Summary
The international C4 rice consortium aims to introduce into rice a high capacity photosynthetic mechanism, the C4 pathway, to increase yield. The C4 pathway is characterised by a complex combination of biochemical and anatomical specialisation that ensures high CO2 partial pressure at RuBisCO sites in bundle sheath (BS) cells. Here we report an update of the progress of the C4 rice project. Since its inception in 2008 there has been an exponential growth in synthetic biology and molecular tools. Golden Gate cloning and synthetic promoter systems have facilitated gene building block approaches allowing multiple enzymes and metabolite transporters to be assembled and expressed from single gene constructs. Photosynthetic functionalisation of the BS in rice remains an important step and there has been some success overexpressing transcription factors in the cytokinin signalling network which influence chloroplast volume. The C4 rice project has rejuvenated the research interest in C4 photosynthesis. Comparative anatomical studies now point to critical features essential for the design. So far little attention has been paid to the energetics. C4 photosynthesis has a greater ATP requirement, which is met by increased cyclic electron transport in BS cells. We hypothesise that changes in energy statues may drive this increased capacity for cyclic electron flow without the need for further modification. Although increasing vein density will ultimately be necessary for high efficiency C4 rice, our modelling shows that small amounts of C4 photosynthesis introduced around existing veins could already provide benefits of increased photosynthesis on the road to C4 rice.
Keywords: C4 photosynthesis, rice, metabolic engineering, bundle sheath cells, plasmodesmata, photosynthetic electron transfer
Significance Statement
Installing a C4 CO2 concentrating mechanism into rice is arguably an ambitious molecular engineering initiative in plant science. This review maps the progress of the C4 rice project since its inception more than 10 years ago. Photosynthetic functionalisation of the bundle sheath compartment and manipulating leaf anatomy remain challenging but rapid progress in installing C4 biochemistry and manipulating multiple traits in a single genetic transformation has been made possible by advances in synthetic biology.
Introduction
The C4 photosynthetic pathway is characterised by a complex combination of biochemical and anatomical specialisation, which provides an elevation of the CO2 partial pressure at the site of ribulose bisphosphate carboxylase oxygenase (RuBisCO) in leaf bundle sheath (BS) cells. CO2 is initially assimilated into C4 acids by phosphoenolpyruvate (PEP) carboxylase in mesophyll (M) cells. These acids then diffuse to and are decarboxylated in BS cells in which CO2 is concentrated. This spatial separation of initial CO2 fixation in M cells and subsequent refixation by RuBisCO in BS cells is mostly associated with Kranz anatomy, but can also occur in single cells (Bowes and Salvucci, 1984; Voznesenskaya et al., 2001). C4 photosynthesis has evolved independently more than 60 times, providing one of the most widespread and effective solutions for overcoming the catalytic inefficiency of RuBisCO (Sage et al., 2012; Christin and Osborne, 2013). Because of the superior nitrogen and water use efficiency of C4 plants, transfer of C4 photosynthetic traits has been an early strategy for improving C3 photosynthesis (Sage, 2004; Miyao et al., 2011). Initial attempts were made to generate hybrids between closely related C3 and C4 plants by conventional crossing (reviewed in Brown and Bouton, 1993). With the advent of transformation technology there were numerous attempts to introduce C4 photosynthetic genes into C3 species (reviewed in Matsuoka et al., 2001; Hausler et al., 2002; Miyao, 2003). These all sought to insert a C4 like pathway into M cells of C3 species following the blue print of the aquatic single cell organism Hydrilla verticillata (Bowes and Salvucci, 1984; von Caemmerer et al., 2014). Four enzymes of the C4 photosynthetic pathway were successfully introduced into rice by Miyao and collaborators and although no photosynthetic gains were observed, Miyao et al. (2011) have elegantly summarised the extensive contribution this endeavour made to our understanding of gene expression and regulation of C4 genes expressed in rice. Current progress on building a two cell C4 pathway in rice are built on these earlier insights.
The current C4 rice project (https://c4rice.com/) which aims to introduce Kranz anatomy into rice was first conceived by John Sheehy (Mitchell and Sheehy, 2006) who invited a group of C4 photosynthesis experts to the international rice research institute (IRRI) in the Philippines to discuss the potential of C4 rice. Engineering the C4 pathway into a C3 plant requires manipulation of both anatomical and biochemical traits and progress of this research has been reviewed a number of times (Hibberd et al., 2008; Hibberd and Covshoff, 2010; Langdale, 2011; Sedelnikova et al., 2018). To introduce Kranz anatomy into rice requires a change of vein spacing patterns so that veins are closer together in the leaf and BS cells need to be ‘functionalised’ for increased photosynthetic capacity, including increased chloroplast content. At this point while there are established candidates for the genes and transcription factors that potentially control vein spacing, the complete transcriptional network remains to be elucidated (for review see Sedelnikova et al., 2018). However, some progress has been made in photosynthetic functionalisation of the rice BS (Wang et al., 2017b) While insertion of C4 biochemistry in rice faces challenges of engineering high level and cell‐specific gene expression (Hibberd and Covshoff, 2010), genes encoding the C4 pathway enzymes and most of the metabolite transporters have now been identified. Here we review current progress and how they have been enabled by technological advances in cloning techniques and highlight future challenges.
Building the biochemistry for C4 rice
In 2008, the year the C4 rice consortium commenced, a remarkable amount of information in regard to the genes encoding the key proteins in the C4 pathway was already known. The cDNA sequences for maize phosphoenolpyruvate carboxylase (PEPC), pyruvate orthophosphate dikinase (PPDK), and NADP‐malate dehydrogenase (MDH) had all been reported and expressed in the M of cells of rice (reviewed in Miyao et al., 2011). The cDNA sequence for NADP‐malic enzyme (NADP‐ME) had also been reported for both the rice endogenous gene and the maize gene (Drincovich et al., 2001). The genes encoding carbonic anhydrase (CA) in maize had also been cloned by this time, but the precise identity of the gene product located in the cytosol of the M cells in C4 leaves was not definitively proven (reviewed in DiMario et al., 2016). Therefore, the key genes encoding the entire biochemical pathway of the NADP‐ME type C4 mechanism as shown in Figure 1 were all available at the commencement of the C4 rice project.
Figure 1.
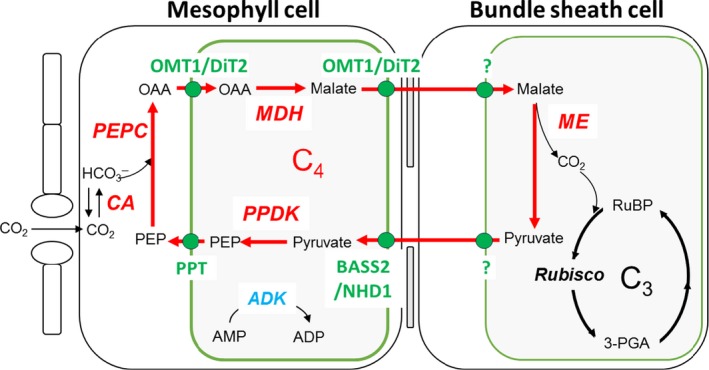
Enzymes and transporters included in the construction of C4 rice. These include carbonic anhydrase (CA), PEP carboxylase (PEPC), malate dehydrogenase (MDH), NADP‐malic enzyme (ME), and pyruvate Pi dikinase (PPDK). Adenylate kinase (ADK) has been added to ensure AMP is converted to ADP. The required transporters are oxoglutarate/malate transporter (OMT1), PEP/phosphate translocator (PPT), dicarboxylate transporter (DiT2), pyruvate/sodium symporter (BASS2), and sodium/proton antiporter (NHD1). Malate importer and pyruvate exporter in bundle sheath chloroplasts are not yet confirmed. Other abbreviations used are OAA for oxaloacetate and PEP for phosphoenolpyruvate.
