Extracellular vesicle Tspan8 levels in pretreatment serum from patients with NSCLC predict distant metastasis–free survival.
Abstract
Non–small cell lung cancer (NSCLC) is the most commonly diagnosed cancer and the leading cause of cancer death worldwide. More than half of patients with NSCLC die after developing distant metastases, so rapid, minimally invasive prognostic biomarkers are needed to reduce mortality. We used proteomics to identify proteins differentially expressed on extracellular vesicles (EVs) of nonmetastatic 393P and metastatic 344SQ NSCLC cell lines and found that tetraspanin-8 (Tspan8) was selectively enriched on 344SQ EVs. NSCLC cell lines treated with EVs overexpressing Tspan8 also exhibited increased Matrigel invasion. Elevated Tspan8 expression on serum EVs of individuals with stage III premetastatic NSCLC tumors was also associated with reduced distant metastasis–free survival, suggesting that Tspan8 levels on serum EVs may predict future metastasis. This result suggests that a minimally invasive blood test to analyze EV expression of Tspan8 may be of potential value to guide therapeutic decisions for patients with NSCLC and merits further study.
INTRODUCTION
Lung cancer is the most common cancer and cause of cancer-related death in men and women worldwide (1). More than 50% of patients diagnosed with lung cancer die within 1 year of diagnosis, and the 5-year survival rate is less than 19% (1). Non–small cell lung cancer (NSCLC) is the most prevalent form of lung cancer, and most patients with NSCLC have metastatic disease at their initial diagnosis (2). However, stage II to III NSCLC tumors are heterogeneous, and patients with the same disease stage can often have very different treatment outcomes. Identification of markers that can be monitored, preferably in a noninvasive manner, to identify which patients are likely to develop metastatic NSCLC would be useful in facilitating treatment choice for such patients to reduce their risk of metastasis development. However, no biomarkers identified to date have adequate sensitivity, specificity, or reproducibility for this purpose (3), and tumor samples can be difficult to obtain, especially from patients with inoperable disease.
Rapid and minimally invasive assay methods hold great potential to improve patient care, and several methods using tumor-derived factors have been attempted to develop such assays, but technical issues have thus far prevented their successful translation to clinical use. Recent studies have shown that circulating tumor cells and circulating tumor DNA can provide useful information on tumor evolution, metastasis, and risk of cancer relapse (4), but low detection rates and high false-positive rates have limited their application (5).
Extracellular vesicles (EVs) are abundantly secreted into circulation by many cells and contain proteins from the plasma and endosomal membranes, as well as proteins, RNAs, and DNAs that reflect the phenotypic state of their cell of origin (6). Unlike conventional tissue biopsies, which are subject to sampling bias from tumor size, EVs are secreted by all cancer cell subsets within tumors and thus can reflect heterogeneous features present in many tumors. EVs can be obtained safely and repeatedly by collecting serial blood samples throughout the course of treatment, unlike conventional monitoring methods, where multiple biopsies are usually not feasible because of prohibitive patient risks or financial costs. Furthermore, the presence of several surface proteins (e.g., CD9, CD63, and CD81) on EVs can be used for EV isolation (7), while additional surface markers can be assayed as potential biomarkers to assess tumor phenotype or fate.
The well-known “seed and soil” theory of tumor metastasis is also supported by recent findings that tumor-derived EVs can migrate to distant organs and tissues long before metastatic tumors form at those sites (8). Lyden’s group has provided direct evidence that tumor cells use EVs to condition sites in distant organs to serve as premetastatic niches for the initiation of metastasis (9). The abundance of such EVs in the bloodstream makes them (or their contents) excellent candidate biomarkers for predicting metastasis or for the early detection of metastasis.
We hypothesized that EVs secreted by metastatic and nonmetastatic NSCLC cells carry proteins that can distinguish these cells and function as diagnostic biomarkers of metastatic NSCLC. To test this hypothesis, we used mass spectrometry (MS) to detect EV proteins that were differentially expressed in two mouse NSCLC cell lines with metastatic and nonmetastatic phenotypes and found that EV expression of tetraspanin-8 (Tspan8) was associated with NSCLC invasiveness, in keeping with findings that its elevated cellular expression is associated with metastasis (10–12). Subsequent analysis found that intersectin-2 (Itsn2) expression, which regulates Tspan8 packaging into EVs (13, 14), was increased in the metastatic NSCLC cell line and that overexpression of Itsn2 increased EV Tspan8 abundance and EV-enhanced NSCLC cell invasion. Finally, analysis of Tspan8 expression in archived serum samples obtained from patients with NSCLC participating in a clinical trial indicated that serum EV-Tspan8 concentration predicted future metastasis. We conclude from these findings that EV-Tspan8 may be an effective, relatively noninvasive biomarker for the early detection of lung cancer metastasis and can facilitate therapeutic decisions for such patients.
RESULTS
Tspan8 in EVs from metastatic and nonmetastatic NSCLC cell lines
We selected two murine NSCLC cell lines, one derived from a primary lung tumor (393P) and one derived from a subcutaneous metastatic lung cancer (344SQ), as a model system to study differences in the EV composition of EVs derived from metastatic and nonmetastatic NSCLC cells (15). We found that 344SQ cells exhibited 10 times the activity of 393P cells to invade through a Matrigel matrix layer, a common surrogate for metastatic activity, confirming the differential phenotype of these two cell lines (P < 0.05; Fig. 1, A and B). Because EVs have been reported to play key roles in regulating metastasis, we isolated EVs from 44SQ and 393P cell cultures by sequential centrifugation and ultracentrifugation to identify proteins that were differentially expressed in the EVs of these cells (fig. S1). Scanning electron microscopy (SEM) and NanoSight particle-tracking analyses revealed smooth, saucer-like vesicles <200 nm in diameter (fig. S2), characteristic of a high-purity EV sample relatively devoid of cell debris (16). Western blot analysis also revealed that the EV fractions expressed the EV markers TSG101, HSP70, and CD9 but did not express the cis-Golgi matrix protein GM130 (Fig. 1C), as expected for an EV preparation without substantial contamination by cytosolic protein (14, 17).
Fig. 1. Characterization of 393P and 344SQ cell phenotype and EV protein expression.
