very close to each other.33 Their position is often cited as the colchicine binding site, which is a pocket for the binding to tubulin of small molecules, such as colchicine, lexibulin, plinabulin, tivantinib, as well as nocodazole, an analogue of carbendazim.22, 24 In contrast, the 16 mutants selected by pyridachlometyl were sensitive to carbendazim, except for one mutant that did not grow on the medium without 1 mg L−1 pyridachlometyl and could not be subjected to the further experiments (Table S4). Twelve of the 17 mutants had a mutation in β‐tubulin (Y222N for nine mutants, Y222S for two mutants, and N219 for one mutant). These positions are far from the colchicine binding site but are included in another pocket for the binding of small molecules to tubulin, which is often called a vinblastine‐binding site23, 25 to which triazolopyrimidines are also reported to bind21, 27 (Fig. 3(a)). The other five did not possess mutations in β‐tubulin, but rather point mutations in the α‐tubulin (P325T for two mutants, P325H for one mutant, P325S for one mutant, and I355F for one mutant, which did not grow on the medium without pyridachlometyl). The proline at 325 and the isoleucine at 355 of α‐tubulin were also reported to mediate the binding of triazolopyrimidines to tubulins in a previous study.26 There has been no report that mutation in α‐tubulin can confer resistance to fungicides as all commercial fungicides, including Oomyceticides, are assumed to bind to the colchicine binding site in β‐tubulin.22 On the contrary, for agrochemicals, tubulin disrupting agents are not only used as fungicides but also as herbicides. Among these herbicides, the binding pocket of carbamates, such as chlorpropham, and benzamides, such as pronamide, are assumed to be the colchicine binding site in β‐tubulin.22 However, dinitroanilines, such as oryzalin, trifluralin, and pendimethalin, have been reported to bind to α‐tubulin.34, 35, 36 Their binding site has been investigated mainly using protozoan organisms.34, 35, 36 In these reports, L136F, S165T, T239I, R243C/K/M/S, V/I252L, and M301T were shown to confer resistance to oryzalin.34, 35, 36 These amino acid positions are located near the interface between α‐tubulin and β‐tubulin forming the dimer.36 In contrast, P325 and I355 of α‐tubulin are located on the other side of α‐tubulin near the interface between the two dimers.26 Therefore, the binding pocket for triazolopyrimidines and pyridachlometyl is also likely different from that of dinitroanilines.
Table 3.
Number of emerged colonies by selection on carbendazim or pyridachlometyl
| Selection | Plated cellsa | Emerged colonies | Frequency | |
|---|---|---|---|---|
| Carbendazim | 1 mg L−1 | 1.1 × 108 | 7 | 6.4 × 10−7 |
| Pyridachlometyl | 1 mg L−1 | 1.1 × 108 | 17 | 1.5 × 10−6 |
Cells exposed to UV light at 30 mJ cm−2 were spread onto plate medium (90 mm diameter) at 1.1 × 107 cells per plate.
Figure 3.
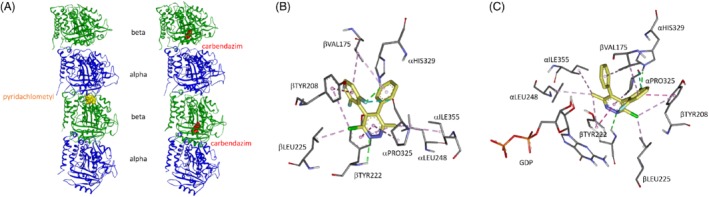
Homology model of Z. tritici tubulin based on X‐ray of Bos taurus, Sus barbatus, and Gallus gallus (PDB 5NJH and 5C1A1) with locations of substitutions leading to resistance against pyridachlometyl and carbendazim, respectively. (a) Overall view of the tubulin dimer unit with putative binding sites of pyridachlometyl and carbendazim. (b) Close‐up view of the putative binding site of pyridachlometyl. (c) Close‐up view of the putative binding site of pyridachlometyl with GDP. See also Fig. S1 for a close‐up view of locations of substitutions leading to resistance to carbendazim.