What set the current C4 rice project apart from previous activities was the desire to engineer a full Kranz two cell‐type mechanism in rice (von Caemmerer et al., 2012). This approach requires not only the cDNA sequences for the relevant photosynthetic proteins but high level expression with suitable promoters in the appropriate cell type of rice. For M cell‐specific expression, the promoters of genes from C4 species encoding PEPC, PPDK, and aspartate aminotransferase (AspAT) had all been tested in C3 species and lead to M‐specific accumulation of the β glucuronidase (GUS) reporter protein (Hibberd and Covshoff, 2010). The PEPC promoter from maize had been the most extensively tested in rice and various truncated versions have been shown to produce M‐specific expression of the reporter gene GUS in rice (Hibberd and Covshoff, 2010).
High level BS cell‐specific expression of proteins such as NADP‐ME has proven to be the greatest challenge for establishing a C4 metabolic pathway in rice. While there are potential anatomical constraints in regard to the photosynthetic competence of the rice BS compartment (Wang et al., 2017b), the paucity of promoters available to drive BS expression in a C3 plant has been a major obstacle. At the commencement of the C4 rice project, the promoters of the genes encoding phosphoenolpyruvate carboxykinase (PEPCK), NADP‐ME, AspAT, small subunit (Engelmann et al.) of RuBisCO, and the P subunit of glycine decarboxylase (GDCP) from C4 species had been tested in C3 plants (Chen et al., 2001; Nomura et al., 2005a; Nomura et al., 2005b; Engelmann et al., 2008). Of these, a version of the Zoysia japonica PEPCK promoter (Nomura et al., 2005a) resulted in BS‐specific GUS accumulation in rice. The Flaveria trinervia GDCP promoter was shown to be BS/ vascular‐specific in Arabidopsis and the promoter of this gene from Flaveria anomala (Chen et al., 2001) was vascular‐specific in rice. Both the PEPCK and GDCP promoters have subsequently been utilised in the C4 rice strategy although promoter strength has been an ongoing issue for engineering efforts and it is notable that in the single cell C4 project (reviewed in Miyao et al., 2011), genomic clones often but not always gave superior expression levels compared with cDNAs driven by their own promoters.
Stacking of genes in transgenic rice in the initial phases of this project required the crossing of homozygous lines harbouring single gene constructs to build a rice plant expressing a complete set of the genes encoding the major enzymes of the C4 pathway. Such a crossing strategy was a hugely time consuming effort as expression levels and cell‐specific expression of the recombinant protein must be checked for each line and a crossing donor identified for each transgene. With segregation of the transgenes inserted at different loci in the stacked lines, many hundreds of individuals had to be genotyped at each cross to obtain a single line harbouring the genes encoding the five key photosynthetic enzymes shown in Figure 1. Indeed, this crossing strategy has taken almost 6 years to achieve in indica rice in the current project. However, five genes are not enough for creating C4 rice and the biochemical pathway is being complemented by a suite of membrane transporters ensuring fast transport of metabolites between cell compartments. Most of the transporters have now been identified and are listed in Figure 1, however there is still some uncertainty about malate import to the BS chloroplast and the export of pyruvate following malate decarboxylation. A recent review of transporters was given by Schuler et al. (2016). Physiological gas exchange techniques exist to allow for the identification of a complete, functioning C4 pathway. These include measurements of CO2 compensation points, reduced oxygen sensitivity of CO2 assimilation and reduced carbon isotope discrimination (Furbank et al., 2009). In the meantime new 13C pulse chase labelling techniques have been developed to analyse the paths of carbon during C3 and C4 photosynthesis (Arrivault et al., 2009) and confirmed that malate is in fact being formed in our current five gene rice transgenics.
Synthetic biology accelerates the pace
A consensus definition drafted by a group of European experts more than a decade ago defined synthetic biology as follows: ‘Synthetic biology is the engineering of biology: the synthesis of complex, biologically based (or inspired) systems, which display functions that do not exist in nature. This engineering perspective may be applied at all levels of the hierarchy of biological structures—from individual molecules to whole cells, tissues and organisms. In essence, synthetic biology will enable the design of ‘biological systems’ in a rational and systematic way’ (Serrano, 2007). Synthetic biology has evolved and adopted many of the commonly used terms in mainstream engineering such as ‘switch’, ‘rewire’, ‘design, test and redesign cycle’. Although in the creation of C4 rice, in which a template or design already existing in nature is being used, the installation of up to 20 genes to completely ‘rewire’ rice metabolism and anatomy surely fits the definition above. A major limitation in the synthetic biology approach however is the cycle time for the design, test and redesign in crops such as rice.
The last 5 years have seen an exponential growth in synthetic biology tools and the cost of gene synthesis has plummeted. This has enabled the C4 rice consortium to adopt a more rapid cycle of design, test and prototype coupled to the adoption of a rapid Agrobacterium‐based rice transformation system in the japonica rice variety ‘Kitaake’. Kitaake is fast flowering, day neutral, small in stature and an established model for functional genomics studies (Li et al., 2017). The obstacle of genetic transformation with a single gene construct at a time and crossing has largely been solved by gene synthesis and Golden Gate cloning or similar ‘gene building block’ approaches (Engler et al., 2014). Gene synthesis allows the ‘domestication’ of coding sequences to remove or insert rare Type IIS restriction enzyme recognition sites while leaving the amino acid sequence unchanged, thus enabling the assembly of gene modules which can be pasted together, often in a ‘one pot cloning’ approach. Assembly of these modules into a T‐DNA suitable for Agrobacterium transformation is therefore greatly accelerated over traditional restriction/ligation approaches (Andreou and Nakayama, 2018). In principle, assembling all the metabolic and transporter components of Figure 1 for rice transformation on a single construct should be readily achievable and this work is currently underway. This approach has so far enabled a 6‐year crossing strategy to be reduced to a 6‐month single transformation experiment in rice.
In a large multigene overexpression construct, it is not desirable to reuse ‘parts’ multiple times due to the possibility of recombination deletion, post‐transcriptional gene silencing or inactivation at the promoter level via methylation (Wassenegger, 2002). Epigenetic promoter silencing is a poorly understood process and can be a major challenge for metabolic engineering. This also presents a challenge for the design of the gene constructs described above for C4 rice. For example, expression of just CA, PEPC, MDH and PPDK would ideally require four heterologous M‐specific promoter sequences. While this may be possible for the M compartment, it is not for the BS compartment (see above).
Synthetic biology has also provided a potential solution to this paucity of promoters in rice. Brückner et al. (2015) described a system compatible with the Golden Gate cloning which utilises multiple promoters (Synthetic TALE Activated Promoters or STAPs) designed to be orthogonal to the genome of the plant to be transformed, which can be activated by a single Transcription Activator‐Like Effector (TALE). This approach provides the opportunity to build multiple transcriptional units driven by different promoters on the same gene construct, trans‐activated by a single transcription factor.
Functionalisation of the bundle sheath: an anatomical roadblock?
