Abstract
Personalized cancer vaccines (PCVs) targeting patient-specific neoantigens are a promising cancer treatment modality; however, neoantigen physicochemical variability can present challenges to manufacturing PCVs in an optimal format for inducing anticancer T cells. Here, we developed a vaccine platform (“SNP-7/8a”) based on charge-modified peptide-TLR-7/8a conjugates that are chemically programmed to self-assemble into nanoparticles of uniform size (~20 nm) irrespective of the peptide antigen composition. This approach provided precise loading of diverse peptide neoantigens linked to TLR-7/8a (adjuvant) in nanoparticles that increased uptake by and activation of antigen-presenting cells that promote T cell immunity. Vaccination of mice with SNP-7/8a using predicted neoantigens (n=179) from three tumor models induced CD8 T cells against ~50% of neoantigens with high predicted MHC-I binding affinity and led to enhanced tumor clearance. SNP-7/8a delivering in silico-designed mock neoantigens also induced CD8 T cells in non-human primates. Altogether, SNP-7/8a is a generalizable approach for co-delivering peptide antigens and adjuvants in nanoparticles for inducing anticancer T cell immunity.
Introduction
T cells that recognize MHC-bound mutant peptides (“neoantigens”)1 are capable of mediating tumor-specific killing and have been shown to promote durable tumor regression and prolonged survival of patients with advanced cancers following adoptive cell therapy2,3. Additionally, improved survival of patients treated with checkpoint inhibitors (CPIs), such as anti-PD-1 and anti-CTLA-4, correlates with tumor mutational load4 and T cell infiltration into tumors5. Based on these findings, personalized cancer vaccines (PCVs) that generate neoantigen-specific T cells are being actively developed as cancer treatments6,7.
The feasibility of using peptide- and RNA-based PCVs has been demonstrated in both mice8,9 and humans10,11. While these studies have established an important proof-of-concept, a major limitation of current peptide- and RNA-based PCV approaches is their efficiency for generating neoantigen-specific CD8 T cells in vivo. For example, less than 10% of 184 predicted neoantigens derived from 3 mouse tumor lines included in either peptide- or RNA-based PCVs induced detectable CD8 T cell responses in mice, even though the putative neoantigens were selected on the basis of high predicted binding affinity for MHC-I9. Moreover, patients who received peptide neoantigens combined with the adjuvant polyICLC had low to undetectable CD8 T responses when assessed directly ex vivo from blood10. Based on current cost and manufacturing constraints that restrict the number of predicted neoantigens that can be included in PCVs6, as well as the limited number of neoantigens in patients with low mutational burden tumors12, more efficient neoantigen prediction and vaccination approaches are likely needed.
The focus of this study was to determine whether the magnitude and breadth of CD8 T cell responses to neoantigens could be improved by optimization of a peptide-based PCV formulation. While numerous formulation approaches have been developed to enhance T cell immunity to peptide antigens13,14, the broad variability of neoantigen physicochemical properties that arise from amino acid sequence variation may limit the translatability of such technologies for use as PCVs15. Indeed, formulating peptide antigens and adjuvants with particle technologies, including poly(lactic-co-glycolic acid) (PLGA)16, liposomes17, lipid nanodiscs18, polymersomes19 and emulsions20, is an empirical process whereby peptide loading and other formulation characteristics may be different for each antigen. An alternative approach is to use conjugate vaccines based on peptide antigens linked to hydrophobic carriers (e.g., lipids21, fatty acids22 and TLRa23–25) that can induce particle assembly or bind albumin for more efficient delivery to lymph nodes21,26. Conjugate vaccines offer the potential advantages that antigen loading is chemically defined and that adjuvants can be covalently attached to ensure co-delivery of both components to antigen presenting cells (APCs), which may be needed for optimal T cell priming27,28. A major limitation of many current particle and conjugate vaccine technologies, however, is that they do not fully account for the broad range of neoantigen properties, which can lead to formulation variability, including the propensity of hydrophobic peptides to form aggregates that complicate manufacturing and form injection-site depots that can lead to sub-optimal CD8 T cell immunity29.
To overcome these limitations, we developed a PCV platform based on charge-modified peptide-TLR-7/8a conjugates that enable reproducible and precise loading of diverse peptide neoantigens with TLR-7/8a in self-assembling nanoparticles (SNP-7/8a) of a defined size (~20 nm, diameter). The data reported here show that SNP-7/8a overcomes several manufacturing and formulation limitations of current peptide-based PCVs and leads to expanded breadth and magnitude of neoantigen-specific CD8 T cells as well as improved tumor clearance.
Results
Peptide physical form is a key determinant of CD8 T cell immunogenicity
Synthetic long peptides (LPs) consisting of 20–40 amino acid sequences, which often include an 8–11 amino acid minimal (“Min”) CD8 T cell epitope, combined with various adjuvants have been widely studied as cancer vaccines30,31. However, it is not well understood how differences in the amino acid composition, which determines the physical form (i.e. hydrodynamic behavior)32 of LPs, affect immunogenicity.
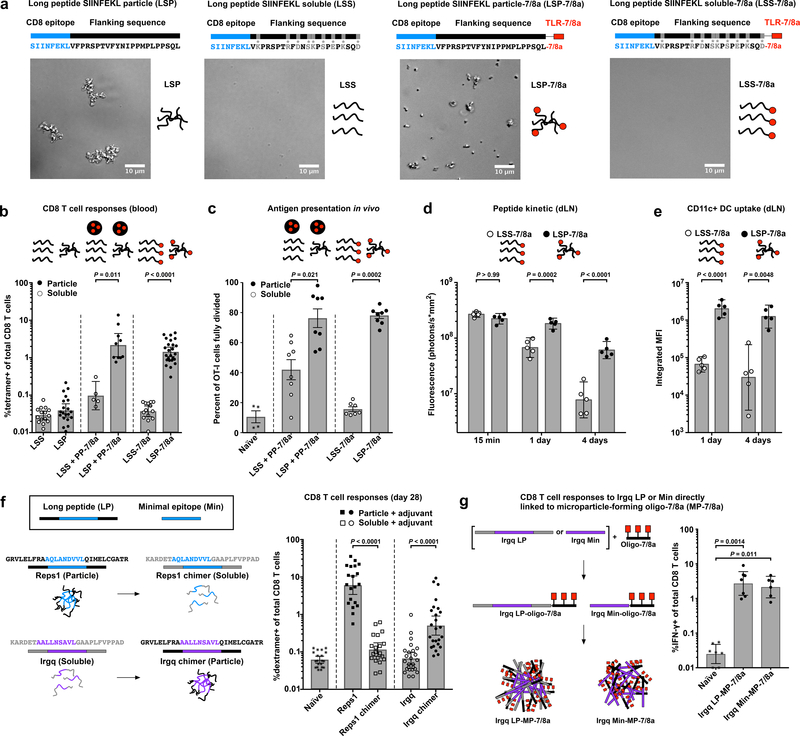
To determine how peptide physical form impacts induction of CD8 T cell responses, the MHC-I epitope from ovalbumin (SIINFEKL) was used as a model immunogen and synthesized as either a hydrophobic 30-amino acid LP that is particulate (“LSP”) or a hydrophilic 30-amino acid LP that is soluble (“LSS”) in aqueous buffer (Fig. 1a). The LPs were then administered to mice alone or in combination with an imidazoquinoline-based TLR-7/8a as a source of adjuvant, which was either covalently attached to the LPs or provided as a particle (PP-7/8a)33,34 admixed with the LPs (Fig. 1a and Supplementary Fig. 1a).
Fig. 1: Peptide antigen physical form is a key determinant of CD8 T cell immunogenicity.
(a) Schematic and brightfield micrographs of the CD8 T cell epitope from Ovalbumin (SIINFEKL) contained within long peptides (LP) that are either particulate (LSP and LSP-7/8a) or soluble (LSS and LSS-7/8a) in PBS. (b) C57BL/6 mice (n = 5–25 per group) were injected subcutaneously with the specified formulations on days 0 and 14, and CD8 T cell responses were assessed by tetramer staining on day 28. (c) C57BL/6 mice (n = 8 per group) were injected subcutaneously with the specified formulations followed by intravenous adoptive transfer of CFSE-labeled OT-I cells. On day 6, cell division of OT-I cells was assessed by flow cytometry. (d,e) C57BL/6 mice (n = 5 per group per time point) were injected subcutaneously with AF647-labeled LSS-7/8a or LSP-7/8a and at various timepoints thereafter draining lymph nodes (dLN) were assessed for (d) total tissue fluorescence and (e) vaccine uptake by CD11c+ DCs. (f) Native and chimeric forms of Reps1 and Irgq LP neoantigens were admixed with an adjuvant (either PP-7/8a, CpG or polyICLC) and administered to C57BL/6 mice (n = 10–25 per group) at days 0 and 14. CD8 T cell responses from blood were determined by dextramer staining on day 28; responses compiled across all adjuvants are shown. (g) C57BL/6 mice (n = 7 per group) were injected subcutaneously with MP-7/8a containing the LP or Min form of the neoantigen Irgq at days 0 and 14 and CD8 T cell responses were assessed by intracellular cytokine staining on day 28. PP-7/8a is a particle-forming polymer-TLR-7/8a adjuvant. Data on log scale are reported as geometric mean with 95% c.i.; data on linear scale are reported as mean ± s.e.m. Statistical significance was determined using Kruskal-Wallis with Dunn’s correction (b,c,f,g) or two-way ANOVA with Bonferroni correction (d,e).
Vaccination with the particle LPs (LSP mixed with PP-7/8a or LSP-7/8a) led to ~20-fold higher CD8 T cell responses as compared with vaccination with the soluble LPs (LSS mixed with PP-7/8a or LSS-7/8a; Fig. 1b). Moreover, the particle LP admixed with PP-7/8a and other TLRa adjuvants known to induce CD8 T cell immunity35, i.e. CpG (TLR-9a) and polyICLC (TLR-3a), induced ~10-fold higher magnitude CD8 T cell responses and improved survival following challenge with ovalbumin-expressing B16 tumor cells (B16.OVA) as compared with LSS combined with the same adjuvants (Supplementary Fig. 1b–d).
As a possible mechanism to account for these findings, we observed that antigen-specific CD8 T cells in mice that received the particle LP expanded for up to 1 week after vaccination and underwent a greater number of cell divisions as compared with CD8 T cells in mice immunized with the soluble LP (Fig. 1c and Supplementary Fig. 2). Additionally, the particle LP was retained longer in draining lymph nodes and had higher uptake by CD11c+ dendritic cells (DCs) compared with the soluble LP (Fig. 1d,e). Together, these data suggest that particle LPs enhance CD8 T cell responses through prolonged antigen presentation by lymph node DCs.
