Stratified assembly of active components drives a tight huddle of proteins on the membrane.
Abstract
Recent in vivo studies reveal that several membrane proteins are driven to form nanoclusters by active contractile flows arising from localized dynamic patterning of F-actin and myosin at the cortex. Since myosin-II assemble as minifilaments with tens of myosin heads, one might worry that steric considerations would obstruct the emergence of nanoclustering. Using coarse-grained, agent-based simulations that account for steric constraints, we find that the patterns exhibited by actomyosin in two dimensions, do not resemble the steady-state patterns in our in vitro reconstitution of actomyosin on a supported bilayer. We perform simulations in a thin rectangular slab, separating the layer of actin filaments from myosin-II minifilaments. This recapitulates the observed features of in vitro patterning. Using super resolution microscopy, we find evidence for such stratification in our in vitro system. Our study suggests that molecular stratification may be an important organizing feature of the cortical cytoskeleton in vivo.
INTRODUCTION
The transient nanoclustering of many cell surface molecules is strongly influenced by interaction with the actomyosin cortex (1–8), a thin layer of actin cytoskeleton and myosin motors (nonmuscle myosin-II) that is juxtaposed with the membrane bilayer. While the thickness of the cortical actin layer has been measured to be around 250 nm [in HeLa cells (9)], its ultrastructure is as yet poorly defined, although evidence suggests that it comprises both dynamic filaments (3) and an extensively branched static meshwork (10). As a consequence, the action of myosin motors on the actin filaments at the cortex drives the local clustering of cell surface proteins that bind to it (1–3).
This layered structure of a multicomponent, asymmetric bilayer juxtaposed with a thin cortical actomyosin layer forms the basis for a model of the cell surface as an active composite (1, 3, 5). In these earlier studies, we had used a coarse-grained description based on active hydrodynamics (11), which is agnostic to molecular details. Recently, it has been shown that the statistics and dynamics of clustering are recapitulated in a minimal in vitro reconstitution of a supported bilayer in contact with a thin layer of short actin filaments and myosin-II minifilaments (both of skeletal muscle origin), driven by the hydrolysis of adenosine triphosphate (ATP) (12, 13). The success of this minimal setup motivates us to revisit our continuum hydrodynamic description from a more molecular, albeit coarse-grained, standpoint.
Nonmuscle myosin-II, which also assemble as minifilaments (14–17) consisting of around 30 myosin heads, are rather bulky structures (16). Hence, a concern could be that steric considerations might obstruct the emergence of membrane protein nanoclustering by this contractile actomyosin mechanism.
To address this, we perform a coarse-grained, agent-based Brownian dynamics simulation of a mixture of polar active filaments built from “actin monomers” and myosin-II minifilaments comprising multiple “myosin heads.” Our coarse-grained model respects the relative sizes of the individual constituents and hence incorporates crucial steric effects, in contrast to most recent in silico studies involving actomyosin assemblies (18–23). We study the phase diagram characterizing actomyosin patterns upon changing F-actin length, myosin minilament, concentration, and the strength of the contractile force exerted by individual myosin-II motors on actin laments. We compare the simulated patterns with the steady-state patterns obtained in an in vitro reconstitution of a thin layer of actomyosin on a supported bilayer first described in (12). We first start with an agent-based model in a strictly two-dimensional (2D) geometry; this has been the geometry of choice in a variety of studies (3, 4, 18–23). We find that in such a strictly 2D setting, steric effects are a serious impediment to the development of the actomyosin patterns (asters) that enable membrane protein clustering seen both in the in vivo (1–3, 5) and in vitro (12) experiments. The patterns obtained do not resemble the steady-state patterns observed in the in vitro reconstitution system. We then perform simulations in a thin rectangular slab, allowing the separation of a layer of actin laments from those of myosin-II minilaments. This simulation recapitulates the in vitro patterning and consequently allows the potential for membrane protein clustering by the contractile platforms. Using super-resolution microscopy, we look for and find direct evidence for a layering or stratification of actin and myosin-II in the in vitro system. As observed in an altogether different in vitro reconstitution of a filament-motor system, a necessary criterion to drive nonequilibrium patterning is the hierarchical assembly of the components (24). Our study suggests the possibility that molecular stratification (25, 26) may be an important organizing feature of the cortical cytoskeleton in vivo.
MODEL
Agent-based simulations of F-actin and myosin-II minifilaments
We model both F-actin and myosin-II minilaments (15, 17) as coarse-grained semiflexible polymers using a bead-spring representation, with steric repulsion (Fig. 1A). We represent the actin filament as a linear polymer, while the myosin-II minifilament as a polymer having a bipolar structure with side branches, which depict a collection of myosin head domains attached to a backbone made up of tail domains that form an inert core (27, 28). We assign the same diameter for all the beads σ for simplicity, irrespective of whether it is a subunit of F-actin or myosin minifilament. Consistency with previously reported dimensions (12, 15, 18, 22, 29) implies that one F-actin bead corresponds to ≈40 G-actin monomers and one myosin head bead corresponds to ≈3 to 4 myosin-II heads. The beads (monomers) are connected by harmonic bonds. For convenience, we use the same stretch spring constant for all bonds. In addition, changing the angle between two consecutive bonds is penalized, effectively introducing a bending stiffness. We choose appropriate values for these elastic constants to ensure that the actin filaments have a persistence length larger than 16 μm (30) and that the myosin minifilaments have flexible heads attached to a very rigid backbone (27). We keep track of the polarity of F-actin by labeling the terminal beads in each filament by a plus or barbed end (+) and minus end (−). The dynamics of the beads are described by overdamped, Brownian dynamics equations (see the Supplementary Materials for details).