Figure 2(a) shows fresh transverse sections of a rice (C3) and Setaria viridis (C4) leaf imaged with the laser confocal microscope using chlorophyll fluorescence overlaid with cell wall fluorescence. This enables visualisation of the chloroplast contents of BS cells in each species. This image clearly indicates that the BS compartment of the C4 leaf is tightly packed with chloroplasts, whereas the rice BS compartment is only sparsely populated by chloroplasts and the cells are highly vacuolated. This has previously been pointed out and ‘photosynthetic functionalisation’ of the BS has been proposed as an early step in evolution of C4 photosynthesis, probably occurring at the C2 or proto‐Kranz stage (Sage, 2004; Wang et al., 2017b). The underlying mechanisms responsible for the proliferation of chloroplasts in BS cells of C4 leaves are largely unknown. However, recently a transgenic approach has been used in rice to ‘recreate’ this key step in evolution by overexpressing the transcription factors GOLDEN2 (G2) or GOLDEN2‐LIKE (GLK) (Wang et al., 2017b) which had been implicated as important in BS cell differentiation in terrestrial plants including Zea mays and more recently in rice (Wang et al., 2013 and references therein). These transcription factors are thought to act in the cytokinin signalling pathway (Wang et al., 2013). Overexpression of these Z. mays transcription factors in rice indeed increased the proportion of vascular cell area occupied by chloroplasts, the mitochondrial population in rice BS cells, and the plasmodesmatal connectivity at the BS/M cell interface (Wang et al., 2017b). The BS chloroplast abundance data from this work are summarised in Figure 3. It is evident from these quantitative data that overexpression of this class of transcription factor can increase BS chloroplast content as a proportion of total leaf chloroplasts by approximately five‐fold (in the case of G2 expressed from the ubiquitin promoter of Z. mays: UBI‐G2). However, another three‐fold increase is required to reach levels equivalent to C2 leaves and potentially another six‐fold to reach C4 levels of chloroplast area in the BS. This can be visualised in Figure 2 which shows a representative image of a leaf transverse (a) and paradermal (b) sections of the UBI‐G2 lines (Wang et al., 2017b) compared with rice and Setaria leaves. Comparison of rice and C4 leaf chloroplast distribution is complicated, however, by the change in vein spacing and reduction in M chloroplast numbers seen in C4 leaves relative to C3 (Stata et al., 2016).
Figure 2.
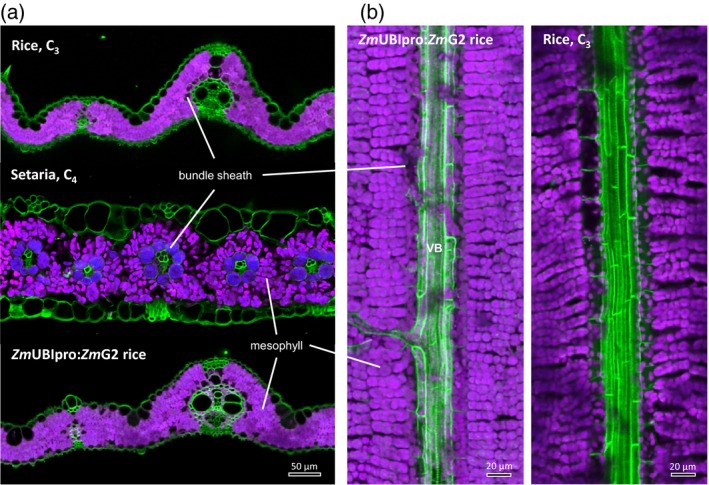
Confocal micrographs of fresh hand‐cut transverse (a) and paradermal (b) leaf sections of Oryza sativa (rice), Setaria viridis (Setaria), and a rice line with constitutively expressed GOLDEN2 transcription factor from Zea mays (ZmUBIpro:ZmG2 rice). Excitation wavelength at 633 nm and dual emission wavelengths at 650–720 nm (magenta for Photosystem II) and 720–800 nm (blue for Photosystem I) were used to visualise chloroplasts. Note that bundle sheath cells of Setaria leaf have chloroplasts appearing pseudo‐blue due to reduced Photosystem II content. Cell wall (green) was visualised at an excitation wavelength of 405 nm and emission wavelength of 420–480 nm. VB, vascular bundle.
Figure 3.
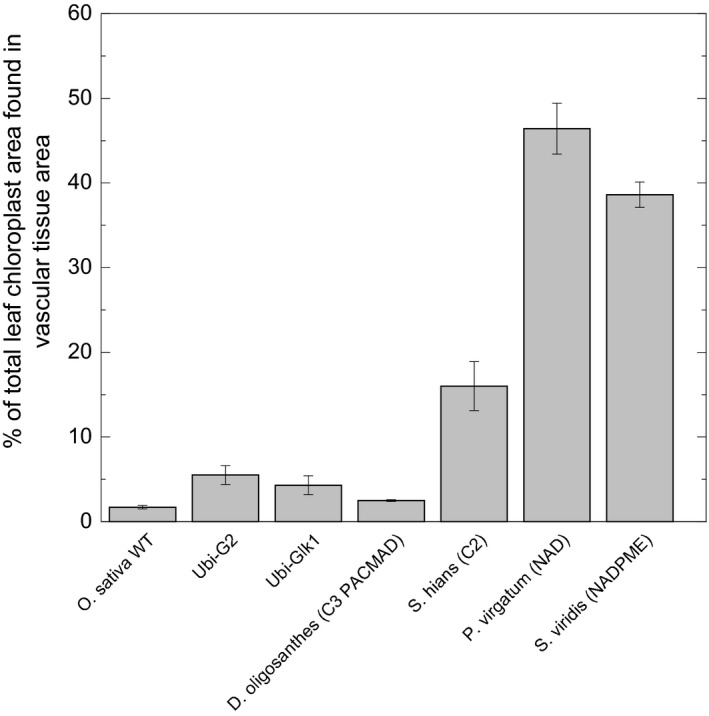
Per cent of total chloroplast area found in vascular tissue area. The data are taken from Table S5 Wang et al. (2017b). Ubi‐G2 and Ubi‐Glk1 are Oryza sativa plants in which the transcription factor GOLDEN2 (G2) or GOLDEN2‐LIKE1 (GLK1) has been overexpressed from the ubiquitin promoter of Zea mays (Wang et al., 2017b). Other monocots shown are Dichanthelium oligosanthes, a C3 from the PACMAD clade; Steinchisma hians, which operates the C2 photosynthetic pathway; Panicum virgatum, a C4 species with the NAD‐ME decarboxylation type; and Setaria viridis, a C4 species with the NADP‐ME decarboxylation type.
Alternative approaches are also under investigation for increasing chloroplast abundance/ volume in the BS compartment of rice (Wang et al., 2017a). The transcription factor CYTOKININ RESPONSIVE GATA FACTOR 1 (CGA1) has also been shown to regulate leaf chloroplast abundance via the cytokinin signalling network (Chiang et al., 2012; Hudson et al., 2013; Wang et al., 2017a) and overexpression of CGA1 in rice resulted in a 30% increase in flag leaf chlorophyll and close to a doubling of chloroplast numbers on a fresh weight basis (Hudson et al., 2013). It has been proposed that regulation of the FtsZ chloroplast division gene by CGA1 is responsible for this phenotype, presenting opportunities for regulating levels of this protein directly or other genes in this pathway controlling chloroplast division (Hudson et al., 2013; Wang et al., 2017a).
It may be that part of the difficulty obtaining high level expression of chloroplast proteins in rice BS cells is the lack of sufficient chloroplast volume to house the recombinant proteins targeted to this compartment. The developmental programme of BS cells in a C3 grass may reflect a more ‘parenchyma‐like’ role in temporary sugar storage and the sugar status of the BS cells may not be conducive to photosynthetic functionalisation. The high level expression of sugar effluxers such as the SWEET13 gene family in the BS of C4 plants has been proposed as evidence that sugar status in the BS or C4 grasses may be quite different from that in C3 grasses (Emms et al., 2016). In addition, partitioning of starch almost exclusively into the BS of C4 grasses is a curious and potentially relevant observation (Lunn and Furbank, 1999).
What can we learn from comparative leaf anatomy between C3 and C4 grass species?
It has previously been proposed that an early step in C4 evolution was ‘inflation’ of BS cell size (Sage, 2004; Sage et al., 2012; Christin et al., 2013). However, our results from anatomical measurements performed in 25 grass species, representing different photosynthetic types and seven independent C4 evolutionary origins (Figure 4a), reveal that C4 leaves do not necessarily have larger BS cells, nor do they always have shorter interveinal distances than C3 leaves (Figure 4b). Rather, a C4 leaf has greater BS surface area per leaf area (Sb), fewer interveinal M cells than a C3 leaf and most notably, more plasmodesmata (PD) at the BS/M cell interfaces (Figure 4; Danila et al., 2016; Danila et al., 2018). For a functional C4 rice, these findings mean that increasing PD connections between M and BS, increasing Sb, and reducing M cells between veins of the rice leaves may be essential for the operation of an efficient photosynthetic pathway.
