Abstract
In this review, we propose a model of PsA as a complex genetically determined autoimmune-mediated disease having a heterogeneous variety of subphenotypes, with each subphenotype under the control of a different susceptibility-associated HLA allele. Since the specific HLA molecules encoded by each susceptibility allele dominantly select a T cell repertoire with the property of recognizing different peptides, we hypothesize each subphenotype reflects a distinct adaptive autoimmune response directed to different target molecules that is mediated by T cells within each selected repertoire. The interaction among the patients’ susceptibility alleles in the selection of their T cell repertoires determines a spectrum of overall clinical disease severity, varying from mild to severe. We further speculate that these different immune responses may result in activation of different immune effector pathways, which might therefore respond differently to various specific biologic agents.
Keywords: HLA allele, genotype, clinical subphenotype, T cell repertoire, autoimmune response
Rheumatology key messages
Different subphenotypes that characterize psoriatic arthritis are strongly associated with different HLA susceptibility alleles.
Each psoriatic arthritis subphenotype may reflect a distinct adaptive autoimmune response.
The interaction of distinct autoimmune responses in psoriatic arthritis may explain differences in disease severity.
Introduction
Heterogeneity in psoriatic disease
PsA is an inflammatory disease that variably affects synovial joints, tendons, entheses and axial sites. Psoriasis is often a feature and PsA develops in about 30% of patients with established skin and/or nail psoriasis. PsA is remarkably heterogeneous in its presentation, disease course and imaging findings compared with, for example, RA. Furthermore, responses to treatment can be quite disparate and divergent.
Clinical heterogeneity
The clinical presentation of PsA often differs considerably between affected patients. While a polyarticular presentation (>4 involved joints) is the most common, an oligo- or monoarticular presentation is not at all uncommon [1]. In some patients, the initial site of inflammation may be at entheses such as the achilles tendon insertion, in others there may be a swollen or dactylitic digit and in yet others, the spine may be the dominant site of inflammation. Throughout the course of the disease, most patients will experience joint inflammation, and enthesitis occurs in 60%, dactylitis in 50% and spinal inflammation may be present, depending on how it is assessed, in up to 30% [2]. Skin and/or nail involvement most commonly precedes musculoskeletal (MSK) inflammation by an average of 10 years but in 10–15%, the MSK inflammation may proceed or occur simultaneously with onset of skin psoriasis [3].
This clinical heterogeneity extends beyond MSK involvement. While plaque psoriasis is most commonly seen, some patients will show other psoriasis patterns including guttate, pustular or inverse forms. Nail changes, too, can be quite varied ranging from the commonly observed pitting and onycholysis to the uncommon but more severe crumbling and complete nail loss. Other organs and tissues are rarely involved including the uveal tract or the gastrointestinal tract. Fifty per cent of patients with established PsA will commonly have three or more features of the metabolic syndrome [4].
Imaging heterogeneity
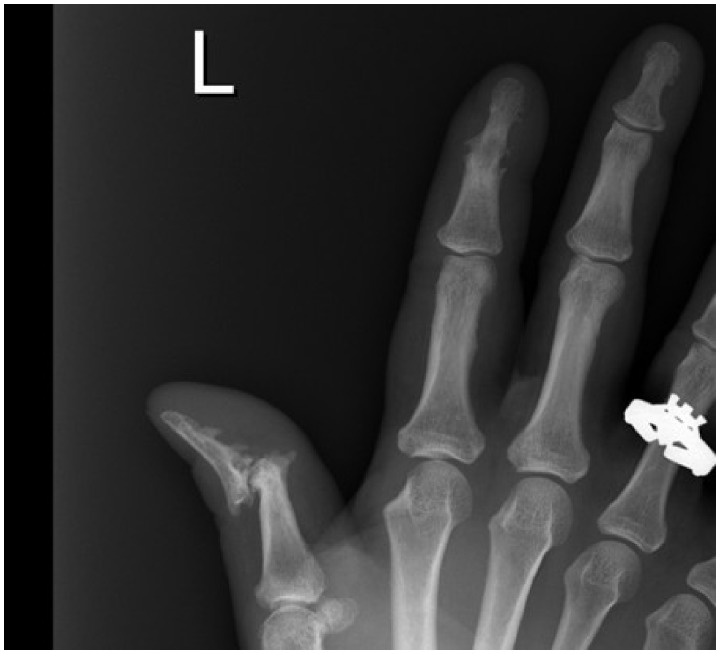
Patients with PsA may show bone destructive (erosions) or resorbing features (osteolysis), they may show features of new bone formation (periostitis, ankylosis), and in some patients both destructive and bone forming processes may occur and even be seen on the same radiograph (Fig. 1). New bone formation is a particular distinguishing feature of PsA with micro-CT studies showing prominent findings in some patients to the extent that it covers the bony surface as in a crown (corona) [5]. Erosions in PsA are also both quantitatively and qualitatively different from those seen in RA, being fewer in number, slower to develop and more Ω-shaped compared with the more U-shaped erosions in RA. Extra-articular bony changes may also be seen in some patients with PsA, including evidence of prominent enthesitis on MRI in about 30% of subjects [6], frequent involvement of tenosynovial structures on ultrasound and MRI [7, 8], and both symmetrical and asymmetrical patterns of involvement on plain radiographs in the sacroiliac joints (75% asymmetrical) and spine [9].