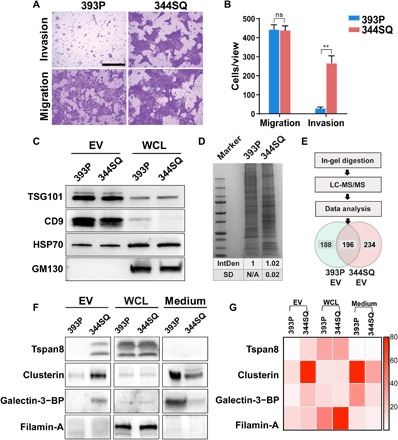
(A) Representative images and (B) graph of the migration and invasion phenotypes of the murine 393P (nonmetastatic) and 344SQ (metastatic) NSCLC cell lines. Scale bar, 100 μm. Data are means ± SEM from three independent experiments; ns, not significant; **P < 0.01. (C) Western blot of EV markers TSG101, HSP70, and CD9 and the Golgi (cytosol) marker GM130 in EVs or whole-cell lysates (WCLs) of 393P and 344SQ cells. (D) Coomassie-stained SDS-PAGE of 393P or 344SQ cell EV isolates; IntDen, relative mean and SD of the integrated lane densitometry from three replicates. N/A, not applicable. (E) Venn diagram of EV proteins identified by LC-MS/MS. (F) Western blot of proteins in EVs, WCLs, and EV-depleted medium. BP, binding protein. (G) Heat map of 393P versus 344SQ EV Western blot expression from low (light red) to high (dark red) optical density.
EV protein lysates were then generated from these samples, and equal amounts of 393P- and 344SQ-derived EV proteins were size-fractionated by SDS–polyacrylamide gel electrophoresis (PAGE) and subjected to in-gel proteolysis, after which eluted peptide fractions were subjected to liquid chromatography–tandem MS (LC-MS/MS), and resulting peptides were queried against the UniProtKB/Swiss-Prot Mus musculus databases to identify proteins differentially expressed in the 393P and 344SQ EV fractions (Fig. 1, D and E). This resulted in the identification of 618 proteins, among which 196 were shared by 393P and 344SQ EVs, 234 were present only in 344SQ EVs, and 188 were present only in 393P EVs. Hypothesizing that EVs from metastatic cells would contain proteins uniquely associated with metastasis, we focused on those proteins that were up-regulated on or unique to the 344SQ-derived EVs and categorized these proteins according to molecular function or biological processes as indicated by their reported UniProt Gene Ontology assignments (fig. S3). EV proteins were also filtered by requiring that they be present in each of the three replicates of this experiment, with at least two identified peptide-spectrum-match sequences. Candidates for EV proteins that were enriched on metastatic 344SQ EVs were required to demonstrate ≥1.5-fold increased expression in 344SQ versus 393P EVs or be uniquely detected in 344SQ EVs, which resulted in 10 candidate proteins. A search of the remaining proteins for those with reported metastasis-related functions led to the final selection of four proteins: Tspan8 (10, 11), clusterin (18), galectin-3–binding protein (19), and filamin-A (20). Western blot analysis of these proteins in 393P and 344SQ cells and their EVs confirmed that Tspan8, clusterin, and galectin-3–binding protein were substantially increased in EVs of 344SQ versus 393P cells, despite no obvious difference in their expression in whole-cell lysates (WCLs) of these cells (Fig. 1, F and G), suggesting that these proteins are selectively packaged into 344SQ EVs. However, filamin-A expression did not detectably differ in the 344SQ and 393P EVs, contrary to our LC-MS/MS results, while both clusterin and galectin-3–binding protein were detected in the EV-depleted supernatants of 344SQ and/or 393P cells in direct opposition to their EV expression, calling into question their utility as EV biomarkers. On the basis of these findings, we selected EV-Tspan8 expression as a potential prognostic biomarker for NSCLC metastasis.
ITSN2 overexpression promotes Tspan8 packaging into EVs of nonmetastatic cells
Internalization of Tspan8 into EVs relies on the association of the Tspan8 N-terminal region with ITSN2, which is known to promote endocytosis and to regulate fission (21). Internalization and recovery of Tspan8 in early endosomes are further promoted by the recruitment of integrin α4 (CD49d) (22). Either or both of these proteins could thus promote Tspan8 enrichment on 344SQ EVs (Fig. 2A). We therefore performed Western blot analyses to evaluate the potential contributions of differential ITSN2 and CD49d expression to Tspan8 recruitment to 344SQ EVs. This analysis found that Itsn2 expression notably increased 344SQ versus 393P cells, while CD49d expression did not change (Fig. 2, B and C), leading us to hypothesize that ITSN2 may play an important role in Tspan8 trafficking to EVs in metastatic tumor cells. To further evaluate the role of ITSN2 in the EV packaging of Tspan8, we established a 393P cell line that overexpressed ITSN2 (393PItsn2+) and observed that increased expression of ITSN2 in cell lysates was matched by a corresponding threefold increase in EV Tspan8 expression (Fig. 2, C and D), while EV ITSN2 levels did not differ with increased ITSN2 expression, indicating its lack of utility as an EV biomarker for metastatic NSCLC. These findings were verified in the human lung cancer cell line A549 (fig. S4), where a cell line with a 6-fold increase in ITSN2 expression (A549ITSN2+) revealed a 2.7-fold increase in EV Tspan8 expression. Collectively, these results suggest that cellular ITSN2 expression plays a key role in recruiting Tspan8 to EVs during metastatic progression.
Fig. 2. Tspan8 is selectively recruited to 344SQ EVs by ITSN2.
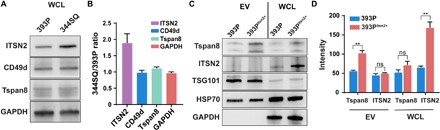
(A) Representative Western blots and (B) quantification of 393P and 344SQ WCL expression of ITSN2, CD49d, Tspan8, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (C) Representative Western blot and (D) quantification of Western blots of Tspan8, ITSN2, TSG101, HSP70, and GAPDH expression in EVs and WCLs of 393P and 393P cells overexpressing ITSN2 (393PItsn2+). Data are means ± SEM from three independent experiments. **P < 0.01.