Figure 4.
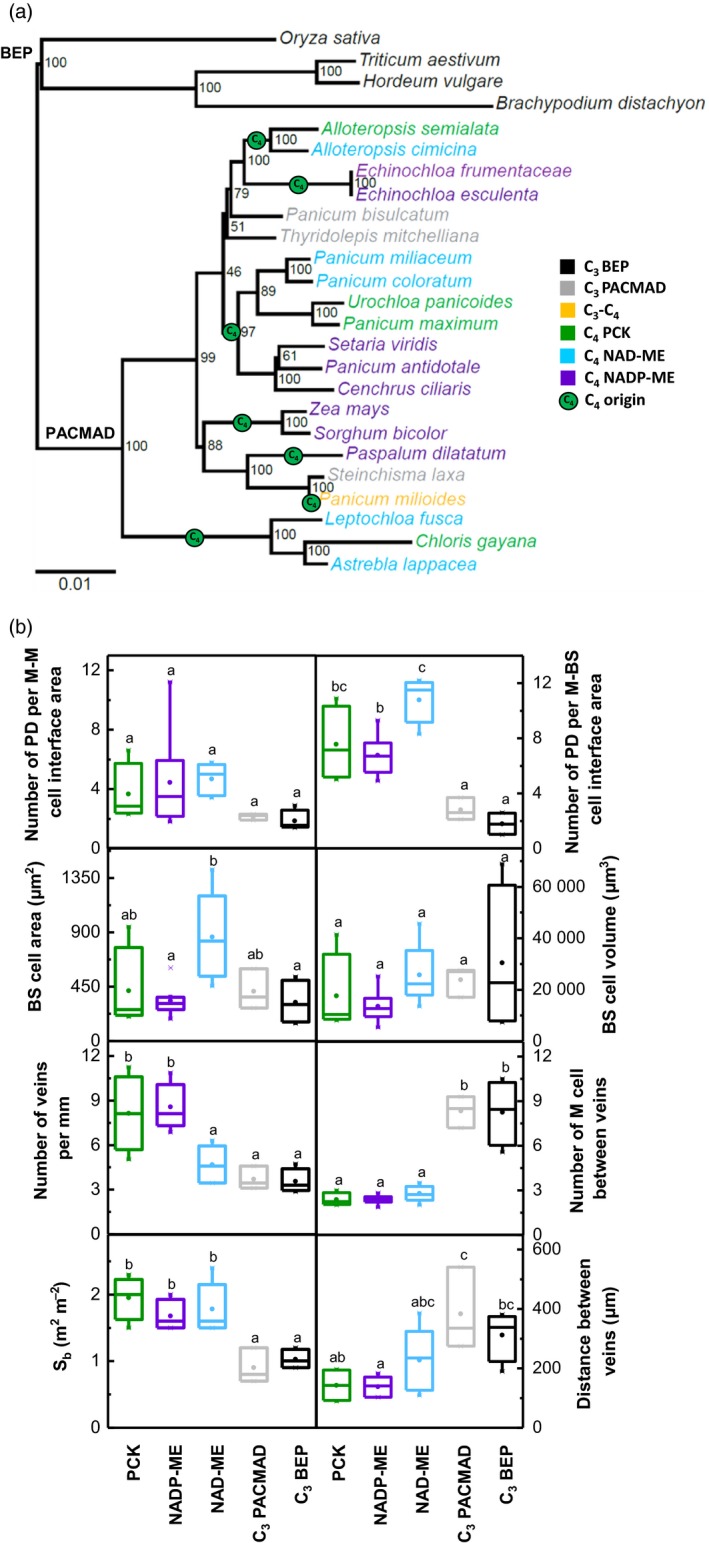
(a) Phylogenetic tree of the C3 and C4 grass species examined generated using sequences from ndhF and rbcL chloroplast genes. Species names are colour‐coded according to photosynthetic types. The seven independent evolutionary origins of C4 photosynthesis according to (GPWGII, 2012) are indicated with green circles at the midpoint of the branches. Note that Panicum milioides, a.k.a. Steinchisma hians, although technically a C3–C4 intermediate (Duvall et al., 2003) was categorised as C4 in GPWGII (2012). Numerical value at internal nodes is the percentage of non‐parametric bootstrap replicates that support the bipartition. Scale bar indicates amino acid substitutions per site.
(b) Distribution of leaf trait values among photosynthetic types in (a) excluding the C3–C4 intermediate type, in which only one species was measured. The distribution of eight variables is summarised by boxplots. Box and whiskers represent the 25 to 75 percentile, and the minimum and maximum distribution. Means are denoted by (•). Letters show the statistical ranking using a post‐hoc Tukey test among photosynthetic types (different letters indicate differences at P‐value < 0.05). BS, bundle sheath; M, mesophyll; PD, plasmodesmata; Sb, bundle sheath surface area per unit leaf area.
It has long been thought that a key feature of C4 leaf anatomy was increased abundance of the symplastic nanochannels (PDs) that facilitate the rapid exchange of metabolites between M and BS cells during C4 photosynthesis (Hatch and Osmond, 1976). Recent work, however, has provided quantitative data on this parameter (Danila et al., 2016; Danila et al., 2018); Figure 4). Enhancement of the PD connections between M and BS of rice to levels observed in closely related C4 species would require increases of at least five‐fold (Danila et al., 2018). While there are few genes known which control PD development and proliferation, it was recently observed that when maize GLK genes were constitutively expressed in rice, increased organelle volume was accompanied by increased M‐BS PD density; although the enhancement is only double that of wild type (Wang et al., 2017b). Coordination of chloroplast and PD development has been suggested previously (Brunkard et al., 2013) and cytokinin has also been implicated in the proliferation of PDs in the shoot apical meristem (Ormenese et al., 2006). These findings offer some hope that a single gene or at least an established transcriptional network could provide a master switch for C4 BS anatomy.
The BS surface area per leaf area (Sb) is a physiological parameter which has proven to be an important feature of modelling the M‐BS interface, including to estimate BS conductance to CO2 diffusion (first estimates ranged between 0.6 m2 m−2 and 3.1 m2 m−2 (Apel and Peisker, 1978; Brown and Byrd, 1993)). Sb was obtained by dividing measurements of BS tissue perimeter from micrographs by interveinal distance (Pengelly et al., 2010). Increasing Sb can be achieved in multiple ways; modification of BS cell size, vasculature size, and the distance between BS. Because BS cell size does not differ substantially between C3 and C4 grass species (Figure 4, Danila et al., 2018), reducing the interveinal distance appears to be the most logical path to increase Sb in rice. Ideally, this would mean upregulation of gene(s) that would promote insertion of additional veins between existing veins of rice, thus reducing interveinal distance and, at the same time, decreasing the number of M cells between veins (Sedelnikova et al., 2018; Hughes et al., 2019).
Paying the energy cost of C4 photosynthesis
The energy cost of C4 photosynthesis is significantly higher compared with C3 as it requires a minimum of two more ATP molecules per one CO2 fixed (Furbank et al., 1990). While high conductance of the M–BS interface to metabolites is essential for the operation of C4 photosynthesis, it also allows a proportion of the CO2 concentrated in the BS to escape back to M (called CO2 leakage; von Caemmerer and Furbank, 2003). This CO2 is either refixed or lost to the intercellular spaces in the M, which increases the cost of C4 photosynthesis (Furbank et al., 1990; von Caemmerer and Furbank, 2003). To sustain higher energy requirements, C4 plants adapt the photosynthetic electron transfer chains of M and BS cells depending on specific needs of the C4 subtype they belong to (Munekage and Taniguchi, 2016). As the efforts of the C4 rice project have targeted NADP‐ME as the decarboxylating enzyme, we focus here on specific energy requirements of the NAPD‐ME subtype of C4 photosynthesis.