Fig. 1.
Hand radiograph from PsA patient demonstrating both new bone forming and resorptive features [12]
Close up of plain radiograph of left hand from patient with PsA showing a destructive mutilating process affecting the interphalangeal joint of the left thumb that has resulted in subluxation. There is also evidence of new bone formation with periostitis along the shaft of the distal phalange of the thumb and ankylosis of the second distal interphalangeal joint.
These diverse features have not yet been found to associate with particular patterns of markers of either bone formation or bone breakdown, but recent genetic studies (reviewed below) suggest that at least some of these imaging features may have a genetic basis.
Heterogeneous treatment responses
The best-designed, phase III randomized controlled trials in patients with PsA have been those conducted with biologic molecules and more recently with small molecules. It is of interest that despite the various cytokine and molecular pathways targeted, the response rates in terms of percentages reaching the primary outcome measure of an ACR 20% (ACR20) response remains at <60%, with 40% and 20% respectively reaching harder targets of ACR50 or ACR70. We might have hoped that non-response to inhibition of one cytokine pathway, such as TNF, might mean an improved response when targeting a different pathway, such as IL-17, but most studies have found reduced responses in those patients previously exposed to anti-TNF therapies [10]. The <60% ACR20 response rate, which is a minimal disease response measure, means of course that >40% do not respond. In addition, discordant responses are not uncommon with, for example, treatment targeting IL-17 resulting in sometimes dramatic improvements in skin psoriasis while features of peripheral arthritis may show little or no response and inflammatory bowel disease features may flair sometimes for the first time [11]. Clinicians have a difficult job trying to identify which drug to prescribe for which patient, often using an individual’s clinical features together with their comorbidity profiles and history of previous drug response as the best guide to treatment choice. This often means that patients cycle through several therapies before finding one that is effective, with this period of non-response contributing to disease progression and poor outcomes, as well as to increasing the societal and individual costs of having this disease.
Genotype determines both susceptibility and subphenotype
Because of the fundamental role played by the HLA alleles in regulating the character and target specificity of each individual’s adaptive immune response, and the increasing evidence that PsA reflects the action of an untoward immune response, the notion of ascertaining if particular HLA alleles could account for the occurrence of PsA and for some of its heterogeneous features was a clear direction to take. It is now evident that the main element responsible for the genetic determinism of the susceptibility to developing PsA and of its phenotypic heterogeneity is the role played by the HLA system.
The first question to be addressed was the nature of the relationship between psoriasis and PsA. Our approach was to compare the HLA genotypes of patients presenting to a rheumatological clinic with PsA and similarly ascertained patients presenting to a dermatological clinic with psoriasis and no clinical evidence of MSK inflammation [13]. While the frequency of the HLA-C*06:02 allele in patients with skin psoriasis and no MSK disease was ∼60% across all age groups, only a subset of under 30% of PsA patients had the HLA-C*06:02 allele. This is a highly significant difference that demonstrates that the psoriasis phenotype is genetically heterogeneous, but these results also provide two other insights: (i) that a subset of PsA patients are indeed genetically related to cutaneous psoriasis, since the frequency of this allele is 17–19% in the matched general population, and (ii) that PsA is genetically heterogeneous since 70% of PsA patients lack this susceptibility allele.
