Abstract
With the emergence of multidrug-resistant and extensively drug-resistant bacterial pathogens, phage therapy and other alternative or additional therapeutic modalities are receiving resurgent attention. One of the major obstacles in developing effective phage therapies is the evolution of phage resistance in the bacterial host. When Pseudomonas aeruginosa was infected with a phage that uses O-antigen as receptor, phage resistances typically achieved through changing or loss of O-antigen structure. In this study, we showed that dsRNA phage phiYY uses core lipopolysaccharide as receptor and therefore efficiently kills the O-antigen deletion mutants. Furthermore, by phage training, we obtained PaoP5-m1, a derivative of dsDNA phage PaoP5, which is able to infect mutants with truncated O-antigen. We then generated a cocktail by mixing phiYY and PaoP5-m1 with additional three wide host range P. aeruginosa phages. The phage cocktail was effective against a diverse selection of clinical isolates of P. aeruginosa, and in the short-term constrained the appearance of the phage-resistant mutants that had beleaguered the effectiveness of single phage. Resistance to the 5-phage cocktail emerges after several days, and requires mutations in both wzy and migA Thus, this study provides an alternative strategy for designing phage cocktail and phage therapy.
Keywords: dsRNA bacteriophage, phage cocktail, Pseudomonas aeruginosa, phage resistance, antibiotic resis
Introduction
Pseudomonas aeruginosa is a common opportunistic pathogen that causes infections of the bloodstream, urinary tract, burn wounds, and is one of the major pathogens infecting the airways of cystic fibrosis patients and P. aeruginosa infections can be life-threatening (Jonckheere et al., 2018; Waters and Grimwood, 2018). Moreover, P. aeruginosa strains are frequently resistant to multiple classes of antibiotics (Lopez-Causape et al., 2018), and P. aeruginosa is a member of the ESKAPE pathogens (Boucher et al., 2009), which include six pathogens with well-recognized abilities to develop antibiotic resistance and cause deadly clinical outbreaks. With the emergence of multidrug-resistant isolates of P. aeruginosa (Sun et al., 2013; Lopez-Causape et al., 2018), phage therapy has received renewed attention (Forde and Hill, 2018; Jault et al., 2018; Kortright et al., 2019), and is a promising alternative approach for treating recalcitrant P. aeruginosa infections (Roach et al., 2017; Waters et al., 2017; Forti et al., 2018; Jault et al., 2018).
An optimal phage therapeutic agent necessitates some features: low immunogenicity, strictly lytic lifestyle, no toxins or antibiotic resistant genes, a wide host range against multiple isolates of the target pathogen, and the ability to constrain the emergence of phage-resistant mutants (Pires et al., 2017; Rohde et al., 2018; Kortright et al., 2019). The host range of a phage treatment can be expanded by incorporating several phages with different host-range specificities within a cocktail. The host range and killing efficiency of phage cocktails targeting P. aeruginosa have been studied in vitro and in vivo (Roach et al., 2017; Forti et al., 2018). Phage resistance is a somewhat more difficult problem to address, as bacteria have evolved a number of different mechanisms to defend themselves against bacteriophages, including prevention of phage adsorption and DNA injection, restriction enzymes, and CRISPR/Cas systems (Hyman and Abedon, 2010). These mechanisms are highly effective and emerge rapidly. For example, a 6-phage cocktail formulated against a broad host range of P. aeruginosa initially killed with great efficiency. As expected, phage-resistant mutants grew to a high density in vitro after only overnight incubation (Forti et al., 2018). However, in the mice and larvae infection model, the phage cocktail resistant mutants are not observed (Forti et al., 2018). When using animal models to evaluate the efficacy of phage therapy, phage resistance is rarely observed, because the resistant mutants usually have a fitness trade-off, or the minor mutants might be cleared by the immune system. On the contrary, phage resistant is quite common in vitro (Kortright et al., 2019). However, in clinics, a growing number of patients infected with multidrug resistant bacteria are immunocompromized, such as AIDS patients, transplant recipients with a suppressed immune system, diabetic patients, et al. These patients might not be able to clear the minor phage resistant bacteria and result in the treatment failure. Thus, the emergence of phage resistance has been reaffirmed by experts in the field as a key issue regarding the feasibility of phage therapy (Rohde et al., 2018; Kortright et al., 2019).
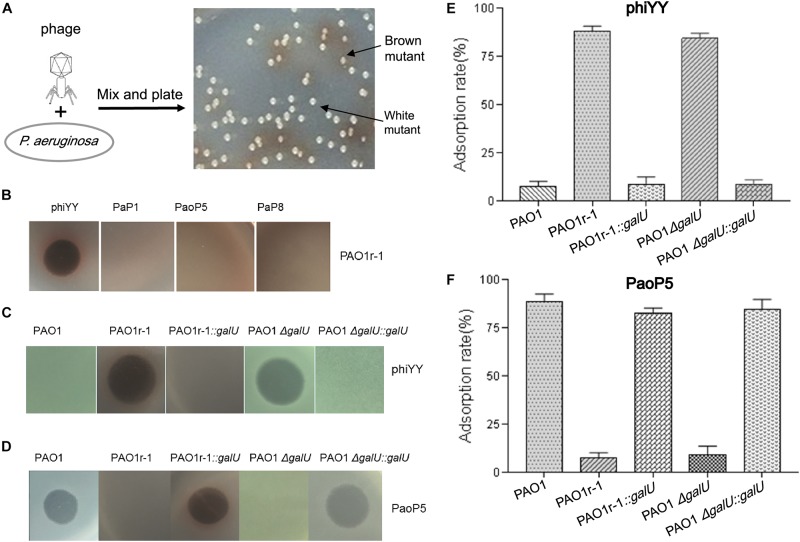
Pseudomonas aeruginosa develops phage resistance through several mechanisms, including modification or loss of the O-antigen component of lipopolysaccharide (LPS), or glycosylation of its type IV pilus (Harvey et al., 2018; Shen et al., 2018). Previously, we identified two types of phage-resistant P. aeruginosa mutants following infection with the dsDNA phages PaP1 or PaoP5, based upon colony pigmentation (Figure 1A). The mutant designated PAO1r-1, with a brown colony phenotype, contained a large chromosomal deletion including the genes galU and hmgA, which respectively, resulted in the complete loss of O-antigen and the accumulation of characteristic brown-colored homogentisic acids, while conferring phage resistance (Le et al., 2014; Shen et al., 2018). Meanwhile, the mutant designated PA1RG, with a white colony phenotype, contained mutations in wzy, an LPS biosynthesis gene, which resulted in a truncated O-antigen structure and also conferred phage resistance (Li et al., 2018). Thus, it seems mutations resulting in changes to LPS structure are likely to be a major source of phage resistance in P. aeruginosa. These phage-resistant mutants arose at a combined high frequency of ∼10–5 in vitro (Shen et al., 2018) and should be considered when designing a phage cocktail for eventual use in the clinical setting.
FIGURE 1.
dsRNA phage phiYY infects PAO1r-1 via exposed LPS core oligosaccharides (A): Colony phenotype of P. aeruginosa phage-resistant mutants, PAO1r-1 (brown) and PAO1w-1 (white). (B): Phage spot assay indicating that dsRNA phage phiYY, but not dsDNA phages PaP1, PaoP5, and PaP8, infects PAO1r-1. Phage spot assays (C,D) and phage adsorption assays (E,F) illustrating that phiYY infects and adsorbs to PAO1-derived strains without O-antigen, including PAO1r-1 and PAO1ΔgalU, while dsDNA phage PaoP5 infects and adsorbs to PAO1-derived strains with full O-antigen, such as PAO1 and the galU complement strains. (P < 0.05, one-way ANOVA, n = 3).
