Abstract
Introduction
Heart failure remains one of the largest clinical challenges in the United States. Researchers have continually searched for more effective heart failure treatments that target the cardiac sarcomere but have found few successes despite numerous expensive cardiovascular clinical trials. Among many reasons, the high failure rate of cardiovascular clinical trials may be partly due to incomplete characterization of a drug candidate’s complex interaction with cardiac physiology.
Areas Covered
In this review, the authors address the issue of preclinical cardiovascular studies. The authors consider inherent tradeoffs made between mechanistic transparency and physiological fidelity for several relevant preclinical techniques at the atomic, molecular, heart muscle fiber, whole heart, and whole-organism levels. Thus, the authors suggest a comprehensive, bottom-up approach to preclinical cardiovascular studies that fosters scientific rigor and hypothesis-driven drug discovery.
Expert opinion
In the authors’ opinion, the implementation of hypothesis-driven drug discovery practices, such as the bottom-up approach to preclinical cardiovascular studies, will be imperative for the successful development of novel heart failure treatments. However, additional changes to clinical definitions of heart failure and current drug discovery culture must accompany the bottom-up approach to maximize the effectiveness of hypothesis-driven drug discovery.
Keywords: drug discovery, sarcomere-based therapy, heart failure, cross-bridge kinetics, in vivo function, solution-based chemistry, biophysical measurements
1. Introduction
Year after year, heart failure (HF) continues to be one of the largest clinical issues faced by the US, accounting for millions of hospitalizations and billions in medical expenses [1]. Furthermore, with approximately 25 million international HF patients, a number that is forecast to rise significantly, the global impact of HF also poses a growing concern [2]. Despite tremendous research effort striving to replace decades-old cardiovascular therapies with modern inotropic agents that target the sarcomere, the root of heart disease, clinical successes remain elusive [3]. This is no more apparent than when considering the ever-growing graveyard of failed cardiovascular clinical trials [3,4]. While clinical trials, in general, are facing low success rates, cardiovascular drugs entering lead-indicated phase I clinical trials had the lowest likelihood of final FDA approval: 8.7%, nearly two times lower than the overall average of all trials [4]. Accompanying the increasing failure rate of clinical trials is ballooning R&D investment in drug discovery; the average drug takes 10–15 years and 1.5–2 billion USD to make it to market [5]. Again, however, the cardiovascular space struggles more than the rest. Among all therapeutic areas considered, the average annual growth rate of cardiovascular clinical trial costs in the US was the highest at 18.65% between 1989 and 2011, almost tripling after the turn of the millennium with no blockbuster cardiovascular drugs to mention [6]. Fortunately, the generally poor state of clinical trials and drug discovery has sparked several discussions about how to reverse many of these worrying trends. Proposed solutions frequently involve improving communication between clinicians, changing pharma business models, and streamlining the clinical trial process. Still, as the statistics suggest, cardiovascular therapies have additional and often more challenging hurdles to overcome.
The base issue lies in the complexity of in vivo heart function and its regulation; biochemistry, mechanics, electrophysiology, hemodynamics, and neurohormonal signaling interface work in tandem unlike any other system in the body. These facets of the heart dynamically regulate cardiac function while maximizing efficiency. The sarcomere hosts the molecular origins of many of these regulatory mechanisms: the molecular motors themselves, myosin II, phosphorylatable accessory proteins such as myosin’s regulatory light chain (RLC), and cardiac myosin binding protein C (cMyBPC), thin-filament calcium handling. Many sarcomeric proteins further enable cooperative ensemble behaviors that result in length, load, and frequency dependencies [7]. In all, the sarcomere is a tightly integrated contractile machine, and disfunction in just one aspect can result in cardiomyopathy and eventual HF. The same fact, however, is what makes the sarcomere such an appealing and logical therapeutic target; precise targeting of just one aspect of the sarcomere could restore normal heart function [8]. But as another has identified, “to be precise one must be comprehensive,” meaning that both the disease and drug mechanism need to be extensively characterized and understood [9]. This is a particularly challenging task for sarcomere targeting therapies due to the multiple levels of sarcomere regulation. Furthermore, a drug’s effect on heart function may not be limited to just the sarcomere. It could elicit slow-onset responses from regulatory mechanisms beyond the sarcomere involving excitation-contraction (EC) coupling, architectural remodeling, or neurohormonal signaling and could make a promising drug candidate eventually fail in clinical trials [3]. In addition, regulatory agencies generally only require a demonstration of safety/efficacy, not detailed knowledge of the mechanism of action – which may contribute to the high failure rate in late-stage clinical trials.
