Abstract
NBS-LRR (nucleotide-binding site and leucine-rich repeat) is one of the largest resistance gene families in plants. The completion of the genome sequencing of wild tomato Solanum pimpinellifolium provided an opportunity to conduct a comprehensive analysis of the NBS-LRR gene superfamily at the genome-wide level. In this study, gene identification, chromosome mapping, and phylogenetic analysis of the NBS-LRR gene family were analyzed using the bioinformatics methods. The results revealed 245 NBS-LRRs in total, similar to that in the cultivated tomato. These genes are unevenly distributed on 12 chromosomes, and ~59.6% of them form gene clusters, most of which are tandem duplications. Phylogenetic analysis divided the NBS-LRRs into 2 subfamilies (CNL-coiled-coil NBS-LRR and TNL-TIR NBS-LRR), and the expansion of the CNL subfamily was more extensive than the TNL subfamily. Novel conserved structures were identified through conserved motif analysis between the CNL and TNL subfamilies. Compared with the NBS-LRR sequences from the model plant Arabidopsis thaliana, wide genetic variation occurred after the divergence of S. pimpinellifolium and A thaliana. Species-specific expansion was also found in the CNL subfamily in S. pimpinellifolium. The results of this study provide the basis for the deeper analysis of NBS-LRR resistance genes and contribute to mapping and isolation of candidate resistance genes in S. pimpinellifolium.
Keywords: Solanum pimpinellifolium, NBS-LRR, duplicated genes, comparative analysis
Introduction
Plants are surrounded by a wide variety of pathogens such as viruses, bacteria, fungi, nematodes, and aphids during their growth and development.1 Some pathogens have successfully invaded crops and have caused severe damage to agricultural production and the quality of crops. To cope with disease attacks, the plants have evolved a series of sophisticated defense mechanisms to defend against various pathogens. Previous studies have shown that disease resistance (R) proteins of plants play an essential role in direct or indirect recognition of corresponding pathogens.2 The plant-pathogen interaction model is regarded as the “gene-for-gene” interaction hypothesis.3 In this hypothesis, an incompatible interaction of host R gene protein products with pathogen Avr proteins produces a defense response termed the hypersensitive response, which impedes pathogen progression via a variety of mechanisms, including localized programmed cell death and correlated immune responses.4 Currently, numerous disease R genes of plants have been cloned, which not only confer resistance to a wide range of pathogens but also play a vital role in resistance to abiotic stress.1,5,6
Presently, researchers had divided R genes into at least 5 diverse classes of families, including NBS-LRR (Nucleotide Binding Site and Leucine-Rich Repeat domains), LRR-TM (Leucine-Rich Repeat plus Transmembrane Receptor), STK (Serine-Threonine Kinase), RLK (Receptor-Like Kinase), and SA-CC (Signal Anchor plus Coiled-Coil).7 Among them, the NBS-LRR family is the largest class of known R proteins in the plant kingdom,1,7 whose encoded proteins are involved in an important part of the plant defense system. In Arabidopsis, researchers have reported that the NBS and LRR domains play different roles in the plant-microbe interaction. The former can bind and hydrolyze ATP or GTP, while the latter is involved in protein-protein interactions.8,9
It was well known that the NBS-LRR protein encodes 3 main domains—N-terminal, NBS, and LRR domains.5,9,10 There are 2 structures in the N-terminal, one is the TIR (Toll/Interleukin-1 receptor) structure, and the other is the non-TIR structure, usually known as CC (coiled-coil). In Arabidopsis, the N-terminal domain has been identified as similar to residues that can enhance gene expression and protein stability.5 The TIR domain of the TNL proteins contain ~175 amino acid residues, and some conserved motifs have been reported in these domains of plant NBS-LRRs (motifs TIR-1, TIR-2, TIR-3, and TIR-4).9,11 The N-terminal CC domain, as a characteristic motif in the N-terminus of the CNL R proteins, plays an important role in protein-protein interactions.12,13 Although the specific mechanism of TIR (toll/interleukin-1 receptor) and CC (coiled-coil) domains’ interactions with pathogens remains unclear, it has been reported that they can stimulate EDS1 (enhanced disease susceptibility 1) and NDR1 (nonrace-specific disease resistance 1), respectively, in the downstream signaling system when the R gene recognizes the pathogen.14,15
The NBS domain, composed of ~300 amino acid sequences, is the main structural domain of the NBS-LRR R genes. Eight distinct conserved motifs—P-loop, RNBS-A, Kinase2, RNBS-B, RNBS-C, GLPL, RNBS-D, and MHDV—have been confirmed in this domain.5,16 However, these 8 motifs are not completely conserved in each subfamily. It has been proved that motifs P-loop, GLPL, Kinase2, and GLPL have high similarity in the TNL and CNL subfamilies, while the similarity levels of the RNBS-A, RNBS-D, and RNBS-C motifs are lower in the TNL and CNL of Arabidopsis.9 Owing to the relatively higher conservativeness than the other 2 domains, the NBS domain is usually used for designing primers to amplify resistance genes.17 The LRR region, with ~24 amino acid sequences, is characterized by leucine-rich repeated C-terminus to the NBS domain in many R genes. Meanwhile, the LRR (leucine-rich repeat) domain has a higher variation than neighboring regions and is assumed to play a key role in the resistance function.18
The NBS-LRR superfamily, accounting for the largest gene family among plant genomes, has become the core of the resistance research field. Currently, NBS-LRR genes have been studied in many monocotyledon and dicotyledon plants, including Arabidopsis, chickpea, potato, rice, cassava, maize, Brassica napus, poplar, Medicago, sorghum, papaya, grape, and lotus.5,18-29 Based on these previous studies, variations (ie, gene number, structural features, and evolutionary pattern) of the NBS-LRR gene family occur in different plants.30 For example, the Arabidopsis genome has approximately 165 NBS-LRRs (112 TNLs and 53 CNLs). A total of 333 NBS-encoding genes have been identified in the Medicago genomic sequence and 92 NBS-LRR genes in B napus.23,25
Tomato (Solanum lycopersicum L.) is one of the most important vegetable crops, whose production, productivity, and quality are adversely affected by abiotic and biotic stresses.31 It is well known that abiotic stresses such as drought, extreme temperature, and high salinity affect almost every stage of a plant’s life cycle.32-40 Depending on the plant stage and duration of the stress, abiotic stress causes serious yield loss.41-46 Furthermore, compared with these gene-related abiotic stresses, which are induced widely by exogenous hormones and abiotic stress factors,47-60 plant NBS-LRR resistance genes are rarely induced. Usually, although several wild tomato species have stress tolerance genes, it is very difficult to transfer them into cultivars due to high genetic distance and crossing barriers. On the contrary, biotic stress causes a significant yield reduction that can be solved using R genes from wild plant species. Currently, a large number of R genes have been found in a wild relative, Solanum pimpinellifolium, which is a critical source of R genes in tomato breeding. The completion of plant genome sequencing provides a rare opportunity to study members of gene families at the whole genome level.61-64 In this study, we identified NBS-LRR-encoding genes in the whole S. pimpinellifolium genome. Similar to the NBS-LRR superfamily of other plants, these genes could be divided into 2 subfamilies—TNL and CNL. All of the NBS-LRR genes were unevenly located in clusters on the chromosomes. We also sought to provide further insights into the diversity or similarity of NBS-LRRs between S. pimpinellifolium and cultivated tomato and Arabidopsis via a genome-wide comparative analysis.
Materials and Methods
Identification of NBS-LRRs in S. pimpinellifolium
The NBS-LRR genes of S. pimpinellifolium were obtained, for the first time, via 3 steps. In the first step, protein sequences of the wild tomato S. pimpinellifolium were downloaded from the Sol Genomics Network (SGN, http://solgenomics.net/) database. In the next step, a local database was created using BioEdit software for the downloaded S. pimpinellifolium protein datasets. In the third step, the protein sequences of the NBS domain of the NBS-encoding sequences from Arabidopsis were used as query sequences to search NBS-LRR candidate genes of the S. pimpinellifolium genome.
