Abstract
This study investigates the immunomodulatory effects of polychromatic polarized light therapy (PLT) on human monocyte cells. While there is some evidence demonstrating a clinical effect in the treatment of certain conditions, there is little research into its mechanism of action. Herein, U937 monocyte cells were cultured and exposed to PLT. The cells were then analyzed for change in expression of genes and cell surface markers relating to inflammation. It was noted that 6 hours of PLT reduced the expression of the CD14, MHC I and CD11b receptors, and increased the expression of CD86. It was also shown that PLT caused downregulation of the genes IL1B, CCL2, NLRP3 and NOD1, and upregulation of NFKBIA and TLR9. These findings imply that PLT has the capacity for immunomodulation in human immune cells, possibly exerting an anti‐inflammatory effect.
Keywords: inflammation, phototherapy, polarized light, polarized light therapy
This study provides evidence for an underlying mechanism for the effects of polarized light therapy on human immune cells. It demonstrates an immunomodulatory effect of polarized light when applied to human monocyte cells, through actions on gene transcription and protein synthesis. These results will help in determining the full action of phototherapies, to provide support for their translation into clinical use.

1. INTRODUCTION
Inflammation is a process heavily implicated in pathological states of all kinds, from allergy and neoplasia, to infection. Despite its central role in the pathophysiology of many common conditions, current methods of managing inflammation are fraught with problems, particularly in a chronic setting. Treatment typically involves pharmacological interventions, many of which exert a range of unwanted effects in addition to their therapeutic action 1. The continuing evolution and development of novel therapeutic approaches to the management of inflammation is essential in order to advance patient care. Evaluation of nonpharmacological anti‐inflammatory treatments is a key, but relatively underrepresented part of this process. A small but growing body of evidence indicates that phototherapies such as polarized light therapy (PLT) are a promising avenue of exploration in this area 2, 3.
The therapeutic benefits of light therapy have been reported since the late 1960s with applications ranging from neonatal jaundice to psoriasis and vitiligo 3, 4. The theoretical basis of phototherapy involves the use of light to induce physiological change within a target tissue 5, with subsequent therapeutic effects. There are several types of phototherapies differentiated by the specific physical qualities of the light used, with the most common being low‐level laser therapy (LLLT) and ultraviolet (UV) therapies. In recent years, however, it has been proposed that a broad light spectrum covering all light colors, and polarization are crucial elements in light therapy 6. Polarized light (PL) is formed by the filtering of light waves so that they are aligned and vibrated in a single plane (Figure 1). Once polarized, light has the ability to penetrate further into tissues than its unpolarized counterpart 7.
Figure 1.

A, Summary figure of different physical properties of phototherapeutic light. B, Summary of the process of light polarization
Broad, visible spectrum PLT differs from other forms of phototherapy as it uses a much wider range of wavelengths than other modalities such as, LLLT or UV. Consequently, the devices used in PLT are generally less expensive and relatively easy to use. PLT has been suggested by advocates and device manufacturers for use in several contexts such as pain, wound healing, skin conditions and inflammatory arthritis. Preliminary clinical evidence indicates efficacy in the management of skin ulcers 8, 9, 10, burns 11, 12, 13, musculoskeletal injuries 14, 15, 16, 17, 18 as well as surgical and nonhealing wounds 6, 19, 20, 21, 22. Despite this, there are little scientific data regarding the physiological mechanisms underlying these changes in vitro or in vivo 2. It has suggested that the interactions of light with the key mitochondrial enzyme cytochrome oxidase C are responsible for the beneficial changes observed in LLLT 23, 24, 25; however, little information has been documented exploring the mechanisms of PLT.
With aging populations, in most developed economies, and the associated increase in demands on health budgets, noninvasive, nonpharmacological avenues of disease treatment will be required to counter the inevitable burden of inflammatory disease. PLT has the potential to be an inexpensive, technically simple and safe phototherapeutic intervention. This study aimed to determine whether PL induces cellular changes to the human monocyte cell line, to provide insight into the possible mechanisms of action underpinning its clinical use. It was hypothesized that a decrease in the inflammatory profile of immune cells may be part of the biological action of PLT. Specifically, this involved the exposure of a cultured U937 monocyte/macrophage cell line to PL, and subsequent assessment of cell surface markers and gene expression relating to inflammation. We showed that exposure to PL decreases the expression of proinflammatory cell surface markers and causes changes in gene expression consistent with a decreased inflammatory profile.