These physiological challenges highlight the need for robust and comprehensive preclinical experiments that consider drug effects at all levels of cardiac physiology. Thus, we suggest a bottom-up approach to preclinical studies that starts with techniques at the atomic and molecular levels, then builds to the cellular and whole organ levels. In this way, the simplest atomic and molecular systems build a strong foundation of highly detailed results that then support findings in more complex and realistic contexts, fostering scientific rigor and hypothesis-driven drug discovery such that only the best drug candidates make it to clinical trials. To this end, we present an overview of relevant computational, physiological, biophysical, and biochemical techniques that can be used to study cardiac sarcomere-targeting drugs.
2. Computational Methods
The first step of the hypothesis-driven drug discovery process entails a detailed understanding of the mode of drug action on molecular structures and its effects on ligand interaction. One traditional method of drug discovery involves testing a large, random number of compounds using high-content or high-throughput screening to identify “lead” compounds more quickly [10]. An abundant amount of experimental data on drug activity, specificity, and/or toxicity can be collected using a diverse array of cellular and chemical assays through this method [11]. In addition, phenotypic screening can identify “first-in-class” drugs that would otherwise be unknown [12]. Recent innovations in high-throughput screenings that use contractile-force based platforms [13,14], functional assays of skeletal muscle [15], and time-resolved fluorescence resonance energy transfer (TR-FRET) [16] have allowed researchers to target sarcomeric proteins. High-throughput screening has also been successful in discovering novel small molecule inhibitors of cardiac hypertrophy [17]. These include various screening systems like induced pluripotent stem cell (iPSC)-derived cardiomyocytes [18], biomimetic cardiac microsystems [19]. High-throughput crystallography techniques can also aid in structure determination [20]. Although there have been advancements in in vitro screening systems thanks to automation and miniaturization, high-throughput screening remains time-consuming, expensive, and far from ideal [21,22]. The test systems used frequently lack the complexity of the target biological system, and chemical compounds are often pursued based on assumptions of structure-function relationships instead of specific biological changes that are relevant to the target pathophysiology [23]. Also, high-throughput screening methods may introduce systematic biases such as batch and edge effects that cause lower screening precision and higher false-positive and -negative rates [24]. Ultimately, drug discovery needs to rely less on luck and more on hypothesis-driven searches.
Therefore, the integration of in silico molecular screening techniques and high-throughput screening offers the drug discovery process to become more knowledge-based and hypothesis-driven. Recent advances in computer-aided high throughput virtual screening using molecular modeling and docking allows for more cost-effective ways to identify lead compounds for treating heart diseases [25]. For example, high throughput screening using human iPSC - cardiomyocytes can be aided by large scale analysis of sarcomere function using the algorithm SarcTrack [26]. Using various initial screening methods such as 1D-3D similarity searches, pharmacophore fingerprints, volume matching, and clustering/partitioning combined with computational filtering based on ADME (absorption, distribution, metabolism, and excretion) properties such as cytochrome P450, solubility, and drug-like characteristics can increase potential hits [27].