An early observation on the anatomy of tropical grasses which predated the discovery of C4 photosynthesis was that chloroplasts in the two cell types of leaves with Kranz anatomy are dimorphic, with the BS chloroplasts often lacking grana stacks (Rhoades and Carvalho, 1944). Subsequently, it was discovered that these grasses such as Z. mays were in fact C4 plants and specifically used the NADP‐ME pathway of C4 photosynthesis (Edwards et al., 1971; Hatch and Kagawa, 1976). As NADP‐ME plants primarily use malate as a C4 acid diffusing from M to BS cells, NADPH produced in the M chloroplasts is consumed for malate synthesis but it is then produced in the BS upon malate decarboxylation to pyruvate (Figure 1). The net transfer of NADPH from M to BS cells means that there is a reduced requirement for linear electron flow to produce NADPH in the BS chloroplast, which have reduced Photosystem II (PSII) content but have highly developed cyclic electron flow (CEF) machinery around Photosystem I (PSI) (Figure 5). Therefore, the observation that BS chloroplasts of NADP‐ME plants are mostly agranal with little or no grana thylakoids functionally reflects the reduced PSII, which would normally be located in the granal stacks (see Munekage and Taniguchi, 2016). However, grana content in BS chloroplasts of NADP‐ME plants is rather variable between species (Ueno et al., 2005) and also within species in response to environmental conditions (Omoto et al., 2009; Danila et al., 2019) and depending on leaf age (Andersen et al., 1972). It has also been shown that aspartate can be used as a transported C4 acid in many NADP‐ME type C4 plants, providing some flexibility in the amount of reducing power transferred to the BS from the M and suggesting that PSII content in the BS cells might in fact respond to the NADPH/NADP+ ratio or the redox state of the BS cells (see Furbank, 2011). This is supported by overexpression of maize NADP‐ME in rice M cells which resulted in agranal chloroplasts, conceivably, due to the increased NADPH/NADP+ ratio depleting PSI of electron acceptor and causing over‐reduction of the electron transfer chain (Takeuchi et al., 2000). While prolonged reduction of PSII acceptors promotes the formation of reactive oxygen species and causes damage and degradation of PSII (Vass, 2012) and thus also grana, there might be also regulatory mechanisms preventing transcription and de novo assembly of PSII polypeptides and grana formation in over‐reduced conditions (Pfannschmidt et al., 1999).
Figure 5.
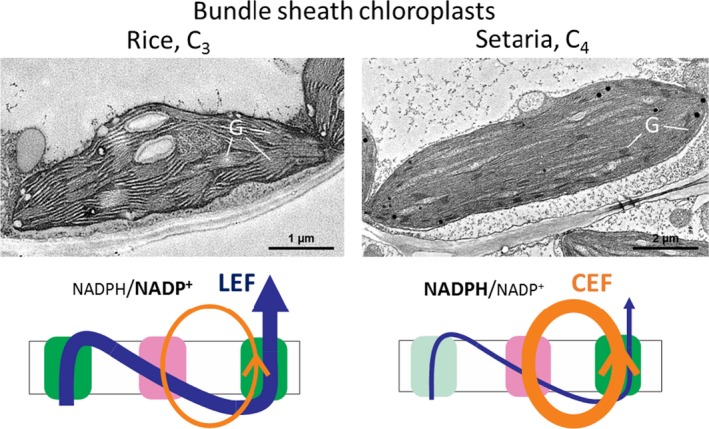
Transmission electron micrographs of the bundle sheath chloroplasts from Oryza sativa (rice) and Setaria viridis (Setaria) and schematic representation of their electron flow pathways. LEF, linear electron flow, is supported by Photosystem II, Cytochrome b 6 f complex and Photosystem I and is dominant in rice bundle sheath chloroplasts. CEF, cyclic electron flow, does not require Photosystem II and is dominant in Setaria bundle sheath chloroplasts. G, thylakoid grana stacks, are abundant in rice but not in Setaria because bundle sheath chloroplasts of NADP‐ME C4 plants require less Photosystem II that is typically localised to grana. NADPH/NADP+ balance in chloroplasts might be a factor defining the ratio between LEF and CEF.
It has been proposed that elevated CEF in BS chloroplasts allows NADP‐ME plants to accommodate the extra costs of C4 photosynthesis by producing ATP without affecting NADPH/NADP+ ratio (Furbank et al., 1990). The chloroplast NADPH dehydrogenase‐like complex and PROTON GRADIENT REGULATION 5 (PGR5) mediate two different CEF routes (Takabayashi et al., 2005; Munekage, 2016). Interestingly, in C3 plants, CEF is promoted in conditions causing over‐reduction of the electron transport chain (Suorsa, 2015) and therefore CEF might be naturally upregulated in BS cells of C4 rice in response to high NADPH/NADP+ ratio.
At present, it is unknown whether alteration of BS chloroplast electron transport components is a strict requirement for C4 rice or only ‘fine tuning’. If the required regulatory mechanisms already exist in rice, BS chloroplasts could conceivably adjust PSII and grana content according to the redox state of cells in C4 rice (Figure 5). Consequently, just the right amount of PSII will be fully assembled in BS to donate electrons for the CEF and compensate for the shortage of NADPH via linear electron transfer. However, the desired adaptation of electron transport requires a strictly coordinated expression and activity of NADP‐ME in rice BS cells and efficient NADPH oxidation by the Calvin cycle to maintain an appropriate NADPH/NADP+ balance. It is worth mentioning that the flexibility of PGA reduction between M and BS cells in C4 plants also contributes to the maintenance of NADPH/NADP+ balance in BS cells, however, it is not clear whether this pathway will be immediately available in C4 rice. The composition of thylakoid protein complexes and energy requirements are similar between C3 and C4 M cells (Munekage and Taniguchi, 2016), but rice BS chloroplasts will require some reorganisation of thylakoid complexes and increased abundance of the NADPH dehydrogenase‐like complex (Majeran et al., 2008; Hernández‐Prieto et al., 2019). This fine tuning will be necessary to run an efficient C4 pathway as our recent research shows that the rate of C4 photosynthesis is strongly dependent on the electron transport capacity of both M and BS cells (Ermakova et al., 2019).
Partial C4: modelling mixed C3 and C4 photosynthesis on the path to C4 rice
The concept of C4 rice has excited photosynthetic modellers and there are a number of photosynthetic models that have tried to evaluate the efficacy of introducing C4 photosynthesis into rice (Bellasio, 2016; Yin and Struik, 2017; Wang et al., 2017c; Bellasio and Farquhar, 2019). Each of these models has a different focus. Wang et al. (2017c) developed a 3D reaction diffusion model of BS and connected M cells with anatomy based on a C3 rice leaf in which C4 photosynthesis was integrated with existing C3 photosynthesis. They concluded that the C4 cycle can operate adjacent to C3 photosynthesis in rice M cells, but that the energy partitioning between the C3 and C4 cycle is an important consideration. In their current model every M cell is adjacent to a BS cell so it is built on a rice leaf with altered vein spacing. Bellasio’s models consider C4 photosynthesis as an addition to C2 photosynthesis and also provides a valuable discussion on energy partitioning. Yin and Struik (2017) consider C4 photosynthesis in rice at the canopy and crop level in different environments.