Subsequently, these candidates were submitted to the online software PFAM (http://pfam.sanger.ac.uk/) to determine whether there are TIR, NBS, and LRR domains. Meanwhile, the CC domain may contain some smaller individual motifs or too divergent proteins; hence, it was further identified using COILS Serve (http://www.ch.embnet.org/software/COILS_form.html). Finally, all NBS-LRR genes were classified according to the presence or absence of TIR, CC, NBS, or LRR domains.
Chromosome mapping of the NBS-LRR genes in S. pimpinellifolium
Information on the physical position on the chromosomes of each NBS-LRR gene from the S. pimpinellifolium genome was searched in the tomato genome database (http://mips.helmholtz-muenchen.de/plant/tomato/searchjsp/index.jsp). The software MapDraw V2.1 was used to draw chromosomal maps of NBS-LRRs.65 Then, gene clusters and tandem duplication were also analyzed based on previously established criteria, respectively. A gene cluster was defined as a region where 2 neighboring homologous genes were less than 200 kb and fewer than 8 non-NBS-encoding genes between NBS-encoding genes.9,26 A tandemly duplicated gene was determined based on Huang’s definition, in which tandem-duplicated candidate gene pairs were defined as a region including 2 or more adjacent homologous genes within 100 kb and the similarity of these genes more than 70%.66
Phylogenetic analysis of the NBS-LRR genes
All NBS-LRR genes with CNL and TNL domains were selected for phylogenetic analysis. The P-loop to GLPL motifs of these chosen NBS-LRR members were extracted to perform multiple sequence alignment using Clustalx 1.83. Then, phylogenetic analysis of the NBS-LRR gene family was performed using the neighbor-joining (NJ) and maximum likelihood (ML) methods. For NJ analysis, MEGA X software was selected.67 The parameters of the phylogenetic tree were set as follows: 1000 bootstrap replications; p-distance model; and pairwise deletion gap. For ML analysis, ProtTest (version 2.4) software was used for model selection68 and PhyML (version 3.0) software was used to construct ML trees with the Whelan and Goldman amino acid substitution model, γ-distribution, and 100 nonparametric bootstrap replicates.69
Homologous comparison and phylogenetic analysis were also investigated between amino acid sequences of S. pimpinellifolium and Arabidopsis NBS-LRR genes. First, NBS-LRR protein sequences of the Arabidopsis genome were obtained from phytozome v12 (https://phytozome.jgi.doe.gov/pz/portal.html). The amino acid sequences of the P-loop to GLPL motifs from CNL and TNL in S. pimpinellifolium and Arabidopsis were applied to construct a phylogenetic tree, respectively. The method and parameter settings for building the phylogenetic tree were the same as described above.
Detection of the conserved motif
The NBS-encoding genes could be divided into CNL and TNL subfamilies based on phylogenetic analysis. To investigate the structural features of these genes, the sequences and distribution of the conserved motifs were analyzed individually using Multiple Expectation Maximization for Motif Elicitation (MEME) (http://meme-suite.org/tools/meme), and the parameters were set as follows: the maximum and minimum lengths of the conserved motif were 50 and 6, respectively, and the largest number of conserved motifs was 20; other parameters used the default settings.70 Conservation or variation of each motif among NBS-LRR members was presented.
Results
Identification and classification of the NBS-LRR gene family in S. pimpinellifolium
Previous studies showed that the NBS-LRR gene family in plants contained different conserved domains, and the NBS-LRR gene family was further divided into 6 different subfamilies according to the domains.5 In this study, we identified a total of 245 NBS-LRR resistance genes in S. pimpinellifolium, and they were classified into 6 NBS-LRR subfamilies: CC-NBS-LRR (CNL), TIR-NBS-LRR (TNL), CC-NBS (CN), TIR-NBS (TN), NBS-LRR (NL), and NBS (N), respectively. The number of genes corresponding to each subfamily is shown in Table 1.
Table 1.
Number and classifications of NBS-LRR genes.
| Predicted Proteins | Letter Code | Solanum Pimpinellifolium | Solanum Lycopersicum 71 | Potato19 | Arabidopsis 5 | Maize22 | Rice20 |
|---|---|---|---|---|---|---|---|
| CC-NBS-LRR | CNL | 78 | 93 | 65 | 51 | 58 | 402 |
| TIR-NBS-LRR | TNL | 15 | 18 | 37 | 92 | 0 | 0 |
| CC-NBS | CN | 54 | 18 | 24 | 6 | 11 | 53 |
| TIR-NBS | TN | 7 | 5 | 12 | 21 | 0 | 0 |
| NBS-LRR | NL | 29 | 66 | 177 | 6 | 31 | 74 |
| NBS | N | 62 | 52 | 104 | 1 | 7 | 16 |
| Total | 245 | 252 | 419 | 177 | 107 | 545 |
Abbreviations: CC, coiled-coil; LRR, leucine-rich repeat; NBS, Nucleotide-binding site; TIR, toll/interleukin-1 receptor.
Among them, both of CNL and TNL were the most typical subfamilies, with 78 and 15 NBS-LRR members, respectively. The numbers of the NBS-LRR genes in the N and CN subfamilies were 62 and 54, respectively. Also, 29 genes were identified as belonging to the NL subfamilies, and only 7 genes were predicted to encode the TIR domain; hence, it belonging to the TN subfamily. Meanwhile, the TN subfamily was the smallest NBS-LRR subfamily among these 6 subfamilies. Based on the above result, we can conclude that these 245 NBS-LRR genes were unevenly located on the 6 subfamilies. Together, high genetic variation was observed in S. pimpinellifolium
Comparative analysis of the NBS-LRR genes between S. pimpinellifolium and S lycopersicum
In addition to comparing the number of genes of different subfamilies in S. pimpinellifolium, we also analyzed the number of genes in these subfamilies in other species (Table 1). The total number of NBS-LRR genes in different plants and the number of genes in each subfamily were significantly different. First, S lycopersicum and S. pimpinellifolium have almost a similar total number of NBS-LRR genes—the former being 25271 and the latter being 245—but there are substantial variations in the number of some NBS-LRR gene subfamilies. For example, the number of CN-type genes in wild tomato was 36 more than that in cultivated tomato, while the number of NL-type genes in wild tomato was 37 less than that in cultivated tomato. Second, 419 NBS-LRR genes were found in potato of the genus Solanum, among which 65 belonged to the CNL subfamily and 37 belonged to the TNL subfamily.
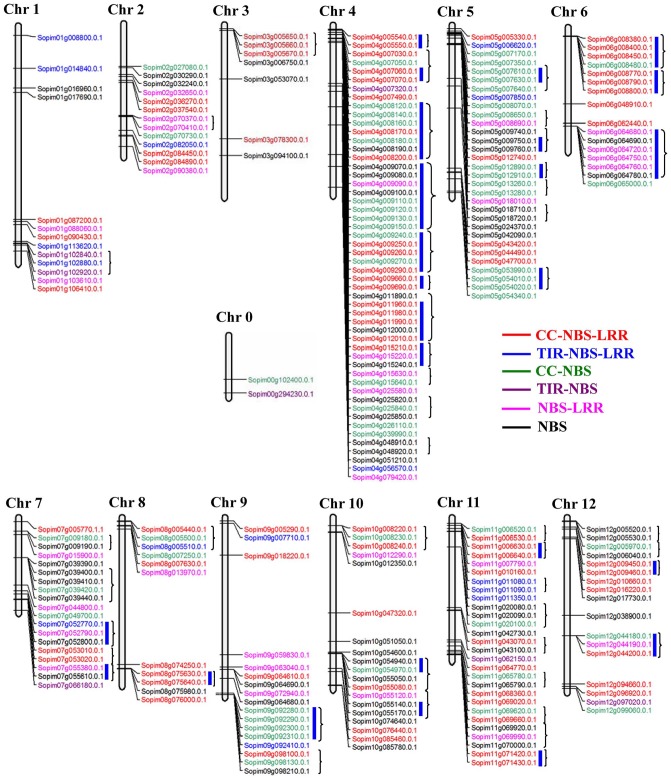
In this context, we further analyzed the number and distribution of the NBS-LRR gene family on the chromosome between S. pimpinellifolium and S lycopersicum. The distribution of genes on different chromosomes in S. pimpinellifolium was mapped (Figure 1), where we found that 243 out of 245 NBS-LRR genes could be located on 12 chromosomes (Figure 1 and Table 2), whereas the remaining 2 NBS-LRR proteins (Sopim00g102400.0.1 and Sopim00g294230.0.1) could not be located on any chromosomes and were assigned to fictitious chromosome 0. Pairwise comparison of NBS-LRR genes on the corresponding chromosomes between wild and cultivated tomatoes is presented in Supplemental Table S1. It is obvious that the NBS-LRRs of these 2 tomato species are unevenly distributed on the chromosomes, which is in accordance with previous reports in Arabidopsis, Medicago truncatula, Populus trichocarpa, and Brassica rapa.5,23-25 Among the 12 chromosomes, the largest number of NBS-LRRs was detected on chromosome 4, containing 52 members. By contrast, chromosome 3 included the smallest number NBS-LRRs, with only 7 genes (Table 2 and Supplemental Table S1).
