Abstract
Cyclins, cyclin-dependent kinases and other components of the core cell cycle machinery drive cell division. Growing evidence indicates that this machinery operates in a distinct fashion in some mammalian stem cell types, such as pluripotent embryonic stem cells. In this review, we discuss our current knowledge of how cell cycle proteins mechanistically link cell proliferation, pluripotency and cell fate specification. We focus on embryonic stem cells, induced pluripotent stem cells, and embryonic neural stem/progenitor cells.
The core cell cycle machinery operating in the cell nucleus orchestrates cell division. The key components of this machinery are proteins called cyclins that bind, activate and provide substrate specificity to their associated catalytic partners, the cyclin-dependent kinases (CDKs)1–4. Cell cycle progression can be divided into four phases: gap 1 (G1), DNA synthesis (S), gap 2 (G2) and mitosis (M). Depending on the mitogenic environment, cells traversing G1 phase either activate a program that will result in cell division, or they enter a quiescent G0 state1–4 (Fig. 1a). At the molecular level, stimulation of cells with growth-promoting factors results in upregulation of the D-type cyclins (D1, D2 and D3), which activate the cyclin-dependent kinase 4 (CDK4) and CDK61–5. In a classical cell cycle model, cyclin D-CDK4/6 complexes, together with E-type cyclins (E1 and E2) and their associated kinases (primarily CDK2, but also CDK1 and CDK3) phosphorylate and functionally inactivate the retinoblastoma protein RB1, and pRB1-related RBL1 and RBL2 proteins1–4. This leads to the activation or de-repression of E2F transcription factors, which then transactivate genes required for the entry and progression of cells into S phase1–4,6,7. This model has been questioned by the demonstration that throughout most of G1 phase, RB1 exists in a mono-phosphosphorylated state, and becomes fully phosphorylated by cyclin E-CDK2 at the end of G1 phase8. In addition to RB1 phosphorylation, inactivation of Cdh1, a substrate recognition subunit of the anaphase promoting complex (APC/C), contributes to an irreversible commitment of cells to cell division9. Inhibition of Cdh1 by cyclin E-CDK2 at the end of G1 phase inactivates APC/C and allows accumulation of S-phase cyclins that are normally degraded by Cdh1-APC/C9–11.
Figure 1 |. Organization of the cell cycle in somatic cells (MEFs) and in different types of ESCs.
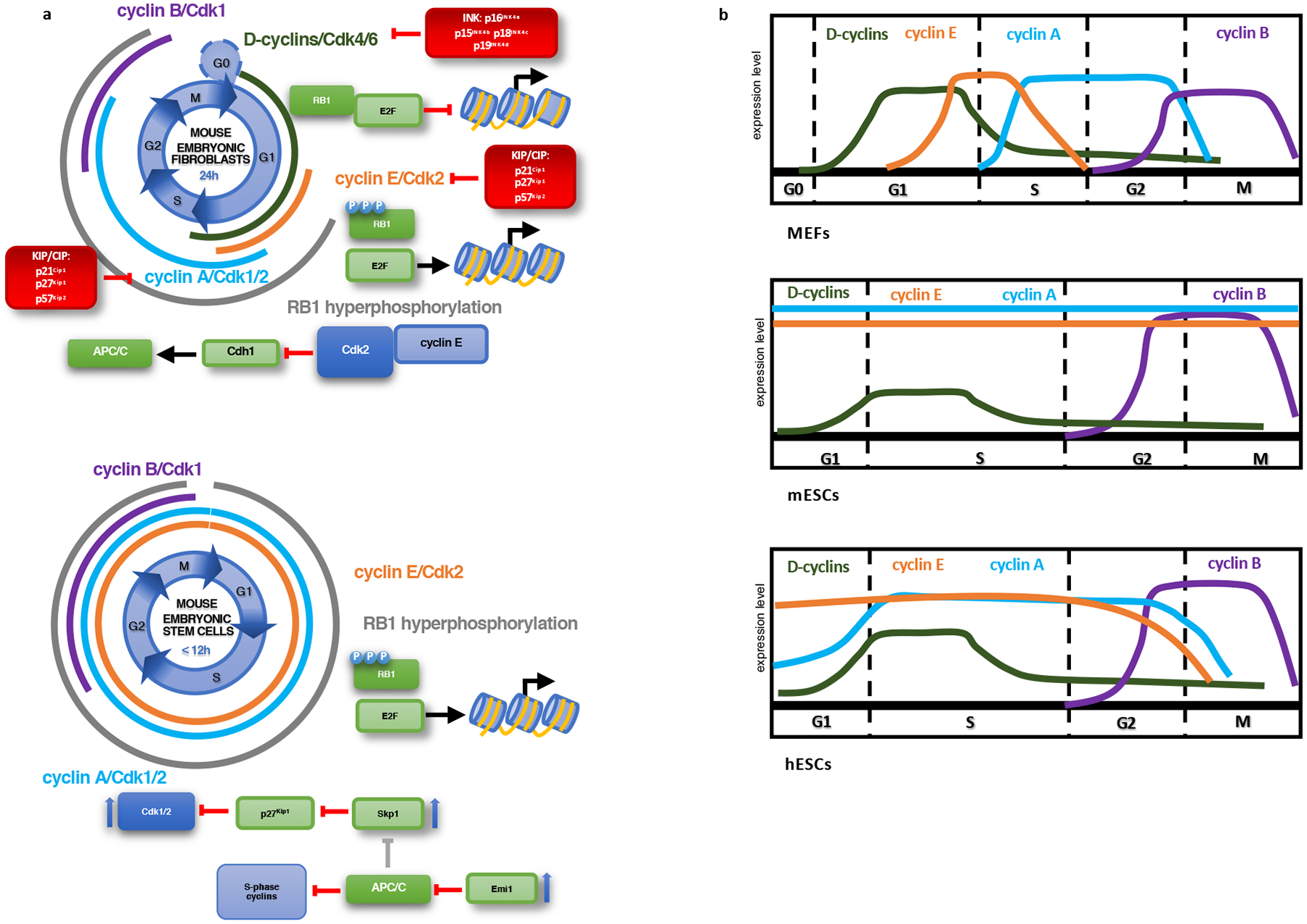
a, Differences in activity and expression of cell cycle components between MEFs and mESCs. In contrast to MEFs, mESCs lack expression of D cyclins and continuously express cyclin A and cyclin E. This allows them to maintain RB1 hyperphosphorylation throughout the cell cycle and results in a very short G1 phase. Cyclin-dependent kinase inhibitors are absent from mESCs. Upward blue arrows indicate increased expression. b, Oscillations of cyclin levels in MEFs, mESCs and hESCs. Abbreviations: MEFs: mouse embryonic fibroblasts; mESCs: mouse embryonic stem cells; hESCs: human embryonic stem cells.
The entry of mammalian cells into S phase is driven by cyclin E-CDK2 acting in concert with the DBF4-associated kinase12–14. Later during S phase, cyclin A2 becomes upregulated, pairs with CDK2 and CDK1 and promotes S-phase progression1–4. Following a second gap phase (G2), cyclin B translocates to the nucleus, activates CDK1, and drives separation of genetic material to daughter cells3,15.
In addition to these positive regulators of the cell cycle, mammalian cells also express two classes of cell cycle inhibitors. The INK proteins (p16INK4a, p15INK4b, p18INK4c, p19INK4d) interact with CDK4 or CDK6 and block their association with D cyclins1–4. The KIP/CIP proteins (p21Cip1, p27Kip1, p57Kip2) form ternary complexes with cyclin-CDK2 and cyclin-CDK1 molecules and inhibit their kinase activities1–4.
