Abstract
Introduction:
Parkinson’s disease is a devastating neurodegenerative disorder preferentially involving loss of dopaminergic neurons in the substantia nigra, leading to typical motor symptoms. While there are still no therapeutics to modify disease course, recent work using induced pluripotent stem cell (iPSC) and 3D brain organoid models have provided further insight into Parkinson’s disease pathogenesis and potential therapeutic targets.
Areas covered:
This review highlights the generation of iPSC neurons and neural organoids as models for studying Parkinson’s disease. It further discusses the recent work using patient-derived neurons from both familial and sporadic forms of Parkinson’s to study disease pathogenic phenotypes and pathways. It additionally provides an evaluation of iPSC neurons and organoid models for therapeutic development in Parkinson’s.
Expert opinion:
The use of Parkinson’s disease patient-derived neurons and organoids provides us with the exciting opportunity to directly investigate pathogenic mechanisms and test drug compounds in human neurons. Future studies will involve generating more sophisticated models of brain organoids, studying neuronal pathways using larger patient cohorts, and routinely assessing therapeutics in these models.
Keywords: Induced pluripotent stem cells, Organoid, Parkinson’s disease, Therapeutics, Dopaminergic neurons, Midbrain, Neurodegeneration, α-synuclein, GBA
1. Introduction
1.1. Induced Pluripotent Stem Cells
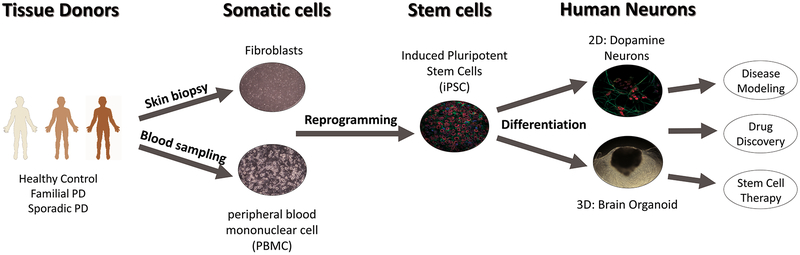
The discovery in 2006 of four transcription factors (Sox2, Oct3/4, c-myc, and Klf4) marked the development of pluripotent stem cells from mouse fibroblasts [1], and was later replicated in human somatic cells [2, 3]. Current research efforts have identified protocols for generating induced pluripotent stem cells (iPSCs) from dermal fibroblasts, hematopoietic stem cells, adipocytes, and peripheral blood mononuclear cells [4, 5, 6, 7]. Together, these findings have opened the field to new advances in patient-specific cell lines and circumvented the need for embryonic stem cells which require gene editing and are linked to ethical concerns [8, 9, 10]. Furthermore, pluripotency now allows researchers to selectively differentiate stem cells into any somatic cell type, resulting in the generation of disease relevant tissues for study. iPSCs additionally offer a strategy for disease modeling using patient-specific cell lines and disease-relevant genetic backgrounds, thus allowing for new opportunities in therapeutic development and drug screening applications (Figure 1).
Figure 1. Application of human induced pluripotent stem cell-derived neurons for disease modeling and drug discovery in Parkinson’s disease.
Human somatic cells such as fibroblasts or peripheral blood mononuclear cell (PBMC) from healthy control, familial and sporadic PD patients are reprogrammed into human iPSCs. Human iPSCs are further differentiated into dopamine neurons or 3D brain organoid depending on the purpose. Differentiated tissues enable replication of Parkinson’s disease in vitro and can be further used for disease modeling, drug discovery and dopamine replacement stem cell therapy.
1.2. IPSC-Derived Neurons
Due to the lack of access to human neuronal tissues [11] and the intrinsic differences in animals models from human pathologies [12, 13], iPSCs provide new methods for modeling disease pathology for multiple neurodegenerative diseases including Alzheimer’s, Parkinson’s, Amyotrophic Lateral Sclerosis (ALS), and Huntington’s disease. Specifically, the identification of neural fate induction by TGFβ antagonists through dual SMAD inhibition [14] has led researchers to further develop protocols for differentiating iPSCs into multiple different neuronal subtypes (cortical, cholinergic, dopaminergic, GABAergic, hippocampal, hypothalamic, motor, serotonergic and Purkinje neurons) as well as glial cells (astrocytes and oligodendrocytes) [15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31]. Furthermore, iPSC differentiation protocols have also been optimized to produce mature electrophysiological neurons supporting basic synaptic functions [32, 33] and have also been transplanted into primates for potential therapeutic applications [34]. For Parkinson’s disease (PD), the ability to generate patient-derived dopaminergic neurons has proved to be particularly insightful, with current differentiation protocols using dual-SMAD inhibition followed by Sonic Hedgehog and FGF8b signaling, and subsequent maintenance in BDNF, GDNF, ascorbic acid, and cAMP [35]. Importantly, multiple studies have been able to recapitulate key PD pathological features and shed light on new mechanistic pathways using patient-derived iPSC dopamine neurons, which will be discussed in further detail in this review.
1.3. IPSC-Derived Neural Organoids
Organoids are derived from stem cells in a 3-dimensional matrix such as Matrigel or animal-derived hydrogels, which allow for efficient cell growth and differentiation. The successful growth of organoids further relies on the innate ability of stem cells to self-organize and form ordered structures and cyto-architecture [36], as well as their cell-cell interactions, and their ability to differentiate into diverse cellular populations [37, 38, 39]. Organoids have been used to model systems ranging from kidneys, liver, intestine, optic cup, cerebral, and midbrain regions [40, 41, 42, 43, 44, 45], reflecting the pluripotent state of iPSCs. Furthermore, organoids have become a critical tool in disease modeling from early stages of development following endogenous temporal stages within cell populations [37, 43], providing readily available models that can replicate disease phenotypes. Indeed, cerebral organoids have been used to model microcephaly, while midbrain organoids have been used to model sporadic PD [43, 46]. Importantly, 3D brain organoids further provide the potential to model complex circuity by generating assembloids (assemblies of different region-specific organoids) to better advance our understanding of the human brain. These include recent studies investigating cell migration in vitro, such as the migration of GABAergic neurons from the ventral forebrain to the dorsal forebrain [47] and neuronal circuity such as cortico-thalamic assembloids which demonstrate projections between deep layer cortical neurons and thalamic neurons [48, 49]. Of note, such organoids may be used to facilitate organ-on-a-chip technology to utilize patient-specific iPSC-derived neurons as an alternative to conventional preclinical models for drugs screening [50]. Thus, brain-region-specific organoids are a valuable asset for studying pathology of diseases during development, and further offer great potential for drug screening in tissue specific environments.
2. IPSC-Derived Neuronal and Organoid Modeling and Drug Discovery in Parkinson’s Disease
2.1. Parkinson’s Disease (PD)
Parkinson’s disease is associated with the progressive loss of A9-dopaminergic neurons in the substantia nigra leading to a loss of dopamine and the dysregulation of fine motor control localized in the basal ganglia. Ultimately, the death of dopaminergic neurons clinically manifests in parkinsonian symptoms including bradykinesia, muscular rigidity, and resting tremors [51] and pathologically involves the presence of Lewy Body aggregates comprised of α-synuclein [52]. PD exists as a multifaceted disease that presents itself clinically with some degree of heterogeneity [53, 54]. Approximately 10% of PD cases are familial with a genetically inherited mutation, while the rest are idiopathic and have an unclear etiology. At the cellular level, PD has been linked to defects in multiple pathways including abnormalities in mitochondrial and lysosomal function, protein accumulation, synaptic and axonal dysfunction, ER stress, and increased oxidative stress [51, 55]. Genetically, multiple genes have been linked to either dominant or recessive familial forms of PD including SCNA, LRRK2, PINK1, PARK2 (parkin) and GBA1, as well as additional genes such as DJ-1, PARK9 (ATP13A2), SJ-1 and VPS35. Thus, iPSC-derived neurons and organoids obtained from PD patients harboring mutations in these genes, as well as from idiopathic PD patients have allowed for new models for understanding PD pathology (Table 1) and testing therapeutics.
