Two Yersinia toxins activate or inactivate Rab proteins involved in vesicle traffic by ADP-ribosylation or glycosylation.
Abstract
We identified a glucosyltransferase (YGT) and an ADP-ribosyltransferase (YART) in Yersinia mollaretii, highly related to glucosylating toxins from Clostridium difficile, the cause of antibiotics-associated enterocolitis. Both Yersinia toxins consist of an amino-terminal enzyme domain, an autoprotease domain activated by inositol hexakisphosphate, and a carboxyl-terminal translocation domain. YGT N-acetylglucosaminylates Rab5 and Rab31 at Thr52 and Thr36, respectively, thereby inactivating the Rab proteins. YART ADP-ribosylates Rab5 and Rab31 at Gln79 and Gln64, respectively. This activates Rab proteins by inhibiting GTP hydrolysis. We determined the crystal structure of the glycosyltransferase domain of YGT (YGTG) in the presence and absence of UDP at 1.9- and 3.4-Å resolution, respectively. Thereby, we identified a previously unknown potassium ion–binding site, which explains potassium ion–dependent enhanced glycosyltransferase activity in clostridial and related toxins. Our findings exhibit a novel type of inverse regulation of Rab proteins by toxins and provide new insights into the structure-function relationship of glycosyltransferase toxins.
INTRODUCTION
Large clostridial glucosylating toxins (CGTs) are major virulence factors and play pivotal roles in many diseases. Prototypes of these exotoxins are toxins A (TcdA) and B (TcdB) of Clostridium (Clostridioides) difficile, which are responsible for antibiotics-associated diarrhea and pseudomembranous colitis (1, 2). TcdA and TcdB are multidomain toxins and consist of an N-terminal glucosyltransferase domain, an autoprocessing cysteine protease domain, a translocation domain, and a C-terminal region called CROPs (combined repetitive oligopeptides) involved in receptor binding (1). After binding, the receptor-toxin complex is endocytosed to reach acidic endosomal compartments from where the toxins (or parts of the toxins) translocate into the cytosol (3). Here, the cysteine protease domain is activated by inositol hexakisphosphate (InsP6), resulting in cleavage of the toxin and release of the glucosyltransferase domain into the cytosol (4, 5). Subsequently, the glucosyltransferase glucosylates Rho proteins at Thr37 (e.g., RhoA, RhoB, and RhoC) or Thr35 of Rac and Cdc42; thereby, the functions of Rho proteins are inhibited (6). Various clostridial toxins share the molecular architecture of TcdA and TcdB, including lethal toxin and hemorrhagic toxins of Clostridium sordellii and the α-toxin of Clostridium novyi (1). The Clostridium perfringens toxin TpeL is another member of this toxin family, which GlcNAcylates Ras and Rac proteins (7). TpeL differs from other toxins because it has no CROP domain.
Yersinia are Gram-negative bacteria. The genus Yersinia has at least 18 species. Only three are known as major human pathogens, including Yersinia pestis, Yersinia enterocolitica, and Yersinia pseudotuberculosis. The pathogens produce numerous virulence factors (8). Well studied are various Yersinia surface adhesins (e.g., YadA, Ail, and Psa) and the large group of Yersinia outer proteins, which are delivered into host cells by type III secretion (8). Another well-characterized pathogen is Yersinia ruckeri, a fish pathogen, which causes the enteric red mouth disease in salmonid fish (9). Recently, AFP18, which is an effector of the type VI secretion-like antifeeding prophage (AFP) complex of Y. ruckeri, has been identified as a tyrosine glycosylating virulence factor (10). Here, we report on two previously unknown toxins produced by Yersinia mollaretii, which largely share the same multidomain architecture with CGTs. While one toxin is a glycosyltransferase, the other toxin has adenosine diphosphate (ADP)–ribosyltransferase activity. Unexpectedly, we found that the substrates of the toxins are Rab proteins, which are bidirectionally (inversely) regulated by the toxins.
RESULTS
C. difficile toxin–related proteins in Y. mollaretii
By comparing the protein sequence of C. difficile toxin B (TcdB) with protein sequences of the genus Yersinia, two Y. mollaretii proteins were found, which shared substantial similarities with TcdB (Fig. 1A). One protein (WP_004876989.1) exhibited similarity with TcdB over the entire protein sequence until amino acid 1834 of TcdB. This protein was called YGT (Yersinia glycosyltransferase). Sequence analysis revealed an N-terminal enzyme domain, followed by a cysteine protease domain and a C-terminal translocation and binding domain with TcdB. However, like in C. perfringens TpeL, the C-terminal CROP domain of TcdB was missing in YGT (fig. S1) (7). The second Yersinia protein (WP_004874725.1) largely differed in its N terminus from YGT and TcdB but shared sequence similarities in the middle and C-terminal parts of the protein with CGTs. This toxin exhibited N-terminal sequence similarity with ADP-ribosyltransferases (see below) and was designated as YART (Yersinia ADP-ribosyltransferase).
Fig. 1. Domain architecture of YART and YGT and analysis of the translocation and protease domains.
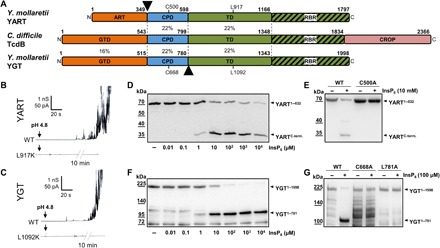
(A) Domain comparison of YART, TcdB, and YGT. The N-terminal domain of YART is an ADP-ribosyltransferase (ART), whereas the N terminus of YGT and TcdB harbors glycosyltransferases (GTD). Like TcdB, YART and YGT have an autocatalytic cysteine protease domain (CPD) and a translocation domain (TD). The C terminus of the TD of TcdB contains a receptor-binding region (RBR). Only TcdB has a CROP domain. Sequence identities of regions indicated by amino acid numbers are in percentage. Arrowheads mark split products of YART and YGT. Cys500 and Leu917 of YART and Cys668 and Leu1092 of YGT indicate the essential cysteine of CPDs and the critical leucine of TDs. (B and C) Membrane activity in lipid bilayer by YART, YGT, and their mutants YART L917K and YGT L1092K. Pore formation was induced by acidification to pH 4.5. The mutants did not induce increase in electric conductance, even after prolonged incubation (10 min). (D and F) In vitro processing of YART1–632 and YGT1–1998 at the indicated concentrations of InsP6, resulting in fragments YARTC-term and YGT1–781. (E and G) Inhibition of autocatalytic cleavage in mutants YGT1–1998 L781A, YGT1–1998 C668A, and YART1–632 C500A in the presence of InsP6. Western blots with anti-His antibody are shown. WT, wild type.
Membrane activity in lipid bilayer
Pore formation is likely involved in translocation of TcdB and related toxins (3, 11). We studied pore formation of the Yersinia toxins in lipid bilayer by monitoring the increase in electric conductance upon acidification of the medium from pH 7.5 to <6 (Fig. 1, B and C) (3). After acidification, YGT and YART exhibited enhanced membrane activity and increased the electrical conductance of lipid membranes but did not induce a defined stepwise increase in conductance. A similar type of interaction with lipid membranes is known for TcdB (3). Notably, Yersinia toxins are highly similar to TcdB (and TcdA) in a peptide region around amino acids 1080 to 1093 of YGT and amino acids 905 to 918 of YART (fig. S1C). This region is essential for translocation of TcdB and is conserved in all CGTs (11). In TcdB, change of Leu1106 to lysine causes inhibition of toxin-induced pore formation (11). Similarly, we observed an inhibition of increased membrane activity with the mutations L917K and L1092K of YART and YGT, respectively (Fig. 1, B and C). These findings suggest highly related translocation mechanisms of the Yersinia toxins as compared with TcdB.
Autoproteolytic activities of YGT and YART
CGTs share an autocatalytic cysteine protease domain, which is allosterically activated by InsP6 (4, 5). Similarly, the autoproteolytic activities of YGT and YART were activated by InsP6 (Fig. 1, D and F). As in all clostridial glucosyltransferase toxins, the cleavage of YART occurred between the cysteine protease domain and the proposed N-terminal enzyme domain (Fig. 1D) (4, 5). Figure 1D shows the InsP6-induced cleavage of the N-terminal YART fragment (amino acids 1 to 632), containing the N-terminal enzyme domain and the proposed upstream protease domain with an additional His tag. This fragment was used because the complete toxin exhibited additional protein bands after purification (fig. S2A), while this was not the case with the N-terminal fragment. Autocleavage in the presence of 1 to 10 μM InsP6 released a ~30-kDa C-terminal peptide. The autoprotease activity of CGTs depends on a catalytic aspartate, histidine, and cysteine (DHC) triad (4), which is conserved in the proposed protease domain of YART with Asp392, His454, and Cys500. As shown in Fig. 1E, YART C500A did not exhibit autocleavage activity in the presence of InsP6 (10 mM).