Fig. 1. Ingredients of agent-based simulations in 2D and in vitro reconstitution setup.
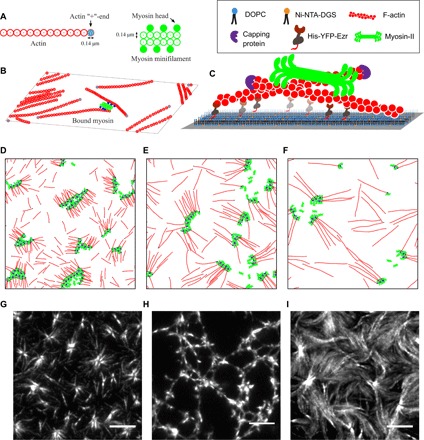
(A) Schematic of actin filaments (red) and myosin minifilaments (green) used in the agent-based model, with real-space dimensions indicated. One F-actin bead corresponds to 40 G-actin monomers, and one myosin bead corresponds to ∼3 to 4 heads. (B) Schematic showing a collection of F-actin and myosin minifilaments in 2D, with myosin motors bound to two actin filaments (bound myosin heads are colored blue). (C) Schematic of the in vitro reconstitution system showing the hierarchical assembly of an SLB, linker protein [His-YFP-Ezrin (HYE)], capped actin filaments, and muscle myosin-II. (D to F) Typical simulation snapshots of different aster configurations at steady state, observed in the strictly 2D simulations, as a function of increasing F-actin length la: (D) la = 2.32 μm, (E) la = 4.84 μm, and (F) la = 719 μm. The (+)-ends of the actin filaments are colored black. (G to I) Typical aster configurations at steady state observed in in vitro experiments: isolated asters, connected asters, and aster bundles, as a function of increasing F-actin length: (G) la = 2 ± 1 μm, (H) la = 3 ± 1 .5 μm, and (I) la = 8 ± 3 μm. Scale bars, 5 μm.
The myosin head beads experience an attractive interaction with the F-actin beads, modeled by a Morse potential where the strength of a myosin-actin bond corresponds to the typical change in free energy associated with hydrolysis of one molecule of ATP (31). In addition, each motor head binds to an actin lament without any geometric restriction, applies an active force fa on actin to displace itself in direction of the (+)-end of actin by sliding the actin in the opposite direction, and eventually unbinds. The binding/unbinding of each motor follows a simple Poisson kinetics (29). Note that we allow a coarse-grained myosin bead to interact with many actin filaments. In addition to this, we allow unbinding of full myosin minifilaments, implemented by allowing the completely detached myosin minifilaments to escape from the medium and to reappear at a different location at a rate km while keeping the mean number of bound myosins fixed.
Our control parameters in the simulation are as follows: (i) length of actin filaments la, which we vary in the range of 2 to 9 μm (we have kept the length of the myosin-II minifilament fixed at ≈1 μm), (ii) magnitude of active force fa (related to the number of phosphorylated heads) for which we use values in the range of 1 to 9 pN per myosin head bead (18, 22, 29), and (iii) concentrations of myosin-II (cm) and F-actin (ca). We have introduced separate unbinding and binding rates ku and kb, respectively, for the myosin head beads (29). All other simulation parameters are held fixed across simulation runs. For other relevant details, see Materials and Methods, and for a full description of the simulation model, the relevant parameters, and their values, see table S1.
In vitro reconstitution experiments
The in vitro reconstitution system consists of a thin actomyosin layer atop a supported lipid bilayer (SLB), as described in our earlier work (12). Briefly, we prepare functionalized lipid membranes, followed by hierarchical addition of actin-binding linker protein [His-YFP-Ezrin (HYE)], prepolymerized actin filaments together with control of actin length by capping proteins, myosin II motors, and limited ATP (100 μM) (Fig. 1C). Actin filaments undergo spatial reorganization in the presence of ATP-fueled myosin activity and reach a static steady state as a consequence of steady ATP depletion (movie S2). We record these steady-state patterns (asters) using total internal reflection fluorescence (TIRF) microscopy (see Materials and Methods). We have systematically tuned the actin filament density, actin filament length, and myosin density and have recorded three distinct actomyosin patterns: isolated asters, connected asters, and aster bundles (Fig. 1, G to I).
RESULTS
Simulations in a strictly 2D layer
We start with the agent-based simulations in a strictly 2D actomyosin system, i.e., when both F-actin filaments and myosin-II motors reside in the same plane. As expected, steady-state configurations displayed by the rigid F-actin filaments alone (with no myosin) fall into three broad classes, determined by an interplay between the filament length la and the filament concentration ca (32, 33): (i) dilute, when the rods are far apart, ; (ii) semi-dilute, when the rods entangle, [the Onsager concentration for isotropic-nematic transition (34)]; and (iii) concentrated, when the rods start to align, c > c*. Here, we restrict ourselves to the dilute and semi-dilute regimes of F-actin concentrations. This corresponds to the regime spanned in the in vitro reconstitution experiment (12). Furthermore, this regime best allows us to probe the fine structure of the individual contractile platforms generated by actomyosin.
The initial configurations for the Brownian dynamics simulation of the full actomyosin system are chosen from an ensemble where the actin and myosin filaments are uniformly distributed, both in center-of-mass position and orientation, at a fixed concentration of filaments. Typically, after a time t ≈ 15 to 30 s, as a result of the binding, active pushing, and unbinding of myosin beads, the actin filaments and myosin are driven to form clusters, consistent with our earlier theoretical studies (3), recent simulations (18–20, 29), and in vitro experiments (12, 35–38).