Tspan8-enriched EVs promote invasiveness of murine and human NSCLC cells
To evaluate whether Tspan8-enriched EVs could promote NSCLC invasion responses, we isolated EVs from 393P and 393PItsn2+ cells and tested the ability to influence the Matrigel invasion of nonmetastatic 393P cells in a modified Transwell assay (fig. S5). EVs isolated from both 393P and 393PItsn2+ cells stimulated the invasion of 393P cells, but the Tspan8-enriched EVs of the 393PItsn2+ cells prompted a 2.6-fold increase in cell invasion (Fig. 3A). Similarly, Tspan8-enriched EVs isolated from an ITSN2-overexpressing human A549 NSCLC cell line (A549ITSN2+) enhanced the Matrigel invasion of A549 cells by 1.5-fold when compared to the response to EVs from unmodified A549 cells (Fig. 3B). Matrigel invasion rates of 393P cells demonstrated EV dose dependence, increasing from 3.3% in untreated cells to 18.9 and 24.9% when these cells were treated with 393PItsn2+ EVs (25 and 50 μg/ml, respectively) (Fig. 3C and fig. S6A). A similar effect was observed with human A549 cells treated with A549ITSN2+ EVs, although untreated A549 cells exhibited a high invasion rate (48.3%), which further increased to 76.2 and 96.3% when these cells were treated with A549ITSN2+ EVs (25 and 50 μg/ml) (Fig. 3D and fig. S6B). No further increases in invasion were observed when either cell was treated with 393PItsn2+ or A549ITSN2+ EVs (100 μg/ml). These findings support the premise that an ITSN2-mediated increase in EV-Tspan8 expression can enhance the invasive behavior of cells in contact with these EVs.
Fig. 3. EV-Tspan8 promotes invasion of mouse 393P and human A549 cells.
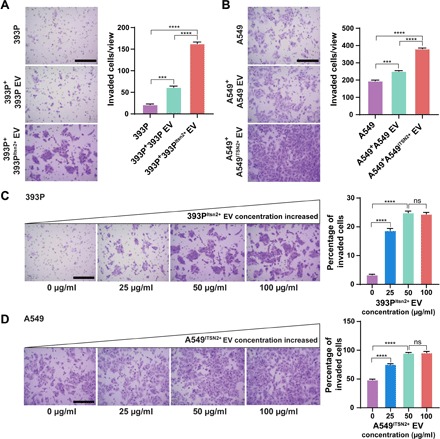
Migration and invasion of (A) 393P cells treated with 393P or 393PItsn2+ EVs (50 μg/ml) for 24 hours and (B) A549 cells treated with A549 or A549ITSN2+ EVs (50 μg/ml) for 24 hours. Scale bars, 100 μm. Representative images and percentage of invasion to migration of (C) 393P and (D) A549 cells in response to increasing concentrations of 393PItsn2+ EVs or A549ITSN2+ EVs. Data indicate means ± SEM from three replicate experiments. Scale bars, 100 μm. ***P < 0.005; ****P < 0.001.
EV-Tspan8 levels predict freedom from distant metastasis in patients with NSCLC
To test the potential diagnostic value of EV-Tspan8 expression as a biomarker of NSCLC metastasis, we analyzed EV-Tspan8 concentrations in 106 pretreatment serum samples collected from individuals diagnosed with locally advanced-stage NSCLC. All patients had received concurrent chemoradiation in a prospective protocol, the primary results of which are reported elsewhere (23). Subjects enrolled in this study had a median follow-up time of 26 months, and 58 of these subjects developed distant metastasis at a median interval of 12.7 months [95% confidence interval (CI), 8.0 to 23.3]. Enrolled subjects had a median age of 66 years and were primary male (60%), white (92%), and diagnosed with stage IIIa tumors (57%) and had a median gross tumor volume of 101 cm3 at initial assay (table S1). No difference was observed in posttreatment freedom from distant metastasis when subjects were analyzed by disease stage [IIIa (n = 61) versus IIIb (n = 45); hazard ratio (HR), 1.1; 95% CI, 0.6 to 1.8; P = 0.77].
EV-Tspan8 expression in these serum samples was measured with a modified sandwich enzyme-linked immunosorbent assay (ELISA) using a Tspan8-specific capture antibody and CD9-specific detection antibody. EV-Tspan8 expression values obtained for these samples ranged from 0.001 to 0.791 (median, 0.120), and low EV-Tspan8 expression was found to predict freedom from distant metastasis. Recursive partitioning analysis identified the optimal cutoff value of EV-Tspan8 to be 0.08, which was used to assign patients to Tspan8low (Tspan8 ≤ 0.08, n = 33) versus Tspan8high (Tspan8 > 0.08, n = 73) groups. Median time to distant metastasis for Tspan8high patients was 8.5 months, while the Tspan8low group did not reach a median during the available follow-up period (Fig. 4). Cox proportional hazard regression analysis of distant metastasis for all 106 patients performed using a multivariate model (disease stage, age, sex, Karnofsky performance status score, smoking status, tumor histology, gross tumor volume, and treatment modality) identified age and Tspan8 expression as independent predictors of freedom from distant metastasis. The HR for those aged >60 years versus those aged ≤60 years was 0.56 (95% CI, 0.31 to 1.00; P = 0.049), and the HR for Tspan8high versus Tspan8low patients was 2.16 (95% CI, 1.06 to 4.40; P = 0.034). These results confirmed our preclinical findings and suggest that low serum EV-Tspan8 level may predict extended time with freedom from distant metastasis after curative chemoradiation for NSCLC.
Fig. 4. EV-Tspan8 level correlates with distant metastasis in patients with NSCLC.
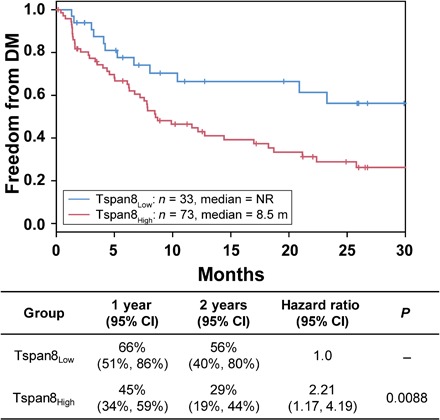
Kaplan-Meier plots of freedom from distant metastasis (DM) in 106 patients with stage III NSCLC segregated by Tspan8Low and Tspan8High serum levels (Tspan8 cutoff = 0.08), with P values calculated with log-rank tests. NR, not reached; m, months.