It is common to compare rice with Z. mays but it is informative to widen this comparison. The analysis of 25 monocot C3 and C4 leaves shows that all C4 leaves have closer vein spacing than C3 species with at most 2–3 M cells between BS (Figure 4). However there are examples in which C4 photosynthesis is naturally supported around widely spaced veins such as in maize husk tissue, albeit at lower rates with little photosynthetic activity in the interveinal M cells (Langdale et al., 1988; Pengelly et al., 2011). Here, in the context of the introduction of C4 metabolism into rice without altered vein spacing, we have asked the simple question: can we detect a small amount of C4 photosynthesis introduced around existing veins using gas exchange techniques? The modelling here uses the Farquhar et al. (1980) C3 model of photosynthesis combined with the enzyme limited C4 model of photosynthesis described in von Caemmerer (2000) with C3 kinetic constants for RuBisCO. Rice leaves have approximately 7 M cells between veins (Figure 4b; Chatterjee et al., 2016). The modelled curves show that partitioning a small amount of RuBisCO to low capacity C4 photosynthesis around veins (i.e. in 2 out of 7 M cells) lowers the compensation point and increase CO2 assimilation rates at all CO2 partial pressures (Figure 6). Hence this should be easily detected with gas exchange techniques. The modelling approach is very basic and has not considered the added energy requirements in M cells running both C3 and C4 photosynthesis given the low capacity C4 photosynthetic rates considered here. Nevertheless it suggests that even this small addition, which would result in the small amount of RuBisCO in BS being more efficient, could have a physiological benefit.
Figure 6.
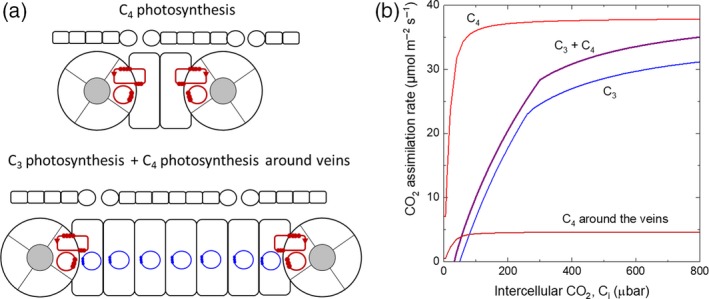
(a) Diagram comparing two cell‐type C4 photosynthesis and a rice leaf with low capacity C4 photosynthesis occurring around the veins. In the latter, C3 photosynthesis occurs in all mesophyll cells (rectangular cells) and in bundle sheath cells (cells inside circles), in addition to C4 photosynthesis occurring only in mesophyll cells adjacent to bundle sheath cells. (b) Modelled CO2 assimilation rates of a rice leaf with low capacity C4 photosynthesis occurring around the veins (purple line). Also shown are rice leaf with C3 photosynthesis (blue line), a standard high capacity C4 photosynthesis (red line), and the C4 photosynthesis around the vein contributing to the enhanced photosynthesis rate of the purple line (dark red line). C3 leaf photosynthesis has been modelled with RuBisCO maximal activity, Vcmax = 90 µmol m−2 sec−1 and electron transport rate, J = 150 µmol m−2 sec−1. The standard C4 photosynthesis was modelled with Vcmax = 40 µmol m−2 sec−1 and PEP carboxylase maximal activity, Vpmax = 300 µmol m−2 sec−1. The low capacity C4 photosynthesis around the veins was modelled with Vcmax = 5 µmol m−2 sec−1 and Vpmax = 25 µmol m−2 sec−1. In the combined C3 and C4 photosynthesis, the total Vcmax was maintained at 90 µmol m−2 sec−1. C3 RuBisCO kinetic constants by von Caemmerer (2000) were used together with the Michaels constant for CO2 of PEPC of 154 µbar (Boyd et al., 2015).
Conclusions
Considerable progress has been made in building C4 rice. The initial concept of assembling a tool box of components and a biochemical, anatomical and molecular blueprint based on evolution and 50 years of study has now become one of the largest synthetic biology projects of all time in plant biology. Discovering that our ‘toolbox’ was often lacking and that new knowledge and technologies would continually reshape the strategy and redefine the blueprint should come as no surprise. ‘Functionalisation’ of the BS compartment is the immediate hurdle. For a fully functional C4 cycle to operate in rice across two cell types, the BS must be functionally capable of housing the photosynthetic machinery necessary for C4 acid decarboxylation and CO2 refixation by RuBisCO. It appears that we have the biochemical and molecular‐genetic components necessary for C4 function; the means to prototype and fine tune them; a promise that C4 photosynthesis around rice leaf veins may be possible and even beneficial, but there is still some distance yet to be travelled on the road to C4 rice.
Author Contributions
All authors contributed to the writing of this review.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
We dedicate this review to the late John Sheehy, the champion of the C4 rice project. The Research was funded by a C4 rice project grant from The Bill & Melinda Gates Foundation to the University of Oxford (2015–2019; OPP1129902) and Australian Research Council Centre of Excellence for Translational Photosynthesis (CE1401000015).
Data Availability
The datasets generated in this paper are available from the corresponding author on request.
References
- Andersen, K.S. , Bain, J.M. , Bishop, D.G. and Smillie, R.M. (1972) Photosystem II activity in agranal bundle sheath chloroplasts from Zea mays . Plant Physiol. 49, 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou, A.I. and Nakayama, N. (2018) Mobius assembly: a versatile golden‐gate framework towards universal DNA assembly. PLoS ONE, 13, e0189892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, P. and Peisker, M. (1978) Influence of high oxygen concentrations on the CO2 compensation concentration in C4 plants. Kulturpflanze, 26, 99–103. [Google Scholar]
- Arrivault, S. , Guenther, M. , Ivakov, A. , Feil, R. , Vosloh, D. , van Dongen, J.T. , Sulpice, R. and Stitt, M. (2009) Use of reverse‐phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 59, 824–839. [DOI] [PubMed] [Google Scholar]
- Bellasio, C. (2016) A generalized stoichiometric model of C3, C2, C2+ C4, and C4 photosynthetic metabolism. J. Exp. Bot. 68, 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellasio, C. and Farquhar, G.D. (2019) A leaf‐level biochemical model simulating the introduction of C2 and C4 photosynthesis in C3 rice: gains, losses and metabolite fluxes. New Phytol. 223, 150–166. [DOI] [PubMed] [Google Scholar]
- Bowes, G. and Salvucci, M.E. (1984) Hydrilla: Inducible C4‐type photosynthesis without Kranz anatomy In Advances in Photosynthesis Research (Sybesma C., ed). The Hague: Martinus Nijhoff/Dr. W Junk Publishers, pp. 829–832. [Google Scholar]
- Boyd, R.A. , Gandin, A. and Cousins, A.B. (2015) Temperature responses of C4 photosynthesis: biochemical analysis of rubisco, phosphoenolpyruvate carboxylase, and carbonic anhydrase in Setaria viridis . Plant Physiol. 169, 1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.H. and Bouton, J.H. (1993) Physiogy and genetics of interspecific hybrids between photosynthetic types [Review]. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 435–456. [Google Scholar]
- Brown, R.H. and Byrd, G.T. (1993) Estimation of bundle sheath cell conductance in C4 species and O2 insensitivity of photosynthesis. Plant Physiol. 103, 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner, K. , Schäfer, P. , Weber, E. , Grützner, R. , Marillonnet, S. and Tissier, A. (2015) A library of synthetic transcription activator‐like effector‐activated promoters for coordinated orthogonal gene expression in plants. Plant J. 82, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard, J.O. , Runkel, A.M. and Zambryski, P.C. (2013) Plasmodesmata dynamics are coordinated by intracellular signaling pathways. Curr. Opin. Plant Biol. 16, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer, S. (2000) Biochemical Models of Leaf Photosynthesis. Collingwood, Australia: CSIRO Publishing. [Google Scholar]