For cleavage studies with YGT, we used the full-length toxin of ~230 kDa, carrying a His and Sumo tag at the N terminus, which largely increased the expression yield. Unexpectedly, in the presence of InsP6, cleavage of YGT occurred downstream of the autocatalytic cysteine protease domain, releasing a fragment of ~110 kDa (Fig. 1F). Accordingly, we did not detect any additional split product, when a toxin fragment, covering the N-terminal enzyme domain and the protease domain only (amino acids 1 to 760), was used (fig. S2B). Sequence analysis suggested that in YGT, cleavage was most likely behind Leu781. This hypothesis was supported by mass spectrometric (MS) analysis. Moreover, when Leu781 of YGT was changed to alanine, cleavage was prevented (Fig. 1G). As found for the protease domain of YART, exchange of the highly conserved cysteine residue (Cys668) of the proposed catalytic triad (Asp562, His625, and Cys668) of YGT resulted in inhibition of autocleavage (Fig. 1G).
Enzyme activity of YART
While the N-terminal region of YART did not exhibit substantial amino acid sequence similarity with CGTs (fig. S1B), we recognized similarity with ADP-ribosyltransferases from the R-S-E type like cholera and pertussis toxins (Fig. 2A) (12).
Fig. 2. ADP-ribosylation by YART and glycosylation by YGT.
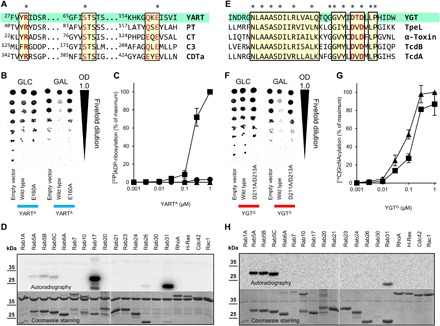
(A) Sequence comparison of ADP-ribosyltransferases [YART, pertussis toxin (PT; P04977), cholera toxin (CT; P01555), Clostridium botulinum C3 toxin (C3; P15879), and C. difficile ADP-ribosyltransferase (CDTa; Q9KH42)]. Conserved amino acids are marked in red; R-S-E amino acid motifs are indicated by asterisk. (B) Spot test of S. cerevisiae growth. Yeast cells, expressing YARTA or mutants under control of a galactose-dependent promoter, were spotted on glucose (GLC) and galactose (GAL) plates in fivefold dilutions. OD, optical density. (C) ADP-ribosylation of Rab proteins. Rab5A (▲), Rab31 (■), and Rab17 (●) (each 5 μg) were incubated at the indicated concentrations of YARTA with 1 μM [32P]NAD+ for 15 min, followed by SDS–polyacrylamide gel electrophoresis (PAGE) and phosphorimaging (data are means ± SD; n = 3). (D) Substrates of YART. Indicated guanosine triphosphatases (GTPases) (Rab31, 0.2 μg; other, 2 μg) were incubated with YARTA (3 μM) and [32P]NAD+ as in (C). Coomassie staining and autoradiogram are shown. (E) Sequence alignment of the DXD region of glucosyltransferase toxins [YGT, C. perfringens TpeL (A2PYQ6), C. novyi α-toxin (Q46149), and C. difficile TcdB (P18177) and TcdA (P16154)]. (F) Growth spot test of YGTG or mutants in yeast cells as in (B). (G) Glycosylation of Rab5A (▲) and Rab31 (■) (each 5 μg) with YGTG and UDP-[14C]GlcNAc (10 μM) for 15 min. Analysis of labeled proteins as in (C). (H) Substrates of YGT. Indicated GTPases (each 2 μg) were incubated with YGTG (0.5 μM) and UDP-[14C]GlcNAc as in (G).
We expressed the ADP-ribosyltransferase domain of YART (amino acids 1 to 349; henceforth called YARTA) in Saccharomyces cerevisiae under the control of a galactose-dependent promoter (Fig. 2B). This resulted in moderate growth inhibition of yeast, when galactose was present. Sequence alignment suggested that Glu160 of YART was the so-called catalytic glutamate, which is conserved in all ADP-ribosyltransferases and essential for enzyme activity (12). Accordingly, expression of YARTA E160A (verified by Western blot; fig. S2C) did not affect yeast growth, supporting the view that YART acts as an ADP-ribosyltransferase. Next, we ADP-ribosylated yeast cell lysate with YARTA in the presence of [32P]NAD+ (nicotinamide adenine dinucleotide). This resulted in radioactive labeling of ~25-, ~35-, and ~40-kDa proteins (fig. S2D). Labeling of the 40-kDa protein band was obviously caused by auto–ADP-ribosylation. MS analysis of the heavily labeled 25-kDa protein band revealed various YPT proteins, including YPT1, YPT6, YPT7, YPT32, YPT11, YPT31, YPT8, and SEC4, but not a direct attachment of ADP-ribose. Because YPT proteins are orthologs of Rab proteins, we tested the ADP-ribosylation of Rab, Rho, and Ras proteins in vitro (Fig. 2D). These studies revealed ADP-ribosylation of Rab31, Rab17, and Rab5 by YARTA. In contrast, Rho proteins (RhoA, Rac, and Cdc42) and H-Ras proteins were not modified. Further analysis of the kinetics of YARTA-induced ADP-ribosylation showed that Rab31 was the preferred substrate of the Yersinia enzyme in vitro (Fig. 2C), showing a >50-fold higher rate in ADP-ribosylation of Rab31 than of Rab17, while Rab5 was only marginally modified under these conditions (low NAD concentration of 1 μM).
Enzyme activity of YGT
The N-terminal part of YGT exhibited substantial sequence similarity with CGTs (Fig. 2E and fig. S1B). Therefore, we studied whether YGT had glycohydrolase activity, a typical property of many glycosyltransferase toxins (13). When radiolabeled uridine diphosphate (UDP)–glucose or UDP–N-acetylglucosamine (GlcNAc) was incubated with the N-terminal domain of YGT (amino acids 1 to 520; henceforth called YGTG), hydrolysis of UDP-GlcNAc but not of UDP-glucose was detected, suggesting that UDP-GlcNAc was the preferred sugar donor (fig. S2E). As for TcdB, the glycohydrolase activity of YGTG depended on the presence of Mn2+ or Mg2+ (fig. S2F).
As found for YARTA, expression of YGTG in S. cerevisiae inhibited yeast growth (Fig. 2F). Growth inhibition was prevented by mutation of the DXD motif (YGTG D211A/D213A), which plays a pivotal role in many glycosyltransferases (13–15). Next, we studied the GlcNAcylation of small guanosine triphosphatases (GTPases) by YGTG in vitro (Fig. 2H), showing that Rab5 and Rab31 were strongly modified by YGTG. By contrast, Rho or Ras proteins, which are the preferred substrates of CGTs like TcdB, were not modified. The concentration-effect curve of YGT-induced GlcNAcylation exhibited a slight preference for Rab5 (Fig. 2G).
Crystal structure analysis of YGT
To gain insights into the architecture and mechanism of the glycosyltransferase domain of YGT, we determined its x-ray structure. A His-tagged construct of YGTG was produced by heterologous expression, purified by affinity chromatography, and the affinity tag was removed by TEV (tobacco etch virus)–protease cleavage. Crystals were obtained in the absence of ligands as well as in the presence of UDP and MnCl2. The structure of YGTG with UDP and Mn2+ bound was solved using native SAD (single-wavelength anomalous diffraction), and it was refined to 1.9-Å resolution (table S1). The ligand-free structure was solved by molecular replacement and refined at 3.4-Å resolution (table S1).
YGTG has a globular structure, which can be divided into two parts. N-terminally, a typical GT-A glycosyltransferase fold comprises a central seven-stranded, mixed β sheet surrounded by 16 α helices in a Rossmann fold–like arrangement (Fig. 3A and fig. S3A). The C-terminal end of the construct comprises a compact antiparallel five-helix bundle (Fig. 3A and fig. S3A). The N-terminal part of YGTG displays high structural similarity with the core glycosyltransferase domain of various toxins. Highest homology was found for clostridial toxins including the lethal toxin from C. sordellii as well as toxins A and B from C. difficile (Fig. 3B and table S2) (14, 16, 17). The N-terminal four-helix bundle characteristic for clostridial glycosylating toxins is absent in YGTG. Substantial homology is also present for the smaller glycosyltransferase domains of the PaTox toxin from Photorhabdus asymbiotica (18) and the Salmonella effectors SseK1 and SseK3 (19, 20). The C-terminal five-helix bundle appears to be specific for YGTG and has no equivalent counterpart in the identified structural homologs. The first helix of the bundle, however, superimposes well with the C-terminal helix of TcdB.
Fig. 3. Structure of YGTG.