Figure 1.
Chromosomal locations and duplications of the paralogous NBS-LRRs on S. pimpinellifolium chromosomes. Chromosome numbers are shown at the top of each bar. Predicted tandem-duplicated genes are indicated by thick blue lines. Gene clusters are marked by black braces. CC indicates coiled-coil; LRR, leucine-rich repeat; NBS, nucleotide-binding site; TIR, toll/interleukin-1 receptor.
Table 2.
Distribution and characteristic of nucleotide-binding site and leucine-rich repeats on different chromosomes between S lycopersicum and S pimpinellifolium.
| Chromosome | Gene Number |
|
|---|---|---|
| S Pimpinellifolium | S Lycopersicum | |
| Chr 00 | 2 | 2 |
| Chr 01 | 13 | 16 |
| Chr 02 | 13 | 14 |
| Chr 03 | 7 | 7 |
| Chr 04 | 52 | 53 |
| Chr 05 | 31 | 34 |
| Chr 06 | 16 | 16 |
| Chr 07 | 19 | 17 |
| Chr 08 | 11 | 10 |
| Chr 09 | 17 | 17 |
| Chr 10 | 19 | 21 |
| Chr 11 | 28 | 27 |
| Chr 12 | 17 | 18 |
| Total number | 245 | 252 |
Gene cluster and tandem duplication of the NBS-LRR genes in S. pimpinellifolium
The distribution of NBS-LRR genes across the chromosomes was used to further analyze the evolutionary patterns of gene expansion (Figure 1). Six different colors represent the genes from the 6 subfamilies of NBS-LRRs in Figure 1. We found that NBS-LRRs of the CNL type were spread across all chromosomes, while TNL genes were selectively distributed on chromosomes 3, 6, 8, 10, and 12. It has been previously proved that most of the NBS-encoding genes are arranged in clusters on chromosomes.5,25 Also, the gene cluster was previously determined by the following criteria: a cluster of NBS-LRR proteins was described as the distance between neighboring homologous genes less than 200 kb and fewer than 8 non-NBS-encoding genes between TNLs and CNLs.5,19,28 Based on the above criteria, NBS-LRR gene clusters were carried out (Figure 1). A total of 49 gene clusters, including 146 NBS-LRR genes, were identified in wild tomato S. pimpinellifolium. In other words, only ~40% of genes did not reside in clusters. Of them, chromosome 4 had 11 gene clusters and 44 genes, occupying ~84.6% (44/52) of the total genes on this chromosome, while chromosomes 1, 2, and 3 each only possessed 1 gene cluster (Figure 1). Meanwhile, chromosome 4 had the most gene clusters (11), including gene members of 3 different subfamilies. Besides, all of these gene clusters consisted of 2 to 8 genes. The gene cluster containing genes from different subfamilies was commonly found in S. pimpinellifolium. For example, 27 out of 49 gene clusters were made up of genes from different subfamilies which were a vital source of gene variation.72
Tandem duplication of NBS-LRRs was analyzed in S. pimpinellifolium (Figure 1). In total, 25 gene pairs were duplicated tandemly, which contained 80 NBS-LRR members. Remarkably, all of the tandem-duplicated NBS-LRRs were anchored in the gene clusters. The biggest gene cluster of chromosome 4 was also the tandem duplication with the largest number of genes. Hence, we speculated that the tandem duplication event was an important factor in the process of NBS-LRR gene cluster evolution. Out of the 25 tandem duplication events, 14 were made up of NBS-LRR members from 1 single subfamily (Figure 1).
Phylogenetic analysis of the NBS-LRR genes in S. pimpinellifolium
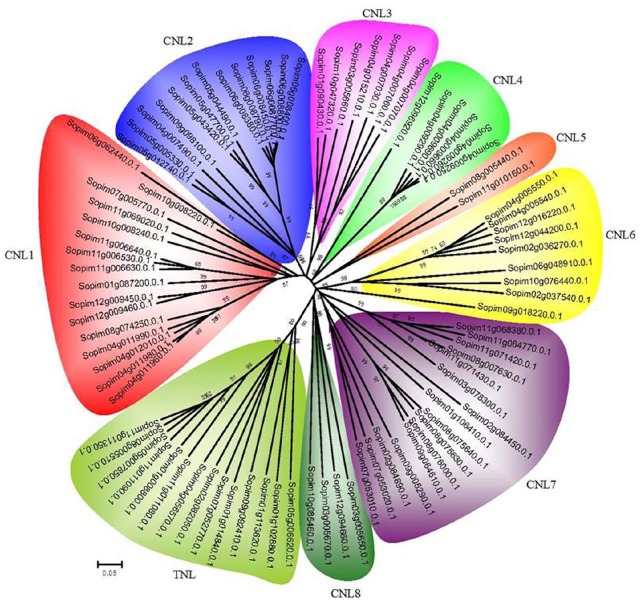
To explore the evolutionary relationships of NBS-LRR genes, we conducted phylogenetic analyses with an alignment of the NBS domain from these 2 species using neighbor-joining (NJ) and maximum likelihood (ML) methods. ML analysis showed that proteins from different species cluster together in clades with high support values (not shown), with support from NJ analysis for most results. Therefore, in the present study, the phylogenetic tree of the NBS-LRR genes from the TNL and CNL subfamilies constructed using the NJ method was selected for analyses. Four members (Sopim04g008170.0.1, Sopim11g069660.0.1, Sopim11g043070.0.1, and Sopim10g055080.0.1) were excluded due to the presence of incomplete NBS domains. Both of the CNL and TNL subfamilies were separated from each other in the phylogenetic tree, and CNL was further divided into eight small branches, namely CNL1 to CNL8, respectively (Figure 2). Inversely, the TNL subfamily remained as one branch owing to fewer gene numbers. Moreover, we found that the CNL1 and CNL7 branches each contained 16 NBS-LRRs, which were from seven different chromosomes, and these 2 branches had the largest gene numbers in the phylogenetic tree. In contrast to CNL1 and CNL7, the CNL5 branch only contained 2 genes from chromosomes 8 and 11, with the lowest number of genes (Figures 1 and 2).
Figure 2.
Phylogenetic relationship of nucleotide-binding site and leucine-rich repeats from CNL and TNL subfamilies in S pimpinellifolium. All of these proteins were grouped into 9 clades (CNL 1-8 and TNL). Numbers above branches represent the support values.
To date, some NBS-LRR-encoding genes have been cloned, like Nrc1, I2, Bs4, Hero, and Rpi-blb1.73-76 The members of the NBS-LRR gene family from S. pimpinellifolium have high sequence similarity with these cloned R genes in Solanaceae based on sequence alignment analysis (Supplemental Table S2). For example, in the CNL7 branch, the NBS-LRR genes were identified as having high homology with the late blight resistance proteins Rpi-bt1, Rpi-blb1/RB, R3a, andRGA2, as well as the R protein I2 to Fusarium oxysporum. Besides, each of the 4 branches (CNL2, CNL3, CNL4, and CNL6) had 1 homologous gene, respectively, with known R proteins (Supplemental Table S2). Both Sopim06g008440.0.1 and Sopim06g008450.0.1 were mapped in a gene cluster on chromosome 6 (Figure 1), and they had high homology with adjacent members of the cluster (Supplemental Table S2). Similarly, the same findings were also uncovered in CNL4. However, no gene clusters were identified in CNL3 and CNL6 (Figure 1). Homologous genes with the R protein of Leaf curl virus and tobacco mosaic virus were also found in the TNL branch, but they exist as single genes. This may explain why the TNL gene family in S. pimpinellifolium had a lower degree of expansion in the process of evolution.