To extend these findings to PCVs, we evaluated the relationship between peptide physical form and immunogenicity using two neoantigens, Reps1 and Irgq, which are both known to bind MHC-I, but were previously reported to be immunogenic and “non-immunogenic,” respectively8. Here, we show that Reps1 LP is particulate in aqueous solution and induces high magnitude CD8 T cell responses when admixed with a variety of adjuvants (i.e. PP-7/8a, CpG or polyICLC), whereas Irgq LP is water-soluble and induces low to undetectable CD8 T cell responses (Fig. 1f and Supplementary Fig. 3). The physical form of each neoantigen was then altered by swapping the amino acid residues flanking each minimal epitope to produce chimeric Reps1, which is soluble, and chimeric Irgq, which is particulate. For each of the adjuvants evaluated, the particulate Irgq chimer induced higher CD8 T cell responses than the native soluble form, whereas the soluble Reps1 chimer did not induce responses significantly above background (Fig. 1f and Supplementary Fig. 3).
To more directly determine how peptide physical form impacts CD8 T cell responses, we directly attached either the native LP (26-mer) or Min (9-mer) Irgq sequence to a hydrophobic oligopeptide-TLR-7/8a to form conjugate vaccines that assemble into microparticles (referred to as “MP-7/8a”) in aqueous conditions and assessed their capacity to induce CD8 T cells in vivo. While the water-soluble native Irgq LP admixed with adjuvants was non-immunogenic (Fig. 1f), both the native LP and Min Irgq sequences induced high magnitude CD8 T cell responses when rendered particulate (Fig. 1g). These data substantiate the finding that particulate delivery of peptide antigens, including neoantigens, is critical for inducing CD8 T cell responses.
Self-assembling nanoparticles based on charged-modified peptide-TLR-7/8a conjugates (SNP-7/8a)
To ensure consistent loading of both peptide neoantigens and adjuvants in particles of a uniform, optimal size (~20 nm, diameter) for delivery to APCs specialized for priming T cell immunity36,37, we developed a vaccine approach that accounts for peptide neoantigen physicochemical variability.
Our approach was to use a modular and chemically tunable vaccine platform based on charge-modified (CM) conjugates comprising peptide antigens linked to both a charge modifying group and a hydrophobic block through enzyme degradable linkers at the N- and C- termini of the peptide, respectively (Fig. 2a). To ensure biocompatibility and manufacturing scalability, biodegradable and chemically-defined compositions of charged amino acids and hydrophobic oligopeptides were used as the charged-modifying groups and hydrophobic blocks, respectively. Finally, to ensure co-delivery of antigen and adjuvant, the oligopeptide-based hydrophobic blocks were linked to a precise number of small molecule imidazoquinoline-based TLR-7/8a, which were specifically selected as adjuvants based on their i) permissibility to chemical conjugation38,39; ii) hydrophobic properties that promote particle self-assembly33,40; and iii) ability to broadly activate human DC subsets to produce key cytokines (i.e. IL-12 and Type-I IFNs) that promote Th1 CD4 and CD8 T cell immunity35,41.
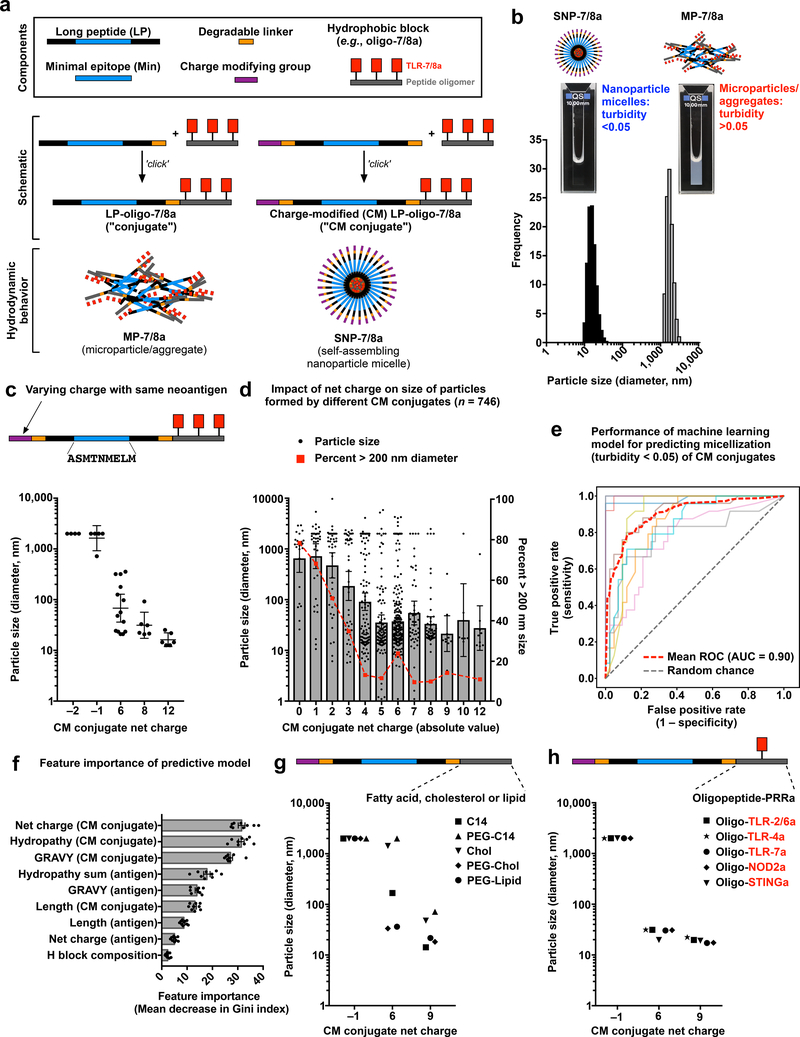
Fig. 2: Self-assembling nanoparticles (SNP-7/8a) based on charge-modified peptide-TLR-7/8a conjugates.
(a) Schematic of modular components comprising peptide-TLR-7/8a conjugate vaccines and charge-modified (CM) peptide-TLR-7/8a conjugate vaccines that form microparticles/aggregates (MP-7/8a) and self-assembling nanoparticle micelles (SNP-7/8a), respectively. (b) Particle size distribution plot for representative SNP-7/8a and MP-7/8a. (c) Particle sizes of CM conjugates (n = 35 unique conjugates) with various charge modifying groups appended to the same peptide antigen sequence. (d) Particle sizes of different CM conjugates (n = 746) with varying net charge (absolute value). (e) Receiver operating characteristic (ROC) curves of a random forest machine learning (ML) model for predicting SNP-7/8a hydrodynamic behavior for any given CM conjugate composition; mean ROC is the average performance of the models based on a 10-fold cross-validated binary classifier (n = 10 runs) trained on data from d. (f) The relative importance of different characteristics of CM conjugates on the performance of the ML model, based on n = 10 cross-validations. (g) Particle size dependency on net charge of CM conjugates containing various hydrophobic blocks. (h) Particle size dependency on net charge of CM conjugates containing hydrophobic blocks based on peptide oligomers linked to various pattern recognition receptor (PRR) agonists. H block = hydrophobic block; C14 = myristic acid; chol = cholesterol. Data on log scale are reported as geometric mean with 95% c.i.; data on linear scale are reported as mean ± s.e.m.
Upon resuspension in aqueous solution, the hydrophobic block promotes multimerization while the charge-modifying group provides a countervailing force that induces formation of ~20 nm, diameter nanoparticle micelles (“SNP-7/8a”) and prevents the formation of large microparticles or aggregates (“MP-7/8a”) that can result from conjugates without charge modification (Fig. 2b).
Net charge of CM conjugates determines SNP formation
It was unknown a priori what magnitude of charge would be required to stabilize self-assembling nanoparticles (SNP) formed by CM conjugates with different neoantigens. Therefore, we systematically investigated how modulating the net charge of CM conjugates impacts particle size. Evaluation of 35 CM conjugates with various charge-modifying groups appended to the N-terminus of the same peptide neoantigen revealed an inverse relationship between the magnitude of CM conjugate net charge and particle size (Fig. 2c). This inverse relationship between magnitude of charge and particle size was confirmed with an additional 746 unique CM conjugates with a variety of antigen sequences, wherein ~90% of CM conjugates having a net absolute charge > 5 assembled into ~20–50 nm nanoparticle micelles (Fig. 2d).
Based on this data set, a random forest machine learning (ML) model42 was used to predict how CM conjugate properties impact the hydrodynamic behavior of SNP-7/8a. This approach used the underlying physical properties of each CM conjugate to predict whether a given composition would form stable nanoparticles (as measured by turbidity < 0.05) or unstable, larger particles (turbidity > 0.05) with a mean ROC AUC of 0.90 (Fig. 2e). Analysis of the underlying ML model revealed that CM conjugate net charge and hydropathy were relatively important features determining nanoparticle formation, whereas peptide antigen length and hydrophobic block composition were less important (Fig. 2f).
To extend these findings, the impact of net charge on the size of particles formed by CM conjugates comprising common hydrophobic carrier molecules (fatty acids, cholesterol and lipids; Fig. 2g) and hydrophobic oligopeptides linked to agonists of other pattern recognition receptors (TLR-2/6, TLR-4, TLR-7, NLR and STING; Fig 2h) was determined. Consistent with the results using CM peptide-TLR-7/8a conjugates, CM conjugates comprising other hydrophobic block compositions and adjuvants also showed an inverse relationship between net charge and particle size, which was independent of the hydrophobic block composition.
SNP-7/8a parameters that affect T cell induction
We next undertook in vivo structure-activity relationship studies to evaluate how various other parameters of SNP-7/8a – including linker composition (cathepsin43 and proteasomal44 processing sites), TLR-7/8a potency and number, and type of charge (i.e. positive versus negative net charge) – impact CD8 T cell responses. Cathepsin degradable linkers placed between the peptide neoantigen and both the charge-modifying group and hydrophobic block increased the efficiency of antigen presentation in vitro and led to higher CD8 T cell responses in vivo (Supplementary Fig. 4). The potency and number of TLR-7/8a linked to each CM conjugate also impacted immunogenicity (Supplementary Fig. 5). Additionally, CM conjugates with net positive charge were more potent in vivo as compared with those bearing net negative charge (Supplementary Fig. 6a–c). Collectively, these data informed the selection of an optimal SNP-7/8a formulation based on CM conjugates with net positive charge (≥ +8), cathepsin degradable linkers and 3 TLR-7/8a, which was found to efficiently prime and boost CD8 T cell responses using a broad range of dosing intervals (Supplementary Fig. 6d).
Manufacturing process for SNP-7/8a as a PCV
A manufacturing process was developed to enable rapid per-patient synthesis of SNP-7/8a (Supplementary Fig. 7). A simple, copper-free “click chemistry” reaction45 was used to link patient-specific, charge-modified peptide neoantigens produced by automated solid-phase synthesis to a pre-built hydrophobic block (oligo-7/8a) (Supplementary Fig. 7a,b). The resulting CM conjugates are chemically defined single molecules, which allow for simple release testing, and can be sterile filtered without material loss (Supplementary Fig. 7c). Addition of aqueous buffer results in the CM conjugates immediately self-assembling to nanoparticles that are stable at room temperature for over 100 hours (Supplementary Fig. 7d).