Cell cycle machinery in embryonic stem cells
Work of several laboratories revealed that the core cell cycle machinery operates differently during early embryonic development16. In developing flies, fish and frogs, first cell cycles are very rapid, lack obvious gap phases and consist of alternating S and M phases16. Analyses of peri-implantation mouse embryos demonstrated that murine embryonic cells display remarkably short division times in vivo (~4.4 – 7.5h)17–19 with a very small fraction of cells residing in G1 phase20,21. Intriguingly, the first two cell cycles are considerably longer, with significant shortening occurring during the third cell division22. Later in development, following gastrulation and formation of the endodermal, mesodermal and ectodermal lineages, the cell cycle length substantially increases, due to extension of the gap phases19.
Murine embryonic stem ES cells (mESCs), derived from inner cell masses of blastocysts and cultured in vitro in conditions that favor retention of their pluripotent state, recapitulate this unique organization of the cell cycle20. They divide very rapidly, although not as fast as their in vivo counterparts (division time ~12h), and have a short G1 phase that lasts only 3h16,23–25 (Table 1). These cells express high levels of cyclins E, A and B, and display elevated levels of Cdk1 and Cdk2 kinase, that greatly exceed those seen in somatic cells20,26,27. Strikingly, high levels of cyclins E and A are observed throughout the cell cycle, in contrast to their periodic expression in somatic cells20,28 (Fig. 1a, b). Consequently, Cdk2, cyclin E- and cyclin A-associated kinases are constitutively active throughout the entire cell cycle, in contrast to somatic cells, where these kinases become transiently activated at specific cell cycle phases16,20,28. The only cell cycle kinase that retains periodicity in mESCs is cyclin B-Cdk1, but its levels are substantially higher than those in somatic cells20,27. As a consequence of hyperactivated Cdk1 and Cdk2 kinases, RB1 is constitutively phosphorylated/inactivated throughout the cell cycle, and E2F activity is constitutively de-repressed16,20,23 (Fig. 1, Table 1).
Table 1 |.
Comparison of cell cycle features in MEFs, mESCs and hESCs.
| MEFs | mESCs | hESCs | |
|---|---|---|---|
| Cell cycle length | 24h | 4.4 – 7.5h in vivo; up to 12h in vitro | 16h |
| G1 phase length | 11h | 3h | 3h |
| CDK1 and CDK2 activity | periodical | very high and constant | very high and periodical |
| D-cyclins expression | ++ | +/− | + |
| RB1 phosphorylation status | hypo- and hyperphosphorylated | hyperphosphorylated | hypo- and hyperphosphorylated |
| KIP/CIP inhibitors expression | present | absent | present |
Abbreviations: MEFs: mouse embryonic fibroblasts; mESCs: mouse embryonic stem cells; hESCs: human embryonic stem cells.
Several mechanisms likely contribute to very high levels of Cdk1 and Cdk2 kinase activity in ESCs. In contrast to somatic cells, ESCs do not express KIP/CIP inhibitors, which normally mitigate the activity of Cdk1/216,20,26. ESCs also express high levels of the APC/C inhibitor Emi1, leading to a decreased activity of APC/C29,30. This mechanism allows cell-cycle-wide accumulation of S-phase cyclins and of another APC/C target Skp211,29. Skp2, in turn, triggers degradation of p27Kip1, thereby further contributing to elevated Cdk1/2 kinase activity16 (Fig. 1a).
Initial reports documented that mESCs express low levels of D cyclins and are refractory to cell cycle inhibition by p16INK4a (ref.31). Consistent with these results, another study32 demonstrated that prior to gastrulation, mouse embryos do not express D cyclins. In contrast to these findings, others reported expression of cyclins D1 and D3 in the pluripotent cells of mouse embryonic epiblast33. Cyclin D3 was also shown to be expressed in ESCs where it complexes with Cdk6; however, these cyclin D3-Cdk6 complexes are refractory to inhibition by p16INK4a (ref.33). A similar cell cycle organization was reported in ESCs derived from rhesus monkeys34. It should be noted that these studies utilized ESCs grown in the presence of serum and leukemia inhibitory factor (LIF). Recent studies indicate that murine cells cultured in a defined medium with inhibitors of MEK and GSK3 kinases resemble true pluripotent stem cells from inner cell masses, and are hence termed to reside in a ‘ground state’35,36. The cell cycle organization of these ground state cells may be different from that of cells cultured in serum/LIF, including a longer G1 phase and the presence of hypo-phosphorylated RB137. This surprising finding calls for more side-by-side comparisons between mESCs cultured in different conditions versus pluripotent cells of early embryos in vivo.
The organization of the cell cycle in human ESCs (hESCs) is slightly different (Fig.1b, Table 1). This likely reflects the fact that hESCs are more similar to ‘primed’ pluripotent cells derived from the late epiblast layer of post-implantation embryos (epiblast-derived stem cells, EpiSCs)19,38,39. It is now appreciated that different forms of pluripotent stem cells exist during early development and that their cell cycle organization might show some differences19. Like mESCs, hESCs cells proliferate rapidly, display a short G1 phase and express high levels of CDK1 and CDK2 kinases40,41. However, these cells express appreciable levels of D cyclins and KIP/CIP inhibitors, show cell-cycle-dependent fluctuations of CDK2 kinase, and contain both hyper- and hypo-phosphorylated RB119,40–42 (Fig. 1b, Table 1).
Molecular links between cell cycle and pluripotency
One of the main questions in the field has been whether the unique cell cycle organization of pluripotent stem cells simply reflects the necessity to rapidly expand this cell population or it plays an active role in enforcing the pluripotent state. Several observations suggested a role for cell cycle proteins in enforcing pluripotency24,27,40,43–46 (Fig. 2). The knock-down of CDK1, CDK2, cyclin E or B1, and treatment with CDK-inhibitors all resulted in the loss of the pluripotent state and triggered differentiation24,27,40,43–46, whereas ectopic overexpression of cyclins E or B1 promoted ESC self-renewal24,44. However, several of these manipulations resulted in cell cycle arrest, apoptosis or perturbed cell cycle progression, which confounded interpretation of the results40,44,45,47. At the mechanistic level, Wang et al.27 proposed that CDK1 kinase maintains the pluripotent state by regulating the PI3K/Akt pathway. Conversely, Kim et al.48 reported that CDK1 inhibits chromatin binding of Oct4 through an indirect mechanism involving Aurkb and PP1. The notion that the core cell cycle machinery actively regulates pluripotency was further supported by genetic studies using ESCs derived from cyclin D-null49 or E-null50 mice. Although ablation of all three D-type (D1−/−D2−/−D3−/−) or both E-type (E1−/−E2−/−) cyclins had no effect on the pluripotent state, a combined acute shutdown of all five G1 cyclins resulted in the strong attenuation of pluripotency25. Importantly, cells lacking all G1 cyclins proliferated in vitro, albeit at a somewhat reduced rate, revealing that D and E cyclins are not essential for the proliferation of mESCs25. At the molecular level, G1 cyclin-CDK kinases were shown to directly phosphorylate core pluripotency factors Oct4, Sox2 and Nanog, resulting in their stabilization25. Ablation of all G1 cyclins strongly diminished phosphorylation of the critical residues of Oct4, Sox2 and Nanog, thereby triggering their proteasomal degradation and attenuation of the pluripotent state25 (Fig. 2).
Figure 2 |. The cell cycle in somatic reprogramming and pluripotency maintenance.