2. MATERIALS AND METHODS
All procedures in this study were performed in PC2 laboratories at the Werribee campus of Victoria University, and the Western Centre for Health Research and Education (Western CHRE) at Sunshine hospital under standard laboratory conditions. All cell culture work was done under aseptic conditions in a class II biosafety cabinet. Trypan blue staining was used before and after illumination to ensure cell viability throughout the experiments. No ethics was required for this research.
2.1. Cell culture and cell differentiation
The U937 monocyte cell line was cultured in Roswell Park Memorial Institute media, supplemented with 10% fetal bovine serum (Interpath Services Pty. Ltd.), 1% penicillin/streptomycin (Sigma‐Aldrich) and 0.1% glutamine (Sigma‐Aldrich), and incubated at 37°C, 5% CO2 as per standard culture protocol. The cells were passaged to 50% to 90% confluency and media replenished every 48 hours until an adequate number of cells were obtained. A stock of experimental cells (cell bank) was then created and stored in liquid nitrogen. One vial from the cell bank was thawed each time the experiment was repeated, to ensure all experimental cells were at the same passage throughout the experiments. U937 cells are promonocytic cells and are differentiated into monocyte/macrophage cells via the addition of vitamin D3 to the media to a final concentration of 100 nм for 72 hours 26.
2.2. Polarized light exposure
The therapeutic PL source employed was using a Bioptron MedAll PAG‐960 lamp (Bioptron, Switzerland) with a wavelength range from 400 to 3400 nm. The light source has an average power density of 40 mW/cm2 evenly distributed across the wavelength spectrum. The entire apparatus, including lamp and tripod, was housed completely within an incubator (Figure 2). The incubator and media temperatures were monitored to ensure there was no overheating of the culture due to the light exposure. This is to ensure that observed effects stemmed from cellular interactions with light, rather than a thermic effect. The light from the lamp was projected perpendicularly onto a 5 cm cell culture plate at a distance of 10 cm, according to manufacturer's instructions, with full light coverage of the sample. The lid of the culture dish containing the experimental cells was removed, to avoid interference of the light passing through. Control cells were kept in the same incubator, separated by a solid, stainless steel partition to ensure no spill‐over light interacted with the control sample. The lid of the control cell plate was also removed to ensure interactions with the atmosphere within the incubator did not account for the changes seen. In initial experiments, cells were exposed for 5 minutes, 15 minutes or 6 hours, followed by incubation for 24 hours before being prepared for flow cytometric analysis of cell surface marker expression. The cells used for analysis of gene expression were exposed to PL for 6 hours, immediately snap frozen, and their RNA isolated prior to their preparation for PCR gene array analysis. All analysis was performed in triplicate as technical replicates.
Figure 2.

Experimental set up in a standard cell culture incubator
2.3. Flow cytometry
The cells were labeled for flow cytometry according to manufacturer's instructions, and standard protocols. All antibodies were titrated prior to the experiment to ensure optimal working concentrations. Briefly, cells were plated in a 96‐well U‐bottom plate and treated with FcR blocking reagent for 10 minutes at room temperature. The cells were then incubated with their specific conjugated antibody (CD86‐Alexa‐Fluor 488, Major Histocompatibility Complex Class [MHC] I, II‐BV510, CD206‐PE‐Cy7, CB11b‐PE or CD14‐BV421) for 45 minutes in the dark at 4°C at a predetermined concentration. Other antibodies against CD40, CD83 and CD209 were negative on this cell line and were subsequently not used. Cells were transported after labeling in the dark on ice prior to analysis. Analysis was performed with a BD FACSCanto II flow cytometer, with three lasers (violet ‐ 405 nm, blue – 488 nm and red – 633 nm). Isotype controls were used to control for background fluorescence. All conjugated antibodies were sourced from BD Biosciences (San Jose, California) to ensure compatibility with the flow cytometer used. Analysis of data was done with BD FACSDiva software (Version 3.0). The monocyte population was gated on forward and side scatter plots and analyzed for fluorescence intensity against unilluminated control cells. Results are reported as flow cytometric dot plots with gating strategies, overlay histograms of control and treated cells, and median fluorescence intensity, as recommended by the International Society for the Advancement of Cytometry data standards task force 27.