Generalizability of SNP-7/8a for neoantigens with extremes of charge and hydropathy
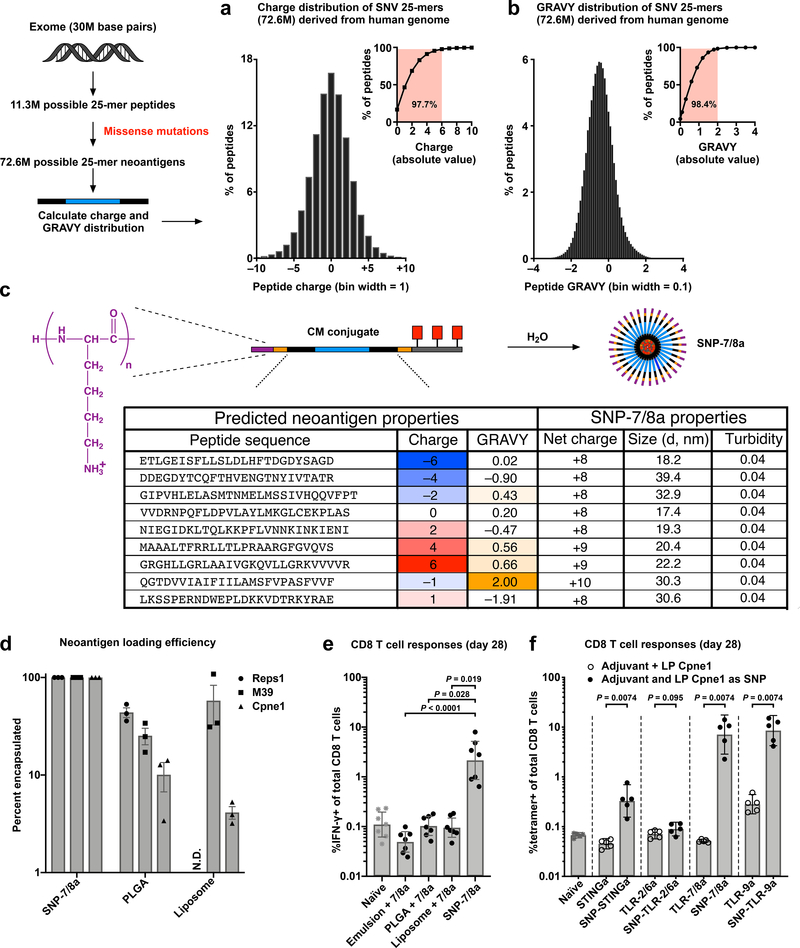
To assess the ability of SNP-7/8a to ensure formulation consistency with heterogeneous peptide neoantigens that may be included in PCVs, we first computed the charge and hydropathy frequency distribution of all 25 amino acid peptides (25-mers) that can be generated from each possible single missense mutation in canonical transcripts from the human genome (n = 72.6M mutant 25-mers). About 98% of mutant 25-mers have a charge between –6 to +6 (at pH 7.4) and a grand average of hydropathy (GRAVY) between –2 and +2 (Fig. 3a,b), which is consistent with the range of properties observed for mouse tumor cell line derived neoantigens (Supplementary Fig. 8a,b). To validate the tolerance of SNP-7/8a for delivering neoantigens at the extremes of charge and hydropathy, 9 different mouse neoantigens with a range of underlying charge (–6 to +6) and GRAVY (–2 to +2) were produced as CM conjugates with net positive charge (≥ +8) fixed by modulating the number of charged amino acids comprising the charge-modifying group, and all formed stable nanoparticles (Fig. 3c).
Fig. 3: Genome-wide analysis of LP neoantigen charge and hydropathy frequency distribution and formulation benchmarking studies validate the generalizability of SNP-7/8a based on CM conjugates.
(a) Charge and (b) hydropathy (GRAVY) frequency distribution of 25 amino acid LPs (n = 72.6M mutant 25-mers) derived from all possible non-synonymous single nucleotide variant (SNV) missense mutations in canonical protein coding transcripts; inset shows the cumulative proportion of mutant 25-mers with a given range of characteristics (i.e. ~98% of possible neoantigen peptides have charge between –6 to +6 and GRAVY between –2 and +2). (c) Predicted neoantigens (mouse-derived) with the indicated charge and GRAVY characteristics were synthesized as CM conjugates with net charge ≥ +8; particle size (diameter, nm) and turbidity were assessed following CM conjugate self-assembly to nanoparticles (SNP-7/8a) in PBS, pH 7.4. Turbidity < 0.05 indicates the absence of aggregates. (d) Different LP neoantigens were synthesized as SNP-7/8a or formulated within PLGA or Liposomal particles and the percentage of peptide neoantigen encapsulated within each was assessed (n = 3 per composition); N.D. indicates that peptide loading was not determined. (e, f) C57BL/6 mice were injected subcutaneously on days 0 and 14 with LP Cpne1 neoantigen using the indicated formulation and CD8 T cell responses were assessed from whole blood on day 28 by either (e) intracellular cytokine staining (n = 7 mice per group) or (f) tetramer staining (n = 5 mice per group). (d) shows the mean ± s.e.m; (e,f) shows the geometric mean with 95% c.i.; statistical significance was determined using Kruskal-Wallis with Dunn’s correction (e) or individual Mann-Whitney U-tests (f).
As PCVs may require multiple predicted neoantigens to be formulated together to maximize T cell breadth, we evaluated the tolerance of SNP-7/8a to formulate multiple CM conjugates with a variety of underlying properties. The SNP-7/8a platform provided consistent particle size, between ~20–30 nm, when the formulation comprised between 5 to 20 different CM conjugates in each particle (Supplementary Fig. 8c,d).
CM conjugates improve peptide antigen loading into particles
To determine whether SNP-7/8a could improve formulation consistency compared with conventional particle delivery systems, we benchmarked peptide neoantigen loading and particle size for SNP-7/8a as compared with PLGA and liposomal particles delivering different neoantigens (Fig. 3d and Supplementary Fig. 8e). As designed, the CM conjugates ensured full conversion (i.e. 100% loading, Fig. 3d) of the peptide antigen into uniformly sized nanoparticles (Supplementary Fig. 8e) with up to 56% peptide neoantigen content by particle mass. While formulations based on the liposomal and PLGA particles resulted in relatively consistent sizes of particles (Supplementary Fig. 8e), peptide loading was highly variable between the different neoantigens, ranging from about 10–50% and about 4–60% for PLGA and liposomes, respectively (Fig. 3d), and resulted in typically low (< 1%) peptide neoantigen content by particle mass.
We next benchmarked the capacity of SNP-7/8a to induce CD8 T cell immunity against a predicted LP neoantigen, Cpne1, as compared with several commonly used particle vaccine formulations. Mice were immunized with the same dose of the LP neoantigen, Cpne1, as either SNP-7/8a; PLGA or liposomal nanoparticles carrying TLR-7/8a; or, a squalene-based oil-in-water emulsion (AddaVax™) admixed with TLR-7/8a, which is representative of a ‘mix-and-shoot’ formulation (i.e. the peptide and emulsion are mixed without any further work-up) that has been used clinically46. SNP-7/8a led to substantially (> 20-fold) higher magnitude CD8 T cell responses as compared with the other particle formulations (Fig. 3e).
CM conjugates of STING, TLR-7/8 and TLR-9 agonists induce T cell immunity
To assess the suitability of the CM conjugate design for accommodating other classes and chemical compositions of immunostimulants, CM conjugates incorporating agonists of STING (cyclic dinucleotides (CDN)), TLR-2/6 (Pam2Cys) and TLR-9 (CpG) were synthesized and evaluated for the capacity to induce CD8 T cell immunity in vivo.
To assess immunogenicity of the different CM conjugate compositions, mice were immunized with either the free agonist admixed with the LP neoantigen, Cpne1, or with Cpne1 as SNP linked to agonist (i.e. SNP-STINGa, SNP-TLR-2/6a, SNP-7/8a and SNP-TLR-9a). The SNPs carrying agonists of STING, TLR-7/8 and TLR-9, but not TLR-2/6, all induced > 5-fold higher magnitude neoantigen-specific CD8 T cell responses as compared with the naïve group (Fig. 3f). These data are consistent with the capacity of STINGa, TLR-7/8a and TLR-9a, but not TLR-2/6a, to induce Type-I IFNs needed to promote cross-priming of exogenously delivered peptide antigens for inducing CD8 T cell immunity35.
SNP-7/8a improves peptide antigen formulation consistency and immunogenicity
To further benchmark immune responses induced by SNP-7/8a against other PCV formulations, 7 MC38-derived neoantigens known to bind MHC-I (Aatf, Adpgk, Cpne1, Dpagt, Irgq, Med12 and Reps1)8 were evaluated. Each of the neoantigens were formulated either as native LPs admixed with adjuvant (i.e. polyICLC + anti-CD40), which is representative of therapeutic cancer vaccines that have been widely used in clinical studies10; as conjugate vaccines based on LPs linked to a hydrophobic molecule that form microparticles/aggregates (MP-7/8a), which is similar to current conjugate vaccine approaches that do not include a charge modifying group22–24; or, as CM conjugates of the LPs, which self-assemble into nanoparticles (SNP-7/8a).
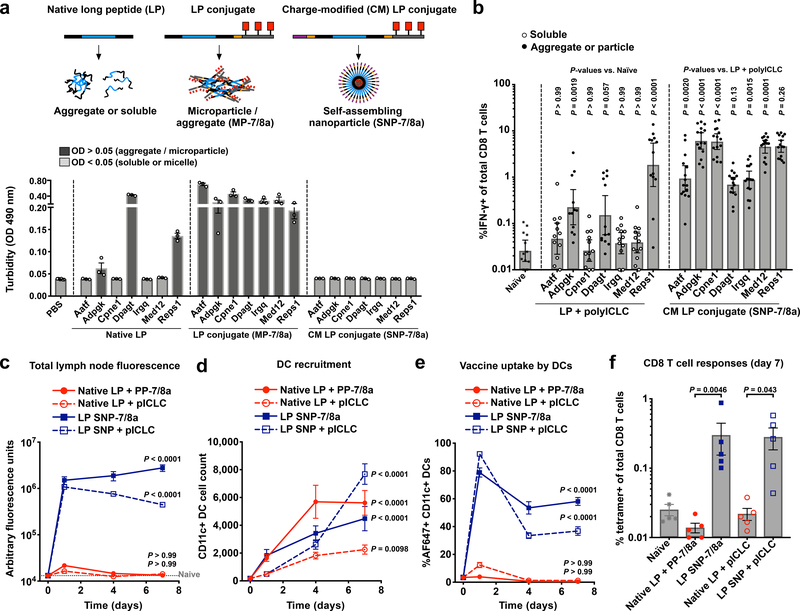
The native LPs showed formulation heterogeneity, with 3 out of 7 LPs (Adpgk, Dpagt and Reps1) forming aggregates, and the remaining 4 occurring as water soluble molecules (Fig. 4a). As designed, all LP neoantigens as MP-7/8a and SNP-7/8a assembled into microparticles/aggregates and nanoparticle micelles, respectively. Consistent with the prior report8, the native LP formulations administered with polyICLC and anti-CD40 induced CD8 T cell responses against 3 of the 7 neoantigens (Adpgk, Dpagt and Reps1; Fig 4b), each of which were particulate, corroborating our findings that peptide physical form is a key determinant of immunogenicity (Fig. 1b–g). Notably, all 7 neoantigens formulated as SNP-7/8a induced high magnitude CD8 T cell responses (Fig. 4b), of which three were associated with delayed tumor growth following challenge (Supplementary Fig. 9).