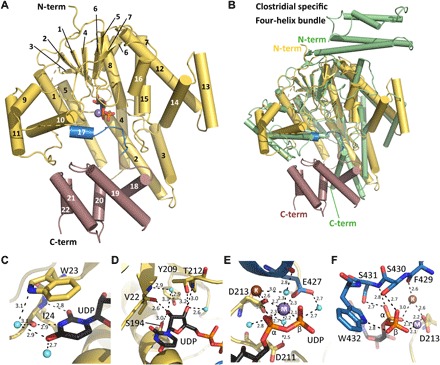
(A) The YGTG structure displays a typical GT-A glycosyltransferase fold (yellow) comprising a seven-stranded β sheet surrounded by α helices and a C-terminal five α-helical bundle (violet). UDP (black sticks) and Mn2+ (purple sphere) are bound centrally. Helix 17 and the loop that encloses the ligand binding site are highlighted in blue. (B) Structural superposition of YGTG [colored as in (A)] with TcdB from C. difficile [Protein Data Bank (PDB) ID: 2BVL, green] showing the GT-A glycosyltransferase fold. While the N-terminal four-helix bundle of TcdB is absent in YGTG, four helices at the C terminus of YGTG are not conserved in clostridial toxins. (C to F) Closeup views of the active site showing the binding mode of UDP (black) with Mn2+ and K+ ions represented as spheres (purple and brown, respectively) and YGTG represented as a cartoon with relevant residues in sticks and colored as in (A). (C) The uridine base moiety is coordinated by a π-stacking interaction with Trp23 and by several direct and water-mediated hydrogen bonds (dashed lines). (D) The ribose moiety is coordinated by a series of hydrogen bonds with its hydroxyl groups. (E) The diphosphate from UDP is stabilized via interactions with the Mn2+ ion. Mn2+ is coordinated with aspartate side chains of the DXD motif, Glu427, and water molecules. Note the presence of a K+ ion in close vicinity. (F) The C-terminal loop of the GT-A fold encloses the nucleotide-binding pocket comprising direct interactions between the diphosphate moiety and Ser430, Ser431, and Trp432.
The 1.9-Å structure of YGTG sheds light on the nucleotide-binding mode (Fig. 3, C to F). The uridine moiety is stabilized by an aromatic π stacking with Trp23 and by four hydrogen bonds, particularly with the backbone atoms of residues Trp23 and Ile24 (Fig. 3C). The ribose moiety is stabilized by several direct or water-mediated hydrogen bonds to either main- or side-chain atoms of Val22, Ser194, Tyr209, and Thr212 (Fig. 3D). The side chains of Asp211 and Asp213, namely, the conserved DXD motif and the adjacent manganese ion (Mn2+), are central for the coordination of the diphosphate moiety (Fig. 3E). The nature of the metal ion was confirmed using the anomalous signal of manganese at 6.7 keV (data not shown). The side chain of Asp213 and one oxygen atom of each α and β-phosphate directly coordinate the Mn2+. Asp211 contributes a water-mediated interaction. The typical octahedral coordination of Mn2+ is completed with a water molecule and with the side chain of Glu427 from helix 17 (Fig. 3E). The side-chain oxygen atom of Asp213 is coordinating not only Mn2+ but also a second atom that was showing a strong peak in the native SAD anomalous Fourier electron density map (fig. S3B). The atom was assigned as potassium ion. It is further coordinated by the main-chain carbonyl groups of residues Glu426 and Phe429 and by the α-phosphate moiety of UDP as well as by several water molecules including one belonging to the first coordination sphere of Mn2+ (Fig. 3, E and F, and fig. S3B). It is well established that the glycohydrolase and glucosyltransferase activities of CGTs are increased by potassium but not by sodium ions (21–23). Similarly, the glycosyltransferase activity of YGT was enhanced by potassium ions (fig. S3C).
Notably, the C terminus of the glycosyltransferase domain comprising the short helix 17 and its consecutive loop (residues Phe429 to Tyr438) encloses the nucleotide-binding site as a lid with multiple interactions to the diphosphate (Ser430 and Trp432 to β-phosphate and Ser431 to α-phosphate) (Fig. 3F). In the ligand-free YGTG structure, which is overall highly similar to the UDP-bound state (1.4-Å root mean square deviation for all atoms), the aforementioned lid is shifted by about 8 Å, including a reorientation of the conserved Trp432 side chain resulting in an open ligand-binding site (fig. S4).
Rab modification by YART and YGT
Next, modification of Rab31 and Rab5 by YART and YGT was analyzed by MS (fig. S5, A to C). These analyses revealed that YARTA ADP-ribosylated Rab31 at Gln64 and Rab5 at the equivalent Gln79, whereas YGTG transferred GlcNAc to Rab5 in position Thr52 (Fig. 4A and fig. S6A). YGTG- and YARTA-induced modifications of Rab proteins were verified by site-directed mutagenesis of Rab5 and Rab31 (Fig. 4B and fig. S6B). The mutant protein Rab5A T52A was not GlcNAcylated by YGTG but slightly ADP-ribosylated by YARTA. On the other hand, the mutant protein Rab5A Q79L was not ADP-ribosylated by YARTA but GlcNAcylated by YGTG. Similar results were obtained with Rab31. YART was not capable to ADP-ribosylate Rab31 Q64L, while YGT caused GlcNAcylation of the mutant Rab31 protein, however, with reduced activity. As expected, GlcNAcylation was blocked in the mutant Rab31 T36A.
Fig. 4. YART- and YGT-induced modifications of Rab5.
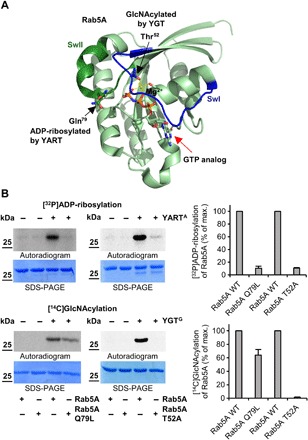
(A) Localization of the sites of Rab5A that are modified by YGT-induced GlcNAcylation and YART-induced ADP-ribosylation. Modification sites are from MS analyses and introduced into the Rab5A structure (PDB ID: 1N6H). Switch-1 (SwI) and switch-2 (SwII) regions are indicated in blue and dark green, respectively. Light green spheres represent the Mg2+ ion, while guanosine triphosphate (GTP) (or a GTP analog) is indicated by red arrow. While YGT modifies Rab5 in Thr52 (black arrow) by GlcNAcylation, YART ADP-ribosylates Gln79 (black arrow). (B) ADP-ribosylation or GlcNAcylation of mutant Rab5 proteins. Wild-type and mutant Q79L or T52A Rab5 proteins (each 2 μg) were incubated with 1 μM YARTA or YGTG, [32P]NAD+, or UDP-[14C]GlcNAc for 1 hour at 21°C. The modifications of proteins were analyzed by SDS-PAGE and phosphorimaging. The amount of modified Rab proteins was normalized to wild-type Rab protein. Means (±SD) of three independent experiments and one representative autoradiogram and Coomassie gel are shown.
Inhibition of GTP hydrolysis by YART-induced ADP-ribosylation
ADP-ribosylation of Rab proteins by YART occurs in a highly conserved glutamine residue that is essential for guanosine triphosphate (GTP) hydrolysis. To study the consequences of ADP-ribosylation on GTP hydrolysis, we used Rab5, although it was not the preferred substrate of YART, because GTPase-activating proteins (GAPs) are available for Rab5 but not for Rab31. Under basal conditions, ~40 to 50% of GTP bound to Rab5 was hydrolyzed after 30 min. Preincubation of Rab5 with YARTA in the presence of high concentrations of NAD+ (400 μM) reduced basal GTP hydrolysis to ~20% (Fig. 5A). Inhibition of GTP hydrolysis was not observed when the inactive mutant YARTA E160A was used. The addition of the GAP (RabGAP5) largely increased hydrolysis of Rab5-bound GTP to >90% (remaining Rab5-bound GTP, ~5%) (Fig. 5A). This effect was strongly inhibited by YARTA-induced ADP-ribosylation (remaining GTP, ~65%). Again, the inactive mutant YARTA E160A was without effect. Thus, ADP-ribosylation blocks GTP hydrolysis and causes activation of Rab5 protein.
Fig. 5. Functional consequences of Rab5-induced modifications by YART and YGT.