However, even without the integration of virtual and in vitro high throughput screening, virtual methods can be helpful in narrowing down drug targets. Structure-based screening methods like computational docking and simulation studies based on experimental X-ray (XR), nuclear magnetic resonance (NMR) structures, or ab initio and template-based homology models can even test protein-ligand dynamics. During docking studies, various stochastic or genetic search algorithms are used to find possible binding properties of ligands and ranked according to a scoring function [28]. Thus, docking studies allow for robust and fast prediction of ligand binding sites. For instance, Figure 1 illustrates an example trial of omecamtiv mecarbil (OM) docked to cardiac myosin S1 fragment crystal structure [29] using AutoDock [30]. Although there are various technical challenges such as imperfect scoring functions, the presence of intrinsically disordered and flexible regions in target proteins, or other complex protein-ligand structural interactions, the field is continually developing. For example, there have been efforts at the multi-scale computational model of hypertrophic cardiomyopathy (HCM) and sarcomeric mechanics of dilated cardiomyopathy (DCM) [31–33]. As a proof of concept, molecular dynamics simulation and docking were done to elucidate the dynamic and mechanistic details of small molecule interaction with the cardiac thin filament in the presence of cardiomyopathic mutations [34]. These techniques explore the energy landscape of the drug-target interactions through analysis of protein conformation, intra- and inter-molecular interactions, and electrostatic properties. The key advantage of the in silico techniques mentioned above is the incredible flexibility that they add to the drug discovery process. Results from computational studies can be refined into leads through iterative pharmacological/activity profiling, and find in-depth, high-resolution atomic information about ligand-target interactions. One important limitation of in silico methods is the maximum size of a system that can be realistically modeled; very large simulations may take too long to simulate properly even with modern computing powers. As a result, simulations must adopt a reductionist approach that may not recapitulate the full complexities of a multi-component system like a muscle fiber or the whole-heart.
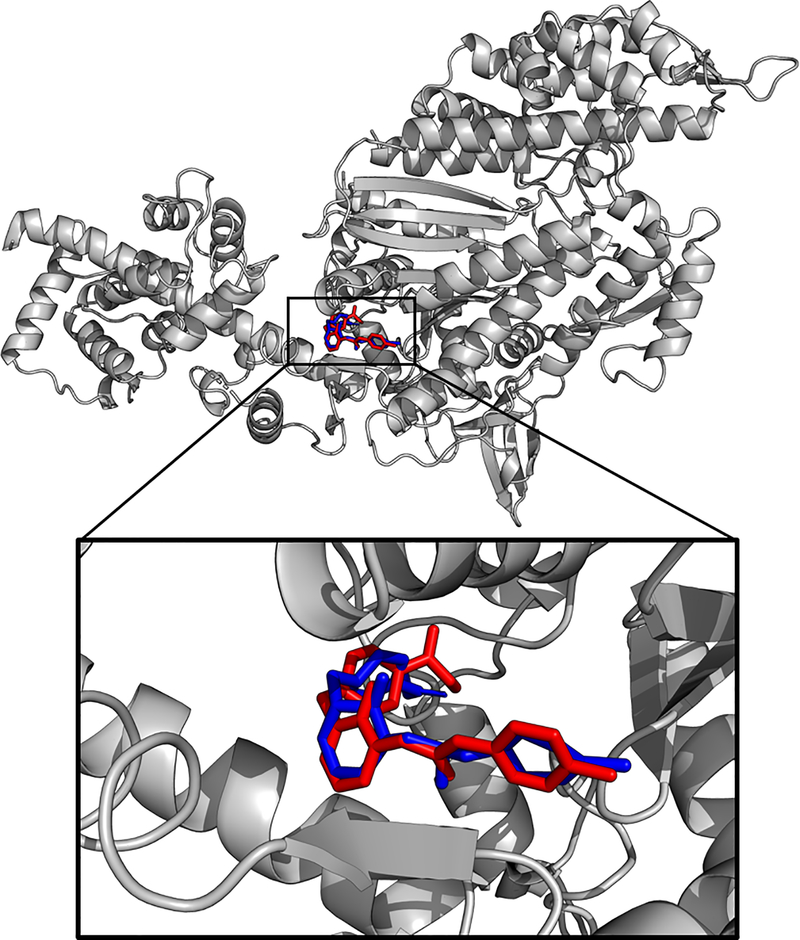
Figure 1.