Figure 1 (D to F) shows typical configurations of clusters observed in simulations, as one increases the length of actin filaments. In comparison, the clusters formed in the in vitro reconstitution experiment (Fig. 1, G to I) go from being isolated asters to connected asters to bundled asters, as the length of actin filaments is increased from 2 to 9 μm, as reported earlier (12). However, note that in terms of the molecular patterning in the isolated clusters (Fig. 2), there are important differences between the configurations obtained in the strict 2D simulations and in vitro experiments.
Fig. 2. Comparison of cluster patterns in the strictly 2D simulations and in vitro experiments.
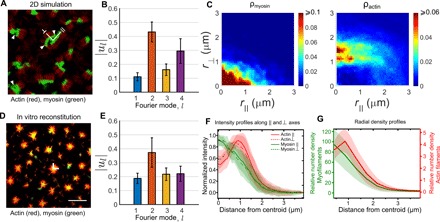
(A) Patterning of actomyosin clusters in the dilute limit obtained from agent-based simulations in the strictly 2D geometry. These patterns have been obtained by coarse graining the density of actin (red) and myosin (green) on a 2D grid with spacing 2σ. The relevant parameters are la = 2.32 μm, ca = 1.2 nM, cm/ca = 0.4, fa = 2 pN, and ku/kb = 0.1. Note that there are no overlapping regions (yellow) as a result of strong steric effects. (B) The shapes of most of the clusters observed in the simulations are strongly anisotropic, almost rectangular, and some even with appearance of bi-stripes [marked by white arrows in (A)], as determined from a Fourier analyses of the shape (Materials and Methods). Note the large amplitude of the l = 2 and l = 4 modes compared to the l = 1 mode, averaged over N = 19,718 clusters. (C) Density profiles of myosin and actin along the ⊥ and ∥ directions of the clusters also show strong anisotropy. Data averaged over N = 24,707 clusters. (D) Patterning of actomyosin clusters in the dilute aster limit obtained from the in vitro experiments, with actin (red), myosin (green), and a distinct region of overlap (yellow). Scale bar, 10 μm. (E) In contrast to the strictly 2D simulations, the shapes of most of the clusters are largely circular as shown by the relatively elevated amplitude of the l = 1 mode compared to the higher modes. Data averaged over N = 561 clusters. (F) Normalized intensity profile (I/Imax) extracted from TIRF images of actin and myosin along orthogonal directions (∥ and ⊥) confirming isotropy of the asters. Data averaged over 20 asters across four different experiments. (G) Circularly averaged radial density profiles of actin and myosin show enrichment of the latter at the core (calibration of TIRF images in fig. S4). Data averaged over 49 asters across four different experiments. Error bars denote SD.
Most of the actomyosin clusters observed under these simulation conditions are anisotropic, many of them forming rectangular bi-stripes, with actin on one side and myosin on the other (Fig. 2A). On the other hand, the clusters in the in vitro experiments are isotropic or circular, with myosin at the core and actin filaments extending radially outward, with the (+)-end oriented toward the core (Fig. 2, D and G). The difference between the strict 2D simulations and in vitro experiments is apparent in the Fourier spectrum of the cluster shape (Fig. 2, B and E) and in the density profiles of myosin and actin (Fig. 2, C and F). When we remove the steric interactions in our simulations, the resulting patterns begin to resemble the experimental patterns (fig. S1). It is clear that the strong steric hindrance of the bulky myosin filaments gets in the way of generating the actomyosin patterns that are potentially responsible for cell surface clustering and bringing membrane proteins in close proximity (3, 12).
Simulations in a stratified actomyosin layer
To circumvent the frustration arising from the unavoidable steric constraints, we perform simulations in a stratified actomyosin layer. Here, we prepare the stratified actomyosin layer by distributing actin filaments along the xy plane at z = 0 while having myosin minifilaments move atop it and within a rectangular slab of thickness zs = 3σ by a harmonic potential centered at z0 = zs/2.
We find that stratification releases the frustration from steric constraints and the steady-state (fig. S2 and movie S1) patterns of actomyosin start resembling the patterns seen in vitro (Fig. 3, A to C). The clusters are now more circular, with myosin concentrated at the cores closely resembling the in vitro patterning (Fig. 3, B and C). Encouraged by this, we study the change in steady-state patterns as a function of actin and myosin concentrations and length of actin filaments. For this, we set fa = 2 pN and ku/kb = 0.2 and work with a large enough kb, the myosin binding rate on actin, so that the eventual remodeling of the aster configurations happens at time scales much larger than typical simulation runs. The resulting phase diagram is qualitatively similar to the one in (12).
Fig. 3. Simulations with stratification.
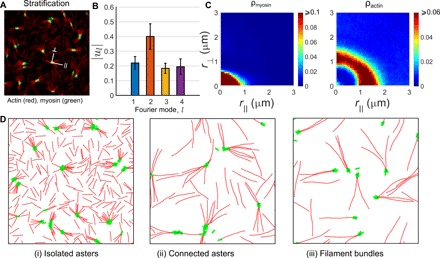
(A) Patterning of actomyosin clusters in the dilute limit obtained from simulations in the stratified geometry. The parameters are la = 2.32 μm, ca = 1.5 nM, cm/ca = 0.2, fa = 2 pN, and ku/kb = 0.2. In this case, we see a colocalization of myosin and actin when projected in the x-y plane. (B) As in the in vitro study, the clusters are more circular, as shown by the relatively elevated amplitude of the l = 1 mode compared to the higher modes. Data averaged over N = 33,518 clusters. (C) Density of myosin and actin along the ⊥ and ∥ directions of the clusters show isotropic profiles, with myosin enriched at the cores and the relative concentration of actin enriched at the periphery. Data averaged over N = 44,537 clusters. (D) Representative snapshots of the different phases obtained on increasing actin filament length, (i) la = 2.32 μm, (ii) la = 6.02 μm, and (iii) la = 8.37 μm, showing isolated asters, connected asters, and filaments bundles.