- von Caemmerer, S. and Furbank, R. (2003) The C4 pathway: an efficient CO2 pump. Photosynth. Res. 77, 191–207. [DOI] [PubMed] [Google Scholar]
- von Caemmerer, S. , Quick, W.P. and Furbank, R.T. (2012) The development of C4 rice: current progress and future challenges. Science, 336, 1671–1672. [DOI] [PubMed] [Google Scholar]
- von Caemmerer, S. , Edwards, G.E. , Koteyeva, N. and Cousins, A.B. (2014) Single cell C4 photosynthesis in aquatic and terrestrial plants: A gas exchange perspective. Aquat. Bot. 118, 71–80. [Google Scholar]
- Chatterjee, J. , Dionora, J. , Elmido‐Mabilangan, A. , Wanchana, S. , Thakur, V. , Bandyopadhyay, A. , Brar, D.S. and Quick, W.P. (2016) The evolutionary basis of naturally diverse rice leaves anatomy. PLoS ONE, 11, e0164532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Qu, N. , Cao, S. , Bauwe, H. , Chen, S. , Tian, W. and Chu, C. (2001) Expression analysis of GDCP promoter from C3–C4 intermediate plant Flaveria anomala in transgenic rice. Chin. Sci. Bull. 46, 1635–1638. [Google Scholar]
- Chiang, Y.‐H. , Zubo, Y.O. , Tapken, W. , Kim, H.J. , Lavanway, A.M. , Howard, L. , Pilon, M. , Kieber, J.J. and Schaller, G.E. (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 160, 332–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin, P.‐A. and Osborne, C.P. (2013) The recurrent assembly of C4 photosynthesis, an evolutionary tale. Photosynth. Res. 1–13. [DOI] [PubMed] [Google Scholar]
- Christin, P.‐A. , Osborne, C.P. , Chatelet, D.S. , Columbus, J.T. , Besnard, G. , Hodkinson, T.R. , Garrison, L.M. , Vorontsova, M.S. and Edwards, E.J. (2013) Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proc. Natl Acad. Sci. USA, 110, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila, F.R. , Quick, W.P. , White, R.G. , Furbank, R.T. and von Caemmerer, S. (2016) The metabolite pathway between bundle sheath and mesophyll: quantification of plasmodesmata in leaves of C3 and C4 monocots. Plant Cell, 28, 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila, F.R. , Quick, W.P. , White, R.G. , Kelly, S. , von Caemmerer, S. and Furbank, R.T. (2018) Multiple mechanisms for enhanced plasmodesmata density in disparate subtypes of C4 grasses. J. Exp. Bot., 69, 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila, F.R. , Quick, W.P. , White, R.G. , von Caemmerer, S. and Furbank, R.T. (2019) Response of plasmodesmata formation in leaves of C4 grasses to growth irradiance. Plant Cell Environ. 42, 24822492. [DOI] [PubMed] [Google Scholar]
- DiMario, R.J. , Quebedeaux, J.C. , Longstreth, D.J. , Dassanayake, M. , Hartman, M.M. and Moroney, J.V. (2016) The cytoplasmic carbonic anhydrases βCA2 and βCA4 are required for optimal plant growth at low CO2 . Plant Physiol. 171, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drincovich, M.F. , Casati, P. and Andreo, C.S. (2001) NADP‐malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 490, 1–6. [DOI] [PubMed] [Google Scholar]
- Duvall, M.R. , Saar, D.E. , Grayburn, W.S. and Holbrook, G.P. (2003) Complex transitions between C3 and C4 photosynthesis during the evolution of paniceae: a phylogenetic case study emphasizing the position of Steinchisma hians (Poaceae), a C3-C4 intermediate. Int. J. Plant Sci., 164 949–958. [Google Scholar]
- Edwards, G.E. , Kanai, R. and Black, C.C. (1971) Phosphoenolpyruvate carboxykinase in leaves of certain plants which fix CO2 by the C4‐dicarboxylic acid cycle of photosynthesis. Biochem. Biophys. Res. Commun. 45, 278–285. [DOI] [PubMed] [Google Scholar]
- Emms, D.M. , Covshoff, S. , Hibberd, J.M. and Kelly, S. (2016) Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol. Biol. Evol. 33, 1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann, S. , Wiludda, C. , Burscheidt, J. , Gowik, U. , Schlue, U. , Koczor, M. , Streubel, M. , Cossu, R. , Bauwe, H. and Westhoff, P. (2008) The gene for the P‐subunit of glycine decarboxylase from the C4 species Flaveria trinervia: Analysis of transcriptional control in transgenic Flaveria bidentis (C4) and Arabidopsis (C3). Plant Physiol. 146, 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, C. , Youles, M. , Gruetzner, R. , Ehnert, T.‐M. , Werner, S. , Jones, J.D.G. , Patron, N.J. and Marillonnet, S. (2014) A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Ermakova, M. , Lopez‐Calcagno, P.E. , Raines, C.A. , Furbank, R.T. and von Caemmerer, S. (2019) Overexpression of the Rieske FeS protein of the Cytochrome b6f complex increases C4 photosynthesis in Setaria viridis . Commun. Biol. 2, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar, G.D. , von Caemmerer, S. and Berry, J.A. (1980) A biochemical‐model of photosynthetic CO2 assimilation in leaves of C3 species. Planta, 149, 78–90. [DOI] [PubMed] [Google Scholar]
- Furbank, R.T. (2011) Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J. Exp. Bot. 62, 3103–3108. [DOI] [PubMed] [Google Scholar]
- Furbank, R. , Jenkins, C. and Hatch, M. (1990) C4 photosynthesis: quantum requirement, C4 acid overcycling and Q‐cycle involvement. Funct. Plant Biol. 17, 1–7. [Google Scholar]
- Furbank, R.T. , von Caemmerer, S. , Sheehy, J. and Edwards, G. (2009) C4 rice: a challenge for plant phenomics. Funct. Plant Biol. 36, 845–856. [DOI] [PubMed] [Google Scholar]
- Hatch, M.D. and Kagawa, T. (1976) Photosynthetic activities of isolated bundle sheath cells in relation to differing mechanisms of C4 pathway photosynthesis. Arch. Biochem. Biophys. 175, 39–53. [DOI] [PubMed] [Google Scholar]
- Hatch, M.D. and Osmond, C.B. (1976) Compartmentation and transport in C4 photosynthesis In Transport in Plants III (Stocking C. R. and Heber U., eds). Berlin, Heidelberg: Springer, pp. 144–184. [Google Scholar]
- Hausler, R.E. , Hirsch, H.J. , Kreuzaler, F. and Peterhansel, C. (2002) Overexpression of C4‐cycle enzymes in transgenic C3 plants: a biotechnological approach to improve C3‐photosynthesis [Review]. J. Exp. Bot. 53, 591–607. [DOI] [PubMed] [Google Scholar]
- Hernández‐Prieto, M.A. , Foster, C. , Watson‐Lazowski, A. , Ghannoum, O. and Chen, M. (2019) Comparative analysis of thylakoid protein complexes in the mesophyll and bundle sheath cells from C3, C4 and C3–C4 Paniceae grasses. Physiol. Plant. 166, 134–147. [DOI] [PubMed] [Google Scholar]
- Hibberd, J.M. and Covshoff, S. (2010) The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Hibberd, J.M. , Sheehy, J.E. and Langdale, J.A. (2008) Using C4 photosynthesis to increase the yield of rice ‐ rationale and feasibility. Curr. Opin. Plant Biol. 11, 228–231. [DOI] [PubMed] [Google Scholar]
- Hudson, D. , Guevara, D.R. , Hand, A.J. , Xu, Z. , Hao, L. , Chen, X. , Zhu, T. , Bi, Y.‐M. and Rothstein, S.J. (2013) Rice cytokinin GATA transcription factor1 regulates chloroplast development and plant architecture. Plant Physiol. 162, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.E. , Sedelnikova, O.V. , Wu, H. , Becraft, P.W. and Langdale, J.A. (2019) Redundant SCARECROW genes pattern distinct cell layers in roots and leaves of maize. Development, 146, dev177543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale, J.A. (2011) C4 cycles: Past, present, and future research on C4 photosynthesis [Review]. Plant Cell, 23, 3879–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale, J.A. , Zelitch, I. , Miller, E. and Nelson, T. (1988) Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. EMBO J. 7, 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Jain, R. , Chern, M. et al. (2017) The sequences of 1504 mutants in the model rice variety Kitaake facilitate rapid functional genomic studies. Plant Cell, 29, 1218–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E. and Furbank, R.T. (1999) Tansley Review No. 105. Sucrose biosynthesis in C4 plants. New Phytol. 143, 221–237. [Google Scholar]