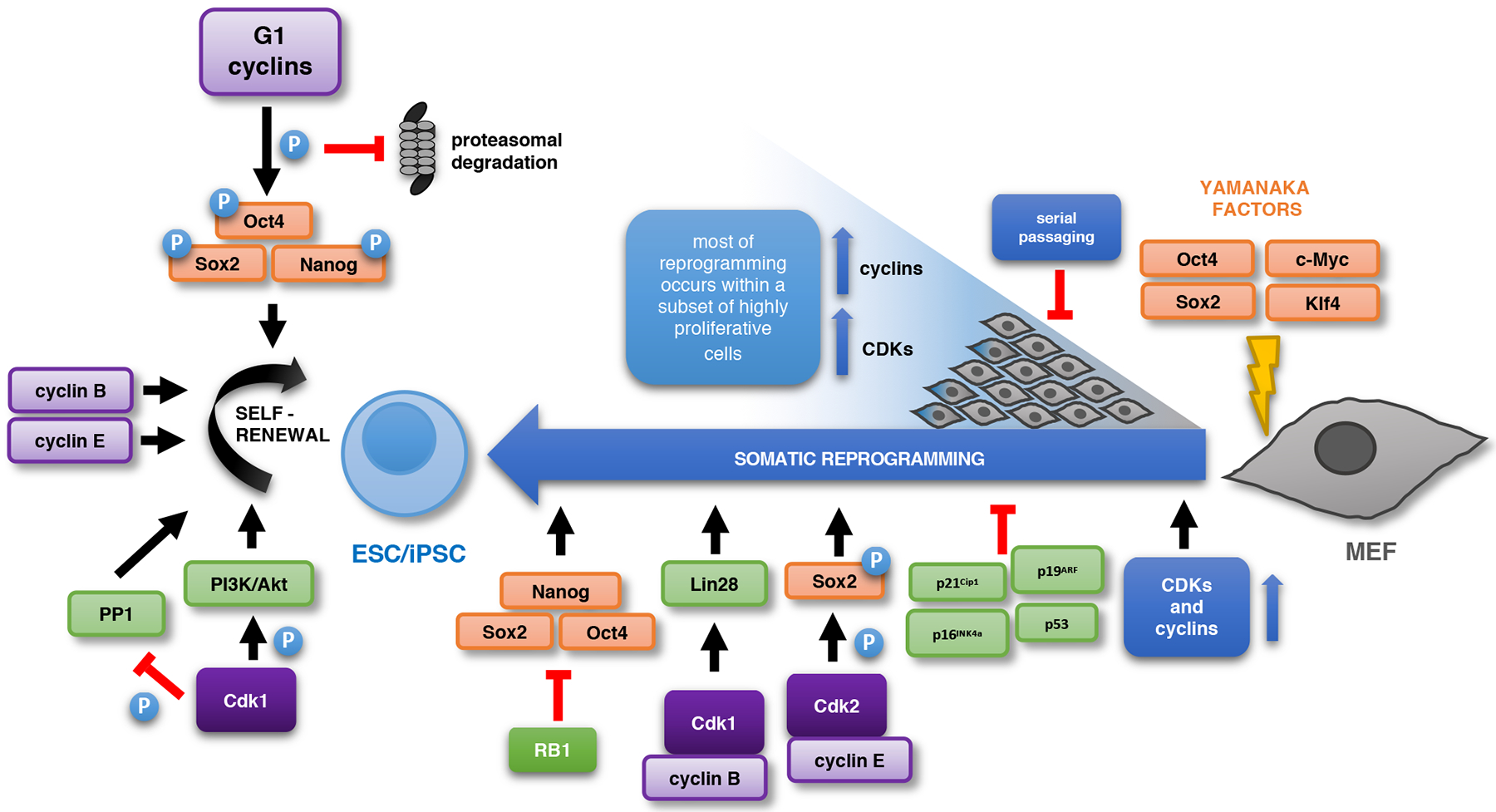
During somatic reprogramming by expression of Oct4, Sox2, Klf4 and c-Myc, somatic cells rapidly accelerate the cell cycle. Ectopic overexpression of cell cycle proteins or inactivation of cell cycle inhibitors increases the efficiency of reprogramming. Conversely, serial passaging leads to a decreased reprogramming rate. CDK2-dependent phosphorylation of Sox2 or cyclin BCDK1-dependent upregulation of LIN28 were postulated to aid reprogramming. RB1 represses expression of core pluripotency factors. The cell cycle machinery is also important for the maintenance of ESC pluripotency. G1 cyclins stabilize core pluripotency factors through phosphorylation, thereby preventing their proteasomal degradation. High levels of cyclins and Cdk1-dependent phosphorylation of PI3K/Akt pathway components likely contribute to the maintenance of pluripotency. Cdk1 also inhibits Oct4 activity during the M phase, acting through PP1 and Aurkb. Upward blue arrows indicate increased expression. Abbreviations: ESC: embryonic stem cell; IPSC: induced pluripotent stem cell.
Although G1 cyclins are dispensable for the proliferation of mESCs, no ESC colonies were observed upon ablation of cyclin A, revealing an essential role for this cyclin in stem cells51. These observations are in stark contrast to the situation seen in fibroblasts, where G1 cyclins are essential, but cyclin A is not (as its function can be carried out by cyclin E)51. Collectively, these observations indicate that, in contrast to differentiated cells, ESCs rely on cyclins A and B for cell cycle progression, whereas G1 cyclins contribute to the maintenance of the pluripotent state.
In addition to cell cycle proteins regulating pluripotency factors, the reverse was also noted, namely the ability of Nanog and Oct4 proteins to affect proliferation52–54. Specifically in hESCs, Nanog was shown to bind the regulatory regions of the CDK6 and CDC25 genes and to upregulate their expression, thereby promoting cell proliferation52. Oct4 was postulated to stimulate cell growth by repressing the expression of p21Cip1, a cell cycle inhibitor gene53. In contrast to these findings, another study reported that Oct4 inhibits cell proliferation by forming a complex with cyclin-Cdk1 and inhibiting Cdk1 activation54. Pluripotency factors can also affect proliferation through indirect mechanisms. For instance, Oct4 and Nanog upregulate c-Myc, which, in turn regulates expression of several cell cycle genes55–57.
Dissolution of pluripotency and cell fate specification
The unusual cell cycle properties of ESCs change upon dissolution of pluripotency, when stem cells undergo cell fate specification and differentiate into different lineages16,. During this process, cell division length increases, mainly due to an expanded G1 phase16,24,28,34,43,58. D cyclins become upregulated, the activity of CDK1 and CDK2 decreases in part due to expression of KIP/CIP inhibitors, and the activation CDK2-, cyclin E- and cyclin A-associated kinases becomes restricted to specific cell cycle phases16,26–29,31,34,45. Downregulation of Emi1 causes an increased APC/C activity, resulting in enhanced degradation of APC/C targets, such as cyclins29,30. All these changes lead to the appearance of hypophosphorylated RB1 during G1 phase, and subject E2F transcriptional activity to a tight cell-cycle-dependent control16,28.
Several studies document that ESCs initiate differentiation in the G1 phase59–62 (Fig. 3). This observation was best demonstrated using a FUCCI system which allows the sorting of cells in defined cell cycle phases24,60,61. One possible mechanistic explanation was provided by the observation that cells traversing the G1 phase express higher levels of developmentally-regulated transcription factors, suggesting that G1 phase ESCs may exist in a ‘lineage-primed’ state61,62. Several developmentally regulated genes in ESCs are marked by overlapping activatory (H3K4me3) and inhibitory (H3K27me3) histone marks63,64. In late G1, the activating H3K4me3 mark increases on many of these ‘bivalent’ developmental genes, resulting in increased levels of their transcripts, which might promote differentiation62. In addition, 5-hydroxymethylation of cytosine increases on some developmental genes during G1 phase, and this mechanism may contribute to their elevated expression61 (Fig. 3).
Fig. 3 |. The cell cycle during dissolution of pluripotency and cell differentiation.