Fig. 4: SNP-7/8a improves formulation consistency, immunogenicity and PK compared to conventional PCV approaches.
(a,b) Seven MC38 neoantigens known to bind MHC-I (Aatf, Adpgk, Cpne1, Dpagt, Irgq, Med12 and Reps1) were produced as either native LPs; conjugates of oligo-7/8a that form microparticles/aggregates (MP-7/8a); or charge-modified (CM) conjugates of oligo-7/8a that self-assemble to nanoparticles (SNP-7/8a). (a) Turbidity of the LP formulations (n = 3 per composition) at 0.5 mg/mL in PBS pH 7.4 was assessed by measuring absorbance (optical density (OD), arbitrary units) at 490 nm; turbidity > 0.05 indicates aggregation. (b) C57BL/6 mice (n = 12–15 per group) were injected subcutaneously with native LPs admixed with polyICLC and anti-CD40 or SNP-7/8a on days 0 and 14, and CD8 T cell responses were assessed from blood by intracellular cytokine staining on day 28. (c–f) C57BL/6 mice (n = 5 per group per time point) were injected subcutaneously with fluorophore (AF647)-labeled Cpne1 neoantigen as either a native LP admixed with a particle adjuvant (polyICLC or PP-7/8a) or as a self-assembling nanoparticle (SNP) admixed with polyICLC or co-delivering TLR-7/8a (SNP-7/8a). Lymph nodes (n = 10 per group per time point) draining the site of immunization were collected at serial time points and assessed for (c) total tissue fluorescence (peptide quantity), (d) total CD11c+ DC count and (e) the percentage of total CD11+ DCs that had taken up vaccine (AF647+). (f) CD8 T cell responses from blood were assessed by tetramer staining on day 7. Data on log scale are reported as geometric mean with 95% c.i.; data on linear scale and all line graphs are reported as mean ± s.e.m. Statistical significance was determined using Kruskal-Wallis with Dunn’s correction (b,f) or two-way ANOVA with Bonferroni correction (c–e).
Though both conjugate vaccines provided increased breadth of CD8 T cell responses as compared with native LPs admixed with adjuvant (e.g., responses against Cpne1; Fig. 4b and Supplementary Fig. 10a), the nanoparticle micelles (SNP-7/8a) based on CM conjugates induced higher magnitude CD8 T cell responses compared with the microparticle/aggregate formulations (MP-7/8a) based on conjugates without charge modification (Supplementary Fig. 10a,b).
Nanoparticles (SNP-7/8a) enhance CD8 T cell responses by increasing APC uptake
We next investigated how the physical form of the peptide antigen affects kinetics and APC uptake in the draining lymph node. The neoantigen Cpne1, which is water-soluble as the native LP, was administered to mice as either the native LP admixed with adjuvant (polyICLC or PP-7/8a) or as a nanoparticle (SNP) admixed with polyICLC or incorporating TLR-7/8a (SNP-7/8a) and then tracked in vivo.
The water-soluble native LP was not detected at levels above background in the draining lymph node (dLN), whereas the nanoparticle LP compositions (SNP and SNP-7/8a) were measured at higher levels at each of the time points evaluated (Fig. 4c). Though all vaccine compositions led to substantial recruitment of total CD11c+ DCs (Fig. 4d), the physical form of the peptide had a major influence on the proportion of those DCs carrying antigen (Fig. 4e). Specifically, whereas greater than 35% of CD11c+ DCs in the dLN of mice vaccinated with nanoparticle LP (SNP or SNP-7/8a) were vaccine+ at 7 days after vaccination, less than 1% were vaccine+ in mice vaccinated with the native LP (Fig. 4e). Moreover, the nanoparticle LP induced higher magnitude CD8 T cell responses as compared with the native LP (Fig. 4f), which did not induce responses above background likely due to insufficient uptake by lymph node DCs.
While both nanoparticle (SNP-7/8a) and microparticle (MP-7/8a) formulations improved lymph node accumulation and uptake by APCs as compared with soluble neoantigen, SNP-7/8a resulted in higher lymph node APC accumulation and induced higher magnitude CD8 T cell responses as compared to MP-7/8a (Supplementary Fig. 10c–f).
SNP-7/8a induces comparable CD8 T cell responses with short and long peptides
Immunization with LPs admixed with adjuvants has been shown to increase the magnitude of CD8 T cell responses as compared with the use of Mins47. However, the influence of peptide length on the efficiency for inducing CD8 T cell immunity when peptide antigens are linked to particle carriers is less well established. Therefore, we evaluated T cell responses induced by Min and LP versions of 7 neoantigens as SNP-7/8a. Both Min and LP versions elicited comparable CD8 T cell responses that were of high magnitude (Supplementary Fig. 11a). As expected, only the LPs elicited CD4 T cell responses (Supplementary Fig. 11b), likely because the Mins are too short to encode an MHC-II epitope, which is contained within some LPs (e.g., Irgq). Of note, the presence of CD4 help did not impact the kinetics, magnitude or phenotype of memory CD8 T cells induced by SNP-7/8a (Supplementary Fig. 11c–e). The results show that both Mins and LPs delivered on SNP-7/8a induce comparable CD8 T cell responses. Nevertheless, LPs may be preferred over Mins based on their ability to also elicit CD4 T cells.
SNP-7/8a validates MHC-I binding prediction algorithms
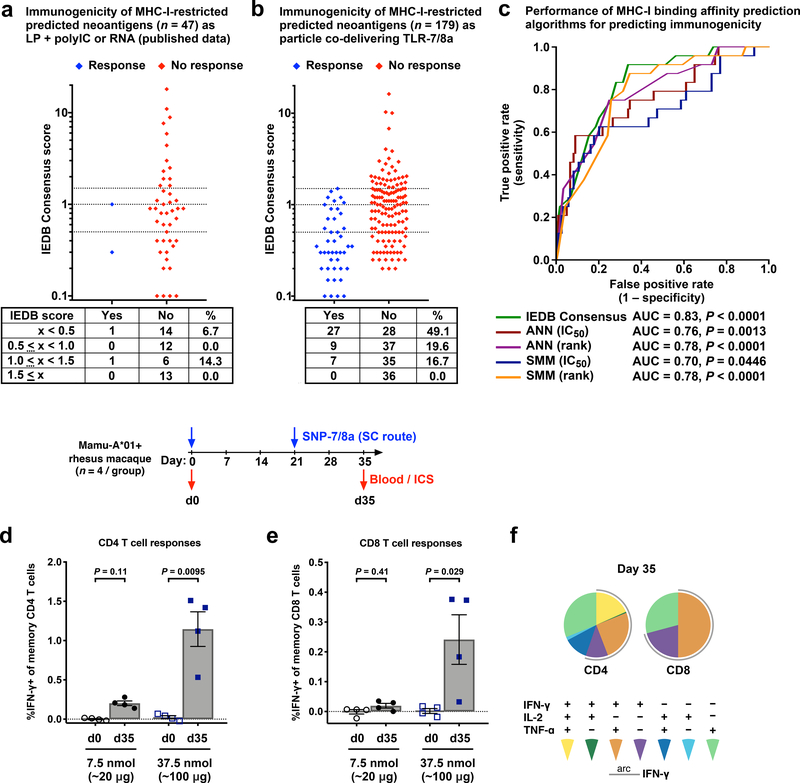
Predicted or measured MHC-I binding affinity is considered an important predictor of immunogenicity48 and is commonly used in the selection of neoantigens to include in PCVs6,49,50. However, a clear correlation between predicted MHC-I binding affinity and immunogenicity is not easily discerned from published data using PCVs based on LP + adjuvant (i.e. polyIC) or RNA (Fig. 5a)9. In contrast, after controlling for the physical form of 179 peptide-based predicted neoantigens from 3 tumor cell lines – by ensuring their delivery in particles co-delivering TLR-7/8a – we observed CD8 T cell responses to approximately 50% of epitopes with high predicted binding affinity (IEDB Consensus score < 0.5) (Fig. 5b), which is about a 5-fold improvement over published rates (~10%) using PCVs based on LP + polyIC or RNA9. The improved efficiency for generating CD8 T cells allowed for the identification of a robust correlation between predicted MHC-I binding affinity and immunogenicity (P < 0.0001) (Fig. 5c) that is consistent with recent reports that many neoantigen-specific CD8 T cells identified in patients recognize epitopes with high predicted MHC-I binding affinity6.
Fig. 5: SNP-7/8a expands the breadth of neoantigen-specific CD8 T cell responses in mice and primates.
(a,b) Induction of neoantigen-specific CD8 T cell responses plotted against in silico predicted MHC-I binding affinity using the immune epitope database (IEDB) consensus algorithm. (a) CD8 T cell responses in mice vaccinated with predicted neoantigens (n = 47) derived from the B16-F10 tumor cell line as LP + polyIC or RNA, as described in ref. < Kreiter S, et al. Nature (2015)>. (b) CD8 T cell responses in mice for predicted neoantigens (n = 179) derived from the B16-F10, MC38 and SB-3123 tumor cell lines as particles co-delivering TLR-7/8a. C57BL/6 mice (n = 5 per group) were injected subcutaneously with up to 4 predicted neoantigens on days 0 and 14, and CD8 T cell responses were assessed from blood by intracellular cytokine staining on day 28. Predicted neoantigens that resulted in CD8 T cell responses that were statistically significantly above background in at least two independent experiments were considered immunogenic. (c) Receiver operating characteristic curve showing performance of different prediction algorithms for classifying neoantigens as immunogenic or non-immunogenic on the basis of predicted MHC-I binding affinity using antigens from b. (d,e) Mamu-A*01-expressing rhesus macaques (n = 4 per dose level) were injected subcutaneously on days 0 and 21 with the indicated doses (7.5 or 37.5 nmol per peptide) of SNP-7/8a containing “mock” neoantigens. Animals were bled on days 0 and 35 (abbreviated d0 and d35) and (d) CD4 and (e) CD8 T cell responses were measured directly from blood (i.e. without a prior expansion step) by intracellular cytokine staining. (f) Proportion of CD4 (left) and CD8 (right) T cells expressing various combinations of IFN-γ, IL-2 and/or TNF-α are shown for the group that received the 37.5 nmol dose at the d35 timepoint. ANN = artificial neural network; SMM = stabilized matrix method. Data on linear scale are reported as mean ± s.e.m. Statistical significance was determined using Kruskal-Wallis with Dunn’s correction (d,e).
SNP-7/8a induces neoantigen-specific CD4 and CD8 T cell responses in primates
We next assessed SNP-7/8a as a PCV in non-human primates (NHPs), which share considerable similarities with the human immune system51. As there are currently no standardized NHP tumor models, we applied an in silico process for identifying “mock” neoantigens (see: Online Methods). Neoantigens with moderate or high predicted binding affinity for the MHC-I allele Mamu-A*01, as determined by the IEDB Consensus algorithm, were evaluated for immunogenicity as SNP-7/8a in Mamu-A*01+ rhesus macaques.