All 16 mutants selected by pyridachlometyl showed cross‐resistance to BAS600F, although the RF for BAS600F was small for two mutants that had the P325T mutation in the α‐tubulin. Among positions in which a single amino acid substitution was observed, our homology model predicted that the amino acid Y222 in β‐tubulin was likely involved in the π‐π stacking with the pyridazine ring of pyridachlometyl, as well as the guanine nucleobase of the GDP nucleotide by the aromatic ring of its side chain (Figs 3(b),(c)). Additionally, this Y222 seems to form a hydrogen bond with the nitrogen atom of the pyridazine ring by its main chain NH group. S221P observed in the mutant of another fungal species, P. digitatum selected by BAS600F (Table 4), will likely interfere with the hydrogen bond between the main chain of Y222 and the pyridazine ring. The role of Y208 for which the Y208S mutation was observed in another mutant of P. digitatum is interesting because this mutant showed hypersensitivity to pyridachlomethyl while showing resistance to BAS600F (Table 4). Using the homology modeling, Y208 seems to mediate π‐π stacking with the 4‐phenyl ring of pyridachlometyl, and also the π‐alkyl interaction with the chlorine atom of the pyridazine ring near its aromatic ring of the side chain. On the contrary, P325 in α‐tubulin may be involved in the hydrophobic interaction with the pyridazine ring of pyridachlometyl and its side chain and also in the interaction with the fluorine atom on the 4‐phenyl ring of pyridachlometyl.
Table 4.
Mutants selected on BAS600F and their sensitivities to pyridachlometyl
| Pyridachlometyl | BAS600F | ||||
|---|---|---|---|---|---|
| Strain | Mutation in β‐tubulin | EC50 (mg L‐1)a | 95% CIb | EC50 (mg L−1) | 95% CI |
| PD5 (wild type) | ‐ | 5.6 | 4.7–7.0 | 0.25 | 0.15–0.31 |
| Mutant 1 | Y208H | 2.2 | 1.5–4.3 | 1.10 | 0.75–1.70 |
| Mutant 2 | S221P | >10 | n.d. | >10 | n.d. |
| Mutant 3 | Y222N | >10 | n.d. | >10 | n.d. |
Each value EC50 is based on two biological replicates.
95% CI: 95% confidence interval of EC50 values (mg L−1).
3.4. Amino acid alignments of tubulin at the positions of mutation
We observed that some mutations in tubulin can confer resistance to pyridachlometyl. However, past studies indicate that mutations observed in laboratory mutants do not always occur in field mutants.37 One reason for this may be the importance of the original amino acid in the intrinsic function of the target proteins. Therefore, we checked the conservation of tubulin primary structures at the positions in which mutations were observed in the laboratory mutants of Z. tritici and P. digitatum. As a result, we confirmed the tyrosine at 222 of the β‐tubulin was highly conserved across all organisms, this tyrosine is replaced with phenylalanine in plants (Fig. 4), indicating the importance of the aromatic ring of the amino acid side chain in this position. Piedra et al. explained the importance of this tyrosine in shielding the guanosine from polymerization contacts by stacking on top of it.38 Additionally, 100% conservation of the proline at 325 of α‐tubulin was confirmed. The change in this amino acid is reported to cause abnormal microtubule arrays and result in helical growth in Arabidopsis thaliana.39 It was also reported that this proline is involved in longitudinal contact in the tubulin protofilament, as well as T221 in β‐tubulin of mammals,40 which is orthologous to N/Q/A219 in fungi (Fig. 4). The S5 mutant of Z. tritici selected by pyridachlometyl harbored the N219K mutation at this position in β‐tubulin and showed relatively lower RF than other mutants (Table S4). As additional variation across different organisms is allowed for this position (Fig. 4), future resistant strains to pyridachlometyl in the field may occur in the form of strains that have a mutation at this position. From another point of view, the amino acid variation in this position may explain the selectivity of pyridachlometyl between fungi, plants, and mammals.
Figure 4.

Alignment of a partial amino acid sequence of α‐tubulin and β‐tubulin in wild types of phytopathogenic fungi, oomycetes, plants, and mammals. Locations within 4.5 Å of pyridachlometyl in the homology model of Z. tritici tubulin dimer‐bound pyridachlometyl are indicated by ‘c’. Locations at which substitutions confer resistance to pyridachlometyl in Z. tritici and P. digitatum are indicated by shading. Amino acids conserved across all organisms are indicated with an asterisk. GenBank accession numbers for the sequences used are listed in Table S5.