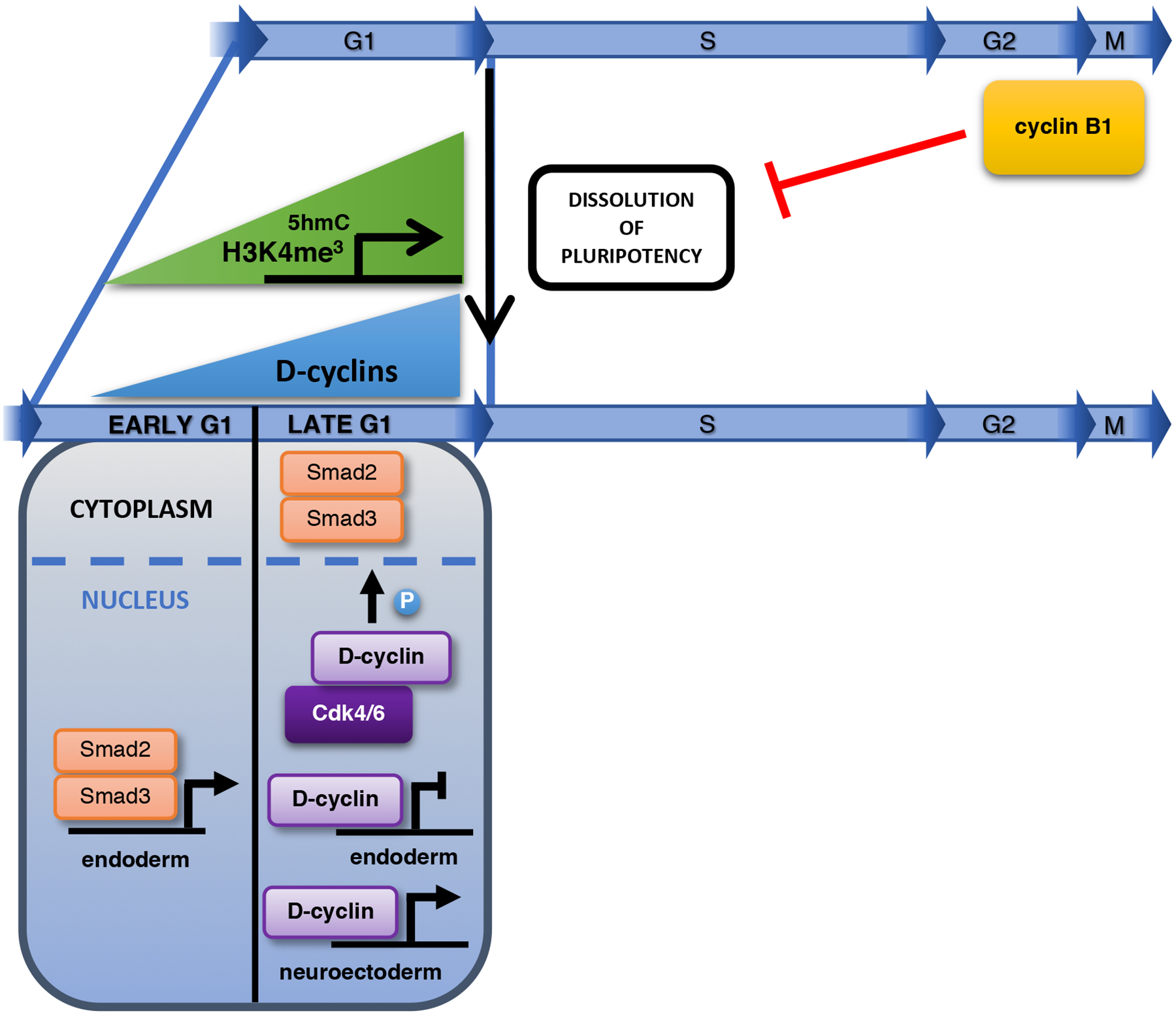
G1 phase may provide a window of opportunity for the dissolution of pluripotency, as during this phase many developmental genes contain permissive epigenetic marks (H3K4me3 and 5-hydroxymethylcytosine). Cyclin B1 actively prevents pluripotency exit. Increased expression of D cyclins in late G1 triggers the phosphorylation and cytoplasmic retention of Smad2/3, thereby inhibiting endodermal differentiation. In addition, D cyclins directly repress the expression of endodermal genes and augment the expression of neuroectodermal genes. Abbreviations: 5hmC: 5-hydroxymethylcytosine.
Collectively, these observations indicate that G1 phase represents a permissive phase for the initiation of cell fate decisions19. In the light of these findings, it has been argued that a long G1 phase may enable the accumulation of factors needed for the dissolution of pluripotency and differentiation and, conversely, a short G1 phase minimizes the exposure to differentiation-promoting signals and helps to maintain the pluripotent state65–67. Work by Pauklin et al. has further extended this model68. The authors reported that during early G1 phase of hESCs, when D cyclins are not yet expressed, Smad2 and Smad3 transcription factors bind and activate endoderm genes, thereby specifying endodermal differentiation. Upregulation of cyclin D-CDK4/6 kinases during late G1 phase leads to phosphorylation of Smad2/3, thereby blocking Smad2/3 entry into the nucleus. This mechanism prevents endodermal cell fate and renders cells susceptible only to neuroectodermal differentiation68. In another study69, these authors reported that in hESCs D cyclins regulate cell fate specification via a CDK4/6- and Smad-independent mechanism. According to this model, cyclin D1 recruits transcriptional coactivators to neuroectoderm genes, thereby promoting neuroectodermal differentiation69. D cyclins also bind endodermal gene promoters, but in this case they recruit transcriptional corepressors, which silence gene expression69 (Fig. 3). Indeed, cyclin D1 was previously shown to interact with gene regulatory regions and regulate gene expression in vivo in a CDK4/6-independent fashion70,71.
These two models68,69 are at odds with several observations. For instance, knockout mice lacking individual D cyclins, all D cyclins (D1−/−D2−/−D3−/−), or Cdk4 and 6 (Cdk4−/−Cdk6−/−) can develop at least until mid-gestation and undergo organogenesis, indicating that specification into different lineages is not significantly affected49,72–75. In addition, mESCs lacking all three D-type cyclins do not upregulate differentiation markers25. A lack of all five G1 cyclins in mESCs increases expression of several neuroectodermal genes and accelerates neural differentiation in vivo in chimeric embryos and teratoma assays25, an opposite outcome to the one described for shRNA-mediated silencing of D cyclins in hESCs68. In agreement with the genetic data, several other reports document that cyclin D-CDK4/6 activity inhibits neuroectodermal differentiation in vivo, whereas its silencing promotes neuroectodermal fate76–80. One possible explanation for these discrepancies would be that the molecular mechanism of lineage specification is fundamentally different between mouse and human cells.
Whereas cells in G1 are prone to differentiation, pathways operating in S and G2 phases may actively repress the dissolution of the pluripotent state44. At the molecular level, it was suggested that cyclin B1 might represent a key component of such pluripotency-promoting pathway during G2 phase44 (Fig. 3). Van Oudenhove et al.81 indicated that ESCs differentiating into mesodermal or endodermal lineages pause during G2 phase due to upregulation of Wee1, a kinase that inhibits CDK182, which may attenuate the pluripotency-promoting mechanism.
Cell cycle alterations during somatic reprogramming
Expression of a defined set of transcription factors, such as Oct4, Sox2, Klf4 and c-Myc, can convert differentiated somatic cells into induced pluripotent stem cells (iPSCs) in a process called somatic reprogramming83–85 (Fig. 2). One of the early events accompanying this process is a strong acceleration of the cell cycle86–89. Guo et al.88 demonstrated that during reprogramming of mouse fibroblasts, a small subpopulation of ‘ultra-fast’ dividing cells (cell division time of ~ 8h) accounts for >99% of the reprogramming. Others have shown that by sorting highly proliferative cells, one can substantially increase the efficiency of reprogramming80. These observations suggest that an acceleration of the cell cycle, which occurs only in a fraction of cells, may represent a rate-limiting factor in reprogramming88. Indeed, ectopic expression of several cyclins and CDKs increased the reprogramming efficiency whereas their depletion had the opposite effect27,89,90. Moreover, the reprogramming potential of primary murine fibroblasts declines after serial passaging, concomitantly with their reduced proliferation rates91. Conversely, genetic lesions in somatic cells that increase cell cycling, such as the inactivation of p19ARF/p16INK4a, p53, or p21Cip1, increased the efficiency of reprogramming91–93.