Table 1:
Key Pathogenic Phenotypes associated with Parkinson’s Disease Mutant iPSC Neuron and Midbrain-like Organoid Models
| Protein/Gene | Mutation | Phenotype | Reference |
|---|---|---|---|
| α-synuclein /SNCA | SNCA Triplication | Increased oxidized dopamine | Burbulla et al 2017 [12] |
| SNCA Triplication | Decreased lysosomal hydrolase trafficking Decreased lysosomal GCase enzyme activity |
Mazzulli et al 2016 [66] | |
| A53T, E46K | Reduced tetramer to monomer ratio of α-synuclein Decreased α-synuclein solubility Increased neurotoxicity |
Dettmer et al 2015 [63] | |
| A53T | Inhibition of MEF2C-PGC1α leading to mitochondrial dysfunction. Increased nitrosative and oxidative stress Increased vulnerability to mitochondrial toxins Aggregation of α-synuclein |
Ryan, SD et al 2013 [61] | |
| A53T, SNCA Triplication | Increased nitrosative stress, ER stress, and UPR GCase accumulation in the ER |
Chung et al 2013 [74] | |
| SNCA Triplication | Aggregation of α-synuclein Overexpression of oxidative stress markers Increased sensitivity to peroxide induced oxidative stress |
Byers et al 2011 [65] | |
| LRRK2/LRRK2 | R1441C/G | Decreased activity dependent synaptic vesicle endocytosis Decreased GDP/GTP cycling Increased auxillin phosphorylation |
Nguyen et al 2018 [94] |
| G2019S | Increased oxidized dopamine | Burbulla et al 2017 [12] | |
| G2019S | Decreased neurite length Upregulation of several autophagic markers |
Borgs et al 2016 [88] | |
| Q456X G2019S/R1441C |
Mitochondrial dysfunction and increase of mitochondrial reactive oxygen species Increased sensitivity to Valinomycin |
Cooper et al 2012 [190] | |
| G2019S | Accumulation of α-synuclein Upregulation of oxidative stress response genes Increased vulnerability to neurotoxins Elevated kinase activity |
Nguyen et al 2011 [93] | |
| Midbrain Organoid: G2019S | Upregulation of TXNIP genes and a-syn accumulation | Kim, H et al 2019 [46] | |
| Midbrain Organoid: G2019S | Increased FOXA2 expression | Smits et al 2019 [158] | |
| Pink1/PINK1 | Exon 3,5 Deletion Exon 3 Deletion |
Increased spontaneous Dopamine release Decreased DA uptake and DAT-binding Increased MAO transcripts and Oxidative stress |
Jiang et al 2012 [112] |
| Q456X; V170G | Impaired parkin recruitment to mitochondria Increased mitochondrial copy number Upregulation of PGC-1α |
Seibler et al 2011 [102] | |
| Parkin/PARK2 | KO | Increased sensitivity to Oxidative stress Deficient glycolysis and lactate metabolism Mitochondrial elongation and enlargement Decreased Neuron Survival |
Bogetofte et al 2019 [191] |
| V324A | Increased oxidized dopamine Decreased GCase activity |
Burbulla et al 2017 [12] | |
| GCase/GBA1 | 84GG/WT | Increased oxidized DA | Burbulla et al 2019 [134] |
| N370S/WT | Reduced protein level and activity of GCase Accumulation of GSL and α-synuclein |
Kim et al 2018 [128] | |
| RecNcil/WT, L444P/WT, N370S/WT | Defective mitochondrial function: Altered cristae morphology Increased mitochondrial diameter Reduced oxygen consumption rate(OCR) Reduced complex I activity Altered NAD+ metabolism Increased ER stress and UPR |
Schondorf et al 2018 [133] | |
| N370S/c0.84dupG | α-synuclein accumulation in cell body and neurites | Mazzuli et al 2016 [132] | |
| N370S/WT | Abnormal GCase post-translation Lipid profile change Upregulation of ER stress Autophagic disturbance Impaired lysosomal degradation capacity Enlargement of lysosomes Increased extracellular α-synuclein |
Fernandes et al 2016 [130] | |
| N370S/N370S N370S/c0.84dupG | Reduced protein level of GCase Reduced DA uptake Elevations in GlcSph, GlcCer Increase α-synuclein protein level but not mRNA level |
Aflaki et al 2016 [127] | |
| PD: RecNcil/WT, L444P/WT, N370S/WT GD: Type 1: N370S/N370S Type 3: L444P/L444P |
Reduction in GCase level and activity Increase in GluCer and a-syn Defected Calcium homeostasis and increased vulnerability to stress response Alteration in the autophagy Increased size and the number of late endosome/lysosome (Lamp1 +) |
Schondorf et al 2014 [129] | |
| N370S/WT | Increased α-synuclein level Elevated mRNA and protein levels of Monoamine oxidase B (MAO-B) and lower DA level |
Woodard et al 2014 [131] | |
| DJ-1/PARK7 | KO | Elevated mitochondrial oxidation Increased oxidized DA Decreased basal respiration Decreased lysosomal proteolysis and GCase enzyme activity α-synuclein accumulation |
Burbulla et al 2017 [12] |
| ATP13A2/PARK9 | 1550 C>T 3176 T>G and 3253 del C |
Disruption of lysosomal Ca2+ homeostasis Reduced lysosomal Ca2+ Storage Increase in cytosolic Ca2+ levels Overall impaired lysosomal exocytosis |
Tsunemi et al 2019 [140] |
| Synaptojanin/SJ1 | R258Q | Accumulation of Atg18a on nascent synaptic autophagosomes Decreased autophagosome maturation Dopaminergic neuron loss |
Vanhauwaert et al 2017 [144] |
| VPS35/VPS35 | D620N | Defective synaptic transmission AMPA-type glutamate receptor (AMPAR) recycling | Munsie et al 2015 [147] |
2.2. IPSC-Derived Neuronal modeling in PD
2.2.1. α-Synuclein (SNCA) models
The SNCA gene encodes the 14kDa monomeric protein α-synuclein, which has been linked to multiple functions including lipid binding and regulation of synaptic vesicles, and is a major component of Lewy Body aggregates in PD patients [52, 56]. Additionally, α-synuclein is a flexible protein that takes on different confirmations dictated by cellular stress and ligand binding [57, 58]. Both N-terminal point mutations (A53E, A53T, A53V, A30P, E46K, H50Q, and G51D) [59] as well as genomic triplication of the SNCA locus [60] lead to autosomal dominant forms of familial PD [56].
iPSC-derived dopamine neurons (iPSC-DA neurons) generated from patients harboring mutant A53T α-synuclein or α-synuclein triplication have highlighted multiple pathways disrupted in patient neurons. These include increased nitrosative stress and mitochondrial dysfunction [61], disrupted synaptic connectivity and transcriptional alterations in synaptic signaling genes [62] and reduced α-synuclein tetramer to monomer ratio [63] in mutant A53T α-synuclein neurons. In addition, iPSC-DA neurons carrying α-synuclein triplication which have elevated levels of α-synuclein [64] demonstrate increased oxidative stress markers [65], decreased lysosomal hydrolase trafficking and lysosomal GCase enzyme activity [66] and increased oxidized dopamine levels [12]. They have also been found to have increased mitochondrial permeability transition pore opening via α-synuclein aggregates that interact with the ATP synthase [67], increased unfolded protein response and ER stress [68], defective ER-mitochondria contacts via VAPB [69] and decreased neurite outgrowth and neuronal activity [70]. Moreover, both A53T and α-synuclein triplication neurons have shown defects in mitochondrial respiration, membrane potential, morphology and expression of genes linked to mitochondrial function [71], as well as abnormal accumulation of Miro on the outer mitochondrial membrane contributing to delayed mitophagy [72]. Additionally, α-synuclein duplication and oligomer forming mutants (E46K and E57K) lead to impaired axonal mitochondrial transport and synaptic degeneration [73]. Interestingly, iPSC-derived cortical neurons harboring A53T or α-synuclein triplication also result in nitrosative stress, accumulation of endoplasmic reticulum (ER)-associated degradation substrates, and ER stress [74], suggesting that certain pathogenic phenotypes may also be present in non-dopaminergic neurons.
2.2.2. LRRK2 models
Leucine-rich repeat Kinase 2 (LRRK2) is a 285 kDa multi-domain protein consisting of Ras-GTPase domain (Roc domain), kinase domain, and protein binding domains with roles in neurite development, phosphorylation of multiple proteins and endocytic sorting via interactions with Rab-GTPases [75, 76, 77, 78, 79, 80]. Mutations in LRRK2 have been identified in the kinase domain (G2019S and I2020T), GTPase domain (R1441G, R1441C, R1441H, and N1437H), and the Carboxy terminal of the Roc domain (Y1699C), leading to autosomal dominant forms of familial PD [81].
LRRK2 G2019S iPSC-DA neurons have been shown to have increased oxidized dopamine [12], increased levels of the lysosomal receptor for chaperone mediated autophagy [82], transcriptome profiles similar to that upon rotenone treatment [83], microRNA alterations [84] and global DNA hyper-methylation [85], and both LRRK2 G2019S and R1441G DA neurons also show impaired NF-κB signaling [86]. iPSC-DA neurons harboring the G2019S and I2020T mutations also show increased levels of apoptosis, reduced neurite outgrowth and length, a disruption of tau and tubulin phosphorylation, an increase in autophagosomes and autophagy genes, increased mitochondrial DNA damage, impaired Miro recruitment to the mitochondria and dysregulation of mitophagy, and upregulation of α-synuclein expression [77, 78, 87, 88, 89, 90, 91, 92, 93]. Interestingly, LRRK2 was also shown to regulate synaptic vesicle recycling, as it phosphorylates auxilin in its clathrin-binding domain at Ser627, leading to disrupted synaptic vesicle endocytosis and decreased synaptic vesicle density in LRRK2 R1441G/C mutant iPSC-DA neurons [94]. Thus, studies in iPSCs investigating LRRK2 function have helped to characterize its role in PD pathogenesis, and may identify additional targets for drug development in future studies.
2.2.3. PINK1 and Parkin models
PINK1 (phosphatase and tensin homolog (PTEN)-induced Putative Kinase1) is a mitochondrial Ser/Thr protein kinase, that is normally cleaved and released into the cytosol for degradation [95]. PINK1 localizes to the mitochondrial membrane upon its depolarization where it phosphorylates Parkin [96, 97, 98]. Parkin is encoded by the PARK2 gene and is an E3 ubiquitin protein ligase which translocates to the mitochondria to ubiquitinate mitochondrial substrates upon PINK1 activation. Together, PINK1 and Parkin regulate mitochondrial health and initiate mitophagy events, and mutations in either gene are associated with autosomal recessive and early onset forms of PD [97, 99, 100, 101].
iPSC-DA neurons from patients expressing PINK1 nonsense (Q456X) or missense (V170G) mutations show mitochondrial defects including impaired parkin recruitment to mitochondria, increased mitochondrial copy number, and upregulation of PGC-1α, a key regulator of mitochondrial biogenesis [102]. PINK1 G411S or Q456X mutant neurons also demonstrate reduced PINK1 kinase activity and mitochondria quality control [103]. In addition, S-nitrosylated PINK1 decreases Parkin translocation to mitochondrial membranes and disrupts mitophagy in iPSC-DA neurons [104]. Of note, partial genetic and pharmacological inhibition of fatty acid synthase was able to decrease toxicity in PINK1 mutant iPSC-DA neurons, potentially by increasing levels of the mitochondrial inner membrane-specific lipid cardiolipin [105].
Parkin mutant iPSC-DA neurons also show mitochondrial alterations [106] which can be rescued by induced expression of the parkin interactor mitochondrial Stomatin-like protein 2 (SLP-2) [107], as well as defective ER-mitochondria contacts and calcium transfer [108] and increased levels of soluble epoxide hydrolase which is involved in inflammation [109]. In addition, both Parkin A324 fsX110 and PINK1 Q456X mutant iPSC-DA neurons show increased α-synuclein accumulation [110] while iPSC-hypothalamic PINK1 and Parkin mutant neurons have excess ER-mitochondria contacts and abnormal lipid trafficking which may deplete phosphatidylserine from the ER to disrupt neuropeptide-containing vesicles production [111].
Interestingly, parkin V324A mutant iPSC-DA neurons also show increased levels of oxidized dopamine and decreased GCase activity [12]. Moreover, iPSC-DA neurons with knockout or mutant parkin have increased expression of monoamine oxidase (MAO) A and B and dysregulation of dopamine release and uptake via dopamine transporter (DAT) and aberrant dopaminergic regulation of presynaptic glutamatergic transmission [112, 113, 114]. Of note, mutant PINK1 Q456X neurons also show increased oxidized dopamine [12], suggesting a further role for both PINK1 and parkin in regulating dopamine homeostasis. Thus, PINK1 and Parkin contribute to a major component of mitochondrial homeostasis and dopamine metabolism, and their loss of function may help drive the loss of DA neurons in the substantial nigra.