DISCUSSION
Lung cancer is the leading cause of cancer-related death worldwide, with a propensity for distant metastasis. Better understanding of the molecular events that regulate the development of lung cancer metastasis and biomarkers that predict which patients will experience metastasis are needed to improve survival rates (24). It has been reported that cancer cells secrete EVs that can be easily isolated from various body fluids (25) and which contain cancer-specific contents. The development of a suitable optical sensor also enables orthogonal analysis of multiple EV samples (16). Mounting evidence indicates that EVs can carry multiple factors that have been validated as surrogate biomarkers of cancer progression and metastasis, suggesting that analysis of circulating EVs could replace some invasive clinical procedures currently used for cancer diagnosis, prognosis, and prediction (26–28). There is substantial interest in the potential of EV biomarkers to serve as less invasive alternatives to tissue biopsy for early diagnosis and prognosis in NSCLC, and a few groups have performed studies to examine the potential of EV proteins to diagnose NSCLC (29). For example, Huang et al. (30) reported that EV expression of epidermal growth factor receptor protein was associated with lung cancer but not with lung inflammation. However, few studies have examined the prognostic potential of EV markers for NSCLC. Sandfeld-Paulsen et al. (31) evaluated the prognostic value of 49 EV membrane proteins in plasma samples from 276 patients with NSCLC and concluded that NY-ESO-1 was correlated with inferior survival. Liu et al. (32) also found that EV miR-10b-5p, miR-23b-3p, and miR-21-5p may be useful as biomarkers in NSCLC, in that elevated levels of these miRNAs in EVs were associated with poor overall survival. However, these studies have limitations with regard to their ability to predict clinical outcomes, especially distant metastasis.
Given that previous studies have reported that circulating EVs can initiate a premetastatic niche at distant sites through the transfer their contents (9), the goals of the current study were to identify candidate EV biomarkers of NSCLC metastasis and evaluate their prognostic potential in a relevant clinical population. In this study, we screened for factors that were differentially enriched in EVs derived from metastatic versus nonmetastatic NSCLC cell lines. Among the factors that met our selection criteria, only Tspan8 exhibited differential EV expression without also revealing appreciable expression in the EV-depleted cell culture medium.
Tspan8 has been reported to modulate invasion of melanoma by affecting integrin-linked kinase signaling and its downstream target protein kinase B (11), to regulate E-cadherin/catenin signaling and metastasis in breast cancer (12), and to promote hepatocellular carcinoma metastasis by increasing expression of A disintegrin and metalloproteinase domain 12 (ADAM12) and mediating astrocyte-elevated gene-1 (AEG-1)–induced invasion (10, 33).
However, while EV expression of Tspan8 differed with metastatic phenotype, its cellular expression levels were not noticeably different between our metastatic and nonmetastatic cell lines, suggesting that this difference resulted from differential EV protein sorting and enrichment in the metastatic cell population. Subsequent studies identified that the multimodular protein ITSN2, which participates in endocytosis with the Tspan8-associating CD49d complex (21, 22), was expressed at a substantially higher level in the metastatic cells, suggesting that increased ITSN2 expression was responsible for the elevated Tspan8 expression and metastatic potential of the EVs of the metastatic cells. This hypothesis was supported by subsequent experiments demonstrating that ITSN2 up-regulation led to both increased EV Tspan8 expression and metastatic promoting activity, without affecting the cellular Tspan8 level. Thus, it appears that any therapeutic approach seeking to attenuate the prometastatic effect of elevated EV-Tspan8 expression may need to target ITSN2 expression rather than Tspan8. We were not able to find any reports linking ITSN2 and metastasis, but our findings from the current study suggest that its dysregulation may play a role in metastatic priming events mediated by EV-Tspan8 expression and may also regulate the sorting of other EV factors that may be involved in this process.
Our findings indicate that the role of EV-Tspan8 in the progression and metastasis of NSCLC may be under the regulation of ITSN2. To the best of our knowledge, a correlation between ITSN2 and metastasis has not previously been reported. It is beyond the scope of the current study, but certainly, our future plan is to determine mechanistically how ITSN2 acts in NSCLC progression, i.e., which signaling pathway or downstream genes are influenced by ITSN2 to result in tumorigenesis.
NSCLC is often diagnosed at advanced stages, and concurrent chemoradiation therapy is considered standard of care for inoperable, locally advanced disease (2). However, stage II to III NSCLC is quite heterogeneous, and standard concurrent chemoradiation often has quite different outcomes in patients at the same apparent disease stage. We found that, among patients with stage III NSCLC, serum EV-Tspan8 expression before treatment distinguished those who developed early distant metastasis and that high serum EV-Tspan8 expression was an independent predictor for distant metastasis, with the optimal serum EV-Tspan8 cut-point differentiating patients with more than twofold difference in median time to distant metastasis. This finding suggests that pretreatment Tspan8 levels may identify patients who could benefit from adjuvant systemic treatment (e.g., chemotherapy and immunotherapy), information that could be used to personalize treatment choices for individual patients based on their risk of distant metastasis.
Standard methods used to diagnose and evaluate cancer metastasis include tissue biopsy and radiographic imaging; however, these methods detect established metastatic sites and cannot predict the likelihood that a tumor will progress to metastasis, limiting the window of opportunity for modifying treatment to slow or prevent the development of distant metastases (34). Serum EV-Tspan8 expression can be analyzed using small, minimally invasive blood samples to identify those patients who might benefit from more aggressive therapeutic interventions. Because of promising data obtained in our NSCLC study cohort, we believe that additional studies are warranted to validate our findings in independent datasets, ideally from large prospective studies. Future studies should also address whether longitudinal analysis of the serum EV-Tspan8 level during the therapeutic intervention can also serve as a biomarker of tumor response and predictive biomarker of metastasis. Such a capability would facilitate the evaluation of treatment responses and potentially allow the rapid adaptation of treatment strategies (35).