At the molecular level, Cdk2-mediated phosphorylation of Sox2 as shown to promote the ability of Sox2 to establish pluripotency during reprogramming, in combination with other factors94, consistent with an observation that cyclin E-Cdk2 stabilizes Sox2 protein25. According to another study27, cyclin B1 and CDK1 play a rate-limiting role in somatic cell reprogramming by upregulating and maintaining the expression of the reprogramming factor LIN2895. Inactivation of RB1 promotes reprogramming via a cell-cycle-independent mechanism, which is related to the ability of RB1 to directly bind pluripotency genes to repress their expression96.
The organization of cell cycle in iPSCs, namely a rapid division time (16–18h) and a very short G1 phase (~2.5h), is similar to that in ESCs89,97. Similarly to ESCs, depletion of CDK1 in reprogrammed iPSCs also attenuates the pluripotent state46. Collectively, these observations suggest that the acquisition of a unique ‘ESC-like’ cell cycle organization in the process of reprogramming is functionally linked to the acquisition of pluripotency97. However, the exact molecular mechanisms by which cell cycle proteins help to acquire and enforce the pluripotent state in iPSCs remain largely unknown.
Cell cycle machinery in neural stem cells
Similarly to pluripotent ESCs, embryonic neural stem/progenitor cells display rapid cell division times (~8h)98–100. Interestingly, a subpopulation of slowly dividing neural progenitors was also identified, which could give rise to adult neural stem cells (NSCs)101. Expression of the cell cycle inhibitor p57Kip2 is essential to maintain this slow dividing pool and is required for the emergence of adult NSCs101. As development proceeds, the cell cycle length of the embryonic neural progenitor cells increases (to up to ~18h), due to a four-fold increase in the length of G180,98,99,102.
Manipulations that prevent the physiological lengthening of G1 phase, such as ectopic overexpression of cyclin D1, E1, or CDK4, increase self-renewal and inhibit neurogenic differentiation76–78. Conversely, depletion of cyclin D1 and CDK4, or treatment with CDK4- or pan-CDK-inhibitors stimulates neurogenesis76,80,103. Likewise, NSCs of Cdk2/Cdk4 double-knockout mice display an increased propensity for neuronal differentiation, resulting in enhanced neurogenic divisions in the brains of these embryos79. Intriguingly, studies of the developing primate cortex revealed that local variations of cyclin E and p27Kip1 levels in neuronal precursors residing in different areas correlate with their cell cycle length (and with G1 length), suggesting that area-specific levels of cyclin-E-associated kinase might locally influence neurogenesis104.
Collectively these analyses strongly suggest that the activity of cyclin-CDK kinases inhibits neuronal differentiation. Lim et al.105 proposed that in self-renewing NSCs, phosphorylation of Sox2 by high levels of cyclin-dependent kinases enhances the ability of Sox2 to inhibit neuronal differentiation105. When the activity of CDKs decreases, the shift towards non-phosphorylated Sox2 allows the proteolytically cleaved, truncated Sox2 species to bind and transactivate proneural genes, thereby promoting neurogenesis105 (Fig. 4). Ali et al.106 postulated that in rapidly proliferating neural stem/progenitor cells, cyclin A- and B-dependent kinases phosphorylate the proneural basic helix-loop-helix transcription factor neurogenin 2 (Ngn2), a master regulator of neuronal differentiation. This phosphorylation inhibits the ability of Ngn2 to bind neurogenic genes and promote neurogenesis106. According to this model, upon lengthening of G1 phase, decreased CDK activity relieves this repression and allows Ngn2 to turn on neurogenesis106. Interestingly, Ngn2 can repress expression of cyclins D1 and E2107, suggesting the presence of a positive feedback loop that enforces the cell cycle exit during neural differentiation.
Fig. 4 |. The cell cycle in neurogenesis.
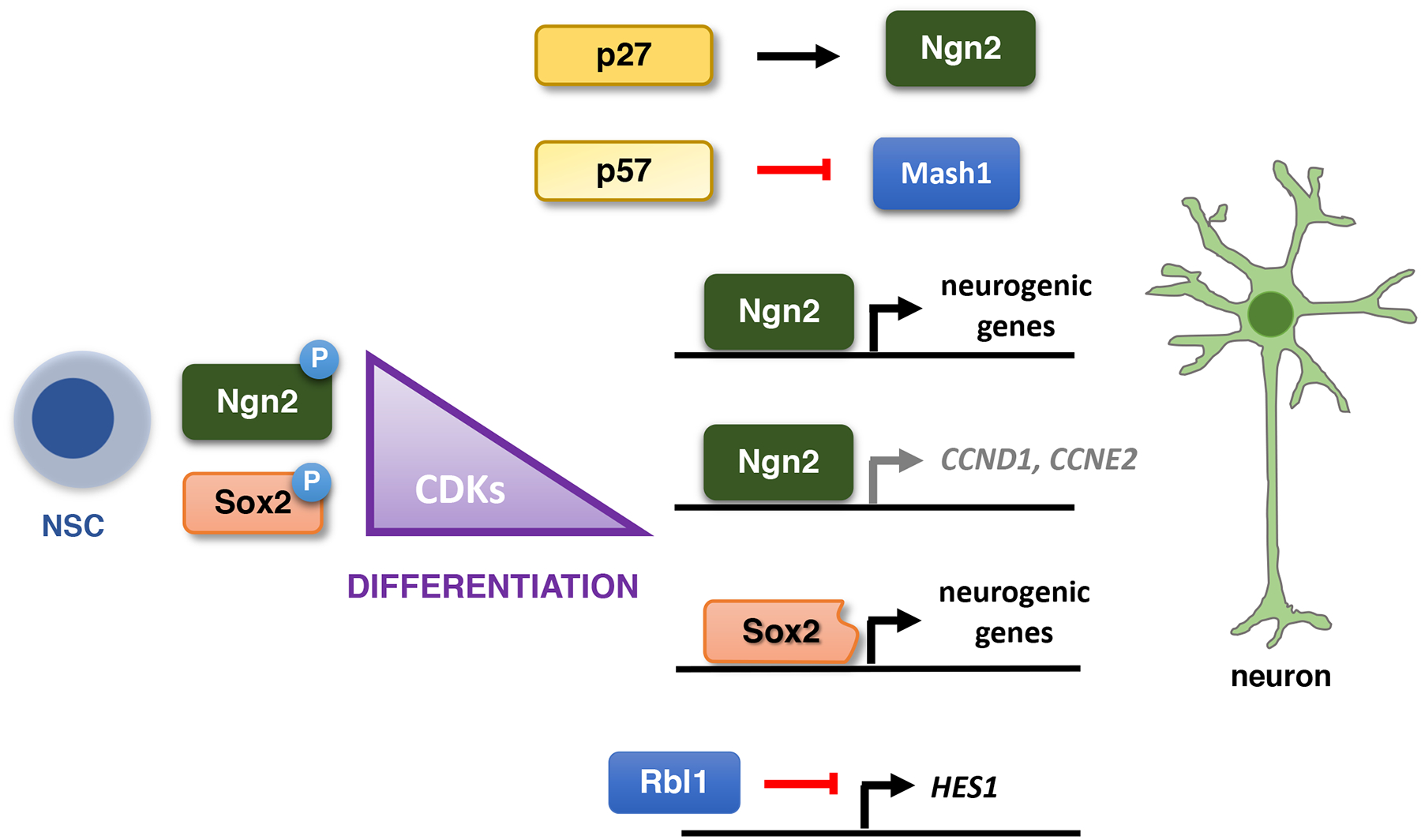
A decrease of CDK activity during neurogenic divisions enables the transactivation of proneural genes by a truncated form of Sox2 and Ngn2. Ngn2, in turn, inhibits the expression of G1 cyclins. A cell cycle inhibitor p27Kip1 stabilizes Ngn2, whereas p57Kip2 interacts with pro-neural factor Mash1 and represses its transcriptional activity. Rbl1 modulates the Notch pathway and affects the expression of its target genes. Abbreviations: NSC: neural stem cell.