2.2.4. GBA models
The GBA1 gene encodes a lysosomal enzyme called glucocerebrosidase (also known as GCase or β-glucosidase) that catalyzes the hydrolysis of glucosylceramide (GlcCer) to glucose and ceramide, as well as the hydrolysis of D-glucosyl-N-acylsphingosine to D-glucose and N-acylsphingosine. Homozygous or compound heterozygote GBA mutations are known to cause Gaucher disease (GD), the most common lysosomal storage disorder. After the first report of GD patients with symptoms of Parkinsonism [115], other studies confirmed the link between PD and GBA mutation including insertion, deletion, frame shift and point mutations in GBA [116, 117, 118, 119]. Approximately 5–10% of PD patients carry GBA1 mutations [120] and the two most common mutations in GBA (N370S and L444P) account for ~3% of GBA-liked PD [115, 121]. Clinically, GBA mutation carriers tend to have an early onset [122, 123] and more cognitive symptoms in addition to severe motor symptoms [124, 125, 126].
Many studies have shown reduced protein levels and GCase activity in mutant GBA patient iPSC-derived neurons across multiple GBA mutations including N370S/N370S, N370S/c0.84dupG, N370S/WT, RecNcil/WT and L444P/WT [127, 128, 129]. In particular, abnormal GCase post-translation has also been observed in iPSC-derived neurons from N370S heterozygous patients [130]. Moreover, in addition to α-synuclein accumulation in GD-linked GBA iPSC-derived neurons [117], increased α-synuclein levels [127, 129, 130, 131] and its aggregation [128, 132] have also been reported in PD-linked GBA iPSC-derived neurons. Accumulation of lipids have also been identified as a major hallmark of mutant GBA iPSC-derived DA neurons including GCase substrates, glycolipids glucosylceramide (GlcCer) and glucosylsphingosine (GlcSph) which are increased in GBA mutant neurons [127, 128, 129, 130].
Additionally, defective function of cellular organelles including lysosomes, mitochondria and the ER have been demonstrated in mutant GBA iPSC-derived neurons. Increased size and number of lysosomes [129] which may result from the reported lysosomal degradation capacity impairment [130] have been clearly observed, along with evidence of autophagic defects [129, 130], potentially rendering neurons more vulnerable to apoptosis. Moreover, mitochondrial dysfunction including decreased oxygen consumption rate (OCR), reduced complex I activity and altered NAD+ metabolism together with altered mitochondrial morphology have also been reported in mutant GBA iPSC-DA neurons [133]. Furthermore, upregulation of ER stress was also observed in multiple studies, leading to defective downstream cellular mechanisms such as calcium homeostasis and the unfolded protein response (UPR) [129, 130, 133]. Lastly, the levels and uptake of dopamine were reduced in GBA N370S iPSC-DA neurons [127, 131], along with upregulated mRNA and protein levels of MAO-B [131], while 84GG/WT iPSC-DA neurons also showed elevated levels of oxidized dopamine [134]. Of note, single-cell transcriptomic analysis of GBA N370S iPSC-DA neurons have highlighted the transcriptional repressor histone deacetylase 4 (HDAC4) as a potential upstream regulator of ER stress and disease pathogenesis [135]. Overall, understanding the role of GBA mutations in PD pathogenesis may further provide possible therapeutic strategies relevant to other forms of PD as discussed below.
2.2.5. Additional genetic PD models
Additional genes linked to familial PD such as DJ-1, PARK9 (ATP13A2), SJ-1 and VPS35 have also been used to model PD pathogenesis in patient-derived iPSC-DA neurons. DJ-1 is involved in regulating mitochondrial oxidant stress and its loss of function mutations result in autosomal recessive PD [136]. iPSC-DA neurons with mutant DJ-1 exhibit elevated mitochondrial oxidation, increased oxidized DA which is exacerbated over time, and decreased basal respiration, in addition to decreased lysosomal proteolysis and GCase enzyme activity and α-synuclein accumulation [12].
PARK9 encodes a lysosomal type 5 P-type ATPase involved in cation homeostasis, ATP13A2. The loss of ATP13A2 function leads to lysosomal dysfunction and the accumulation of α-synuclein seen in Kufor-Rakeb syndrome (KRS), a rare form of juvenile-onset PD and familial PD [137, 138]. Recently, loss of function mutations of ATP13A2 in iPSC-DA neurons have exhibited α-synuclein accumulation as well as impaired lysosomal exocytosis due to the disruption of calcium homeostasis [139, 140]. Interestingly, ATP13A2 overexpression or activators of the lysosomal calcium channel TRPML1 (transient receptor potential mucolipin 1) were able to increase α-synuclein secretion and lysosomal exocytosis, and prevent neuronal toxicity [140]. ATP13A2 thus offers potential as a therapeutic target for PD-related synucleinopathies by increasing lysosomal exocytosis and neuronal secretion of intracellular α-synuclein.
In addition, the homozygous missense R258Q mutation in the Sac domain of Synaptojanin (SJ-1) results in autosomal recessive early onset PD [141, 142, 143]. iPSC neurons with the SJ-1 mutation exhibit accumulation of Atg18a on nascent synaptic autophagosomes, thus blocking autophagosome maturation and contributing to dopaminergic neuron loss [144], further suggesting a role for SJ-1 in synaptic macroautophagy. Finally, the D620N mutation in the retromer protein VPS35 leads to autosomal dominant PD [145, 146], and iPSC-DA patient neurons show defective synaptic transmission AMPA-type glutamate receptor (AMPAR) recycling [147], which may further contribute to dopaminergic neuron loss.
2.2.6. Idiopathic PD
The generation of iPSC-derived DA neurons from patients has for the first time, allowed for the possibility to model non-genetic forms of PD and provide important insights into sporadic etiology, which account for the majority of PD patients. Interestingly, idiopathic PD patient-derived DA neurons have been shown to have decreased basal mitochondrial respiration, increased oxidized dopamine levels and oxidized DJ-1, and decreased GCase enzyme activity [12] and maturation [132]. In addition, idiopathic DA neurons also demonstrate microRNA alterations [84], global DNA hyper-methylation [85], as well as impaired Miro degradation and mitochondrial motility [89], suggesting that pathways disrupted in genetic forms of PD may be similarly affected in idiopathic forms.
Of note, environmental factors have also been proposed to play a key role in driving PD pathogenesis [148, 149, 150, 151]. These include previous studies reporting the association between smoking and monoamine oxidase B (MAO-B) polymorphism [152], agricultural insecticides and polymorphisms in the Acetylcholinesterase/paraoxonase locus [153], pesticide exposure and the CYP2D6 polymorphism [154, 155], and mitochondrial toxin-induced defects in α-synuclein A53T iPSC-derived neurons [61]. Thus, although the role of gene-environmental interactions (GxEs) in PD are still not completely understood, the use of patient neurons with mutant genetic backgrounds together with environmental risk factors such as toxins may be useful in further testing the two-hit (or double hit) hypothesis in PD progression and further used for drug discovery models.
2.3. Neural Organoid Modeling in PD
Midbrain-like organoids were first characterized in 2016 and have been gaining interest with their potential to model PD [42]. In particular, organoids offer the advantage of providing a more appropriate 3D niche environment which can consist of multiple cell types, special organizational structure, and enhanced cellular maturity [156]. Further characterization of midbrain-like organoids have shown the presence of neuromelanin-like granules and the ability to form functional neural networks [42, 157]. Thus far, studies have found that midbrain-like organoids derived from LRRK2 G2019S iPSCs show upregulation of the thiol-oxidoreductase TXNIP [46] and an increase in the floor plate marker FOXA2 during organoid development [158]. Consequently, midbrain-like organoids need to be further investigated in studies of PD, as they have great potential for identifying neurodevelopmental and novel characteristics of PD that cannot be recapitulated in 2D neuronal systems.
2.4. PD Drug Discovery in iPSC Neurons
The overall drug discovery process using iPSCs has been reviewed recently [21] and our focus will be on discoveries made using iPSC models of PD (Table 2). Therapeutic strategies have currently shown potential in inhibiting overactive proteins, rescuing phenotypes with wildtype proteins, or using agonists to rescue the activity of associated proteins.
Table 2:
Drug Discovery in Parkinson’s Disease iPSC DA Neurons
| Tested drug | Target | Mode of action | Reference |
|---|---|---|---|
| NCGC758 NCGC607 | β-Glucocerebrosidase (GCase) | Small-molecule chaperone | Aflaki et al 2016; Mazzulli et al 2016) [127, 132] |
| Quinazoline inhibitors | β-Glucocerebrosidase (GCase) | Selective stabilization of GCase | Zheng et al 2019 [159] |
| Carmofur | Acid ceramidase | Inhibitor | Kim et al 2018 [160] |
| S-181 | β-Glucocerebrosidase (GCase) | Increased wild-type GCase activity | Burbulla et al 2019 [134] |
| mito-TEMPO NAC | Mitochondria | Antioxidants | Burbulla et al. 2017 [12] |
| Isoxasole | MEF2C | Activator | Ryan et al. 2013 [61] |
| PD0325901 | MEK | Inhibitor | Reinhardt et al 2013 [78] |
| NPT100–18A NPT100–14A ELN48228 | α-synuclein | Interfering oligomerization | Kouroupi et al 2017 [62] |
In particular, multiple studies have highlighted the potential use for targeting the GCase pathway in PD human iPSC-DA models. GBA chaperones NCGC758 and NCGC607 were found to restore GCase activity and reduce substrate accumulation in the lysosome in multiple PD models of iPSC-DA neurons [127, 132]. In addition, studies on GCase have identified quinazoline inhibitors that can be derived into activators that stabilize GCase activity within iPSC-DA and fibroblasts [159]. Moreover, inhibition of acid ceramidase using carmofur was shown to reduce α-synuclein accumulation in GBA-1 mutant iPSC-DA neurons [160], while reducing glycosphingolipids in GBA mutant (N370S/c.84dupG) or α-synuclein triplication neurons diminished pathology and restored physiological α-synuclein conformers that associated with synapses [161]. Finally, recent work has identified a novel chemical series of GCase activators, including a new small-molecule modulator (S-181) that increased wild-type GCase activity in iPSC-derived dopaminergic neurons from patients with 84GG-GBA1, as well as in LRRK2-, Parkin-, DJ-1-linked and sporadic PD [134]. Thus, GCase activity represents a major target for PD therapeutic treatment that is associated with multiple forms of PD, including both genetic and idiopathic cases.