Omecamtiv mecarbil (OM) was docked to the cardiac myosin S1 fragment in the pre-powerstroke state crystal structure (PDB ID 5N69) using the Genetic Lamarckian algorithm in AutoDock. The OM pose from the crystal structure (blue) and the predicted pose from the example docking experiment (red) differs by RMSD = 6.364 Å, calculated using PyMOL’s rms_cur matchmaker command (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). The predicted affinity via docking was 0.23 μM.
Unfortunately, both in vitro and virtual screening methods are limited by the number of compounds in the compound libraries and are not able to create unique and novel ligands. Therefore, hypothesis-driven drug discovery may benefit from structure-based de novo rational drug design to develop purpose-built drug molecules from scratch. Because de novo drug design provides an unlimited search space, it poses the problem of combinatorial explosion. Avoiding such an explosion requires careful and systematic selection of primary target constraints using rule- or grid-based approaches and identification of interaction sites using techniques such as Multiple Copy Simultaneous Search or docking [35]. In this method, the assembly of a complete, bound ligand is performed through atom-by-atom design, fragment linking/growing, or scaffold hopping techniques, which is then assessed by various scoring functions [28]. In ligand-based methods, a 3D ligand pharmacophore model can be used to develop a target-specific quantitative structure-activity relationship (QSAR) model using various regression and pattern-recognition techniques [36]. Two examples of sarcomere-specific drugs that have been developed using rational drug design and QSAR for optimization include bepridil and levosimendan [37]. While structure-based rational drug design improves the success rate and gives researchers a basis for further development of drugs [38], it can be more time consuming than virtual screening.
After the screening process identifies promising chemical compounds, additional computational tools can optimize the ligand-receptor binding free energies to augment ligand selectivity/potency to the desired target. Some of these methods include force-field based Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA), cheminformatics-based QSAR, quantum mechanics-based Coupled Cluster calculations, or enhanced molecular dynamics sampling methods including weighted histogram analysis method (WHAM), the potential of mean force (PMF), metadynamics, and convex-PL scoring function [39]. A more comprehensive and detailed review of these and other computational methods in drug discovery and cardiology can be found elsewhere [40,41].
In summary, computational methods allow researchers to quickly and efficiently prioritize promising drug candidates. It is also worth noting that some in vitro experiments can also be simulated in silico. For example, steered or force-probe molecular dynamics simulation, in place of atomic force microscopy, can be conducted to study conformation or energy landscape of sarcomeric proteins like titin and troponin [42,43] However, due to constraints in time, complexity, and computational power, a physiologically realistic pulling speed is not possible. Novel techniques and models are continually being developed, such as the virtual heart model to screen for cardiotoxicity [44], improving the efficiency of the computational drug discovery process. The use of various artificial intelligence methods like machine learning, deep learning, and neural networks can further accelerate and automate the drug discovery process, resulting in more accurate lead generation and prediction of molecular level protein dynamics in a reasonable time frame [45,46]. However, computational methods are still theoretical, mathematical models with various underlying assumptions that may limit their utility. Thus, the molecular knowledge gained through docking and simulations should be the first step in designing more complex in vitro experiments in real biological systems.
3. Solution-based and Single-Molecule Techniques
While the flexibility of molecular in silico simulation affords investigators many irreplaceable tools, they do not serve as replacements for biochemical, biophysical, and physiological experiments. Current computational tools are not equipped to capture the full complexity of real biological systems. While a range of possible biological testbeds exists, the bottom-up approach suggests that early experiments should focus on studying drug effects in the simplest systems possible. Doing so helps to confirm and extend any in silico results while maintaining a high level of experimental control, flexibility, and detail. Such experiments naturally lend themselves to techniques that utilize purified or isolated proteins to characterize molecular behavior and interactions. An added advantage of these experiments is that they can easily utilize recombinant proteins to test drug behavior in the context of a specific mutation or disease [47–50]. This is essential for inotropic therapies intended to treat cardiomyopathies that are caused by particular mutations.