SNP-7/8a induced dose-dependent neoantigen-specific CD4 and CD8 T cell responses that were directly measurable from the blood of primates following prime and boost vaccinations (Fig. 5d,e). Moreover, the neoantigen-specific T cells induced were relatively high quality as indicated by their polyfunctionality (Fig. 5f). Assessment of the response to each individual antigen revealed that the CD8 T cell response was directly measurable against 5, 4, 5 and 2 unique antigens for the four primates assessed (Supplementary Fig. 12), indicating that SNP-7/8a induced CD8 T cells to a wide breadth of antigens.
Optimization of therapeutic regimen
Consistent with recent reports showing improved therapeutic efficacy of vaccines in combination with checkpoint inhibitors (e.g., anti-PD-1/PDL-1 and anti-CTLA-4)52, SNP-7/8a combined with anti-PD-L1 resulted in a modest improvement in tumor control as compared with either treatment used alone (Supplementary Fig. 13a,b). Thus, SNP-7/8a was used in combination with anti-PD-L1 for all subsequent studies assessing vaccine efficacy in the therapeutic setting.
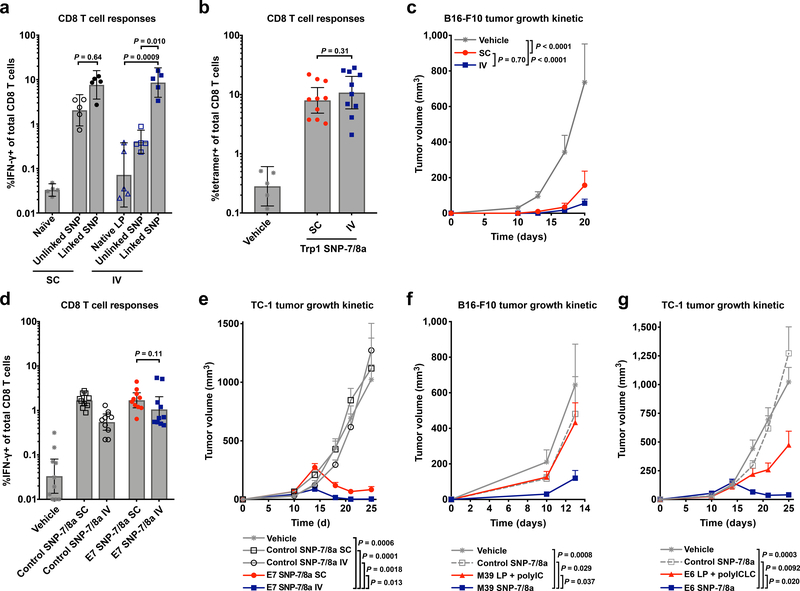
As the impact of co-delivery (i.e. physical linkage) of peptide antigen and adjuvant on immunogenicity following different routes of vaccination has not been closely studied, we also evaluated CD8 T cell responses in mice immunized with a neoantigen and TLR-7/8a either in separate particles (“unlinked SNP”) or together in the same particle (“linked SNP”), by the subcutaneous (SC) or IV route. For the SC route, mice that received the linked SNP had ~3-fold higher, albeit not significantly different, CD8 T cell responses as compared with mice that received unlinked SNP (Fig. 6a). In contrast, the differences in responses were more striking by the IV route. Mice administered particles co-delivering peptide antigen and adjuvant (i.e. linked SNP) by the IV route had CD8 T cell responses that were ~20- or ~50-fold higher than the responses in mice that received antigen and adjuvant in separate particles (i.e. unlinked SNP) or as native LP admixed with particle adjuvant, respectively (Fig. 6a).
Fig. 6: IV administration of SNP-7/8a induces CD8 T cells that mediate tumor regression.
(a) C57BL/6 mice (n = 5 per group) were vaccinated with TLR-7/8a and the neoantigen Reps1 in separate particles (“unlinked SNP”) or together in the same particle (“linked SNP”) by either the subcutaneous (SC) or intravenous (IV) route on days 0 and 14. An additional group of mice received the native LP admixed with a particulate TLR-7/8a adjuvant by the IV route. CD8 T cell responses were assessed on day 21 by intracellular cytokine staining. (b,c) C57BL/6 mice (n = 10 per group) implanted subcutaneously with 1.0 × 105 B16-F10 tumor cells were treated with either SNP-7/8a delivering the self-antigen Trp1 by the SC or IV route, or vehicle control (DMSO/PBS) by the IV route, which were each given along with anti-PD-L1 by the IP route on days 3, 10 and 17; (b) CD8 T cell responses were assessed from blood by tetramer staining on day 12 and (c) tumor growth was monitored at various time points. (d,e) C57BL/6 mice (n = 10 per group) implanted subcutaneously with 1.0 × 105 TC-1 tumor cells were treated with either vehicle control (DMSO/PBS) by the IV route, or SNP-7/8a delivering the virus-associated tumor antigen HPV E7 or an irrelevant antigen (Adpgk neoantigen from MC38) as an inflammation control (“control SNP-7/8a”) by the SC or IV route. Treatments were given along with anti-PD-L1 delivered by the IP route on days 7 and 14. (d) CD8 T cell responses were assessed from blood by intracellular cytokine staining on day 17; (e) tumor growth was monitored at various time points. (f) C57BL/6 mice (n = 10 per group) implanted subcutaneously with 1.0 × 105 B16-F10 tumor cells were treated with either the M39 neoantigen as LP admixed with polyIC by the SC route, M39 as SNP-7/8a by the IV route, an irrelevant neoantigen (Adpgk) as SNP-7/8a (“control SNP-7/8a”) by the IV route or vehicle control (DMSO/PBS) by the IV route. Treatments were given along with anti-PD-L1 delivered by the IP route on days 1,8 and 15. Tumor growth curves are shown. (g) C57BL/6 mice (n = 10 per group) implanted subcutaneously with 1.0 × 105 TC-1 tumor cells were treated with either the virus-associated tumor antigen HPV E6 as an LP admixed with polyICLC by the SC route, E6 as SNP-7/8a by the IV route, an irrelevant neoantigen (Adpgk) as SNP-7/8a by the IV route (“control SNP-7/8a”); or, vehicle control (DMSO/PBS) by the IV route. Treatments were given along with anti-PD-L1 delivered by the IP route on days 7 and 14. Tumor growth was monitored at various time points. Data on log scale are reported as geometric mean with 95% c.i.; data on linear scale are reported as mean ± s.e.m. Statistical significance was determined using Kruskal-Wallis with Dunn’s correction (a), Mann Whitney U-test (b,d), or two-way ANOVA with Bonferroni correction (c,e–g).
While SNP-7/8a induced similar magnitude of T cells by both local (SC) and systemic (IV) routes of vaccination (Fig. 6a,b,d), vaccination by the IV route provided a trend toward higher efficacy (Fig. 6c and Supplementary Fig. 14a) or significantly (P = 0.013) higher efficacy (Fig. 6e and Supplementary Fig. 14b) compared to vaccination by the SC route.
Therapeutic vaccination with SNP-7/8a enhances tumor regression
After identifying a preferred therapeutic regimen, we sought to determine if the improved efficiency of SNP-7/8a could lead to a greater breadth of CD8 T cells that mediate tumor clearance in vivo. We first screened the immunogenicity of 24 predicted neoantigens from the B16-F10 tumor cell line as LPs delivered on SNP-7/8a (Supplementary Fig. 15a,b). Among the 24 predicted neoantigens screened, 10 induced CD8 T cell responses as SNP-7/8a (Supplementary Fig. 15b and Supplementary Table 1), of which only 1 or 2 were previously reported to induce CD8 T cells using LP + polyIC or RNA, respectively9. Though the 24 predicted neoantigens screened were selected based on MHC-I binding, 10 were also found to induce CD4 T cell responses as SNP-7/8a (Supplementary Table 2).
We then assessed whether 4 of the previously reported non-immunogenic neoantigens (i.e. M01, M07, M21 and M39) that induced a CD8 T cell response but no significant CD4 T cell response as SNP-7/8a could limit tumor growth when used as a therapeutic vaccine administered intravenously (IV). Vaccination with SNP-7/8a delivering either of the neoantigens M07 or M21 led to improved control of tumor growth as compared with untreated animals (Supplementary Fig. 15c). Moreover, vaccination with SNP-7/8a delivering the neoantigen M39 also resulted in improved tumor control that was CD8 T cell dependent (Supplementary Fig. 15d).
Finally, we benchmarked anti-tumor efficacy with SNP-7/8a against other peptide vaccines administered by their preferred route. In a first study, we treated tumor-bearing animals with M39 neoantigen as SNP-7/8a by the IV route or as LP + polyIC administered by the SC route. Consistent with the previously reported lack of immunogenicity of M39 delivered as LP + polyIC, there was no efficacy observed with LP + polyIC, whereas there was significant efficacy when M39 was delivered as SNP-7/8a (Fig. 6f and Supplementary Fig. 15e). In a second study, animals bearing TC-1 tumors treated with the HPV E6 antigen as SNP-7/8a by the IV route had improved tumor control as compared with animals treated with E6 as LP + polyICLC by the SC route (Fig. 6g and Supplementary Fig. 15f).
Altogether, these results demonstrate the broad therapeutic potential of SNP-7/8a as a vaccine platform for targeting diverse tumor antigens (including self-antigens, neoantigens and viral antigens) and that improved T cell priming efficiency with SNP-7/8a leads to a greater breadth of neoantigen-specific CD8 T cells capable of mediating tumor control.
Discussion
Herein we report the rationale for and systematic development of charge-modified (CM) conjugates as a generalizable vaccine platform for co-delivering any peptide-based tumor antigen with molecularly-defined adjuvants (e.g., TLR-7/8a, TLR-9a and STINGa) in self-assembling nanoparticles that efficiently induce anticancer T cell immunity. The major findings were that conjugate vaccines can be chemically programmed (via charge modification) to account for peptide antigen physicochemical heterogeneity to provide consistent formulations optimized for T cell priming.
A broad variety of particle vaccine technologies (e.g., liposomes, PLGA particles, etc.) have been developed to enhance the immunogenicity of peptide-based cancer vaccines; however, many such approaches rely on empirical formulation processes that can lead to variability (e.g., inconsistent material loading) arising from differences in antigen properties. While several emerging technologies have demonstrated promise for improving the consistency of peptide neoantigen incorporation into particles53,54, these and many other particle vaccine technologies also face challenges during sterile filtration using ≤0.2 μm pore membranes, which can be clogged by particles that exceed the filter membrane pore size and result in significant material loss55. These challenges may limit the use of such technologies for personalized, on-demand therapies that require consistent formulations with short production timelines. In contrast, the CM conjugates described herein are chemically-defined single molecules that allow for sterile filtration without material loss and ensure consistent formulations (i.e. precise material loading in uniformly sized particles) through a controlled nanoparticle self-assembly process.