The next question of which HLA alleles were operating in the development of PsA in those lacking HLA-C*06:02 yielded the intriguing result that several HLA-B alleles are significantly increased in PsA but not in the psoriasis cohort [14]. These HLA-B alleles included B*08:01, B*27:05:02, B*38:01 and B*39:01. Indeed, the frequency of B*08:01 is significantly decreased in the psoriasis cohort. Moreover, in the PsA cohort several HLA-B alleles were found at a significantly decreased frequency, such as B*40:01 and B*44:01, suggesting they were ‘protective’ against the development of PsA. The notable feature is that all of these alleles are alternative genes at the HLA-B locus, although because of linkage disequilibrium, it is possible, and indeed perhaps likely, that some HLA-C alleles also contribute to susceptibility.
Since the role of the each different HLA molecule is to bind and present certain different types of peptides to the TCR, examining the known preferences of the HLA molecules for different peptides showed the HLA molecules associated with PsA susceptibility tended to be characterized by the property of binding peptides containing a positively charged amino acid at position 2 or 3 in the peptide. In contrast, those significantly decreased in frequency, such as B*40:01 and B*44:01, suggesting they were somewhat protective against the development of PsA, were characterized by a preference for binding peptides that had negatively charged amino acids at this position. These associations with HLA alleles, and the peptide-binding characteristics of the molecules they encode, provided additional evidence for the role of the recognition properties of the individual’s T cell repertoires in the development of PsA.
The finding of a single, well-etched disease entity that is mediated by elements of the immune system exhibiting a highly pluralistic association between disease susceptibility and multiple different HLA alleles was puzzling, but even in the early studies evidence emerged that different alleles were associated with quantitatively different traits such as whether the MSK features appeared more contemporaneously with the onset of psoriasis, as was the case with HLA-B*27:05:02 and B*39:01:01 patients, or whether MSK disease appeared only a decade or more after the onset of psoriasis, as was known to be the situation in the psoriasis cohorts characterized by HLA-C*06:02. This finding indicated that the action of the HLA-C*06:02 allele led to a highly penetrant cutaneous phenotype and a less penetrant, delayed development of MSK features, while the HLA-B*27:05:02 allele specified a more equivalently penetrant cutaneous and MSK phenotype. Patients with C*06:02 had more severe psoriasis and a delayed mean onset of milder PsA features of over 11 years after the onset of skin disease, accounting for their presence in a dermatology clinic, while those with B*27:05:02 had significantly milder skin disease that appeared more contemporaneously with the MSK features that brought them to the attention of the rheumatology clinic.
In further studies, specific additional elements of the heterogeneous clinical subphenotypes were preferentially associated with different susceptibility alleles in univariate analysis, summarized in Table 1 for three of the principal susceptibility alleles. B*08:01 was strongly associated with the characteristic asymmetric sacroiliitis of PsA (accounting for 75% of those with sacroiliitis), with dactylitis and with nail disease. B*27:05:02 was associated with symmetrical sacroiliitis (accounting for 25% of those with sacroiliitis), enthesitis and dactylitis. Reciprocally, there was a trend towards negative associations for B*27:05:02 with asymmetric sacroiliitis and B*08:01 with symmetrical sacroiliitis. Of interest, individuals with C*06:02 exhibited a trend towards a negative association with asymmetric sacroiliitis and were significantly less likely to have dactylitis and nail disease.
Table 1.
The influence of HLA genotype on clinical subphenotype
| HLA Genotype | ||||||
|---|---|---|---|---|---|---|
| B*08:01:01 | B*27:05:02 | C*06:02 | ||||
| Clinical subphenotype | OR | P | OR | P | OR | P |
| Asymmetric sacroiliitis | 1.97 | 0.008 | 0.49 | 0.07 | 0.54 | 0.36 |
| Symmetric sacroiliitis | 0.049 | 0.07 | 4.16 | <0.001 | 0.47 | 0.053 |
| Enthesitis | 0.81 | 0.33 | 3.65 | <0.001 | 1.09 | 0.68 |
| Dactylitis | 1.57 | 0.019 | 1.97 | 0.014 | 0.67 | 0.04 |
| Nail disease | 2 | 0.003 | 0.95 | 0.86 | 0.52 | 0.002 |
OR: odds ratio. Results were considered significant (bold) when P<0.05.
What does the finding of an HLA association tell us about the underlying autoimmune response?