3.5. In vitro analysis of fitness cost by each mutation
We tested the speed of growth in three different temperatures versus that of the wild type because some mutations in β‐tubulin were reported to confer negative effects on the growth of fungi at high temperature.41, 42 No difference was observed in their multiplication speed in liquid medium at all three temperatures, even though most of them had a mutation in tubulin at the positions for which original amino acids are highly conserved and thus original amino acids should be important for the intrinsic function of tubulin (Table 5). Although some of these showed slightly reduced multiplying speeds, the difference from that of the wild type was less than 15% in any case and no correlation with types of mutations was observed. That is, the S15 mutant showed slightly reduced multiplying speed as compared with the parental strain at 18 °C, but the S8 mutant, which possesses the same mutation, showed a slight increase in multiplying speed compared with that of the parental strain. In filamentous fungi, tubulin is known to be essential for polarized growth, such that the mutants may show abnormal growth in the filamentous phase in planta, although Z. tritici multiply in liquid culture as yeast‐like blastospores.43 As previously mentioned, Hawking et al. discussed that mutations observed in laboratory mutants do not always occur in the field.37 In fact, two mutation types obtained as carbendazim‐resistant mutants in our study (compound mutation of Q134K with F265L and single mutation E198D) were not reported in field isolates of plant pathogens.22, 37 Although we could not find a deficiency in growth or pathogenicity of mutants selected by pyridachlometyl, it is not certain if resistant strains with mutations observed in this study will emerge after future adoption of pyridachlometyl in commercial fields. However, it would be better to assume that the risk of resistance exists for pyridachlometyl and to have a concrete strategy to control the risk of resistance development should it be established.
Table 5.
Growth rate at three different temperatures
| Growth Rate | ||||||||
|---|---|---|---|---|---|---|---|---|
| Mutation in tubulin | 12 °C | 18 °C | 27 °C | |||||
| Mutant | α‐tubulin | β‐tubulin | μ[h−1] a | t d[h] b | μ[h−1] | t d[h] | μ[h−1] | t d[h] |
| Set1 (wild type) | WT | WT | 1.0 | 0.70 | 1.3 | 0.54 | 1.5 | 0.46 |
| S13 | P325H | WT | 1.0 | 0.73 | 1.2 | 0.55 | 1.5 | 0.48 |
| S17 | P325S | WT | 1.0 | 0.69 | 1.4 | 0.49 | 1.8 | 0.40 |
| S8 | P325T | WT | 1.0 | 0.70 | 1.7 | 0.41 | 1.8 | 0.38 |
| S15 | P325T | WT | 1.0 | 0.69 | 1.1 | 0.60 | 1.5 | 0.47 |
| S5 | WT | N219K | 1.0 | 0.72 | 1.5 | 0.48 | 1.6 | 0.43 |
| S1 | WT | Y222N | 1.0 | 0.69 | 1.3 | 0.54 | 1.6 | 0.44 |
| S2 | WT | Y222N | 1.0 | 0.70 | 1.5 | 0.46 | 1.7 | 0.40 |
| S3 | WT | Y222N | 1.0 | 0.69 | 1.4 | 0.51 | 1.7 | 0.42 |
| S6 | WT | Y222N | 1.0 | 0.70 | 1.3 | 0.52 | 1.5 | 0.47 |
| S7 | WT | Y222N | 1.0 | 0.69 | 1.5 | 0.45 | 1.6 | 0.43 |
| S10 | WT | Y222N | 1.0 | 0.70 | 1.2 | 0.59 | 1.5 | 0.47 |
| S11 | WT | Y222N | 1.0 | 0.70 | 1.4 | 0.49 | 1.7 | 0.40 |
| S12 | WT | Y222N | 1.0 | 0.69 | 1.2 | 0.56 | 1.4 | 0.50 |
| S16 | WT | Y222N | 1.0 | 0.69 | 1.3 | 0.53 | 1.6 | 0.45 |
| S4 | WT | Y222S | 1.0 | 0.69 | 1.5 | 0.46 | 2.0 | 0.35 |
| S9 | WT | Y222S | 1.0 | 0.72 | 1.8 | 0.40 | 1.8 | 0.38 |
Specific growth rate μ[h−1] was calculated by linear approximation of ΔOD at the log phase (0 to 96 h for 12 °C, 0 to 72 h for 18 °C, 0 to 72 h for 27 °C) with the least squares method. Under these conditions, the R‐squared values of the calculated formula were always more than 0.9 for each wild type and mutant strain.