Apart from the members of the CNL and TNL subfamilies, NBS-LRR genes from other subfamilies also showed high homology with known functional genes. As shown in Supplemental Table S2, all subfamilies had homologous genes with known resistance genes, except for the TN subfamily. Twenty-one known resistance genes, involving multiple resistances to various pathogens, had as high as 80% similarity with the NBS-LRR genes in S. pimpinellifolium. It is worth mentioning that the similarity of 2 pairs of analogous genes, Sopim01g090430.0.1 and Nrc1, Sopim05g007850.0.1 and Bs4, was as high as 100%. All of the NBS-LRR genes, which had homology with the known resistance genes in S. pimpinellifolium, were mapped on 10 chromosomes of the wild tomato, except chromosomes 3 and 10. Only 6 genes existed alone, and all of the others were located in gene clusters.
Conserved motif analysis of the NBS-LRR genes
To uncover the structural characteristics of the NBS-LRR gene family, MEME was applied to analyze the structure and distribution of the conserved motifs among the TNL and CNL subfamilies. Twenty distinct motifs were determined in each subfamily (Tables 3 and 4). All of the conserved motifs displayed a diversity distribution in their respective subfamilies (Supplemental Figures S1-TNL and S2-CNL1-4, respectively).
Table 3.
Conserved motifs of the TNL subfamily in S pimpinellifolium.
| Domain | Motif Number | Motif | Width | E-Value | Motif Sequence |
|---|---|---|---|---|---|
| TIR | Motif01 | T-1 | 40 | 1.20E-236 | xkYDVFLSFRGEDTRxtFtxHLYxaLxnrGIxTFxDdkrL |
| Motif03 | T-2 | 29 | 2.00E-197 | AIeeSxxaxvIFSkNYAxSrWCLxELVkI | |
| Motif02 | T-3 | 29 | 7.70E-198 | qxViPvFYdVDPShVRxQxesfxeaFxkH | |
| Motif08 | T-4 | 21 | 9.30E-104 | VxrWRxALxxAAdlxGxDxxn | |
| NBS | Motif04 | P-loop | 30 | 2.40E-192 | dVRixGIwGxGGIGKTTiAkAxFdxlxxxF |
| Motif05 | Kinase-2/RNBS-B | 40 | 1.80E-246 | kKVLiVLDDvDhxdqLdyLagxxxWFGxGSRIIxTTRdKH | |
| Motif09 | RNBS-C | 26 | 1.10E-96 | AxxLFnxhAFkxxxPxxxFxxlsxeV | |
| Motif12 | GLPL | 15 | 7.90E-80 | VxhAxGLPLALKVlG | |
| Motif20 | RNBS-D | 22 | 1.50E-39 | sxLhkrxxxxWrxtvxxlKxxp | |
| Motif13 | TNBS-1 | 15 | 5.80E-68 | dqxiFLDIACFfrGk | |
| Motif18 | TNBS-2 | 29 | 2.90E-65 | VxqILesCdFgAexGlxVLIdkSLVfISx | |
| LRR | Motif07 | L-1 | 21 | 8.00E-120 | xnxixMHdLIqeMGxxiVrxe |
| Motif10 | L-2 | 29 | 6.90E-92 | gkxSRlWxxeDxxxVlxxntgTxavEgIw | |
| Motif17 | L-3 | 21 | 3.80E-58 | LPENWYVsDNFLGFAVCYSGn | |
| Motif14 | L-4 | 21 | 1.00E-84 | yLPnxLRWlxWxxyPlxSlPx | |
| Motif06 | L-5 | 29 | 5.40E-153 | TPDFsgmPnLExLxLxxCxnLxEVHxSlG | |
| Motif15 | L-6 | 29 | 4.00E-97 | PxSIcxLkxLxxLxxsxCxkLexlPexiG | |
| Motif16 | L-7 | 29 | 6.60E-90 | dxxxPxDigxLSsLxxLxLxgNNFxxLPx | |
| Motif19 | L-8 | 40 | 5.30E-52 | TqLPEFPxQLDTIxADWSNDxICNSLFQNISsfQHDISAS | |
| Motif11 | L-9 | 29 | 6.70E-83 | aIHFFLVPLAGLWdTSkANGkTPNDYglI |
Abbreviations: LRR, leucine-rich repeat; NBS, nucleotide-binding site; TIR, toll/interleukin-1 receptor.
Table 4.
Conserved motifs of the CNL subfamily in S pimpinellifolium.
| Domain | Motif Number | Motif | Width | E-Value | Motif Sequence |
|---|---|---|---|---|---|
| CC | Motif19 | C-1 | 40 | 4.40E-177 | QYElLQNVcGNlRDFHgLIVNGCikhEtvEnVLPlFQLMA |
| Motif20 | C-2 | 40 | 1.30E-193 | VMHICyTNLKASTSaEVGrFIKkLLETSPDILREYlIhLQ | |
| Motif16 | C-3 | 40 | 2.40E-223 | kLxxxLxxxqxfLxDAExKQxxdxxvxxWlxelxxxaxxA | |
| Motif14 | C-4 | 29 | 6.40E-267 | xxxxxxxvGxxxexxxixxxLxxxxxxxx | |
| NBS | Motif01 | P-loop | 26 | 3.3e-969 | vixIxGMgGxGKTTLAxkxyxxxxxx |
| Motif07 | RNBS-A | 29 | 1.5e-509 | FxxxaWxxVSqxxxxxxllxxixxxxxxx | |
| Motif04 | Kinase-2 | 15 | 3.7e-433 | ryLiVlDDVWxxxxx | |
| Motif06 | RNBS-B | 15 | 4.4e-405 | xGsRIIxTTRxxxVa | |
| Motif08 | RNBS-C | 21 | 8.9e-414 | xxxlxxLxxeeSWxLfxxkxF | |
| Motif02 | GLPL | 29 | 1.1e-929 | xxxeLxxxgkxIaxkCxGLPLaixxxaGx | |
| Motif15 | CNBS-1 | 15 | 4.70E-254 | xxxxxlxLSYxxLpx | |
| Motif03 | CNBS-2 | 15 | 5.7e-477 | xLKxCFLYxxxfPeD | |
| Motif10 | RNBS-D | 21 | 1.7e-375 | xxixxxxLixLWiAEGfvxxx | |
| LRR | Motif11 | CNBS-3 | 21 | 9.20E-297 | xxExvaexylxdLixRsLvxx |
| Motif09 | MHDV | 15 | 1.8e-404 | cxxHDlxxdxxxxxa | |
| Motif13 | L-1 | 21 | 5.90E-274 | xxxlpxxixxLxhLRyLxxxx | |
| Motif12 | L-2 | 15 | 1.60E-291 | lPxsxxxLxnLqtLx | |
| Motif17 | L-3 | 15 | 1.50E-197 | xxxlxxLpxLexLxl | |
| Motif18 | L-4 | 29 | 1.10E-212 | xxexxFxxLKxLxlxxxxLxxWeaxxxxF | |
| Motif05 | L-5 | 31 | 4.7e-444 | xLxxLxlxxCxxLxxiPxxxxxxxxLxxxxx |
Abbreviations: CC, coiled-coil; LRR, leucine-rich repeat; NBS, nucleotide-binding site.