The recognition properties of the T cells comprising the adaptive immune system of each person are determined by the individual’s particular HLA alleles. Each of the HLA molecules encoded by all of these HLA alleles in a person acts in the thymus to positively select the individual’s T cell repertoire, and does so by binding a self-peptide. The most overtly autoreactive T cells are eliminated by negative selection, but since a highly efficient negative selection process would remove all T cells, each of us is left with a T cell repertoire composed of T cells selected on self-peptides and self MHC. It will be the task of these T cells to recognize pathogen-related peptides that resemble the self-peptides that initially selected the T cell. These peptides are brought to the lymph node by antigen presenting cells, such as dendritic cells and macrophages, and initiate the adaptive immune response to the pathogen. Since different alleles encode HLA molecules that bind different peptides distinguished by their strong preferences for different anchoring amino acids, the repertoires selected by these alternative alleles recognize different self-peptides. According to the peptide composition of the pathogen, some individuals will have an advantage in recognizing certain pathogens. The large number of alternative HLA alleles at each HLA class I and class II locus make it likely that the recognition properties of each individual in terms of the specific peptides recognized are quite different from those of the next person. This heterogeneity in the recognition properties of the adaptive immune response is of great selective advantage to the species in responding to epidemics of infectious disease, and indeed, it is this selective advantage that drives the considerable diversity of the HLA system.
The fact that each individual’s T cell repertoire is selected on self-peptides lays the foundation for the development of a T cell response directed to one’s own proteins that results in autoimmunity and autoimmune diseases. In fact, the clonal TCR cannot usually distinguish between self-peptides and pathogen peptides. It is the inflammatory environment provided by the dendritic cell or other antigen presenting cells that activates the T cell. In the absence of pro-inflammatory signals, this presentation of self-peptides results in tolerization of the T cell to self. However, environmental factors, or polymorphisms of other genes that enhance the activation state of the antigen presenting cell, can create an inflammatory microenvironment that triggers the T cell to respond to a self-peptide, initiating the autoimmune response. Because the T cells of different individuals are selected on different peptides bound to different HLA molecules, the potential target of the autoimmune response accordingly differs.
We consider that this relationship between genotype and subphenotype suggests that a subphenotype, e.g. enthesitis, reflects a specific autoimmune attack on the molecules comprising the enthesis by T cells that were selected on HLA-B*27:05:02 molecules in the thymus, and among this repertoire of T cells might well have been some that were positively selected on peptides from self-molecules identical to those found in the enthesis, whereas B*08:01 molecules do not appear to select a T cell repertoire that mediates an autoimmune response to this target. In contrast, the subphenotype of dactylitis includes enthesitis, but involves other target structures, and here the association with both B*08:01 and B*27:05:02 alleles suggests that the T cell repertoires of individuals with either of these alleles can mediate a response to target molecules that manifests itself clinically as the development of dactylitis. Since each HLA molecule selects its own repertoire of T cells on self-peptides, the major implication of these observations is that different adaptive immune responses targeting different molecules contribute to the development of each of the clinical subphenotypes.
The perplexing difference in distribution between symmetric and asymmetric sacroiliitis, which appear otherwise radiographically identical, is supported by the strongly contrasting HLA association of symmetric and asymmetric sacroiliitis [9]. This genotypic difference in these two forms of axial spondyloarthritis appears consistent with the different patterns of axial involvement that predominate in PsA and in ankylosing spondylitis and suggests that different autoimmune responses targeting a different molecule in the SI joint underlie each pattern of involvement. A further possible implication of this concept would be that the spinal involvement in a HLA-B*27:05:02 PsA patient would be more analogous to that of ankylosing spondylitis in terms of natural history and perhaps therapeutic response than that of a HLA-B*08:01 PsA patient. The response to mechanical stress is a component of PsA, and one possibility to be explored is that the B*08:01 molecule selects a T cell repertoire that responds to peptides from a molecule induced by MSK stress.
The significantly decreased frequency of HLA-C*06:02 in PsA compared with patients with skin psoriasis only (30% vs 60%) found in the above studies and since confirmed by others [15], together with the negative association of HLA-C*06:02 with certain PsA severity features such as sacroiliitis and dactylitis used in the CASPAR criteria, may contribute to difficulty in the diagnosis of PsA in individuals with psoriasis that have HLA-C*06:02. Additionally, since HLA-C*06:02 is positively associated with more severe skin psoriasis, the overwhelming nature of the skin involvement may lead to failure to appreciate the milder MSK symptoms and findings. This may in part be the explanation also for the common clinical observation that patients presenting first to rheumatologists with their MSK symptoms tend to have mild or undiagnosed psoriasis.
