Abstract
This study investigates the feasibility of in vivo quantitative optical coherence tomography (OCT) of human brain tissue during glioma resection surgery in six patients. High‐resolution detection of glioma tissue may allow precise and thorough tumor resection while preserving functional brain areas, and improving overall survival. In this study, in vivo 3D OCT datasets were collected during standard surgical procedure, before and after partial resection of the tumor, both from glioma tissue and normal parenchyma. Subsequently, the attenuation coefficient was extracted from the OCT datasets using an automated and validated algorithm. The cortical measurements yield a mean attenuation coefficient of 3.8 ± 1.2 mm−1 for normal brain tissue and 3.6 ± 1.1 mm−1 for glioma tissue. The subcortical measurements yield a mean attenuation coefficient of 5.7 ± 2.1 and 4.5 ± 1.6 mm−1 for, respectively, normal brain tissue and glioma. Although the results are inconclusive with respect to trends in attenuation coefficient between normal and glioma tissue due to the small sample size, the results are in the range of previously reported values. Therefore, we conclude that the proposed method for quantitative in vivo OCT of human brain tissue is feasible during glioma resection surgery.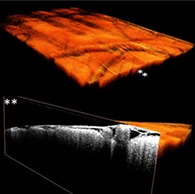
Keywords: attenuation coefficient, glioma, in vivo human brain tissue, intraoperative, optical coherence tomography
This study shows the feasibility of in vivo OCT imaging of the human brain tissue during glioma resection surgery combined with quantitative analysis by means of the OCT attenuation coefficient. The attenuation coefficient is extracted from the data using an automated and validated algorithm.
1. INTRODUCTION
The recurrence and survival benefit for glioma patients can be increased by more effective tumor resection 1, 2. The gold standard for glioma diagnosis is histological and immunohistochemical analyses. In daily practice preoperative MRI is used to image the patient's brain as a visual guidance for the surgeon. However, due to the limited resolution of MRI (approximately 1‐1.5 mm), there is a need for a high‐resolution intraoperative imaging modality, which has led to the development and study of various imaging techniques for surgical navigation, such as: intraoperative CT and MRI, fluorescence imaging, Raman spectroscopy and ultrasound 3.
Optical coherence tomography (OCT) allows non‐contact label‐free volumetric visualization of tissue with a resolution of approximately 10 to 20 micrometer and a depth of view of 1 to 2 mm which is typical for commercially available clinical OCT systems. OCT has previously been applied to image ex vivo and in vivo tissues in various human organs, including the retina, coronary artery, gastric tract, urinary tract, prostate, breast and also the human brain 3, 4, 5, 6, 7, 8, 9. The label‐free and real‐time imaging capabilities make OCT a suitable tool for intraoperative use 10, 11. In addition to tissue visualization, extraction of quantitative parameters from the OCT signal is proposed as a tool to discriminate between healthy and diseased tissue. The microstructure and organization of tissue is altered due to disease and reflected in its optical properties 12, which are accessible through quantitative OCT parameters. A frequently used quantitative OCT parameter is the optical attenuation coefficient (μOCT). At commonly used OCT wavelengths, at which absorption in most biological tissues is much lower than scattering, μOCT estimates the scattering properties of tissue. Within the single scattering approximation, μOCT can be determined by fitting a single exponential model to the OCT data after correction for system‐dependent parameters 13. Multiple studies have shown promising results for the use of μOCT in order to enhance contrast between healthy and diseased tissue to offer visual guidance to clinicians 14, 15, 16, 17, 18, 19. In addition to μOCT, the speckle distribution of OCT images has been shown to contain information on sub‐resolution scattering particles and is studied as a quantitative parameter for tissue characterization 20, 21, 22. In a study on rat liver and brain tissue, speckle distribution analysis of the OCT images has shown to provide for added contrast based on tissue heterogeneity 21. In general, OCT speckle is expected to follow a Rayleigh distribution for static homogenous tissues 22, 23. Sub‐resolution tissue inhomogeneities may therefore be detected through OCT speckle analysis 21.
Imaging of human brain tissue using OCT has been investigated in multiple studies 3, 4, 5, 6, 7, 8, 9. Most of these studies have examined the difference between cancerous and normal tissue in ex vivo brain samples 3, 4, 5, 6, 8, 9. Assayag et al 6 show that brain tissue microstructures are clearly identified using full‐field OCT with a high lateral resolution of 1 μm in ex vivo fresh samples of human brain. The authors conclude that high‐grade glioma, but not low‐grade glioma, could be distinguished in the OCT images. A disadvantage of high‐resolution OCT is the limited imaging depth (from 1 to 2 mm for conventional resolution systems to approximately 200 μm for 1 μm resolution systems) and the increase in acquisition time and data volume. Quantitative analysis of conventional resolution OCT images may provide an alternative real‐time visual guidance by discriminating normal from glioma brain tissue based on the difference in tissue optical properties.