It should be noted that an opposite conclusion has also been reported108, namely that in embryonic chick spinal cord, cyclin D1 serves to promote neurogenesis through a cell cycle-independent function mediated by Hes6. As mentioned before, Pauklin et al. noted that in hESCs, cyclin D1 and CDK4 promote neurogenic differentiation68,69.
In addition to cyclins and CDKs, other cell cycle proteins were shown to play roles in neurogenesis. For instance, p27Kip1 can promote neuronal differentiation by stabilizing Ngn2 protein, independently of its ability to inhibit CDKs109. In contrast, a related inhibitor, p57Kip1, interacts with pro-neural basic helix-loop-helix factor Mash1 and represses its transcriptional activity, thereby inhibiting neurogenesis110. In addition, Rbl1 protein regulates neural precursor cells in the mammalian brain by repressing the Notch1 pathway111. Lastly, CDK inhibitors p27Kip1, p57Kip2 and RB1 play cell-cycle-independent roles in regulating neuronal cell migration109,112–117.
In contrast to highly proliferative ESCs, which are the focus of this Review, tissue-specific stem cells of adult organisms are largely quiescent65,66. It was proposed that their G0 arrest serves the same purpose as G1 phase truncation in ESCs, namely to limit the window of opportunity for cell differentiation66. Several cell cycle proteins, including CDK inhibitors, can enforce a quiescent state118–124, as detailed in excellent Reviews on this subject published elsewhere125,126.
Conclusions and outlook
One of the major questions in the field is whether the unusual cell cycle organization seen in ESCs and iPSCs plays a direct role in maintaining pluripotency. In addition, it remains to be resolved whether the reorganization of the cell cycle upon pluripotency dissolution represents the cause or consequence of cell differentiation. The combined evidence to date suggests that cell cycle proteins are causally involved in these processes, however, the mechanistic underpinnings are only beginning to emerge. The physiological role of very high Cdk1 and Cdk2 kinase activities in pluripotent ESCs remains a mystery. These kinases might likely phosphorylate additional substrates in stem cells that are different from those in differentiated cells, and this might contribute to pluripotency through some currently unknown mechanisms. Proteome-wide identification of cyclin-CDK substrates in pluripotent cells and during pluripotency dissolution will help to decipher the full repertoire of cyclin-CDK functions in these processes. The role of cell cycle proteins in cell fate specification and differentiation remains largely unclear, with different reports proposing different mechanisms and outcomes. Combinations of genome- and proteome-wide approaches might help to address this point. The contribution of 3D chromatin conformation, promoter-enhancer looping, insulated neighborhoods and topologically associating domains, as well as enhancer repertoire and decommissioning has recently been acknowledged in pluripotency and differentiation127–132, however it remains unclear whether these processes are linked to cell cycle proteins. Another largely unaddressed issue is the heterogeneity within the stem cell compartments. Single-cell approaches such as live single-cell microscopy9,133–135 or single-cell RNA sequencing136–139 will help to ascribe specific molecular functions to cell cycle proteins at discreet stages of fate specification and cell differentiation.
An improved understanding of the cell cycle machinery in stem cell compartments will likely have important implications for regenerative medicine. For example, a transient expression of cyclin D1 and Cdk4 in NSCs in vivo resulted in increased neurogenesis and improved neuronal function140. Other work revealed the utility of inhibiting cell cycle proteins to promote hepatic and pancreatic differentiation68,141. Given the growing armamentarium of specific cell cycle inhibitors82, these studies offer opportunities to specifically promote cell differentiation into a lineage of interest. Conversely, activation of the cell cycle machinery can be used as a mean to augment somatic reprogramming27,89,90. For these reasons, the elucidation of the full range of molecular functions of cell cycle proteins in stem cell self-renewal, differentiation and reprogramming should provide important insights to help increase the efficiency of these processes in a therapeutically meaningful way.
Acknowledgements
This work was supported by a grant R01CA202634 (to PS).
Footnotes
Competing interests
PS has been a consultant at Novartis, Genovis, Guidepoint, The Planning Shop, ORIC Pharmaceuticals and Exo Therapeutics; his laboratory receives research funding from Novartis. WM is currently an employee of Cedilla Therapeutics.
References
- 1.Malumbres M & Barbacid M To cycle or not to cycle: a critical decision in cancer. Nature reviews. Cancer 1, 222–231 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ & Roberts JM Living with or without cyclins and cyclin-dependent kinases. Genes & development 18, 2699–2711 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Morgan DO The Cell Cycle: Principles of Control OUP/New Science Press (2007). [Google Scholar]
- 4.Satyanarayana A & Kaldis P Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28, 2925–2939 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ Growth factor-regulated G1 cyclins. Stem Cells 1, 47–55 (1994). [PubMed] [Google Scholar]
- 6.Geng Y et al. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97, 767–777 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Hwang HC & Clurman BE Cyclin E in normal and neoplastic cell cycles. Oncogene 24, 2776–2786 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Narasimha AM et al. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife 3, doi: 10.7554/eLife.02872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cappell SD, Chung M, Jaimovich A, Spencer SL & Meyer T Irreversible APC(Cdh1) Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell 166, 167–180, doi: 10.1016/j.cell.2016.05.077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manchado E, Eguren M & Malumbres M The anaphase-promoting complex/cyclosome (APC/C): cell-cycle-dependent and -independent functions. Biochem Soc Trans 38, 65–71, doi: 10.1042/BST0380065 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Pines J Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol 12, 427–438, doi: 10.1038/nrm3132 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Heller RC et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146, 80–91, doi: 10.1016/j.cell.2011.06.012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter JC Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem 275, 39773–39778, doi: 10.1074/jbc.M008107200 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Yeeles JT, Deegan TD, Janska A, Early A & Diffley JF Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435, doi: 10.1038/nature14285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavet O & Pines J Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Developmental cell 18, 533–543, doi: 10.1016/j.devcel.2010.02.013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boward B, Wu T & Dalton S Concise Review: Control of Cell Fate Through Cell Cycle and Pluripotency Networks. Stem Cells 34, 1427–1436, doi: 10.1002/stem.2345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson KA, Meneses JJ & Pedersen RA Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891–911 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Snow MHL Gastrulation in the mouse: growth and regionalization of the epiblast. J Embryol Exp Morphol 42, 293–303 (1977). [Google Scholar]
- 19.Dalton S Linking the Cell Cycle to Cell Fate Decisions. Trends Cell Biol 25, 592–600, doi: 10.1016/j.tcb.2015.07.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stead E et al. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 21, 8320–8333, doi: 10.1038/sj.onc.1206015 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Solter D, Skreb N & Damjanov I Cell cycle analysis in the mouse EGG-cylinder. Exp Cell Res 64, 331–334 (1971). [DOI] [PubMed] [Google Scholar]
- 22.Smith RK & Johnson MH Analysis of the third and fourth cell cycles of mouse early development. Journal of reproduction and fertility 76, 393–399 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Savatier P, Huang S, Szekely L, Wiman KG & Samarut J Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene 9, 809–818 (1994). [PubMed] [Google Scholar]
- 24.Coronado D et al. A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res 10, 118–131, doi: 10.1016/j.scr.2012.10.004 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Liu L et al. G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat Cell Biol 19, 177–188, doi: 10.1038/ncb3474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujii-Yamamoto H, Kim JM, Arai K & Masai H Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J Biol Chem 280, 12976–12987, doi: 10.1074/jbc.M412224200 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Wang XQ et al. CDK1-PDK1-PI3K/Akt signaling pathway regulates embryonic and induced pluripotency. Cell death and differentiation 24, 38–48, doi: 10.1038/cdd.2016.84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White J et al. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Molecular biology of the cell 16, 2018–2027, doi: 10.1091/mbc.e04-12-1056 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bar-On O, Shapira M, Skorecki K, Hershko A & Hershko DD Regulation of APC/C (Cdh1) ubiquitin ligase in differentiation of human embryonic stem cells. Cell Cycle 9, 1986–1989, doi: 10.4161/cc.9.10.11727 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Ballabeni A et al. Cell cycle adaptations of embryonic stem cells. Proc Natl Acad Sci U S A 108, 19252–19257, doi: 10.1073/pnas.1116794108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB & Samarut J Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene 12, 309–322 (1996). [PubMed] [Google Scholar]