While other conjugate vaccine technologies have been shown to be effective for inducing anticancer T cell immunity21,22,26, a major limitation is their propensity to form aggregates that complicate manufacturing and lead to injection-site depots that can cause T cell exhaustion29. To address this challenge, we introduced a charge-modifying group to the N-terminus of peptide antigens that i) improves the solubility of hydrophobic peptide antigens during synthesis and purification and ii) induces conjugates to self-assemble to nanoparticle micelles of a small, optimal size (~20 nm) for targeting APCs that promote T cell immunity36,37. Importantly, SNP-7/8a based on CM conjugates enhanced uptake by APCs and led to superior CD8 T cell induction over conjugates without charge modification.
Consistent with recent studies showing that the route of administration can impact efficacy of cancer vaccines in mouse models56,57, we observed improved efficacy with SNP-7/8a by the IV route as compared with the SC route and are undertaking studies to understand the mechanistic basis for these differences. Successful IV vaccination strategies will likely require the physical association of antigen and adjuvant in particles to ensure that both components are co-delivered to APCs for efficient T cell priming (e.g., see: Fig. 6a) and to prevent antigen presentation without innate immune stimulation, which can lead to tolerance47. In addition, small, ~10–30 nm, nanoparticles that can passively accumulate in tumors following IV administration may provide additional benefit by enabling the adjuvant to access and alter the tumor microenvironment58.
Finally, maximizing efficacy will also require identification of the optimal combination with complementary immunotherapies52, chemotherapeutics59 and/or radiation treatment60 to promote efficient tumor killing while maintaining acceptable safety profiles. Peptide-based vaccine platforms such as SNP-7/8a can be used to prime T cells that, at a minimum, should be combined with checkpoint inhibitors. The pool of vaccine primed T cells may be expanded to higher numbers in vivo using heterologous prime-boost immunization with viral or RNA vaccines, or cytokines, such as IL-2, that promote T cell expansion. Vaccine primed T cells may also be isolated and manipulated ex vivo to increase their number and alter their quality for use in ACT or sequenced to identify T cell receptors (TCRs) of interest. Indeed, vaccines that efficiently prime anticancer T cells have the potential to play a central role in many promising combination immunotherapies.
In conclusion, the results presented herein show how a peptide-based PCV can be systematically optimized to enhance the magnitude and breadth of neoantigen-specific T cell responses while addressing manufacturing challenges of a personalized therapy.
Online Methods
Animal protocols
Animal experiments were conducted at the Vaccine Research Center (VRC) at the National Institutes of Health (Bethesda, MD) and the Institut Curie (Paris, France). Animal protocols underwent review and were approved by the respective ACUCs prior to the start of experiments. Animal experiments complied with the respective ethical guidelines as set by each ACUC.
Animals
Female C57BL/6 (B6) mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained under specific-pathogen-free conditions. B6 mice were 8–12 weeks of age at the start of experiments. Animals were randomly assigned to either control or experimental groups. Eight healthy female and male rhesus macaques (“NHPs”) of Indian origin (Macaca mulatta) with a mean (s.d.) age and weight of 3.0 (0.7) years and 4.3 (0.9) kg, respectively, were pair housed in animal biosafety level 2 facilities and were monitored throughout the study for physical health, food consumption, body weight, and temperature. Study groups were balanced with respect to age, weight, and gender. Sample size was based on prior NHP immunogenicity studies and calculated using Prism (GraphPad) and JMP Design of Experiment functionality (SAS).
Peptide antigens
Native peptide antigens and modified peptide antigens were custom synthesized by Genscript (Piscataway, NJ) using standard solid phase peptide synthesis and purified (> 90%) by HPLC.
TLR-7/8 agonists and hydrophobic blocks (e.g., oligo-7/8a)
Imidazoquinoline-based TLR-7/8 agonists were produced by Avidea Technologies, Inc. (Baltimore, MD) as previously described33. Detailed chemical schematics and descriptions of the methods used to synthesize and characterize the TLR-7/8a and hydrophobic blocks is provided in the Supplementary Materials and Methods.
Conjugate vaccine synthesis
Conjugate vaccines were produced by linking peptide antigens to hydrophobic blocks (e.g., oligo-7/8a) using a copper-free strain-promoted azide-alkyne cycloaddition click chemistry reaction. A detailed description of the methods used to synthesize and characterize the conjugate vaccines is provided in the Supplementary Materials and Methods.
Immunizations & treatment with checkpoint inhibitors
Vaccines were prepared in sterile, endotoxin-free (<0.05 EU/mL) PBS (Gibco). For mice, vaccines were administered either subcutaneously in a volume of 50 μL in each hind footpad or intravenously via the tail vein in a volume of 200 μL. Adjuvants were either prepared in-house as previously described33 and summarized in the Supplementary Materials and Methods or were acquired from commercial sources: polyIC (InvivoGen, San Diego, CA), anti-CD40 agonist (clone FGK4.5, BioXCell cat #BE0016–2, West Lebanon, NH), and CpG 1826 (InvivoGen). PolyICLC (Hiltonol) was a kind gift of A. M. Salazar (Oncovir). Animals treated with checkpoint inhibitor (CPI), anti-PD-L1 (clone 10F.9G2, BioXCell cat #BE0101), received 200 μg administered by the IP route in 100 μL PBS. For NHP, SNP-7/8a was formulated in 1 mL PBS for each of 4 SC sites (left and right deltoid; left and right thigh). Immunizations and blood sampling occurred with the NHP under anesthesia (10 mg per kg weight ketamine HCl).
Tumor cell lines
B16-F10 was acquired from ATCC (CRL-6475), MC38 was a kind gift from Lélia Delamarre (Genentech), TC-1 was a kind gift from T.C. Wu (Johns Hopkins University), and B16.OVA was a kind gift from H. Levitsky (Juno Therapeutics). Working cell banks (passage 4) were generated immediately upon receipt and used for tumor experiments. Cells were determined to be mycoplasma free upon establishment of each working cell bank.
Tumor implantations
B16-F10, TC-1, and B16.OVA were cultured in RPMI-1640 media (GE Life Sciences), and MC38 was cultured in DMEM media (Gibco), each supplemented with 10% v/v heat-inactivated FCS (Atlanta Biologicals), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco), 1× non-essential amino acids (GE Life Sciences), and 1 mM sodium pyruvate (GE Life Sciences). B16.OVA media was supplemented with 0.5 mg/mL G418. For each tumor cell implantation, a frozen aliquot was thawed, passaged once, and harvested using trypsin EDTA (Gibco), quenched with HI-FCS, washed in PBS, and implanted subcutaneously in sterile PBS. Tumors were measured using digital calipers twice per week, and tumor volume was estimated using the formula [tumor volume = short × short × long / 2]. Animals were euthanized when tumors reached size criteria (1000 mm3 or 2000 mm3).
CD8 depletion
Mice were depleted for CD8 T cells by intraperitoneal injection of 250 μg of anti-CD8 (clone 2.43, BioXCell cat #BE0061) in 100 μL PBS 1 d prior to tumor implantation, and 1 d and 7 d after tumor implantation. CD8 T cell depletion in the blood was confirmed using flow cytometry methods as described below.
Measurement of mouse T cell responses by intracellular cytokine staining
Measurement of antigen-specific CD8 and CD4 T cell responses by intracellular cytokine staining was performed as previously described61. Briefly, 200 μL heparin-treated whole blood was lysed with ACK lysis buffer (Quality Biologicals), filtered, and cultured in complete RPMI in 96-well plates with 2 μg/mL anti-CD28 (clone 37.51, BD cat #553294) in combination with 2 μg/mL of the native (unmodified) peptide antigen (Genscript). Brefeldin A (BFA, BD) was added to a final concentration of 10 μg/mL 2 h after purified peptides were added, and cells were incubated for an additional 4 h. After washing with PBS, cells were stained with UV Blue Live-Dead Dye (Life Technologies), washed, and blocked with anti-CD16/CD32 (clone 2.4G2, BD cat #553142) for 10 minutes at room temperature. After blocking, cells were surface stained for 30 minutes at room temperature with BUV805-anti-CD8 (clone 53–6.7, BD cat #564920) and BUV395-anti-CD4 (clone RM4–4, BD cat #740209). Cells were then fixed and permeabilized using Fix / Perm solution (BD) and incubated at 4°C for 30 minutes. Cells were washed and then suspended in Perm / Wash buffer containing AlexaFluor700-anti-CD3 (clone 17A2, BioLegend cat #100216), APC-anti-IFN-γ (clone XMG1.2, BD cat #554413), PE-anti-IL-2 (clone JES6–5H4, BD cat #554428) and BV650-anti-TNF-α (clone MP6-XT22, BD cat #563943) at 4°C for 30 minutes. Cells were washed and suspended in 0.5% paraformaldehyde (Electron Microscopy Sciences) in PBS and then evaluated by flow cytometry.
Multimer (tetramer/dextramer) staining of CD8 T cells from whole blood
Tetramer+ or dextramer+ CD8 T cell responses were characterized from whole blood as previously described62. Briefly, 200 μL heparin-treated whole blood was lysed with ACK lysis buffer, filtered, and plated in 96-well plates in PBS. Cells were stained with the viability dye Live/Dead Fixable Orange (OrViD, Life Technologies) for 10 minutes at room temperature. After washing, cells were stained for 15 minutes at 4°C with multimers (PE-H2-Kb OVA (SIINFEKL) tetramer (Beckman Coulter, Brea, California), PE-H2-Db Reps1 (AQLANDVVL) or Irgq (AALLNSAVL) dextramers (Immudex, Copenhagen, Denmark), or PE-H2-Db Cpne1 (SSPYSLHYL) or Trp1 (TAPDNLGYM) tetramers (kind gift of John Finnigan and Nina Bhardwaj, Mt. Sinai Icahn School of Medicine). Cells were then blocked with anti-CD16/CD32 (clone 2.4G2, BD cat #553142) for 10 minutes, followed by the addition of APC-Cy7-anti-CD8 (clone 53–6.7, Biolegend cat #100714), PE-Cy7-anti-CD62L (clone MEL-14, Abcam cat #ab25569, Cambridge, England), eFluor-660-anti-CD127 (clone A7R34, eBioscience cat #50–1271-82) and FITC-anti-KLRG1 (clone 2F1, Southern Biotech cat #1807–02, Birmingham, Alabama). After incubating for 20 minutes at room temperature, cells were washed and then incubated with Fix / Perm solution (BD) for 20 minutes at 4°C. After washing, cells were suspended in Perm / Wash buffer containing PerCP-Cy5.5-anti-CD3 (clone 145–2C11, BD cat #551163) and incubated at 4°C for 30 minutes. Cells were washed and suspended in Perm / Wash buffer and then evaluated by flow cytometry.
Measurement of NHP T cell responses by intracellular cytokine staining
Measurement of antigen-specific CD8 and CD4 T cell responses by intracellular cytokine staining was performed as previously described63, with modifications as noted below. PBMCs were isolated by density-gradient centrifugation from acid-citrate-dextrose–anti-coagulated whole blood. Without a prior expansion step, PBMCs were stimulated with peptides corresponding to the native (unmodified) neoantigens at 2 μg/mL for 2 hours followed by 10 hours in the presence of BFA at 10 μg/mL. Antigen-specific responses are reported after background subtraction of identical gates from the same sample incubated with the control antigen stimulation (irrelevant neoantigens). The staining protocol and staining panel were as previously described63.