Doubling time (t d[h]) was calculated by formula t d[h] = loge 2/μ[h−1].
4. CONCLUSION
Pyridachlometyl is a new tool to control crop pathogenic fungi that have developed resistance to the existing mode of action. Our experiments demonstrated a broad spectrum of fungicidal activity for pyridachlometyl and also an absence of cross‐resistance between other major agricultural fungicides. The target site of pyridachlometyl is likely to be orthologous to the target site of triazolopyrimidines in mammalian tubulins. This target pocket between α‐ and β‐tubulin is clearly distinct from that of the known tubulin‐disrupting fungicide carbendazim. Some patterns of artificial mutation in tubulin were indicated and could be able to confer resistance to pyridachlometyl. Their relevance and the risk for emergence as field‐resistant mutants in the future should be further investigated.
Supporting information
Table S1. Origin of Zymoseptoria tritici strains in this study.
Table S2. Incubation conditions for antifungal activity tests†.
Table S3. Primer nucleotides used in this study.
Table S4. Sensitivity of each mutant against pyridachlometyl and other fungicides.
Table S5. GenBank accession numbers of protein sequences used in Fig. 4.
Figure S1. Close‐up view of locations of substitutions leading to resistance to carbendazim.
ACKNOWLEDGEMENTS
The authors would like to thank Yukie Uda and Akio Manabe of Sumitomo Chemical who provided technical assistance and helped with sample preparation, respectively.
REFERENCES
- 1. Balmford A, Amano T, Bartlett H, Chadwick D, Collins A, Edwards D et al, The environmental costs and benefits of high‐yield farming. Nat Sustainability 1:477–485 (2018). [PMC free article] [PubMed] [Google Scholar]
- 2. Popp J, Peto K and Nagy J, Pesticide productivity and food security. A review. Agron Sustain Dev 33:243–255 (2013). [Google Scholar]
- 3. Brent K, Historical perspectives of fungicide resistance, in Fungicide Resistance in Crop Protection: Risk and Management, ed. by Thind TS. CABI, Wallingford, pp. 141–154 (2012). [Google Scholar]
- 4. Hollomon DW, Fungicide resistance: 40 years on and still a major problem, in Fungicide Resistance in Plant Pathogens, ed. by Ishii H. and Hollomon DW. Springer, Tokyo, pp. 3–11 (2015). [Google Scholar]
- 5. Leadbeater A, Recent developments and challenges in chemical disease control. Plant Protect Sci 51:163–169 (2015). [Google Scholar]
- 6. Rehfus A, Strobel D, Bryson R and Stammler G, Mutations in sdh genes in field isolates of Zymoseptoria tritici and impact on the sensitivity to various succinate dehydrogenase inhibitors. Plant Pathol 67:175–180 (2018). [Google Scholar]
- 7. Kirikyali N, Diez P, Luo J, Hawkins N and Fraaije BA, Azole and SDHI sensitivity status of Zymoseptoria tritici field populations sampled in France, Germany and the UK during 2015, in Modern Fungicides and Antifungal Compounds, Vol. VIII, ed. by Deising HB, Fraaije B, Mehl A, Oerke EC, Sierotzki H. and Stammler G. Deutsche Phytomedizinische Gesellschaft, Braunschweig, pp. 153–158 (2017). [Google Scholar]
- 8. Cherrad S, Hernandez C, Vacher S and Steva H, First detection of boscalid‐resistant strains of Erysiphe necator in French vineyards: biological and molecular characterization, in Modern Fungicides and Antifungal Compounds, Vol. VIII, ed. by Deising HB, Fraaije B, Mehl A, Oerke EC, Sierotzki H. and Stammler G. Deutsche Phytomedizinische Gesellschaft, Braunschweig, pp. 211–216 (2017). [Google Scholar]
- 9. Rallos L and Baudoin A, Co‐occurrence of two allelic variants of CYP51 in Erysiphe necator and their correlation with over‐expression for DMI resistance. PLoS One 11:e0148025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rallos LEE, Johnson NG, Schmale DG, Prussin JA and Baudoin AB, Fitness of Erysiphe necator with G143A‐based resistance to quinone outside inhibitors. Plant Dis 98:1494–1502 (2014). [DOI] [PubMed] [Google Scholar]