- 32.Wianny F et al. G1-phase regulators, cyclin D1, cyclin D2, and cyclin D3: up-regulation at gastrulation and dynamic expression during neurulation. Dev Dyn 212, 49–62 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Faast R et al. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a). Oncogene 23, 491–502, doi: 10.1038/sj.onc.1207133 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Fluckiger AC et al. Cell cycle features of primate embryonic stem cells. Stem Cells 24, 547–556, doi: 10.1634/stemcells.2005-0194 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols J & Smith A Naive and primed pluripotent states. Cell Stem Cell 4, 487–492, doi: 10.1016/j.stem.2009.05.015 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Ying QL et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ter Huurne M, Chappell J, Dalton S & Stunnenberg HG Distinct Cell-Cycle Control in Two Different States of Mouse Pluripotency. Cell Stem Cell 21, 449–455 e444, doi: 10.1016/j.stem.2017.09.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brons IG et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195, doi: 10.1038/nature05950 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Tesar PJ et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199, doi: 10.1038/nature05972 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Neganova I, Zhang X, Atkinson S & Lako M Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene 28, 20–30 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Becker KA et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol 209, 883–893, doi: 10.1002/jcp.20776 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Conklin JF, Baker J & Sage J The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat Commun 3, 1244, doi: 10.1038/ncomms2254 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Filipczyk AA, Laslett AL, Mummery C & Pera MF Differentiation is coupled to changes in the cell cycle regulatory apparatus of human embryonic stem cells. Stem Cell Res 1, 45–60, doi: 10.1016/j.scr.2007.09.002 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Gonzales KA et al. Deterministic Restriction on Pluripotent State Dissolution by Cell-Cycle Pathways. Cell 162, 564–579, doi: 10.1016/j.cell.2015.07.001 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Zhang WW et al. Cdk1 is required for the self-renewal of mouse embryonic stem cells. J Cell Biochem 112, 942–948, doi: 10.1002/jcb.23010 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Neganova I et al. CDK1 plays an important role in the maintenance of pluripotency and genomic stability in human pluripotent stem cells. Cell death & disease 5, e1508, doi: 10.1038/cddis.2014.464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huskey NE et al. CDK1 inhibition targets the p53-NOXA-MCL1 axis, selectively kills embryonic stem cells, and prevents teratoma formation. Stem Cell Reports 4, 374–389, doi: 10.1016/j.stemcr.2015.01.019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HJ et al. Cyclin-dependent kinase 1 activity coordinates the chromatin associated state of Oct4 during cell cycle in embryonic stem cells. Nucleic Acids Res 46, 6544–6560, doi: 10.1093/nar/gky371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozar K et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118, 477–491, doi: 10.1016/j.cell.2004.07.025 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Geng Y et al. Cyclin E ablation in the mouse. Cell 114, 431–443 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Kalaszczynska I et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell 138, 352–365, doi: 10.1016/j.cell.2009.04.062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X et al. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol 184, 67–82, doi: 10.1083/jcb.200801009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J, Go Y, Kang I, Han YM & Kim J Oct-4 controls cell-cycle progression of embryonic stem cells. The Biochemical journal 426, 171–181, doi: 10.1042/BJ20091439 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao R et al. A nontranscriptional role for Oct4 in the regulation of mitotic entry. Proc Natl Acad Sci U S A 111, 15768–15773, doi: 10.1073/pnas.1417518111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kareta MS, Sage J & Wernig M Crosstalk between stem cell and cell cycle machineries. Curr Opin Cell Biol 37, 68–74, doi: 10.1016/j.ceb.2015.10.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varlakhanova NV et al. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation 80, 9–19, doi: 10.1016/j.diff.2010.05.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amati B, Alevizopoulos K & Vlach J Myc and the cell cycle. Front Biosci 3, d250–268 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Becker KA, Stein JL, Lian JB, van Wijnen AJ & Stein GS Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. J Cell Physiol 222, 103–110, doi: 10.1002/jcp.21925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sela Y, Molotski N, Golan S, Itskovitz-Eldor J & Soen Y Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of retinoblastoma protein. Stem Cells 30, 1097–1108, doi: 10.1002/stem.1078 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Calder A et al. Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells Dev 22, 279–295, doi: 10.1089/scd.2012.0168 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Singh AM et al. Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Reports 1, 532–544, doi: 10.1016/j.stemcr.2013.10.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh AM et al. Cell-Cycle Control of Bivalent Epigenetic Domains Regulates the Exit from Pluripotency. Stem Cell Reports 5, 323–336, doi: 10.1016/j.stemcr.2015.07.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernstein BE et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326, doi: 10.1016/j.cell.2006.02.041 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Mikkelsen TS et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560, doi: 10.1038/nature06008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lange C & Calegari F Cdks and cyclins link G1 length and differentiation of embryonic, neural and hematopoietic stem cells. Cell Cycle 9, 1893–1900, doi: 10.4161/cc.9.10.11598 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Orford KW & Scadden DT Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet 9, 115–128, doi: 10.1038/nrg2269 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Singh AM & Dalton S The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell 5, 141–149, doi: 10.1016/j.stem.2009.07.003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pauklin S & Vallier L The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pauklin S, Madrigal P, Bertero A & Vallier L Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev 30, 421–433, doi: 10.1101/gad.271452.115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bienvenu F et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature 463, 374–378, doi: 10.1038/nature08684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casimiro MC et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. The Journal of clinical investigation 122, 833–843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malumbres M et al. Mammalian cells cycle without the D-type cyclin-dependent kinases CDK4 and CDK6. Cell 118 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Sicinska E et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer cell 4, 451–461 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Sicinski P et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 384, 470–474, doi: 10.1038/384470a0 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Sicinski P et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82, 621–630 (1995). [DOI] [PubMed] [Google Scholar]
- 76.Lange C, Huttner WB & Calegari F Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5, 320–331 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Artegiani B, Lindemann D & Calegari F Overexpression of cdk4 and cyclinD1 triggers greater expansion of neural stem cells in the adult mouse brain. J Exp Med 208, 937–948, doi: 10.1084/jem.20102167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pilaz LJ et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci U S A 106, 21924–21929, doi: 10.1073/pnas.0909894106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lim S & Kaldis P Loss of Cdk2 and Cdk4 induces a switch from proliferation to differentiation in neural stem cells. Stem Cells 30, 1509–1520 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Roccio M et al. Predicting stem cell fate changes by differential cell cycle progression patterns. Development 140, 459–470, doi: 10.1242/dev.086215 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Van Oudenhove JJ et al. Lineage-Specific Early Differentiation of Human Embryonic Stem Cells Requires a G2 Cell Cycle Pause. Stem Cells 34, 1765–1775, doi: 10.1002/stem.2352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Otto T & Sicinski P Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 17, 93–115, doi: 10.1038/nrc.2016.138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park IH et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146, doi: 10.1038/nature06534 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Takahashi K & Yamanaka S Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676, doi: 10.1016/j.cell.2006.07.024 (2006). [DOI] [PubMed] [Google Scholar]