Additionally, other drugs have also been tested in iPSC-DA models which have been shown to help ameliorate several phenotypes. Mitochondrial antioxidants, mito-TEMPO and NAC, show reduction of oxidized dopamine leading to reduced insoluble α-synuclein levels and increased GCase activity in patient iPSC-DA neurons [12]. Activation of MEF2C using isoxasole was sufficient to rescue α-synuclein A53T iPSC-DA neurons from nitrosative stress via the MEF2C-PGC1α pathway by increasing mitochondria respiration and biogenesis [61]. In addition, LRRK2 G2019S iPSCs treated with the MEK inhibitor PD0325901 showed protection from oxidative stress [78], while knockdown of α-synuclein showed rescue of the phenotype and survival of DA neurons [92]. Interfering with α-synuclein oligomerization in iPSC-DA neurons with NPT100–18A, NPT100–14A, and ELN48228 also rescued PD pathology [62]. Moreover, upregulation of Synoflavin or the ubiquitin ligase NEDD4 which it affects were shown to reverse accumulation of GCase and other glycoproteins in the ER in α-synuclein A53T and triplication iPSC-derived cortical neurons [74], while inhibition of stearoyl-CoA-desaturase was also shown to reduce α-synuclein inclusion formation caused by excess oleic acid in iPSC neurons [162]. Finally, treatment with coenzyme Q(10), rapamycin, or the LRRK2 kinase inhibitor GW5074 were previously shown to ameliorate mitochondrial dysfunction in iPSC neurons from multiple PD patients (LRRK2 R1441C; LRRK2 G2019S; PINK1 Q456X mutants) [163].
Of note, human iPSC-DA neurons have also been transplanted into in vivo models of PD to determine their ability to survive and function in future potential dopamine cell replacement therapy strategies for PD patients [164]. Initial studies found that human non-iPSC-derived DA neurons could efficiently engraft into PD rodent models [20, 165, 166], while later work showed that iPSC-derived DA neurons were also successful in integrating into PD rodent models [167, 168], including neurons derived from PD patients [169]. More recently, both human and non-human primate iPSC-derived DA neurons could function properly following transplantation back into PD non-human primate models [34, 170], highlighting the potential for success in future dopamine replacement studies based on iPSC-derived neurons.
3. Conclusion
iPSC-derived neurons and organoids generated from both familial and idiopathic patients have replicated key PD pathogenic phenotypes and are an important model for studying and identifying novel neuronal pathways involved in disease. While the majority of studies thus far have involved 2D neuronal cultures, the use of midbrain-like 3D organoids will be important for investigating pathologies and neuronal complexity that are not reflected in 2D models. Importantly, these technologies have added unique approaches to drug screens, and additionally provide new models to reassess current neuroprotective and neurotoxic compounds that are under consideration. Patient-derived cell cultures can thus play a key role in identifying disease mechanisms that can be therapeutic targets for multifaceted diseases such as PD. In addition, drug screens in iPSC-DA neurons thus far have been capable of identifying neuroprotective effects, and may additionally provide insight into the efficacy of compounds in human neurons. Finally, the use of organoids for drug discovery allow the possibility for studying how therapeutics modulate multiple cell types in a more physiological 3D model.
4. Expert Opinion
4.1. IPSC Derived Neurons for Parkinson’s Disease Modeling
Since 2006 and the discovery of the factors capable of rendering cells into a state of pluripotency, many advancements have been made in Parkinson’s disease research. One of the main benefits to using iPSCs to model disease is the genetic background of the patient cells, which allows for direct study of relevant disease mutations. Moreover, pluripotency allows for the generation of disease specific cell types, allowing for the study of disease mechanisms in human dopaminergic neurons. In addition, the ability to generate isogenic controls from patient lines using gene corrective technologies has proven to be essential for identifying which phenotypes arise from specific disease mutations.
Importantly, human neurons have also provided a unique opportunity for screening and testing novel therapeutics that could not have been revealed by traditional cell culture or animal model experiments. Indeed, human PD iPSC-DA neurons in long-term culture show time-dependent onset of PD phenotypes such as early defects in lysosomal dysfunction followed by subsequent α-synuclein accumulation [140]. Patient-derived PD iPSC-DA neurons also exhibit multiple pathogenic phenotypes that are not observed in mouse models of PD, which display negligible levels of oxidized dopamine. In particular, human DJ-1 KO iPSC-derived neurons demonstrate decreased lysosomal GCase activity and reduced tyrosine hydroxylase (TH) in the SNc which are not observed in DJ-1 KO mice [12]. Thus, patient-derived iPSC-based PD modeling may allow for the study of distinct pathogenic phenotypes arising over time which were not previously found in animal or traditional cell models.
Of note, one important factor in studying neurodegenerative diseases is understanding the role of aging. Indeed, age represents the greatest risk factor for PD, with the incidence of PD at ~0.5 to 1 percent in people 65 to 69 years of age rising sharply to 1 to 3 percent in people over 80 years of age [171, 172]. Thus, studying aging in the context of iPSC-derived neuronal models has represented another key angle for PD research. Previous studies have used long term cultures [90, 173], artificial cellular aging induced by genotoxic stress [174] or expression of progerin [175] to induce age-related features of PD. Indeed, long-term cultures grown for hundreds of days have been used to study progressive PD-linked disease phenotypes in human iPSC-derived neurons [173]. Interestingly, age-related PD phenotype such as gradual loss of tyrosine hydroxylase (TH), mitochondrial dysfunction, dendrite degeneration, Lewy body formation, and accumulation of neuromelanin were accelerated in iPSC-derived dopaminergic neurons upon progerin-induced artificial aging [175].
Another angle used to study aging phenotypes has been the generation of induced neurons (iNs) which involve the direct conversion from somatic cells to functional neurons. Importantly, this strategy overcomes the problem of rejuvenation during iPSC reprogramming from somatic cells [176, 177, 178] which makes it hard for iPSCs to reflect a donor’s age. Compared to iPSC-derived neurons, iNs maintain epigenetic features and aging phenotypes of donors which drive age-related pathologies in the disease of interest. Previous studies on PD iNs have shown that PINK1 Q456X iNs do not demonstrate pS65-ubiquitin accumulation upon mitochondrial damage [179] and that both p.G411S and p.Q456X PINK1 heterozygote iNs have reduced PINK1 kinase activity and pS65-ubiquitin level [103]. However, the use of iNs also has several limitations including the fact that while iPSCs are self-renewing, the somatic cells from which iNS are derived become senescent after multiple passages and are thus limited in their ability to be expanded over long periods [180]. Thus, taken together, multiple approaches may be required to enable better modeling of aging in PD drug discovery.
In addition, there are currently several key points that must be considered when using iPSC technology. First, there exists significant batch to batch variation across patient lines and even within identical clonal populations taken from the same patient, which must be taken into consideration when deciphering and interpreting results from drug screens aimed at targeting pathogenic phenotypes in patient neurons. Multiple factors account for this variability, including health of the neuronal culture, as well as the formation of gradients within media which can lead to differences in 2D neuronal differentiations. Furthermore, comparative studies using patient-derived and healthy donor-derived iPSCs often do not distinguish between general individual variations caused by different genetic background [181]. To address this issue, multiple studies have now used genetic editing to create isogenic controls by specifically correcting the known PD-linked mutation in patient-derived iPSCs [61, 129, 134, 175]. Addtionally, to overcome batch by batch differences, more than 3 independent batches of differentiation are normally required for iPSC-based disease studies. Thus, given small sample sizes and high sample variability, there is a need to recruit multiple patient and healthy donors, and create extensive biobanks (ex. skin and/or blood samples) with multiple clonal lines with isogenic controls for each patient line that will aid in reducing the effect of patient to patient variability in drug screens and aid in the discovery of specific phenotypes.
Secondly, there is a need for standardizing differentiation protocols across researchers and strict guidelines for characterizing healthy and successfully differentiated neuronal cultures. One of the challenges of iPSC-based PD modeling is the efficient generation of high quality A9 DA neurons and their subsequent validation. Currently, common markers used for validation of iPSC-derived DA neurons are immunostaining or immunoblot levels of TH, FOXA2, LMX1 and NURR1 [20]. In addition, electrophysiological recordings have also been used to further support the functional specific characterization of DA neurons including oscillation of membrane potential (2~5Hz) [20] or voltage sag and short rebound delay upon injection of a hyperpolarizing current (−250 pA) [12]. Thus, standardizing the methods for validating the generation of DA neurons and further characterizing the genetic, functional and/or molecular similarities and differences between iPSC-derived DA neurons and actual DA neurons from human brains will be important for understanding both the effectiveness and limitations of this model in future PD drug discovery.
Thirdly, iPSC-derived neuronal cultures are extremely costly, due to the sheer cost of media, peptides, small molecules, supplements, and consumables that are required for iPSC maintenance and throughout the duration of neuronal differentiation. This is further exacerbated in studies on neurodegenerative and age-related phenotypes which may require even longer culture durations. Thus, identifying ways to decrease these costs will be important for moving forward. Practically, this may involve purchasing expensive media and growth factors in bulk at lower costs to be shared across multiple labs, or identifying companies which are able to generate these reagents at reduced prices. It may also be important to plan experiments in advance to ensure that all neurons from each differentiation are efficiently used to avoid wastage of cells, materials and/or incubator space. Alternatively, the generation of iNeurons via direct differentiation of cultured patient fibroblasts into neurons which bypasses the need to culture iPSCs may also help to reduce time and cell culture costs for patient-derived neurons [182, 183]. Of note, these difficulties further present a challenge for conducting high-throughput screens using iPSC-derived neurons, where there is a need to produce large-scale expansion of neuronal cells, which is further limited by both time and cost, as well as incubator space for storing large batches of neurons needed for screening.
Thus, future studies and additional models may be required to further study the role of aging in patient neurons, how different time points in 2D cultures map to an aging patient cohort, and the role of epigenetic components in mediating PD onset and progression. Finally, further work using genome-scale networks in iPSC-derived neurons may help to further identify possible targets for therapies in PD [184].
4.2. Neural Organoids for Parkinson’s Disease Modeling
The use of midbrain organoids thus far for studying PD pathology has been mainly restricted to only a few studies. However, organoids can provide key insights into developmental characteristics that might be implicated in disease progression. Current studies have shown that organoids cultured up to two years give rise to functionally mature neurons, have increased cell diversity, and demonstrate time points which parallel in vivo development at postnatal stages [185]. Moreover, organoids may be capable of representing key time points in development, as seen in cerebral organoids with the appearance of astroglial cells during differentiation [38]. Additionally, unlike mouse models, midbrain-like organoids are able to produce neuromelanin [12, 42], and thus allow for possible testing of therapeutic compounds on multiple cell types, including both those affected in PD and those that are spared. In addition, the use of CRISPR technology in organoids [186] is highly relevant for studying the role of PD familial genes and generating isogenic controls to which to compare pathogenic phenotypes, as well as for the application of organoids in personalized medicine.
Importantly, neuronal organoids have previously shown drug discovery potential in other diseases, such as the discovery of pharmaceutical modulators of the mutated L-type calcium channel associated with Timothy Syndrome which can restore the migration of cortical interneurons [47]. Furthermore, 3D brain organoids have demonstrated promise by recapitulating key β-amyloid and tau pathology phenotypes seen in Alzheimer’s disease [187]. Finally, neuronal organoids have been able to recapitulate aspects of the blood brain barrier in co-cultures with endothelial cells and thus provide the potential for screening compounds for CNS drug targeting [188]. As assembloids offer an interesting tool for modeling the connections between different brain regions, this may be relevant for PD pathogenesis in elucidating how patient neurons might exhibit dysfunctional circuitry between midbrain dopaminergic and striatal neurons.