In this study, a phage cocktail designed to constrain the emergence of these phage-resistant phenotypes in P. aeruginosa was developed. The phage cocktail was successful in that it had a broader host range against a panel of P. aeruginosa clinical isolates compared to any of the tested single phages, and constrained the emergence of the previously described phage resistance phenotypes.
Results
The dsRNA Phage phiYY Infects Phage-Resistant PAO1r-1 With Exposed Core LPS
Previously, in P. aeruginosa PAO1, we identified two types of mutants that resist infection by the dsDNA phage, PaoP5 (Figure 1A). One of these, PAO1r-1 has lost the O-antigen and can be easily identified via a brown colony phenotype caused by the accumulation of homogentisic acids (Shen et al., 2018). To attempt to identify a phage that bypasses this resistance mechanism, and was able to kill PAO1r-1, a panel of lytic phages was examined (Table 1). While PAO1r-1 was resistant to dsDNA phages, including PaP1 (Lu et al., 2013), PaoP5 (Shen et al., 2016), and PaP8 (Table 1), the dsRNA phage, phiYY, was able to lyse PAOr-1 and form clear plaques (Figure 1B).
TABLE 1.
Bacterial strains and phages used in this study.
| Strain or phage | Description | Sources |
| PAO1 | Wild type P. aeruginosa strain | Jacobs et al., 2003 |
| PA1 | Wild type P. aeruginosa strain | Lu et al., 2015 |
| PAO1r-1 | Phage-resistant mutant with brown pigment | Shen et al., 2018 |
| PAO1r-1::galU | PAO1r-1 complemented with galU | This study |
| PAO1ΔgalU | PAO1ΔgalU from the transposon library | Jacobs et al., 2003 |
| PAO1ΔgalU::galU | PAO1ΔgalU complemented with galU | This study |
| PAO1w-1 | Phage-resistant mutant with a C1075T mutation in wzy | This study |
| PAO1w-1::wzy | PAO1w-1 complemented with wzy | This study |
| PAO1Δwzy | Knock out Δwzy in PAO1 background | This study |
| PAO1Δwzy::wzy | PAO1Δwzy complemented with wzy | This study |
| PAO1ΔmigA | PAO1ΔmigA from the transposon library | Jacobs et al., 2003 |
| PAO1ΔmigAΔwzy | Knock out wzy in PAO1ΔmigA | This study |
| phiYY | dsRNA bacteriophage isolated from Southwest hospital sewage | Yang et al., 2016 |
| PaP1 | dsDNA bacteriophage isolated from Southwest hospital sewage | Le et al., 2013 |
| PaP8 | dsDNA bacteriophage isolated from Southwest hospital sewage | This study |
| PaoP5 | dsDNA bacteriophage isolated from Southwest hospital sewage | Shen et al., 2016 |
| PaoP5-m1 | PaoP5 mutant phage with a A715C mutation in orf75 | This study |
As stated above, PAO1r-1 carries a deletion in galU and therefore lacks the O-antigen component of LPS, leaving the core oligosaccharides of LPS exposed on the cell surface (Choudhury et al., 2005; Le et al., 2014). Accordingly, phiYY is likely to utilize the LPS core oligosaccharide instead of O-antigen as the receptor to initiate infection (Figure 2). Because PAO1r-1 contains a large chromosomal deletion, a mutant containing a single-gene disruption of galU (PAO1ΔgalU), as well as complement strains restoring galU in both genetic backgrounds (PAO1r-1::galU and PAO1ΔgalU::galU), were examined for susceptibility to phiYY and PaoP5. As shown in Figure 1C, phiYY lysed PAO1r-1 and PAO1ΔgalU, but did not kill the galU complemented strains, indicating that loss of galU allows for phiYY infection. On the contrary, PaoP5 infected PAO1, PAO1r-1::galU and PAO1ΔgalU::galU, but not the PAO1ΔgalU mutant (Figure 1D), indicating that loss of galU prevents infection by PaoP5.
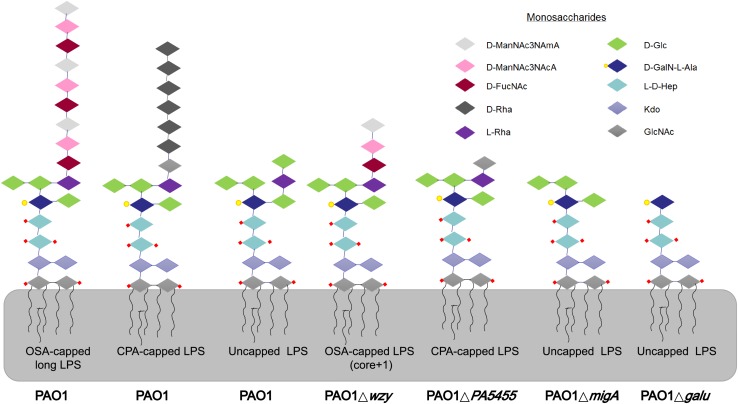
FIGURE 2.
LPS glycoforms on the surface of P. aeruginosa strains. Wild-type PAO1 forms OSA-capped LPS, CPA-capped, and uncapped LPS. PAO1Δwzy has only one copy of the typically repeating O-antigen sugar unit. PAO1ΔgalU has no O-antigen, exposing the uncapped core. PAO1ΔPA5455 has only GlcNAc, instead of the full-length CPA. PAO1ΔmigA lacks the L-Rha and D-Glc normally observed on the uncapped core. d-ManNAc3NAcA:di-N-acetylated mannuronic acid; D-FucNAc: d-2-amino-2,6-dideoxy-galactose-acetamido; D-Rha: D-rhamnose; L-Rha: L-rhamnose; D-Glc: D-glucose; D-GalN-L-Ala: d-galactosamine-l-alanyl; L-D-Hep: L-glycero-d-manno-heptose; Kdo: 3-deoxy-d-manno-oct-2-ulosonic acid; GlcNAc: glucose-acetamido.
A phage adsorption assay was used to further investigate the binding of these phages to the bacterial strains. phiYY adsorbs to PAO1r-1 and PAO1ΔgalU with high efficiency, but cannot bind to PAO1 or the galU complement strains (Figure 1E). Meanwhile, phage PaoP5 efficiently adsorbs to PAO1 and the galU complement strains, but not PAO1r-1 or PAO1ΔgalU (Figure 1F). Together, these data strongly indicate that the receptor for phiYY is the core oligosaccharide, while the receptor for PaoP5 is O-antigen. Therefore, phiYY may infect P. aeruginosa mutants lacking an O-antigen that arise following infection by O-antigen-binding phages, such as PaoP5.
Training the dsDNA Phage PaoP5 to Infect White Mutants With Truncated LPS
We previously identified PA1RG, a mutant of P. aeruginosa PA1, resistant to the dsDNA phage, PaP1, with a white colony phenotype (Li et al., 2018). This strain carries a C595T mutation in wzy, the B-band O-antigen polymerase, which results in a truncated O-antigen with only one copy of the normally-repeating sugar unit (Figure 2).