2.4. Gene arrays
All reagents and consumables used in RNA extraction, cDNA synthesis and gene expression analysis were acquired from QIAGEN (Hilden, Germany). The array used was the RT2 Profiler PCR array “Human Innate and Adaptive Immune Responses” which analyses 84 genes related to immune and inflammatory responses, including key cytokines, chemokines and immune active surface membrane receptors. RNA quality and concentration were assessed via Agilent 2100 Bioanalyzer, Qubit fluorimeter (Invitrogen) and spectrophotometer (DeNovix). RNA extraction was performed using the RNeasy Mini kit according to the manufacturer's instructions. Briefly, cells were lysed in RLT buffer containing β‐mercaptoethanol (10 μL/mL) and centrifuged through a QIAshredder column. Lysates were passed through RNeasy spin columns, treated on‐column with DNase, following which the RNA was collected in RNase free water for preparation of cDNA. cDNA was prepared in a thermal cycler using the Qiagen RT2 First Strand kit. Briefly, RNA (500 ng) was incubated at 42°C for 2 minutes in the presence of GE buffer (containing DNase) to eliminate any remaining genomic DNA, before being immediately transferred to ice. cDNA was reverse transcribed for 15 minutes at 42°C, and at 92°C for 3 minutes to terminate the reactions. cDNA was then added to the RT2 SYBR green qPCR master mix and dispensed to the 96‐well gene array plate as per manufacturer's instructions. The gene array was run in Biorad CFX96 real‐time PCR cycler with an initial denaturation step of 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Once the RT‐PCR was complete, melt curve analysis was performed to ensure the presence of a single PCR product for each gene. Gene array data were analyzed using CFX Maestro (BioRad). An unpaired t test was used to compare groups; a P value ≤.05 and a 2‐fold change in expression were set as thresholds for significance. Genes were excluded from further analysis if the crossing point (Cq) value was greater than 34 and/or a unique melting temperature was not observed.
3. RESULTS
3.1. Polarized light decreases the expression of cell surface markers related to inflammation
Gating used for analysis was performed against appropriate isotype controls to account for background fluorescence. Example gating strategy is shown in Figure 3. No change to cell surface marker expression was seen after 5 or 30 minutes of PL exposure. The cell surface marker expression change after 6 hours exposure to PL is shown (Figure 4). Six hours of exposure to PL caused a mean decrease in the median fluorescence intensity of 23% for CD11b, 39% for CD14, 27% in MHC I and 35% in MHC II, though MHC II expression was low at baseline (Figure 4). Conversely, there was a mean increase in the median fluorescence of 20% in CD86. There were no consistent changes seen in CD206 expression.
Figure 3.

Example gating strategy. Left‐hand panel, doublet discrimination strategy; middle panel, monocytes gated using size and density; right‐hand panel, fluorescence intensity of the given antibody with quadrants for visual inspection. FSC‐A, forward scatter area; FFC‐H, forward scatter height; SSC‐A: Side scatter area
Figure 4.
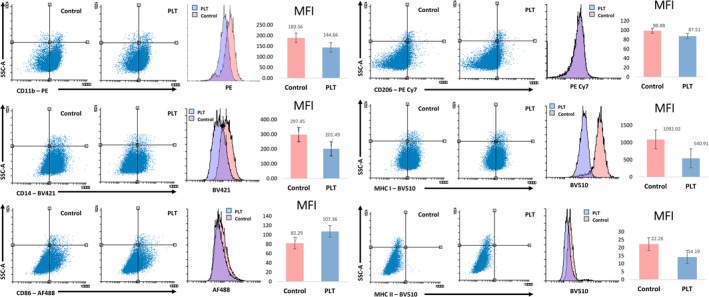
Changes in cell surface marker expression as assessed by flow cytometry following 6 hours exposure to polarized light therapy. Live cells were gated on FSC vs SSC profile and isotype control antibodies were used as background control. Shown are values above the background isotype controls. Experiments were performed in triplicate; representative samples are displayed in dot plots and histograms. MFI, median fluorescence intensity
3.2. Polarized light decreases genes related to inflammation
All experimental and control samples passed the inbuilt quality control measures in the gene array. Normalization was performed against the two most stable housekeeping genes—GAPDH and RPLP0. Cutoff points were set at 2‐fold up or downregulation, and P values calculated with significance level set at P < .05. The results are summarized in Figures 5 and 6. Figure 5 shows genes with significant change in regulation, and more than 2‐fold regulation, and the total changes for all expressed genes are presented as a scatter plot and heat map. Figure 6 shows the specific change in expression of the six genes that reached greater than 2‐fold up‐ or downregulation, and statistical significance. Six hours of PLT caused upregulation of NFKBIA and TLR9, and downregulation of IL1B, CCL2, NLRP3 and NOD1. NFKBIA is a key inflammatory inhibitor of cytokine production, and TLR9 codes for the production of the toll‐like receptor 9, a key membrane receptor involved in pathogen recognition and immune activation. IL1B and CCL2 are important cytokines, while NLRP3 and NOD1 are membrane receptors involved in inflammatory signaling. All other genes assessed in the array did not reach the significance and fold regulation cutoffs.