- 11. Miyamoto T, Ishii H and Tomita Y, Occurrence of boscalid resistance in cucumber powdery mildew in Japan and molecular characterization of the iron‐sulfur protein of succinate dehydrogenase of the causal fungus. J Gen Plant Pathol 76:261–267 (2010). [Google Scholar]
- 12. Perez‐Garcia A, Romero D, Fernández‐Ortuño D, López‐Ruiz F, De Vicente A and Torés JA, The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol Plant Pathol 10:153–160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walker AS, Auclair C, Gredt M and Leroux P, First occurrence of resistance to strobilurin fungicides in Microdochium nivale and Microdochium majus from French naturally infected wheat grains. Pest Manag Sci 65:906–915 (2009). [DOI] [PubMed] [Google Scholar]
- 14. Kozawa T, Occurrence of QoI fungicide‐resistant strains of Microcohium nivale in Hokkaido, in Proceedings of the 24th Symposium of Research Committee on Fungicide Resistance; 2014 June 5. Sapporo; Tokyo, The Phytopathological Society of Japan: (2014). [Google Scholar]
- 15. Shimizu M, Decreased sensitivity to DMI‐fungicides in Cercospora beticola, the causal fungus of Cercospora leaf spot on sugar beet. Shokubutu Boueki 61:421–425 (2007). [Google Scholar]
- 16. Bolton MD, Rivera V and Secor G, Identification of the G143A mutation associated with QoI resistance in Cercospora beticola field isolates from Michigan, United States. Pest Manag Sci 69:35–39 (2013). [DOI] [PubMed] [Google Scholar]
- 17. Serey RA, Torres R and Latorre BA, Pre‐ and post‐infection activity of new fungicides against Botrytis cinerea and other fungi causing decay of table grapes. Cienc Inv Agr 34:215–224 (2007). [Google Scholar]
- 18. Pees K‐J and Albert G, Triazolopyrimidine derivatives with fungicidal activity. European Patent 550113 (1993).
- 19. Lamberth C, Trah S, Wendeborn S, Dumeunier R, Courbot M, Godwin J et al, Synthesis and fungicidal activity of tubulin polymerisation promoters. Part 2: Pyridazines. Bioorg Med Chem 20:2803–2810 (2012). [DOI] [PubMed] [Google Scholar]
- 20. Zhang N, Ayral‐Kaloustian S, Nguyen T, Afragola J, Hernandez R, Lucas J et al, Synthesis and SAR of [1,2,4]triazolo[1,5‐a]pyrimidines, a class of anticancer agents with a unique mechanism of tubulin inhibition. J Med Chem 50:319–327 (2007). [DOI] [PubMed] [Google Scholar]
- 21. Beyer CF, Zhang N, Hernandez R, Vitale D, Lucas J, Nguyen T et al, TTI‐237: a novel microtubule‐active compound with in vivo antitumor activity. Cancer Res 68:2292–2300 (2008). [DOI] [PubMed] [Google Scholar]
- 22. Young DH, Anti‐tubulin agents, in Fungicide Resistance in Plant Pathogens, ed. by Ishii H. and Hollomon DW. Springer, Tokyo, pp. 93–103 (2015). [Google Scholar]
- 23. Tangutur AD, Kumar D, Krishna KV and Kantevari S, Microtubule targeting agents as cancer chemotherapeutics: an overview of molecular hybrids as stabilizing and destabilizing agents. Curr Top Med Chem 17:2523–2527 (2017). [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Zhang H, Gigant B, Yu Y, Wu Y, Chen X et al, Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery. FEBS J 283:102–111 (2016). [DOI] [PubMed] [Google Scholar]
- 25. Hyams J. and Lloyd CW. eds, Microtubules. Wiley‐Liss, New York, NY: (1994). [Google Scholar]
- 26. Rai SS and Wolff J, Localization of the vinblastine‐binding ste on β‐tubulin. J Biol Chem 271:14707–14711 (1996). [DOI] [PubMed] [Google Scholar]
- 27. Sáez‐Calvo G, Sharma A, Balaguer FA, Barasoain I, Rodríguez‐Salarichs J, Olieric N et al, Triazolopyrimidines are microtubule‐stabilizing agents that bind the Vinca inhibitor site of tubulin. Cell Chem Biol 24:737–750 (2017). [DOI] [PubMed] [Google Scholar]
- 28. Morishita H and Manabe A, Pyridazine compound and use of thereof. JP Patent 4747680 (2005).