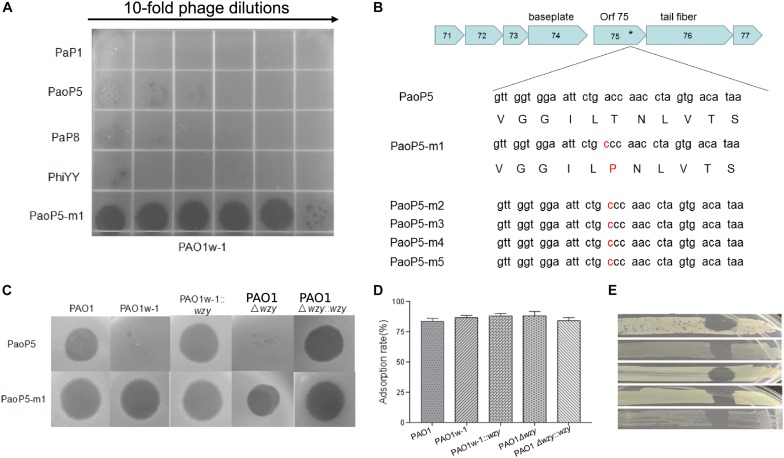
In this study, we isolated 5-phage-resistant PAO1 mutants with a white colony phenotype. Three out of these five isolates had mutations in wzy as detected by Sanger sequencing of the PCR fragment amplified from the isolates. Two mutants had a wzy C736G mutation and the third, designated PAO1w-1, contained a wzy C1075T mutation, which resulted in a premature stop codon.
PAO1w-1 was highly resistant to the phages PaP1, PaoP5, PaP8, and phiYY (Figure 3A). To attempt to isolate a mutant phage that is able to effectively infect PAO1w-1, 107 plaque forming units (pfu) of PaoP5 was mixed with PAO1w-1, and dozens of mutant phages that overcame resistance and lysed PAO1w-1 were observed. Five mutant phages were selected, purified and sequenced (Table 2). All 5-phage isolates that broke resistance of PAO1w-1 contained a A715C mutation in orf75, which was likely to be responsible for the expanded host range (Figure 3B). orf75 is located between the tail fiber gene and the baseplate gene. Though it is not annotated by blastp, Orf75 was identified as a structural protein by SDS-PAGE and high-performance liquid chromatography – mass spectrometry (Shen et al., 2016). Thus, the product of orf75 is highly likely to be a receptor-binding protein that is associated with host range.
FIGURE 3.
Phage PaoP5-m1 is able to infect P. aeruginosa phage-resistant mutants with truncated O-antigen structures. (A) Phage spot assay that indicates PaoP5-m1, but not PaP1, PaoP5, PaP8, and phiYY, efficiently infects PAO1w-1. (B) PaoP5-m1-5 contain an A715C mutation in orf75. PaoP5-m1 efficiently infects (C) and adsorbs (D) to all the PAO1 derived strains with full or truncated O-antigen structure. (E) Phage spot assay indicating five additional P. aeruginosa mutants selected by resistance to dsDNA phage PaoP5 are sensitive to PaoP-m1.
TABLE 2.
Primers used in this study.
| Primers | Sequence (5′–3′) |
| Validating wzy mutation | |
| Wzy-F1 | TCACTGAAACAGGTCGGTAT |
| Wzy-R1 | TACCTATAACAACAGCAATCG |
| Knockout of wzy | |
| Wzy-K-F(EcoRI) | gGAATTCCTCGCCGTGCACTTGCTCGT |
| Wzy-K-R(BamHI) | cgGGATCCGCCAAACACCTCATGTTCCA |
| Complementation of wzy | |
| Wzy –C-F(KpnI) | ggGGTACCCTTGCCGTCACTTTCTCCGA |
| Wzy –C-R(PstI) | aaCTGCAGGGAGTTGGCGCATATGCATA |
| Complementation of galU | |
| galU –C-F(BamHI) | cgGGATCCTATGATCAAGAAATGTC TTTTCCCG |
| galU –C-R(PstI) | aaCTGCAGTCAGTGAGCCTTGCC GGTCTTGT |
| Mapping mutations in PaoP5-m1 | |
| 062orf-F1 (structural protein-F1) | TTTTGGAAGACCTACCCCTC |
| 062orf-R1 (structural protein-R1) | GCCTTCCTGAACAACACTGA |
| 063orf-F1 | GACATGGCAATGGCGATTAC |
| 063orf-R1 | GTCGTAGTTGTAAAGAGCAG |
| 064orf-067orf-F1 | CGTCTGTGGTTCCGCATGGC |
| 064orf-067orf-R1 | AATTTAAGTTCCCCAGTAAG |
| 068orf-F1 (tape measure protein-F1) | CGGTTTTTTCTCTAATCGGT |
| 068orf-R1 (tape measure protein-R1) | CAAATCTAGACGTTGTGATAACA |
| 069orf-073orf-F1 | TAGCGACAGACTTCTAGCTC |
| 069orf-073orf-R1 | CTGGTAGGGCGAGCCGGATA |
| 074orf-F1 | TGGTAGACATTGTTGCTCAA |
| 074orf-R1 | TTCCAGCGAATCCAAAATAG |
| 075orf-F1 | ATAGAGATCAAGATCAGTGTAACA |
| 075orf-R1 | TCGCTACCTGTATGGGTACG |
| 076orf-F1 (tailfiber1-F1) | TCGAAGCTTATAAATTTGTATTCG |
| 076orf-R1 (tailfiber1-R1) | TCGATAGCAATCTCTACTTGACG |
| 077orf-F1 | AAACTGACGTAATGACTGATGC |
| 077orf-R1 | GGAATCTTGTCGGAATGGGT |
| 078orf-F1 (tailfiber2-F1) | TAGAGGGCTGTCCCACATCT |
| 078orf-R1 (tailfiber2-R1) | CAGTCTCTGTCCATGTTGACTG |
| 079orf-F1 (endolysin-F1) | GCCAATCGGATATTTCCAGA |
| 079orf-R1 (endolysin-R1) | TTGCGGCAGCTACTACGAG |
| 080-088orf-F1 | AGACTGCGCCTTGCAAAG |
| 080-088orf-R1 | CAAGCCAGAATACTACGTCTATGA |
One of the 5 mutant phages, designated PaoP5-m1, was selected for further study. Interestingly, unlike its parent strain, PaoP5, which did not infect PAO1w-1 or a single-gene inactivation of wzy (PAO1Δwzy), PaoP5-m1 was able to adsorb and infect all of the PAO1-derived strains, regardless of the presence of full-length or truncated O-antigen (Figures 3C,D). To test the host range of PaoP5-m1 against a larger panel of PaoP5-resistant mutants, we selected 30 white colonies surviving PaoP5 infection across 3 biological replicates. Strikingly, PaoP5-m1 infected all white mutants collected (Figure 3E), indicating that PaoP5-m1 is an excellent phage to eliminate P. aeruginosa mutants with truncated O-antigen structures.
Interestingly, similar phage training was unable to generate a mutant phage that infected PAO1r-1, bearing a complete lack of O-antigen (data not shown). This is likely due to the fact that the structures of the O-antigen and core oligosaccharides are quite different, compared to the structural differences between the full-length and truncated O-antigen (Lam et al., 2011). Thus, although phage mutants can adapt rapidly to a truncated receptor structure, they cannot adapt to a completely novel receptor structure in such a short period of time (Schwartz and Lindell, 2017).