For use with sarcomere specific proteins, most in vitro techniques fall into four main categories: binding affinity assays, structural studies, biochemical kinetic assays, and functional assays. Since binding affinity assays and structural studies are extensively reviewed and discussed elsewhere [39,51,52] and are not unique to the study of sarcomeric proteins [29,53] the rest of this section will focus on the kinetic and functional assays that are more specific to studying the sarcomere. Furthermore, because most of the kinetic and functional assays available characterize myosin itself, the discussion will primarily be in the context of myosin-targeting drugs.
For myosin-interacting drug candidates, solution-based kinetic assays are often the first performed. Kinetic assays provide standard methods for determining effects on different steps of the myosin-ATPase cycle, which is the biochemical engine of contractility [50,54]. The simplest and easiest of these techniques measures the steady-state rate of actin-activated myosin-ATPase. Various actin concentrations can be used, but the maximal actin-activated myosin-ATPase rate (kcat) is most commonly reported. Because of its relative simplicity, kcat is an indicator sometimes used for high throughput screening of small molecules [55,56]. However, kcat alone is not specific enough to parse the fine-grained details of a drug mechanism, so a significant change in kcat mainly warrants more thorough mechanistic investigations. To further isolate a drug’s effect on specific states and transitions within the ATPase cycle, a myriad of well-established transient kinetic techniques are available [54]. For example, transient kinetic assays have been used to isolate the effects of many small molecules to the inorganic phosphate (Pi) release step. High profile drug candidates like OM, mavacamten, and several other research molecules such as blebbistatin and 2,3,-butanedione monoxime (BDM) all modulate the rate of actin-activated Pi release [53,56–58]. These similar drug actions make sense considering that Pi release is the rate-limiting step of the actin-activated ATPase activity assay used to identify many of these molecules [59]. Thus, any changes in Pi release rate would alter kcat most significantly and be easiest to detect.
Of course, drug-induced changes in other steps of the myosin-ATPase cycle are possible and could be responsible for secondary drug mechanisms. For example, while the primary action of mavacamten was first attributed to the slowing of Pi release, later transient kinetic assays found that it also slowed the actomyosin association rate [60]. This discovery later helped explain mavacamten’s effect on data from skinned cardiac fibers [61]. Several other solution-based assays can offer additional insights into the mechanism of myosin-targeting drug candidates. For instance, FRET and TR-FRET can be used to probe the mechanical movements associated with the myosin power and recovery strokes [62–64].
One significant limitation that virtually all solution-based myosin assays share is that they cannot sufficiently characterize strain-dependent transitions such as ADP release. The strain dependence of ADP release is particularly important to consider because it forms the molecular basis of muscle’s force-velocity and power-velocity relationships, which are strong predictors of whole-heart function [47,50]. Fortunately, a class of functional assays that test in vitro motility and single myosin performance compensates for this particular shortcoming of solution-based assays. The simplest of these measures the in vitro motility of fluorescent F-actin as it is moved by myosin molecules fixed to the floor of a flow-cell [65]. An actin-binding protein, such as α -actinin, can then be added to increase the force that the myosin works against [66]. Repeating the motility assay with varying α-actinin concentrations eventually produces a force-normalized force-velocity curve. More sophisticated assays utilizing optical trapping techniques allow for actual quantification of parameters like the single-molecule force, its power-stroke step size, actin-myosin binding lifetime, and the force dependence of ADP-release [67,68]. Applying these functional assays to OM illustrates their experimental importance. While transient kinetic assay showed that OM increased the rate of Pi release, in vitro motility assays and optical trap techniques suggested that OM also inhibits the power stroke and causes ADP release to be independent of load, thus slowing detachment [47,69]. Further experiments with optical trap techniques went on to suggest that the latter may be another common drug mechanism in addition to affecting Pi release [47,70,71].