In the TNL subfamily, there were 4, 7, and 9 motifs identified in the TIR, NBS, and LRR domains, respectively (Table 3 and Supplemental Table S3). The motifs in the TIR domain were named T-1 to T-4, and the motifs in the LRR domain were named L-1 to L-9. The motifs of the NBS domain were named following previous studies.5 All of the 14 TNL members contained 4 motifs of the TIR domain, except that Sopim09g092410.0.1 lacked motif T-1 (Supplemental Figure S1 and Supplemental Table S3). The motif RNBS-A was not found in the NBS domain. Two novel motifs (TNBS-1 and TNBS-2) were identified in most of the TNL subfamily. Both of Kinase-2 and RNBS-B existed in Motif 5, as these 2 proteins were so close to each other. The motifs of the NBS domain had higher conservation in the 14 TNL genes, except TNBS-2, which was missed in some of the genes. Also, the motif compositions of the NBS-LRR genes provided further support for the grouping of phylogenetic branches. For example, 3 NBS-LRR genes (Sopim09g092410.0.1, Sopim04g056570.0.1, and Sopim07g052770.0.1) were locat-ed in the adjacent branches of the phylogenetic tree, and they did not contain the motif TNBS-2.
In the CNL subfamily, there were 4, 11, and 5 motifs in the CC, NBS and LRR domains, respectively (Table 4 and Supplemental Table S4). A low degree of conservation of motifs was observed from CC to LRR. Of them, 3 out of the 4 conserved motifs (C-1, C-2, and C-3) had lower conservation in the CC domain compared with the NBS and LRR domains (Supplemental Figure S2). In other words, most of the genes in the CNL subfamily lost motifs C-1, C-2, and C-3. Besides, most of the NBS-LRRs from the CNL4 to 8 branches lacked motif L-4 (Supplemental Table S4). In the NBS domain, most of the genes from CNL 6 missed the RNBS-D and CNBS-3 motifs. The remaining conserved motifs were detected in most of the NBS-LRR genes. Overall, the motifs of the NBS domain were relatively conservative compared with that in the N-terminal domain (Supplemental Figure S2).
When compared with the TNL and CNL subfamilies, some differences regarding the motif compositions were observed. For example, the MHDV motif was unique to the CNL family (Table 4). Besides, 2 conserved motifs (TNBS-1 and TNBS-2) were identified as novel members in the TNL subfamily, while 3 unique motifs (CNBS-1, CNBS-2, and CNBS-3) were only found in the CNL subfamily, with more diversity (Supplemental Table S3 and Table 4). In the TNL and CNL subfamilies, the conserved motifs of the LRR domain had relatively high diversity.
Evolutionary comparison of the NBS-LRR genes between S. pimpinellifolium and Arabidopsis
Previous findings have revealed that the NBS-LRR genes of Arabidopsis could be definitively divided into 2 subfamilies (TNL and non-TNL), and the CNL subfamily was the main member of the non-TNL group.9 In this current study, to investigate gene distribution and expansion of the NBS-LRR gene family between S. pimpinellifolium and Arabidopsis, genes from the CNL and TNL subfamilies were selected for phylogenetic analysis. The NBS-LRRs from S. pimpinellifolium and Arabidopsis fell into 2 distinct branches (TNL and CNL) (Figures 3 and 4). Then, the TNL and CNL subfamilies were separately analyzed, and the results are as follows.
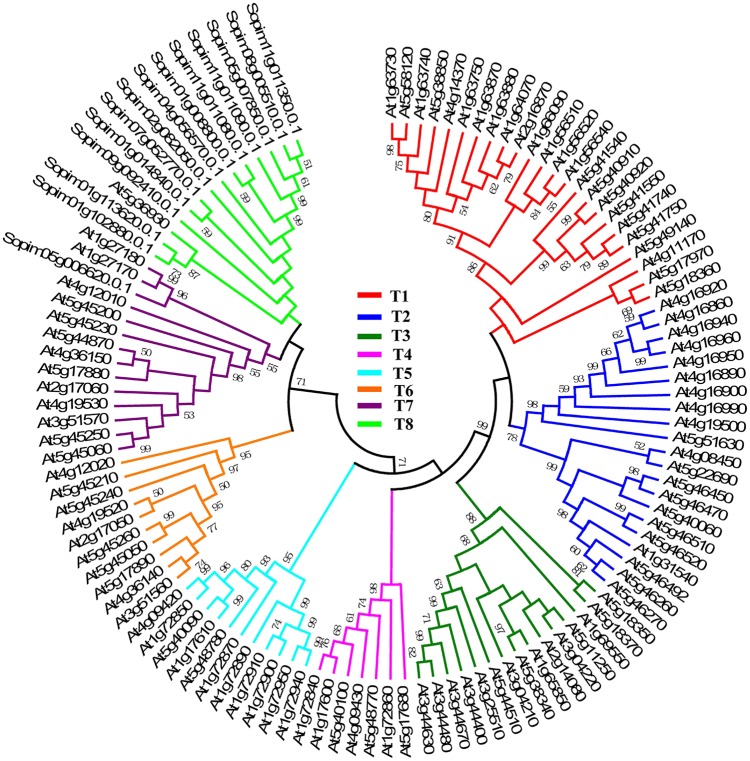
Figure 3.
Phylogenetic relationship of the TNL subfamily between Arabidopsis and S pimpinellifolium. Nucleotide-binding site and leucine-rich repeats are classified into 8 distinct branches (T1 T8), and these branches are shown in different colors in the phylogenetic tree, respectively. Numbers above branches represent the support values.
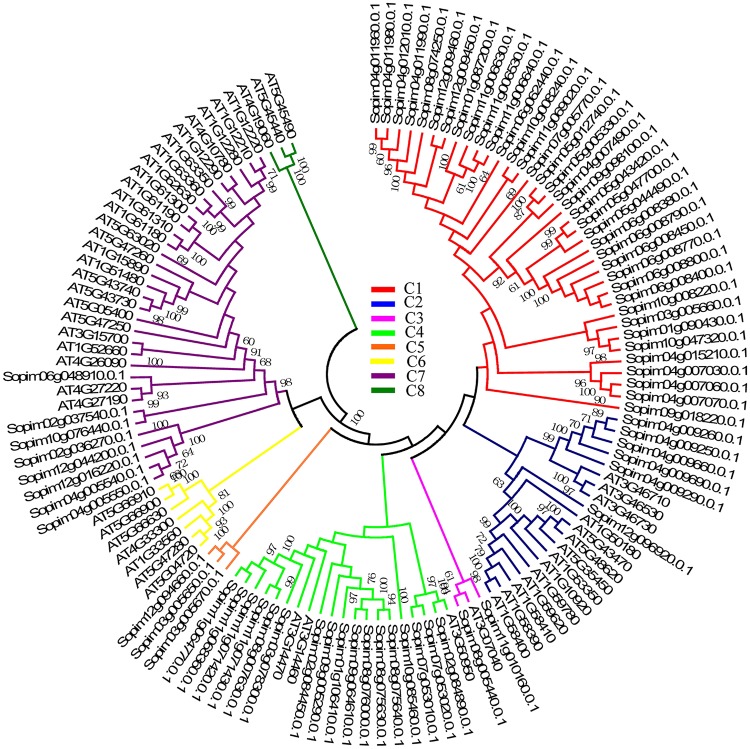
Figure 4.
Phylogenetic relationship of the CNL subfamily between Arabidopsis and S pimpinellifolium. Nucleotide-binding site and leucine-rich repeats are classified into 8 distinct branches (C1 C8), and these branches are shown in different colors in the phylogenetic tree, respectively. Numbers above branches represent the support values.
The phylogenetic tree of the TNL genes from the Arabidopsis and S. pimpinellifolium genomes is shown in Figure 3. The phylogenetic tree showed that the TNL genes were classified into 8 clades, namely T1 to T8. Among them, T1 to T6 only contained NBS-LRR genes from Arabidopsis, and the existence of multiple NBS-LRR gene copies across Arabidopsis suggests that the expansion of the NBS-LRR gene family occurred after differentiation of Arabidopsis and S. pimpinellifolium. One gene of S. pimpinellifolium was distributed in T7, while one gene of Arabidopsis was distributed in T8. Accordingly, 2 pairs of orthologous genes (Sopim05g006620.0.1 and At1g27170/At1g27180, and Sopim01g113620.0.1/Sopim01g102880.0.1 and At5g36930) were detected between the S. pimpinellifolium and Arabidopsis genomes. The phylogeny showed that multiple rounds of duplication events occurred in Arabidopsis.