Characterization of innate immune cells and cytokines from lymph nodes
The uptake of AlexaFluor647 (AF647)-labeled vaccines by antigen presenting cells (APCs) in the vaccine-draining popliteal lymph nodes (dLN) were evaluated as previously described33. Briefly, dLN of vaccinated mice were harvested at specified time points and mechanically disrupted in BioMasher tubes (Nippi Inc, Japan) containing PBS. Resulting cell suspensions were filtered through a 40 μm nylon mesh filter plate (EMD Millipore). Half of each dLN cell suspension was incubated at 37°C in complete RPMI for 12 hours. Supernatants were harvested and IL-12p40 concentration was determined by quantitative ELISA (Peprotech). The other half of each dLN cell suspension was transferred to V-bottom 96-well plates (Sigma Aldrich) for staining. Cells were stained for 10 minutes at room temperature with Live/Dead Fixable Aqua (Life Technologies), washed, and blocked with anti-CD16/CD32 (clone 2.4G2, BD cat #553142) for 10 minutes at room temperature. After blocking, cells were surface stained with BV510-anti-CD3 (clone 145–2C11, BD cat #563024), BV421-anti-CD19 (clone 1D3, BD cat #562701), BV605-anti-Ly-6G (clone 1A8, BD cat #563005), Cy7APC-anti-Ly-6C (clone HK1.4, BioLegend cat #128026), BV711-anti-CD103 (clone 2E7, BioLegend cat #121435), BV786-anti-CD8 (clone 53–6.7, BD cat #563332), BV510-anti-NK-1.1 (clone PK136, BD cat #563096), Cy7-PE-anti-B220 (clone RA3–6B2, BD cat #552772), AlexaFluor488-anti-IA/I-E (clone M5/114.15.2, Biolegend cat #107616), PE-anti-CD11c (clone HL3, BD cat #553802), AlexaFluor700-anti-CD11b (clone M1/70, BioLegend cat #101222), Cy5-PE-anti-F4/80 (clone BM8, eBioscience cat #15–4801-82), and CF594-PE-anti-CD80 (clone 16–10A1, BD cat #562504). Cells were washed, fixed in 0.5% paraformaldehyde in PBS, and analyzed by flow cytometry. The integrated median fluorescence intensity (iMFI) was calculated by multiplying the total number of cells positive for the vaccine (AF647+ cells) and the Median Fluorescence Intensity of the AF647+ cells.
Quantification of vaccine in draining lymph nodes by fluorescence measurements
Single cell suspensions of lymph nodes were added to black-walled 96-well plates and quantified for AF647-labeled vaccines by performing epifluorescence imaging (excitation = 650 nm; emission = 700 nm; 0.50 second exposure) using a Bruker In Vivo Xtreme (Bruker, Billerica, MA); fluorescence was determined by placing identically-sized regions of interest over each well.
Evaluation of OT-I expansion in vivo
The duration of antigen presentation following vaccination was assessed by measuring the expansion of OT-I cells adoptively transferred into vaccinated mice. OT-I cells were prepared by isolating total CD8+ T cells from spleen and lymph nodes of OT-I transgenic CD45.1 mice using Miltenyi beads (Bergisch Gladbach, Germany). The OT-I CD8 T cells were then labeled with 5 μM CFSE (carboxyfluorescein succinimidyl ester, Life Technologies) in PBS containing 0.1% BSA (Bovine Serum Albumin, Sigma-Aldrich) for 8 min at 37°C. CFSE-labeled OT-I cells (1×106) were injected intravenously in 100 μL PBS 0.1% BSA at different time points (day 0, 3, or 7) after C57BL/6 CD45.2 mice were injected subcutaneously with the indicated vaccines. Analysis of the in vivo expansion was performed 6 days after adoptive transfer by enumerating the number of CFSE-diluted CD8+ CD45.1+ OT-I cells from draining lymph nodes of vaccinated mice.
Flow cytometry
Samples were acquired on a modified BD LSR Fortessa X-50 flow cytometer running BD FACSDiva software v8.0.1. Results were analyzed using FlowJo v9.9.6 (TreeStar), Pestle v1.8, and SPICE v6.0 (ref.64).
Prediction of MHC-I binding affinity and immunogenicity screens
Mutations resulting from a single-nucleotide polymorphism that were also transcribed in a mouse tumor were selected from MC38 (ref.8), B16-F10 (Nina Bhardwaj, personal communication), and SB-3123 cell line (Nicholas Restifo, personal communication) without confirmation of MHC-I binding. Binding affinity for each peptide was predicted using Immune Epitope Database (IEDB) Consensus algorithm v2013–02-22 (ref.65), the ANN method66, and the SMM method67. Predictions were made for both H2-Kb and H2-Db MHC-I alleles, and the higher predicted binding affinity of the two alleles was selected for subsequent regression analysis. Each epitope (n = 179) selected for analysis was prepared as a peptide antigen linked to oligo-7/8a. Mice (n = 5 per group) were vaccinated subcutaneously in two sites with a pool of four antigens (1 nmol per antigen per site) on days 0 and 14. CD8 T cell responses were assessed 7 days after final vaccination for each antigen individually by ICS as described above. If any antigen in a pool was positive, each of the four antigens were then tested separately to confirm immunogenicity.
Hydropathy and charge frequency distribution of human neoantigens
All available human protein sequences were downloaded from Ensembl 2017 (ref.68). Canonical transcripts were identified as the longest sequence for each unique Ensembl protein-coding gene identifier. All possible wild type 25-mer peptides (n = 11.3M) were extracted from canonical protein sequences using a bespoke Python script. All possible single nucleotide mutations resulting in a missense substitution were identified using the database for non-synonymous functional prediction (dbNSFP; ref.69). All possible 25-mer peptides incorporating a single missense mutation were then extracted (n = 72.6M). The net charge (K, R = +1; E, D = –1; all other amino acids = 0) and grand average of hydropathy (GRAVY)70 were calculated for each wild type and mutant peptide.
Selection of rhesus macaque antigens
Rhesus neoantigens were selected from ‘hotspot’ mutations or ‘random’ mutations. Hotspot mutations were selected from the most prevalent (n = 100) mutations in human cancers from the COSMIC (Catalogue of Somatic Mutations in Cancer; ref.71) database. For the hotspot to be selected, the reference rhesus genome had to have exact homology to the wildtype human sequence where the mutation occurs. Random mutations were generated by introducing random mutations in the rhesus exome in silico. Non-synonymous mutations (n = 100) were selected. Neoantigens with high predicted binding affinity for Mamu-A*01 (IEDB Consensus score < 0.5) and moderate predicted binding affinity (IEDB Consensus score = 0.5 to 1.0) were selected for vaccination.
Machine learning model
A random forest machine learning model42 was used to predict whether a given conjugate vaccine would form nanoparticles (as assessed by turbidity < 0.05) or larger particles (turbidity > 0.05) based on measured and derived properties of the underlying composition (net charge, hydropathy, lengths of the constituent structural elements). 10-fold cross-validated models were derived to avoid overfitting the data. In each of these cross-validations, the random forest hyperparameters (the number of trees and the number of variables considered at each split) were tuned via Gaussian process optimization (scikit-optimize: Sequential model-based optimization, GitHub, 2018). To avoid overfitting the hyperparameters, their tuning was performed with 5-fold cross-validation, in 100 iterations (including 10 initial steps where the hyperparameters were set randomly), controlled by the log-loss. The resulting 10-fold cross-validated out-of-sample ROC curves and average ROC were reported. The total decrease in node impurity weighted by the probability of reaching that node (Gini Index) was calculated for each derived property of the underlying compositions.
Statistics and graphs
Sample sizes for biological studies were chosen based on calculations using JMP statistical analysis software (SAS, Cary, NC); standard deviations and pre-specified differences in groups (“differences to detect”) were based on historical data, and type I and type II error rates were set at 0.05 and 0.2, respectively. All relevant statistical tests were two-sided. Unless stated otherwise, data on linear axes are reported as mean ± s.e.m. Data on log scale are reported as geometric mean with 95% c.i. Statistical analyses were carried out using Prism software (GraphPad) or JMP. Unless stated otherwise within the figure legends, Kruskal-Wallis one-way ANOVA with Dunn’s post-test correction for multiple comparisons was used to calculate P-values for comparisons between > 2 groups; two-way ANOVA with Bonferroni correction was used to calculate P-values for comparisons between groups over multiple time points; and log rank test was used to compare survival differences for Kaplan-Meier plots.
Code availability
Scripts used to determine hydropathy and charge frequency distribution of human neoantigens and to conduct machine learning analyses of charge-modified conjugates are available from the corresponding authors upon request.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgements
The authors wish to acknowledge: M. Dillon, G. Salbador, L. Gilliam, S. Rush, M. Carballo, E. McCarthy, J. Noor, C. Chiedi, JP Todd, and D. Scorpio of the Translational Research Program (VRC) as well as B. Klaunberg and V. Diaz at the Mouse Imaging Facility (MIF, NIH) for their valuable support with the animal studies; T. Abram and A. Mullen (VRC) for administrative support; S. McWhirter and C. Ndubaku (Aduro BioTech) for providing CDN-based reagents; Cytometry Platform and the Animal facilities of the Institut Curie; INSERM; Agence Nationale de la Recherche (ANR-11-LABX-0043 and ANR-10-IDEX-0001-02 PSL); Institut Curie (CIC IGR-Curie 1428); La Ligue Nationale Contre le Cancer program; SiRIC grant INCa-DGOS-12554; the Czech Science Foundation project 19-08176S; and the Ministry of Education, Youth and Sports of the Czech Republic within the National Sustainability Program II (project BIOCEV-FAR LQ1604). This work was supported by the Intramural Research Program of the U.S. National Institutes of Health as well as NIH grant # R01EB027143. CJ is an employee of the VA Maryland Health Care System. The views reported in this paper do not reflect the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Competing Financial Interests
GL, YZ, VC, RL, LS, AI, and RS are listed as inventors on patents describing polymer-based vaccines. GL, YZ, VC, SN, YI, BD, JB, and AI are employees/consultants of Avidea Technologies, Inc. (Baltimore, MD), which is commercializing polymer-based drug delivery technologies for immunotherapeutic applications. CJ has an equity position in Cellth Systems, LLC. JB is a member of the POCKiT Diagnostics Board of Scientific Advisors.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.