Subcortical brain tissue (white matter) contains myelinated nerve fibers, which give the subcortical tissue its white color. The myelinated nerve fibers are strongly scattering and visible as bright white lines in microscopic OCT images 2. Due to its strong scattering and abundance in subcortical brain tissue, myelin is expected to be the main contributor to scattering in subcortical brain tissue. Cortical brain tissue (gray matter) contains a minimal amount of myelin, therefore a lower μOCT is expected for normal cortical brain tissue compared to normal subcortical brain tissue. This hypothesis is substantiated by quantitative OCT studies which found a lower μOCT for cortical tissue (gray matter) compared to subcortical tissue (white matter) in fixated 7 and fresh ex vivo samples of human brain tissue 3. Changes in tissue caused by glioma are expected to change the scattering properties of the affected brain tissue and therefore measureable by means of quantitative OCT. Low‐grade glioma is expected to show an increase interstitial water content and potentially a minor increase in cell density compared to normal brain tissue 24, 25. High‐grade glioma shows an increase in cell density, vascularization and necrosis compared to normal tissue. Furthermore, high‐grade glioma degrades the myelin content in subcortical tissue 3, 24. These changes in tissue composition have contrary effects on tissue optical properties, which make it hard to predict the expected trend in μOCT. Experimentally an increase of μOCT for both low‐grade and high‐grade glioma compared to normal cortical brain tissue was reported based on a study on ex vivo samples collected in 32 patients 3. For subcortical tissue a decrease in μOCT for both low‐grade and high‐grade glioma tissue compared to normal tissue was reported. The only in vivo OCT study on glioma detection in human brain tissue to our knowledge reports on OCT images in nine patients during brain tumor resection using a TD‐OCT system 5. After semi‐quantitative analysis, the authors conclude that analysis of visible microstructure and light attenuation in OCT images discriminates between normal brain tissue, infiltrative zone, solid cancer and necrosis. The promising results of these studies suggest that μOCT could be used to differentiate glioma from normal brain tissue, and therefore serve as motivation for continued investigations, particularly in in vivo and intraoperative settings. Intraoperative in vivo measurements introduce challenges in data collection due to time pressure and motion artifacts caused by, for example, heartbeat and respiration. Quantitative data analysis of these datasets is not trivial. Challenges such as unequal tissue edges, the presence of blood vessels and the accumulation of liquid and blood need to be addressed. Moreover, automated analysis is required to provide for objective, unsupervised and robust analysis of such large datasets.
In this pilot study, we investigate the feasibility of in vivo quantitative OCT during brain surgery in six patients. We present OCT data collected during standard surgical glioma resection procedure from normal and glioma tissue in cortical and subcortical brain regions. A validated and automated custom‐written software was developed for data analysis. Using this software, we extract μOCT and speckle contrast, which can be used as a potential tool to discriminate between normal and glioma tissue. Additionally, we use the obtained speckle contrast as a quality check for the edge detection method to provide for a robust and unbiased exclusion of data containing incorrect edge detection. We show that the obtained μOCT values are in the same range as reported in literature and thereby confirm the validity and feasibly of our approach. Based on our results we recommend further studies in order to assess the ability of quantitative OCT to differentiate glioma from normal brain tissue. This is the first intraoperative in vivo study of the use of quantitative OCT during glioma resection.
2. MATERIALS AND METHODS
2.1. Study design
The feasibility of the use of OCT imaging during glioma resection surgery, followed by quantitative analysis of the collected data, was studied in six patients. The preoperative MRI scans of these six patients asserted that the tumor location was likely to enable successful measurements. This study was carried out at the Radboud University (Nijmegen, Netherlands) according to the Declaration of Helsinki for experiments involving humans. An informed consent was obtained from each patient and the privacy of the patients was ensured by anonymization of the data. The OCT data were collected during the standard surgery protocol after craniotomy and opening of the dura. Glioma and normal brain tissue were scanned cortically (gray matter) before resection and subcortically (white matter) after partial resection of the tumor. Tissue areas were labeled as glioma and normal based on visual inspection of the brain tissue as well as MRI based neuro‐navigation by the neurosurgeon. 3D OCT datasets of (x,y,z) 1024, 1024, 600 pixels corresponding to 10,10, 3.75 mm, respectively, were collected using a commercial 50 kHz Santec IVS 2000 swept source OCT systems, operating at a center wavelength of approximately1300 nm with a axial and lateral resolution, experimentally determined as approximately 13 and 25 μm FWHM, respectively. The OCT systems' signal to noise was determined at 118 dB 26. All depth distances on OCT images throughout this paper are given in physical length (mm), using a refractive index of 1.4 for brain tissue 3. In accordance to standard surgical procedure, a sample of the tumor was collected for histopathological analysis. Based on the histopathological outcome we have grouped the patients as low grade (grade 2) and high grade (grade 4). Afterwards, the collected OCT datasets were analyzed by extracting μOCT and speckle contrast from the OCT datasets.