However, there still exists several limitations to the use of neuronal organoids as a model for drug discovery. Similar to 2D cultures, 3D neuronal organoids are also highly variable even if generated from the same clonal cell line. Thus, recent studies have sought to reduce the variability between organoids [37, 189], in addition to controlling the nutrient flow and cell gradient, and further reducing the spatial and temporal variability of the cell culture environment, as seen in organ-on-a-chip technology. Moreover, organoids present additional costs compared to 2D cultures, as they often require spinning bioreactors and may require even longer differentiation times to develop age-associated phenotypes. Additionally, as much of the current progress in organoid technology has been developed using cortical and forebrain organoids, further work needs to be conducted characterizing and standardizing the differentiation of midbrain organoids which will be relevant for PD research.
Moving forward, it will thus be critical to further develop brain organoid models by improving vascularization to avoid neuronal death on the inside of the organoid, and to test the ability of compounds to cross the blood brain barrier. As organoids often represent multi-cell type cultures, it will also be important to adapt assays from 2D cultures which are used to screen singular cell types to multi-cell type cultures, such as the use of fluorescence-activated cell sorting to examine phenotypes in individual cell types after organoid dissociation. In addition, efforts to further optimize organoid differentiation protocols from patient cells may provide new insights into PD pathology.
4.3. Future of Stem Cell modeling in Parkinson’s Disease
Future work using iPSC-DA neuron and organoid modeling in PD will be highly valuable for elucidating neuronal pathways and identifying relevant therapeutic targets, as well as providing important models for testing future therapeutics. Moving forward, it will be imperative to develop biobanks that host iPSCs from specific patient lines and disease linked mutations, as well as to establish standardized differentiation protocols and markers for midbrain DA neuron generation. More sophisticated brain modeling including the incorporation of different neuronal subtypes, as well as glial cells into organoid systems will be important. Moreover, studies involving multiple clones and patient lines, possibly comparing different PD genes and mutations, will help advance our understanding of PD pathogenesis. Finally, further studies involving idiopathic PD patient-derived iPSC neurons will be crucial for shedding light on the pathways contributing to PD onset in sporadic patients which represent the vast majority of PD cases and aid in the development of future therapeutics.
Article Highlights box.
iPSC-derived dopamine neurons represent an important model for studying familial and idiopathic Parkinson’s disease mechanisms.
Parkinson’s patient-derived dopamine neurons recapitulate key pathogenic phenotypes including α-synuclein accumulation, decreased GCase activity and organelle dysfunction.
iPSC-derived midbrain-like organoids may offer more sophisticated and physiologically relevant 3D models for studying Parkinson’s.
iPSC-derived neurons and organoids are useful models for drug discovery, identifying therapeutic targets, and compound screening and may also be relevant for future dopamine replacement studies.
Several studies have used iPSC-derived neurons as models for assessing the efficacy of Parkinson’s disease therapeutics.
Funding:
This manuscript was funded by the National Institutes of Health (NIH) via grant R01 NS076054.
Footnotes
Declaration of Interest:
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures:
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006. August 25;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]; ** Initial paper identifying factors used for the derivation of iPSCs in mice
- 2.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]; * Hallmark paper on the derivation of iPSCs from human somatic cells
- 3.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008. January;26(1):101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 4.Gu H, Huang X, Xu J, et al. Optimizing the method for generation of integration-free induced pluripotent stem cells from human peripheral blood. Stem Cell Res Ther. 2018. June 15;9(1):163. doi: 10.1186/s13287-018-0908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu X, Liu T, Song K, et al. Induced pluripotent stem cells generated from human adipose-derived stem cells using a non-viral polycistronic plasmid in feeder-free conditions. PLoS One. 2012;7(10):e48161. doi: 10.1371/journal.pone.0048161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takenaka C, Nishishita N, Takada N, et al. Effective generation of iPS cells from CD34+ cord blood cells by inhibition of p53. Exp Hematol. 2010. February;38(2):154–62. doi: 10.1016/j.exphem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Wiedemann A, Hemmer K, Bernemann I, et al. Induced pluripotent stem cells generated from adult bone marrow-derived cells of the nonhuman primate (Callithrix jacchus) using a novel quad-cistronic and excisable lentiviral vector. Cell Reprogram. 2012. December;14(6):485–96. doi: 10.1089/cell.2012.0036. [DOI] [PubMed] [Google Scholar]
- 8.Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22(4):635–41. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- 9.Robertson JA. Human embryonic stem cell research: ethical and legal issues. Nature Reviews Genetics. 2001. 2001/01/01;2(1):74–78. doi: 10.1038/35047594. [DOI] [PubMed] [Google Scholar]
- 10.Barker RA, de Beaufort I. Scientific and ethical issues related to stem cell research and interventions in neurodegenerative disorders of the brain. Prog Neurobiol. 2013. November;110:63–73. doi: 10.1016/j.pneurobio.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Arthur KC, Calvo A, Price TR, et al. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016. August 11;7:12408. doi: 10.1038/ncomms12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burbulla LF, Song P, Mazzulli JR, et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science. 2017. September 22;357(6357):1255–1261. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Paper identifying elevated oxidized dopamine in iPSC-derived DA neurons from both familial and idiopathic Parkinson’s patients and downstream pathogenic phenotypes; Also demonstrated key differences in dopamine metabolism between human and mouse Parkinson’s models
- 13.Gitler AD, Dhillon P, Shorter J. Neurodegenerative disease: models, mechanisms, and a new hope. Dis Model Mech. 2017. May 1;10(5):499–502. doi: 10.1242/dmm.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009. March;27(3):275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abud EM, Ramirez RN, Martinez ES, et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017. April 19;94(2):278–293 e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emdad L, D’Souza SL, Kothari HP, et al. Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev. 2012. February 10;21(3):404–10. doi: 10.1089/scd.2010.0560. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y, Qu ZY, Cao SY, et al. Directed differentiation of basal forebrain cholinergic neurons from human pluripotent stem cells. J Neurosci Methods. 2016. June 15;266:42–9. doi: 10.1016/j.jneumeth.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Kirkeby A, Grealish S, Wolf DA, et al. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012. June 28;1(6):703–14. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Kirkeby A, Nolbrant S, Tiklova K, et al. Predictive Markers Guide Differentiation to Improve Graft Outcome in Clinical Translation of hESC-Based Therapy for Parkinson’s Disease. Cell Stem Cell. 2017. January 5;20(1):135–148. doi: 10.1016/j.stem.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriks S, Shim JW, Piao J, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011. November 6;480(7378):547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little D, Ketteler R, Gissen P, et al. Using stem cell–derived neurons in drug screening for neurological diseases. Neurobiology of Aging. 2019. 2019/06/01/;78:130–141. doi: 10.1016/j.neurobiolaging.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Lu J, Zhong X, Liu H, et al. Generation of serotonin neurons from human pluripotent stem cells. Nat Biotechnol. 2016. January;34(1):89–94. doi: 10.1038/nbt.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maroof AM, Keros S, Tyson JA, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013. May 2;12(5):559–72. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merkle FT, Maroof A, Wataya T, et al. Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development. 2015. February 15;142(4):633–43. doi: 10.1242/dev.117978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas CR, Chen J, Tang Y, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013. May 2;12(5):573–86. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nistor GI, Totoiu MO, Haque N, et al. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia. 2005. February;49(3):385–96. doi: 10.1002/glia.20127. [DOI] [PubMed] [Google Scholar]
- 27.Nizzardo M, Simone C, Falcone M, et al. Human motor neuron generation from embryonic stem cells and induced pluripotent stem cells. Cell Mol Life Sci. 2010. November;67(22):3837–47. doi: 10.1007/s00018-010-0463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012. October;7(10):1836–46. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Wang B, Pan N, et al. Differentiation of human induced pluripotent stem cells to mature functional Purkinje neurons. Sci Rep. 2015. March 18;5:9232. doi: 10.1038/srep09232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang N, Chanda S, Marro S, et al. Generation of pure GABAergic neurons by transcription factor programming. Nat Methods. 2017. June;14(6):621–628. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Diana X, Di Giorgio Francesco P, Yao J, et al. Modeling Hippocampal Neurogenesis Using Human Pluripotent Stem Cells. Stem Cell Reports. 2014. 2014/03/11/;2(3):295–310. doi: 10.1016/j.stemcr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunhanlar N, Shpak G, van der Kroeg M, et al. A simplified protocol for differentiation of electrophysiologically mature neuronal networks from human induced pluripotent stem cells. Mol Psychiatry. 2018. May;23(5):1336–1344. doi: 10.1038/mp.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardy C, van den Hurk M, Eames T, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A. 2015. May 19;112(20):E2725–34. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi T, Morizane A, Doi D, et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature. 2017. August 30;548(7669):592–596. doi: 10.1038/nature23664. [DOI] [PubMed] [Google Scholar]; * This study found that human iPSC dopamine neurons could be transplanted and functionally integrate into a primate model
- 35.Garcia-Leon JA, Vitorica J, Gutierrez A. Use of human pluripotent stem cell-derived cells for neurodegenerative disease modeling and drug screening platform. Future Med Chem. 2019. June;11(11):1305–1322. doi: 10.4155/fmc-2018-0520. [DOI] [PubMed] [Google Scholar]
- 36.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012. June 14;10(6):771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Velasco S, Kedaigle AJ, Simmons SK, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019. 2019/06/01;570(7762):523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper showed that organoids contain diverse cell population that change overtime, including the rise of astroglial cells from three months to six months, allowing for the potential to study development and aging in vitro
- 38.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. 2018. November;19(11):671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]; * This review provides a detailed analysis on the state of organoid research and the possibilities for their application
- 39.Sasai Y Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013. January 17;493(7432):318–26. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- 40.Assawachananont J, Mandai M, Okamoto S, et al. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Reports. 2014. May 6;2(5):662–74. doi: 10.1016/j.stemcr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freedman BS, Brooks CR, Lam AQ, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun. 2015. October 23;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo J, Xiao Y, Sun AX, et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016. August 4;19(2):248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes the first protocol for differentiating midbrain-like organoids, and established the finding that human but not mouse midbrain-like organoids were capable of producing neuromelanin
- 43.Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013. September 19;501(7467):373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper pioneered the derivation of cerebral organoids with multiple brain regions and is one of the first to use organoids in modeling disease pathology
- 44.Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011. February 3;470(7332):105–9. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013. July 25;499(7459):481–4. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 46.Kim H, Park HJ, Choi H, et al. Modeling G2019S-LRRK2 Sporadic Parkinson’s Disease in 3D Midbrain Organoids. Stem Cell Reports. 2019. March 5;12(3):518–531. doi: 10.1016/j.stemcr.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birey F, Andersen J, Makinson CD, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017. May 4;545(7652):54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolsey TA, Van der Loos H. The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. The description of a cortical field composed of discrete cytoarchitectonic units. Brain Res. 1970. January 20;17(2):205–42. doi: 10.1016/0006-8993(70)90079-x. [DOI] [PubMed] [Google Scholar]
- 49.Pasca SP. The rise of three-dimensional human brain cultures. Nature. 2018. January 24;553(7689):437–445. doi: 10.1038/nature25032. [DOI] [PubMed] [Google Scholar]; **Excellent review on the current use of brain organoids and other 3D Neuronal cultures detailing the progress that they have contributed to understanding disease pathogenesis and technology advancement.
