Abstract
Contamination of soils with heavy metals, such as nickel (Ni), is a major environmental concern due to increasing pollution from industrial activities, burning of fossil fuels, incorrect disposal of sewage sludge, excessive manure application and the use of fertilizers and pesticides in agriculture. Excess Ni induces leaf chlorosis and inhibits plant growth, but the mechanisms underlying growth inhibition remain largely unknown. A detailed analysis of root development in Arabidopsis thaliana in the presence of Ni revealed that this heavy metal induces gravitropic defects and locally inhibits root growth by suppressing cell elongation without significantly disrupting the integrity of the stem cell niche. The analysis of auxin-responsive reporters revealed that excess Ni inhibits shootward auxin distribution. Furthermore, we found that PIN2 is very sensitive to Ni, as the presence of this heavy metal rapidly reduced PIN2 levels in roots. A transcriptome analysis also showed that Ni affects the expression of many genes associated with plant cell walls and that Ni-induced transcriptional changes are largely independent of iron (Fe). In addition, we raised evidence that excess Ni increases the accumulation of reactive oxygen species and disturbs the integrity and orientation of microtubules. Together, our results highlight which processes are primarily targeted by Ni to alter root growth and development.
Keywords: Arabidopsis thaliana, Heavy metals, Polar auxin transport, Reactive oxygen species, Root development, Transcriptome
Introduction
The transition metal nickel (Ni) is an essential element for many organisms, including plants, bacteria and fungi. In microorganisms, several metalloenzymes rely on Ni as a cofactor, including [NiFe]-hydrogenases, ureases, carbon monoxide dehydrogenases, methyl-coenzyme M reductase and acireductone dioxygenases (reviewed in Li and Zamble 2009). In plants, the only Ni-dependent enzyme identified so far is urease, which catalyzes the hydrolysis of urea into ammonia and carbon dioxide (Dixon et�al. 1975). Therefore, Ni is required in very low amounts by most plants with critical levels in shoots ranging between 0.01 and 10 �g Ni per g dry weight depending on the plant species (Marschner 2012).
Under aerobic conditions, Ni is present in soils mainly in its divalent cationic form Ni2+, which is the form most available to plants (Cempel and Nikel 2006, Kabata-Pendias 2011). Nickel concentrations vary considerably depending on the soil type (Kabata-Pendias 2011), ranging from 10 to 40 ppm in most soils while even surpassing 1,000 ppm in serpentine soils or soils enriched with Ni-bearing ores (Mizuno and Kirihata 2015, Mizuno et�al. 2018). Importantly, anthropogenic activities such as combustion of fossil fuels, mining, smelting, welding, electroplating and the production of alloys have dramatically increased the amount of Ni released in the environment (Cox and Hutchinson 1981, Michaeli et�al. 2012). In addition to these sources of industrial origin, soil levels of Ni can be increased by sewage sludge disposal, irrigation with polluted water and excessive fertilization with animal manure (Chang et�al. 1984). Increased Ni levels in soils impose large environmental concerns, due to the toxic effects of this heavy metal on all living organisms. Especially in intensive agricultural systems, plants are more likely to be exposed to Ni levels that can induce toxicity rather than Ni deficiency. Symptoms of Ni toxicity in plants include chlorosis, wilting and necrosis of leaves, stunted growth and inhibited root elongation (Seregin and Kozhevnikova 2006, Leškov� et�al. 2017). Other physiological and biochemical changes reported in Ni-stressed plants include disturbed water balance (Pandey and Sharma 2002, Gajewska et�al. 2006), inhibition of photosynthesis (Carlson et�al. 1975, Sheoran et�al. 1990) and increased oxidative stress (Boominathan and Doran 2002, Gajewska et�al. 2006, Hao et�al. 2006). Not being a redox-active metal, Ni-induced oxidative stress is usually associated with altered synthesis and activity of various antioxidant enzymes (Gajewska and Skłodowska 2005, Zhao et�al. 2008, Ghasemi et�al. 2009).
The induction of oxidative stress by Ni can be considered as a direct mechanism of its toxicity, while it has been proposed that Ni toxicity can arise from indirect mechanisms as well, such as the interference of Ni with other essential metals. Due to its similarity to other divalent metals, Ni2+ can compete with these metals during transport and in many biochemical and physiological processes. This is the case for iron (Fe), as high Ni levels induce the appearance of symptoms that resemble Fe deficiency, such as leaf chlorosis, and evoke some transcriptional and physiological responses in roots that are typical of Fe-deficient plants (Nishida et�al. 2011, Leškov� et�al. 2017). The presence of high concentrations of Ni in the growth media results in decreased Fe accumulation in shoots (Ghasemi et�al. 2009, Nishida et�al. 2011, Leškov� et�al. 2017). Many transporters involved with Fe2+ uptake and distribution are also capable of mediating Ni2+ transport across membranes (Schaaf et�al. 2004, Gendre et�al. 2006, Nishida et�al. 2011) suggesting the existence of competition between these two metals at the level of transport. In Arabidopsis thaliana, the transporter IRON-REGULATED TRANSPORTER 1 (IRT1), which is the major transporter for Fe2+ uptake from soils (Vert et�al. 2002), greatly facilitates Ni accumulation in plants (Nishida et�al. 2011). Furthermore, the nonproteinogenic amino acid nicotianamine can form strong complexes with transition metals, such as Fe and Ni (Beneš et�al. 1983). Interestingly, increasing nicotianamine levels can significantly enhance plant tolerance to high Ni, as shown by the overexpression of a nicotianamine synthase from barley or from the metal hyperaccumulator Noccaea caerulescens in tobacco or Arabidopsis, respectively (Kim et�al. 2005, Pianelli et�al. 2005). However, not all responses triggered by Ni are directly associated with altered Fe homeostasis. Previously, we have shown that while Ni-induced leaf chlorosis and root ferric-chelate reductase activity could be reverted by foliar supply of Fe, the strong primary root inhibition and root branching induced by high Ni could not (Leškov� et�al. 2017). In addition, the inhibited primary root elongation and increased lateral root density were still observed even when IRT1 was knocked out. Thus, these results indicate that the strong impact of Ni on root system architecture is due to Fe-independent toxic effects triggered by this heavy metal. However, it remains unresolved which developmental processes are altered by Ni to induce the short- and highly branched root phenotype.
Some studies have indicated that Ni can inhibit cell division in different plant species (L'Huillier et�al. 1996, Knasm�ller et�al. 1998, Demchenko et�al. 2005, Kozhevnikova et�al. 2009, Pavlova 2017). This negative effect of Ni on mitotic activity likely results from Ni-induced damages of nucleolar structure, aberrations in chromosome integrity and abnormalities during mitosis (Fiskesj� 1988, Liu et�al. 1994, Sresty and Madhava Rao 1999, Pavlova 2017). Ni may also inhibit cell elongation. The presence of polysaccharides with functional groups in the cell wall of plants offers binding sites for divalent and trivalent metals. Ni2+ ions bind preferentially to carboxyl groups of polygalacturonic acids and hydroxycinnamic acids, the proportion depending on the pH of the root apoplast and the plant species (Meychik et�al. 2014). In some plant species, the cell wall even represents the major compartment for Ni sequestration (Kr�mer et�al. 2000, Redjala et�al. 2010). Although enhancing plant tolerance to certain metals, binding of metals to cell walls can ultimately increase cell wall rigidity and result in cell rupture, hence inhibiting cell elongation as shown for copper (Cu) and aluminum (Al) (Jones et�al. 2006, Kopittke et�al. 2008, Kopittke et�al. 2009). However, it remains unclear whether Ni-induced inhibition of root elongation is due to altered cell elongation or meristematic activity.
In our previous study, we showed that excess Ni evokes a transient upregulation of several Fe deficiency-regulated genes (Leškov� et�al. 2017). However, we also found that many phenotypical changes triggered by Ni cannot be explained by the interference of this heavy metal with Fe homeostasis. In this study, we combined transcriptomics and microscopic analyses of several reporters to elucidate which developmental and cell biological processes are targeted by Ni toxicity in roots. Our transcriptome analysis revealed that many Fe deficiency-independent processes are altered by excess Ni and indicate that genes associated with the cell wall are negatively affected by this heavy metal. We also found that high Ni rapidly inhibits primary root elongation and that this effect is largely confined to the roots directly exposed to this heavy metal. Furthermore, we show that Ni disturbs auxin response and transport in the root apical meristem and inhibits a specific set of auxin transporters, in particular PIN2. We also raised evidence that high Ni induces reactive oxygen species (ROS) accumulation in roots and disturbs the integrity and orientation of cortical microtubules in cells of the elongation zone. Altogether, our study unveiled a set of targets that respond sensitively to Ni to impair root development in the presence of this heavy metal.
Results
Transcriptome analysis reveals that Ni affects many iron- and cell wall-associated genes
To gain further insights into a larger number of processes potentially altered by high Ni, we performed a transcriptional profiling of roots of A. thaliana accession Columbia-0 (Col-0) exposed to 100 �M Ni, a concentration that induces slight chlorosis and intermediate inhibition of root and shoot growth (Leškov� et�al. 2017).
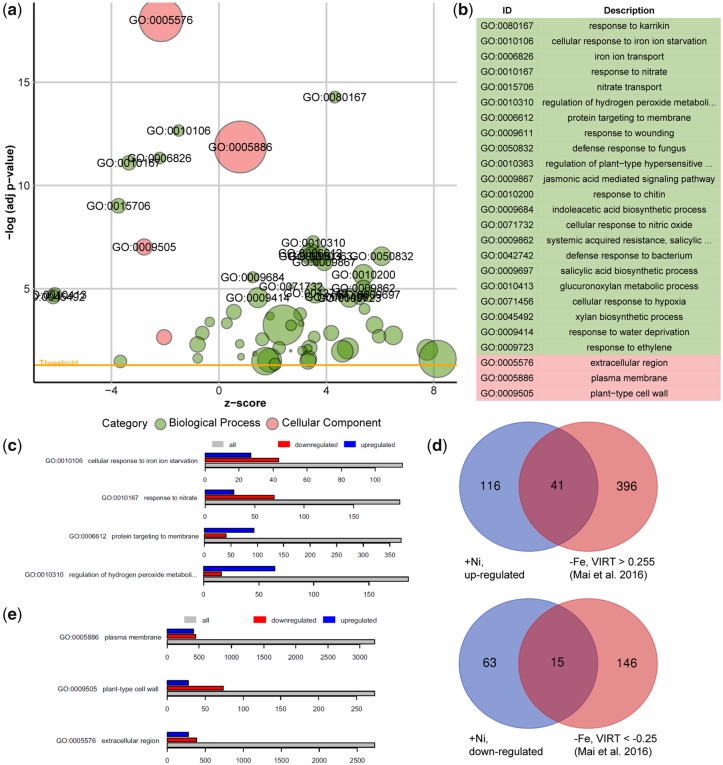
A total of 5,694 genes responded significantly (adjusted P-value <0.05) to high Ni. By setting the log2 fold change cutoff at |1.0|, we detected 235 responsive genes, of which 156 genes were upregulated and 78 genes were downregulated by 100 �M Ni for 4 d. To identify biological processes significantly altered in response to high Ni, we performed a gene ontology (GO) enrichment analysis including all genes significantly (i.e. adjusted P-value <0.05) altered in Ni-treated roots (Fig.�1a, b). The top significant GO terms included response to karrikin (GO:0080167), cellular response to Fe ion starvation (GO:0010106), Fe ion transport (GO:0006826), response to nitrate (GO:0010167) and nitrate transport (GO:0015706). Other biological processes also significantly enriched in response to high Ni were related to different hormones, defense responses, protein targeting to membrane and regulation of hydrogen peroxide (H2O2) metabolism (Fig.�1a, b). Whereas a larger number of genes belonging to GO terms ‘cellular response to Fe ion starvation’ and ‘nitrate transport’ were downregulated by Ni, genes of GO terms ‘protein targeting to membrane’ and ‘regulation of H2O2 metabolism’ were mainly upregulated (Fig.�1c). The significant enrichment of genes related with cellular response to Fe starvation and Fe transport in Ni-exposed roots is in line with the short-term responses detected by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) in our previous study (Leškov� et�al. 2017). Although a larger number of genes associated with GO term ‘cellular response to Fe ion starvation’ was downregulated in Ni-exposed roots (Fig.�1c), many genes directly related with cellular Fe homeostasis and transport were induced by the presence of Ni (Table�1).