2.2. Data selection for quantitative analysis
A total of 19 cortical 3D OCT datasets were recorded in five patients and a total of eight subcortical 3D datasets were recorded in three patients. Due to the surgical settings and time restrictions we were not able to record subcortical scans in two patients.
After visual inspection a region of approximately 2.4 by 2.4 mm suitable for quantitative analysis was selected from each OCT dataset. This selection was made in order to exclude parts from quantitative analysis that contain image artifacts caused by out of focus, mirror images, large blood vessels and accumulation of blood and fluids on the tissue surface. In total nine cortical and three subcortical OCT datasets were fully excluded due to low quality and image artifacts. An overview of patient and data inclusion is given in Supporting Information Figure S1.
2.3. Quantitative analysis
2.3.1. Attenuation coefficient
Automated quantitative analysis of the OCT data included determination of μOCT and OCT speckle contrast 22. μOCT was determined by a nonlinear least squares (NLS) fit using the following model 13:
| (1) |
where, 〈A(z)〉 is the averaged OCT amplitude in depth, z is the position in depth, z0 is the position of the tissue boundary, A and μOCT are free running parameters (amplitude and attenuation coefficient, respectively). The system‐dependent parameter t(z) (describing the point spread function) is defined as: , where zf is the position of the focus in depth, ZR0 is the Rayleigh length in air and n is the refractive index of the medium 27. The sensitivity roll‐off h(z) is defined as: 13, 28. Here, zmax is the maximal imaging depth of the OCT system and s is the ratio between the spectral resolution and the sampling interval. The contribution of the point spread function and the sensitivity roll‐off were determined by fitting the equation for 〈A(z)〉 to the OCT amplitude of a sample with a low concentration of Intralipid (0.003%), for which an scattering coefficient of 0.15 mm−1 was assumed, and ZR0 and s were the free running parameters.
2.3.2. Speckle contrast
The speckle contrast (C), which is the ratio of standard deviation over mean of the OCT amplitude values, was calculated from each region of interest (ROI) 22.
| (2) |
To explore the use of speckle contrast as a tool to discriminate between normal and glioma tissue the mean and standard deviation of speckle contrast from each scan was calculated after excluding the ROI's for which did not meet the R 2 and residual criteria of the μOCT fit.
2.4. Edge detection and ROI selection
Quantitative analysis of the OCT data was performed using custom‐written code (Matlab 7.11.0 R2010b; The Mathworks Inc., Natick, Massachusetts). Prior to edge detection, the uppermost 28 pixels of all A‐lines were removed to exclude system‐related signal artifacts. Detection of the brain tissue surface in the dB‐scale data was achieved by first excluding low SNR regions by means of a pixel value threshold after which we applied a 3 × 3 median filter and then a vertical edge filter. The thus detected tissue edge was then smoothed further by carrying out local regression using weighted linear least‐squares and a second degree polynomial model. Selecting all data below this smoothed edge and converting it to linear scale then yielded raw OCT amplitude data of the brain tissue. Finally, this data was straightened and then divided into laterally overlapping regions of interest (ROIs): 10 × 10 × 30 pixels (0.1 mm × 0.1 mm × 0.2 mm) for the determination of μOCT and 10 × 10 × 5 pixels (0.1 mm × 0.1 mm × 0.03 mm) for the determination of speckle contrast. These ROIs were positioned below the detected tissue edge to exclude structures on the tissue surface from the fit analysis. In the cortical scans the ROIs were set between 10 and 30 pixels below the tissue surface while for the subcortical scans the ROIs were positioned 10 pixels below the tissue edge. The A‐lines of the individual ROIs for the μOCT determination were then averaged, yielding a single average A‐line for each ROI. Subsequently, a ROI‐specific μOCT per pixel was determined by non‐linear least squares fitting Equation (1) to this average A‐line. The noise was defined as the average of the last 20 pixels of the average A‐line. Speckle contrast was calculated from the un‐averaged OCT amplitude data from the speckle contrast ROIs, for which the depth of the ROI was limited to 5 pixels (0.03 mm) 22 in order to avoid influence of attenuation of the OCT signal in depth on the speckle distribution within the ROI.