At the concentrations of actin under study, the steady-state patterns are determined by a competition between the nematic alignment, parametrized by the Frank splay elastic constant K1, and a splay instability induced by myosin contractility (39). The splay elastic constant depends linearly on the filament concentration and filament aspect ratio (40); thus, when the filament concentration is low and the filament lengths are small, ≤6 μm , the splay instability due to myosin contractility dominates. As shown in (39), this leads to the formation of an isolated aster phase, with the (+)-ends of actin filaments in each aster pointing inward.
As one increases the length or concentration of actin filaments, the system transitions from an isolated aster phase in the dilute regime to a semi-dilute regime. To understand the structures that form in the semi-dilute regime, we first note that because of the active contractile stresses, there emerge two new length scales—the aster size and the aster separation 𝓁*—both functions of the filament length, concentration, and contractility. As we increase the actin filament length (or concentration) while keeping the other control parameters fixed, we obtain configurations where the long filaments span across the asters. Upon increasing myosin density, these spanning filaments via myosin result in connected asters with altered polarity patterns, which we will discuss below.
Upon increasing the length or concentration of actin filaments further, the aligning tendency of the filaments starts to dominate over the contractility-induced splay instability. This is because the splay elastic constant K1 depends linearly on the filament aspect ratio and filament concentration (40). This leads to the formation of filament bundles of actin beyond la > 6 μm, amid a dilute dispersion of bundled asters. This is the active analog of the entangled phase in passive mixtures of filaments and crosslinkers (41).
To display these changes in filament-motor configurations as a phase diagram, we need to arrive at a suitable order parameter. Because the various aster configurations are accompanied by filament polarity sorting, an appropriate order parameter can be extracted from the behavior of the pair correlation function gm−(r) between the bound myosin heads and the (−)-end of actin filaments (Fig. 4A and fig. S3) (see Materials and Methods for definition). In the isolated aster phase, myosin heads are proximal to the (+)-ends of actin filaments; consequently, the correlation between the myosin heads and the (−)-end of the filaments will peak at a separation r = la. In the connected aster phase, a fraction of the myosin heads that connect across asters is proximal to the (−)-end of actin; this will give rise to a new peak at r = σ, the minimum separation of two beads. Last, in the filament bundle phase, the peak height at r = σ relative to that at r = la decreases. Defining R ≡ gm−(r = σ), we record a nonmonotonic variation as one sweeps across the three phases upon tuning la and cm/ca (fig. S3). This allows us to plot the phase diagrams shown in Fig. 4 (B and C).
Fig. 4. Characterization of the phases observed in our simulations of stratified actin filaments and myosin minifilaments.
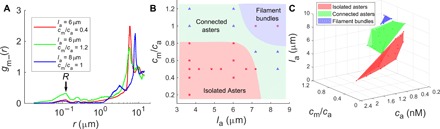
(A) Plots of gm–(r) for three different cases depicting three different phases observed. (B) A 2D phase diagram showing three different phases observed in our simulations on the cm/ca versus la plane. The squares correspond to cases where R ≤ Rc, and triangles correspond to cases with R > Rc, where Rc is a suitable value (0.15) fixed to draw the phase boundaries. The bundle phase is confirmed by calculating the orientational alignment of the bound actin filaments around the myosin core, defined by ϕ (see Materials and Methods), which changes from 0 in a disordered configuration to 1 in a fully aligned configuration. (C) Full phase diagram of the different phases in the space of length of actin (la), concentration of actin (ca), and ratio of myosin to actin concentrations (cm/ca). The phase points have been assigned on the basis of plots as in (B).
The phase diagram as a function of actin filament length and concentration of actin and myosin (Fig. 4C) bears a qualitative resemblance to the in vitro phase diagram in (12). This resemblance and the similarity of the actomyosin patterning in asters suggest that stratification releases the frustration imposed by steric constraints. We next look for evidence of such a release in the in vitro experiments, especially at the cores of the asters observed in Fig. 2D.
Stratification revealed by STED microscopy
To obtain evidence of stratification in our in vitro setup, we need to probe the configurations of actin and myosin transverse to the plane of the actomyosin layer. To this end, we perform 3D stimulated emission depletion (STED) microscopy on the in vitro system, prepared with Star-635–labeled actin and Star-580–labeled myosin-II, with a resolution of 70 nm (100 nm) in the lateral xy plane and 150 nm (180 nm) in the transverse z direction; for actin-635 (myosin-II-580), see the Supplementary Materials. Analysis of the actin and myosin-II signal along the z axis shows distinct stratified organization of actin and myosin.
We see that the periphery of the aster along the xy plane is relatively enriched in actin filaments, with few myosins attached (Fig. 5, A to D). The separation between the actin and myosin peak intensities at the periphery ranges from 0 to 100 nm, with a mean at 29 nm. On the other hand, the aster core is relatively enriched in myosin-II; the actin laments appear to protrude into the z direction, with myosin-II forming a cap over it. The separation between the actin protruding column and myosin-II at the core ranges from 150 to 800 nm, with a mean at 393 nm (Fig. 5, D and E).