MATERIALS AND METHODS
Study design
The primary objective of this translational study was to search for EV biomarkers to predict NSCLC distant metastasis with a long-term goal to develop rapid, minimally invasive markers to guide precision treatment decisions. The research subjects and units of investigation were mouse-derived cultured cells and serum samples from human donors. We first used comparative proteomics to identify EV proteins with known metastasis associations that were selectively enriched on EVs of higher metastatic murine lung cancer cell line as candidates for a predictor of distant metastasis. To explore the role of Itsn2 in EV-Tspan8 recruitment, EVs were isolated from Itsn2-overexpressing cell lines and evaluated for the influence on cell invasion. We then adapted sandwich ELISA to develop a serum EV capture and detection system, where a capture antibody recognizing Tspan8 was used to selectively enrich EVs, while antibody recognizing EV membrane protein CD9 served as detection probe. The predictive value of serum EV-Tspan8 level for freedom from distant metastasis was assessed in 106 patients with stage III NSCLC from a prospective randomized trial. For these studies, randomization or blinding was not applicable.
Cell cultures
The murine lung cancer cell lines 393P (nonmetastatic) and 344SQ (metastatic) were provided by J. M. Kurie (Department of Thoracic/Head and Neck Medical Oncology at MD Anderson). The human lung cancer cell line A549 was purchased from the American Type Culture Collection. The 393P, 344SQ, and A549 cells were cultured in RPMI 1640 (HyClone) containing 10% fetal bovine serum (Gibco) for growth or maintenance or in serum-free medium for EV collection. Cultures were incubated at 37°C in a humidified 5% CO2 incubator and supplemented with penicillin (100 U) and streptomycin (100 μg/ml; Invitrogen).
EV isolation
The protocol for isolating EVs from conditioned cell medium was described previously (36) and depicted schematically in fig. S1. Briefly, eight to ten 15-cm dishes of cells grown to 70% confluence were washed three times with phosphate-buffered saline (PBS) to remove residual serum contamination and then incubated for 48 hours in serum-free media. The cell culture supernatant was then collected, and cell debris was removed by centrifugation at 800g for 5 min. The supernatant was then centrifuged at 2000g for 20 min, followed by filtration through a 0.45-μm pore filter (10,040 to 466, VWR), and concentrated by retention and collection with 100-kDa cutoff filters (Amicon Ultra-15, Millipore). After another centrifugation at 12,000g for 30 min, the EVs were pelleted by ultracentrifugation at 100,000g for 2 hours with a Ti 100.2 rotor (Beckman Instruments), after which the EV pellets were washed with ice-cold PBS and pelleted again at 100,000g for 2 hours. All sequential centrifugations were done at 4°C. Pellets were resuspended in PBS for functional assays and used immediately or stored at −80°C.
EVs were isolated from serum samples by using the ExoQuick exosome precipitation solution (EXOQ20A-1, System Biosciences) according to the manufacturer’s instructions. Briefly, serum was centrifuged at 3000g for 15 min to remove cells and cell debris, and 15 μl of ExoQuick exosome precipitation solution was added to 45 μl of serum supernatant, which was mixed gently and incubated at 4°C for 30 min, followed by centrifugation at 1500g for 30 min. The supernatant was then aspirated, and the EV pellet was suspended in 50 μl of sterile PBS for immediate use.
Scanning electron microscopy
Pellets containing EVs isolated from 393P and 344SQ cells were vortexed and suspended in 0.2- to 1-ml PBS and serially diluted in distilled water. Vesicle mixtures were then added at 1 to 5 μl to cleaned silicon chips, which were sonicated in acetone, ethanol, and distilled water for 5 min in each solvent, flushed with water, air-dried, and immobilized after drying under a ventilation hood. Samples on silicon chips were mounted on an SEM stage with carbon paste and imaged by a Hitachi S-4700 SEM under low beam energies (5.0 to 10.0 kV). Freshly isolated EVs were immobilized on silicon immediately after isolation and imaged within 3 days.
Nanoparticle tracking analysis
Nanoparticle tracking analysis (NanoSight) was performed as described elsewhere (37), with minor modifications as follows. Three 30-s videos were taken under controlled fluid flow with the pump speed set to 80, and the videos were analyzed with the batch analysis tool of NTA 2.3 software (version 2.3 build 2.3.5.0033.7-Beta7), where minimum particle size, track length, and blur were set to “automatic.” Representative vesicle size and concentrations given were recorded over three tracking runs.
Protein concentration measurement
WCLs, purified EVs, and EV-depleted cell culture media (medium from which EVs were removed by overnight ultracentrifugation) were lysed in Mammalian Protein Extraction Reagent in the presence of a protease inhibitor cocktail (Pierce). Concentrations of proteins in WCLs and EVs were measured with a bicinchoninic acid assay kit (Pierce) according to the manufacturer’s instructions. Protein concentrations in medium were measured with a Bradford assay (Pierce) according to the manufacturer’s instructions.
Western blot analyses
Lysed samples were loaded onto two 4 to 20% gradient SDS-PAGE gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad) by using standard methods. Gels were blocked with 5% bovine serum albumin in PBS with 0.05% Tween 20 (PBST). Antibodies used for Western blotting included 1:500 Tspan8 (sc-292058, Santa Cruz Biotechnology Inc.), 1:1000 TSG101 (ab83, Abcam), 1:1000 CD9 (ab92726, Abcam), 1:1000 GM130 (sc-55591, Santa Cruz Biotechnology Inc.), 1:1000 HSP70 (sc-32239, Santa Cruz Biotechnology Inc.), 1:1000 galectin-3–binding protein (sc-74970, Santa Cruz Biotechnology Inc.), 1:1000 clusterin (sc-8354, Santa Cruz Biotechnology Inc.), 1:1000 filamin-A (sc-28284, Santa Cruz Biotechnology Inc.), 1:1000 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (2118L, Cell Signaling Technology), 1:1000 Itsn2 (ab133854, Abcam), and 1:1000 CD49d (PA520595, Invitrogen).