Fig. 1.
Genome-wide transcriptional changes in response to high Ni. (a) GO enrichment analysis of overrepresented categories of genes significantly altered by high Ni. Each circle corresponds to a significantly enriched GO category (adjusted P-value <0.05 indicated by the ‘threshold’ line). Size of circles reflects the number of genes that are associated with each respective GO category, and colors reflect the ontology, where green color indicates biological processes and red color indicates cellular component. z-score indicates whether genes from GO terms were primarily upregulated (positive z-score) or downregulated (negative z-score). (b) List of most significant GO terms. (c) Analysis of differentially expressed genes of selected biological processes in Ni-exposed roots. Shown is the total number of genes belonging to the indicated GO clusters and the number of genes significantly up- or downregulated by Ni. (d) Venn diagrams showing the overlap between genes significantly up- or downregulated in response to Ni [−1.0 > log2FC (+Ni/control) > 1.0; adjusted P-value <0.05] and previously shown to robustly respond to Fe deficiency (−Fe). Control, no Ni added; +Ni, 100 �M Ni added to one-half-strength MS media. (e) Analysis of differentially expressed genes of selected cellular compartments.
Table 1.
Expression of genes related with Fe homeostasis in response to high Ni
| Involvement in Fe homeostasis | AGI ID | Annotation | log2�FC +Ni/control | Adj. P-value |
|---|---|---|---|---|
| Fe acquisition | AT3G56970 | BASIC HELIX-LOOP-HELIX 38 (BHLH38) | 2.45 | 0.0001 |
| � | AT3G12900 | SCOPOLETIN 8-HYDROXYLASE (S8H) | 2.17 | 0.0006 |
| � | AT4G31940 | CYTOCHROME P450, FAMILY 82, SUBFAMILY C, POLYPEPTIDE 4 (CYP82C4) | 1.31 | 0.0004 |
| � | AT3G56980 | BASIC HELIX-LOOP-HELIX 39 (BHLH39) | −0.95 | 0.0015 |
| � | AT1G01580 | FERRIC REDUCTION OXIDASE 2 (FRO2) | −0.94 | 0.0008 |
| � | AT4G19690 | IRON-REGULATED TRANSPORTER 1 (IRT1) | −0.87 | 0.0003 |
| � | AT3G13610 | FERULOYL-COA 6′-HYDROXYLASE (F6'H1) | −0.71 | 0.0004 |
| � | AT2G28160 | FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR (FIT) | −0.04 | 0.8062 * |
| � | AT4G16770 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | −0.02 | 0.8207 |
| Fe acquisition/Fe allocation | AT1G56160 | MYB DOMAIN PROTEIN 72 (MYB72) | 1.93 | 0.0002 |
| AT3G12820 | MYB DOMAIN PROTEIN 10 (MYB10) | 1.89 | 0.0001 | |
| Fe allocation | AT2G41240 | BASIC HELIX-LOOP-HELIX PROTEIN 100 (bHLH100) | 2.93 | 0.0002 |
| � | AT1G56430 | NICOTIANAMINE SYNTHASE 4 (NAS4) | 1.16 | 0.0003 |
| � | AT5G24380 | YELLOW STRIPE LIKE 2 (YSL2) | 0.25 | 0.0056 |
| � | AT5G53550 | YELLOW STRIPE LIKE 3 (YSL3) | 0.16 | 0.0575 |
| � | AT5G56080 | NICOTIANAMINE SYNTHASE 2 (NAS2) | −0.80 | 0.0025 |
| � | AT3G47640 | POPEYE (PYE) | −0.61 | 0.0002 |
| � | AT5G04950 | NICOTIANAMINE SYNTHASE 1 (NAS1) | −0.49 | 0.0036 |
| � | AT3G08040 | FERRIC REDUCTASE DEFECTIVE 3 (FRD3) | −0.35 | 0.0359 |
| � | AT4G24120 | YELLOW STRIPE LIKE 1 (YSL1) | −0.01 | 0.9206 |
| Intracellular Fe homeostasis/Fe transport | AT4G19680 | IRON REGULATED TRANSPORTER 2 (IRT2) | 1.32 | 0.0008 |
| � | AT1G23020 | FERRIC REDUCTION OXIDASE 3 (FRO3) | 1.21 | 0.0001 |
| � | AT2G40300 | FERRITIN 4 (FER4) | 0.98 | 0.0001 |
| � | AT1G21140 | VACUOLAR IRON TRANSPORTER-LIKE 1 (VTL1) | 0.80 | 0.0041 |
| � | AT3G56090 | FERRITIN 3 (FER3) | 0.79 | 0.0001 |
| � | AT3G25190 | VACUOLAR IRON TRANSPORTER-LIKE 5 (VTL5) | 0.61 | 0.0022 |
| � | AT3G11050 | FERRITIN2 (FER2) | 0.57 | 0.0007 |
| � | AT4G27860 | VACUOLAR IRON TRANSPORTER (VIT) | 0.32 | 0.0018 |
| � | AT1G10500 | CHLOROPLAST-LOCALIZED ISCA-LIKE PROTEIN (CPISCA) | 0.15 | 0.3599 |
| � | AT1G76800 | VACUOLAR IRON TRANSPORTER-LIKE 2 (VTL2) | −1.07 | 0.0004 |
| � | AT5G67330 | NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN 4 (NRAMP4) | −0.41 | 0.0006 |
| � | AT2G23150 | NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 3 (NRAMP3) | −0.14 | 0.3470 |
| � | AT4G32590 | 2Fe-2S ferredoxin-like superfamily protein | −0.01 | 0.9571 |
| Regulators of Fe deficiency responses | AT5G19700 | EARLY LEAF SENESCENCE 1 (ELS1) | 1.13 | 0.0005 |
| AT1G74770 | BRUTUS LIKE 1 (BTSL1) | 1.01 | 0.0001 | |
| � | AT3G20770 | ETHYLENE-INSENSITIVE3 (EIN3) | 0.08 | 0.3469 |
| � | AT3G18290 | BRUTUS (BTS) | −0.28 | 0.0024 |
| � | AT2G27050 | ETHYLENE-INSENSITIVE3-LIKE 1 (EIL1) | −0.21 | 0.0139 |
| Fe cluster assembly | AT5G51720 | NEET GROUP PROTEIN (NEET) | 0.43 | 0.3219 |
| � | AT4G25910 | NFU DOMAIN PROTEIN 3 (NFU3) | 0.25 | 0.0050 |
| � | AT2G16710 | Iron-sulfur cluster biosynthesis family protein | 0.04 | 0.6154 |
| � | AT4G01940 | NFU DOMAIN PROTEIN 1 (NFU1) | 0.01 | 0.9386 |
| � | AT3G12260 | LYR family of Fe/S cluster biogenesis protein | 0.00 | 0.9823 |
| � | AT2G26060 | CYTOSOLIC IRON-SULFUR PROTEIN ASSEMBLY 1 (CIA1) | −0.21 | 0.0329 |
| � | AT4G04080 | ISCU-LIKE 3 (ISU3) | −0.09 | 0.5026 |
| � | AT1G51390 | NFU DOMAIN PROTEIN 5 (NFU5) | −0.08 | 0.3304 |
| � | AT4G22220 | IRON-SULFUR CLUSTER ASSEMBLY PROTEIN 1 (ISU1) | −0.07 | 0.2100 |
| AT1G60990 | Chloroplast-localized COG0354 protein | −0.03 | 0.6810 | |
| � | AT3G01020 | ISCU-LIKE 2 (ISU2) | −0.02 | 0.6982 |
| � | AT3G20970 | NFU DOMAIN PROTEIN 4 (NFU4) | −0.02 | 0.8424 |
Adjusted P-values >0.05 are in italic.
To further assess the degree of interplay between Ni and Fe deficiency, we compared the list of Ni-responsive genes with genes that are robustly regulated by Fe, as determined by Mai et�al. (2016) using data from 10 distinct transcriptome studies. This comparison indicated that approximately 26% and 19% of all genes significantly upregulated and downregulated by high Ni, respectively, are also typically responsive to Fe deficiency (Fig.�1d). Despite the substantial overlap, this result demonstrates that most Ni-responsive genes actually responded to Ni toxicity and not to Ni-induced Fe deficiency.
By assessing GO terms for cellular compartments, our analysis also provided evidence for the most likely cellular targets of Ni. We found that genes belonging to the terms extracellular region (GO:0005576), plasma membrane (GO:0005886) and plant-type cell wall (GO:0009505) were significantly enriched among Ni-responsive genes (Fig.�1a, b). Interestingly, genes belonging to the term ‘plant-type cell wall’, including BETA-XYLOSIDASE 1 (BXL1), BXL2, ENDOXYLOGLUCAN TRANSFERASE A1 (EXGT-A1), PECTIN METHYLESTERASE 2 (PME2) and GLYCOSYL HYDROLASE 9B4 (GH9B4), were mainly downregulated by high Ni (Fig.�1e and Supplementary Dataset 1).
To validate our microarray analysis, we assessed the expression of selected genes by qPCR. This additional analysis confirmed that SCOPOLETIN 8-HYDROXYLASE (S8H), CYTOCHROME 82C4 (CYP82C4), MYB DOMAIN PROTEIN 72 (MYB72), BASIC HELIX-LOOP-HELIX PROTEIN 100 (bHLH100), COPPER/ZINC SUPEROXIDE DISMUTASE 1 (CSD1) and LOB DOMAIN-CONTAINING PROTEIN 41 (LBD41) were significantly upregulated, whereas IRON SUPEROXIDE DISMUTASE 1 (FSD1), ARABINOGALACTAN PROTEIN 22 (AGP22), BETA-XYLOSIDASE 2 (BXL2) and a member of the peroxidase superfamily (ATG2G37130) were significantly downregulated by high Ni (Supplementary Fig. S1). Therefore, although restricted to a single time-point, our transcriptome analysis provided an overview of which root-related processes respond sensitively to high external Ni availability.
Nickel induces root gravitropic defects and localized inhibition of root elongation
Previously, we have shown that excess Ni inhibits root elongation in an Fe-independent manner (Leškov� et�al. 2017). To understand the dynamics of this repressive action, we assessed time-course changes in root length and lateral root density in response to increasing Ni concentrations. We observed that primary roots were highly sensitive to external Ni, with concentrations 100 and 150 �M strongly repressing elongation as soon as plants were transferred to treatments (Fig.�2a, b). Average lateral root length was also significantly inhibited by high Ni, but only when 150 �M Ni was supplied to plants (Fig.�2a, c). Following primary root repression, lateral root density increased significantly in plants exposed to 100 and 150 �M Ni, especially in the first 6 d after transfer (Fig.�2d). A closer inspection of the developmental stage of all lateral root primordia in roots exposed to two Ni concentrations that caused differential repression of primary root elongation revealed that 100 �M Ni stimulated significantly the proportion of emerged lateral roots (Supplementary Fig. S2a).