Fig. 5. Stratified organization of actin and myosin in asters.
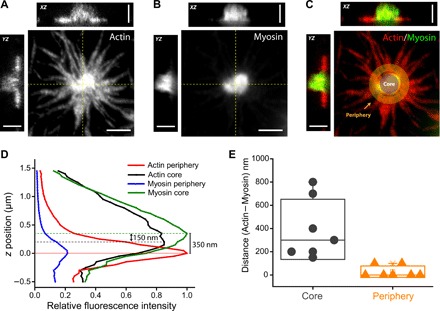
(A to C) 3D STED microscopy of an actomyosin aster showing average intensity projection and XZ/YZ intensity profile of actin (A), myosin (B), and the merge (C) demarcating the core and the periphery used for subsequent measurements. Scale bars, 2 μm (XY) and 1.5 μm (XZ/YZ). (D) Intensity plot showing the relative intensities (I/Imax) of actin and myosin along the z axis in the core (solid line) and periphery (dashed lines). Actin intensity peak in the periphery was arbitrarily chosen as position 0. (E) Distance between actin and myosin intensity peaks [as shown in (D)] in the core and the periphery, quantified across seven different asters.
Our STED observations demonstrate a relative separation between the actin and myosin, both at the periphery and at the core. We also observe that the myosin enriched at the core draws in the actin laments, resulting in a protrusion in the transverse direction (with respect to the bilayer). This can only arise in a stratified configuration, where myosin is layered atop the actin layer. This coincides with our operational definition of stratification in the earlier section. The results obtained from the STED measurements are an experimental confirmation of the stratification of the actomyosin layer.
DISCUSSION
We have presented results of detailed agent-based simulations of actomyosin assemblies that account for realistic steric restrictions, both in strict 2D and in stratified geometries. A key observation is that steric effects, often ignored in these simulations, can markedly affect the steady-state configurations at short scales. We find that to recover the steady-state configurations observed in in vitro experiments (12), it is necessary to stratify the F-actin and myosin-II minifilaments. We have constructed a phase diagram as a function of the concentrations of F-actin, myosin motors, and the length of actin filaments in this actively driven system, which bears a satisfying resemblance to the steady-state phase diagram reported in (12). The generation of this phase diagram, resembling the in vitro system, is a major achievement of the simulations developed here. This reveals many of the key ingredients at the microscopic level and emergent mesoscale, which recapitulate the in vitro system.
Motivated by our agent-based simulations, we next look for evidence of stratification in an in vitro reconstitution of actin and myosin on a supported bilayer. Using super-resolution microscopy, we find direct evidence for stratification. We believe that these observations have a bearing on the possible role of molecular stratification within the active cortex at the cell surface.
Molecular stratification and its regulation have been invoked as an organizing principle in the elaboration of the focal adhesion complex (25, 26) at the cell surface. In the context of the lateral nanoclustering of the outer cell surface glycosylphosphatidylinositol (GPI)–anchored proteins, we demonstrated that the lipid-tethered protein—in conjunction with cholesterol, sphingolipids, and inner leaflet phosphatidylserine—forms a molecular bridge transverse to the asymmetric bilayer membrane, which helps connect the outer cell surface to the actomyosin cortex (42). Here, we argue that a transverse organization or stratification continues into the actomyosin cortex, thereby facilitating lateral nanoscale proximity of cell membrane proteins, which engage with the cortex. Our study shows that in the absence of stratification, steric effects would preclude this lateral nanoscale proximity. This has functional implications, because, for example, cholesterol-dependent GPI-anchored protein nanoclustering is necessary for promoting integrin function (43, 44). We take up some of these issues in a later study.
The molecular stratification that we observe is a consequence of steric effects. In the cellular context, this is likely to be a key organizing element in many types of hierarchical self-assembly and organization processes (25, 26). Stratification of the cortex opens up a new dimension, allowing the cortex more room to display its fine structure in terms of a diversity of cortical elements with a variety of structural motifs and modes of stress generation.
MATERIALS AND METHODS
Simulation details and units
In the agent-based model, we measure energy in units of kBT at the physiological temperature of 310 K, length in units of bead diameter σ ≈ 0.14 μm, and time in units of τ = ζσ2/kBT ≈ 10−2 s, where ζ is the friction coefficient of the medium associated with the drag on a monomer bead in the model (29). All the parameter values used are listed in table S1.
Equations of motion (see the Supplementary Materials) are integrated using standard Euler-Maruyama algorithm with the time step δt = 1.5 × 10−4 τ. The simulations are carried out up to 108 time steps (≈104 τ). The simulations took 1 × 107 to 2 × 107 steps to reach the steady state, which was identified by monitoring the autocorrelation functions of density and local orientation of the filaments (fig. S2 and movie S1). We perform simulations in 2D with all polymers restricted to the xy plane and also in a quasi-3D geometry, with a stratified layer of myosin minifilaments placed “on top” of the plane of actin filaments. Dimensions of the 2D box in which all the actin filaments are restricted are Lx = Ly = 28 μm. Our simulations, done with the SAMoS program (45), include steric hindrance among all polymers via a truncated Lennard-Jones potential.
Calculation of the order parameters for different phases
The pair correlation function gm−(r) is a two-point density-density correlation function defined as follows
where gm−(r) represents the probability to find a myosin head located at position rm and an actin (−)-end located at position r− within a thin annular region of radius r and thickness Δr. Nm and N− are the number of myosin heads and number of actin (−)-ends, respectively.