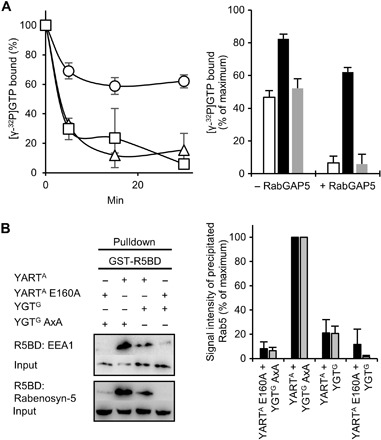
(A) YART effects on GTP hydrolysis by Rab5. Rab5A was ADP-ribosylated by active YARTA or inactive YARTA E160A and loaded with [γ-32P]GTP. Left: RabGAP5-stimulated GTP hydrolysis (time course) by Rab5A pretreated with YARTA (○), with YARTA E160A (□) or untreated control (△). Right: GTP hydrolysis after 30 min with or without RabGAP5 (untreated control, white column; YARTA pretreatment, black column; YARTA E160A pretreatment, gray column). Data are means ± SD (n = 3). (B) Pulldown of Rab5A by the R5BDs of EEA1 and Rabenosyn-5. Rab5A (Flag-tagged) was coexpressed overnight in HeLa cell with active YARTA and YGTG or with the inactive mutants YARTA E160A and YGTG AXA. Cell lysates were incubated with GST-R5BD–coupled beads (30 min). Thereafter, beads were precipitated and washed, and interacting proteins were analyzed by SDS-PAGE and Western blot. Immunoblots with anti-Flag antibody before (input) and after pulldown are shown. Left: One representative experiment. Right: Results of three independent experiments with EEA1 (black column) or with Rabenosyn-5 (gray column) (data are means ± SD). The amount of precipitated Rab5 was normalized to the pulldown of Rab5 after coexpression with YARTA and YGTG AXA.
Effects of YART and YGT expression on Rab5 effector interaction
Next, we studied the effects of YART and YGT in intact cells. We coexpressed active or inactive YARTA with active or inactive YGTG in HeLa cells together with Flag-tagged Rab5. To analyze the consequences of ADP-ribosylation and/or glycosylation of Rab5 on the interaction with its effectors, we performed pull-down assays of Flag-tagged Rab5 with the Rab5-binding domains (R5BDs) of the effectors EEA1 (early endosome antigen 1) and Rabenosyn-5. The amount of precipitated Rab5 was detected by an anti-Flag antibody. As shown in Fig. 5B, expression of active YARTA together with inactive YGTG largely increased the interaction of R5BDs with Rab5 as compared with controls (e.g., expression of inactive YARTA and inactive YGTG), indicating strong activation of Rab5. When both YARTA and YGTG were expressed as active proteins, the R5BD pulldown of Rab5 was significantly reduced compared with active YARTA and inactive YGTG. Last, expression of active YGTG in the presence of inactive YARTA resulted in minor pulldown of Rab5. Thus, all these data are in agreement with an activation of Rab5 by YARTA and its inactivation by YGTG.
It has been reported that overexpression of Rab5 or expression of constitutively active Rab5 induces enlargement of early endosomes (24, 25). Therefore, we studied the effects of YART and YGT on the size of vesicles associated with Rab5. To this end, mCherry-tagged Rab5A (mCherry-Rab5) was expressed in HeLa cells. In addition, the constructs contained a Flag tag for detection by Western blotting. When mCherry-Rab5A was coexpressed with the active ADP-ribosyltransferase domain of YART [green fluorescent protein (GFP)–YARTA], the immunofluorescence microscopic analysis revealed that the area of mCherry-Rab5–associated vesicles was significantly increased as compared with coexpression with the inactive YART mutant (GFP-YARTA E160A) (Fig. 6, A and C). Next, we tested the influence of YGT on Rab5-associated vesicles in the presence of active and inactive YART (Fig. 6, B and D). When the active YART domain (GFP-YARTA) was expressed together with the inactive glycosyltransferase domain of YGT (GFP-YGTG AXA), vesicle size was increased. However, when activated glycosyltransferase domain (GFP-YGTG) was expressed, no labeled vesicles were detected, and mCherry-Rab5 was equally distributed in the cytosol. The same result was obtained after expression of active YGTG together with inactive YART mutant (GFP-YARTA E160A). Western blot analysis revealed efficient coexpression of the various constructs under the conditions used (Fig. 6E). Again, these findings supported the notion that YART activates Rab5, thereby increasing the fusion and enlargement of Rab5-associated vesicles, whereas YGT inactivates Rab5, which appears to dominantly block Rab5 interaction with vesicles.
Fig. 6. Effect from YART and YGT on the Rab5A distribution in HeLa cells.
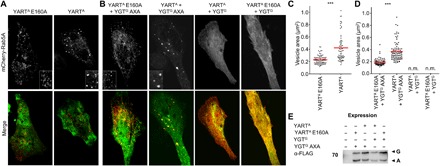
HeLa cells were transiently transfected overnight with mCherry-C1-Rab5A (top; red) together with the indicated pEGFP-C1-toxin constructs (green). Afterward, cells were washed and incubated at 21°C for an additional 3 hours. Insets show in detail Rab5A-vesicle structure. Pictures are representative of three independent experiments. Scale bar, 10 μm. (A) Effects of YARTA or YARTA E160A on the distribution of overexpressed Rab5A. (B) Effects of active YARTA and YGTG and inactive YARTA E160A and YGTG AXA on the distribution of overexpressed mCherry-C1-Rab5A. (C) Quantification by MetaMorph imaging software of the size of mCherry-C1-Rab5A vesicles in the presence of the active (YARTA) or inactive ADP-ribosyltransferase domain (YARTA E160A). Cells of view are ≥20, and n = 3. Unpaired two-sample t test was used (***P < 0.001). (D) Quantification of (B) as described in (C). n.m., not measurable. (E) For analysis of the expression of pEGFP-C1-toxin constructs, HeLa cells were lysed after transfection and expression. Proteins were analyzed by SDS-PAGE, Western blotting, and immunostaining with anti-FLAG tag antibody. G indicates GFP-YGTG and GFP-YGTG AXA; A indicates GFP-YARTA and GFP-YARTA E160A.
DISCUSSION
Here, we detected in Y. mollaretii two Yersinia toxins called YGT and YART that were largely related to CGTs. While YGT was only found in Y. mollaretii (strain ATCC 43969), YART-like proteins (~95% sequence identity) were also detected in a Yersinia massiliensis and a Yersinia intermedia strain. Both proteins (YGT and YART) comprised the typical polymodular structure of CGTs with an N-terminal enzyme domain, followed by an autoprotease domain and a C-terminal translocation and binding domain (1). In contrast to most CGTs, YGT and YART have no C-terminal CROP domains. Thus, the putative Yersinia toxins resemble the structure of C. perfringens toxin TpeL, which also lacks CROPs (7). Similar to TcdB (3), both Yersinia proteins increased electric conductance in black lipid membranes at low pH, suggesting pore-forming activity involved in membrane translocation. Both YART and YGT share amino acid sequences (amino acids 1080 to 1093 of YGT and amino acids 905 to 918 of YART) with C. difficile toxins, which appear to be essential for pore formation of TcdB (11). Exchange of Leu917 and Leu1092 of YART and YGT, respectively, which are equivalent to Leu1106 in TcdB, blocked membrane activity. Thus, YART and YGT are probably translocated across membranes by the same molecular mechanism as CGTs. Further studies are necessary to reveal the typical low pH–dependent uptake of the toxins from endosomes and to exclude delivery mechanisms directly involving bacterial secretion systems.
Similar to CGTs (4, 5), YGT and YART have autocleavage activity in the presence of InsP6. While YART cleaves behind the ADP-ribosyltransferase domain, YGT cuts downstream of the autoprotease domain after Leu781. Change of this residue to alanine blocked cleavage. These findings suggest that the glycosyltransferase is released into the cytosol together with the autoprotease domain. Whether this is of functional importance remains to be studied.
The N-terminal enzyme domain of YART (YARTA) has ADP-ribosyltransferase activity. This finding shows that the toxin activity of CGT-like toxins is not coupled to only one type of enzyme activity (e.g., glycosyltransferase activity). We found that YART specifically modifies Rab proteins. Rab31 was the highly preferred substrate followed by Rab17 and Rab5. Notably, Ras and Rho proteins were not ADP-ribosylated by YART. The identification of Rab31 protein as target of a bacterial toxin is unique. So far, only Pseudomonas aeruginosa exoenzyme S (ExoS), which is a type III secretion effector, has been reported to ADP-ribosylate Rab5 (26). However, ExoS ADP-ribosylates multiple target proteins, including various low–molecular mass GTPases (Rho, Ras, and Rab), ERM (ezrin-radixin-moesin) proteins, and vimentin (26). Moreover, modification of Rab proteins by ExoS occurs at multiple sites. By contrast, ADP-ribosylation of Rab31 and Rab5 by YART is restricted to Gln64 and Gln79, respectively, which are functionally equivalent glutamine residues. This glutamine residue is conserved in nearly all small GTPases and involved in GTP hydrolysis. The function of the glutamine residue is the optimal positioning of the nucleophilic water molecule for attacking the γ phosphate of GTP and to stabilize the transition state of GTP hydrolysis (27). Therefore, ADP-ribosylation inhibits hydrolysis of GTP even after stimulation of the GTPase activity by RabGAP5.