2.5. Inclusion criteria for μOCT based on fit statistics and speckle contrast
The residual and R 2 of the NLS fits were calculated to serve as a threshold to only include the outcome of fits for which the fitted curve has a good overlap with the experimental data. The thresholds (residual < 0.4, R 2 > 0.8) were chosen based on visual inspection of the overlap between model‐based fitted curves and experimental data from a selection of the datasets. In addition, only μOCT values from ROI's with speckle contrast between 0.47 and 0.57 were included, which is assumed to correspond to a homogenous tissue region. The speckle gate was chosen at ±10% of the theoretical speckle contrast value of 0.52 after visual inspection of multiple randomly selected datasets. Finally, μOCT values for each included ROI were obtained after correction of the depth axis for the refractive index of brain tissue 3, 1.4, yielding values per mm.
3. RESULTS
3.1. Validation of analysis software
Intralipid is commonly used as a calibration sample in scattering‐based experiments. To validate our custom‐written automated algorithm presented in this paper, μOCT and speckle contrast were extracted from OCT data of a concentration series of Intralipid (Figure 1). The obtained μOCT values using our custom‐written software were in good agreement with in literature reported experimental values 29.
Figure 1.
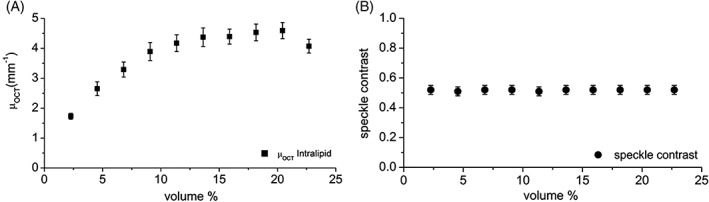
A, OCT attenuation coefficient (μOCT) and B, speckle contrast of a concentration series of Intralipid. μOCT values per are obtained using the refractive index of 1.34 for water
3.2. Histopathology
Cancer type and grade were determined by histopathology and are summarized in the Table 1. Patients 1 and 4 were diagnosed with a low‐grade tumor, patients 2, 3 and 5 were diagnosed with a high‐grade tumor.
Table 1.
Histopathological results per patient
| Patient number | Histopathological outcome |
|---|---|
| Pt 1 | Astrocytoma (grade 2) |
| Pt 2 | Glioblastoma (grade 4) |
| Pt 3 | Glioblastoma (grade 4) |
| Pt 4 | Oligodendroglioma (grade 2) |
| Pt 5 | Glioblastoma (grade 4) |
3.3. Qualitative analysis of OCT datasets
3D OCT datasets were successfully collected from cortical brain tissue in five patients and from subcortical brain tissue in three patients. Cross‐sections (B‐scans) from normal brain and glioma areas of cortical and subcortical brain tissue are shown in Figure 2. In the cortical scans superficial blood vessels were clearly visible next to the structure of the arachnoid space. No visual features could be identified to differentiate between glioma and normal brain tissue. Figure 3 shows a volumetric view of a subcortical OCT dataset together with a cross‐sectional view at two locations on the y‐plane (Figure 3).
Figure 2.
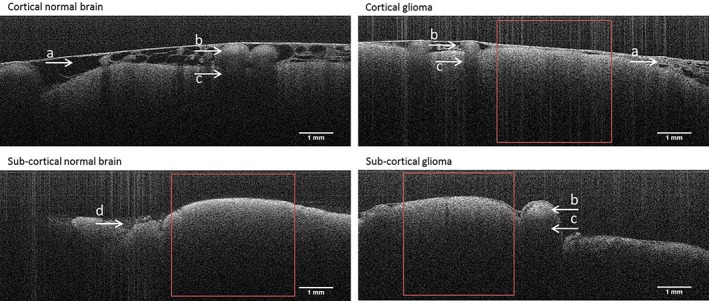
Cortical and subcortical gray scale cross‐sectional OCT scans of glioma and normal brain tissue. A) arachnoid space, B) blood vessel, C) shadowing due to blood vessel, D) blood or fluid pool. The red squares show a typical region selected for quantitative analysis
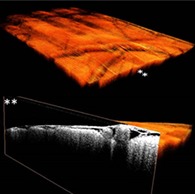
Figure 3.