Figure 5.

Gene expression changes following 6 hours of PLT. Top Left, fold regulation andsignificance of genes; top right, scatter plot of fold regulation; bottom, clustered heat map. Experiments were repeated three times and mean of three repeats are shown
Figure 6.

Bar graphs of relative normalized fold regulation of significant genes compared to reference point of 1.0. *P < .05; **P < .01; ***P < 0.001
4. DISCUSSION
This study demonstrates that PLT can exert a measurable effect on the human immune system, giving much needed mechanistic evidence for its clinical effects. While phototherapies such as low‐level laser have been previously shown to have a suppressive effect on inflammation and immune response 2, very limited evidence exists investigating whether PLT has the same capacity. The results of this study demonstrate changes in gene expression and cell surface marker expression in differentiated U937 cells following 6 hours exposure of PL. This suggests that PLT has the capacity to cause modulation of the behavior of activated monocytes in vitro. This could aid in explaining the demonstrated effects of PLT in improving wound healing and decreasing pain and inflammation in vivo.
The cell surface markers studied here are known to be involved in a range of processes vital to the immune and inflammatory response, including recognition of self, T cell activation, recognition and phagocytosis of pathogens and immune cell infiltration and accumulation. It is difficult to predict how the observed changes demonstrated in this research translate into the infinitely more complex cellular environment surrounding an inflammatory or healing process, however possible in vivo effects may be inferred. The MHC I and MHC II receptors are involved in antigen presentation 28 and CD11b receptor in inflammatory cell accumulation 29 and the CD14 receptor is associated with the detection and phagocytosis of bacterial lipopolysaccharides, and other proteinaceous debris by macrophages 30. Their suppression may signify a decrease in the inflammatory activity of these cells and have the net effect of dampening the intensity and hastening the resolution of inflammation in vivo. Similar changes in immune cell marker expression have been shown in other cell types following exposure to LLLT, suggesting the possibility of common mechanisms for the actions of phototherapies 31, 32. CD86 has been shown to play an important role in the amplification and maintenance of an inflammatory state in vivo 33, 34. This could result in faster clearance of debris and pathogens and a hastening of the resolution of the inflammatory response, leading to enhanced healing times, correlating with the observed clinical effects of PLT.
While the reduction in cell surface markers suggests a functional immunomodulatory effect, changes in gene expression provide a mechanistic insight into the cellular response to PLT. The downregulation of IL1B, CCL2, NLRP3 and NOD1 suggests a dampening of the immune and inflammatory response. These genes and their associated products are implicated in lymphocyte differentiation and proliferation, inflammasome activation 35, systemic inflammation 36, immune cell migration and accumulation 37, antigen recognition 38, 39 and phagocyte activity 39. Others have described decreases in IL1B in murine wound models and aortic smooth muscle cells following exposure to LLLT 40, 41 and LED therapy 42. Additionally, LLLT has been shown to increase or decrease the expression of CCL2 at different intensities in the THP‐1 monocyte cell line, suggesting a dose‐dependent effect 43. The findings in this study provide a theoretical mechanism for their in vivo findings. Decreasing the expression of these genes in activated monocyte/macrophage cells is likely to cause a functional reduction in inflammation. The observed increase in NFKBIA fits within the picture of immune suppression, as this gene has been shown to modulate and decrease the effects of malignant 44, inflammatory 45 and auto‐immune disease 46. The outcome of the PL mediated increase in TLR9 expression might play is less clear. It is involved in the recognition and binding of bacterial and viral DNA and has been associated with auto‐immune disease and inflammation 47, but how an upregulation of these processes might exert an anti‐inflammatory or increased healing state is unclear.