Similar to CGTs, the YGTG has glycosyltransferase activity by using UDP-GlcNAc as sugar donor. Both glucosylation and GlcNAcylation are observed within the family of CGTs (13). While C. difficile toxins (TcdA and TcdB) use UDP-glucose, C. novyi α-toxin and C. perfringens TpeL prefer UDP-GlcNAc like YGT (13). Similarly, as found for YART, YGT targets Rab5 and Rab31, which were modified in Thr52 and Thr36, respectively. These functionally equivalent residues are located in the switch I region of the GTPases and have a crucial role in Mg2+ ion coordination and nucleotide binding. Modification of Rab at this site is unique for YGT. Notably, CGTs (e.g., TcdA and TcdB) modify the equivalent residue (Thr37 or Thr35) in Rho proteins (6). Other CGTs modify Ras proteins like C. sordellii lethal toxin (TcsL) and C. perfringens toxin TpeL (7, 13).
In line with the functional similarity of YGTG with CGTs, the YGTG structure has a typical GT-A glycosyltransferase fold and shows highest structural similarity to clostridial toxins (14–16) including lethal toxin, TcdA and TcdB, and α-toxin. In contrast to these, YGTG includes a C-terminal five-helix bundle of yet unknown function. The canonical core GT-A glycosyltransferase, which harbors the Rossmann fold–like and its nucleotide-binding site, is also structurally similar to the smaller glycosyltransferase domain of PaTox (18) from P. asymbiotica as well as to the arginine glycosylating effector proteins SseK1, SseK3, and NleB (19, 20). The structure of UDP-bound YGTG sheds light on the nucleotide-binding mode reflecting the situation after the glycosyltransferase reaction has occurred. The binding mode ensures nucleoside specificity. The conserved DXD motif and a divalent cation, here referred to as a manganese ion, position the diphosphate moiety. Overall, the binding mode is similar to those reported for clostridial toxins (14).
The structures of ligand-free and ligand-bound YGTG reveal two conformations of the terminal loop, which covers the diphosphate moiety when the nucleotide is bound and leaves the binding pocket open in ligand-free state. The closed conformation is similar to those in glycosyltransferase structures from TcdA (15) and lethal toxin (16) in the presence of UDP-glucose. Consistent with the ligand-free YGTG structure, in nucleotide-free structures of α-toxin from C. novyi and of TcdA from C. difficile (15, 16, 23), the loop is displaced, and the binding site is open. Together, clostridial and YGTG structures show a consistent conformational change of the so-called lid loop from open conformation in the nucleotide-free state to closed conformation when the nucleotide is present.
A notable feature of the UDP-bound YGTG structure is the presence of a potassium ion at the ligand-binding site. Potassium strongly enhances the activity of CGTs (21–23) and YGT. Yet so far, the relevant ion-binding site was not structurally resolved. Because the potassium ion in YGTG is directly coordinated by the DXD motif’s Asp213 side chain, which also coordinates the manganese ion, the potassium ion is likely to affect the charge equilibration in the binding site. The ion also interacts with residues of the lid loop in the closed state and may help to stabilize ligand binding. We suggest that the identified potassium ion–binding site is the basis for enhanced glycosyltransferase activity in clostridial and related toxins.
Rab proteins are master regulators of endosome fusions (28). Canonically, the presence of Rab5 and its effectors like EEA1 defines prototypic early endosomes (29). The GTPase controls the maturation of phagosomes to phagolysosomes, which have major antimicrobial activity (30). Accordingly, various studies report on a role of Rab5 in host-pathogen interactions, including pathogens such as Listeria monocytogenes (31), P. aeruginosa (32), Salmonella (33), and Yersinia (34). Rab31 (also called Rab22B) belongs to the Rab5 subfamily. It mainly localizes to the trans-Golgi network and is probably involved in the anterograde exit from the trans-Golgi network to the cell membrane (35). Considering that many Yersinia species (e.g., Y. pestis, Y. pseudotuberculosis, Y. enterocolitica, and Y. ruckeri) are facultative intracellular pathogens (36, 37), the same is probably true for Y. mollaretii. Our coexpression studies show that YART and YGT regulate Rab proteins in a bidirectional (inverse) manner. Expression of YARTA largely increased the level of activated Rab5. Similarly, expression of YART increased the size of Rab5-associated vesicles, which is a consequence of constitutive activation of Rab5 (24, 25). These effects were inhibited by YGT. Moreover, YGT caused equal distribution of labeled Rab5 in the cytosol without any vesicle association, indicating inactivation of Rab5. Rab5 and Rab31 are likely involved in invasion and intracellular infection of Yersinia. Thus, we speculate that YART- and YGT-induced activation and/or inhibition of Rab proteins occur in a time- and space-dependent manner to optimize the survival of the pathogen.
Y. mollaretii have been isolated from human feces, fresh water, animal feces, and intestines and foods (38). The bacteria are also found in the oral microbiome of children (39). So far, however, a pathogenic role of Y. mollaretii for humans is not known. Specific Yersinia (e.g., Yersinia entomophaga) are pathogenic for insects (40). This is also true for Y. mollaretii (41). It remains to be studied whether YART and YGT are virulence factors, which are important for host-pathogen interaction in insects.
MATERIALS AND METHODS
Cell culture, preparation of mammalian cell lysates, and immunostaining
HeLa cells were grown in Dulbecco’s modified Eagle’s medium in a humidified atmosphere of 5% CO2 at 37°C. The media contained 10% fetal calf serum, penicillin (4 mM), and streptomycin (4 mM). For preparation of cell lysate, cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in the indicated buffer for 5 min on ice. Afterward, the cell lysate was transferred into a 1.5-ml tube and cleared by centrifugation (10 min, 4°C, 20,000g). The supernatants were used for the following experiments. After separation of proteins by SDS–polyacrylamide gel electrophoresis (PAGE) and Western blotting, the proteins were detected by specific antibodies. Horseradish peroxidase–conjugated donkey anti-mouse immunoglobulin G (H&L) (610-703-124; Rockland) or goat anti-rabbit immunoglobulin G (H&L) (no. 7074, Cell Signaling) antibody was used as a secondary antibody. Antibody signals were detected by enhanced chemiluminescence using the LAS-4000 reader (Fujifilm) and quantified with MultiGauge software (FujiFilm).
Yeast culture conditions
Yeast strains were cultivated on minimal YNB medium (0.67% yeast nitrogen base without amino acids; Difco) containing 2% glucose or galactose and adapted from the specific S. cerevisiae strain requirements, with the suitable supplements (uracil, tryptophan, adenine, or leucine) completed. For successful yeast transformation, the lithium acetate method (42) was used.
Preparation of yeast lysates and membrane fractions
Yeast cells were cultivated in YPD (yeast extract–peptone–dextrose) medium or in selective SD (supplemental dextrose) medium on a shaker at 30°C; afterward, the cells were harvested by centrifugation at 4000 rpm for 10 min at 4°C. The pellet was washed once with ice-cold water. Then, the pellet was resuspended in a minimal volume of lysis buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, and Protease Inhibitor Cocktail (Roche Diagnostics GmbH, Mannheim, Germany)], and the cells were disrupted using acid-washed glass beads (425 to 600 μm, Sigma-Aldrich) by vortexing of the 1:1 cell pellet–beads mixture for 1 min at maximal power. After 1 min on ice, the procedure was repeated six times. Last, the whole mixture was centrifuged at 20,000g for 15 min at 4°C, and the protein content of the supernatant was determined by using the bicinchoninic acid Protein Assay Kit (Life Technologies, Waltham) according to the manufacturer’s protocol. For separating the membrane fraction, the supernatant was additionally ultracentrifuged for 1 hour at 100,000g at 4°C, and the membrane pellet was resuspended in buffer [25 mM tris-HCl (pH 7.5) and 1 mM EDTA].
Cloning, expression, and purification of proteins
Cloning and recombinant protein production were performed in Escherichia coli TG1 and BL21, respectively. S. cerevisiae MH272-3fα (ura3, leu2, his3, trp1, and ade2) and pESC-Ura galactose-inducible expression vectors were obtained from Stratagene (Agilent, Frankfurt, Germany). The various mutants were constructed by site-directed mutagenesis using QuikChange kit (Stratagene) according to the manufacturer’s instructions, with the exception that the Velocity DNA Polymerase (Bioline) was used. The constructs were checked by DNA sequencing (Eurofins GATC Biotech GmbH, Konstanz, Germany). Primers (Biomers, Ulm; Apara Bioscience, Denzlingen) and gBlock Gene fragments (Integrated DNA Technologies, USA) used for cloning are listed in table S3. For purification, the E. coli strain BL21 was transformed with the corresponding plasmids and was grown at 37°C in Luria-Bertani broth supplemented with the respective antibiotics [ampicillin (100 μg/ml) or kanamycin (30 μg/ml)] until an optical density of A600 = 0.5 to 0.8. The expression of the proteins was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) (Roth, Karlsruhe), and bacteria were grown for indicated times at the given temperatures (table S4). The cells were harvested by centrifugation with 6000 rpm for 15 min and resuspended in the indicated lysis buffer with DNAse (deoxyribonuclease; 5 μg/ml), lysozyme (1 mg/ml), and 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich). The bacterial cells were lysed by using an ultrasonic homogenizer and centrifuged for 45 min (14,000 rpm, 4°C). The proteins were purified from the supernatant by nickel affinity purification or Glutathione Sepharose 4B (GE Healthcare) according to the tag of the protein (table S4). Proteins were stored in the indicated buffers at −80°C (table S4).