Volumetric view of a cortical OCT dataset (A) together with a cross‐sectional view at two locations (B,C) on the y‐plane
3.4. Quantitative analysis of OCT datasets
The regions selected for quantitative analysis were analyzed using our custom‐written automated software to extract μOCT and speckle contrast from the OCT data. The obtained μOCT and speckle contrast per scan are shown in a color‐coded map in Figures 4 and 5 for the cortical and subcortical datasets, respectively. In patients with high‐grade glioma (n = 3) we found a mean and standard deviation μOCT of 3.7 ± 1.5 mm−1 for normal cortical tissue and 3.9 ± 1.4 mm−1 for glioma tissue. In patients with low‐grade glioma (n = 2) we found a μOCT of 4.0 ± 1.1 mm−1 for normal cortical tissue and 3.1 ± 0.5 mm−1 for glioma tissue. For the subcortical datasets (all high‐grade tumors) a μOCT of 5.7 ± 2.1 mm−1 for normal tissue and 4.5 ± 1.6 mm−1 for glioma tissue was found.
Figure 4.
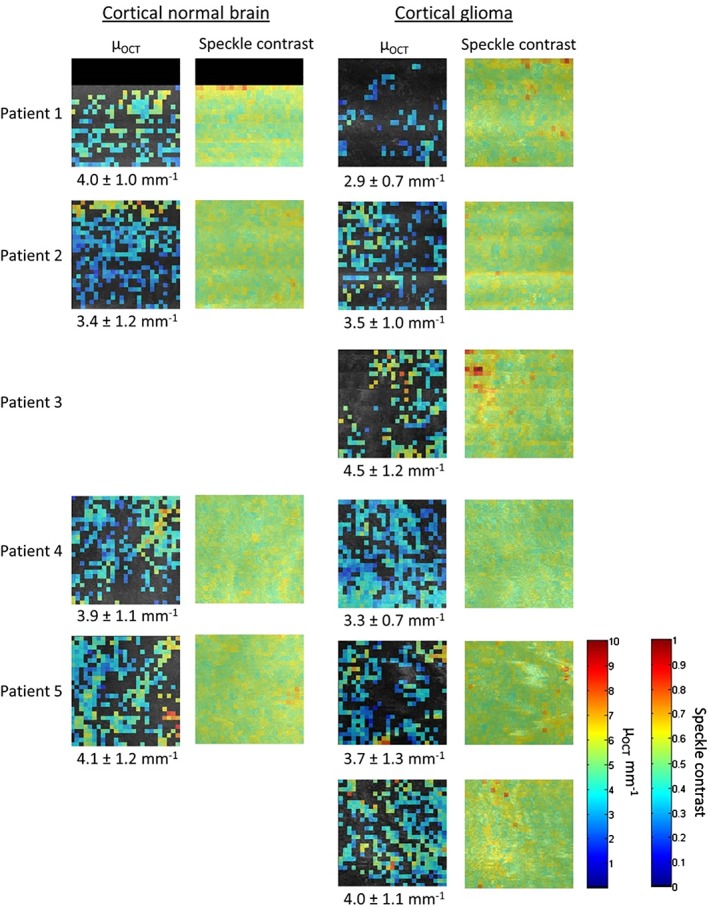
Color map of μOCT and speckle contrast of the cortical OCT datasets. The μOCT color scale bar runs from 0 to 10 mm−1 and the color scale bar of the speckle contrast runs from 0 to 1. The μOCT mean and standard deviation are given below the color maps
Figure 5.
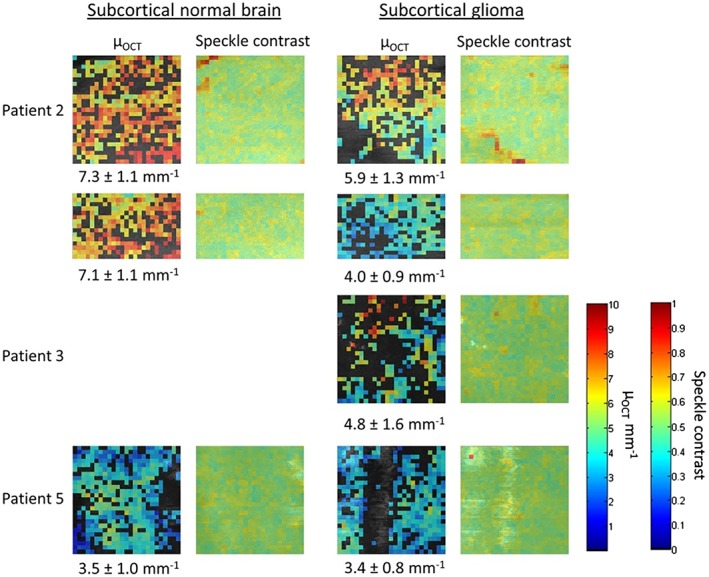
Color map of speckle contrast and μOCT of the subcortical OCT datasets. The μOCT color scale bar runs from 0 to 10 mm−1 and the color scale bar of the speckle contrast runs from 0 to 1. The μOCT mean and standard deviation are given below the color maps
From the speckle contrast analysis we obtained a mean and standard deviation of 0.54 ± 0.19 for cortical and 0.55 ± 0.19 for subcortical brain tissue. Moreover, no difference was found in speckle contrast values for normal and glioma tissue.