PsA susceptibility alleles interact to result in more severe disease
Since each of us receives one HLA-B and HLA-C locus allele from each parent, an individual patient could receive different combinations of susceptibility or resistance alleles. One PsA patient might inherit two B*27:05:02 alleles, while another inherits a B*27:05:02 allele and a B*08:01 allele. A genetic risk score based on the combination of only six HLA haplotypes or alleles involved in PsA susceptibility was developed by Jon Giles [14]. To determine the influence of these combinations of susceptibility genes on PsA disease severity, a disease severity score was developed based on the presence of enthesitis, sacroiliitis, dactylitis, joint deformity, joint fusion, erosions or osteolysis. The greatest probability of being in the highest severity tertile was best modelled by a genetic risk score based on the combination of these six HLA haplotypes or alleles involved in PsA susceptibility. This analysis showed that the clinical severity of PsA is best explained by an additive model in which the positive and negative risk alleles participate in the final composite phenotype of more or less severe disease. For example, homozygosity for two HLA-B*27:05:02 alleles places one at the highest probability of being in the severe PsA disease tertile, while having one HLA-B*27:05:02 allele together with B*44:02 greatly reduces the likelihood of having severe disease.
Mechanistically, this additivity of the effects of positive and negative risk alleles resulting in a composite phenotype can best be interpreted by their mutual effect on the recognition and response properties of the patient’s overall T cell repertoire. Each allomorphic molecule acts independently, and thus dominantly, to select a T cell repertoire with the property of recognizing particular peptides bound to the HLA molecule. Since the allele score indicates that a patient with a B*27:05:02 allele and a B*44:02 allele has a lower propensity for severe disease than a patient with two B*27:05:02 alleles, one implication of this observation is that the presence of self-peptides presented by the B*44:02 molecule subtracts from the T cell repertoire generated by the B*27:05:02 molecule certain T cell clones that if they were otherwise present would mediate more severe disease. Clearly, larger series of patients are required to more precisely map the interaction among susceptibility and other potentially modifying alleles. Understanding the differences contributed by the individual’s entire HLA genotype, as well as the many other genes influencing the immune response, underlies the concept of personalized medicine, and such differences should explain some of the heterogeneity in therapeutic response.
This leads to the model of PsA as a complex autoimmune-mediated disease with a variety of subphenotypes, each subphenotype under the control of a different HLA allele that is associated positively or negatively with susceptibility through its action on the T cell repertoire [16]. We hypothesize each subphenotype reflects a distinct adaptive autoimmune response directed to different target molecules, but that act in concert to result in the spectrum of more or less severe PsA.
CD8+ T cells recognize antigen presented by HLA class 1 molecules, with studies showing that CD8+ T cells predominate in PsA synovial fluid [17]. CD8+ T cells obtained from PsA synovial tissue and fluid are clonally expanded, and following MTX therapy, a reduction in non-specific polyclonal T cell infiltration has been demonstrated [18, 19]. In a more recent study, PsA genetic risk variants were shown to co-localize with epigenetic markers of open chromatin preferentially in CD8+ memory T cells [20]. Furthermore, the association of human immunodeficiency virus infection with PsA suggests that when CD4+ T cells are depleted, persisting memory-effector CD8+ T cells can drive the development of PsA [21]. Given the above evidence, it can be proposed that in PsA, self-peptides derived perhaps from entheseal or synovial proteins are presented by antigen presenting cells expressing certain HLA class I alleles to CD8+ T cells. The CD8+ T cells that are specific for these self-peptide–MHC complexes thus become activated, with the activated state perpetuated by the continual supply of self-peptides.
We speculate that these different immune responses might be mediated by different effector T cells and accordingly respond differently to various newer pathway-specific therapeutic agents. In support of this speculation, there are some data to suggest that the heterogeneous treatment response in PsA might have a basis in the HLA genotype of the patient. This was shown to be clearly the case in psoriasis patients in which those patients who were HLA-C*06:02 positive had a significantly superior response to the IL-12–IL-23 blocking biologic ustekinumab, while HLA-C*06:02 negative patients were significantly more likely to respond to the TNF inhibitor adalimumab than to ustekinumab [22, 23]. Whether HLA genotype might also explain some of the heterogeneous responses in PsA has not yet been studied; however, because of the diversity and interactions among the susceptibility and phenotype-associated HLA alleles, and the possibility that only one facet of the disease might substantively respond, testing this hypothesis in a randomized clinical trial will be challenging and large numbers of patients may be required to show effect. It is possible, though, that association with one genotype, for example, will result in activation of a particular inflammatory pathway. In this regard, in unpublished work from our own laboratories, in B cell clones from PsA patients selected for disease-associated mutations (rs33980500 and rs13190832) in the TRAF3IP gene, those with single mutations in the TRAF3IP gene had no major effects on cytokine production (IL-6, IL-8), whereas cell lines with double mutations resulted in highly significant pro-inflammatory cytokine production.