Black lipid bilayer experiments
Membrane insertion and pore activity were measured using black lipid membranes composed of diphytanoyl phosphatidylcholine (DiPhPC). Black lipid bilayer experiments were performed as described previously (3). The instrumentation consisted of a Teflon chamber divided into two compartments by a thin wall and connected by a small circular hole (∼0.4 mm2). The aqueous solutions of both chambers contained 1 M KCl and were buffered with 20 mM tris-HCl (pH 7.4). After painting a 1% solution of DiPhPC (Avanti Polar Lipids, Alabaster, AL) dissolved in n-decane across the hole to form a membrane, the toxins (3.5 nM) were added to one side of the membrane (the cis side). After 5 min, by the addition of 1.85% HCl into the same compartment containing the toxins, the pH drop to pH 4.8 was obtained. The current of the membrane was measured by using Ag/AgCl electrodes (with salt bridges) switched in series with a voltage source and a homemade current amplifier. The applied membrane potential was −50 mV at room temperature. The signal was recorded on a strip chart recorder (Rikadenki, Freiburg, Germany).
In vitro autocleavage assay
For the in vitro autocleavage assay, 2 μg of full-length toxins or toxin fragments was incubated in cleavage buffer [100 mM tris-HCl (pH 7.4)] with the indicated concentrations of InsP6. After incubation for 1 hour at 37°C, the reaction was stopped by adding Laemmli buffer and boiling for 5 min at 95°C. The visualization of the cleavage products was done by SDS-PAGE, subsequent Western blotting, and anti-His (no. 2365S, Cell Signaling) antibody staining.
Spot test assay
For testing the toxicity of YART and YGT in S. cerevisiae, yeast growth on agar plates was analyzed. Therefore, fivefold serial dilutions of yeast cultures were spotted onto YNB agar supplemented with glucose or galactose and with adenine, tryptophan, leucine, and uracil. Yeast was incubated for 3 days at 30°C.
Hydrolysis of UDP-glucose and UDP-GlcNAc by YGT
For determination of UDP-glucose hydrolysis, YGTG was incubated with UDP-[14C]Glc or UDP-[14C]GlcNAc (at indicated concentrations) in the reaction buffer, containing 50 mM Hepes (pH 7.5), 100 mM KCl, 2 mM MgCl2, and 1 mM MnCl2. One-microliter samples were taken at the indicated time points and subjected to PEI (polyethyleneimine)–cellulose thin-layer chromatography (Merck, Darmstadt, Germany). To separate hydrolyzed [14C]glucose from UDP-[14C]glucose, 0.2 mM LiCl was used as mobile phase, and the samples were allowed to run for 8 min. Eventually, the plates were dried and analyzed with the phosphorimager (Typhoon FLA 7000, GE Healthcare, Freiburg, Germany), and radioactive samples were quantified by using MultiGauge software.
Glycosylation assay
For the glycosylation assay, cell lysate or recombinant GTPases were incubated with recombinant YGTG or its mutants and 10 μM UDP-[14C]GlcNac in a buffer containing 50 mM Hepes (pH 7.5), 100 mM KCl, 2 mM MgCl2, 1 mM MnCl2, and BSA (0.1 mg/ml) at 21°C for the indicated time. Analyses of modified proteins were done by SDS-PAGE and afterward by phosphorimaging (Typhoon FLA 7000, GE Healthcare, Freiburg, Germany).
ADP-ribosylation reaction
Cell lysate or recombinant GTPases were incubated for the indicated time at 21°C with 1 μM [32P]NAD+ and YARTA in ADP-ribosylation buffer [20 mM tris-HCl (pH 7.4), 1 mM EDTA, and 5 mM MgCl2]. All radiolabeled proteins were detected by SDS-PAGE and subsequent phosphorimaging (Typhoon FLA 7000, GE Healthcare, Freiburg, Germany).
GTP hydrolysis assay
First, Rab5A (10 μM) was ADP-ribosylated by active YARTA or inactive YARTA E160A (each 1 μM) in the presence of 400 μM NAD+ in a buffer containing 50 mM Hepes (pH 7.5), 100 mM KCl, 2 mM MgCl2, 1 mM MnCl2, and BSA (0.1 mg/ml) for 60 min at 24°C.Afterward, for loading of Rab proteins, 5 μl of 10-fold buffer A [500 mM Hepes (pH 7.4), BSA (10 mg/ml), 10 mM dithiothreitol (DTT), and 10 mM adenosine triphosphate], 1 μl of 50 mM EDTA, 1 μl of 25 mM MgCl2, 25 μl of 10 μM [γ-32P]GTP [in 50 mM tris-HCl (pH 8.0), 1 mM DTT, 1 mM ATP, and 10 μM GTP], 13 μl of H2O, and 5 μl of 10 μM pretreated Rab5A were mixed on ice and then incubated for 45 min at 24°C. Then, 10 μl of the preloaded Rab was added to a 90-μl reaction mix (buffer A plus 5 mM MgCl2, and 1 mM GTP). GAP reaction was started by the addition of 1 pmol of the MBP (maltose-binding protein)–tagged RabGAP5. GTP hydrolysis was performed at 16°C. At indicated times, 10-μl samples were removed and added to 950-μl ice-cold stopping buffer [20 mM tris-HCl (pH 8.0), 100 mM NaCl, and 5 mM MgCl2]. Bound radioactively labeled GTP was measured using a nitrocellulose filter-binding assay (10).
Effector pull-down assay
The effector pull-down was performed as described previously (43) with the exception that the HeLa cells were transfected with human Flag-Rab5A [a gift from Q. Zhong (plasmid no. 28043, Addgene; http://n2t.net/addgene:28043; RRID: Addgene_28043)] and in addition with the different toxin constructs. The transfection was done according to the manufacturer’s instructions with Lipofectamine 2000 (Thermo Fisher Scientific, Karlsruhe, Germany). Data were obtained by SDS-PAGE, subsequent Western blotting, and anti-Flag (no. 14793S, Cell Signaling) antibody staining.
Expression of Rab5A, YART, and YGT in HeLa cells
HeLa cells were seeded at a density of 3.5 × 105 cells per well in six-well tissue culture plates. After settling down, cells were transfected with the mCherry-C1-Rab5A construct together with indicated pEGFP-C1-toxin constructs. The transfection was done by using Lipofectamine 2000 (Thermo Fisher Scientific, Karlsruhe, Germany) according to the manufacturer’s protocol. After overnight culture, HeLa cells were washed with PBS and Dulbecco’s modified Eagle’s medium containing the described additives. Afterward, HeLa cells were incubated in a humidified atmosphere of 5% CO2 at 21°C for 3 hours. Then, the cells were washed with PBS, fixed for 15 min with 4% paraformaldehyde in PBS, and washed again with PBS (all was done at room temperature). Last, cells were dried and embedded with Mowiol (Merck, Darmstadt, Germany) for microscopy.
Microscopy and imaging acquisition
Confocal fluorescence microscopy was performed with an Axiovert 200M microscope (Carl Zeiss, Jena, Germany), driven by the VisiView (Visitron, Puchheim, Germany) imaging software with plan-apochromat objectives, a Yokogawa CSU-X1 spinning disk confocal head with emission filter wheel, and a CoolSNAP HQ2 digital camera with 488- and 561-nm laser lines. Images were processed with MetaMorph imaging software (Universal Imaging, NY).