Fig. 2.
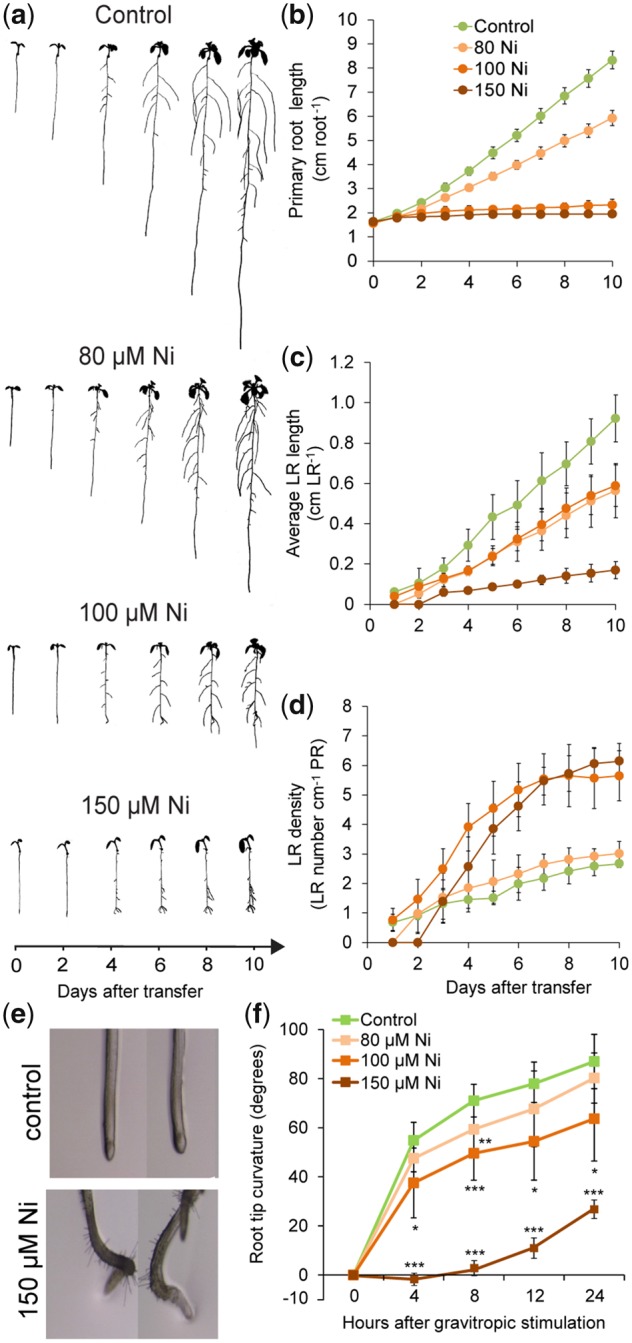
Ni inhibits root elongation and induces gravitropic defects. Seedlings of A. thaliana accession Col-0 were pre-cultured for 7 d on one-half-strength MS agar medium and then transferred to fresh medium without added Ni (control) or containing the indicated concentrations of Ni. Plants were imaged daily with a scanner. (a) Images of representative plants at the indicated time-points after transfer to treatments. Daily changes in primary root length (b), average lateral root length (c) and lateral root density (d) in response to Ni. Bars represent mean � SD (n = 7 plants). High Ni induces agravitropic root growth (e) and reduces the root response to gravistimulation (f). Seven-day-old seedlings pre-cultured on solid one-half-strength MS medium transferred to control medium or medium containing the indicated concentrations of Ni. Bars represent mean � SD (n = 7 plants). *P < 0.05; **P < 0.01; ***P < 0.001 according to Student’s t-test (pairwise comparison with control at each time-point). LR, lateral root; PR, primary root.
In addition to inhibiting root elongation, high Ni induced agravitropic primary root growth (Fig.�2e). We then submitted Ni-treated roots to gravistimulation and observed that they gradually lost their ability to grow toward the gravity vector with each increase in external Ni levels (Fig.�2f). We wondered whether the loss of gravitropism could also be linked to Ni-elicited changes in the differentiation and starch accumulation of columella cells. Therefore, we performed Lugol’s staining of starch granules. The number of stained layers was decreased after exposure to high Ni for 4 d (Supplementary Fig. S2b), indicating that Ni has a strong negative impact on the number of mature columella cells.
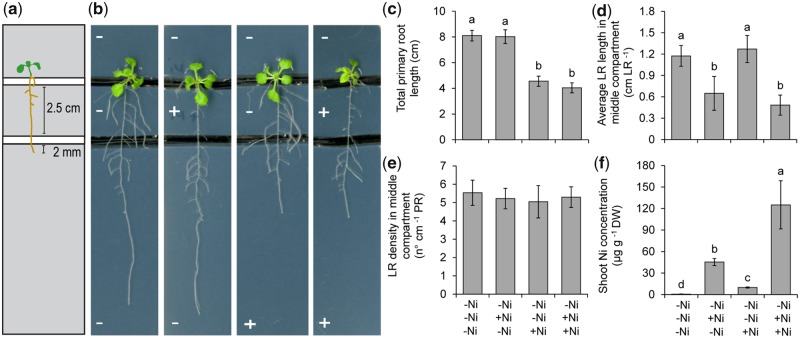
Our time-course experiments revealed that Ni induces rapid changes in root developmental processes, suggesting that Ni exerts a local effect on these processes. To test this hypothesis, we grew plants on horizontally split agar plates with three spatially separated compartments (Fig.�3a). Seedlings precultured without supplemented Ni were transferred to split agar plates so that shoots were placed on the upper, Ni-free compartment, and roots exposed to two compartments with or without added Ni. Primary root length was only significantly inhibited when the root tips but not the other root parts were directly exposed to Ni (Fig.�3a, b). This effect was also observed when plants were grown in two-compartment agar plates (Supplementary Fig. S3). Ni had also a strong local effect on average lateral root length, as only lateral roots directly exposed to Ni were inhibited (Fig.�3a, c). In contrast, lateral root density did not directly respond to localized Ni supply (Fig.�3d). In shoots, Ni concentrations were the highest in plants grown on agar plates supplied with Ni in both middle and lower compartments (Fig.�3e). Plants grown on this condition also exhibited significantly lower concentrations of macronutrients and of the micronutrient manganese as compared with those grown on a split agar without Ni supplementation in any of the compartments (Supplementary Fig. S4). However, in all cases, shoot levels were either just at critical deficiency values or well above these values. Further support to the localized effect of Ni was the fact that Ni supply only to the lowest agar segment evoked a similar inhibitory effect on primary root elongation as the presence of Ni on two compartments despite inducing an almost 13-fold lower Ni accumulation in leaves (Fig.�3a, b, e). Taken together, these results indicate that Ni has a rapid, local inhibitory effect on root elongation.
Fig. 3.
Ni exerts a local inhibitory effect on root elongation. (a) Schematic representation of the split-agar setup. Plants were pre-cultured on one-half-strength MS agar medium and transferred to agar plates in which three compartments were spatially separated with a gap. Ten-day-old seedlings were placed in a way that only approximately 2 mm of the apical root zone was in contact with the lowest segment, while shoots were placed on the uppermost, Ni-free compartment. (b) Appearance of A. thaliana plants accession Col-0 on horizontally split agar plates with differential Ni supply to roots. − and + indicate medium without added Ni and medium containing 150 �M Ni, respectively. Plants were photographed after 7 d and representative plants are shown. Total primary root length along three compartments (c), average lateral root length (d) and lateral root density in the middle compartment (e) and Ni concentrations in whole shoots (f). Bars represent mean � SD (n = 15 for root traits and n = 6 replicates containing five shoots each for elemental 1analysis). Different letters indicate significant differences according to Tukey’s multiple test at P < 0.05. DW, dry weight; LR, lateral root; PR, primary root.
Nickel preferentially targets cell elongation
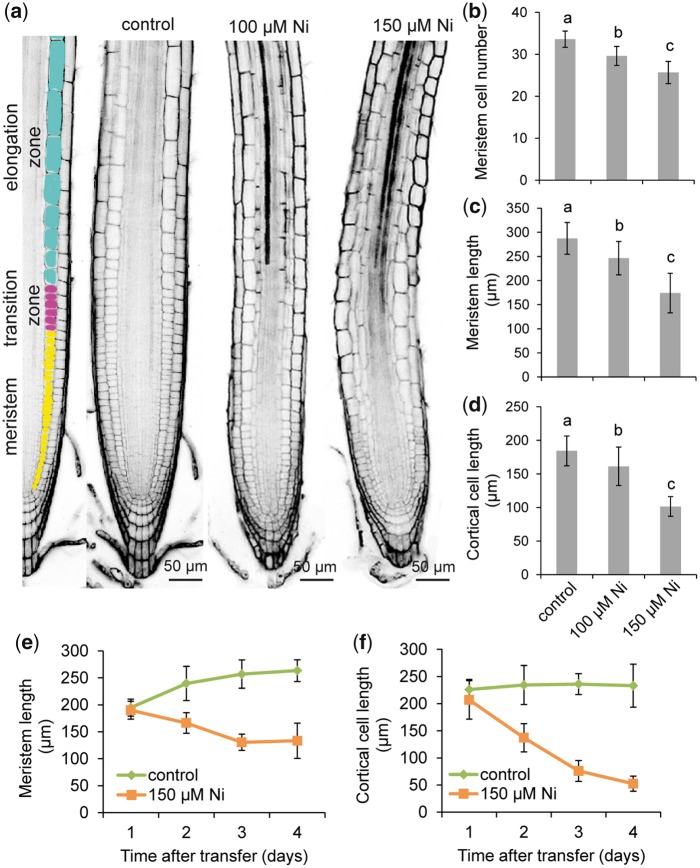
Root elongation is largely dependent on the rate of cell division in the apical meristem and cell expansion in the elongation zone (Kong et�al. 2018). Disturbances in any of these processes can significantly alter the length of roots. To determine root meristem size, we assessed the number of cortex cells in a file extending from the quiescent center (QC) to the first elongated cell. Our analysis revealed that Ni negatively affected the meristems by decreasing the number of meristematic cells and the meristem length (Fig.�4a–c). Excess Ni also inhibited the elongation of mature cells (Fig.�4d) and altered their overall morphology, as many cells within the elongation zone also expanded more radially (Fig.�4a). For all parameters assessed, we observed that the deleterious effects of Ni became more prominent as the external levels of this heavy metal were increased. A temporal analysis further revealed that both meristem length and mature cortical cell length were inhibited by high Ni as early as 2 d after transfer (Fig.�4e, f). In addition, we observed that while initially inhibiting longitudinal cell expansion in the elongation zone, prolonged exposure to Ni gradually increased radial expansion and deformations of cells near the transition zone (Supplementary Fig. S5).
Fig. 4.
Effect of Ni on cell elongation. (a) Longitudinal view of representative roots of A. thaliana accession Col-0 exposed to different Ni treatments showing tissue organization and developmental zones. Seven-day-old seedlings pre-cultured on one-half-strength MS agar medium were transferred to fresh medium without added Ni (control) or containing the indicated concentrations of Ni. After 4 d, roots were stained with propidium iodide and imaged under a confocal laser scanning microscope. Cells highlighted in different colors belong to the indicated root zones. Meristem cell number (b), meristem length (c) and average cell size (length) of mature cells of the cortical cell layer (d) of roots exposed for 4 d to control or the indicated Ni concentrations, as detailed in (a). Different letters indicate significant differences according to Tukey’s multiple test at P < 0.05. Time-course changes in meristem length (e) and cell length of mature cells of the cortical cell layer (f) under control or 150 �M Ni. Values represent the mean � SD of 15 measurements.