Summary and next steps
Familial aggregation in PsA has certainly been demonstrated, with the recurrence ratio in first degree relatives of PsA patients reported at between 30 and 55, compared with between 4 and 10 for patients with psoriasis [24]. What has not been explored, however, is whether this familial aggregation relates to specific genetic associations such as those in the MHC region. Neither has it been explored whether clinical phenotypes or treatment responses tend to aggregate in PsA families. In addition, the genetic associations of PsA extend beyond the MHC region and there is no information yet available as to whether expression of non-MHC genotypes, either alone or in combination with the MHC, relates either to disease phenotype or to treatment responses.
The HLA associations described above are only a preliminary road map to the next phase of delineating the specific pathways through which the adaptive and innate immune system mediates each of the subphenotypes that comprise the many faces of PsA. In the class I-associated autoimmune diseases, we lack the development of sentinal autoantibodies that signal the preclinical phase of autoimmunity in lupus or RA, and this knowledge is particularly needed to address the potential to thwart the onset of the final stages of the immune response that result in PsA. Perhaps some combinations of HLA alleles will be found that have a highly predictive value for the development of PsA. It will be challenging to define whether certain therapeutic agents will be more effective in a specific subphenotype, or in a patient with a particular susceptibility allele, in view of the interactions among the alleles contributing to the composite phenotype. The allelic interactions will involve contributions from many additional alleles, beyond those associated with clinical disease. Already the relationship between genetic heterogeneity and clinical heterogeneity promises to be a significant chapter in personalized medicine. However, it is likely that additional alleles, well beyond the key six alleles identified in our studies, will be involved in this determination. Moreover, these additional HLA alleles, along with non-HLA alleles, may be found to account for some of the individual personalized variations in disease course and therapeutic response. An important caution is that this knowledge largely reflects studies performed in Caucasians, and needs to be replicated to other races and ethnicities, where it surely will provide novel insights.
Funding: This paper was published as part of a supplement funded by an educational grant from Novartis.
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1. Kane D, Stafford L, Bresnihan B, FitzGerald O.. A classification study of clinical subsets in an inception cohort of early psoriatic peripheral arthritis—‘DIP or not DIP revisited’. Rheumatology (Oxford) 2003;42:1469–76. [DOI] [PubMed] [Google Scholar]
- 2. Kane D, Stafford L, Bresnihan B, FitzGerald O.. A prospective, clinical and radiological study of early psoriatic arthritis: an early synovitis clinic experience. Rheumatology (Oxford) 2003;42:1460–8. [DOI] [PubMed] [Google Scholar]
- 3. Gladman DD, Shuckett R, Russell ML, Thorne JC, Schachter RK.. Psoriatic arthritis (PSA)—an analysis of 220 patients. Q J Med 1987;62:127–41. [PubMed] [Google Scholar]
- 4. Haroon M, Gallagher P, Heffernan E, FitzGerald O.. High prevalence of metabolic syndrome and of insulin resistance in psoriatic arthritis is associated with the severity of underlying disease. J Rheumatol 2014;41:1357–65. [DOI] [PubMed] [Google Scholar]
- 5. Finzel S, Englbrecht M, Engelke K. et al. A comparative study of periarticular bone lesions in rheumatoid arthritis and psoriatic arthritis. Ann Rheum Dis 2011;70:122–7. [DOI] [PubMed] [Google Scholar]
- 6. Marzo-Ortega H, Tanner SF, Rhodes LA. et al. Magnetic resonance imaging in the assessment of metacarpophalangeal joint disease in early psoriatic and rheumatoid arthritis. Scand J Rheumatol 2009;38:79–83. [DOI] [PubMed] [Google Scholar]