4. DISCUSSION
In this study we have demonstrated the feasibility of in vivo quantitative OCT of human brain tissue during glioma resection surgery. OCT datasets of glioma and normal tissue were successfully collected in the cortical and subcortical brain areas in respectively five and three patients. The quality of 15 out of 27 of these OCT datasets was sufficient for automated quantitative analysis. The quality of the excluded datasets was hampered by motion artifacts and specular reflections caused by fluid accumulation on the tissue surface. Data collection was especially challenging for subcortical areas due to the limited size of the resection site compared to the size of the OCT probe. In future research, the quality of the OCT datasets would strongly benefit from a dedicated probe design 3. To our knowledge, this is the second study to collect in vivo OCT images of normal and glioma human brain tissue. In addition, in this study, the OCT datasets were analyzed with an automated custom‐written software to extract μOCT and speckle contrast applying a validated and robust approach.
4.1. Qualitative analysis
Böhringer et al 5 report OCT images recorded in nine patients during glioma resection (in vivo and ex vivo fresh tissue on ice) and conclude that qualitative analysis of the microstructure and optical attenuation in OCT images discriminates between normal brain tissue, infiltrated brain tissue, solid cancer and necrosis. While the arachnoid space and blood vessels were clearly visible in our OCT images (Figure 2), the pathological microstructures of glioma tissue described by Böhringer et al were not apparent.
4.2. Quantitative analysis
4.2.1. OCT attenuation coefficient (μOCT)
The analysis yields a mean and standard deviation of μOCT of 3.8 ± 1.2 mm−1 for normal gray matter (cortical) and 5.7 ± 2.1 mm−1 and white matter (subcortical). The higher μOCT for white matter is in agreement with literature and is attributed of the high myelin content in white matter 3, 6, 7. Our results are very similar to the values reported by Kut et al, that is, 2.8 ± 0.9 mm−1 for normal cortical tissue and 6.2 ± 0.8 mm−1 for normal subcortical tissue.
In cortical brain tissue an increase in μOCT is reported in literature for both low‐grade and high‐grade glioma compared to normal tissue. Therefore, grouping low‐grade and high‐grade glioma together is justified, which yields a mean of 3.8 ± 1.2 mm−1 for normal tissue and 3.6 ± 1.1 mm−1 for glioma tissue. In subcortical brain tissue we have only collected datasets from patients with high‐grade glioma, which yield a mean μOCT of 5.7 ± 2.1 mm−1 for normal tissue and 4.5 ± 1.6 mm−1 for glioma tissue. A decrease in μOCT for glioma compared to normal brain tissue is in agreement with literature, and is commonly attributed to the decay of myelin 3, 5. The obtained μOCT results here, however, are inconclusive with respect to trends in μOCT between normal and glioma tissue. The small number of patients restricts any meaningful statistical analysis. Nevertheless, we observe that the obtained μOCT values are in the range of values reported by Kut et al for ex vivo brain samples 3. Any differences in μOCT values between this study and the study by Kut et al can be explained by the difference between in vivo and ex vivo tissue optical properties, and the lack of on to one match of in vivo OCT data with histology in this study.
4.2.2. Speckle contrast
The distribution of OCT speckle is expected to be Rayleigh distributed for a homogenous distribution of scattering particles 22, 23. For such Rayleigh distribution, a speckle contrast of 0.52 is expected. The speckle contrast within a ROI is influenced by tissue inhomogeneities both smaller and larger than the imaging resolution. In this study, speckle contrast from each ROI was calculated for two purposes. First, speckle contrast was analyzed to explore its use to discriminate between normal and glioma tissue. Therefore, the mean and standard deviation speckle contrast of each scan was calculated after excluding the ROI's for which the μOCT fits did not met the R 2 > 0.8 and residual < 0.4 criteria. We found that the mean speckle contrast was equal for normal and glioma tissue in both cortical and subcortical brain tissue. Second, a speckle contrast gate (0.47–0.57) was applied to the extracted μOCT values to only include the outcome of fits from ROIs containing more or less homogenous tissue and correct edge detection. The speckle gate boundaries were chosen at 10% of the theoretical value of 0.52 for homogenous tissue, based on visual inspection of a number of randomly selected datasets. This approach allowed for a more robust method for edge detection.