While this research provides proof of concept for a physiological mechanism by which PLT may exert its clinical effects, many questions remain. Firstly, whilst monocyte/macrophage cells are important contributors to inflammation and healing processes, there are a host of other cells involved in the natural setting. Further studies to evaluate whether PLT exerts a change on other important immune, connective tissues and stem cells are necessary to give a more integrated picture of the effects of PLT on tissue healing. Additionally, while cell lines are a convenient and useful research tools, their behavior may not perfectly reflect that of their in vivo counterparts. Consequently, evaluation of how PLT may affect the diverse range of native cellular interactions involved in healing and inflammation should be examined through animal model research. The effect of light penetrance must also be evaluated to identify whether the effects shown in this study are equivalent to those that might occur in a clinical setting where the incident light is obstructed by overlying tissues. While PL has been shown to penetrate to a depth of up to 5 centimeters 7, it is currently unclear if its ability to exert a physiological change is affected by distance from the light source or the light transmitting properties of the tissue being treated. Finally, the dose response of PLT must be evaluated further. This study identified 6 hours of PLT as being able to generate a cellular response, but for translation into clinical use, minimum effective timeframes must be identified, and the effects of repeated dosages must also be explored further.
5. CONCLUSION
Noninvasive, nonpharmacological interventions to treat inflammation and to assist in the process of tissue healing have the potential to revolutionize the treatment of a wide range of conditions. This study provides preliminary evidence suggesting that PLT could be a candidate to fill this gap in therapeutic approaches.
This study provides key mechanistic evidence for the capacity of PLT to impart a physiological change. The changes in gene and cell surface marker expression observed after 6 hours of PLT were suggestive of immune and inflammatory suppression, providing potential for clinical use in a host of illnesses and injuries. These findings support the reported results of PLT seen in clinical practice, where it is used to assist in the healing of wounds and injuries. Future research on other cell types should be performed to establish a clear mechanism, and dose response experiments done to identify optimal treatment practices. Further, studies comparing the effect of the individual wavelengths that make up PLT could allow for more targeted therapeutic approaches.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
J.F., N.T. and K.M. performed the experiments under the supervision of V.A., S.F. and M.H.; J.F., D.K. and V.A. wrote the article and J.F., V.A., S.F., M.H., D.K. and A.T. edited and reviewed the article.
ACKNOWLEDGMENTS
The authors would like to thank the Immunology and Translational Group within the Institute for Health and Sport, Victoria University, for support and discussions. V.A. would like to thank the Victoria University College of Health and Biomedicine start‐up funds for financial support. J.F. was supported by the Australian Government Research Training Program and MK by the Victoria University Vice‐Chancellors Postgraduate Scholarship.
Feehan J, Tripodi N, Fraser S, et al. Polarized light therapy: Shining a light on the mechanism underlying its immunomodulatory effects. J. Biophotonics. 2020;13:e201960177 10.1002/jbio.201960177
Contributor Information
Jack Feehan, Email: jfeehan@student.unimelb.edu.au.
Vasso Apostolopoulos, Email: vasso.apostolopoulos@vu.edu.au.