We calculate the local polar order parameter ϕ to quantify the orientational alignment of the bound actin filaments around a myosin core. We define ϕ to be the net orientation of actin filament segments (two consecutive actin beads bound by harmonic bond) found within 1 μm of the centroids of bound myosin minifilaments. The formal definition of ϕ for Ns actin segments is as follows (46)
where the angular brackets represent time averages and θk represents the orientation of the kth actin segment. In the isolated aster phase, ϕ is small (≤0.5) because bound actins nearly form a closed loop of orientations around a bound myosin. However, ϕ slowly increases in the connected aster phase because of nonuniform populations of actin (+)- and (−)-ends around bound myosin. Last, ϕ approaches unity in the phase dominated by actin bundles. This variation of ϕ has been used to confirm the phase boundary between the connected aster phase and the filament bundle phase of actin.
In vitro reconstitution system
Our reconstitution system consisted of thin actomyosin layers coupled to SLBs via linker proteins, as described in our earlier work (12). Briefly, actin and myosin were purified from fresh chicken skeletal muscles. HYE and capping protein were expressed in BL21EDE3* bacterial cells and purified by Ni2+ affinity and gel filtration chromatography. SLBs were prepared by fusing small unilamellar vesicles (SUVs) dioleoylphosphatidylcholine (DOPC) [98 mole percent (mol %)] and Ni-NTA-DGS 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (2 mol %) inside small artificial chambers glued on top of clean coverslips (purchased from Avanti Polar). Actin was polymerized by mixing dark and Atto 633 maleimide (AttoTech)–labeled G-actin in a 90:10 ratio, along with capping protein, ATP (Sigma-Aldrich), and 1× KMEH buffer to generate filaments of controlled average length. Atto 565 maleimide–labeled myosin minifilaments were added along with ATP on SLB-tethered actin filaments to study its effect on actin filament organization. The three distinct actomyosin patterns were obtained as a function of myosin density, F-actin density, and F-actin length, as previously reported.
Fourier analysis of shapes
To analyze the shapes of myosin clouds, we compute the Fourier spectrum of the shape boundaries (47). The outline contour (r) of any given shape with a closed boundary can be expanded in terms of Fourier series as follows
where r0 is the mean distance of any point on the boundary of the outline from the curve centroid, ul is the amplitude of the lth mode, and θ is the angle made by the radius vector of such a point with the reference axis. From such an expansion, we can determine the different mode amplitudes by taking a Fourier transform of r(θ) as follows
where Nc is the total number of points on the contour and , rj and θj being the length of radius vector and orientation, respectively, of the jth point on the contour. These modes capture specific features, for instance, l = 1 mode reports on the circular behavior, l = 2 captures the elliptic nature, l = 3 describes the triangular character, l = 4 describes the quadrateness, and so forth.
We use the above analysis to calculate the Fourier modes of the shapes observed in our experimental images. We work with images of isolated actomyosin clusters, which are subjected to a Gaussian filter to smoothen the boundaries. The properties of the shapes are computed using the regionprops function and their boundaries using bwboundaries function in MATLAB. We perform the Fourier analysis on the shapes within a certain size range (100 to 500 boundary pixels) and look at absolute values of first four harmonics of ul. We repeat the procedure of shape analysis on our simulated actomyosin structures. Note that the ul values (heights of bars) presented in Figs. 2 and 3 are mean amplitudes from samples collected over simulated snapshots or experimental regions of interest (ROIs), as the case may be, and the associated error bars represent the SDs across the samples.
TIRF imaging
Imaging was performed on a Nikon Ti Eclipse microscope quipped with a TIRF module coupled to Agilent monolithic laser combiner MLC400 (laser lines: 405, 488, 561, and 653 nm; Agilent Technologies). Images were collected with a 100 × 1.49 numerical aperture (NA) objective with an electron multiplying charge-coupled device camera (Evolve 512, Photometrics), yielding a pixel size of 103 nm. Multicolor, time lapse, and stream acquisition were controlled and recorded using the software μManager. Image analysis was performed in ImageJ (imagej.nih.gov/ij/).
3D STED nanoscopy
STED microscopy was performed on STAR-635–labeled F-actin and STAR-580–labeled myosin-II on an Abberior Expert Line 775 nm STED system. A pulsed 775-nm depletion laser and two pulsed excitation lasers (561 and 640 nm) were used to image myosin and actin, respectively. Spatial light modulator technology on the system allowed us to tune the STED beam between lateral and axial directions to improve xy and z resolutions. We characterized our fluorophores by measuring their full width at half maximum at different 775-nm power densities. We could achieve a resolution as good as ≈30 nm in xy and 60 nm in z. Actomyosin asters were imaged at 50% 3D settings, with a measured resolution of 70 and 100 nm in xy and 120 and 150 nm in z for actin-635 and myosin-II-580, respectively (fig. S5). Signal was collected with a 100 × 1.4 NA Olympus objective with an avalanche photodiode (APD) by Excelitas Technologies. Images were analyzed using ImageJ.