LC-MS/MS analysis
EVs were isolated from three independent cell cultures of each cell line as described above; 20 μg of protein was loaded on SDS-PAGE precast gradient gels (Bio-Rad), and proteins were separated at 125 V. In preparation for MS analysis, the gels were stained with Coomassie EZBlue and destained overnight in water before band excision. For in-gel digestion, each sample lane was excised into 12 equal bands approximately 5 mm in height. Gel bands were dehydrated in acetonitrile for 5 min, rehydrated in 10 mM dithiothreitol in 50 mM ammonium bicarbonate, and incubated at 56°C for 1 hour. After subsequent dehydration with acetonitrile, the gel bands were treated with 55 mM iodoacetic acid in 50 mM ammonium bicarbonate in the dark at room temperature for 1 hour. Bands were dehydrated once more with acetonitrile and treated with 50 μl of a solution of MS grade trypsin (10 ng/μl) in 50 mM ammonium bicarbonate. After overnight digestion at 37°C, the gel band supernatant was transferred to a new tube, and the tryptic peptides were extracted three times with 50% acetonitrile containing 5% acetic acid. The combined extracts were dried by vacuum centrifugation and suspended to ~20 μl with 0.1% formic acid/2% acetonitrile before injection. Subsequently, the digests were separated by using UltiMate 3000 Nano LC (Dionex Corporation) with an nC18 enrichment column (Dionex C18 Pepmap 100, 5-μm particles, 100-Å pores, 300 μm internal diameter × 5 mm) and a Dionex nC18 analytical column (C18 Pepmap 100, 3-μm particles, 100-Å pores, 75 μm internal diameter × 150 mm). Flow rates of 20 μl/min and 300 nl/min were used for the loading and analytical columns, respectively. Eluted peptides were analyzed on a linear ion trap LTQ Velos Pro mass spectrometer (Thermo Fisher Scientific). One MS scan was followed by eight MS/MS scans, and MS/MS spectra were queried against the UniProt database (www.uniprot.org) by using in-house Mascot 2.3.0 (www.matrixscience.com) software and a mass tolerance of 0.5 Da.
ITSN2 overexpression
To overexpress Tspan8 in 393P cells, myc-DDK–tagged mouse Itsn2, transcript variant 3, was purchased from OriGene (MR230794); to overexpress Tspan8 in A549 cells, Homo sapiens ITSN2 in pENTR223 gateway donor was purchased from the DNASU Plasmid Repository, and a plasmid vector pLenti-CMV-Puro-DEST (w118-1) was purchased from Addgene (17452). Transfections were performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA), typically with 500 ng of plasmid unless otherwise noted. Cells were seeded at a density of 40,000 cells per well in 24-well plates and transfected the next day by using Lipofectamine 2000 (Invitrogen). Cells were then incubated with puromycin (10 μg/ml; Invitrogen) for 2 days to select for transduced cells, and Itsn2 expression was confirmed by Western blotting.
Migration and invasion assay
Migration and invasion assays were performed as described elsewhere (38). A total of 20,000 393P or A549 cells were seeded in serum-free medium in Transwell inserts (8-μm pores) in 24-well plates (Corning). To measure invasion, the inserts were precoated with Matrigel (BD Biosciences) overnight at room temperature. Complete medium containing 10% fetal bovine serum with or without EVs from the same cell line was used as the chemoattractant in the lower compartment of the well. After 24 hours of incubation at 37°C, the cells on the upper compartment were removed by using cotton swabs, and the membrane surface was stained with 0.1% crystal violet. Permeating (invaded) cells were visualized under a microscope and quantified with ImageJ.
Patient and clinical samples
Serum samples analyzed from this study were obtained from archived samples of patients enrolled in a completed prospective randomized trial (NCT00915005), the primary outcomes of which were reported by Liao et al. (23). Serum samples were obtained from 106 patients [64 men and 42 women; median age, 66 years (range, 42 to 81 years); table S1] under an Institutional Review Board–approved protocol before chemoradiation therapy was begun. Demographic information was collected as part of the standard protocol, and the occurrence of distant metastasis was determined by routine follow-up workup. All patients included in this analysis provided written informed consent for the primary protocol and the serum sample collection.
Enzyme-linked immunosorbent assay
EVs isolated from serum samples of patients with NSCLC were assayed for Tspan8 expression by sandwich ELISA according to the following procedure, which was modified to measure EV membrane protein. Briefly, 50-μl isolated serum EV samples were applied onto the 96-well microplate, which was precoated with an anti-Tspan8 murine monoclonal antibody (1:500; sc-293317, Santa Cruz Biotechnology Inc.) and subsequently blocked with 5% bovine serum albumin in PBST for 2 hours at room temperature. After incubation at 4°C overnight, wells were gently washed four times with PBST and then incubated at room temperature for 2 hours with 50 ng of anti–CD9-biotin rabbit polyclonal antibody (1:2000; MA119485, Invitrogen) in 100 μl of PBS. Wells were then washed four times with PBST and incubated for 1 hour at room temperature with 25 ng of streptavidin-conjugated horseradish peroxidase in 100 μl of PBS (1:5000; N100, Thermo Fisher Scientific). Wells were then incubated at room temperature for 20 min with substrate after the addition of 50 μl of tetramethylbenzidine solution (34022, Thermo Fisher Scientific) and then supplemented with 50 μl of 2 M H2SO4 solution to each well to terminate the reaction, and the absorbance at 450 nm was measured with a plate reader (SK601, Seikagaku Corporation). No cross-reactivity was observed between the Tspan8 antibody and CD9-biotin antibody with the absence of EV samples. Each sample was assayed with analytical runs that were performed in duplicate.