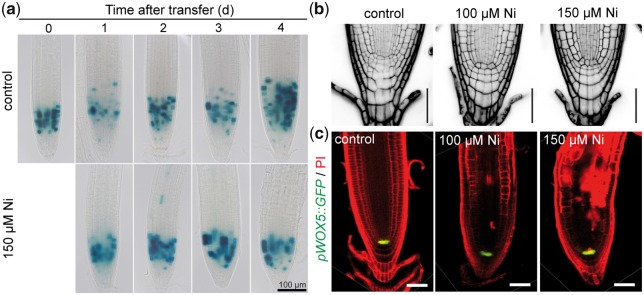
We next addressed whether the smaller meristems under high Ni were associated with decreased mitotic activity. Thus, we assessed CYCLIN B1;1 promoter (pCYCB1;1)--driven �-glucuronidase (GUS) activity in response to high Ni. Although meristems of Ni-exposed roots were shorter, mitotic activity could still be detected in these roots (Fig.�5a). Despite the severe defects observed in the elongation and differentiation zones of Ni-treated roots (Fig.�4a and Supplementary Fig. S5), the structure of the apical part of the meristem also remained surprisingly unaffected 5 d after transferring plants to treatments (Fig.�5b). At the same time, the expression of the QC-specific marker WUSCHEL RELATED HOMEOBOX 5 (WOX5) also indicated that QC identity was maintained even when such severe Ni toxicity was imposed to plants (Fig.�5c).
Fig. 5.
Effect of Ni on mitotic activity and the integrity of the root apical meristem. (a) Time-course changes in cell division activity in primary roots as revealed by pCYCB1;1-dependent GUS assay. Seven-day-old seedlings pre-cultured on one-half-strength MS agar medium were transferred to fresh medium without added Ni (control) or containing 150 �M Ni. GUS activity was assessed at the indicated time-points, and representative images of 15 plants are shown. Close-up view of apical meristems (b) and pWOX5::GFP in the quiescent center (c) in response to external Ni. Seedlings pre-cultured for 7 d on one-half-strength MS medium were transferred to control medium or medium supplemented with 100 and 150 �M Ni. Representative images of eight plants are shown. Scale bars: 50 �m.
Taken together, our results demonstrate that Ni preferentially targets cell expansion and has comparatively minor effects on mitotic activity and cell identity in root meristems of Arabidopsis. The fact that Ni can decrease the size of root meristems without evoking pronounced changes in cell division might suggest that this heavy metal accelerates the rate at which cells progress through the meristem to the elongation zone.
Nickel alters auxin distribution by rapidly inhibiting the auxin transporter PIN2
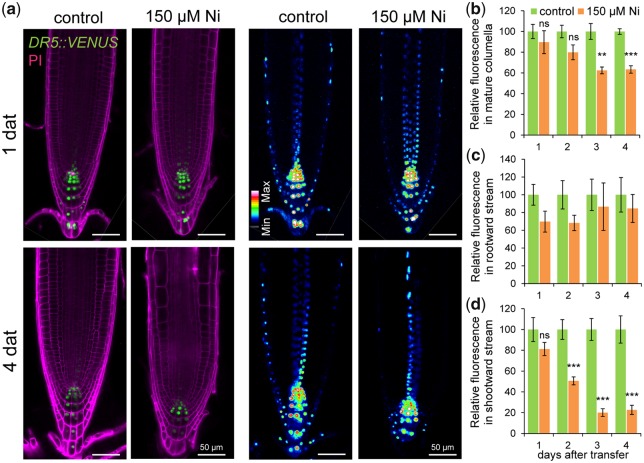
The observed Ni-induced gravitropic defects, arrested cell elongation and stimulated lateral root emergence resemble phenotypes associated with disturbed auxin response and distribution in plants (Benkov� et�al. 2003, Blilou et�al. 2005, Sato et�al. 2015). We first investigated this possibility with the auxin-responsive DR5::VENUS reporter in a time-course experiment. Our analysis revealed that high Ni-altered Venus-dependent fluorescence intensity in the root apex (Fig.�6a). DR5::VENUS signals in mature columella cells and in cells engaged with rootward and shootward auxin flows were then quantified. We found that high Ni significantly decreased DR5::VENUS signals in mature columella cells 3 d after transfer (Fig.�6b). Notably, auxin response in the stele (rootward stream) and lateral root cap + epidermis (shootward stream) was differentially affected by high Ni. While DR5::VENUS signals were not significantly altered in the stele over the course of the experiment, high Ni quickly inhibited auxin response in outer cell layers (Fig.�6c, d). The inhibited auxin response in columella cells and outer cells was also accompanied by decreased auxin levels in these cells as revealed by the analysis of the DII::VENUS reporter line (Supplementary Fig. S6).
Fig. 6.
Ni inhibits auxin responses in outer cell layers. (a) Localization of DR5::VENUS expression in root tips of plants grown for 1 or 4 d on one-half-strength MS agar medium without (control) or with 150 �M Ni. Shown are representative images of DR5::VENUS distribution in propidium iodide-stained roots and pseudo-colored confocal images of DR5::VENUS expression intensity. Quantification of DR5::VENUS signal intensities in mature columella cells (b), in inner cell layers where rootward auxin stream takes place (c) and in outer cell layers engaged with shootward auxin flow (d). Values represent the mean � SE of 10 measurements per condition and time-point. **P < 0.01; ***P < 0.001; ns, not significant (P > 0.05) according to Student’s t-test (pairwise comparison with control at each time-point).
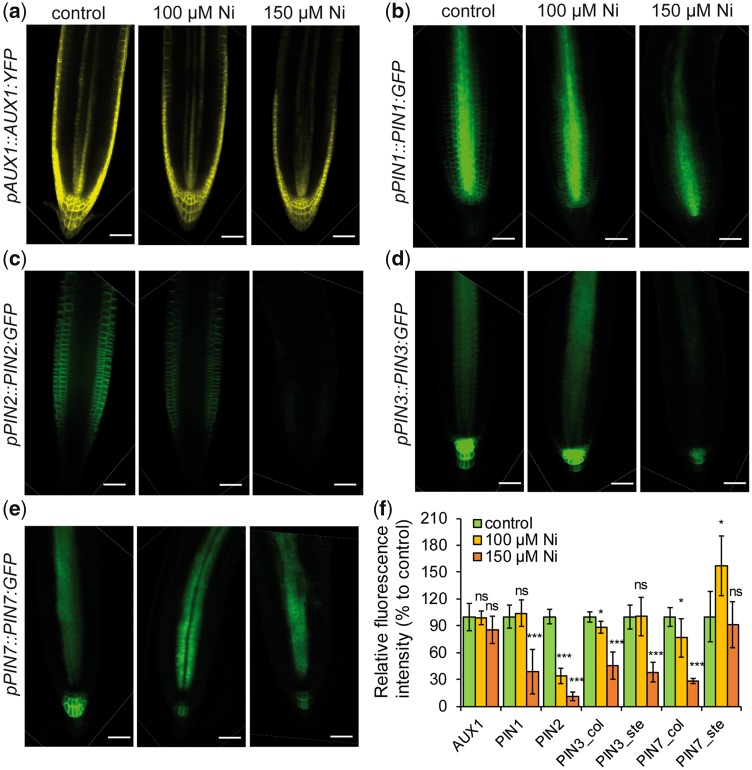
Auxin distribution is highly dependent on the action of plasma membrane-associated auxin influx carriers, such as AUXIN RESISTANT 1 (AUX1), and polarly localized auxin efflux carriers of the PIN-FORMED (PIN) family (Bennett et�al. 1996, Petr�šek et�al. 2006). The altered auxin response and distribution in Ni-treated roots prompted us to investigate whether Ni affected the expression and/or localization of these transporters. No marked change in AUX1:GFP and PIN1:GFP accumulation was observed at 100 �M Ni, whereas 150 �M Ni significantly repressed PIN1 but not AUX1 (Fig.�7a, b, f). In columella cells, high Ni decreased PIN3:GFP and PIN7:GFP expressions (Fig.�7c, f). In the stele, whereas PIN7:GFP was not substantially affected by Ni, PIN3:GFP accumulation was strongly inhibited by the highest Ni levels (Fig.�7c, e, f). However, Ni exerted the strongest overall inhibitory effect on PIN2, as PIN2:GFP-derived signal was almost completely abolished already by 100 �M Ni (Fig.�7d, f), indicating that PIN2 is particularly sensitive to the presence of external Ni.
Fig. 7.
Ni affects differentially the protein levels of different auxin transporters. Distribution of AUX1 (a), PIN1 (b), PIN2 (c), PIN3 (d) and PIN7 (e) in response to Ni. Translational fusion lines of the indicated auxin transporters were pre-cultured on one-half-strength MS agar medium for 7 d and then transferred for 5 d to fresh medium containing the indicated treatments. Scale bars: 50 �m. (f) Relative quantification of the fluorescence signals. Data represent the mean � SD (n = 7–9 roots). *P < 0.05; ***P < 0.001; ns, not significant (P > 0.05) according to Student’s t-test (pairwise comparison with control). Col, fluorescence in columella cells; ste, fluorescence in root stele.
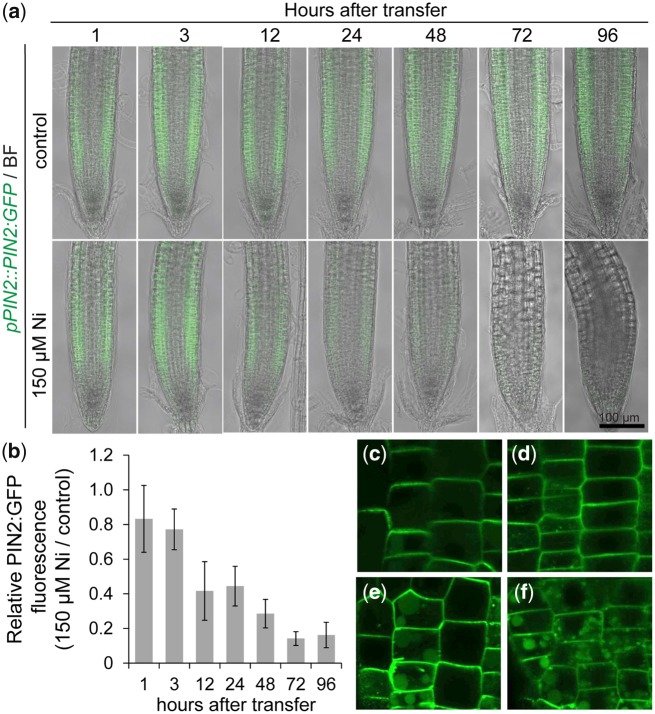
The strong repression of PIN2, the main PIN-type transporter responsible for shootward auxin flows in roots (Wiśniewska et�al. 2006), could explain the decrease in auxin response that we detected in outer cells of the root meristem using the DR5::VENUS reporter (Fig.�6). To investigate in more details the dynamics of Ni-induced PIN2 inhibition, we carried out a time-course experiment. We found that PIN2:GFP signals in outer cells were repressed by Ni as quickly as 12 h after transferring plants to treatments (Fig.�8a, b). Interestingly, PIN2 transcript levels were not significantly altered by Ni even after 9 d of exposure to this heavy metal (Supplementary Fig. S7), suggesting that Ni targets PIN2 at the post-transcriptional level. We therefore investigated the effect of Ni on PIN2 protein distribution by incubating roots with one-half-strength Murashige and Skoog (MS) solution containing increasing concentrations of Ni. Without added Ni, PIN2 was correctly localized on the apical side (i.e. toward the shoot) of the epidermal cells of root tips (Fig.�8c). In the presence of Ni, PIN2 polar localization in plasma membranes became less obvious (Fig.�8d–f). At concentrations ≥100 �M, Ni also induced PIN2 intracellular localization because an increasing number of PIN2:GFP-containing agglomerates began to accumulate within epidermal cells (Fig.�8e, f). Taken together, these results suggest that PIN2 protein accumulation and polar distribution are rapidly and sensitively altered by Ni.