- 85.Wernig M et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324, doi: 10.1038/nature05944 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Cacchiarelli D et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell 162, 412–424, doi: 10.1016/j.cell.2015.06.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mikkelsen TS et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55, doi: 10.1038/nature07056 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo S et al. Nonstochastic reprogramming from a privileged somatic cell state. Cell 156, 649–662, doi: 10.1016/j.cell.2014.01.020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ruiz S et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Current biology : CB 21, 45–52, doi: 10.1016/j.cub.2010.11.049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Edel MJ et al. Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev 24, 561–573, doi: 10.1101/gad.1876710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Utikal J et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460, 1145–1148, doi: 10.1038/nature08285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hong H et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–1135, doi: 10.1038/nature08235 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kawamura T et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144, doi: 10.1038/nature08311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ouyang J et al. Cyclin-dependent kinase-mediated Sox2 phosphorylation enhances the ability of Sox2 to establish the pluripotent state. The Journal of biological chemistry 290, 22782–22794, doi: 10.1074/jbc.M115.658195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tanabe K, Nakamura M, Narita M, Takahashi K & Yamanaka S Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc Natl Acad Sci U S A 110, 12172–12179, doi: 10.1073/pnas.1310291110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kareta MS et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell 16, 39–50, doi: 10.1016/j.stem.2014.10.019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghule PN et al. Reprogramming the pluripotent cell cycle: restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J Cell Physiol 226, 1149–1156, doi: 10.1002/jcp.22440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Salomoni P & Calegari F Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol 20, 233–243, doi: 10.1016/j.tcb.2010.01.006 (2010). [DOI] [PubMed] [Google Scholar]
- 99.Caviness VS Jr., Takahashi T & Nowakowski RS Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci 18, 379–383 (1995). [DOI] [PubMed] [Google Scholar]
- 100.Takahashi T, Nowakowski RS & Caviness VS Jr. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci 15, 6046–6057 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Furutachi S et al. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat Neurosci 18, 657–665, doi: 10.1038/nn.3989 (2015). [DOI] [PubMed] [Google Scholar]
- 102.Arai Y et al. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat Commun 2, 154, doi: 10.1038/ncomms1155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Calegari F & Huttner WB An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J Cell Sci 116, 4947–4955, doi: 10.1242/jcs.00825 (2003). [DOI] [PubMed] [Google Scholar]
- 104.Lukaszewicz A et al. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47, 353–364, doi: 10.1016/j.neuron.2005.06.032 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lim S et al. Cyclin-Dependent Kinase-Dependent Phosphorylation of Sox2 at Serine 39 Regulates Neurogenesis. Mol Cell Biol 37, doi: 10.1128/MCB.00201-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ali F et al. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138, 4267–4277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lacomme M, Liaubet L, Pituello F & Bel-Vialar S NEUROG2 drives cell cycle exit of neuronal precursors by specifically repressing a subset of cyclins acting at the G1 and S phases of the cell cycle. Mol Cell Biol 32, 2596–2607, doi: 10.1128/MCB.06745-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lukaszewicz AI & Anderson DJ Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proceedings of the National Academy of Sciences of the United States of America 108, 11632–11637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nguyen L et al. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes & development 20, 1511–1524 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Joseph B et al. p57Kip2 is a repressor of Mash1 activity and neuronal differentiation in neural stem cells. Cell Death Differ 16, 1256–1265, doi: 10.1038/cdd.2009.72 (2009). [DOI] [PubMed] [Google Scholar]
- 111.Vanderluit JL et al. p107 regulates neural precursor cells in the mammalian brain. J Cell Biol 166, 853–863, doi: 10.1083/jcb.200403156 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ferguson KL et al. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J 24, 4381–4391, doi: 10.1038/sj.emboj.7600887 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Godin JD et al. p27(Kip1) is a microtubule-associated protein that promotes microtubule polymerization during neuron migration. Dev Cell 23, 729–744, doi: 10.1016/j.devcel.2012.08.006 (2012). [DOI] [PubMed] [Google Scholar]
- 114.Itoh Y, Masuyama N, Nakayama K, Nakayama KI & Gotoh Y The cyclin-dependent kinase inhibitors p57 and p27 regulate neuronal migration in the developing mouse neocortex. The Journal of biological chemistry 282, 390–396 (2007). [DOI] [PubMed] [Google Scholar]
- 115.Kawauchi T, Chihama K, Nabeshima Y & Hoshino M Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nature cell biology 8, 17–26 (2006). [DOI] [PubMed] [Google Scholar]
- 116.Kawauchi T, Shikanai M & Kosodo Y Extra-cell cycle regulatory functions of cyclin-dependent kinases (CDK) and CDK inhibitor proteins contribute to brain development and neurological disorders. Genes Cells 18, 176–194, doi: 10.1111/gtc.12029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McClellan KA et al. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol Cell Biol 27, 4825–4843, doi: 10.1128/MCB.02100-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng T, Rodrigues N, Dombkowski D, Stier S & Scadden DT Stem cell repopulation efficiency but not pool size is governed by p27(kip1). Nature medicine 6, 1235–1240 (2000). [DOI] [PubMed] [Google Scholar]
- 119.Cheng T et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808 (2000). [DOI] [PubMed] [Google Scholar]
- 120.Furutachi S, Matsumoto A, Nakayama KI & Gotoh Y p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J 32, 970–981, doi: 10.1038/emboj.2013.50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kippin TE, Martens DJ & van der Kooy D p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev 19, 756–767, doi: 10.1101/gad.1272305 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Matsumoto A et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell 9, 262–271, doi: 10.1016/j.stem.2011.06.014 (2011). [DOI] [PubMed] [Google Scholar]
- 123.Viatour P et al. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell 3, 416–428, doi: 10.1016/j.stem.2008.07.009 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yuan Y, Shen H, Franklin DS, Scadden DT & Cheng T In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18INK4C. Nat Cell Biol 6, 436–442, doi: 10.1038/ncb1126 (2004). [DOI] [PubMed] [Google Scholar]
- 125.Cheung TH & Rando TA Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14, 329–340, doi: 10.1038/nrm3591 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Velthoven CTJ & Rando TA Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell 24, 213–225, doi: 10.1016/j.stem.2019.01.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Barakat TS et al. Functional Dissection of the Enhancer Repertoire in Human Embryonic Stem Cells. Cell Stem Cell 23, 276–288 e278, doi: 10.1016/j.stem.2018.06.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dowen JM et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387, doi: 10.1016/j.cell.2014.09.030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hnisz D, Day DS & Young RA Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell 167, 1188–1200, doi: 10.1016/j.cell.2016.10.024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ji X et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell 18, 262–275, doi: 10.1016/j.stem.2015.11.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kishi Y & Gotoh Y Regulation of Chromatin Structure During Neural Development. Frontiers in neuroscience 12, 874, doi: 10.3389/fnins.2018.00874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Whyte WA et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cappell SD et al. EMI1 switches from being a substrate to an inhibitor of APC/C(CDH1) to start the cell cycle. Nature 558, 313–317, doi: 10.1038/s41586-018-0199-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sakaue-Sawano A et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498, doi: 10.1016/j.cell.2007.12.033 (2008). [DOI] [PubMed] [Google Scholar]
- 135.Spencer SL et al. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cao J et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502, doi: 10.1038/s41586-019-0969-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guo H et al. Single-Cell RNA Sequencing of Human Embryonic Stem Cell Differentiation Delineates Adverse Effects of Nicotine on Embryonic Development. Stem Cell Reports, doi: 10.1016/j.stemcr.2019.01.022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pijuan-Sala B et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495, doi: 10.1038/s41586-019-0933-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao T et al. Single-Cell RNA-Seq Reveals Dynamic Early Embryonic-like Programs during Chemical Reprogramming. Cell Stem Cell 23, 31–45 e37, doi: 10.1016/j.stem.2018.05.025 (2018). [DOI] [PubMed] [Google Scholar]
- 140.Bragado Alonso S et al. An increase in neural stem cells and olfactory bulb adult neurogenesis improves discrimination of highly similar odorants. EMBO J 38, doi: 10.15252/embj.201798791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Azzarelli R et al. Multi-site Neurogenin3 Phosphorylation Controls Pancreatic Endocrine Differentiation. Dev Cell 41, 274–286 e275, doi: 10.1016/j.devcel.2017.04.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