Using a combination of all the above techniques, one can get a more complete picture of the molecular effects of myosin-targeting cardiac drug candidates. Unfortunately, cardiac myosin never operates alone; the sarcomere organizes ensembles of myosin into an intricate hexameric lattice structure with several additional thick and thin filament regulatory proteins (e.g. RLC, cMyBPC, the troponin complex, tropomyosin, etc.) that augment myosin’s function [7,72]. That being said, it is possible to test the effects of regulatory proteins incorporated into some of the above assays. The most common method involves adding the troponin complex and tropomyosin to F-actin, creating regulated thin filaments. Then, the Ca2+ dependence of some in vitro parameters such as kcat and motility can be quantified [48,50,73]. Regulated thin filaments can serve as a high throughput screening method for troponin targeting drugs [55]. However, aside from the Ca2+ handling proteins, regulatory proteins are rarely considered in solution. One of the only other examples is a study that showed how adding purified cMyBPC to myosin in solution yields cooperative behavior even without thin filaments [74]. Such a technique could hypothetically screen for drugs targeting cMyBPC, which may offer a novel therapeutic target, but no such assay exists to date [75,76]. Even if solutions-based assays incorporating cMyBPC or other accessory proteins gained popularity, like all other solution-based and single-molecule assays, they would still fail to capture ensemble behaviors and sarcomere geometry that introduce additional complexities like cooperative effects [7].
Another aspect to consider is that the asynchronous action of multiple myosin molecules during sarcomere contraction potentially uncouples the chemical and mechanical transitions of the actomyosin-ATPase cycle, including that of Pi release [77–79]. While the in vitro motility assay and optical trapping techniques can be modified to test the behaviors of small ensembles of myosin (5–10 molecules), the effects are not the same because of the missing lattice structure [80]. Nevertheless, mathematical modeling efforts have found that single myosin behavior can sufficiently predict ensemble behavior [81]. But these were not models of diseased states, and little modeling work exists to back the predictive power of single-molecule studies with altered function [81]. As such, drug effects on ensemble behavior may be unpredictable; sarcomere-level phenomena could presumably amplify, mask, or even alter the drug’s molecular effects which can be tested using detergent-skinned fiber preparations that harbor an intact sarcomere lattice.
4. Muscle Fiber-Based Techniques
While in silico computational screening, solution-based in vitro motility assays, and other avenues reviewed above have distinct advantages like elucidating the drug effects at an atomic scale and discerning protein-protein interactions to quickly screen a large number of potential drug candidates, those techniques do not factor in the complexity of higher-order assemblies characteristic of multicellular fiber preparations. Moreover, impaired sarcomeric contractile function underlies impaired whole-heart function, suggesting the need for protocols that allow testing the impact of drug candidates that target the cardiac sarcomere. Detergent-skinning of myocardial preparations with Triton-X 100 is widely used for this purpose and involves permeabilizing the preparations to facilitate rapid equilibration of ions and drug molecules between the sarcomere’s contractile apparatus and the external medium (Figure 2). Importantly, skinned fiber experiments can serve as a nice middle ground in the bottom-up approach for drug testing: the preparations are not too complex but at the same time capture most of the fundamental behaviors underlying muscle contraction.