YGT crystallization and x-ray diffraction data collection
Before crystallization, YGTG [final product after TEV (tobacco etch virus)–cleavage Gly-YGTG 2–520] was concentrated to 15 mg/ml as estimated by Bradford assay. Vapor diffusion crystallization assays were performed at 4°C and at 20°C mixing 150 μl of YGT or YGT supplemented with 2 mM UDP and 2 mM MnCl2 and an identical volume of precipitant solution. Within a few days, needle-shaped crystals appeared in several conditions at 20°C. At 4°C, the crystallization process took up to a month, but the nucleation was lower, and thus larger crystals could be obtained. Large crystals of YGT supplemented with 2 mM UDP and 2 mM MnCl2 were grown in 100 mM tris-HCl (pH 8.5), 200 mM sodium acetate, and 15% polyethylene glycol (PEG) 4000. Ligand-free YGT crystals grew as very thin needles in a condition containing 200 mM sodium formate and 20% PEG 3350. Crystals were cryoprotected by the stepwise addition of ethylene glycol–supplemented reservoir solution to a final concentration of about 15% of ethylene glycol before transfer in liquid nitrogen. All data collections were carried out at 100 K. Initial diffraction data were collected at beamline ID23-1 (ESRF, Grenoble, France). A dataset at 1.9-Å resolution was collected from a crystal of UDP and MnCl2-supplemented YGTG at beamline ID30A-3 (MASSIF3, ESRF) at an energy of 12.8 keV. A native SAD dataset at 6-keV energy was collected at beamline ID29 (ESRF) (table S1). Data from YGT crystals were collected at 12.4 keV on beamline PXI (SLS, Villigen, Switzerland) (table S1).
Data processing and structure determination using native SAD
X-ray diffraction data were processed with XDS and scaled with XSCALE (44). Phenix.AutoSol (45) was used to solve the phase problem using the native SAD method, and the data were collected at 6 keV. A total of 17 sites were identified corresponding after structure refinement to 13 sulfur atoms, 2 phosphor atoms, 1 manganese atom, and 1 potassium atom. About 400 residues were built automatically, and the structure was finalized by manual modeling in Coot (46) and refinement cycles using Phenix (45). The 1.9-Å resolution structure of YGTG with bound UDP and Mn2+ was refined to final Rwork and Rfree values of 18.6 and 22.3%, respectively. About 97.4 and 2.6% of the residues are in the favored and in the allowed areas of the Ramachandran plot, respectively. No Ramachandran outlier is present in the structure. The latter includes one YGTG molecule (resolved residues 12 to 150, 153 to 230, and 233 to 518) with 1 UDP molecule, 1 manganese ion, 1 potassium ion, 8 ethylene glycol molecules, and 408 water molecules. This high-resolution structure was used as a molecular replacement search model to solve the phase problem of the ligand-free YGTG structure using the program Phaser (47). The crystal belongs to another space group and has two monomers in the asymmetric unit. Several cycles of manual modeling and refinement using the program Buster (48) were required to obtain the final structure. Noncrystallographic symmetry was used in initial refinement steps, and the 1.9-Å resolution structure was used as reference model. The final structure shows Rwork and Rfree of 22.7 and 24.9%, respectively, with 95.8 and 3.9% of the residues in the most favored and allowed regions of the Ramachandran plot. Only four residues are in the disallowed regions of the Ramachandran plot. In the structure, one monomer comprises residues 12 to 430 and 435 to 519, while the second monomer has a few disordered short peptides and comprises residues 12 to 150, 153 to 301, 304 to 340, 344 to 364, 366 to 473, and 475 to 516. Final coordinates were deposited in the Protein Data Bank with accession codes 6RTG and 6RTH for the UDP-bound and ligand-free YGTG, respectively. The DALI server (49) was used to analyze structural homology. The PyMOL Molecular Graphics System, version 2.3.0, Schrödinger LLC was used to prepare the figures.
NanoLC-MS/MS analysis
Ten micrograms of Rab5A or 8 μg of Rab31 was incubated for 4 hours at 21°C, followed by overnight incubation at 4°C with 400 μM NAD+ and 8.5 μM YARTA. Afterward, the sample was reduced with 50 mM DTT and alkylated with 120 mM iodoacetamide before SDS-PAGE. For YGT, 10 μg of Rab5A was incubated for 4 hours at 21°C. Then, the reaction mix was incubated overnight at 4°C with 10 μM UDP-GlcNAc and 500 nM YGTG. For cleavage analysis, YART1–632 and YGT1–1998 (each 4 μg of protein) were incubated for 1 hour at 37°C with 10 mM InsP6, followed by SDS-PAGE.
The excised gel bands were destained with 30% acetonitrile in 0.1 M NH4HCO3 (pH 8), shrunk with 100% acetonitrile, and dried in a vacuum concentrator (Concentrator 5301, Eppendorf, Germany). Digests were performed with 0.1 μg of trypsin respectively elastase per gel band overnight at 37°C in 0.1 M NH4HCO3 (pH 8). After removing the supernatant, peptides were extracted from the gel slices with 5% formic acid, and extracted peptides were pooled with the supernatant.
Nano liquid chromatograyphy–tandem MS (NanoLC-MS/MS) analyses were performed on an Orbitrap Fusion (Thermo Fisher Scientific) equipped with an EASY-Spray Ion Source and coupled to an EASY-nLC 1000 (Thermo Fisher Scientific). Peptides were loaded on a trapping column (2 cm by 75 μm ID, PepMap C18, 3-μm particles, 100-Å pore size) and separated on an EASY-Spray column (50 cm by 75 μm ID, PepMap C18, 2-μm particles, 100-Å pore size) with a 30-min linear gradient from 3 to 40% acetonitrile and 0.1% formic acid.
Both MS and MS/MS scans were acquired in the Orbitrap analyzer with a resolution of 60,000 for MS scans and 15,000 for MS/MS scans. A mixed ETD (electron transfer dissociation)/HCD (high-energy collision dissociation) method was used. HCD fragmentation was applied with 35% normalized collision energy. For ETD, calibrated charge-dependent ETD parameter was applied. A top-speed data-dependent MS/MS method with a fixed cycle time of 3 s was used. Dynamic exclusion was applied with a repeat count of 1 and an exclusion duration of 10 s; singly charged precursors were excluded from selection. Minimum signal threshold for precursor selection was set to 50,000. Predictive AGC (automatic gain control) was used with a target value of 2 × 105 for MS scans and 5 × 104 for MS/MS scans. EASYIC was used for internal calibration.
Mascot Distiller 2.5 was used for raw data processing and generating peak lists, essentially with standard settings for the Orbitrap (high/high settings). Mascot Server 2.5 was used for database searching with the following parameters: peptide mass tolerance, 10 parts per million; MS/MS mass tolerance, 0.02 Da; enzyme, “trypsin” with three missed cleavage sites allowed for trypsin or “none” for elastase; fixed modification, carbamidomethyl (C); variable modifications, Gln->pyroGlu (N-terminal Q), oxidation (M), HexNAc (ST), and ADP-ribosylation (NQ). Database searching was performed against a custom database containing the protein sequences of interest.
Statistics
Results are presented as means ± SD of at least three independent experiments (unless otherwise stated). Student’s t test was applied when two groups with normal distribution had to be compared. Statistical analyses were performed using GraphPad Prism version 5.04. P < 0.05 was considered statistically significant and marked with an asterisk (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
We thank O. Wunderlich and V. Kreiner for excellent technical assistance and Y. Belyi for the support in yeast work. Funding: This study was supported by the Deutsche Forschungsgemeinschaft (DFG) AK 784 6/16-4 (to K.A.), CRC 746 (to K.A. and C.H.), and the DFG under Germany’s Excellence Strategy (CIBSS–EXC-785 2189–Project ID 390939984 to C.H.). Author contributions: G.S.O. designed and performed the experiments, analyzed the data, and wrote the paper; C.W., X.B., W.-C.K., and C.H. performed the structure determination and analysis; C.W. and C.H. contributed to the writing of the manuscript; B.S. and P.J.K.A. designed and performed the experiments and analyzed the data; P.P., T.J., and C.S. designed the experiments and analyzed the data; A.S. performed the MS analyses and analyzed the data; and K.A. supervised the study, designed the experiments, analyzed the data, and wrote the manuscript. All authors discussed the results and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaaz2094/DC1
Fig. S1. Model of the domain architecture of YART and YGT, in comparison to C. difficile TcdB and C. perfringens TpeL.
Fig. S2. Processing, expression, and enzyme activity of YART and YGT.
Fig. S3. Topological representation of YGTG and potassium ion coordination in the ligand-bound YGTG structure.
Fig. S4. C-terminal loop of the glycosyltransferase domain encloses the nucleotide-binding site in the UDP-bound state and leaves it open in the ligand-free state.
Fig. S5. ETD MS/MS spectra.
Fig. S6. YART- and YGT-induced modifications of Rab31.
Table S1. Data collection and refinement statistics.
Table S2. Proteins structurally homologous to YGTG as identified by DALI and showing a Z score higher than 10.
Table S3. Genomic DNA, primer, gblocks, plasmids, and vectors.
Table S4. Buffer and conditions of protein purification.