References
- 1.Lennerz V et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A 102, 16013–16018, doi: 10.1073/pnas.0500090102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran E et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645, doi: 10.1126/science.1251102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran E et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med 375, 2255–2262, doi: 10.1056/NEJMoa1609279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371, 2189–2199, doi: 10.1056/NEJMoa1406498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg JE et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920, doi: 10.1016/S0140-6736(16)00561-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin U & Tureci O Personalized vaccines for cancer immunotherapy. Science 359, 1355–1360, doi: 10.1126/science.aar7112 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Hu Z, Ott PA & Wu CJ Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat Rev Immunol 18, 168–182, doi: 10.1038/nri.2017.131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav M et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576, doi: 10.1038/nature14001 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Kreiter S et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696, doi: 10.1038/nature14426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ott PA et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221, doi: 10.1038/nature22991 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226, doi: 10.1038/nature23003 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74, doi: 10.1126/science.aaa4971 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Mehta NK, Moynihan KD & Irvine DJ Engineering New Approaches to Cancer Vaccines. Cancer Immunol Res 3, 836–843, doi: 10.1158/2326-6066.CIR-15-0112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bookstaver ML, Tsai SJ, Bromberg JS & Jewell CM Improving Vaccine and Immunotherapy Design Using Biomaterials. Trends Immunol 39, 135–150, doi: 10.1016/j.it.2017.10.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheetz L et al. Engineering patient-specific cancer immunotherapies. Nat Biomed Eng 3, 768–782, doi: 10.1038/s41551-019-0436-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilyinskii PO et al. Adjuvant-carrying synthetic vaccine particles augment the immune response to encapsulated antigen and exhibit strong local immune activation without inducing systemic cytokine release. Vaccine 32, 2882–2895, doi: 10.1016/j.vaccine.2014.02.027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varypataki EM et al. Synthetic long peptide-based vaccine formulations for induction of cell mediated immunity: A comparative study of cationic liposomes and PLGA nanoparticles. Journal of controlled release : official journal of the Controlled Release Society 226, 98–106, doi: 10.1016/j.jconrel.2016.02.018 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A & Moon JJ Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat Mater 16, 489–496, doi: 10.1038/nmat4822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott EA et al. Dendritic cell activation and T cell priming with adjuvant- and antigen-loaded oxidation-sensitive polymersomes. Biomaterials 33, 6211–6219, doi: 10.1016/j.biomaterials.2012.04.060 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Fox CB & Haensler J An update on safety and immunogenicity of vaccines containing emulsion-based adjuvants. Expert review of vaccines 12, 747–758, doi: 10.1586/14760584.2013.811188 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Liu H et al. Structure-based programming of lymph-node targeting in molecular vaccines. Nature 507, 519–522, doi: 10.1038/nature12978 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho HI, Barrios K, Lee YR, Linowski AK & Celis E BiVax: a peptide/poly-IC subunit vaccine that mimics an acute infection elicits vast and effective anti-tumor CD8 T-cell responses. Cancer Immunol Immunother 62, 787–799, doi: 10.1007/s00262-012-1382-6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ignacio BJ, Albin TJ, Esser-Kahn AP & Verdoes M Toll-like Receptor Agonist Conjugation: A Chemical Perspective. Bioconjug Chem 29, 587–603, doi: 10.1021/acs.bioconjchem.7b00808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zom GG et al. Efficient induction of antitumor immunity by synthetic toll-like receptor ligand-peptide conjugates. Cancer Immunol Res 2, 756–764, doi: 10.1158/2326-6066.CIR-13-0223 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Lu BL, Williams GM, Verdon DJ, Dunbar PR & Brimble MA Synthesis and Evaluation of Novel TLR2 Agonists as Potential Adjuvants for Cancer Vaccines. J Med Chem, doi: 10.1021/acs.jmedchem.9b01044 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Zhu G et al. Albumin/vaccine nanocomplexes that assemble in vivo for combination cancer immunotherapy. Nat Commun 8, 1954, doi: 10.1038/s41467-017-02191-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair-Gupta P et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell 158, 506–521, doi: 10.1016/j.cell.2014.04.054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wille-Reece U et al. HIV Gag protein conjugated to a Toll-like receptor 7/8 agonist improves the magnitude and quality of Th1 and CD8+ T cell responses in nonhuman primates. Proc Natl Acad Sci U S A 102, 15190–15194, doi:0507484102 [pii] 10.1073/pnas.0507484102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hailemichael Y et al. Persistent antigen at vaccination sites induces tumor-specific CD8(+) T cell sequestration, dysfunction and deletion. Nature medicine 19, 465–472, doi: 10.1038/nm.3105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenter GG et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N Engl J Med 361, 1838–1847, doi: 10.1056/NEJMoa0810097 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Melief C Peptide-Based Therapeutic Cancer Vaccines. (2018).
- 32.Cavalli S, Albericio F & Kros A Amphiphilic peptides and their cross-disciplinary role as building blocks for nanoscience. Chem Soc Rev 39, 241–263, doi: 10.1039/b906701a (2010). [DOI] [PubMed] [Google Scholar]
- 33.Lynn GM et al. In vivo characterization of the physicochemical properties of polymer-linked TLR agonists that enhance vaccine immunogenicity. Nat Biotechnol 33, 1201–1210, doi: 10.1038/nbt.3371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynn GM et al. Impact of Polymer-TLR-7/8 Agonist (Adjuvant) Morphology on the Potency and Mechanism of CD8 T Cell Induction. Biomacromolecules 20, 854–870, doi: 10.1021/acs.biomac.8b01473 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Coffman RL, Sher A & Seder RA Vaccine adjuvants: putting innate immunity to work. Immunity 33, 492–503, doi:S1074-7613(10)00362-6 [pii] 10.1016/j.immuni.2010.10.002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy ST, Rehor A, Schmoekel HG, Hubbell JA & Swartz MA In vivo targeting of dendritic cells in lymph nodes with poly(propylene sulfide) nanoparticles. Journal of controlled release : official journal of the Controlled Release Society 112, 26–34, doi: 10.1016/j.jconrel.2006.01.006 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Manolova V et al. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol 38, 1404–1413, doi: 10.1002/eji.200737984 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Nuhn L et al. pH-degradable imidazoquinoline-ligated nanogels for lymph node-focused immune activation. Proc Natl Acad Sci U S A 113, 8098–8103, doi: 10.1073/pnas.1600816113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla NM, Malladi SS, Mutz CA, Balakrishna R & David SA Structure-activity relationships in human toll-like receptor 7-active imidazoquinoline analogues. J Med Chem 53, 4450–4465, doi: 10.1021/jm100358c (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson DS et al. Antigens reversibly conjugated to a polymeric glyco-adjuvant induce protective humoral and cellular immunity. Nat Mater 18, 175–185, doi: 10.1038/s41563-018-0256-5 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Vasilakos JP & Tomai MA The use of Toll-like receptor 7/8 agonists as vaccine adjuvants. Expert review of vaccines 12, 809–819, doi: 10.1586/14760584.2013.811208 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Liaw A & Wiener M Classification and Regression by randomForest. R News 2, 18–22, doi:citeulike-article-id:1121494 (2002). [Google Scholar]
- 43.Choe Y et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem 281, 12824–12832, doi: 10.1074/jbc.M513331200 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Kloetzel PM Antigen processing by the proteasome. Nat Rev Mol Cell Biol 2, 179–187, doi: 10.1038/35056572 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Jewett JC & Bertozzi CR Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev 39, 1272–1279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Poelgeest MI et al. Vaccination against Oncoproteins of HPV16 for Noninvasive Vulvar/Vaginal Lesions: Lesion Clearance Is Related to the Strength of the T-Cell Response. Clin Cancer Res 22, 2342–2350, doi: 10.1158/1078-0432.CCR-15-2594 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Bijker MS et al. Superior induction of anti-tumor CTL immunity by extended peptide vaccines involves prolonged, DC-focused antigen presentation. Eur J Immunol 38, 1033–1042, doi: 10.1002/eji.200737995 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Moutaftsi M et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol 24, 817–819, doi: 10.1038/nbt1215 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Rubinsteyn A et al. Computational Pipeline for the PGV-001 Neoantigen Vaccine Trial. Frontiers in immunology 8, 1807, doi: 10.3389/fimmu.2017.01807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abelin JG et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 46, 315–326, doi: 10.1016/j.immuni.2017.02.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson EA & Lore K Non-human primates as a model for understanding the mechanism of action of toll-like receptor-based vaccine adjuvants. Curr Opin Immunol 47, 1–7, doi: 10.1016/j.coi.2017.06.006 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Moynihan KD et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nature medicine 22, 1402–1410, doi: 10.1038/nm.4200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu F et al. Poly(propylacrylic acid)-peptide nanoplexes as a platform for enhancing the immunogenicity of neoantigen cancer vaccines. Biomaterials 182, 82–91, doi: 10.1016/j.biomaterials.2018.07.052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li AW et al. A facile approach to enhance antigen response for personalized cancer vaccination. Nat Mater 17, 528–534, doi: 10.1038/s41563-018-0028-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wendorf J et al. A practical approach to the use of nanoparticles for vaccine delivery. J Pharm Sci 95, 2738–2750, doi: 10.1002/jps.20728 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Kranz LM et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401, doi: 10.1038/nature18300 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Sultan H et al. Designing therapeutic cancer vaccines by mimicking viral infections. Cancer Immunol Immunother 66, 203–213, doi: 10.1007/s00262-016-1834-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lynn GM, Laga R & Jewell CM Induction of anti-cancer T cell immunity by in situ vaccination using systemically administered nanomedicines. Cancer Lett 459, 192–203, doi: 10.1016/j.canlet.2019.114427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welters MJ et al. Vaccination during myeloid cell depletion by cancer chemotherapy fosters robust T cell responses. Sci Transl Med 8, 334ra352, doi: 10.1126/scitranslmed.aad8307 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Cadena A et al. Radiation and Anti-Cancer Vaccines: A Winning Combination. Vaccines (Basel) 6, doi: 10.3390/vaccines6010009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darrah PA et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nature medicine 13, 843–850, doi: 10.1038/nm1592 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Quinn KM et al. Comparative analysis of the magnitude, quality, phenotype, and protective capacity of simian immunodeficiency virus gag-specific CD8+ T cells following human-, simian-, and chimpanzee-derived recombinant adenoviral vector immunization. J Immunol 190, 2720–2735, doi: 10.4049/jimmunol.1202861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishizuka AS et al. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nature medicine 22, 614–623, doi: 10.1038/nm.4110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roederer M, Nozzi JL & Nason MC SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry. Part A : the journal of the International Society for Analytical Cytology 79, 167–174, doi: 10.1002/cyto.a.21015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim Y et al. Immune epitope database analysis resource. Nucleic Acids Res 40, W525–530, doi: 10.1093/nar/gks438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen M et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12, 1007–1017, doi: 10.1110/ps.0239403 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peters B & Sette A Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics 6, 132, doi: 10.1186/1471-2105-6-132 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aken BL et al. Ensembl 2017. Nucleic Acids Res 45, D635–D642, doi: 10.1093/nar/gkw1104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, Wu C, Li C & Boerwinkle E dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat 37, 235–241, doi: 10.1002/humu.22932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kyte J & Doolittle RF A simple method for displaying the hydropathic character of a protein. J Mol Biol 157, 105–132 (1982). [DOI] [PubMed] [Google Scholar]
- 71.Tate JG et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research 47, D941–D947, doi: 10.1093/nar/gky1015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.