- Majeran, W. , Zybailov, B. , Ytterberg, A.J. , Dunsmore, J. , Sun, Q. and van Wijk, K.J. (2008) Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell. Proteomics, 7, 1609–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, M. , Furbank, R.T. , Fukayama, H. and Miyao, M. (2001) Molecular engineering of C4 photosynthesis [Review]. Ann. Rev. Plant Physiol. Plant Mol. Biol. 52, 297–314. [DOI] [PubMed] [Google Scholar]
- Mitchell, P.L. and Sheehy, J.E. (2006) Supercharging rice photosynthesis to increase yield. New Phytol. 171, 688–693. [DOI] [PubMed] [Google Scholar]
- Miyao, M. (2003) Molecular evolution and genetic engineering of C4 photosynthetic enzymes [Review]. J. Exp. Bot. 54, 179–189. [DOI] [PubMed] [Google Scholar]
- Miyao, M. , Masumoto, C. , Miyazawa, S.‐I. and Fukayama, H. (2011) Lessons from engineering a single‐cell C4 photosynthetic pathway into rice. J. Exp. Bot. 62, 3021–3029. [DOI] [PubMed] [Google Scholar]
- Munekage, Y.N. (2016) Light harvesting and chloroplast electron transport in NADP‐malic enzyme type C4 plants. Curr. Opin. Plant Biol. 31, 9–15. [DOI] [PubMed] [Google Scholar]
- Munekage, Y.N. and Taniguchi, Y.Y. (2016) Promotion of cyclic electron transport around photosystem I with the development of C4 photosynthesis. Plant Cell Physiol. 57, 897–903. [DOI] [PubMed] [Google Scholar]
- Nomura, M. , Higuchi, T. , Ishida, Y. , Ohta, S. , Komari, T. , Imaizumi, N. , Miyao‐Tokutomi, M. , Matsuoka, M. and Tajima, S. (2005a) Differential Expression Pattern of C4 Bundle Sheath Expression Genes in Rice, a C3 Plant. Plant Cell Physiol. 46, 754–761. [DOI] [PubMed] [Google Scholar]
- Nomura, M. , Higuchi, T. , Katayama, K. , Taniguchi, M. , Miyao‐Tokutomi, M. , Matsuoka, M. and Tajima, S. (2005b) The promoter for C4‐type mitochondrial aspartate aminotransferase does not direct bundle sheath‐specific expression in transgenic rice plants. Plant Cell Physiol. 46, 743–753. [DOI] [PubMed] [Google Scholar]
- Omoto, E. , Kawasaki, M. , Taniguchi, M. and Miyake, H. (2009) Salinity Induces Granal Development in Bundle Sheath Chloroplasts of NADP‐Malic Enzyme Type C4 Plants. Plant Prod. Sci. 12, 199–207. [Google Scholar]
- Ormenese, S. , Bernier, G. and Périlleux, C. (2006) Cytokinin application to the shoot apical meristem of Sinapis alba enhances secondary plasmodesmata formation. Planta, 224, 1481–1484. [DOI] [PubMed] [Google Scholar]
- Pengelly, J.J.L. , Sirault, X.R.R. , Tazoe, Y. , Evans, J.R. , Furbank, R.T. and von Caemmerer, S. (2010) Growth of the C4 dicot Flaveria bidentis: photosynthetic acclimation to low light through shifts in leaf anatomy and biochemistry. J. Exp. Bot. 61, 4109–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly, J.J.L. , Kwasny, S. , Bala, S. , Evans, J.R. , Voznesenskaya, E.V. , Koteyeva, N.K. , Edwards, G.E. , Furbank, R.T. and von Caemmerer, S. (2011) Functional analysis of corn husk photosynthesis. Plant Physiol. 156, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt, T. , Nilsson, A. and Allen, J.F. (1999) Photosynthetic control of chloroplast gene expression. Nature, 397, 625. [Google Scholar]
- Rhoades, M.M. and Carvalho, A. (1944) The function and structure of the parenchyma sheath plastids of the maize leaf. Bull. Torrey Botanical Club, 71, 335–346. [Google Scholar]
- Sage, R.F. (2004) The evolution of C4 photosynthesis. New Phytol. 161, 341–370. [DOI] [PubMed] [Google Scholar]
- Sage, R.F. , Sage, T.L. and Kocacinar, F. (2012) Photorespiration and the evolution of C4 photosynthesis. Ann. Rev. Plant Biol. 63, 19–47. [DOI] [PubMed] [Google Scholar]
- Schuler, M.L. , Mantegazza, O. and Weber, A.P.M. (2016) Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 87, 51–65. [DOI] [PubMed] [Google Scholar]
- Sedelnikova, O.V. , Hughes, T.E. and Langdale, J.A. (2018) Understanding the genetic basis of C4 kranz anatomy with a view to engineering C3 crops. Ann. Rev. Genet. 52, 249–270. [DOI] [PubMed] [Google Scholar]
- Serrano, L. (2007) Synthetic biology: promises and challenges. Mol. Syst. Biol. 3, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stata, M. , Sage, T.L. , Hoffmann, N. , Covshoff, S. , Ka‐Shu Wong, G. and Sage, R.F. (2016) Mesophyll chloroplast investment in C3, C4 and C2 species of the genus Flaveria. Plant Cell Physiol. 57, 904–918. [DOI] [PubMed] [Google Scholar]
- Suorsa, M. (2015) Cyclic electron flow provides acclimatory plasticity for the photosynthetic machinery under various environmental conditions and developmental stages. Front Plant Sci. 6, 800–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi, A. , Kishine, M. , Asada, K. , Endo, T. and Sato, F. (2005) Differential use of two cyclic electron flows around photosystem I for driving CO2‐concentration mechanism in C4 photosynthesis. Proc. Natl Acad. Sci. USA, 102, 16898–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, Y. , Akagi, H. , Kamasawa, N. , Osumi, M. and Honda, H. (2000) Aberrant chloroplasts in transgenic rice plants expressing a high level of maize NADP‐dependent malic enzyme. Planta, 211, 265–274. [DOI] [PubMed] [Google Scholar]
- Ueno, O. , Yoshimura, Y. and Sentoku, N. (2005) Variation in the activity of some enzymes of photorespiratory metabolism in C4 grasses. Ann. Bot. 96, 863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass, I. (2012) Molecular mechanisms of photodamage in the photosystem II complex. Biochimica et Biophysica Acta (BBA) ‐ . Bioenergetics, 1817, 209–217. [DOI] [PubMed] [Google Scholar]
- Voznesenskaya, E.V. , Franceschi, V.R. , Kiirats, O. , Freitag, H. and Edwards, G.E. (2001) Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nature, 414, 543–546. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Fouracre, J. , Kelly, S. et al . (2013) Evolution of GOLDEN2‐LIKE gene function in C3 and C4 plants. Planta, 237, 481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Hendron, R.‐W. and Kelly, S. (2017a) Transcriptional control of photosynthetic capacity: conservation and divergence from Arabidopsis to rice. New Phytol. 216, 32–45. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Khoshravesh, R. , Karki, S. , Tapia, R. , Balahadia, C.P. , Bandyopadhyay, A. , Quick, W.P. , Furbank, R. , Sage, T.L. and Langdale, J.A. (2017b) Re‐creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Current Biol. 27, 3278–3287.e3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Tholen, D. and Zhu, X.‐G. (2017c) C4 photosynthesis in C3 rice: a theoretical analysis of biochemical and anatomical factors. Plant Cell Environ. 40, 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M. (2002) Gene silencing In International Review of Cytology (Jeon K. W., ed). New York: Academic Press, pp. 61–113. [DOI] [PubMed] [Google Scholar]
- Yin, X. and Struik, P.C. (2017) Can increased leaf photosynthesis be converted into higher crop mass production? A simulation study for rice using the crop model GECROS. J. Exp. Bot. 68, 2345–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated in this paper are available from the corresponding author on request.