Generation of a Five-Phage Cocktail With Broad Host Range
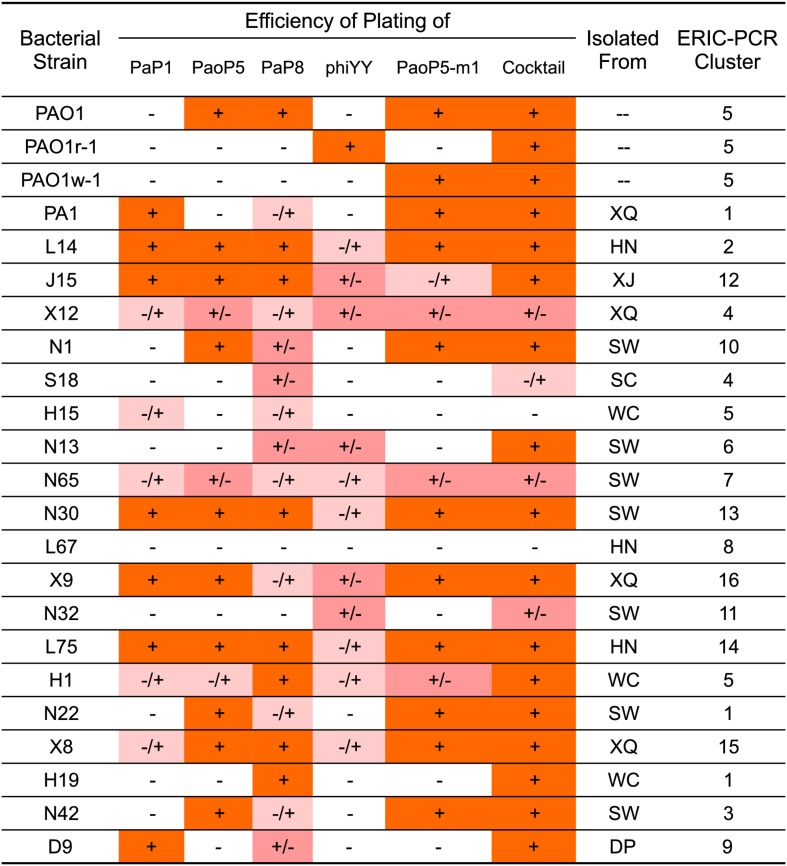
As the genotypes and phenotypes exemplified by PAO1r-1 and PAO1w-1 represented the two major types of resistant mutants which emerged following dsDNA phage treatment of P. aeruginosa, phiYY and PaoP5-m1 were included in a 5-phage cocktail we formulated to constrain the emergence of resistance during phage therapy of P. aeruginosa. In addition to phiYY and PaoP5-m1, the parent phage of PaoP5-m1, PaoP5, and 2 additional dsDNA phages with a wide host range (PaP1 and PaP8) were included in the phage cocktail. The effectiveness of this cocktail at killing P. aeruginosa was examined against a panel of strains that included the type strains PAO1 and PA1, the phage-resistant mutants PAO1-r-1 and PAO1-w-1, as well as 19 P. aeruginosa clinical isolates from seven Chinese hospitals (Yang et al., 2016). To maximize the diversity of the panel, the 19 clinical isolates were selected based on diverse Eric-PCR typing (Khosravi et al., 2016; Yang et al., 2016): two strains were selected from clusters 1, 4, and 5, while one strain was selected from each of the remaining 13 typing clusters (Figure 4).
FIGURE 4.
EOP of single phages or a 5-phage cocktail on a panel of 23 P. aeruginosa strains. The killing efficiency of each phage strain and cocktail were determined by an EOP assay. (+) = EOP 1; (±) = EOP 0.1–0.01; (–/+) = EOP 0.001; (–) = EOP < 0.0001. For clinical isolates of P. aeruginosa, the “Isolated From” column lists the hospital where the strain was isolated: SW, SouthWest Hospital; DP, DaPing Hospital; XQ, XinQiao Hospital; WC, West China Hospital; SC, SiChuan Provincial People’s Hospital; XJ, XiJing Hospital; HN, HeNan Provincial People’s Hospital.
The efficiency of plating (EOP) of each phage and the 5-phage cocktail was measured across the panel of 23 P. aeruginosa strains (Figure 4). EOP is more accurate than a simple dot plaque assay in determining phage sensitivity and host range, as the phage lysate is serially diluted to minimize the impact of other antimicrobial agents, such as lysin, within the lysate (Forti et al., 2018). Interestingly, phiYY was able to kill 12 of the strains on the panel, however the EOP was low. Each phage had a distinct host range, and no individual phage lysed all strains in the panel. As expected, the 5-phage cocktail had by far the widest host range and high EOP values.
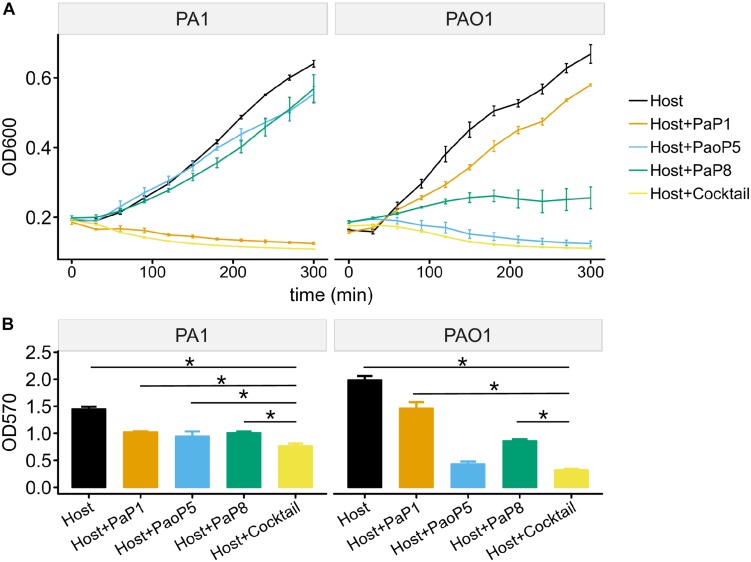
The Phage Cocktail Inhibits Bacterial Growth in Liquid Cultures and in Biofilms
To test the effects of the individual phages or the phage cocktail on a liquid culture of P. aeruginosa, and provide more detail regarding the lysis kinetics of each treatment, cultures of PAO1 or PA1 were infected with each phage or the cocktail, and the optical density (OD600) was monitored over 5 h (Figure 5A). For PA1, which is sensitive to PaP1, the addition of PaP1 resulted in a decrease in the OD600 of PA1 within 1 h after infection. Meanwhile, challenge with PaP8 did not significantly affect OD600 compared to the no phage negative control, which is likely due to the low infection efficiency of PaP8 against PA1, as seen above in Figure 4. In PAO1, phage PaoP5 infection caused a decrease in OD600 1–2 h post-infection, and PaP8 could kill and inhibit PAO1 growth, as indicated by the relatively stable OD600. When treated with the 5-phage cocktail, the OD600 of both the PAO1 and PA1 cultures decreased about 1 h after phage infection, indicating that the phage cocktail was effective in lysing liquid cultures of both P. aeruginosa strains, and it did so in a similar timeframe as the challenge with the single, effective phages.