Fig. 8.
Ni rapidly inhibits PIN2 accumulation and alters PIN2 membrane distribution. (A and B) Time-course analysis of pPIN2::PIN2:GFP accumulation in response to elevated Ni. Representative images (a) and quantification of the GFP fluorescence signal (b). Values are mean � SD of 10 measurements. Seven-day-old seedlings pre-cultured on one-half-strength MS agar medium were transferred to fresh medium without added Ni (control) or containing 150 �M Ni. (c–f) Close-up view of PIN2:GFP protein localization in cell membranes and in intracellular agglomerates. Pictures were taken after 12 h incubation in 0 (c), 50 (d), 100 (E) or 200 �M Ni (f). BF, bright-field.
Nickel interferes with microtubule organization and induces oxidative stress in roots
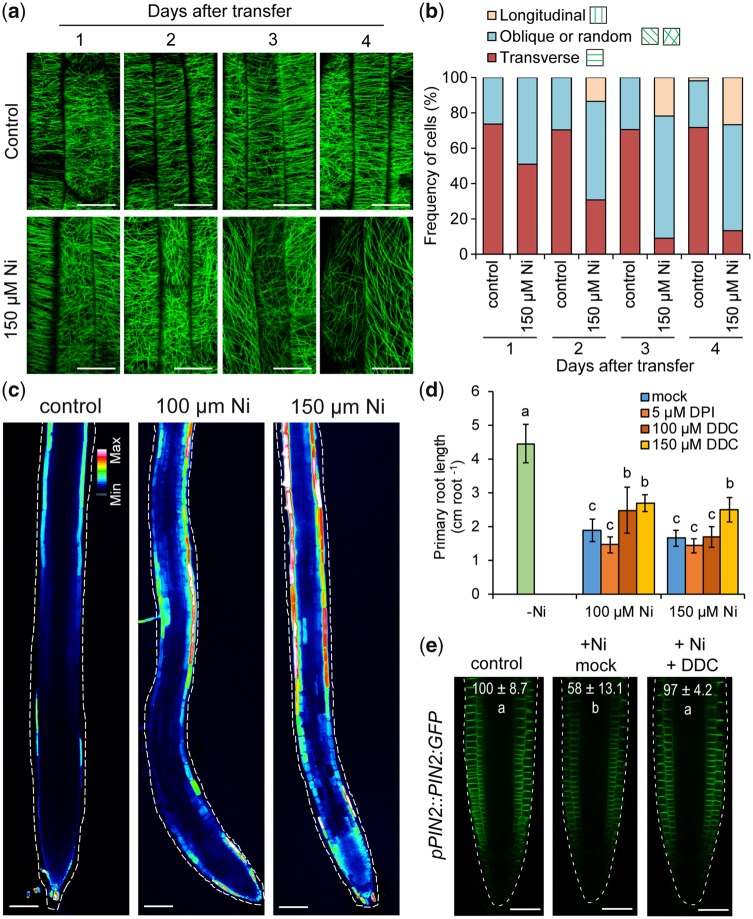
The aberrant cell expansion (Fig.�4a) and inhibited PIN2 accumulation (Figs.�7e, f, 8) of Ni-exposed roots prompted us to investigate the orientation and integrity of cortical microtubules in cells of the elongation zone. We observed cortical microtubules with a transgenic line constitutively expressing the MICROTUBULE-ASSOCIATED PROTEIN4 (MAP4) fused to the green fluorescence protein (GFP) under the control of cauliflower mosaic virus 35S promoter (Marc et�al. 1998). In roots of control plants, microtubules were mainly transverse throughout the experiment (Fig.�9a, b). However, the exposure of roots to high Ni induced re-orientation of microtubules, especially after 2 d. In Ni-exposed roots, a higher proportion of microtubules underwent random, oblique and longitudinal realignments (Fig.�9a, b). In addition to re-orientation, high Ni gradually decreased the density of microtubules (Fig.�9a), suggesting that Ni may induce microtubule depolymerization.
Fig. 9.
Ni induces microtubule re-orientation and ROS accumulation. (a) Ni-induced changes in microtubule integrity and orientation in epidermal cells of the elongation zone assessed with the microtubular marker line 35S::GFP:MAP4. Confocal images were taken at the indicated time-points after transfer to treatments. Note that after 3 d and especially 4 d of exposure to high Ni, microtubule density is decreased. (b) Frequency of different microtubule orientation patterns in epidermal cells of the elongation zone (n > 20 cells per condition and time-point). (c) Carboxy-H2DCFDA staining of ROS in roots exposed to the indicated Ni concentrations for 4 d. Representative false color confocal images are shown. To allow comparison, images were taken with the same microscopic settings. Effect of the NADPH oxidase and superoxide dismutase inhibitors DPI and DDC, respectively, on Ni-induced primary root arrest (d) and PIN2 accumulation in root tips (e). Seven-day-old seedlings pre-cultured on one-half-strength MS agar medium were transferred for 4 d to fresh media without added Ni (control) or containing the indicated concentrations of Ni and ROS inhibitors. In (d), data represent mean � SD (n = 12 roots). In (e), values indicate relative PIN2-GFP signal intensities (% compared with control) after 12 h treatment. Data are mean � SD (n = 7–8 roots). +Ni, 150 �M Ni; +DDC, 150 �M. Different letters indicate significant differences according to Tukey’s multiple test at P < 0.05. Scale bars: 20 �m (a), 100 �m (c) and 50 �m (e).
Recently, it has been shown that H2O2 modulates cytoskeleton function and PIN2 levels at the plasma membrane (Zwiewka et�al. 2019). To investigate whether Ni can also alter the accumulation and distribution of ROS, we stained roots with carboxylated 2′,7′-dichlorodihydrofluorescein diacetate (C-H2DCFDA) as described previously (M�ller et�al. 2015). We observed that Ni increased C-H2DCFDA signals along the whole root and was especially concentrated in outer cells (Fig.�9c). Our transcriptome analysis had indicated a significant enrichment of genes involved in the regulation of H2O2 metabolism in response to high Ni (Fig.�1b, c). Among these, we found a large number of peroxidases and superoxide dismutases, of which most were significantly upregulated by excess Ni (Supplementary Table S1). We then grew Ni-exposed plants in the presence of diphenyleneiodonium (DPI) and diethyldithiocarbamic acid (DDC), which inhibit the activity of NADPH oxidases and superoxide dismutases, respectively. In our growth conditions, DPI was not able to rescue the root growth of Ni-exposed plants (Fig.�9d). However, the supply of DDC with high Ni was able to significantly increase primary root length. Notably, we also observed that DDC could partially restore PIN2:GFP in root tips as soon as 12 h after transferring plants to high Ni (Fig.�9e). The effect of DDC was still persistent after 4 d (Supplementary Fig. S8a, b). Thus, these results indicate that there might be a link between Ni-induced changes in ROS accumulation and the observed changes in PIN2 distribution.
Discussion
Transcriptional changes in response to Ni are largely independent of Ni-induced Fe deficiency
Previously, we found that Ni-induced Fe deficiency is the main reason for the appearance of leaf chlorosis and for the upregulation of many genes related with Fe homeostasis in Arabidopsis (Leškov� et�al. 2017). However, since only the expression of a selected number of genes was assessed by qRT-PCR in our previous study, it remained unclear to what extent transcriptional changes induced by high Ni overlap with those triggered by Fe deficiency. Our transcriptome analysis now showed that approximately one quarter of all genes differentially regulated by Ni are also typically up- or downregulated by Fe deficiency (Fig.�1d). Interference with Fe is not restricted to Ni, since also high levels of cadmium (Cd) and Zn have been shown to significantly alter the expression of Fe-related genes (Becher et�al. 2004, van de Mortel et�al. 2006, van de Mortel et�al. 2008, Besson-Bard et�al. 2009, Landa et�al. 2015). We found that excess Ni can alter the expression of several Fe deficiency-induced genes involved with Fe acquisition and internal chelation, such as IRT2, S8H, CYP82C4 and NAS4 (Table�1 and Supplementary Fig. S1). Interestingly, in line with our earlier findings (Leškov� et�al. 2017), upregulation of these genes was not accompanied by a significant change in the expression of FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR (FIT; Table�1). Instead, high Ni specifically induced the expression of other Fe-related transcription factors, such as bHLH038, bHLH100 and bHLH101, MYB72 and MYB10 (Table�1 and Supplementary Fig. S1). In agreement with these findings, expression of the root stele-specific gene NAS4 (Dinneny et�al. 2008, Palmer et�al. 2013), which is regulated by MYB10 and MYB72 (Palmer et�al. 2013) was upregulated by high Ni, while the expression of NAS1 (Dinneny et�al. 2008), which is controlled by FIT (Klatte et�al. 2009), was slightly downregulated (Table�1). These observations are consistent with the fact that high Ni evokes selective responses resembling Fe deficiency, many of which can be reverted by extra Fe supply via the leaves (Ghasemi et�al. 2009, Leškov� et�al. 2017).
Despite the significant overlap with Fe deficiency, most transcriptional changes induced by high Ni were not related to disturbed Fe homeostasis (Fig.�1d). We identified several processes that are affected by Ni toxicity, including response to nitrate, the biosynthesis of hemicellulosic components and the regulation of H2O2 metabolism (Fig.�1a, b). With regard to the latter, our C-H2DCFDA staining confirmed that high Ni induces the accumulation of ROS in roots (Fig.�9c). As shown in the study of Gajewska and Skłodowska (2005) with wheat plants, superoxide and H2O2 levels are particularly increased upon exposure to high Ni. At least part of the Ni-induced oxidative stress results from the inhibitory effect of this heavy metal on the activity of a number of antioxidant enzymes, such as catalases and superoxide dismutases (Gajewska and Skłodowska 2005, Zhao et�al. 2008, Ghasemi et�al. 2009). Thus, it is likely that the altered expression of genes associated with regulation of H2O2 metabolism in Ni-exposed roots reflects a response induced by the increased ROS accumulation.
Nickel modifies root system architecture by targeting PIN2-mediated shootward auxin flow
By using a split-agar experimental setup, we showed that Ni has a local toxic effect in roots, characterized by a specific inhibition of the elongation of roots directly exposed to Ni (Fig.�3). In addition, root inhibition was largely independent of Ni levels accumulating in shoots. This agrees with the fact that high Ni also inhibits the elongation of irt1 roots (Leškov� et�al. 2017). Similar local toxicity has been reported for Arabidopsis plants exposed to high Cu in a split-root system (Lequeux et�al. 2010). However, unlike Ni, excess Cu strongly inhibited the mitotic activity in the root apical meristem (Yuan et�al. 2013, Lequeux et�al. 2010) and induced premature cell death (Lequeux et�al. 2010). We found that cell division and the integrity of the stem cell niche were maintained even after prolonged exposure to Ni levels when primary root elongation was completely inhibited (Fig.�5). Instead, high Ni strongly repressed cell expansion in the root elongation zone (Fig.�4). These results indicate that Ni and Cu interfere with specific root developmental processes to inhibit primary root elongation.