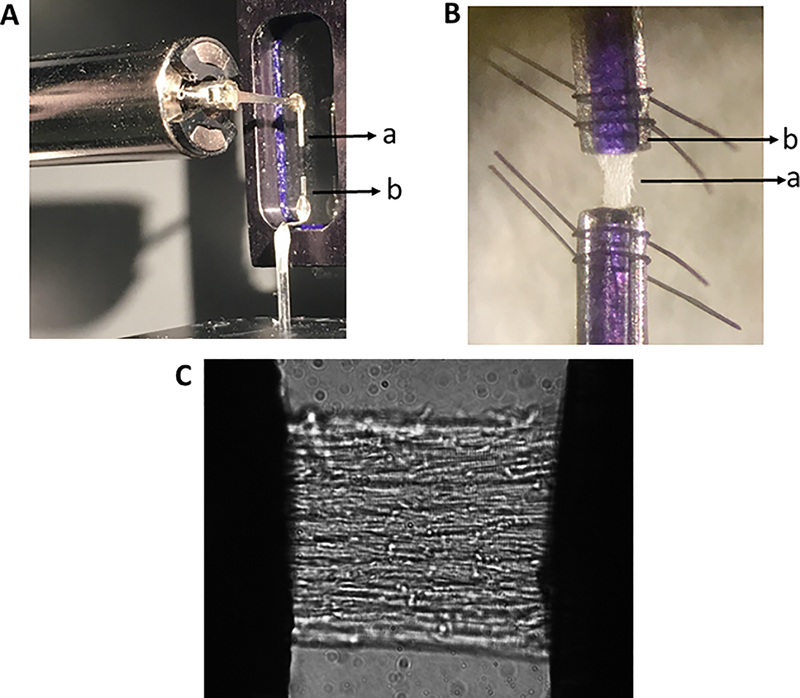
Figure 2:
(A) Experimental setup consisting of a motor arm (a) and force transducer (b) placed within the chamber in which solutions containing various amounts of Ca2+ and drugs can be loaded. (B) A multicellular detergent-skinned cardiac muscle preparation (a) is fixed in between the motor arm and a force transducer troughs (b) and secured using polydioxanone pins (violet-colored) and nylon loops (black colored). (C) Shown is a magnified image (40x) of a detergent-skinned cardiac muscle preparation secured on either end as shown in panel B.
There are several advantages to testing novel drug compounds on cardiac skinned fibers. One key advantage is that drugs can be tested using myocardial tissue samples collected from donor and HF patients. Skinned preparations can easily be mounted on a variety of experimental setups to monitor trends in force generation, optical properties, and ionic fluxes [82]. Importantly, skinned preparations permit the study of drug effects on a multitude of sarcomeric contractile mechanisms that ultimately shape the whole-heart function: cooperativity of force development, cooperative cross-bridge (XB) recruitment to actin, force responses to intracellular [Ca2+] fluctuations, sensitivity of myofilaments to Ca2+, dynamic XB behavior [83], tension cost of force production [84], lattice spacing [85], and length-dependent activation [86]. For details on translating cardiac muscle mechanics data from skinned preparations to whole-heart function, please refer to a recent review [87]. Skinned preparations can be utilized to perform various techniques like stretch activation (SA) [88,89], ktr [76], loaded shortening and power output [90], and XR diffraction studies [91]. For example, SA experiments allow the experimenter to further dissect the dynamic XB behavior into XB on and off rates as well as assess XB recruitment and measure the magnitudes of XB detachment and XB recruitment [92]. Apart from elucidating the drug effects on various sarcomere-based contractile mechanisms, skinned preparations allow us to test the drug effects following altered phosphorylation status of contractile proteins, an important consideration due to its functional implications in HF [93]. Further, the effect of pharmacological agents on genetic models like cMyBPC knock out (KO) [76], pathological HF [93], mutated, and phospho-ablated samples can also be evaluated.