- 29. Nakaune R and Nakano M, Benomyl resistance of Colletotrichum acutatum is caused by enhanced expression of beta‐tubulin 1 gene regulated by putative leucine zipper protein CaBEN1. Fungal Genet Biol 44:1324–1235 (2007). [DOI] [PubMed] [Google Scholar]
- 30. Zhao Z, Liu H, Luo Y, Zhou S, An L, Wang C et al, Molecular evolution and functional divergence of tubulin superfamily in the fungal tree of life. Sci Rep 4:1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leroux P, Walker AS, Albertini C and Gredt M, Resistance to fungicides in French populations of Septoria tritici, the causal agent of wheat leaf blotch, in Proceedings of the Association of Applied Biologists Meeting, "Fungicide Resistance: Are we Winning the Battle but Losing the War?". Edinburgh, Wellesbourne, Association of Applied Biologists: (2006). [Google Scholar]
- 32. Edwards SG, Pirgozliev SR, Hare MC and Jenkinson P, Quantification of trichothecene‐producing Fusarium species in harvested grain by competitive PCR to determine efficacies of fungicides against Fusarium head blight of winter wheat. Appl Environ Microbiol 67:1575–1580 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oakley CE, Tubulins in Aspergillus nidulans . Fungal Genet Biol 41:420–427 (2004). [DOI] [PubMed] [Google Scholar]
- 34. Morrissette NS, Mitra A, Sept D and Sibley LD, Dinitroanilines bind α‐tubulin to disrupt microtubules. Mol Biol Cell 15:1960–1968 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lyons‐Abbott S, Sackett DL, Wloga D, Gaertig J and Morgan RE, Werbovetz KA and Morrissette NS, α‐tubulin mutations alter oryzalin affinity and microtubule assembly properties to confer dinitroaniline resistance. Eukaryot Cell 12:1825–1834 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu Z, Chen J, Nyporko A, Han H, Yu Q and Powles S, Novel α‐tubulin mutations conferring resistance to dinitroaniline herbicides in Lolium rigidum . Front Plant Sci 9:97 (2018). 10.3389/fpls.2018.00097. eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hawkins NJ and Fraaije B, Predicting resistance by mutagenesis: lessons from 45 years of MBC resistance. Front Microbiol 7:1814 eCollection (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Piedra FA, Kim T, Garza ES, Geyer EA, Burns A, Ye X et al, GDP‐to‐GTP exchange on the microtubule end can contribute to the frequency of catastrophe. Mol Biol Cell 27:3515–3525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ishida T, Kaneko Y, Iwano M and Hashimoto T, Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana . Proc Natl Acad Sci USA 104:8544–8549 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nogales E, Whittaker M, Milligan RA and Downing KH, High‐resolution model of the microtubule. Cell 96:79–88 (1999). [DOI] [PubMed] [Google Scholar]
- 41. Koenraadt H, Somerville SC and Jones AL, Characterization of mutations in the beta‐tubulin gene of benomyl‐resistant field strains of Venturia inaequalis and other plant pathogenic fungi. Phytopathology 82:1348–1354 (1992). [Google Scholar]
- 42. Takeshita N, Coordinated process of polarized growth in filamentous fungi. Biosci Biotechnol Biochem 80:1693–1699 (2016). [DOI] [PubMed] [Google Scholar]
- 43. Quaedvlieg W, Kema GH, Groenewald JZ, Verkley GJ, Seifbarghi S, Razavi M et al, Zymoseptoria gen. Nov.: a new genus to accommodate Septoria‐like species occurring on graminicolous hosts. Persoonia 26:57–69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Origin of Zymoseptoria tritici strains in this study.
Table S2. Incubation conditions for antifungal activity tests†.
Table S3. Primer nucleotides used in this study.
Table S4. Sensitivity of each mutant against pyridachlometyl and other fungicides.
Table S5. GenBank accession numbers of protein sequences used in Fig. 4.
Figure S1. Close‐up view of locations of substitutions leading to resistance to carbendazim.