- 50.Zhang BY, Korolj A, Lai BFL, et al. Advances in organ-on-a-chip engineering. Nat Rev Mater. 2018. August;3(8):257–278. doi: 10.1038/s41578-018-0034-7. [DOI] [Google Scholar]
- 51.Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015. August 29;386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]; * A review on the complexities of Parkinson’s disease and the interplay of genetic and environmental factors that affect neuronal physiology
- 52.Spillantini MG, Schmidt ML, Lee VM, et al. Alpha-synuclein in Lewy bodies. Nature. 1997. August 28;388(6645):839–40. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]; * This paper details the discovery of α-synuclein as a primary component in Lewy bodies, a pathological hallmark of Parkinson’s disease
- 53.Lewis SJ, Foltynie T, Blackwell AD, et al. Heterogeneity of Parkinson’s disease in the early clinical stages using a data driven approach. J Neurol Neurosurg Psychiatry. 2005. March;76(3):343–8. doi: 10.1136/jnnp.2003.033530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidailhet M [Heterogeneity of Parkinson’s disease]. Bull Acad Natl Med. 2003;187(2):259–75; discussion 275–6. [PubMed] [Google Scholar]
- 55.Nguyen M, Wong YC, Ysselstein D, et al. Synaptic, Mitochondrial, and Lysosomal Dysfunction in Parkinson’s Disease. Trends Neurosci. 2018. November 30. doi: 10.1016/j.tins.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong YC, Krainc D. alpha-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nat Med. 2017. February 7;23(2):1–13. doi: 10.1038/nm.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lashuel HA, Overk CR, Oueslati A, et al. The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nat Rev Neurosci. 2013. January;14(1):38–48. doi: 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theillet FX, Binolfi A, Bekei B, et al. Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature. 2016. February 4;530(7588):45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- 59.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997. June 27;276(5321):2045–7. [DOI] [PubMed] [Google Scholar]
- 60.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003. October 31;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 61.Ryan SD, Dolatabadi N, Chan SF, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell. 2013. December 5;155(6):1351–64. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kouroupi G, Taoufik E, Vlachos IS, et al. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’s disease. Proceedings of the National Academy of Sciences. 2017;114(18):E3679. doi: 10.1073/pnas.1617259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dettmer U, Newman AJ, Soldner F, et al. Parkinson-causing alpha-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nat Commun. 2015. June 16;6:7314. doi: 10.1038/ncomms8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Devine MJ, Ryten M, Vodicka P, et al. Parkinson’s disease induced pluripotent stem cells with triplication of the alpha-synuclein locus. Nature Communications. 2011. August;2. doi: ARTN 440 10.1038/ncomms1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Byers B, Cord B, Nguyen HN, et al. SNCA triplication Parkinson’s patient’s iPSC-derived DA neurons accumulate alpha-synuclein and are susceptible to oxidative stress. PLoS One. 2011;6(11):e26159. doi: 10.1371/journal.pone.0026159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mazzulli JR, Zunke F, Isacson O, et al. alpha-Synuclein-induced lysosomal dysfunction occurs through disruptions in protein trafficking in human midbrain synucleinopathy models. Proc Natl Acad Sci U S A. 2016. February 16;113(7):1931–6. doi: 10.1073/pnas.1520335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ludtmann MHR, Angelova PR, Horrocks MH, et al. alpha-synuclein oligomers interact with ATP synthase and open the permeability transition pore in Parkinson’s disease. Nature Communications. 2018. June 12;9. doi: ARTN 2293 10.1038/s41467-018-04422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heman-Ackah SM, Manzano R, Hoozemans JJM, et al. Alpha-synuclein induces the unfolded protein response in Parkinson’s disease SNCA triplication iPSC-derived neurons. Human Molecular Genetics. 2017. November 15;26(22):4441–4450. doi: 10.1093/hmg/ddx331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paillusson S, Gomez-Suaga P, Stoica R, et al. alpha-Synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca2+ homeostasis and mitochondrial ATP production. Acta Neuropathol. 2017. July;134(1):129–149. doi: 10.1007/s00401-017-1704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliveira LMA, Falomir-Lockhart LJ, Botelho MG, et al. Elevated alpha-synuclein caused by SNCA gene triplication impairs neuronal differentiation and maturation in Parkinson’s patient-derived induced pluripotent stem cells. Cell Death Dis. 2015. November;6. doi: ARTN e1994 10.1038/cddis.2015.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zambon F, Cherubini M, Fernandes HJR, et al. Cellular alpha-synuclein pathology is associated with bioenergetic dysfunction in Parkinson’s iPSC-derived dopamine neurons. Human Molecular Genetics. 2019. June 15;28(12):2001–2013. doi: 10.1093/hmg/ddz038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaltouki A, Hsieh CH, Kim MJ, et al. Alpha-synuclein delays mitophagy and targeting Miro rescues neuron loss in Parkinson’s models. Acta Neuropathol. 2018. October;136(4):607–620. doi: 10.1007/s00401-018-1873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prots I, Grosch J, Brazdis RM, et al. alpha-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. P Natl Acad Sci USA. 2018. July 24;115(30):7813–7818. doi: 10.1073/pnas.1713129115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung CY, Khurana V, Auluck PK, et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013. November 22;342(6161):983–7. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guaitoli G, Raimondi F, Gilsbach BK, et al. Structural model of the dimeric Parkinson’s protein LRRK2 reveals a compact architecture involving distant interdomain contacts. Proceedings of the National Academy of Sciences. 2016;113(30):E4357. doi: 10.1073/pnas.1523708113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kawakami F, Yabata T, Ohta E, et al. LRRK2 phosphorylates tubulin-associated tau but not the free molecule: LRRK2-mediated regulation of the tau-tubulin association and neurite outgrowth. PLoS One. 2012;7(1):e30834. doi: 10.1371/journal.pone.0030834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohta E, Kawakami F, Kubo M, et al. Dominant-negative effects of LRRK2 heterodimers: a possible mechanism of neurodegeneration in Parkinson’s disease caused by LRRK2 I2020T mutation. Biochem Biophys Res Commun. 2013. January 11;430(2):560–6. doi: 10.1016/j.bbrc.2012.11.113. [DOI] [PubMed] [Google Scholar]
- 78.Reinhardt P, Schmid B, Burbulla LF, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013. March 7;12(3):354–67. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 79.Steger M, Tonelli F, Ito G, et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife. 2016. January 29;5. doi: 10.7554/eLife.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parisiadou L, Xie CS, Cho HJ, et al. Phosphorylation of Ezrin/Radixin/Moesin Proteins by LRRK2 Promotes the Rearrangement of Actin Cytoskeleton in Neuronal Morphogenesis. J Neurosci. 2009. November 4;29(44):13971–13980. doi: 10.1523/Jneurosci.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci. 2010. December;11(12):791–7. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orenstein SJ, Kuo SH, Tasset I, et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013. April;16(4):394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sandor C, Robertson P, Lang C, et al. Transcriptomic profiling of purified patient-derived dopamine neurons identifies convergent perturbations and therapeutics for Parkinson’s disease. Human Molecular Genetics. 2017. February 1;26(3):552–566. doi: 10.1093/hmg/ddw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tolosa E, Botta-Orfila T, Morato X, et al. MicroRNA alterations in iPSC-derived dopaminergic neurons from Parkinson disease patients. Neurobiol Aging. 2018. September;69:283–291. doi: 10.1016/j.neurobiolaging.2018.05.032. [DOI] [PubMed] [Google Scholar]
- 85.Fernandez-Santiago R, Merkel A, Castellano G, et al. Whole-genome DNA hyper-methylation in iPSC-derived dopaminergic neurons from Parkinson’s disease patients. Clin Epigenetics. 2019. July 23;11. doi: ARTN 108 10.1186/s13148-019-0701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Maturana RL, Lang V, Zubiarrain A, et al. Mutations in LRRK2 impair NF-kappa B pathway in iPSC-derived neurons. J Neuroinflamm. 2016. November 18;13. doi: ARTN 295 10.1186/s12974-016-0761-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin L, Goke J, Cukuroglu E, et al. Molecular Features Underlying Neurodegeneration Identified through In Vitro Modeling of Genetically Diverse Parkinson’s Disease Patients. Cell Rep. 2016. June 14;15(11):2411–26. doi: 10.1016/j.celrep.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 88.Borgs L, Peyre E, Alix P, et al. Dopaminergic neurons differentiating from LRRK2 G2019S induced pluripotent stem cells show early neuritic branching defects. Sci Rep. 2016. September 19;6:33377. doi: 10.1038/srep33377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsieh CH, Shaltouki A, Gonzalez AE, et al. Functional Impairment in Miro Degradation and Mitophagy Is a Shared Feature in Familial and Sporadic Parkinson’s Disease. Cell Stem Cell. 2016. December 1;19(6):709–724. doi: 10.1016/j.stem.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol Med. 2012. May;4(5):380–95. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanders LH, Laganiere J, Cooper O, et al. LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson’s disease patients: reversal by gene correction. Neurobiol Dis. 2014. February;62:381–6. doi: 10.1016/j.nbd.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Skibinski G, Nakamura K, Cookson MR, et al. Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J Neurosci. 2014. January 8;34(2):418–33. doi: 10.1523/JNEUROSCI.2712-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen HN, Byers B, Cord B, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011. March 4;8(3):267–80. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nguyen M, Krainc D. LRRK2 phosphorylation of auxilin mediates synaptic defects in dopaminergic neurons from patients with Parkinson’s disease. Proc Natl Acad Sci U S A. 2018. May 22;115(21):5576–5581. doi: 10.1073/pnas.1717590115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Greene AW, Grenier K, Aguileta MA, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012. April;13(4):378–85. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010. June 10;66(5):646–61. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015. January 21;85(2):257–73. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zeng XS, Geng WS, Jia JJ, et al. Cellular and Molecular Basis of Neurodegeneration in Parkinson Disease. Front Aging Neurosci. 