REFERENCES AND NOTES
- 1.Aktories K., Schwan C., Jank T., Clostridium difficile toxin biology. Annu. Rev. Microbiol. 71, 281–307 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Voth D. E., Ballard J. D., Clostridium difficile toxins: Mechanism of action and role in disease. Clin. Microbiol. Rev. 18, 247–263 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth H., Pfeifer G., Hofmann F., Maier E., Benz R., Aktories K., Low pH-induced formation of ion channels by Clostridium difficile toxin B in target cells. J. Biol. Chem. 276, 10670–10676 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Egerer M., Giesemann T., Jank T., Satchell K. J., Aktories K., Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 282, 25314–25321 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Reineke J., Tenzer S., Rupnik M., Koschinski A., Hasselmayer O., Schrattenholz A., Schild H., von Eichel-Streiber C., Autocatalytic cleavage of Clostridium difficile toxin B. Nature 446, 415–419 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K., Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375, 500–503 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Nagahama M., Ohkubo A., Oda M., Kobayashi K., Amimoto K., Miyamoto K., Sakurai J., Clostridium perfringens TpeL glycosylates the Rac and Ras subfamily proteins. Infect. Immun. 79, 905–910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson S., Williams P., Yersinia virulence factors - a sophisticated arsenal for combating host defences. F1000Res 5, 10.12688/f1000research.8466.1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rucker R. R., Redmouth disease of rainbow trout (Salmo gairdneri). Bull. Off Int. Epizoot 65, 825–830 (1966). [PubMed] [Google Scholar]
- 10.Jank T., Eckerle S., Steinemann M., Trillhaase C., Schimpl M., Wiese S., van Aalten D. M. F., Driever W., Aktories K., Tyrosine glycosylation of Rho by Yersinia toxin impairs blastomere cell behaviour in zebrafish embryos. Nat. Commun. 6, 7807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Park M., Tam J., Auger A., Beilhartz G. L., Lacy D. B., Melnyk R. A., Translocation domain mutations affecting cellular toxicity identify the Clostridium difficile toxin B pore. Proc. Natl. Acad. Sci. U.S.A. 111, 3721–3726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lüscher B., Bütepage M., Eckei L., Krieg S., Verheugd P., Shilton B. H., ADP-Ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 118, 1092–1136 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Jank T., Belyi Y., Aktories K., Bacterial glycosyltransferase toxins. Cell. Microbiol. 17, 1752–1765 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Reinert D. J., Jank T., Aktories K., Schulz G. E., Structural basis for the function of Clostridium difficile toxin B. J. Mol. Biol. 351, 973–981 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Pruitt R. N., Chumbler N. M., Rutherford S. A., Farrow M. A., Friedman D. B., Spiller B., Lacy D. B., Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J. Biol. Chem. 287, 8013–8020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziegler M. O., Jank T., Aktories K., Schulz G. E., Conformational changes and reaction of clostridial glycosylating toxins. J. Mol. Biol. 377, 1346–1356 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Chumbler N. M., Rutherford S. A., Zhang Z., Farrow M. A., Lisher J. P., Farquhar E., Giedroc D. P., Spiller B. W., Melnyk R. A., Lacy D. B., Crystal structure of Clostridium difficile toxin A. Nat. Microbiol. 1, 15002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jank T., Bogdanović X., Wirth C., Haaf E., Spoerner M., Böhmer K. E., Steinemann M., Orth J. H. C., Kalbitzer H. R., Warscheid B., Hunte C., Aktories K., A bacterial toxin catalyzing tyrosine glycosylation of Rho and deamidation of Gq and Gi proteins. Nat. Struct. Mol. Biol. 20, 1273–1280 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Park J. B., Kim Y. H., Yoo Y., Kim J., Jun S.-H., Cho J. W., El Qaidi S., Walpole S., Monaco S., García-García A. A., Wu M., Hays M. P., Hurtado-Guerrero R., Angulo J., Hardwidge P. R., Shin J.-S., Cho H.-S., Structural basis for arginine glycosylation of host substrates by bacterial effector proteins. Nat. Commun. 9, 4283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esposito D., Günster R. A., Martino L., Omari K. E., Wagner A., Thurston T. L. M., Rittinger K., Structural basis for the glycosyltransferase activity of the Salmonella effector SseK3. J. Biol. Chem. 293, 5064–5078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciesla W. P. Jr., Bobak D. A., Clostridium difficile toxins A and B are cation-dependent UDP-glucose hydrolases with differing catalytic activities. J. Biol. Chem. 273, 16021–16026 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Genth H., Schelle I., Just I., Metal ion activation of Clostridium sordellii lethal toxin and Clostridium difficile toxin B. Toxins 8, 109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Urzo N., Malito E., Biancucci M., Bottomley M. J., Maione D., Scarselli M., Martinelli M., The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release. FEBS J. 279, 3085–3097 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Bucci C., Parton R. G., Mather I. H., Stunnenberg H., Simons K., Hoflack B., Zerial M., The small GTPase Rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70, 715–728 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Stenmark H., Parton R. G., Steele-Mortimer O., Lütcke A., Gruenberg J., Zerial M., Inhibition of Rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13, 1287–1296 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon N. C., Barbieri J. T., Exoenzyme S ADP-ribosylates Rab5 effector sites to uncouple intracellular trafficking. Infect. Immun. 82, 21–28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittinghofer A., Vetter I. R., Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Zerial M., McBride H., Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Villaseñor R., Kalaidzidis Y., Zerial M., Signal processing by the endosomal system. Curr. Opin. Cell Biol. 39, 53–60 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Flannagan R. S., Cosío G., Grinstein S., Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Ireton K., Rigano L. A., Dowd G. C., Role of host GTPases in infection by Listeria monocytogenes. Cell. Microbiol. 16, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbieri A. M., Sha Q., Bette-Bobillo P., Stahl P. D., Vidal M., ADP-ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect. Immun. 69, 5329–5334 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madan R., Krishnamurthy G., Mukhopadhyay A., SopE-mediated recruitment of host Rab5 on phagosomes inhibits Salmonella transport to lysosomes. Methods Mol. Biol. 445, 417–437 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Sarantis H., Balkin D. M., De Camilli P., Isberg R. R., Brumell J. H., Grinstein S., Yersinia entry into host cells requires Rab5-dependent dephosphorylation of PI(4,5)P2 and membrane scission. Cell Host Microbe 11, 117–128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Gabin A. G., Cammer M., Almazan G., Charron M., Larocca J. N., Role of rRAB22b, an oligodendrocyte protein, in regulation of transport of vesicles from trans Golgi to endocytic compartments. J. Neurosci. Res. 66, 1149–1160 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Dhar M. S., Virdi J. S., Strategies used by Yersinia enterocolitica to evade killing by the host: Thinking beyond Yops. Microbes Infect. 16, 87–95 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Ryckaert J., Bossier P., D’Herde K., Diez-Fraile A., Sorgeloos P., Haesebrouck F., Pasmans F., Persistence of Yersinia ruckeri in trout macrophages. Fish Shellfish Immunol. 29, 648–655 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Sulakvelidze A., Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: The ignored species. Microbes Infect. 2, 497–513 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Ribeiro A. A., Azcarate-Peril M. A., Cadenas M. B., Butz N., Paster B. J., Chen T., Bair E., Arnold R. R., The oral bacterial microbiome of occlusal surfaces in children and its association with diet and caries. PLOS ONE 12, e0180621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurst M. R., Jones S. A., Binglin T., Harper L. A., Jackson T. A., Glare T. R., The main virulence determinant of Yersinia entomophaga MH96 is a broad-host-range toxin complex active against insects. J. Bacteriol. 193, 1966–1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuchs T. M., Bresolin G., Marcinowski L., Schachtner J., Scherer S., Insecticidal genes of Yersinia spp.: Taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol. 8, 214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavallius J., Merrick W. C., Site-directed mutagenesis of yeast eEF1A. Viable mutants with altered nucleotide specificity. J. Biol. Chem. 273, 28752–28758 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Qi Y., Liang Z., Wang Z., Lu G., Li G., Determination of Rab5 activity in the cell by effector pull-down assay. Methods Mol. Biol. 1298, 259–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.G. Bricogne, E. Blanc, M. Brandl, C. Flensburg, P. Keller, P. Paciorek, P. Roversi, A. Sharff, O. Smart, C. Vonrhein, T. Womack, BUSTER version 2.10.2 (Cambridge, United Kingdom: Global Phasing Ltd., 2011).
- 49.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaaz2094/DC1
Fig. S1. Model of the domain architecture of YART and YGT, in comparison to C. difficile TcdB and C. perfringens TpeL.
Fig. S2. Processing, expression, and enzyme activity of YART and YGT.
Fig. S3. Topological representation of YGTG and potassium ion coordination in the ligand-bound YGTG structure.
Fig. S4. C-terminal loop of the glycosyltransferase domain encloses the nucleotide-binding site in the UDP-bound state and leaves it open in the ligand-free state.
Fig. S5. ETD MS/MS spectra.
Fig. S6. YART- and YGT-induced modifications of Rab31.
Table S1. Data collection and refinement statistics.
Table S2. Proteins structurally homologous to YGTG as identified by DALI and showing a Z score higher than 10.
Table S3. Genomic DNA, primer, gblocks, plasmids, and vectors.
Table S4. Buffer and conditions of protein purification.