REFERENCES
- 1. Cascorbi I., Clin. Pharmacol. Ther. 2017, 102(4), 564. [DOI] [PubMed] [Google Scholar]
- 2. Feehan J., Burrows S. P., Cornelius L., Cook A. M., Mikkelsen K., Apostolopoulos V., Husaric M., Kiatos D., Maturitas 2018, 116, 11. [DOI] [PubMed] [Google Scholar]
- 3. Bjordal J. M., Johnson M. I., Iversen V., Aimbire F., Lopes‐Martins R. A. B., Photomed. Laser Ther. 2006, 24(2), 158. [DOI] [PubMed] [Google Scholar]
- 4. Gambichler T., Terras S., Kreuter A., Clin. Dermatol. 2013, 31(4), 438. [DOI] [PubMed] [Google Scholar]
- 5. Jahanshahifard S., Ahmadpour‐Kacho M., Pasha Y. Z., J. Clin. Neonatol. 2012, 1(3), 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Monstrey S., Hoeksema H., Depuydt K., Maele V., Van Landuyt K., Blondeel P., Eur. J. Plast. Surg. 2002, 24(8), 377. [DOI] [PubMed] [Google Scholar]
- 7. Da Silva A., Deumié C., Vanzetta I., Biomed. Opt. Express 2012, 3(11), 2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iordanou P., Baltopoulos G., Giannakopoulou M., Bellou P., Ktenas E., Int. J. Nurs. Pract. 2002, 8(1), 49. [DOI] [PubMed] [Google Scholar]
- 9. Durovic A., Maric D., Brdareski Z., Jevtic M., Durdevic S., Vojnosanit. Pregl. 2008, 65(12), 906. [DOI] [PubMed] [Google Scholar]
- 10. Medenica L., Lens M., J. Wound Care 2003, 12(1), 37. [DOI] [PubMed] [Google Scholar]
- 11. Monstrey S., Hoeksema H., Saelens H., Depuydt K., Hamdi M., Van Landuyt K., Blondeel P., Br. J. Plast. Surg. 2002, 55(5), 420. [DOI] [PubMed] [Google Scholar]
- 12. Karadag C. A., Birtane M., Aygit A. C., Uzunca K., Doganay L., J. Burn Care Res. 2007, 28(2), 291. [DOI] [PubMed] [Google Scholar]
- 13. Oliveira P. C., Pinheiro A. L. B., de Castro I. C., Reis Junior J. A., Noia M. P., Gurgel C., Teixeira Cangussú M. C., Pedreira Ramalho L. M., Photomed. Laser Surg. 2011, 29(9), 619. [DOI] [PubMed] [Google Scholar]
- 14. Stasinopoulos D., Photomed. Laser Ther. 2005, 23(1), 66. [DOI] [PubMed] [Google Scholar]
- 15. Stasinopoulos D., Papadopoulos C., Lamnisos D., Stasinopoulos I., Disabil. Rehabil. 2017, 39(5), 450. [DOI] [PubMed] [Google Scholar]
- 16. Stasinopoulos D., Stasinopoulos I., Clin. Rehabil. 2006, 20(1), 12. [DOI] [PubMed] [Google Scholar]
- 17. Stasinopoulos D., Stasinopoulos I., Johnson M., Photomed. Laser Ther. 2005, 23(2), 225. [DOI] [PubMed] [Google Scholar]
- 18. Stasinopoulos D., Stasinopoulos I., Pantelis M., Stasinopoulou K., Photomed. Laser Surg. 2009, 27(3), 513. [DOI] [PubMed] [Google Scholar]
- 19. Fenyo M., Theoretical and experimental basis of biostimulation by laser irradiation. Opt. Laser Technol. 1984, 16(4), 209. [Google Scholar]
- 20. Pinheiro A. L. B., Pozza D. H., Oliveira M. G. D., Weissmann R., Ramalho L. M. P., Photomed. Laser Ther. 2005, 23(5), 485. [DOI] [PubMed] [Google Scholar]
- 21. Iordanou P., Lykoudis E. G., Athanasiou A., Koniaris E., Papaevangelou M., Fatsea T., Bellou P., Photomed. Laser Surg. 2009, 27(2), 261. [DOI] [PubMed] [Google Scholar]
- 22. Tada K., Ikeda K., Tomita K., J. Trauma 2009, 67(5), 1073. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Huang Y.‐Y., Wang Y., Lyu P., Hamblin M. R., Biochim. Biophys. Acta Gen. Subj. 2017, 1861(2), 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X., Tian F., Soni S. S., Gonzalez‐Lima F., Liu H., Sci. Rep. 2016, 6, 30540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X., Tian F., Reddy D. D., Nalawade S. S., Barrett D. W., Gonzalez‐Lima F., Liu H., J. Cereb. Blood Flow Metab. 2017, 37(12), 3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rots N. Y., Iavarone A., Bromleigh V., Freedman L. P., Blood 1999, 93(8), 2721. [PubMed] [Google Scholar]