In the CNL subfamily, 53 NBS-LRRs in Arabidopsis and 73 NBS-LRRs in S. pimpinellifolium were classified into 8 branches in the phylogenetic tree, namely, C1 to C8 (Figure 4). Genes from the same species tend to cluster together, some of them directly forming a single branch. For example, both the C1 and C5 branches were NBS-LRR proteins from S. pimpinellifolium, while NBS-LRR members of C6 and C8 belonged to the Arabidopsis genome.C1 was composed of 37 members, and C5 only contained 3 members. The C6 and C8 branches had 7 and 3 NBS-LRR genes, respectively. The remaining 4 branches (C2, C3, C4, and C7) harbored genes from these 2 species, but genes from different species tended to cluster. C4 contained a large number of S. pimpinellifolium NBS-LRRs (16) and only 3 members from Arabidopsis. C2 and C7 had similar patterns that contained genes from both species: C2 included 20 genes, 14 members from Arabidopsis and 6 from S. pimpinellifolium; C7 included 33 genes, 25 from Arabidopsis, and 8 from S. pimpinellifolium. In these branches, C1 was the largest among the 8 branches, while the C3, C5, and C8 branches were the smallest. Based on the phylogeny, a mass of paralogous genes were observed in both species, and there were 3 pairs of orthologous genes (At3g07040 and Sopim08g05440.0.1, At3g50950 and Sopim02g084890.0.1, and At4g27220/At4g27190 and Sopim06g048910.0.1).
Discussion
Wild species, as an important component of germplasm resources, contain resistance to disease and abiotic stress genes and play a key role in the hereditary improvement of the cultivated species. In disease-resistant tomato cultivars, a crowd of resistance genes was derived from the wild species.77 Among them, the wild tomato S. pimpinellifolium was one of the important wild types. Several NBS-LRR disease-resistant proteins from S. pimpinellifolium have been cloned.78 Therefore, an in-depth analysis of the NBS-LRR genes in S. pimpinellifolium will help promote the progress of disease resistance breeding in tomato.
In previous studies, the numbers of NBS-LRR gene families and members of each subfamily were different in distinct plants. In this study, a total of 245 NBS-LRR genes were identified, among which 78 genes belonged to the CNL subfamily and 15 genes belonged to the TNL subfamily (the ratio of CNL/TNL was about 5:1). A similar phenomenon has been found in other plants. For example, the number of CNL subfamilies in potato is 4.7 times that of that in the TNL subfamily,19 being the closest ratio with S. pimpinellifolium, and the ratio in grape and poplar are 3.8 and 2.0, respectively.28 Research has shown that the number of TNL subfamily genes is dominant in Arabidopsis and Brassica crops compared with other higher plants, which is due to the resistance of plants to the main pathological mechanism of long-term evolution.79 Likewise, a large number of CNL genes in S. pimpinellifolium were also used for resistance development to major diseases in Solanaceae. Also, it was found that there were distinctive characteristics of NBS-LRR disease resistance genes between monocotyledon and dicotyledon. Rice, as a typical monocotyledon, contained 402 CNL subfamily genes but TNL-type genes were missing.20 Similarly, the TNL-type genes were not detected in monocotyledon maize.22
There was no significant difference in the NBS-LRR disease resistance genes between S. pimpinellifolium and S lycopersicum (Table 1). However, the members of the NBS-LRR genes in each subfamily were varied, which implies that there were different patterns of expansion in the evolution from wild species to cultivated species. Interestingly, there was no significant difference in the number of NBS-LRRs on each chromosome in both species, indicating that they have high homology. Also, some NBS-LRR disease resistance genes may be from other wild relatives of tomato, rather than from S. pimpinellifolium.
The identified 245 NBS-LRR genes in S. pimpinellifolium were unevenly distributed across 12 chromosomes, most of which tended to form gene clusters of different sizes (Figure 1). The results are consistent with the distribution of NBS-LRR disease resistance genes of other plants.5,23,28 It was reported that the expansion of the NBS-LRR gene family on plant chromosomes can occur via gene recombination, transform, duplication, and selection.72,79,80 Tandem duplication genes with high similarity have common ancestors, and these duplicated genes were usually located in a homologous cluster. In S. pimpinellifolium, a total of 24 tandem duplication events, containing 80 NBS-LRR genes, were found in gene clusters.
In addition, there were lots of singleton genes in these 2 plant species. Approximately 42.9% (12/28) of singleton NBS-LRR genes shared a homologous relationship with the cloned resistance genes (Supplemental Table S2). Several singletons were homologous genes on other chromosomes, such as singletons Sopim08g074250.0.1, Sopim03g005660.0.1, and Sopim04g056570.0.1, and some of them seem to have evolved independently.
Two subfamilies, CNL and TNL, were identified through phylogenetic tree construction. The conserved motifs were used to distinguish the difference of protein sequences of N-terminal and NBS domains between these 2 subfamilies (Tables 3 and 4). Most of the conserved motifs were selectively distributed within a subfamily in the phylogenetic tree, implying that structural and functional similarities existed among NBS-LRRs within the same clade. In the TNL subfamily, all motifs were detected in all of the analyzed genes, except that the novel identified the TNBS-2 motif of the NBS domain was missed in 3 genes (Sopim09g092410.0.1, Sopim07g052770.0.1, and Sopim04g056570.0.1) (Supplemental Table S3 and Supplemental Figure S1). In contrast to the TNL subfamily, motifs of the CNL subfamily had much higher diversity (Supplemental Table S4 and Supplemental Figure S2). Some motifs were specific to each branch within the CNL subfamily, such as Motifs 03, 10, and 11 found in CNL4, while these motifs were not observed in CNL6. Whether this discovery reflected the more ancient origin of the CNL subfamily during plant evolution was unclear. We also found that some conserved motifs only existed in a particular clade, for example, Motif 19 and Motif 20 existed in the CNL2 and CNL3 branches, respectively. The motif analysis of the NBS-LRR genes in S. pimpinellifolium provided evidence of the complex evolutionary relationship among gene members in this family, which was also supported by the results of phylogenetic analysis.
Although the functions of most of these motifs have not been identified, it is plausible that some probably involved a crucial role. For instance, previous reports demonstrated that 3 domains (LRR, TIR, and CC) regulated downstream signaling events through intramolecular interactions.81-83 Proteins homologous to plant NBS-LRR proteins play a role in mammalian defense responses. In these mammalian proteins, the N-terminal domain is involved in downstream signaling partners through protein-protein interactions, the NBS hydrolyzes ATP functions as a regulatory domain, and LRR binds to upstream regulatory factors.84,85
When compared with the TNL and CNL subfamilies between S. pimpinellifolium and Arabidopsis, a large number of species-specific expansions of the TNL subfamily were detected in the Arabidopsis lineage after the divergence of these 2 species (Figure 3). By contrast, the number of genes in the CNL subfamily relatively decreased in Arabidopsis. Moreover, a species-specific expansion of NBS-LRR genes was also observed in Cucurbitaceae.86 These findings probably hint that these genes might have originated from NBS-encoding genes or resistance gene homologs. Comparative analysis of the CNL subfamily revealed that only 3 CNL genes were from Arabidopsis in the C4 branch, and the other members all belonged to S. pimpinellifolium which were abundant in this branch (Figure 4), suggesting that these 3 Arabidopsis genes were orthologous genes with those from S. pimpinellifolium. Also, the T8 branch belongs to this kind of clade (Figure 3).
Conclusions
In this study, a comprehensive and systematic analysis of the NBS-LRR genes of S. pimpinellifolium was conducted by the integration of chromosome distribution, phylogenetic relationships, and conserved motifs. The results revealed that a large number of NBS-LRR genes were present in gene clusters, and tandem duplication events were observed in these clusters. Phylogenetic analysis divided NBS-LRR genes into 2 distinct subfamilies, which was further supported by conservation or variation in motif compositions and functional divergence among clades. Moreover, a comparative analysis was performed in S. pimpinellifolium and Arabidopsis to reveal evolutionary relationships and functional characterization of the NBS-LRR gene family. This will provide a basis of information regarding the function research of NBS-LRR genes in S. pimpinellifolium, and possibly in other plant species.