FIGURE 5.
Effects of the 5-phage cocktail on liquid cultures and biofilms of P. aeruginosa. (A) Growth kinetics of P. aeruginosa cells in liquid culture in presence or absence of single phage strains or the phage cocktail. (B) Disruption of P. aeruginosa biofilms by single phage strains or the phage cocktail. (P < 0.05, one-way ANOVA, n = 3). The asterisks mark P-value of <0.05 as calculated by Student’s t-test between two groups.
The capability of phages to reduce biofilms formed by P. aeruginosa PA1 or PAO1 was examined using crystal violet staining. Following 24 h of biofilm formation by P. aeruginosa strains on 96-well plates, phages were applied individually or in the 5-phage cocktail. Plates were incubated for 4 h, at which point the biofilms were stained with crystal violet. Challenge with the phage cocktail caused a significant reduction in the biofilm biomass of both PA1 and PAO1 (Figure 5B), and eliminated biofilms with a slightly higher efficacy compared to that of challenge using the single, effective phage.
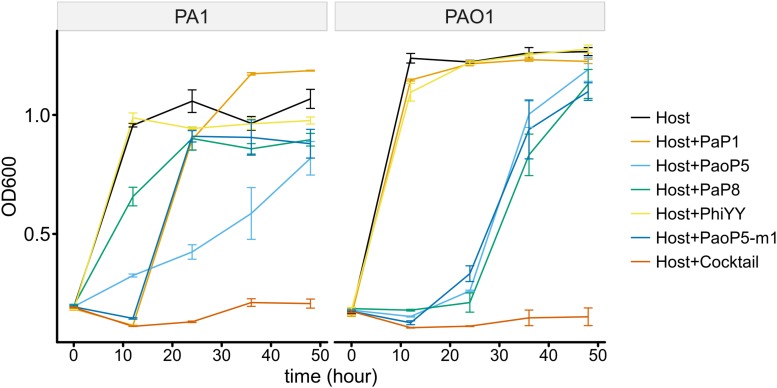
The Phage Cocktail Inhibits the Development of Phage-Resistant Bacteria in the Short-Term
To determine whether the 5-phage cocktail inhibits the emergence of phage-resistant mutants, P. aeruginosa PAO1 or PA1 was mixed with a single phage or the phage cocktail. When treated with a single phage, the OD600 of PAO1 and PA1 suffered an initial decrease during the first few hours. However, both strains re-grew to high densities after 12–24 h of incubation, most likely due to the growth of phage-resistant mutants. On the other hand, when treated with the 5-phage cocktail, the OD600 of both strains remained low even after 48 h of incubation (Figure 6). This data strongly indicates that the 5-phage cocktail not only efficiently kills P. aeruginosa, but also prevents the emergence of phage-resistant mutants in vitro.
FIGURE 6.
The 5-phage cocktail constrains phage-resistant mutants in the short-term. Growth kinetics of P. aeruginosa PA1 and PAO1 in liquid cultures in the presence or absence of single phage strains or the phage cocktail for 48 h.
After Extended Exposure to the Phage Cocktail, Resistant Isolates Emerge With Mutations in migA and wzy
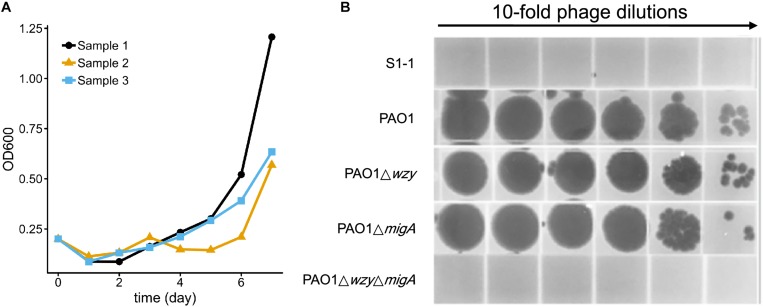
To test whether resistance to the 5-phage cocktail emerged after a longer period of time, the P. aeruginosa PAO1 culture containing the phage cocktail was incubated for an additional 7 days without the addition of fresh growth media (Figure 7A). In all three replicates, bacterial density finally began to increase after around 5 days (OD600 = 0.5∼1.1), indicating that resistance to the phage cocktail had developed. These cultures were plated on LB agar and two colonies were isolated from each plate (Table 3). The EOP of each selected mutant was examined, and all six isolates were indeed resistant to all phages in the cocktail as no plaques were formed. To determine a genetic cause of the phage resistance, these six isolates, and wild type PAO1, were subjected to whole genome sequencing. The sequencing data revealed that all 6 isolates contained mutations in both wzy and migA (Table 3).
FIGURE 7.
Resistance to the 5-phage cocktail emerges after 7 days, and requires mutations in wzy and migA. (A) Growth kinetics of P. aeruginosa PAO1 in liquid culture in the presence of the phage cocktail for 7 days. Three biological repeats were performed. (B) Phage spot assay illustrating that the phage cocktail resistant mutant S1-1 and PAO1ΔwzyΔmigA double mutant are resistant to infection by PaoP5-m1, while single-gene disruptions of PAO1Δwzy and PAO1ΔmigA are sensitive to PaoP5-m1.
TABLE 3.
Mutations found in phage cocktail resistant mutants.
| Sample | Nucleotide change | Amino acid change | Affected gene | Affected protein | Function |
| S1-1/S1-2 | T→G | H123P | migA | alpha-1,6-rhamnosyltransferase | Uncapped core |
| (T)7→9 | Shift (130/1317 nt) | wzy | B-band O-antigen polymerase | OSA | |
| S2-1 | G→A | Q42* | migA | alpha-1,6-rhamnosyltransferase | Uncapped core |
| (CTGGCC)2→3 | coding (1353/1689 nt) | fimL | hypothetical protein | Unkonwn | |
| G→A | P73S | wzy | B-band O-antigen polymerase | OSA | |
| (CCA)2→3 | coding (95/972 nt) | gmd | GDP-mannose 4,6-dehydratase | CPA | |
| S2-2 | G→A | Q42* | migA | Alpha-1,6-rhamnosyltransferase | Uncapped core |
| (CTGGCC)2→3 | coding (1353/1689 nt) | fimL | hypothetical protein | Unkonwn | |
| G→A | P73S | wzy | B-band O-antigen polymerase | OSA | |
| C→A | G31C | gmd | GDP-mannose 4,6-dehydratase | CPA | |
| S3-1 | C→T | R217Q | migA | Alpha-1,6-rhamnosyltransferase | Uncapped core |
| G→A | P73S | wzy | B-band O-antigen polymerase | OSA | |
| C→T | W151* | rmd | oxidoreductase Rmd | CPA | |
| S3-2 | C→T | R217Q | migA | Alpha-1,6-rhamnosyltransferase | Uncapped core |
| G→A | P73S | wzy | B-band O-antigen polymerase | OSA | |
| G→A | W100* | PA5455 | hypothetical protein | CPA |
PAO1 was infected with the phage cocktail in three biological repeats. Two phage cocktail resistant mutants S1-1 and S1-2 selected from sample 1 share the same mutation sites in migA and wzy. The other four mutants have additional mutations in gmd, rmd or PA5455, which are involved in CPA synthesis. * indicates nonsense mutation.