Statistical analysis
Statistical analyses were conducted with two-tailed unpaired Student’s t tests, or one- or two-way analysis of variance tests using GraphPad Prism software (version 7.0 for Windows, GraphPad, CA). Patient or treatment data were expressed as means ± SEM. For survival analyses, the Kaplan-Meier method was used to estimate the distribution of freedom from distant metastasis over time. Groups were compared with HRs, and 95% CIs were computed by Cox proportional hazard regression analysis. An optimal cut-point for Tspan8 was identified with recursive partitioning analysis. Differences with a P value less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank J. M. Kurie (The University of Texas MD Anderson Cancer Center) for providing the murine lung cancer cell lines 393P and 344SQ and L. Song (Arizona State University) for sharing technical expertise on Itsn2 overexpression. Funding: The work was primarily supported by research funding provided by the National Institutes of Health (U01CA214254, R01HD090927, R01AI122932, R01AI113725, R03AI140977, and R21Al126361-01), the Fred Hutchinson Cancer Research Center (0000917241), and an Arizona Biomedical Research Commission (ABRC) young investigator award. The clinical correlative studies were supported in part by the National Cancer Institute of the National Institutes of Health, 5P01CA021239 and P30CA016672. Author contributions: Y.L., J.F., and T.Y.H. conceived the study, generated the hypotheses, and designed the experiments. Y.L. performed all the experiments and analyzed the data. J.F. provided technical expertise and contributed to the LC-MS/MS experiments. Y.L. and B.N. wrote the manuscript. Z.L., T.X., J.Z., and S.H.L. conceived and designed the clinical study. Z.L., T.X., N.A., X.L., L.L., and K.H. analyzed and interpreted the clinical data. All authors contributed to the manuscript review, revision, and finalization. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Clinical samples from patients with NSCLC were provided by The University of Texas MD Anderson Cancer Center under a fully executed material transfer agreement (MTA no. 21959). All data needed to evaluate the conclusions in the paper are presented in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Materials described in this paper can be provided by The University of Texas MD Anderson Cancer Center pending scientific review and a completed material transfer agreement. Please send communications to Z. Liao to initiate material transfer requests.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaaz6162/DC1
Fig. S1. Schematic of the procedures for isolating EVs from conditioned cell culture medium.
Fig. S2. Characterization of EVs isolated from murine NSCLC cell lines.
Fig. S3. Gene Ontology analysis of proteins up-regulated or uniquely detected in EVs from 344SQ cells.
Fig. S4. Tspan8 enriched in EVs derived from A549ITSN2+ cells.
Fig. S5. Schematic of the procedures for analyzing the influence of EV-Tspan8 on cell metastasis.
Fig. S6. EV-Tspan8 induced invasion in a concentration-dependent manner.
Table S1. Patient and treatment characteristics.
REFERENCES AND NOTES
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Walters S., Maringe C., Coleman M. P., Peake M. D., Butler J., Young N., Bergström S., Hanna L., Jakobsen E., Kölbeck K., Sundstrøm S., Engholm G., Gavin A., Gjerstorff M. L., Hatcher J., Johannesen T. B., Linklater K. M., McGahan C. E., Steward J., Tracey E., Turner D., Richards M. A., Rachet B.; ICBP Module 1 Working Group , Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: A population-based study, 2004-2007. Thorax 68, 551–564 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Mamdani H., Ahmed S., Armstrong S., Mok T., Jalal S. I., Blood-based tumor biomarkers in lung cancer for detection and treatment. Transl. Lung Cancer Res. 6, 648–660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merker J. D., Oxnard G. R., Compton C., Diehn M., Hurley P., Lazar A. J., Lindeman N., Lockwood C. M., Rai A. J., Schilsky R. L., Tsimberidou A. M., Vasalos P., Billman B. L., Oliver T. K., Bruinooge S. S., Hayes D. F., Turner N. C., Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. Arch. Pathol. Lab. Med. 142, 1242–1253 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Alix-Panabières C., Pantel K., Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–631 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R., The biology and function of exosomes in cancer. J. Clin. Invest. 126, 1208–1215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halvaei S., Daryani S., Eslami-S Z., Samadi T., Jafarbeik-Iravani N., Bakhshayesh T. O., Majidzadeh-A K., Esmaeili R., Exosomes in cancer liquid biopsy: A focus on breast cancer. Mol. Ther. Nucleic Acids 10, 131–141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langley R. R., Fidler I. J., The seed and soil hypothesis revisited—The role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 128, 2527–2535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Mark M. T., Molina H., Kohsaka S., Di Giannatale A., Ceder S., Singh S., Williams C., Soplop N., Uryu K., Pharmer L., King T., Bojmar L., Davies A. E., Ararso Y., Zhang T., Zhang H., Hernandez J., Weiss J. M., Dumont-Cole V. D., Kramer K., Wexler L. H., Narendran A., Schwartz G. K., Healey J. H., Sandstrom P., Labori K. J., Kure E. H., Grandgenett P. M., Hollingsworth M. A., de Sousa M., Kaur S., Jain M., Mallya K., Batra S. K., Jarnagin W. R., Brady M. S., Fodstad O., Muller V., Pantel K., Minn A. J., Bissell M. J., Garcia B. A., Kang Y., Rajasekhar V. K., Ghajar C. M., Matei I., Peinado H., Bromberg J., Lyden D., Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiel M. A., Santhekadur P. K., Mendoza R. G., Siddiq A., Fisher P. B., Sarkar D., Tetraspanin 8 mediates AEG-1-induced invasion and metastasis in hepatocellular carcinoma cells. FEBS Lett. 590, 2700–2708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Kharbili M., Agaësse G., Barbollat-Boutrand L., Pommier R. M., de la Fouchardière A., Larue L., Caramel J., Puisieux A., Berthier-Vergnes O., Masse I., Tspan8-β-catenin positive feedback loop promotes melanoma invasion. Oncogene 38, 3781–3793 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Voglstaetter M., Thomsen A. R., Nouvel J., Koch A., Jank P., Navarro E. G., Gainey-Schleicher T., Khanduri R., Groß A., Rossner F., Blaue C., Franz C. M., Veil M., Puetz G., Hippe A., Dindorf J., Kashef J., Thiele W., Homey B., Greco C., Boucheix C., Baur A., Erbes T., Waller C. F., Follo M., Hossein G., Sers C., Sleeman J., Nazarenko I., Tspan8 is expressed in breast cancer and regulates E-cadherin/catenin signalling and metastasis accompanied by increased circulating extracellular vesicles. J. Pathol. 