2018;10:109. doi: 10.3389/fnagi.2018.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Durcan TM, Fon EA. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015. May 15;29(10):989–99. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998. April 9;392(6676):605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 101.Shimura H, Hattori N, Kubo S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000. July;25(3):302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 102.Seibler P, Graziotto J, Jeong H, et al. Mitochondrial Parkin Recruitment Is Impaired in Neurons Derived from Mutant PINK1 Induced Pluripotent Stem Cells. Journal of Neuroscience. 2011. April 20;31(16):5970–5976. doi: 10.1523/Jneurosci.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Puschmann A, Fiesel FC, Caulfield TR, et al. Heterozygous PINK1 p.G411S increases risk of Parkinson’s disease via a dominant-negative mechanism. Brain. 2017. January;140(1):98–117. doi: 10.1093/brain/aww261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oh CK, Sultan A, Platzer J, et al. S-Nitrosylation of PINK1 Attenuates PINK1/Parkin-Dependent Mitophagy in hiPSC-Based Parkinson’s Disease Models. Cell Reports. 2017. November 21;21(8):2171–2182. doi: 10.1016/j.celrep.2017.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vos M, Geens A, Bohm C, et al. Cardiolipin promotes electron transport between ubiquinone and complex I to rescue PINK1 deficiency. Journal of Cell Biology. 2017. March;216(3):695–708. doi: 10.1083/jcb.201511044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shaltouki A, Sivapatham R, Pei Y, et al. Mitochondrial Alterations by PARKIN in Dopaminergic Neurons Using PARK2 Patient-Specific and PARK2 Knockout Isogenic iPSC Lines. Stem Cell Rep. 2015. May 12;4(5):847–859. doi: 10.1016/j.stemcr.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zanon A, Kalvakuri S, Rakovic A, et al. SLP-2 interacts with Parkin in mitochondria and prevents mitochondrial dysfunction in Parkin-deficient human iPSC-derived neurons and Drosophila. Human Molecular Genetics. 2017. July 1;26(13):2412–2425. doi: 10.1093/hmg/ddx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gautier CA, Erpapazoglou Z, Mouton-Liger F, et al. The endoplasmic reticulum-mitochondria interface is perturbed in PARK2 knockout mice and patients with PARK2 mutations. Human Molecular Genetics. 2016. July 15;25(14):2972–2984. doi: 10.1093/hmg/ddw148. [DOI] [PubMed] [Google Scholar]
- 109.Ren Q, Ma M, Yang J, et al. Soluble epoxide hydrolase plays a key role in the pathogenesis of Parkinson’s disease. P Natl Acad Sci USA. 2018. June 19;115(25):E5815–E5823. doi: 10.1073/pnas.1802179115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chung SY, Kishinevsky S, Mazzulli JR, et al. Parkin and PINK1 Patient iPSC-Derived Midbrain Dopamine Neurons Exhibit Mitochondrial Dysfunction and alpha-Synuclein Accumulation. Stem Cell Rep. 2016. October 11;7(4):664–677. doi: 10.1016/j.stemcr.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Valadas JS, Esposito G, Vandekerkhove D, et al. ER Lipid Defects in Neuropeptidergic Neurons Impair Sleep Patterns in Parkinson’s Disease. Neuron. 2018. June 27;98(6):1155.-+. doi: 10.1016/j.neuron.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 112.Jiang H, Ren Y, Yuen EY, et al. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat Commun. 2012. February 7;3:668. doi: 10.1038/ncomms1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ren Y, Jiang H, Ma D, et al. Parkin degrades estrogen-related receptors to limit the expression of monoamine oxidases. Hum Mol Genet. 2011. March 15;20(6):1074–83. doi: 10.1093/hmg/ddq550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhong P, Hu Z, Jiang H, et al. Dopamine Induces Oscillatory Activities in Human Midbrain Neurons with Parkin Mutations. Cell Rep. 2017. May 2;19(5):1033–1044. doi: 10.1016/j.celrep.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Neumann J, Bras J, Deas E, et al. Glucocerebrosidase mutations in clinical and pathologically proven Parkinson’s disease. Brain. 2009. July;132(Pt 7):1783–94. doi: 10.1093/brain/awp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gegg ME, Burke D, Heales SJ, et al. Glucocerebrosidase deficiency in substantia nigra of parkinson disease brains. Ann Neurol. 2012. September;72(3):455–63. doi: 10.1002/ana.23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mazzulli JR, Xu YH, Sun Y, et al. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011. July 8;146(1):37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murphy KE, Gysbers AM, Abbott SK, et al. Reduced glucocerebrosidase is associated with increased alpha-synuclein in sporadic Parkinson’s disease. Brain. 2014. March;137(Pt 3):834–48. doi: 10.1093/brain/awt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wong YC, Krainc D. Lysosomal trafficking defects link Parkinson’s disease with Gaucher’s disease. Mov Disord. 2016. November;31(11):1610–1618. doi: 10.1002/mds.26802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schapira AH. Glucocerebrosidase and Parkinson disease: Recent advances. Mol Cell Neurosci. 2015. May;66(Pt A):37–42. doi: 10.1016/j.mcn.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sidransky E, Nalls MA, Aasly JO, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009. October 22;361(17):1651–61. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gan-Or Z, Bar-Shira A, Mirelman A, et al. LRRK2 and GBA mutations differentially affect the initial presentation of Parkinson disease. Neurogenetics. 2010. February;11(1):121–5. doi: 10.1007/s10048-009-0198-9. [DOI] [PubMed] [Google Scholar]
- 123.Lesage S, Anheim M, Condroyer C, et al. Large-scale screening of the Gaucher’s disease-related glucocerebrosidase gene in Europeans with Parkinson’s disease. Hum Mol Genet. 2011. January 1;20(1):202–10. doi: 10.1093/hmg/ddq454. [DOI] [PubMed] [Google Scholar]
- 124.Cilia R, Tunesi S, Marotta G, et al. Survival and dementia in GBA-associated Parkinson’s disease: The mutation matters. Ann Neurol. 2016. November;80(5):662–673. doi: 10.1002/ana.24777. [DOI] [PubMed] [Google Scholar]
- 125.Oeda T, Umemura A, Mori Y, et al. Impact of glucocerebrosidase mutations on motor and nonmotor complications in Parkinson’s disease. Neurobiol Aging. 2015. December;36(12):3306–3313. doi: 10.1016/j.neurobiolaging.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 126.Thaler A, Gurevich T, Bar Shira A, et al. A “dose” effect of mutations in the GBA gene on Parkinson’s disease phenotype. Parkinsonism Relat Disord. 2017. March;36:47–51. doi: 10.1016/j.parkreldis.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 127.Aflaki E, Borger DK, Moaven N, et al. A New Glucocerebrosidase Chaperone Reduces alpha-Synuclein and Glycolipid Levels in iPSC-Derived Dopaminergic Neurons from Patients with Gaucher Disease and Parkinsonism. J Neurosci. 2016. July 13;36(28):7441–52. doi: 10.1523/JNEUROSCI.0636-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper found that the GCase chaperone NCGC607 could increase GCase activity and decrease α-synuclein levels in patient iPSC-DA neurons
- 128.Kim S, Yun SP, Lee S, et al. GBA1 deficiency negatively affects physiological alpha-synuclein tetramers and related multimers. Proc Natl Acad Sci U S A. 2018. January 23;115(4):798–803. doi: 10.1073/pnas.1700465115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schondorf DC, Aureli M, McAllister FE, et al. iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun. 2014. June 6;5:4028. doi: 10.1038/ncomms5028. [DOI] [PubMed] [Google Scholar]
- 130.Fernandes HJ, Hartfield EM, Christian HC, et al. ER Stress and Autophagic Perturbations Lead to Elevated Extracellular alpha-Synuclein in GBA-N370S Parkinson’s iPSC-Derived Dopamine Neurons. Stem Cell Reports. 2016. March 8;6(3):342–56. doi: 10.1016/j.stemcr.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woodard CM, Campos BA, Kuo SH, et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep. 2014. November 20;9(4):1173–82. doi: 10.1016/j.celrep.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mazzulli JR, Zunke F, Tsunemi T, et al. Activation of beta-Glucocerebrosidase Reduces Pathological alpha-Synuclein and Restores Lysosomal Function in Parkinson’s Patient Midbrain Neurons. J Neurosci. 2016. July 20;36(29):7693–706. doi: 10.1523/JNEUROSCI.0628-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper found that the GCase chaperone NCGC578 could increase GCase activity and decrease α-synuclein levels in patient iPSC-DA neurons
- 133.Schondorf DC, Ivanyuk D, Baden P, et al. The NAD+ Precursor Nicotinamide Riboside Rescues Mitochondrial Defects and Neuronal Loss in iPSC and Fly Models of Parkinson’s Disease. Cell Rep. 2018. June 5;23(10):2976–2988. doi: 10.1016/j.celrep.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 134.Burbulla LF, Jeon S, Zheng J, et al. A modulator of wild-type glucocerebrosidase improves pathogenic phenotypes in dopaminergic neuronal models of Parkinson’s disease. Sci Transl Med. 2019. October 16;11(514). doi: 10.1126/scitranslmed.aau6870. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study found that increasing GCase activity ameliorated phenotypes in both Parkinson’s disease familial and idiopathic patient iPSC-DA neurons
- 135.Lang C, Campbell KR, Ryan BJ, et al. Single-Cell Sequencing of iPSC-Dopamine Neurons Reconstructs Disease Progression and Identifies HDAC4 as a Regulator of Parkinson Cell Phenotypes. Cell Stem Cell. 2019. January 3;24(1):93.-+. doi: 10.1016/j.stem.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bonifati V, Rizzu P, van Baren MJ, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003. January 10;299(5604):256–9. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 137.Ramirez A, Heimbach A, Grundemann J, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006. October;38(10):1184–91. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 138.Mc Donald JM, Krainc D. Lysosomal Proteins as a Therapeutic Target in Neurodegeneration. Annu Rev Med. 2017. January 14;68:445–458. doi: 10.1146/annurev-med-050715-104432. [DOI] [PubMed] [Google Scholar]
- 139.Tsunemi T, Hamada K, Krainc D. ATP13A2/PARK9 regulates secretion of exosomes and alpha-synuclein. J Neurosci. 2014. November 12;34(46):15281–7. doi: 10.1523/JNEUROSCI.1629-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tsunemi T, Perez-Rosello T, Ishiguro Y, et al. Increased Lysosomal Exocytosis Induced by Lysosomal Ca(2+) Channel Agonists Protects Human Dopaminergic Neurons from alpha-Synuclein Toxicity. J Neurosci. 2019. July 17;39(29):5760–5772. doi: 10.1523/JNEUROSCI.3085-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Krebs CE, Karkheiran S, Powell JC, et al. The Sac1 Domain of SYNJ1 Identified Mutated in a Family with Early-Onset Progressive Parkinsonism with Generalized Seizures. Hum Mutat. 2013. September;34(9):1200–1207. doi: 10.1002/humu.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Olgiati S, De Rosa A, Quadri M, et al. PARK20 caused by SYNJ1 homozygous Arg258Gln mutation in a new Italian family. Neurogenetics. 2014. August;15(3):183–188. doi: 10.1007/s10048-014-0406-0. [DOI] [PubMed] [Google Scholar]