Supplementary Material
Acknowledgments
We thank S. Henkes and K. Weijer for many illuminating discussions. A.D. thanks School of Life Sciences, University of Dundee, UK, for hospitality and the Simons Foundation for funding. M.R. and S.M. thank J. Spudich for drawing attention to the role of steric interactions. We thank CIFF Imaging Facility at C-Camp, NCBS, for the use of the STED microscope. Funding: R.S. thanks UK BBSRC (award no. BB/N009789/1) for funding. S.M. and M.R. acknowledge a JC Bose Fellowship from DST (Government of India) and support from the NCBS–Max Planck Lipid Centre. S.M. acknowledges HFSP (RGP0027/2012) and Margadarshi Fellowship (IA/M/15/1/502018) from the Wellcome Trust–DBT India Alliance. Author contributions: M.R. and S.M. conceived the idea. A.D., R.S., and M.R. designed the simulation strategy. A.B., D.K., M.R., and S.M. designed the experimental strategy. A.D. and R.S. executed the simulations. A.B. and D.K. executed the experiments. All authors contributed to the analysis of the data and the writing of the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaay6093/DC1
Section S1. Simulation methods
Section S2. In vitro experiments
Table S1. Parameter values in real units with dimensionless values used in simulations.
Fig. S1. Description of agent-based model, active force versus velocity plot for a single myosin-II minifilament, and snapshots from simulation without steric interactions.
Fig. S2. Time series of aster density and aster strength showing fluctuations about steady state.
Fig. S3. Relative populations of F-actin (−)-ends near myosin-II heads help determine different aster and bundle phases.
Fig. S4. Density measurements of actin and myosin in our in vitro experiments.
Fig. S5. Characterization of the fluorophores on Abberior 775 STED nanoscope.
Movie S1. A representative time course from the simulation of myosin-II minifilaments and F-actin in the stratified geometry.
Movie S2. Time-lapse images from steady-state asters in our in vitro experiments.
REFERENCES AND NOTES
- 1.Rao M., Mayor S., Active organization of membrane constituents in living cells. Curr. Opin. Cell Biol. 29, 126–132 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., Mayor S., Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135, 1085–1097 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gowrishankar K., Ghosh S., Saha S., Mayor S., Rao M., Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 149, 1353–1367 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri A., Bhattacharya B., Gowrishankar K., Mayor S., Rao M., Spatiotemporal regulation of chemical reactions by active cytoskeletal remodeling. Proc. Natl. Acad. Sci. U.S.A. 108, 14825–14830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha S., Lee I.-H., Polley A., Groves J. T., Rao M., Mayor S., Diffusion of GPI-anchored proteins is influenced by the activity of dynamic cortical actin. Mol. Biol. Cell 26, 4033–4045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plowman S. J., Muncke C., Parton R. G., Hancock J. F., H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 102, 15500–15505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lingwood D., Simons K., Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010). [DOI] [PubMed] [Google Scholar]
- 8.van Zanten T. S., Cambi A., Koopman M., Joosten B., Figdor C. G., Garcia-Parajo M. F., Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 106, 18557–18562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chugh P., Clark A. G., Smith M. B., Cassani D. A. D., Dierkes K., Ragab A., Roux P. P., Charras G., Salbreux G., Paluch E. K., Actin cortex architecture regulates cell surface tension. Nat. Cell Biol. 19, 689–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morone N., Fujiwara T., Murase K., Kasai R. S., Ike H., Yuasa S., Usukura J., Kusumi A., Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J. Cell Biol. 174, 851–862 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchetti M. C., Joanny J. F., Ramaswamy S., Liverpool T. B., Prost J., Rao M., Simha R. A., Hydrodynamics of soft active matter. Rev. Mod. Phys. 85, 1143–1189 (2013). [Google Scholar]
- 12.Köster D. V., Husain K., Iljazi E., Bhat A., Bieling P., Mullins R. D., Rao M., Mayor S., Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc. Natl. Acad. Sci. U.S.A. 113, E1645–E1654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murrell M., Oakes P. W., Lenz M., Gardel M. L., Forcing cells into shape: The mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol. 16, 486–498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beach J. R., Shao L., Remmert K., Li D., Betzig E., Hammer J. A. III, Nonmuscle myosin II isoforms coassemble in living cells. Curr. Biol. 24, 1160–1166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonn-Segev A., Bernheim-Groswasser A., Roichman Y., Dynamics in steady state in vitro acto-myosin networks. J. Phys. Condens. Matter 29, 163002 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Billington N., Wang A., Mao J., Adelstein R. S., Sellers J. R., Characterization of three full-length human nonmuscle myosin II paralogs. J. Biol. Chem. 288, 33398–33410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finer J. T., Simmons R. M., Spudich J. A., Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature 368, 113–119 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Hiraiwa T., Salbreux G., Role of turnover in active stress generation in a filament network. Phys. Rev. Lett. 116, 188101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Q., Li J., Murrell M. P., Kim T., Balance between force generation and relaxation leads to pulsed contraction of actomyosin networks. Biophys. J. 115, 2003–2013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freedman S. L., Hocky G. M., Banerjee S., Dinner A. R., Nonequilibrium phase diagrams for actomyosin networks. Soft Matter 14, 7740–7747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo W., Yu C.