248, 421–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zöller M., Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9, 40–55 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Nazarenko I., Rana S., Baumann A., McAlear J., Hellwig A., Trendelenburg M., Lochnit G., Preissner K. T., Zöller M., Cell surface tetraspanin tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 70, 1668–1678 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Gibbons D. L., Lin W., Creighton C. J., Rizvi Z. H., Gregory P. A., Goodall G. J., Thilaganathan N., Du L., Zhang Y., Pertsemlidis A., Kurie J. M., Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 23, 2140–2151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyu Y., Cui D., Huang J., Fan W., Miao Y., Pu K., Near-infrared afterglow semiconducting nano-polycomplexes for the multiplex differentiation of cancer exosomes. Angew. Chem. Int. Ed. Engl. 58, 4983–4987 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Plebanek M. P., Angeloni N. L., Vinokour E., Li J., Henkin A., Martinez-Marin D., Filleur S., Bhowmick R., Henkin J., Miller S. D., Ifergan I., Lee Y., Osman I., Thaxton C. S., Volpert O. V., Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat. Commun. 8, 1319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau S. H., Sham J. S., Xie D., Tzang C.-H., Tang D., Ma N., Hu L., Wang Y., Wen J.-M., Xiao G., Zhang W.-M., Lau G. K. K., Yang M., Guan X.-Y., Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene 25, 1242–1250 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Lin T.-W., Chang H.-T., Chen C.-H., Chen C.-H., Lin S.-W., Hsu T.-L., Wong C.-H., Galectin-3 binding protein and galectin-1 interaction in breast cancer cell aggregation and metastasis. J. Am. Chem. Soc. 137, 9685–9693 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Jiang X., Yue J., Lu H., Campbell N., Yang Q., Lan S., Haffty B. G., Yuan C., Shen Z., Inhibition of Filamin-A reduces cancer metastatic potential. Int. J. Biol. Sci. 9, 67–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koh T.-W., Verstreken P., Bellen H. J., Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron 43, 193–205 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Rana S., Claas C., Kretz C. C., Nazarenko I., Zoeller M., Activation-induced internalization differs for the tetraspanins CD9 and Tspan8: Impact on tumor cell motility. Int. J. Biochem. Cell Biol. 43, 106–119 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Liao Z., Lee J. J., Komaki R., Gomez D. R., O'Reilly M. S., Fossella F. V., Blumenschein G. R. Jr., Heymach J. V., Vaporciyan A. A., Swisher S. G., Allen P. K., Choi N. C., DeLaney T. F., Hahn S. M., Cox J. D., Lu C. S., Mohan R., Bayesian adaptive randomization trial of passive scattering proton therapy and intensity-modulated photon radiotherapy for locally advanced non-small-cell lung cancer. J. Clin. Oncol. 36, 1813–1822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dopeso H., Jiao H.-K., Cuesta A. M., Henze A.-T., Jurida L., Kracht M., Acker-Palmer A., Garvalov B. K., Acker T., PHD3 controls lung cancer metastasis and resistance to EGFR inhibitors through TGFα. Cancer Res. 78, 1805–1819 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Wang W., Luo J., Wang S., Recent progress in isolation and detection of extracellular vesicles for cancer diagnostics. Adv. Healthc. Mater. 7, e1800484 (2018). [DOI] [PubMed] [Google Scholar]
- 26.An T., Qin S., Xu Y., Tang Y., Huang Y., Situ B., Inal J. M., Zheng L., Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J. Extracell. Vesicles 4, 27522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alix-Panabières C., Pantel K., Clinical prospects of liquid biopsies. Nat. Biomed. Eng. 1, 0065 (2017). [Google Scholar]
- 28.Cui S., Cheng Z., Qin W., Jiang L., Exosomes as a liquid biopsy for lung cancer. Lung Cancer 116, 46–54 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Jakobsen K. R., Paulsen B. S., Bæk R., Varming K., Sorensen B. S., Jørgensen M. M., Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J. Extracell. Vesicles 4, 26659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S.-H., Li Y., Zhang J., Rong J., Ye S., Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. 31, 330–335 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Sandfeld-Paulsen B., Aggerholm-Pedersen N., Bæk R., Jakobsen K. R., Meldgaard P., Folkersen B. H., Rasmussen T. R., Varming K., Jørgensen M. M., Sorensen B. S., Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 10, 1595–1602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q., Yu Z., Yuan S., Xie W., Li C., Hu Z., Xiang Y., Wu N., Wu L., Bai L., Li Y., Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 8, 13048–13058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang T., Lin J., Wang Y., Chen G., Huang J., Chen J., Zhao Y., Sun R., Liang C., Liu B., Tetraspanin-8 promotes hepatocellular carcinoma metastasis by increasing ADAM12m expression. Oncotarget 7, 40630–40643 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gourin C. G., Watts T. L., Williams H. T., Patel V. S., Bilodeau P. A., Coleman T. A., Identification of distant metastases with positron-emission tomography-computed tomography in patients with previously untreated head and neck cancer. Laryngoscope 118, 671–675 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Pi C., Zhang M.-F., Peng X.-X., Zhang Y.-C., Xu C.-R., Zhou Q., Liquid biopsy in non-small cell lung cancer: A key role in the future of personalized medicine? Expert Rev. Mol. Diagn. 17, 1089–1096 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Fan J., Wei Q., Koay E. J., Liu Y., Ning B., Bernard P. W., Zhang N., Han H., Katz M. H., Zhao Z., Hu Y., Chemoresistance transmission via exosome-mediated EphA2 transfer in pancreatic cancer. Theranostics 8, 5986–5994 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerritzen M. J. H., Martens D. E., Wijffels R. H., Stork M., High throughput nanoparticle tracking analysis for monitoring outer membrane vesicle production. J. Extracell. Vesicles 6, 1333883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan J., Sun L., Xu F., Liu L., Hu F., Song D., Hou Z., Wu W., Luo X., Wang J., Yuan X., Hu J., Wang G., M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 79, 146–158 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/11/eaaz6162/DC1
Fig. S1. Schematic of the procedures for isolating EVs from conditioned cell culture medium.
Fig. S2. Characterization of EVs isolated from murine NSCLC cell lines.
Fig. S3. Gene Ontology analysis of proteins up-regulated or uniquely detected in EVs from 344SQ cells.
Fig. S4. Tspan8 enriched in EVs derived from A549ITSN2+ cells.
Fig. S5. Schematic of the procedures for analyzing the influence of EV-Tspan8 on cell metastasis.
Fig. S6. EV-Tspan8 induced invasion in a concentration-dependent manner.
Table S1. Patient and treatment characteristics.