Many P. aeruginosa simultaneously synthesize multiple forms of lipopolysaccharide (Figure 2). These include the capped core glycoforms O-antigen-specific (OSA) and common polysaccharide antigen (CPA), as well as the uncapped core. The enzyme encoded by migA is responsible for the transfer of an L-rhamnose to glucose II of the core polysaccharide, and is required for the synthesis of uncapped core. Thus, the migA mutant lacks the L-rhamnose residue on the uncapped cores (Kocincova et al., 2012), and the wzy mutant lacks the OSA and produces “core + 1” LPS (Li et al., 2018). The sequencing data presented here implies that mutations in both wzy and migA are required for phage cocktail resistance. As expected, although both PAO1Δwzy and PAO1ΔmigA were sensitive to PaoP5-m1, a PAO1ΔwzyΔmigA double mutant was resistant to PaoP5-m1 (Figure 7B). This data implies that PaoP5-m1 may use either core antigen or OSA as receptors.
Among the six selected isolates resistant to the 5-phage cocktail, four had additional mutations. These mutations were located in the genes gmd, rmd or PA5455 (Table 3), all of three of which are involved in CPA synthesis (Lam et al., 2011; Hao et al., 2013). These mutations are less likely to be involved in the resistance to the phage cocktail seen in these strains, as the PAO1ΔwzyΔmigA double mutation was sufficient to confer resistance to the phage cocktail.
Discussion
Pseudomonas aeruginosa is an opportunistic pathogen which poses a serious health threat, particularly to patients with cystic fibrosis or traumatic burns, as well as the immunocompromised. P. aeruginosa is a master of antibiotic resistance, with intrinsic resistance to several drugs via low membrane permeability and expression of efflux pumps, and has a remarkable capacity to mutate and horizontally acquire additional traits leading to multidrug-resistant (MDR) and extensively-resistant (XDR) strains (Lopez-Causape et al., 2018). With the emergence of these phenotypes, which severely limit current treatment options, phage therapy is once again being explored as a potential therapeutic. However, phage therapy is still not widely used due to legislation issues and the lack of proper clinical trials and efficacy studies. As to the phage agents, one of the major barriers to successful phage therapy is the rapid emergence of phage-resistant mutants (Rohde et al., 2018).
Co-evolution of phages and their hosts is common in the natural environment, and the mutation and modification of receptors and receptor binding proteins by both phage and their hosts is well-understood as a major mechanism for this classic Red Queen phenomenon (Paterson et al., 2010). Indeed, one of the major mechanisms of phage resistance in P. aeruginosa is modifications to the LPS molecules exposed on the cell surface. As mutant phages with an expanded or altered host range can be isolated through phage training, and serve as complementary components of a therapeutic phage cocktail to prevent emergence of phage-resistant mutants, the goal of this study was to further characterize phage-resistant mutants of P. aeruginosa, perform phage training to isolate phages that break these resistant phenotypes, and develop a phage cocktail effective at constraining the development of phage-resistant P. aeruginosa during treatment.
The mutant strain PAO1r-1, derived from P. aeruginosa PAO1, was resistant to dsDNA phages, but is susceptible to infection by the dsRNA phage, phiYY (Figure 1). This is due to the fact that PAO1r-1 contains a large deletion, including galU, which leads to truncated LPS, with core oligosaccharides exposed. phiYY appears to utilize these core oligosaccharides as the receptor for infection (Figure 1). Further credence for this hypothesis was provided by the fact that phiYY did not infect or adsorb the parent strain with a complete LPS, or a the complement strains PAO1r-1::galU or PAO1ΔgalU::galU (Figure 1).
dsRNA phages are a unique group of phages, which are classified in the Cystoviridae family (Mantynen et al., 2018). Currently, there are only 7 sequenced dsRNA phages, and phiYY is the only one that infects human pathogen (Yang et al., 2016), while other 6 dsRNA phages all infects Pseudomonas syringae. phi6 is the first isolated dsRNA phage, and serves as an excellent model to study the biology of dsRNA phage (Bamford et al., 1987), while our study demonstrated the antimicrobial potential of dsRNA phage phiYY, which eliminates the brown mutants after infection with dsDNA phages. Moreover, brown mutants are also frequently detected from cystic fibrosis patients with chronic P. aeruginosa infections, suggesting the brown mutants can better survive in chronic infection (Smith et al., 2006; Hocquet et al., 2016). Thus, dsRNA phage phiYY might be an efficient agent to kill the brown mutants in chronic P. aeruginosa infections, which needs further investigation.
Meanwhile, mutations in another locus involved in LPS production, wzy, generated phage-resistant mutants in P. aeruginosa PA1, which express a truncated “core + 1” LPS with only one repeat of the O-antigen trisaccharide moiety. Similar phage-resistant wzy mutants were obtained in this study from a PAO1 genetic background (Figure 3). Phage training of PaoP5 generated mutant phages capable of breaking the resistance afforded to PAO1 by the wzy mutation harbored by PAO1w-1, including the phage isolate, PaoP5-m1 (Figure 3). PaoP5-m1 contains a mutation in orf75, and was capable of adsorbing to and infecting PAO1 and all tested wzy mutants that arose following infection of PAO1 with PaoP5 (Figure 3). Phage-host co-evolution could expand the host range of the phages and increase the resistance of evolved bacteria (Paterson et al., 2010). Thus, mutant phages with expanded or altered host range can be isolated through phage training, and serve as auxiliary components of a phage cocktail to eliminate the phage resistant bacteria. However, a similar phage training approach failed to produce phage that could break the resistance phenotype of PAO1r-1, likely because the exposed LPS core oligosaccharides of PAO1r-1 are too structurally disparate from the full-length LPS.
A 5-phage cocktail was generated containing three P. aeruginosa phages with a broad host range (PaP1, PaoP5, PaP8), as well as the two phages capable of breaking P. aeruginosa phage resistance mechanism described above (phiYY and PaoP5-m1). This phage cocktail was examined against a panel of P. aeruginosa strains including two laboratory type strains relevant to this study (PAO1 and PA1), the phage-resistant mutants described above (PAO1r-1 and PAO1w-1), and 19 clinical isolates which were selected to represent the 15 Eric-PCR typing clusters. The phage cocktail was able to infect and kill ∼90% of the strains tested, higher than each of the components of the cocktail individually, and did so with increased EOP values (Figure 4). The phage cocktail was effective in killing P. aeruginosa in liquid cultures and biofilms, in addition to solid media (Figure 5). While the phage cocktail was effective in constraining the emergence of phage-resistant mutants during the time frames utilized to develop the phage-resistant mutants described above, extended incubation out to 5–7 days led to growth of mutants resistant to the cocktail. All isolated mutants which were resistant to the cocktail had mutations in both wzy and migA. This development of resistance requiring mutations in multiple loci is a classic illustration of a Red Queen dynamic.
In the natural environment, phage-host co-evolution dynamics have been described as arms race fluctuation selection. It is intriguing to see that this study, while the inclusion of phiYY and PaoP5-m1, the two phages that target two main types of phage resistant mutants in P. aeruginosa, significantly constrained the emergence of phage resistant mutants, mutants carrying double or triple gene mutations, including wzy and migA eventually appeared after 5 days’ incubation. Thus, a better understanding of the mechanism of how these mutants evolved even after long term incubation is important in designing better phage-therapy strategy, and warrant further investigation.