4.3. Limitations
Tumor tissue was identified using neuro‐navigation on preoperative MRI scans. Identified areas were part of the bigger tissue sample that was sent for histopathological examination and all were positive for glioma. However, obviously, normal brain tissue was not sent for histopathological examination, so we cannot rule out tumor infiltration in the areas that were labeled as normal brain. Furthermore, the depth of analysis was limited, which could have hampered detection of glioma tissue in the cortical scans.
To extract reliable μOCT values from OCT data we have fitted a well‐studied and validated model of the OCT signal to the data. In this model system‐dependent parameters are taken into account. Moreover, we have used fit statistics to only include the outcome of the NLS fits that are a good representation of the data. Furthermore, the model assumes single scattering from a homogeneous region of tissue. To satisfy this assumption we have areas for quantitative analysis (250 by 250 pixels) without large blood vessel based on visual inspection of the datasets. Moreover, we applied a speckle gate as a quality check for homogeneity and valid edge detection.
In summary, quantitative OCT has been suggested in literature as a tool with great potential for intraoperative imaging during brain tumor resection as a visual guide to discriminate glioma from normal brain tissue. Studies on ex vivo human brain tissue have shown promising results, motivating this pilot study to investigate the feasibility of OCT imaging on in vivo human brain during glioma resection surgery. We have proposed and validated a methodology for an automated quantitative analysis to extract μOCT and speckle contrast from the OCT data. We successfully collected OCT images during glioma resection surgery, from which the quantitative parameters μOCT and speckle contrast were extracted. From the results of this pilot study we conclude, that the proposed method for quantitative in vivo OCT of the brain cortex is feasible during glioma resection surgery. However, applicability for subcortical areas is limited due to surgical restrictions and current probe size, which has resulted in low data quality and exclusion of data from quantitative analysis. While the obtained μOCT values are in the same range as reported values of ex vivo brain tissue samples, further studies on a larger group of patients together with one to one matching between in vivo OCT scans and gold standard histology are needed to investigate the ability of μOCT to discriminate between glioma and normal brain tissue. For successful translation to the clinical, real‐time and fully automated extraction of quantitative parameters is needed 8. However, this is challenging due to irregular tissue structures present in in vivo collected OCT data. At the moment analysis steps such as ROI selection and edge detection optimization require supervision. Therefore, additional image analysis steps are required to provide fully unsupervised quantitative analysis of in vivo OCT data.
Supporting information
FIGURE S1. Overview of patient and data inclusion
FIGURE S2. Flow chart of quantitative data analysis
ACKNOWLEDGMENTS
This research was funded by the Netherlands Organization for Scientific Research as a part of iMIT, IOP Photonic Devices Project number IPD 12020, managed by the Netherlands Enterprise Agency and was performed within the platform of Institute Quantivision.
Almasian M, Wilk LS, Bloemen PR, van Leeuwen TG, ter Laan M, Aalders MCG. Pilot feasibility study of in vivo intraoperative quantitative optical coherence tomography of human brain tissue during glioma resection. J. Biophotonics. 2019;12:e201900037 10.1002/jbio.201900037
Funding information IOP photonic devices , Grant/Award Number: IPD 12020; NWO, iMIT, Grant/Award Number: Optical Biopsy; the Netherlands Organization for Scientific Research
REFERENCES
- 1. Smith J. S., Chang E. F., Lamborn K. R., Chang S. M., Prados M. D., Cha S., Tihan T., VandenBerg S., McDermott M. W., Berger M. S., J. Clin. Oncol. 2008, 26, 1338. [DOI] [PubMed] [Google Scholar]
- 2. Sanai N., Polley M. Y., McDermott M. W., Parsa A. T., Berger M. S., J. Neurosurg. 2011, 115, 3 10.3171/2011.2.JNS10998. [DOI] [PubMed] [Google Scholar]