Plant hormones, especially auxin, play an important role in the regulation of root cell elongation and tropic growth (Vanneste and Friml 2009). Changes in auxin-related processes are frequently observed when plants are exposed to metals. For example, it has been shown that auxin responses were increased in the stele and attenuated in the columella cells when A. thaliana plants were exposed to high Cu (Yuan et�al. 2013).
Our transcriptome analysis indeed showed that high Ni alters the expression of genes involved with the biosynthesis or signaling of salicylic acid, jasmonic acid and ethylene (Fig.�1a, b), three hormones typically associated with stress responses and plant defense signaling (Li et�al. 2019). Interestingly, we found no significant enrichment for terms associated with auxin. However, we observed that high levels of this heavy metal inhibited auxin accumulation and responses in columella cells and in outer cell layers involved in shootward auxin flow (Fig.�6 and Supplementary Fig. S6). Since the strong inhibitory effect of Ni on PIN2 protein accumulation was not due to changes in PIN2 expression levels (Fig.�8 and Supplementary Fig. S7), it is possible that Ni interfered with auxin-related processes mainly at the post-transcriptional level. These rather specific changes were associated with the fact that excess Ni preferentially targeted the auxin transporters PIN7, PIN3 and especially PIN2 (Fig.�7). In contrast to Ni, excess Cu results in a specific repression of PIN1 (Yuan et�al. 2013). Also, Cd toxicity affects more strongly PIN1 than PIN2 in roots (Bruno et�al. 2017). These results further support the notion that different heavy metals target specific components of the auxin transport system in roots to remodel root growth.
Although PIN-dependent auxin transport is also required for cell elongation, significant reduction of root growth due to repressed cell expansion is only observed in the pin3 pin7 double mutant but not in pin3, pin7 or pin2 single mutants (Blilou et�al. 2005). Apart from reduced root elongation, we found that high Ni also impaired root gravitropic responses and reduced starch accumulation in columella cells (Fig.�2f and Supplementary Fig. S2b). Also, here the negative impact of Ni on PIN2, PIN3 and PIN7 was likely involved, since strong agravitropic phenotypes are observed in the pin2 single and pin3 pin7 double mutants (Luschnig et�al. 1998, Friml et�al. 2002, Kleine-Vehn et�al. 2010), while the pin2 pin3 pin7 triple mutant also contains less starch granules in columella cells (Blilou et�al. 2005). The strong and rapid inhibition of PIN2 protein accumulation and polar distribution by high Ni (Fig.�7) may explain the lateral root phenotype of Ni-exposed roots. As shown before, lateral roots of pin2 mutant plants emerge at a faster rate as compared with wild-type plants (Benkov� et�al. 2003), a phenotype that resembles exposure to high Ni (Supplementary Fig. S2a). Therefore, altogether our findings indicate that Ni perturbs gravitropic responses, cell elongation and lateral root development, at least in part, by negatively affecting a subset of auxin transporters in roots.
Recently, it has been shown that H2O2 triggers a dose-dependent inhibition of PIN transporters (Zwiewka et�al. 2019). In the case of PIN2, it was found that H2O2 induces the accumulation of PIN2:GFP-containing intracellular agglomerates and inhibits PIN2 recycling. Since our results demonstrate that excess Ni induces the accumulation of ROS in roots (Fig.�9c), it is tempting to speculate that an oxidative burst triggered by Ni was involved with the observed changes in PIN2 accumulation and cellular distribution. When the activity of superoxide dismutases was inhibited by the drug DDC, PIN2:GFP levels in Ni-exposed roots were partially restored (Fig.�9e). Although DDC also has been shown to act as a chelator for Ni (Smirnova et�al. 2004), we only detected small decreases in shoot Ni levels when plants were supplied with DDC (Supplementary Fig. S8c). This suggests that Ni entry into the plants was not significantly inhibited by this drug. However, we cannot completely exclude that the partial restoration of primary root elongation and of PIN2 levels by DDC was, at least in part, due to increased Ni chelation.
Another possible explanation for the disturbed PIN2 accumulation in Ni-exposed roots comes from the link between membrane targeting of auxin transporters and microtubule stability. The application of the microtubule depolymerization inhibitor, oryzalin, increases the accumulation of PIN2 in the cytosol (Kleine-Vehn et�al. 2008, Ambrose et�al. 2013). However, we found that Ni stimulated PIN2 degradation within 12 h (Fig.�8a, c), whereas significant changes in microtubule stability and orientation were detected only after 2 d (Fig.�9a, b). Furthermore, the altered polar distribution of PIN2 upon Ni supply (Fig.�8c–f) seems unlikely to be caused by changes in microtubule properties, as disturbance of microtubules by oryzalin does not alter the polar localization of PIN2 in the plasma membrane of epidermal cells (Kleine-Vehn et�al. 2008). Instead, polar distribution of PIN2 appears to be regulated by other factors, such as cell wall integrity, as shown by genetically or chemically disturbing cellulose synthesis or by removing cell walls by protoplasting (Feraru et�al. 2011). Therefore, possible alterations in cell wall properties induced by the presence of Ni, as suggested by our transcriptome analysis, might have contributed to the disturbed polar distribution and altered lateral diffusion of PIN2 in epidermal cells.
Putative inhibitory action of Ni on cell wall-associated processes
Our microarray data indicated that Ni evoked significant changes in the expression of genes associated with cell walls and extracellular region (Fig.�1e). Interestingly, in each case, the majority of these differentially expressed genes were downregulated by high Ni. Carboxyl and hydroxyl groups of cell wall polysaccharides as well as protein-derived sulfhydryl and histidyl groups play a central role in the immobilization of metal ions in cell walls (reviewed in Krzesłowska 2011). Many metals ions, including Al3+, Cu2+, Cd2+ and Ni2+, can bind to de-esterified pectins (Dronnet et�al. 1996, Meychik et�al. 2011, Meychik et�al. 2014). Direct effects of Ni in root cell walls could also explain why primary root inhibition is largely confined to roots directly exposed to this heavy metal (Fig.�3). In line with this assumption, we have previously shown that Ni also inhibits root elongation of irt1 plants (Leškov� et�al. 2017), which have greatly reduced Ni uptake (Nishida et�al. 2011). It has been demonstrated that polygalacturonic and hydroxycinnamic acids of root cell walls can effectively bind Ni2+ (Meychik et�al. 2014). As a consequence, cell wall rigidity and integrity can be altered, which may explain the strong inhibition of cell elongation in Ni-exposed roots (Fig.�4). Noteworthy, our transcriptome data demonstrated that many genes involved in cell wall modifications were significantly repressed by high Ni (Fig.�1e and Supplementary Dataset 1). Hence, we propose a scenario in which Ni disturbs growth by initially directly interacting with cell wall components. The resulting inhibition of cell expansion then initiates a signaling response that is also manifested at the transcriptional level.
We also observed that among all genes differentially regulated by high Ni with |log2FC|�≥�1, seven genes encoding for peroxidases were present (Supplementary Table S1). Interestingly, all peroxidases differentially expressed in response to Ni are predicted to reside in the extracellular region, indicating that they belong to class III (Passardi et�al. 2004). Depending on whether class III peroxidases mediate hydroxylic or peroxidative cycles, they can either generate ROS to stimulate cell wall loosening or promote the cross-linking of phenolics to induce cell wall stiffening (Passardi et�al. 2004). In maize, it has been shown that the rapid Al-induced ROS accumulation is associated with cell wall rigidification and the consecutive root growth repression (Jones et�al. 2006). However, it remains to be tested whether the increased ROS accumulation and differential expression of peroxidases in response to high Ni are responsible for the observed inhibition of cell elongation and compromised anisotropic expansion.
In addition to inhibiting cell elongation along the longitudinal axis, high Ni partially impaired the anisotropic expansion of epidermal and cortical cells in the elongation zone (Fig.�4a). Defective anisotropic growth is also induced in roots of maize and rice plants exposed to toxic Al levels (Blancaflor et�al. 1998, Jones et�al. 2006, Wu et�al. 2014). By studying the mechanical properties of Al-stressed rice roots, it was found that these defects are related with decreased cell extensibility and increased cell rigidity (Wu et�al. 2014). The directionality of expansion has been proposed to be dependent on the orientation of cortical microtubules (Baskin 2005), which directs the deposition of cellulose microfibrils (Gutierrez et�al. 2009). We found that the integrity and orientation of cortical microtubules in cells of the elongation zone were disturbed by high Ni (Fig.�9a, b). As soon as 2 d after transferring plants to treatments, we detected a decreased number of cells displaying transverse arrangement of cortical microtubules relative to the elongation axis (Fig.�9b). Such Ni-induced disruption of microtubule orientation could, in part, underlie the root growth inhibition and compromised anisotropy of Ni-exposed roots. It is also possible that the hierarchy of the processes affected by Ni is reversed. Indeed, the disruption of cellulose synthesis can, conversely, also alter the organization of cortical microtubules, as shown previously by Fisher and Cyr (1998). In such scenario, a putative interference of Ni with cellulose synthesis could result in disorganized cortical microtubules. Furthermore, it has been recently shown that root epidermal and cortical cells undergo two rapid modes of cell elongation, the first involving endoreplication and the second involving dynamic actin reorganization (Takatsuka et�al. 2018). Future research will be necessary to investigate whether Ni does indeed disrupt microfibril orientation. Another important aspect that should be assessed is whether the presence of Ni in the cell wall of elongating cells can alter cell wall composition and actin organization.
Materials and Methods
Plant material and growth conditions
Arabidopsis (A. thaliana) ecotype Col-0 was used as wild type throughout the study. All transgenic lines used in this study have been previously described: pCYCB1;1::GUS (Ferreira et�al. 1994), pWOX5::GFP (Blilou et�al. 2005), DR5::VENUS (Heisler et�al. 2005), pAUX1::AUX1:YFP (Swarup et�al. 2004), pPIN1::PIN1:GFP (Benkov� et�al. 2003), pPIN2::PIN2:GFP (Xu and Scheres 2005), pPIN3::PIN3:GFP (Dello Ioio et�al. 2007), pPIN7::PIN7:GFP (Blilou et�al. 2005), 35S::GFP:MAP4 (Marc et�al. 1998) and DII::VENUS (Brunoud et�al. 2012).
Arabidopsis seeds were surface sterilized in 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 and sown on solid media containing half-strength MS medium supplemented with 0.5% (w/v) sucrose, 1% (w/v) Difco agar (Becton Dickinson, USA) and 2.5 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.7). After a 2-d stratification at 4�C, agar plates were placed vertically in a growth cabinet and plants were grown in conditions of 10 h of light (120 �mol m−2 s−1) at 22�C and 14 h of dark at 19�C for 7 d. Afterward, plants were transferred to fresh solid agar prepared as described above with or without the supplementation of nickel sulfate (NiSO4�6H2O) at concentrations as indicated in the legends of figures. For transcriptome analysis, seedlings pre-cultured for 7 d on solid half-strength MS agar media without added Ni were transferred to fresh media containing no added Ni or 100 �M Ni, a concentration that induces slight chlorosis and intermediate inhibition of root and shoot growth (Leškov� et�al. 2017). Root samples for microarray analysis (four biological replicates consisting of 16 whole roots per condition) were harvested 4 d after plants were transferred to control and 100 �M Ni conditions.