-h., Lieu Z. Z., Allard J., Mogilner A., Sheetz M. P., Bershadsky A. D., Analysis of the local organization and dynamics of cellular actin networks. J. Cell Biol. 202, 1057–1073 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller C. J., Harris D., Weaver R., Ermentrout G. B., Davidson L. A., Emergent mechanics of actomyosin drive punctuated contractions and shape network morphology in the cell cortex. PLOS Comput. Biol. 14, e1006344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S., Wolynes P. G., Active contractility in actomyosin networks. Proc. Natl. Acad. Sci. U.S.A. 109, 6446–6451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez T., Chen D. T. N., DeCamp S. J., Heymann M., Dogic Z., Spontaneous motion in hierarchically assembled active matter. Nature 491, 431–434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F., Waterman C. M., Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Case L. B., Waterman C. M., Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melli L., Billington N., Sun S. A., Bird J. E., Nagy A., Friedman T. B., Takagi Y., Sellers J. R., Bipolar filaments of human nonmuscle myosin 2-A and 2-B have distinct motile and mechanical properties. eLife 7, e32871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spudich J., Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 106, 1236–1249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mak M., Zaman M. H., Kamm R. D., Kim T., Interplay of active processes modulates tension and drives phase transition in self-renewing, motor-driven cytoskeletal networks. Nat. Commun. 7, 10323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott A., Magnasco M., Simon A., Libchaber A., Measurement of the persistence length of polymerized actin using fluorescence microscopy. Phys. Rev. E 48, R1642–R1645 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Tran Q. H., Unden G., Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur. J. Biochem. 251, 538–543 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Käs J., Strey H., Tang J. X., Finger D., Ezzell R., Sackmann E., Janmey P. A., F-actin, a model polymer for semiflexible chains in dilute, semidilute, and liquid crystalline solutions. Biophys. J. 70, 609–625 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Höfling F., Munk T., Frey E., Franosch T., Entangled dynamics of a stiff polymer. Phys. Rev. E 77, 060904 (2008). [DOI] [PubMed] [Google Scholar]
- 34.P. G. De Gennes, J. Prost, The Physics of Liquid Crystals (Clarendon Press, 1993). [Google Scholar]
- 35.Alvarado J., Sheinman M., Sharma A., MacKintosh F. C., Koenderink G. H., Molecular motors robustly drive active gels to a critically connected state. Nat. Phys. 9, 591–597 (2013). [Google Scholar]
- 36.Linsmeier I., Banerjee S., Oakes P. W., Jung W., Kim T., Murrell M. P., Disordered actomyosin networks are sufficient to produce cooperative and telescopic contractility. Nat. Commun. 7, 12615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber L., Suzuki R., Krüger T., Frey E., Bausch A. R., Emergence of coexisting ordered states in active matter systems. Science 361, 255–258 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Sonal, Ganzinger K. A., Vogel S. K., Mücksch J., Blumhardt P., Schwille P., Myosin-II activity generates a dynamic steady state with continuous actin turnover in a minimal actin cortex. J. Cell Sci. 132, jcs219899 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Husain K., Rao M., Emergent structures in an active polar fluid: Dynamics of shape, scattering, and merger. Phys. Rev. Lett. 118, 078104 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Odijk T., Elastic constants of nematic solutions of rod-like and semi-flexible polymers. Liq. Cryst. 1, 553–559 (1986). [Google Scholar]
- 41.Borukhov I., Bruinsma R. F., Gelbart W. M., Liu A. J., Structural polymorphism of the cytoskeleton: A model of linker-assisted filament aggregation. Proc. Natl. Acad. Sci. U.S.A. 102, 3673–3678 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raghupathy R., Anilkumar A.A., Polley A., Singh P.P., Yadav M., Johnson C., Suryawanshi S., Saikam V., Sawant S. D., Panda A., Guo Z., Vishwakarma R. A., Rao M., Mayor S., Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 161, 581–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Zanten T. S., Cambi A., Koopman M., Joosten B., Figdor C. G., Garcia-Parajo M. F., Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 106, 18557–18562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalappurakkal J. M., Anilkumar A. A., Patra C., van Zanten T. S., Sheetz M. P., Mayor S., Integrin mechano-chemical signaling generates plasma membrane nanodomains that promote cell spreading. Cell 177, 1738–1756.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soft Active Matter on Surfaces (SAMoS); https://github.com/sknepneklab/SAMoS.
- 46.Weitz S., Deutsch A., Peruani F., Self-propelled rods exhibit a phase-separated state characterized by the presence of active stresses and the ejection of polar clusters. Phys. Rev. E Stat. Nonlin. Soft Matter. Phys. 92, 012322 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Chu F.-Y., Haley S. C., Zidovska A., On the origin of shape fluctuations of the cell nucleus. Proc. Natl. Acad. Sci. U.S.A. 114, 10338–10343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borau C., Kim T., Bidone T., García-Aznar J. M., Kamm R. D., Dynamic mechanisms of cell rigidity sensing: Insights from a computational model of actomyosin networks. PLOS ONE 7, e49174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weeks J. D., Chandler D., Andersen H. C., Role of repulsive forces in determining the equilibrium structure of simple liquids. J. Chem. Phys. 54, 5237–5247 (1971). [Google Scholar]
- 50.Erdmann T., Schwarz U. S., Stochastic force generation by small ensembles of myosin II motors. Phys. Rev. Lett. 108, 188101 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaay6093/DC1
Section S1. Simulation methods
Section S2. In vitro experiments
Table S1. Parameter values in real units with dimensionless values used in simulations.
Fig. S1. Description of agent-based model, active force versus velocity plot for a single myosin-II minifilament, and snapshots from simulation without steric interactions.
Fig. S2. Time series of aster density and aster strength showing fluctuations about steady state.
Fig. S3. Relative populations of F-actin (−)-ends near myosin-II heads help determine different aster and bundle phases.
Fig. S4. Density measurements of actin and myosin in our in vitro experiments.
Fig. S5. Characterization of the fluorophores on Abberior 775 STED nanoscope.
Movie S1. A representative time course from the simulation of myosin-II minifilaments and F-actin in the stratified geometry.
Movie S2. Time-lapse images from steady-state asters in our in vitro experiments.