Experimental Procedures
Bacterial Strains, Phages and Culture Conditions
The bacterial strains and phages in this work are listed in Table 1. P. aeruginosa strains were grown on Luria-Bertani (LB) broth at 37°C. When required, gentamicin (50 μg/ml), or tetracycline (35 μg/ml) were used at a final concentrations in LB broth as described below. To completely remove bacteria, dsDNA phages were treated with chloroform, while dsRNA phage was filtered multiple times through 0.22 μm filters (Millipore), because phiYY is chloroform sensitive. To generate the phage cocktail, 5-phages with the same titer were mixed to a final titer of 109pfu/ml.
Selection of Phage-Resistant Mutants
The process of isolating phage-resistant mutants and calculating the frequency of resistance has been described in detail previously (Shen et al., 2018). PAO1r-1 was previously isolated, tested and sequenced (Shen et al., 2018). To isolate a PAO1-derived white-phenotype mutant PAO1w-1, PAO1 was infected with PaoP5 and immediately plated on LB agar plates for 24 h. Then, the white mutants were isolated. The wzy gene was PCR amplified with primers listed in Table 2, and the PCR products were sequenced to identify the mutation site in wzy.
Construction of P. aeruginosa Strains
The knockout and complementation of galU and wzy were performed as previously described (Le et al., 2014; Li et al., 2018). Briefly, to complement galU in PAO1r-1 and PAO1ΔgalU, the galU gene was PCR amplified using primers galU–C-F and galU–C-R, and the PCR product was digested by BamHI/PstI, and cloned into the BamHI/PstI digested plasmid pUCP24 to generate pucp-galU. Then, pucp-galU was electroporated into PAO1r-1 or PAO1ΔgalU. The sample approach was applied to complement wzy in PAO1w-1 and PAO1Δwzy with primers Wzy–C-F and Wzy–C-R.
To make insertional deletion of wzy in PAO1 and PAO1ΔmigA, a fragment within wzy was amplified with primers Wzy-K-F and Wzy-K-R, and the PCR product was digested by BamHI/EcoRI, and cloned into the BamHI/EcoRI digested plasmid pEX18Gm to generate plasmid pEX-wzy. Then, pEX-wzy was electroporated into PAO1 or PAO1ΔmigA to generate insertional mutant.
Bacteriophage Adsorption Assay
A bacteriophage adsorption assay with various P. aeruginosa strains was performed according to a previously described protocol (Le et al., 2014). Briefly, the log phase bacterial cultures were harvested and resuspended in LB broth with an OD600 of 0.5. Phage was then added at an MOI of 0.01, and the adsorption proceeded at 37°C for 5 min. Then, 1 ml samples were collected and centrifuged at 16,000 × g for 1 min. The supernatant was filtrated using 0.22-μm-pore-size filters (Millipore). Then, the phage titer in the supernatant (t1) and the original phage stock (t0) were determined using double-agar plating assays. The phage adsorption rate was calculated as (t0−t1)/t0. The values of adsorption rate shown in the bar graph are the means and standard deviations are the error bars from three biological replicates.
EOP Assay and Phage Spot Assay
The EOP of each phage isolate and the 5-phage cocktail on P. aeruginosa strains was determined according to a previously described protocol (Forti et al., 2018). 5 μl of serial 10-fold dilutions of phage were spotted on double layer agar plates, on which a specific bacterial host was spread in the upper soft agar. The number of plaques observed after overnight incubation were compared to the number obtained on sensitive strain PAO1, PA1 or PAO1r-1. The phage spot assay is similar to the EOP assay. Mix 200 μl of bacterial culture with 4 ml of soft agar and pour in the agar plate. Then drop 1 μl of phage on the soft agar and observe the formation of plaque after overnight incubation.
Isolation and Sequencing of Mutant PaoP5 Phages
The process of phage training to break resistance has been described previously (Le et al., 2013). Briefly, ∼107 pfu of phage PaoP5 was mixed with 200 μl of log phase P. aeruginosa strain PAO1 (OD600 = 0.5) and plated in a double-layer plate. Dozens of plaques appeared after overnight culture. Five clear plaques were randomly selected and transferred into the LB medium. After serial 10- fold dilutions, the plaque assay was repeated with 10 μl of each dilution to obtain well-separated plaques. The same procedure was repeated twice to purify each mutant phage. After purification of mutant phage, genomic DNA was extracted as previously described (Le et al., 2013). Subsequently, the whole region encoding the structural proteins (Orf62 to Orf 88) was amplified by PCR using the primers indicated in Table 2. PCR amplicons were then purified and sequenced using Sanger technology. The mutation site was determined by blast the sequencing data with PaoP5 genome (GeneBank accession number KU297675).
Biofilm Disruption Experiment
Crystal violet staining was used to monitor biofilm disruption (Forti et al., 2018). An overnight culture of P. aeruginosa was diluted to 0.02 OD600 in LB broth in 96-well polystyrene microtiter plates, which were then incubated at 37°C for 24 h. The broth was removed and the wells were washed with 200 μl of LB. Then, 200 μl of LB or LB containing phage or phage cocktail was added, and incubated for 4 h at 37°C. Then, crystal violet staining was applied as previously described (Forti et al., 2018). Three biological replicates were performed.
Selection of Mutants Resistant to the Phage Cocktail
100 μl of phage cocktail was added into 5 ml exponential phase PAO1 (OD600 = 0.2). The culture was incubated at 37°C for 7 days with shaking, and the OD600 was monitored.
Bacterial Genome Sequencing
Bacterial genomic DNA was extracted from six mutants which were resistant to the phage cocktail and wild type PAO1 using the UNlQ-10 Column Bacterial Genomic DNA Isolation Kit (Sangon Biotec), and then sequenced using an Illumina Hiseq 2500 platform (∼1 Gbp/sample). Fastp (Chen et al., 2018) was used for adapter trimming and quality filtering after de-multiplexing the raw reads. Mutations were identified in clean reads using Breseq (Barrick et al., 2014) with PAO1 (GenBank accession: NC_002516.2) as the reference genome. The sequence data is available in the NCBI Sequence Read Archive under SRA accession number PRJNA517283.
Statistical Analysis
The statistical analysis was performed using One-way ANOVA or student’s t test. A P value < 0.05 was considered as statistically significant.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: NCBI Sequence Read Archive under SRA accession number PRJNA517283.
Author Contributions
SL and FH conceived the study. YY, WS, and QZ performed the experiments. QC, KX, XJ, and JW analyzed the data. SL, XH, and JB wrote the manuscript. All authors read and approved the final manuscript for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was supported by the National Natural Science Foundation of China (NSFC, 31870167 to SL) and by NIH/NIDCR F32-DE026947 (JB).