Supplemental Material
Supplemental material, Figure_S1_xyz330564316c3b6 for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Figure_S2_xyz33056e5992d7c for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S1_xyz33056b5977b4d for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S2_xyz33056ee822a92 for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S3_xyz330562220ba64 for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S4_xyz33056bbc1857b for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was financially supported by the National Key Research and Development Program of China (2017YFE0114500), the State Key Laboratory Breeding Base for the Zhejiang Sustainable Pest and Disease Control (2010DS700124-ZZ1903 and 2010DS700124-ZZ1807), the National Natural Science Foundation of China (31772294), the Zhejiang Provincial major Agricultural Science and Technology Projects of New Varieties Breeding (2016C02051), the Zhejiang Provincial Natural Science Foundation of China (LY18C150008), the General Program from the National key research and development program (2017YFD0101902, 2018YFD1000800), and the earmarked fund for China Agriculture Research System (CARS-23-G-44).
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: HJW and HWW conceived and designed the research. JL, QWG, LZP, SLC, YC, MYR, QJY, and RQW performed the experiments. ZPY and GZZ analyzed the data and wrote the manuscript. HJW revised the manuscript. All authors read and approved the final manuscript.
ORCID iD: Hongjian Wan  https://orcid.org/0000-0001-7426-498X
https://orcid.org/0000-0001-7426-498X
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826-833. [DOI] [PubMed] [Google Scholar]
- 2. van der Hoorn RA, Kamoun S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Flor H. The complementary genic systems in flax and flax rust. Adv Genet. 1956;8:29-54. [Google Scholar]
- 4. Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323-329. [DOI] [PubMed] [Google Scholar]
- 5. Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schulze-Lefert P, Panstruga R. A molecular evolutionary concept connecting nonhost resistance pathogen host range and pathogen speciation. Trends Plant Sci. 2011;16:117-125. [DOI] [PubMed] [Google Scholar]
- 7. Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285-312. [DOI] [PubMed] [Google Scholar]
- 8. Lukasik E, Takken FLW. STANDing strong resistance proteins instigators of plant defence. Curr Opin Plant Biol. 2009;12:427-436. [DOI] [PubMed] [Google Scholar]
- 9. Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999;20:317-332. [DOI] [PubMed] [Google Scholar]
- 10. Pan Q, Wendel J, Fluhr R. Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol. 2000;50:203-213. [DOI] [PubMed] [Google Scholar]
- 11. Meyers BC, Morgante M, Michelmore RW. TIR-X andTIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002;32:77-92. [DOI] [PubMed] [Google Scholar]
- 12. Nooren IM, Kaptein R, Sauer RT, Boelens R. The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils. Nat Struct Biol. 1999;6:755-759. [DOI] [PubMed] [Google Scholar]
- 13. Liu J, Liu X, Dai L, Wang G. Recent progress in elucidating the structure function and evolution of disease resistance genes in plants. J Genet Genomics. 2007;34:765-776. [DOI] [PubMed] [Google Scholar]
- 14. Bai J, Pennill LA, Ning J, et al. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002;12:1871-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306-10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao YL, Ying C, Wei M, Yang Z, Hai HJ, Ming Z. Identification and characterization of NBS-encoding disease resistance genes in Lotus japonicus. Plant Syst Evol. 2010;289:101-110. [Google Scholar]
- 17. Panwar P, Jha AK, Pandey P, Gupta AK, Kumar A. Functional markers based molecular characterization and cloning of resistance gene analogs encoding NBS-LRR disease resistance proteins in finger millet (Eleusinecoracana). Mol Biol Rep. 2011;38:3427-3436. [DOI] [PubMed] [Google Scholar]
- 18. Sagi MS, Deokar AA, Tar’an B. Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to ascochyta blight infection. Front Plant Sci. 2017;8:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jupe F, Pritchard L, Etherington Mackenzie K, et al. Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics. 2012;13:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou T, Wang Y, Chen JQ, et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genomics. 2004;271:402-415. [DOI] [PubMed] [Google Scholar]
- 21. Lozano R, Hamblin MT, Prochnik S, Jannink JL. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics. 2015;16:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng Y, Li X, Jiang H, et al. Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J. 2012;279:2431-2443. [DOI] [PubMed] [Google Scholar]
- 23. Mun JH, Yu HJ, Park S, Park BS. Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol Genet Genomics. 2009;282:617-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kohler A, Rinaldi C, Duplessis S, et al. Genome-wide identification of NBS resistance genes in Populus trichocarp. Plant Mol Biol. 2008;66:619-636. [DOI] [PubMed] [Google Scholar]
- 25. Ameline-Torregrosa C, Wang BB, O’Bleness MS, et al. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008;146:5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paterson AH, Bowers JE, Bruggmann R, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551-556. [DOI] [PubMed] [Google Scholar]
- 27. Porter BW, Paidi M, Ming Alam M, Nishijima WT, Zhu YJ. Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genomics. 2009;281:609-626. [DOI] [PubMed] [Google Scholar]
- 28. Yang S, Zhang X, Yue JX, Tian D, Chen JQ. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics. 2008;280:187-198. [DOI] [PubMed] [Google Scholar]
- 29. Lozano R, Ponce O, Ramirez M, Mostajo N, Orjeda G. Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS ONE. 2012;7:e34775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cannon SB, Zhu H, Baumgarten AM, et al. Diversity distribution and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol. 2002;54:548-562. [DOI] [PubMed] [Google Scholar]
- 31. Ercolano MR, Sanseverino W, Carli P, Ferriello F, Frusciante L. Genetic and genomic approaches for mediated disease resistance in tomato: retrospects and prospects. Plant Cell Rep. 2012;31:973-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qi J, Song CP, Wang B, et al. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol. 2018;60:805-826. [DOI] [PubMed] [Google Scholar]
- 33. Li K, Yang FB, Miao YC, Song CP. Abscisic acid signaling is involved in regulating the mitogen-activated protein kinase cascade module AIK1-MKK5-MPK6. BMC Plant Biol. 2017;12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu LY, Li N, Yao CP, Meng SS, Song CP. Functional analysis of the ABA-responsive protein family in ABA and stress signal transduction in Arabidopsis. Chin Sci Bull. 2013;58:3721-3730. [Google Scholar]
- 35. Hao F, Zhao S, Dong H, Zhang H, Sun L, Miao C. Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J Integr Plant Biol. 2010;52:298-307. [DOI] [PubMed] [Google Scholar]
- 36. Song Y, Xiang F, Zhang G, Miao Y, Miao C, Song CP. Abscisic acid as an internal integrator of multiple physiological processes modulates leaf senescence onset in Arabidopsis thaliana. Front Plant Sci. 2016;7:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pang Y, Li J, Qi B, et al. Aquaporin AtTIP51 as an essential target of gibberellins promotes hypocotyl cell elongation in Arabidopsis thaliana under excess boron stress. Func Plant Biol. 2017;45:305-314. [DOI] [PubMed] [Google Scholar]
- 38. Sun L, Ma L, He S, Hao F. AtrbohD functions downstream of ROP2 and positively regulates waterlogging response in Arabidopsis. Plant Signal Behav. 2018;13:e1513300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu F, Liu H, Xu Y, et al. Heterogeneous expression of the cotton R2R3-MYB transcription factor GbMYB60 increases salt sensitivity in transgenic Arabidopsis. Plant Cell Tiss Organ Cul. 2018;133:15-25. [Google Scholar]
- 40. Lv S, Yu D, Sun Q, et al. Activation of gibberellin 20-oxidase 2 undermines auxin-dependent root and root hair growth in NaCl-stressed Arabidopsis seedlings. Plant Growth Regul. 2018;84:225-236. [Google Scholar]
- 41. Li L, Hou M, Cao L, et al. Glutathione S-transferases modulate Cu tolerance in Oryza sativa. Environ Exp Bot. 2018;155:313-320. [Google Scholar]