- 7. Kaeley GS. Use of ultrasound in psoriatic arthritis In: FitzGerald O, Gladman D, eds. Oxford textbook of psoriatic arthritis. Oxford, UK: Oxford University Press, 2018, 155–61. [Google Scholar]
- 8. McGonagle D, Eshed I.. MRI In: FitzGerald O, Gladman D, eds. Oxford textbook of psoriatic arthritis. Oxford, UK: Oxford University Press, 2018, 165–75. [Google Scholar]
- 9. Haroon M, Winchester R, Giles JT, Heffernan E, FitzGerald O.. Clinical and genetic associations of radiographic sacroiliitis and its different patterns in psoriatic arthritis. Clin Exp Rheumatol 2017;35:270–6. [PubMed] [Google Scholar]
- 10. Mease P. Biologic treatments for psoriatic arthritis apart from TNF inhibition In: FitzGerald O, Gladman D, eds. Oxford textbook of psoriatic arthritis. Oxford, UK: Oxford University Press, 2018, 281–6. [Google Scholar]
- 11. Coates LC, Kavanaugh A, Mease PJ. et al. Group for research and assessment of psoriasis and psoriatic arthritis 2015 treatment recommendations for psoriatic arthritis. Arthritis Rheumatol 2016;68:1060–71. [DOI] [PubMed] [Google Scholar]
- 12. FitzGerald O, Gladman D. editors, Oxford textbook of psoriatic arthritis. Oxford: Oxford University Press, 2018. [Google Scholar]
- 13. Winchester R, Minevich G, Steshenko V. et al. HLA associations reveal genetic heterogeneity in psoriatic arthritis and in the psoriasis phenotype. Arthritis Rheum 2012;64:1134–44. [DOI] [PubMed] [Google Scholar]
- 14. Haroon M, Winchester R, Giles JT. et al. Certain class I HLA alleles and haplotypes implicated in susceptibility play a role in determining specific features of the psoriatic arthritis phenotype. Ann Rheum Dis 2016;75:155–62. [DOI] [PubMed] [Google Scholar]
- 15. Eder L, Chandran V, Pellet F. et al. Human leucocyte antigen risk alleles for psoriatic arthritis among patients with psoriasis. Ann Rheum Dis 2012;71:50–5. [DOI] [PubMed] [Google Scholar]
- 16. FitzGerald O, Haroon M, Giles JT, Winchester R.. Concepts of pathogenesis in psoriatic arthritis: genotype determines clinical phenotype. Arthritis Res Ther 2015;17:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costello P, Bresnihan B, O'Farrelly C, FitzGerald O.. Predominance of CD8+ T lymphocytes in psoriatic arthritis. J Rheumatol 1999;26:1117–24. [PubMed] [Google Scholar]
- 18. Costello PJ, Winchester RJ, Curran SA. et al. Psoriatic arthritis joint fluids are characterized by CD8 and CD4 T cell clonal expansions appear antigen driven. J Immunol 2001;166:2878–86. [DOI] [PubMed] [Google Scholar]
- 19. Curran SA, FitzGerald OM, Costello PJ. et al. Nucleotide sequencing of psoriatic arthritis tissue before and during methotrexate administration reveals a complex inflammatory T cell infiltrate with very few clones exhibiting features that suggest they drive the inflammatory process by recognizing autoantigens. J Immunol 2004;172:1935–44. [DOI] [PubMed] [Google Scholar]
- 20. Bowes J, Budu-Aggrey A, Huffmeier U. et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat Commun 2015;6:6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winchester R, Bernstein DH, Fischer HD. et al. The co-occurrence of Reiter’s syndrome and acquired immunodeficiency. Ann Intern Med 1987;106:19–26. [DOI] [PubMed] [Google Scholar]
- 22. Talamonti M, Botti E, Galluzzo M. et al. Pharmacogenetics of psoriasis: hLA-Cw6 but not LCE3B/3C deletion nor TNFAIP3 polymorphism predisposes to clinical response to interleukin 12/23 blocker ustekinumab. Br J Dermatol 2013;169:458–63. [DOI] [PubMed] [Google Scholar]
- 23. Dand N, Duckworth M, Baudry D. et al. HLA-C*06:02 genotype is a predictive biomarker of biologic treatment response in psoriasis. J Allergy Clin Immunol 2019;143:2120–30. [DOI] [PubMed] [Google Scholar]
- 24. Winchester R, Rahman P.. Genetics of psoriatic arthritis In: FitzGerald O, Gladman D, eds. Oxford textbook of psoriatic arthritis. Oxford, UK: Oxford University Press, 2018, 57–68. [Google Scholar]