Transcriptome analysis
All preprocessing and statistical analyses of microarray data were performed using R statistical computing environment (R Development Core Team 2008). RNA amplification, labeling and hybridization to Agilent microarrays [Arabidopsis (V4, 021169) Gene Expression Microarray] were conducted following the manufacturer’s protocol (Agilent Technologies, USA). Analysis of microarray data was performed with the R package ‘limma’ (Ritchie et�al. 2015). Raw feature intensities were background corrected using ‘normexp’, and ‘quantile’ normalization method was used to normalize between arrays. As the next step in preprocessing, we filtered out control and low expressed probes. Differential expression was performed by fitting a linear model to log2-transformed data by an empirical Bayes method (Smyth 2004). Resulting P-values were tested for false discoveries due to multiple hypotheses testing via the Benjamini and Hochberg procedure (adjusted P-values). To extract genes with significant expression differences, a cutoff of adjusted P < 0.05 and |log2FC|�>�1 was applied. Microarray and sample annotation data were deposited in the Gene Expression Omnibus database under the accession number GSE138859.
GO enrichment analysis
The GO terms enrichment analysis was conducted by ‘topGO’ package (Alexa and Rahnenfuhrer 2019) with the gene annotation database of A. thaliana (v3.8.2; Carlson 2019). Fisher’s exact test was performed to compare the frequency of GO terms in the given gene sets with the background frequency. The enriched GO terms were considered to be significant with adjusted P-value <0.05 according to the Benjamini–Hochberg P-value correction method. The results of the enrichment analyses were visualized using ‘GOplot’ package (Walter et�al. 2015).
Quantitative real-time PCR
Root tissues were collected by excision and immediately frozen in liquid N. Total RNA was extracted using the RNeasy Plant Mini Kit (Macherey-Nagel GmbH & Co KG, Germany). Quantitative reverse transcriptase PCRs were conducted with the CFX38TM Real-Time System (Biorad, Germany) and the Go Taq qPCR Master Mix SybrGreen I (Promega, Germany) using the primers listed in Supplementary Table S2. Since we have previously determined that UBQ2 has the highest stability under different heavy metal supplies (Leškov� et�al. 2017), we used this reference gene to normalize relative expression levels of all tested genes. Relative expression was calculated according to Pfaffl (2004).
Root system architecture and gravistimulation
Plants were scanned at the times indicated in the legends of figures with an Epson Expression 10000XL scanner (Seiko Epson, Germany) at a resolution of 300 dots per inch using settings described by Gruber et�al. (2013). Primary root lengths, total lateral root lengths, and lateral root numbers were analyzed using WinRhizo Pro version 2009c (Reagent Instrument, Canada). Average lateral root length was calculated by dividing the total length of lateral roots by the number of lateral roots. Lateral root density was calculated by dividing the number of lateral roots by the length of the primary root.
The gravistimulation assay was carried out with seedlings previously treated or not with high Ni for 3 d. The plates with the seedlings were reoriented by 90� relative to the gravity vector. The plants were scanned every 4 h after gravistimulation up to 24 h, and the angle of curvature was analyzed using ImageJ software.
Microscopy and image acquisition
Images of transgenic lines carrying GFP, yellow fluorescent protein (YFP) or VENUS reporters were acquired with a confocal laser-scanning microscope (LSM 780; Zeiss, Germany) equipped with 20�/0.8 M27 objective. GFP signal was excited with 488-nm argon laser, and the emission was captured at 505–560-nm wavelengths, while the fluorescence of YFP and VENUS was excited with a 514-nm laser and the emission was detected at 518–575 nm. Propidium iodide (PI) staining was visualized with a 561-nm laser and detected at 550–650 emission range. A 40�/1.20 water immersion objective was used to image GFP-tagged microtubules. For comparative analyses, all recordings of each reporter line were made with the same settings. The images were recorded and adjusted with ZEN (Zeiss) software.
ROS staining
The presence of ROS in roots was determined as described in M�ller et�al. (2015) with slight modifications. Whole seedlings were stained with 10 μM Carboxy-H2DCFDA (Invitrogen, Germany) for 20 min in darkness, briefly rinsed to remove the excess of extracellular dye and subsequently mounted on microscope slides for imaging under confocal microscope (excitation 493 nm, emission 517–527 nm).
Histochemical staining
For counterstaining cell walls during reporter analyses and for the measurements of meristem size and mature cell length, roots were incubated for 10 min in aqueous solution of 10 mg l−1 PI in dark, rinsed twice with distilled water and imaged immediately.
Starch granules were stained by incubating roots in Lugol solution (Sigma, Germany) for 10 min, followed by double washout in distilled water. GUS activity of the proCYCB1;1::GUS plant roots was visualized by overnight incubation of roots at 37�C in GUS staining solution containing 0.4 g l−1 5-bromo-4-chloro-3-indolyl-beta-d-glucuronic acid, 100 mM sodium phosphate buffer (pH 7.0), 10 mM EDTA, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide and 0.1% Triton-X100. Samples were mounted in chloral hydrate solution (2 mg l−1 prepared in 25% glycerol) and imaged under a light microscope (Axio Imager2, Zeiss).
Measurement of cell size and meristem length
The length of root apical meristems and mature cortical cells was measured from PI-stained images with ZEN software. Meristem size was determined in a single-cell file, starting from the QC until the first elongating cell. The number of cells in root apical meristems was counted manually.
Elemental analysis
Whole shoots were dried at 65�C and digested in HNO3 solution in polytetrafluoroethylene tubes under pressurized system (UltraCLAVE IV, Germany; MLS). Elemental analysis was performed by sector field high-resolution inductively coupled plasma-mass spectrometry (ELEMENT 2; Thermo Fisher Scientific, Germany). Element standards were prepared from certified reference materials from CPI International.
Funding
Vedeck� Grantov� Agent�ra [VEGA 2/0115/17] to A.L. and Deutsche Forschungsgemeinschaft, Bonn, Germany [HE 8362/1-1] to R.F.H.G.
Supplementary Material
Acknowledgments
We thank Elis Fraust and Yudelsy A. Tandron Moya (Leibniz Institute of Plant Genetics and Crop Plant Research) for excellent technical assistance and Kenneth D. Birnbaum (New York University), Jiř� Friml (Institute of Science and Technology Austria), Ivan Paponov (Albert-Ludwigs-University of Freiburg) and Malcolm J. Bennett (University of Nottingham) for providing mutants and reporter lines. We thank Nicolaus von Wir�n (Leibniz Institute of Plant Genetics and Crop Plant Research) for critically reading the article.
Disclosures
No conflicts of interest declared.
References
- Alexa A., Rahnenfuhrer J. (2019) topGO: Enrichment Analysis for Gene Ontology. R Package Version 2.36.0.
- Ambrose C., Ruan Y., Gardiner J., Tamblyn L.M., Catching A., Kirik V. (2013) CLASP interacts with Sorting Nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 24: 649–659. [DOI] [PubMed] [Google Scholar]
- Baskin T.I. (2005) Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21: 203–222. [DOI] [PubMed] [Google Scholar]
- Becher M., Talke I.N., Krall L., Kr�mer U. (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 37: 251–268. [DOI] [PubMed] [Google Scholar]
- Beneš I., Schreiber K., Ripperger H., Kircheiss A. (1983) Metal complex formation by nicotianamine, a possible phytosiderophore. Experientia 39: 261–262. [Google Scholar]
- Benkov� E., Michniewicz M., Sauer M., Teichmann T., Seifertov� D., J�rgens G., et al. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett M.J., Marchant A., Green H.G., May S.T., Ward S.P., Millner P.A., et al. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950. [DOI] [PubMed] [Google Scholar]
- Besson-Bard A., Gravot A., Richaud P., Auroy P., Duc C., Gaymard F., et al. (2009) Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol. 149: 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancaflor E.B., Jones D.L., Gilroy S. (1998) Alterations in the cytoskeleton accompany aluminum-induced growth inhibition and morphological changes in primary roots of maize. Plant Physiol. 118: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., et al. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Boominathan R., Doran P.M. (2002) Ni-induced oxidative stress in roots of the Ni hyperaccumulator, Alyssum bertolonii. New Phytol. 156: 205–215. [DOI] [PubMed] [Google Scholar]
- Bruno L., Pacenza M., Forgione I., Lamerton L.R., Greco M., Chiappetta A., et al. (2017) In Arabidopsis thaliana cadmium impact on the growth of primary root by altering SCR expression and auxin-cytokinin cross-talk. Front Plant Sci 8: 1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103. [DOI] [PubMed] [Google Scholar]
- Carlson M. (2019) org.At.tair.db: Genome Wide Annotation for Arabidopsis. R Package Version 3.8.2.