- 42. Guo S, Dai S, Singh PK, et al. A membrane-bound NAC-like transcription factor OsNTL5 represses the flowering in Oryza sativa. Front Plant Sci. 2018;9:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu LH, Wang WY, Guo JJ, et al. Zinc improves salt tolerance by increasing reactive oxygen species scavenging and reducing Na+ accumulation in wheat seedlings. Biol Plantarum. 2014;58:751-757. [Google Scholar]
- 44. Gao W, Xu FC, Guo DD, et al. Calcium-dependent protein kinases in cotton: insights into early plant responses to salt stress. BMC Plant Biol. 2018;18:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li W, Zhao F, Fang W, et al. Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L) employing iTRAQ-based proteomic technique. Front Plant Sci. 2015;6:732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang G, Lu T, Miao W, et al. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ. 2017;5:e4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao X, Wang YJ, Wang YL, Wang XL, Zhang X. Extracellular Ca2+ alleviates NaCl-induced stomatal opening through a pathway involving H2O2-blocked Na+ influx in Vicia guard cells. J Plant Physiol. 2011;168:903-910. [DOI] [PubMed] [Google Scholar]
- 48. Ma L, Zhang H, Sun L, et al. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J Exp Bot. 2012;63:305-317. [DOI] [PubMed] [Google Scholar]
- 49. Zhang J, Wang F, Zhang C, et al. A novel VIGS method by agroinoculation of cotton seeds and application for elucidating functions of GhBI-1 in salt-stress response. Plant Cell Rep. 2018;37:1091-1100. [DOI] [PubMed] [Google Scholar]
- 50. Zhao Q, Chen W, Bian J, et al. Proteomics and phosphoproteomics of heat stress-responsive mechanisms in spinach. Front Plant Sci. 2018;9:800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang PT, Liu H, Hua HJ, Wang L, Song CP. A vacuole localized beta-glucosidase contributes to drought tolerance in Arabidopsis. Chin Sci Bull. 2011;56:3538-3546. [Google Scholar]
- 52. Lü D, Wang W, Miao C. ATHK1 acts downstream of hydrogen peroxide to mediate ABA signaling through regulation of calcium channel activity in Arabidopsis guard cells. Chin Sci Bull. 2013;58:336-343. [Google Scholar]
- 53. Ma XN, Zhang XR, Yang L, et al. Hydrogen peroxide plays an important role in PERK4-mediated abscisic acid-regulated root growth in Arabidopsis. Func Plant Biol. 2018;46:165-174. [DOI] [PubMed] [Google Scholar]
- 54. Liang JY, Xia JY, Liu LL, Wan SQ. Global patterns of the responses of leaf-level photosynthesis and respiration in terrestrial plants to experimental warming. J Plant Ecol. 2013;6:437-447. [Google Scholar]
- 55. Zhao X, Wang YL, Qiao XR, et al. Phototropins function in high-intensity blue light-induced hypocotyl phototropism in Arabidopsis by altering cytosolic calcium. Plant Physiol. 2013;162:1539-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Han YJ, Wang XH, Chen WC, et al. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in flower petal of Osmanthus fragrans. Tree Genet Genomes. 2014;10:329-338. [Google Scholar]
- 57. Han Y, Wu M, Cao L, et al. Characterization of OfWRKY3, a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene OfCCD4 in Osmanthus fragrans. Plant Mol Biol. 2016;91:485-496. [DOI] [PubMed] [Google Scholar]
- 58. Zhao X, Li YY, Xiao HL, et al. Nitric oxide blocks blue light-induced K(+) influx by elevating the cytosolic Ca2+ concentration in Vicia faba L. guard cells. J Integr Plant Biol. 2013;55:527-536. [DOI] [PubMed] [Google Scholar]
- 59. Wang K, He JN, Zhao Y, et al. EAR1 negatively regulates ABA signaling by enhancing 2C protein phosphatase activity. Plant Cell. 2018;30:815-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li W, de Ollas C, Dodd IC. Long-distance ABA transport can mediate distal tissue responses by affecting local ABA concentrations. J Integr Plant Biol. 2018;60:16-33. [DOI] [PubMed] [Google Scholar]
- 61. Lu T, Zhang G, Sun L, Wang J, Hao F. Genome-wide identification of CBL family and expression analysis of CBLs in response to potassium deficiency in cotton. PeerJ. 2017;5:e3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen MY, Li K, Li HP, Song CP, Miao Y. The glutathione peroxidase gene family in Gossypium hirsutum: genome-wide identification classification gene expression and functional analysis. Sci Rep. 2017;7:44743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun Q, Wang GH, Zhang X, et al. Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci Rep. 2017;7:42418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Guo YW, Guo HL, Li X, et al. Two type III polyketide synthases from Polygonum cuspidatum: gene structure evolutionary route and metabolites. Plant Biotech Rep. 2013;7:371-381. [Google Scholar]
- 65. Liu RH, Meng JL. MapDraw: a Microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas. 2003;25:317-321. [PubMed] [Google Scholar]
- 66. Huang S, Gao Y, Liu J, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Genet Genomics. 2012;287:495-513. [DOI] [PubMed] [Google Scholar]
- 67. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 69. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696-704. [DOI] [PubMed] [Google Scholar]
- 70. Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202-W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Andolfo G, Sanseverino W, Rombauts S, et al. Overview of tomato (S lycopersicum) candidate pathogen recognition genes reveals important R locus dynamics. New Phytol. 2013;197:223-237. [DOI] [PubMed] [Google Scholar]
- 72. Friedman AR, Baker BJ. The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007;17:493-499. [DOI] [PubMed] [Google Scholar]
- 73. Schornack S, Ballvora A, Gurlebeck D, et al. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004;37:46-60. [DOI] [PubMed] [Google Scholar]
- 74. Ernst K, Kumar A, Kriseleit D, Kloos DU, Phillips MS, Ganal MW. The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 2002;31:127-136. [DOI] [PubMed] [Google Scholar]
- 75. Ori N, Eshed Y, Paran I, et al. The I2C family from the wilt disease resistance locusI2 belongs to the nucleotide binding leucine-rich repeat superfamily of plant resistance genes. Plant Cell. 1997;9:521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van der Vossen E, Sikkema A, Hekkert Bt, et al. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867-882. [DOI] [PubMed] [Google Scholar]
- 77. Stevens MA, Rick CM. Genetics and breeding. In: Atherton JG, Rudich J, eds. The Tomato Crop: A Scientific Basis for Improvement. New York, NY: Springer; 1987:35-109. [Google Scholar]
- 78. Rasul I, Zafar F, Ali MA, et al. Genetic basis for biotic stress resistance in plants from Solanaceae family: a review. Intl J Agric Biol. 2019;22:178-194. [Google Scholar]
- 79. Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004;20:116-122. [DOI] [PubMed] [Google Scholar]
- 80. Baumgarten A, Cannon S, Spangler R, May G. Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics. 2003;165:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hwang CF, Bhakta AV, Truesdell GM, Pudlo WM, Williamson VM. Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell. 2000;12:1319-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Luck JE, Lawrence GJ, Dodds PN, Shepherd KW, Ellis JG. Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell. 2000;12:1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moffett P, Farnham G, Peart J, Baulcombe DC. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 2002;21:4511-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hu Y, Benedict MA, Ding L, Nunez G. Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. EMBO J. 1999;18:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang L, Manji GA, Grenier JM, et al. PYPAF7 a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874-29880. [DOI] [PubMed] [Google Scholar]
- 86. Wan H, Yuan W, Bo K, Shen J, Pang X, Chen J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genomics. 2013;14:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Figure_S1_xyz330564316c3b6 for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Figure_S2_xyz33056e5992d7c for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S1_xyz33056b5977b4d for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S2_xyz33056ee822a92 for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S3_xyz330562220ba64 for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics
Supplemental material, Table_S4_xyz33056bbc1857b for Genomic Organization and Comparative Phylogenic Analysis of NBS-LRR Resistance Gene Family in Solanum pimpinellifolium and Arabidopsis thaliana by Huawei Wei, Jia Liu, Qinwei Guo, Luzhao Pan, Songlin Chai, Yuan Cheng, Meiying Ruan, Qingjing Ye, Rongqing Wang, Zhuping Yao, Guozhi Zhou and Hongjian Wan in Evolutionary Bioinformatics