- 27. Lee J. A., Spidlen J., Boyce K., Cai J., Crosbie N., Dalphin M., Furlong J., Gasparetto M., Goldberg M., Goralczyk E. M., Hyun B., Jansen K., Kollmann T., Kong M., Leif R., McWeeney S., Moloshok T. D., Moore W., Nolan G., Nolan J., Nikolich‐Zugich J., Parrish D., Purcell B., Qian Y., Selvaraj B., Smith C., Tchuvatkina O., Wertheimer A., Wilkinson P., Wilson C., Wood J., Zigon R., International Society for Advancement of Cytometry Data Standards Task Force , Scheuermann R. H., Brinkman R. R., Cytometry A 2008, 73(10), 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gazi U., Martinez‐Pomares L., Immunobiology 2009, 214(7), 554. [DOI] [PubMed] [Google Scholar]
- 29. Solovjov D. A., Pluskota E., Plow E. F., J. Biol. Chem. 2005, 280(2), 1336. [DOI] [PubMed] [Google Scholar]
- 30. Kitchens R. L., Chem. Immunol. 2000, 74(1), 61. [DOI] [PubMed] [Google Scholar]
- 31. Chen A. C., Huang Y. Y., Sharma S. K., Hamblin M. R., Photomed. Laser Surg. 2011, 29(6), 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. King D. E., Jiang H., Simkin G. O., Obochi M. O., Levy J. G., Hunt D. W., Scand. J. Immunol. 1999, 49(2), 184. [DOI] [PubMed] [Google Scholar]
- 33. Sansom D. M., Manzotti C. N., Zheng Y., Trends Immunol. 2003, 24(6), 313. [DOI] [PubMed] [Google Scholar]
- 34. Rutkowski R., Moniuszko T., Stasiak‐Barmuta A., Kosztyla‐Hojna B., Alifier M., Rutkowski K., Tatarczuk‐Krawiel A., Arch. Immunol. Ther. Exp. 2003, 51(6), 421. [PubMed] [Google Scholar]
- 35. Vandanmagsar B., Youm Y.‐H., Ravussin A., Galgani J. E., Stadler K., Mynatt R. L., Ravussin E., Stephens J. M., Dixit V. D., Nat. Med. 2011, 17(2), 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haukeland J. W., Damås J. K., Konopski Z., Løberg E. M., Haaland T., Goverud I., Torjesen P. A., Birkeland K., Bjøro K., Aukrust P., J. Hepatol. 2006, 44(6), 1167. [DOI] [PubMed] [Google Scholar]
- 37. Qian B.‐Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L. R., Kaiser E. A., Snyder L. A., Pollard J. W., Nature 2011, 475(7355), 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., Modlin R. L., Akira S., J. Immunol. 2002, 169(1), 10. [DOI] [PubMed] [Google Scholar]
- 39. Mahla R. S., Reddy C. M., Prasad D., Kumar H., Front. Immunol. 2013, 4, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sayed I. S., Saafan A., Abdel‐Gawad F. K., Harhash T. A., Abdel‐Rahman M. A., J. Dent. Lasers 2015, 1(9), 23. [Google Scholar]
- 41. Gavish L., Perez L., Gertz S. D., Lasers Surg. Med. 2006, 38(8), 779. [DOI] [PubMed] [Google Scholar]
- 42. Martins D. F., Turnes B. L., Cidral‐Filho F. J., Bobinski F., Rosas R. F., Danielski L. G., Petronilho F., Santos A. R., Neuroscience 2016, 324, 485. [DOI] [PubMed] [Google Scholar]
- 43. Chen C. H., Wang C. Z., Wang Y. H., Liao W. T., Chen Y. J., Kuo C. H., Kuo H. F., Hung C. H., Mediators Inflamm. 2014, 2014, 625048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bredel M., Scholtens D. M., Yadav A. K., Alvarez A. A., Renfrow J. J., Chandler J. P., Yu I. L., Carro M. S., Dai F., Tagge M. J., N. Engl. J. Med. 2011, 364(7), 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoshioka T., Nishikomori R., Hara J., Okada K., Hashii Y., Okafuji I., Nodomi S., Kawai T., Izawa K., Ohnishi H., J. Clin. Immunol. 2013, 33(7), 1165. [DOI] [PubMed] [Google Scholar]
- 46. Klein W., Tromm A., Folwaczny C., Hagedorn M., Duerig N., Epplen J. T., Schmiegel W. H., Griga T., Int. J. Colorectal Dis. 2004, 19(2), 153. [DOI] [PubMed] [Google Scholar]
- 47. Wong F. S., Hu C., Zhang L., Du W., Alexopoulou L., Flavell R. A., Wen L., Ann. N. Y. Acad. Sci. 2008, 1150(1), 146. [DOI] [PubMed] [Google Scholar]