- 143.Quadri M, Fang M, Picillo M, et al. Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism. Hum Mutat. 2013. September;34(9):1208–15. doi: 10.1002/humu.22373. [DOI] [PubMed] [Google Scholar]
- 144.Vanhauwaert R, Kuenen S, Masius R, et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. Embo Journal. 2017. May 15;36(10):1392–1411. doi: 10.15252/embj.201695773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vilarino-Guell C, Wider C, Ross OA, et al. VPS35 Mutations in Parkinson Disease. American Journal of Human Genetics. 2011. July 15;89(1):162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zimprich A, Benet-Pages A, Struhal W, et al. A Mutation in VPS35, Encoding a Subunit of the Retromer Complex, Causes Late-Onset Parkinson Disease. American Journal of Human Genetics. 2011. July 15;89(1):168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Munsie LN, Milnerwood AJ, Seibler P, et al. Retromer-dependent neurotransmitter receptor trafficking to synapses is altered by the Parkinson’s disease VPS35 mutation p.D620N. Human Molecular Genetics. 2015. March 15;24(6):1691–1703. doi: 10.1093/hmg/ddu582. [DOI] [PubMed] [Google Scholar]
- 148.Cannon JR, Greenamyre JT. Gene-environment interactions in Parkinson’s disease: specific evidence in humans and mammalian models. Neurobiol Dis. 2013. September;57:38–46. doi: 10.1016/j.nbd.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chai C, Lim KL. Genetic insights into sporadic Parkinson’s disease pathogenesis. Curr Genomics. 2013. December;14(8):486–501. doi: 10.2174/1389202914666131210195808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Horowitz MP, Greenamyre JT. Gene-environment interactions in Parkinson’s disease: the importance of animal modeling. Clin Pharmacol Ther. 2010. October;88(4):467–74. doi: 10.1038/clpt.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vance JM, Ali S, Bradley WG, et al. Gene-environment interactions in Parkinson’s disease and other forms of parkinsonism. Neurotoxicology. 2010. September;31(5):598–602. doi: 10.1016/j.neuro.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 152.Checkoway H, Franklin GM, Costa-Mallen P, et al. A genetic polymorphism of MAO-B modifies the association of cigarette smoking and Parkinson’s disease. Neurology. 1998. May;50(5):1458–61. doi: 10.1212/wnl.50.5.1458. [DOI] [PubMed] [Google Scholar]
- 153.Benmoyal-Segal L, Vander T, Shifman S, et al. Acetylcholinesterase/paraoxonase interactions increase the risk of insecticide-induced Parkinson’s disease. FASEB J. 2005. March;19(3):452–4. doi: 10.1096/fj.04-2106fje. [DOI] [PubMed] [Google Scholar]
- 154.Deng Y, Newman B, Dunne MP, et al. Further evidence that interactions between CYP2D6 and pesticide exposure increase risk for Parkinson’s disease. Ann Neurol. 2004. June;55(6):897. doi: 10.1002/ana.20143. [DOI] [PubMed] [Google Scholar]
- 155.Elbaz A, Levecque C, Clavel J, et al. CYP2D6 polymorphism, pesticide exposure, and Parkinson’s disease. Ann Neurol. 2004. March;55(3):430–4. doi: 10.1002/ana.20051. [DOI] [PubMed] [Google Scholar]
- 156.Kelava I, Lancaster MA. Dishing out mini-brains: Current progress and future prospects in brain organoid research. Dev Biol. 2016. December 15;420(2):199–209. doi: 10.1016/j.ydbio.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Monzel AS, Smits LM, Hemmer K, et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports. 2017. May 9;8(5):1144–1154. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Smits LM, Reinhardt L, Reinhardt P, et al. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis. 2019;5:5. doi: 10.1038/s41531-019-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zheng J, Jeon S, Jiang W, et al. Conversion of Quinazoline Modulators from Inhibitors to Activators of beta-Glucocerebrosidase. J Med Chem. 2019. February 14;62(3):1218–1230. doi: 10.1021/acs.jmedchem.8b01294. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study was able to modify current compounds that are inhibitors to an activator as a potential therapeutic drug for increasing GCase activity
- 160.Kim MJ, Jeon S, Burbulla LF, et al. Acid ceramidase inhibition ameliorates alpha-synuclein accumulation upon loss of GBA1 function. Hum Mol Genet. 2018. June 1;27(11):1972–1988. doi: 10.1093/hmg/ddy105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zunke F, Moise AC, Belur NR, et al. Reversible Conformational Conversion of alpha-Synuclein into Toxic Assemblies by Glucosylceramide. Neuron. 2018. January 3;97(1):92.-+. doi: 10.1016/j.neuron.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fanning S, Haque A, Imberdis T, et al. Lipidomic Analysis of alpha-Synuclein Neurotoxicity Identifies Stearoyl CoA Desaturase as a Target for Parkinson Treatment. Molecular Cell. 2019. March 7;73(5):1001.-+. doi: 10.1016/j.molcel.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cooper O Pharmacological Rescue of Mitochondrial Deficits in iPSC-Derived Neural Cells from Patients with Familial Parkinson’s Disease. Sci Transl Med. 2012. July 4;4(141). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Barker RA, Farrell K, Guzman NV, et al. Designing stem-cell-based dopamine cell replacement trials for Parkinson’s disease. Nature Medicine. 2019. July;25(7):1045–1053. doi: 10.1038/s41591-019-0507-2. [DOI] [PubMed] [Google Scholar]
- 165.Rhee YH, Ko JY, Chang MY, et al. Protein-based human iPS cells efficiently generate functional dopamine neurons and can treat a rat model of Parkinson disease. Journal of Clinical Investigation. 2011. June;121(6):2326–2335. doi: 10.1172/Jci45794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gonzalez R, Garitaonandia I, Poustovoitov M, et al. Neural Stem Cells Derived From Human Parthenogenetic Stem Cells Engraft and Promote Recovery in a Nonhuman Primate Model of Parkinson’s Disease. Cell Transplant. 2016;25(11):1945–1966. doi: 10.3727/096368916x691682. [DOI] [PubMed] [Google Scholar]
- 167.Cai JL, Yang M, Poremsky E, et al. Dopaminergic Neurons Derived from Human Induced Pluripotent Stem Cells Survive and Integrate into 6-OHDA-Lesioned Rats. Stem Cells Dev. 2010. July;19(7):1017–1023. doi: 10.1089/scd.2009.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wernig M, Zhao JP, Pruszak J, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. P Natl Acad Sci USA. 2008. April 15;105(15):5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hargus G, Cooper O, Deleidi M, et al. Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. P Natl Acad Sci USA. 2010. September 7;107(36):15921–15926. doi: 10.1073/pnas.1010209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hallett PJ, Deleidi M, Astradsson A, et al. Successful Function of Autologous iPSC-Derived Dopamine Neurons following Transplantation in a Non-Human Primate Model of Parkinson’s Disease. Cell Stem Cell. 2015. March 5;16(3):269–274. doi: 10.1016/j.stem.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nussbaum RL, Ellis CE. Alzheimer’s disease and Parkinson’s disease. N Engl J Med. 2003. April 3;348(14):1356–64. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- 172.Tanner CM, Goldman SM. Epidemiology of Parkinson’s disease. Neurol Clin. 1996. May;14(2):317–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Burbulla LF, Beaumont KG, Mrksich M, et al. Micropatterning Facilitates the Long-Term Growth and Analysis of iPSC-Derived Individual Human Neurons and Neuronal Networks. Adv Healthc Mater. 2016. August;5(15):1894–903. doi: 10.1002/adhm.201500900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhu L, Sun C, Ren J, et al. Stress-induced precocious aging in PD-patient iPSC-derived NSCs may underlie the pathophysiology of Parkinson’s disease. Cell Death & Disease. 2019. 2019/02/04;10(2):105. doi: 10.1038/s41419-019-1313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Miller JD, Ganat YM, Kishinevsky S, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013. December 5;13(6):691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lapasset L, Milhavet O, Prieur A, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011. November 1;25(21):2248–53. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Marion RM, Strati K, Li H, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009. February 6;4(2):141–54. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 178.Suhr ST, Chang EA, Tjong J, et al. Mitochondrial rejuvenation after induced pluripotency. PLoS One. 2010. November 23;5(11):e14095. doi: 10.1371/journal.pone.0014095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fiesel FC, Ando M, Hudec R, et al. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep. 2015. September;16(9):1114–30. doi: 10.15252/embr.201540514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Drouin-Ouellet J, Lau S, Brattas PL, et al. REST suppression mediates neural conversion of adult human fibroblasts via microRNA-dependent and -independent pathways. EMBO Mol Med. 2017. August;9(8):1117–1131. doi: 10.15252/emmm.201607471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.International Parkinson Disease Genomics C, Nalls MA, Plagnol V, et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011. February 19;377(9766):641–9. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010. February 25;463(7284):1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang Y, Pak C, Han Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013. June 5;78(5):785–98. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Khurana V, Peng J, Chung CY, et al. Genome-Scale Networks Link Neurodegenerative Disease Genes to alpha-Synuclein through Specific Molecular Pathways. Cell Syst. 2017. February 22;4(2):157.-+. doi: 10.1016/j.cels.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mariani J, Simonini MV, Palejev D, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012. July 31;109(31):12770–5. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schwank G, Koo B-K, Sasselli V, et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell. 2013. 2013/12/05/;13(6):653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 187.Choi SH, Kim YH, Hebisch M, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014. November 13;515(7526):274–8. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Phan DT, Bender RHF, Andrejecsk JW, et al. Blood-brain barrier-on-a-chip: Microphysiological systems that capture the complexity of the blood-central nervous system interface. Exp Biol Med (Maywood). 2017. November;242(17):1669–1678. doi: 10.1177/1535370217694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yoon SJ, Elahi LS, Pasca AM, et al. Reliability of human cortical organoid generation. Nature Methods. 2019. January;16(1):75.-+. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cooper O, Seo H, Andrabi S, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med. 2012. July 4;4(141):141ra90. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bogetofte H, Jensen P, Ryding M, et al. PARK2 Mutation Causes Metabolic Disturbances and Impaired Survival of Human iPSC-Derived Neurons. Front Cell Neurosci. 2019;13:297–297. doi: 10.3389/fncel.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]