- Carlson R.W., Bazzaz F.A., Rolfe G.L. (1975) The effect of heavy metals on plants: II. Net photosynthesis and transpiration of whole corn and sunflower plants treated with Pb, Cd, Ni, and Tl. Environ. Res. 10: 113–120. [DOI] [PubMed] [Google Scholar]
- Cempel M., Nikel G. (2006) Nickel: a review of its sources and environmental toxicology. Pol. J. Environ. Stud. 15: 375–382. [Google Scholar]
- Chang A.C., Warneke J.E., Page A.L., Lund L.J. (1984) Accumulation of heavy metals in sewage sludge-treated soils. J. Environ. Qual. 13: 87–91. [Google Scholar]
- Cox R.M., Hutchinson T.C. (1981) Environmental factors influencing the rate of spread of the grass Deschampsia caespitosa invading areas around the sudbury nickel-copper smelter. Water Air Soil Pollut. 16: 83–106. [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., et al. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Demchenko N.P., Kalimova I.B., Demchenko K.N. (2005) Effect of nickel on growth, proliferation, and differentiation of root cells in Triticum aestivum seedlings. Russ. J. Plant Physiol. 52: 220–228. [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., et al. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945. [DOI] [PubMed] [Google Scholar]
- Dixon N.E., Gazzola C., Blakeley R.L., Zerner B. (1975) Letter: Jack bean urease (EC 3.5.1.5). A metalloenzyme. A simple biological role for nickel? J. Am. Chem. Soc. 97: 4131–4133. [DOI] [PubMed] [Google Scholar]
- Dronnet V.M., Renard C.M.G.C., Axelos M.A.V., Thibault J.F. (1996) Characterisation and selectivity of divalent metal ions binding by citrus and sugar-beet pectins. Carbohydr. Polym. 30: 253–263. [Google Scholar]
- Feraru E., Feraru M.I., Kleine-Vehn J., Martini�re A., Mouille G., Vanneste S., et al. (2011) PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21: 338–343. [DOI] [PubMed] [Google Scholar]
- Ferreira P.C., Hemerly A.S., Engler J.D., van Montagu M., Engler G., Inz� D. (1994) Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell 6: 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D.D., Cyr R.J. (1998) Extending the microtubule/microfibril paradigm. Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol. 116: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskesj� G. (1988) The Allium test—an alternative in environmental studies: the relative toxicity of metal ions. Mutat. Res. Fund. Mol. Mech. Mut. 197: 243–260. [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benkov� E., Mendgen K., Palme K. (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809. [DOI] [PubMed] [Google Scholar]
- Gajewska E., Skłodowska M. (2005) Antioxidative responses and proline level in leaves and roots of pea plants subjected to nickel stress. Acta Physiol. Plant. 27: 329–340. [Google Scholar]
- Gajewska E., Skłodowska M., Słaba M., Mazur J. (2006) Effect of nickel on antioxidative enzyme activities, proline and chlorophyll contents in wheat shoots. Biol. Plant. 50: 653–659. [Google Scholar]
- Gendre D., Czernic P., Con�j�ro G., Pianelli K., Briat J.-F., Lebrun M., et al. (2006) TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J. 49: 1–15. [DOI] [PubMed] [Google Scholar]
- Ghasemi R., Ghaderian S.M., Kr�mer U. (2009) Interference of nickel with copper and iron homeostasis contributes to metal toxicity symptoms in the nickel hyperaccumulator plant Alyssum inflatum. New Phytol. 184: 566–580. [DOI] [PubMed] [Google Scholar]
- Gruber B., Giehl R.F.H., Friedel S., von Wir�n N. (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163: 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M.C., Ehrhardt D.W. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797. [DOI] [PubMed] [Google Scholar]
- Hao F.S., Wang X.C., Chen J. (2006) Involvement of plasma-membrane NADPH oxidase in nickel-induced oxidative stress in roots of wheat seedlings. Plant Sci. 170: 151–158. [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., et al. (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Jones D.L., Blancaflor E.B., Kochian L.V., Gilroy S. (2006) Spatial coordination of aluminium uptake, production of reactive oxygen species, callose production and wall rigidification in maize roots. Plant Cell Environ. 29: 1309–1318. [DOI] [PubMed] [Google Scholar]
- Kabata-Pendias A. (2011) Trace Elements in Soils and Plants. p. 505 CRC Press, Florida. [Google Scholar]
- Kim S., Takahashi M., Higuchi K., Tsunoda K., Nakanishi H., Yoshimura E., et al. (2005) Increased nicotianamine biosynthesis confers enhanced tolerance of high levels of metals, in particular nickel, to plants. Plant Cell Physiol. 46: 1809–1818. [DOI] [PubMed] [Google Scholar]
- Klatte M., Schuler M., Wirtz M., Fink-Straube C., Hell R., Bauer P. (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150: 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Ding Z., Jones A.R., Tasaka M., Morita M.T., Friml J. (2010) Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl. Acad. Sci. USA 107: 22344–22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Łangowski Ł., Wiśniewska J., Dhonukshe P., Brewer P.B., Friml J. (2008) Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Knasm�ller S., Gottmann E., Steinkellner H., Fomin A., Pickl C., Paschke A., et al. (1998) Detection of genotoxic effects of heavy metal contaminated soils with plant bioassays. Mutat. Res. Genet. Toxicol. Environ. 420: 37–48. [DOI] [PubMed] [Google Scholar]
- Kong X., Liu G., Liu J., Ding Z. (2018) The root transition zone: a hot spot for signal crosstalk. Trends Plant Sci. 23: 403–409. [DOI] [PubMed] [Google Scholar]
- Kopittke P.M., Blamey F.P.C., Menzies N.W. (2008) Toxicities of soluble Al, Cu, and La include ruptures to rhizodermal and root cortical cells of cowpea. Plant Soil 303: 217–227. [Google Scholar]
- Kopittke P.M., McKenna B.A., Blamey F.P.C., Wehr J.B., Menzies N.W. (2009) Metal-induced cell rupture in elongating roots is associated with metal ion binding strengths. Plant Soil 322: 303–315. [Google Scholar]
- Kozhevnikova A.D., Seregin I.V., Bystrova E.I., Belyaeva A.I., Kataeva M.N., Ivanov V.B. (2009) The effects of lead, nickel, and strontium nitrates on cell division and elongation in maize roots. Russ. J. Plant Physiol. 56: 242–250. [Google Scholar]
- Kr�mer U., Pickering I.J., Prince R.C., Raskin I., Salt D.E. (2000) Subcellular localization and speciation of nickel in hyperaccumulator and non-accumulator Thlaspi species. Plant Physiol. 122: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzesłowska M. (2011) The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiol. Plant. 33: 35–51. [Google Scholar]
- Landa P., Prerostova S., Petrova S., Knirsch V., Vankova R., Vanek T. (2015) The transcriptomic response of Arabidopsis thaliana to zinc oxide: a comparison of the impact of nanoparticle, bulk, and ionic zinc. Environ. Sci. Technol. 49: 14537–14545. [DOI] [PubMed] [Google Scholar]
- Lequeux H., Hermans C., Lutts S., Verbruggen N. (2010) Response to copper excess in Arabidopsis thaliana: impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 48: 673–682. [DOI] [PubMed] [Google Scholar]
- Leškov� A., Giehl R.F.H., Hartmann A., Fargašov� A., von Wir�n N. (2017) Heavy metals induce iron deficiency responses at different hierarchic and regulatory levels. Plant Physiol. 174: 1648–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Huillier L., d'Auzac J., Durand M., Michaud-Ferri�re N. (1996) Nickel effects on two maize (Zea mays) cultivars: growth, structure, Ni concentration, and localization. Can. J. Bot. 74: 1547–1554. [Google Scholar]
- Li N., Han X., Feng D., Yuan D., Huang L.-J. (2019) Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20: 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zamble D.B. (2009) Nickel homeostasis and nickel regulation: an overview. Chem. Rev. 109: 4617–4643. [DOI] [PubMed] [Google Scholar]
- Liu D., Jiang W., Guo L., Hao Y., Lu C., Zhao F. (1994) Effects of nickel sulfate on root growth and nucleoli in root tip cells of Allium cepa. Isr. J. Plant Sci. 42: 143–148. [Google Scholar]
- Luschnig C., Gaxiola R.A., Grisafi P., Fink G.R. (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Gene Dev. 12: 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai H.-J., Pateyron S., Bauer P. (2016) Iron homeostasis in Arabidopsis thaliana: transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biol. 16: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao T-h., McCubbin A.G., et al. (1998) A GFP–MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P. (2012) Marschner's Mineral Nutrition of Higher Plants. p. 651 Academic Press, San Diego. [Google Scholar]
- Meychik N., Nikolaeva Y., Kushunina M., Yermakov I. (2014) Are the carboxyl groups of pectin polymers the only metal-binding sites in plant cell walls? Plant Soil 381: 25–34. [Google Scholar]
- Meychik N.R., Nikolaeva Y.I., Komarynets O.V., Ermakov I.P. (2011) Barrier function of the cell wall during uptake of nickel ions. Russ. J. Plant Physiol. 58: 409–414. [Google Scholar]
- Michaeli E., Boltižiar M., Sol�r V., Ivanov� M. (2012) The landfill of industrial waste—l�ženec near the former nickel smelter at Sereď town as an example of environmental load. Životn� Prostredie 46: 63–68. [Google Scholar]
- Mizuno T., Kirihata Y. (2015) Elemental composition of plants from the serpentine soil of Sugashima Island, Japan. Aust. J. Bot. 63: 252–260. [Google Scholar]
- Mizuno T., Nakahara Y., Fujimori T., Yoshida H. (2018) Natural revegetation potential of Japanese wild thyme (Thymus quinquecostatus Celak.) on serpentine quarries. Ecol. Res. 33: 777–788. [Google Scholar]
- M�ller J., Toev T., Heisters M., Teller J., Moore K.�L., Hause G., et al. (2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 33: 216–230. [DOI] [PubMed] [Google Scholar]
- Nishida S., Tsuzuki C., Kato A., Aisu A., Yoshida J., Mizuno T. (2011) AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol. 52: 1433–1442. [DOI] [PubMed] [Google Scholar]
- Pandey N., Sharma C.P. (2002) Effect of heavy metals Co2+, Ni2+ and Cd2+ on growth and metabolism of cabbage. Plant Sci. 163: 753–758. [Google Scholar]
- Palmer C.M., Hindt M.N., Schmidt H., Clemens S., Guerinot M.L. (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F., Penel C., Dunand C. (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9: 534–540. [DOI] [PubMed] [Google Scholar]
- Pavlova D. (2017) Nickel effect on root-meristem cell division in Plantago lanceolata (Plantaginaceae) seedlings. Aust. J. Bot. 65: 446–452. [Google Scholar]
- Petr�šek J., Mravec J., Bouchard R., Blakeslee J.J., Abas M., Seifertov� D., et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2004) A-Z of Quantitative PCR. pp. 87–112. International University Line (IUL; ), La Jolla. [Google Scholar]
- Pianelli K., Mari S., Marqu�s L., Lebrun M., Czernic P. (2005) Nicotianamine over-accumulation confers resistance to nickel in Arabidopsis thaliana. Transgenic Res. 14: 739–748. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2008) A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- Redjala T., Sterckeman T., Skiker S., Echevarria G. (2010) Contribution of apoplast and symplast to short term nickel uptake by maize and Leptoplax emarginata roots. Environ. Exp. Bot. 68: 99–106. [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato E.M., Hijazi H., Bennett M.J., Vissenberg K., Swarup R. (2015) New insights into root gravitropic signalling. J. Exp. Bot. 66: 2155–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G., Ludewig U., Erenoglu B.E., Mori S., Kitahara T., von Wir�n N. (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J. Biol. Chem. 279: 9091–9096. [DOI] [PubMed] [Google Scholar]
- Seregin I.V., Kozhevnikova A.D. (2006) Physiological role of nickel and its toxic effects on higher plants. Russ. J. Plant Physiol. 53: 257–277. [Google Scholar]
- Sheoran I.S., Singal H.R., Singh R. (1990) Effect of cadmium and nickel on photosynthesis and the enzymes of the photosynthetic carbon reduction cycle in pigeonpea (Cajanus cajan L.). Photosynth. Res. 23: 345–351. [DOI] [PubMed] [Google Scholar]
- Smirnova A.P., Chernysh V.V., Proskurnin M.A. (2004) Investigations of cobalt, nickel, and copper diethyldithiocarbamates using thermal lens spectrometry. J. Anal. Chem. 59: 424–432. [Google Scholar]
- Smyth G.K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: 1. [DOI] [PubMed] [Google Scholar]
- Sresty T.V.S., Madhava Rao K.V. (1999) Ultrastructural alterations in response to zinc and nickel stress in the root cells of pigeonpea. Environ. Exp. Bot. 41: 3–13. [Google Scholar]
- Swarup R., Kargul J., Marchant A., Zadik D., Rahman A., Mills R., et al. (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka H., Higaki T., Umeda M. (2018) Actin reorganization triggers rapid cell elongation in roots. Plant Physiol. 178: 1130–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel J.E., Schat H., Moerland P.D., Van Themaat E.V.L., Van Der Ent S., Blankestijn H., et al. (2008) Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 31: 301–324. [DOI] [PubMed] [Google Scholar]
- van de Mortel J.E., Villanueva L.A., Schat H., Kwekkeboom J., Coughlan S., Moerland P.D., et al. (2006) Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 142: 1127–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dedaldechamp F., Gaymard F., Guerinot M.L., Briat J.F., et al. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter W., S�nchez-Cabo F., Ricote M. (2015) GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31: 2912–2914. [DOI] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertov� D., Brewer P.B., Růžička K., Blilou I., et al. (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883–883. [DOI] [PubMed] [Google Scholar]
- Wu D., Shen H., Yokawa K., Baluška F. (2014) Alleviation of aluminium-induced cell rigidity by overexpression of OsPIN2 in rice roots. J. Exp. Bot. 65: 5305–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005) Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.M., Xu H.H., Liu W.C., Lu Y.T. (2013) Copper regulates primary root elongation through PIN1-mediated auxin redistribution. Plant Cell Physiol. 54: 766–778. [DOI] [PubMed] [Google Scholar]
- Zhao J., Shi G., Yuan Q. (2008) Polyamines content and physiological and biochemical responses to ladder concentration of nickel stress in Hydrocharis dubia (Bl.) Backer leaves. Biometals 21: 665. [DOI] [PubMed] [Google Scholar]
- Zwiewka M., Bielach A., Tamizhselvan P., Madhavan S., Ryad E.E., Tan S., et al. (2019) Root adaptation to H2O2-induced oxidative stress by ARF-GEF BEN1- and cytoskeleton-mediated PIN2 trafficking. Plant Cell Physiol. 60: 255–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.