- 3. Kut C., Chaichana K. L., Xi J., Raza S. M., Ye X., McVeigh E. R., Li X., Sci. Transl. Med. 2015, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Böhringer H. J., Boller D., Leppert J., Knopp U., Lankenau E., Reusche E., Bo H. J., Hu G., Giese A., Lasers Surg. Med. 2006, 38, 588. [DOI] [PubMed] [Google Scholar]
- 5. Böhringer H. J., Lankenau E., Stellmacher F., Reusche E., Hüttmann G., Giese A., Neurosurg. Concepts 2009, 151, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Assayag O., Grieve K., Devaux B., Harms F., Pallud J., Chretien F., Boccara C., Varlet P., Neuroimage 2013, 2, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H., Magnain C., Sakadžić S., Fischl B., Boas D., Biomed. Opt. Express 2017, 8, 5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan W., Kut C., Liang W., Li X., Sci. Rep. 2017, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bizheva K., Unterhuber A., Hermann B., Povaz B., Sattmann H., Fercher A. F., Drexler W., J. Biomed. Opt. 2005, 10, 011006‐1. [DOI] [PubMed] [Google Scholar]
- 10. Carrasco‐Zevallos O. M., Viehland C., Keller B., Draelos M., Kuo A. N., Toth C. A., Izatt J. A., Biomed. Opt. Express 2017, 8, 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jansen S. M., Almasian M., Wilk L. S., De Bruin D. M., Henegouwen M. I. V. B., Strackee S. D., Id P. R. B., Meijer S. L., Id S. S. G., Id T. G. V. L., Sensors 2018, 18, 1. [Google Scholar]
- 12. Cheong W. F., Prahl S. A., Welch A. J., IEEE J. Quantum Electron. 1990, 26, 2166. [Google Scholar]
- 13. Almasian M., Bosschaart N., van Leeuwen T. G., Faber D. J., J. Biomed. Opt. 2015, 20, 121314‐1. [DOI] [PubMed] [Google Scholar]
- 14. Swager A., Faber D. J., De Bruin D. M., Meijer S. L., Bergman J. J., Curvers W. L., J. Biomed. Opt. 2017, 22, 086001. [DOI] [PubMed] [Google Scholar]
- 15. Scolaro L., McLaughlin R. a., Klyen B. R., Wood B. a., Robbins P. D., Saunders C. M., Jacques S. L., Sampson D. D., Biomed. Opt. Express 2012, 3, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wessels R., de Bruin D. M., Faber D. J., van Boven H. H., Vincent A. D., van Leeuwen T. G., van Beurden M., Ruers T. J. M., J. Biomed. Opt. 2012, 17, 116022‐1. [DOI] [PubMed] [Google Scholar]
- 17. McLaughlin R. A., Scolaro L., Robbins P., Saunders C., Jacques S. L., Sampson D. D., J. Biomed. Opt. 2010, 15, 46029. [DOI] [PubMed] [Google Scholar]
- 18. Cauberg E. C. C., de Bruin D. M., Faber D. J., de Reijke T. M., Visser M., de la Rosette J. J. M. C. H., van Leeuwen T. G., J. Biomed. Opt. 2010, 15, 066013‐1. [DOI] [PubMed] [Google Scholar]
- 19. McLaughlin R. A., Scolaro L., Robbins P., Saunders C. M., Jacques S. L., Sampson D. D., Med. Image Comput. Comput. Assist. Interv. 2009, 12, 657. [DOI] [PubMed] [Google Scholar]
- 20. Lindenmaier A. A., Conroy L., Farhat G., Dacosta R. S., Flueraru C., Vitkin I. A., Opt. Lett. 2013, 38, 1280. [DOI] [PubMed] [Google Scholar]
- 21. Sugita M., Brown R. A., Popov I., Vitkin A., J. Biophotonics 2018, 11, e201700055. [DOI] [PubMed] [Google Scholar]
- 22. Almasian M., van Leeuwen T. G., Faber D. J., Sci. Rep. 2017, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hillman T. R., Adie S. G., Seemann V., Armstrong J. J., Jacques S. L., Sampson D. D., Opt. Lett. 2006, 31, 190. [DOI] [PubMed] [Google Scholar]
- 24. DeAngelis L. M., Brain Tumors English J. 2010, 344, 114. [DOI] [PubMed] [Google Scholar]
- 25. Lee E. J., Lee S. K., Agid R., Bae J. M., Keller A., Terbrugge K., Am. J. Neuroradiol. 2008, 29, 1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Bruin D. M., Bremmer R. H., Kodach V. M., de Kinkelder R., van Marle J., van Leeuwen T. G., Faber D. J., J. Biomed. Opt. 2010, 15, 25001. [DOI] [PubMed] [Google Scholar]
- 27. Van Leeuwen T. G., Faber D. J., Aalders M. C., IEEE J. Sel. Top. Quantum Electron. 2003, 9, 227. [Google Scholar]
- 28. Nassif N., Cense B., Park B., Pierce M., Yun S., Bouma B., Tearney G., Chen T., de Boer J., Opt. Express 2004, 12, 367. [DOI] [PubMed] [Google Scholar]
- 29. Kodach V. M., Kalkman J., Faber D. J., van Leeuwen T. G., Biomed. Opt. Express 2010, 1, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. Overview of patient and data inclusion
FIGURE S2. Flow chart of quantitative data analysis