References
- Bamford D. H., Romantschuk M., Somerharju P. J. (1987). Membrane fusion in prokaryotes: bacteriophage phi 6 membrane fuses with the Pseudomonas syringae outer membrane. The EMBO J. 6 1467–1473. 10.1002/j.1460-2075.1987.tb02388.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Colburn G., Deatherage D. E., Traverse C. C., Strand M. D., Borges J. J. (2014). Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. 10.1186/1471-2164-15-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B. (2009). Bad bugs, no drugs: no ESKAPE! an update from the infectious diseases society of America. Clin. Infect. Dis. 48 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury B., Carlson R. W., Goldberg J. B. (2005). The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohyd.Res. 340 2761–2772. 10.1016/j.carres.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Forde A., Hill C. (2018). Phages of life - the path to pharma. Br.J.Pharmacol. 175 412–418. 10.1111/bph.14106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti F., Roach D. R., Cafora M., Pasini M. E., Horner D. S., Fiscarelli E. V. (2018). Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 62:e02573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., King J. D., Huszczynski S., Kocincova D., Lam J. S. (2013). Five new genes are important for common polysaccharide antigen biosynthesis in Pseudomonas aeruginosa. mBio 4 e631–e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey H., Bondy-Denomy J., Marquis H., Sztanko K. M., Davidson A. R., Burrows L. L. (2018). Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation. Nat.Microbiol. 3 47–52. 10.1038/s41564-017-0061-y [DOI] [PubMed] [Google Scholar]
- Hocquet D., Petitjean M., Rohmer L., Valot B., Kulasekara H. D., Bedel E. (2016). Pyomelanin-producing Pseudomonas aeruginosa selected during chronic infections have a large chromosomal deletion which confers resistance to pyocins. Environ. Microbiol. 18 3482–3493. 10.1111/1462-2920.13336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman P., Abedon S. T. (2010). Bacteriophage host range and bacterial resistance. Adv.Appl.Microbiol. 70 217–248. 10.1016/S0065-2164(10)70007-1 [DOI] [PubMed] [Google Scholar]
- Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S. (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100 14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jault P., Leclerc T., Jennes S., Pirnay J. P., Que Y. A., Resch G. (2018). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. LancetInfect.Dis. 19 35–45. 10.1016/s1473-3099(18)30482-1 [DOI] [PubMed] [Google Scholar]
- Jonckheere L., Schelstraete P., Van Simaey L., Van Braeckel E., Willekens J., Van Daele S. F. (2018). Establishing the diagnosis of chronic colonization with Pseudomonas aeruginosa of cystic fibrosis patients: comparison of the European consensus criteria with genotyping of P. aeruginosa isolates. J.Cyst.Fibrosis 17 729–735. 10.1016/j.jcf.2018.03.004 [DOI] [PubMed] [Google Scholar]
- Khosravi A. D., Hoveizavi H., Mohammadian A., Farahani A., Jenabi A. (2016). Genotyping of multidrug-resistant strains of Pseudomonas aeruginosa isolated from burn and wound infections by ERIC-PCR. Acta Cir.Bras. 31 206–211. 10.1590/S0102-865020160030000009 [DOI] [PubMed] [Google Scholar]
- Kocincova D., Ostler S. L., Anderson E. M., Lam J. S. (2012). Rhamnosyltransferase genes migA and wapR are regulated in a differential manner to modulate the quantities of core oligosaccharide glycoforms produced by Pseudomonas aeruginosa. J.Bacteriol. 194 4295–4300. 10.1128/JB.05741-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortright K. E., Chan B. K., Koff J. L., Turner P. E. (2019). Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25 219–232. 10.1016/j.chom.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Lam J. S., Taylor V. L., Islam S. T., Hao Y., Kocincova D. (2011). Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front.Microbiol. 2:118. 10.3389/fmicb.2011.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S., He X. S., Tan Y. L., Huang G. T., Zhang L., Lux R., et al. (2013). Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PloS one 8:8. 10.1371/journal.pone.0068562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S., Yao X., Lu S., Tan Y., Rao X., Li M. (2014). Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci.Rep. 4 4738. 10.1038/srep04738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Shen M., Yang Y., Le S., Li M., Wang J. (2018). Adaptation of Pseudomonas aeruginosa to Phage PaP1 Predation via O-Antigen polymerase mutation. Front.Microbiol. 9:1170. 10.3389/fmicb.2018.01170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Causape C., Cabot G., Del Barrio-Tofino E., Oliver A. (2018). The versatile mutational resistome of Pseudomonas aeruginosa. Front.Microbiol. 9:685. 10.3389/fmicb.2018.00685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. G., Le S., Tan Y. L., Zhu J. M., Li M., Rao X. C. (2013). Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa Phage PaP1: establishment of genus PaP1-Like Phages. PloS One 8:13. 10.1371/journal.pone.0062933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Le S., Li G., Shen M., Tan Y., Zhao X. (2015). Complete genome sequence of Pseudomonas aeruginosa PA1, Isolated from a patient with a respiratory tract infection. Genome Announc. 3:e01453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantynen S., Sundberg L. R., Poranen M. M. (2018). Recognition of six additional cystoviruses: Pseudomonas virus phi6 is no longer the sole species of the family cystoviridae. Arch. Virol. 163 1117–1124. 10.1007/s00705-017-3679-4 [DOI] [PubMed] [Google Scholar]
- Paterson S., Vogwill T., Buckling A., Benmayor R., Spiers A. J., Thomson N. R. (2010). Antagonistic coevolution accelerates molecular evolution. Nature 464 275–278. 10.1038/nature08798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. P., Melo L., Vilas Boas D., Sillankorva S., Azeredo J. (2017). Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 39 48–56. 10.1016/j.mib.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Roach D. R., Leung C. Y., Henry M., Morello E., Singh D., Di Santo J. P. (2017). Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22 38.e–47.e. [DOI] [PubMed] [Google Scholar]
- Rohde C., Resch G., Pirnay J. P., Blasdel B. G., Debarbieux L., Gelman D. (2018). Expert opinion on three phage therapy related topics: bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 10:E178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. A., Lindell D. (2017). Genetic hurdles limit the arms race between Prochlorococcus and the T7-like podoviruses infecting them. Isme J. 11 1836–1851. 10.1038/ismej.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Zhang H., Shen W., Zou Z., Lu S., Li G. (2018). Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic Acids Res. 46 4505–4514. 10.1093/nar/gky160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M. Y., Le S., Jin X. L., Li G., Tan Y. L., Li M. (2016). Characterization and comparative genomic analyses of Pseudomonas aeruginosa Phage PaoP5: new members assigned to PAK_P1-like viruses. Sci.Rep. 6 6. 10.1038/srep34067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. E., Buckley D. G., Wu Z. N., Saenphimmachak C., Hoffman L. R., D’Argenio D. A. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 103 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. J., Shi H. Q., Zhang X. B., Fang Y. D., Chen Y. C., Chen J. H. (2013). Detection of carbapenemase-encoding genes among clinical Isolates of Pseudomonas aeruginosa in a chinese burn unit. J. Burn Care Res. 34 453–458. 10.1097/BCR.0b013e3182700afd [DOI] [PubMed] [Google Scholar]
- Waters E. M., Neill D. R., Kaman B., Sahota J. S., Clokie M. R. J., Winstanley C., et al. (2017). Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 72 666–667. 10.1136/thoraxjnl-2016-209265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters V., Grimwood K. (2018). Defining chronic Pseudomonas aeruginosa infection in cystic fibrosis. J.Cyst.Fibrosis 17 292–293. 10.1016/j.jcf.2018.03.007 [DOI] [PubMed] [Google Scholar]
- Yang Y. H., Lu S. G., Shen W., Zhao X., Shen M. Y., Tan Y. L. (2016). Characterization of the first double-stranded RNA bacteriophage infecting Pseudomonas aeruginosa. Sci.Rep. 6 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: NCBI Sequence Read Archive under SRA accession number PRJNA517283.