In contrast to in vitro motility and solution-based assays, skinned fiber preparations retain a well-organized sarcomeric lattice with essential regulatory and structural protein constituents and thus preserve near-normal structure-function relationships. Thus, testing newly identified drug candidates on skinned cardiac preparations may yield additional insights that might even differ from findings obtained using lattice-free systems. For instance, initial solution based stopped-flow and transient-state kinetic experiments with OM showed an increased rate of actin-dependent phosphate release from cardiac myosin-S1 leading to the idea that the sarcomeric modulator OM accelerates myosin XB transition rate from weakly- to a strongly-bound force-generating state [53] – which would predict faster rates of force generation in cardiac muscle. However, contrary to this notion, recent skinned cardiac fiber experiments revealed that OM acts to slow overall force generation by slowing the rate of XB detachment (krel), rate of cooperative XB recruitment (kdf), and rate of tension redevelopment (ktr) [76,94]. Data from skinned fiber studies suggest that the OM-induced slowing of krel acts to extend the amount of time that OM-bound myosin XBs spend in their actin-bound state [76]. The prolonged binding-times of OM-bound myosins enhances cooperative thin filament activation via nearest-neighbor effects, allowing non-OM-bound myosins to bind to actin, and results in an OM-induced increase in net force production [69]. Additionally, OM-induced slowing of kdf and ktr also suggests that the drug prolongs the total time course of OM-bound XB transitions to force-bearing states, an effect that potentially spreads cooperative activation along the thin filaments during OM-induced force enhancement [93].
Skinned preparations are a good experimental system to test drug impact on the cardiac sarcomere because these preparations provide a versatile testbed to precisely control factors such as intracellular [Ca2+], temperature, degree of sarcomere length changes, and to equilibrate drug doses around the contractile apparatus. Fiber preparations also facilitate performing structure-function studies by enabling selective extraction of endogenous sarcomeric proteins/complexes and reconstitution with exogenous proteins/complexes [95,96], a valuable method to test drug impact on contractile function by simulating conditions like reduced or altered protein content within the sarcomere. Skinned preparations are ideal to test a range of direct sarcomere targeting drugs and their dose-dependent effects [61,93] to understand the cellular basis of functional data collected from whole-heart studies. For example, the impact of cardiac thin filament modulators like levosimendan, bepridil and thick filament modulators like OM, EMD57033, and mavacamten [97] can be evaluated in skinned cardiac preparations to help draw meaningful conclusions about the drug effects on in vivo function. For instance, using skinned fibers, we recently showed that OM significantly enhances myocardial force at submaximal [Ca2+] but significantly slows XB behavior by its effect on both XB recruitment and detachment phases [76], suggesting that OM prolongs XB duty cycle time to further enhance cooperative XB recruitment to open actin sites –findings that support OM-mediated prolongations of in vivo systolic ejection time (SET) observed in human clinical trials [98,99]. Thus, inferences from fiber studies provide a mechanistic basis for drug-mediated contractile modulation and can have direct implications for the design and development of new cardiac sarcomere-based therapies.
Despite the advantages, some inherent limitations prevent a direct one-to-one translation of fiber studies to whole-heart function. Skinned preparations lack the architecture to mimic the complex fiber orientation that is characteristic of whole-heart, and it is difficult to resolve individual sarcomere lengths within a large fiber [100]. Fiber experiments are also performed in a static Ca2+ environment, whereas Ca2+ levels that underlie in vivo function transiently change. Furthermore, the whole-heart functions by integrating a complex interplay of sarcomeric function with multiple upstream and downstream cellular/neurohormonal signaling cascades, Ca2+ cycling mechanisms, EC coupling, and myocardial electrical conduction pathways which cannot be simulated using skinned fibers.
As an example of these differences, skinned fibers from murine cMyBPC KO hearts exhibit faster krel and kdf when compared with wild-type mouse fibers, suggesting that force relaxation and development are sped at the fiber level [88,101]. Such effects would be expected to enhance diastolic and systolic functions at the whole-heart level. Intriguingly, KO hearts display impaired diastolic and systolic functions as indicated by increased isovolumic relaxation time and reduced fractional shortening suggesting that in vivo ventricular function is severely affected in the KO hearts [102]. This functional disparity between fiber and whole-heart experiments may be related to altered Ca2+ handling due to secondary adaptations and functional remodeling that occurs in the heart in response to cMyBPC ablation [103]. Magnetic resonance diffusion spectrum imaging studies revealed that these differences can also be attributable to the fact that cMyBPC ablation causes substantial myoarchitectural disarray, especially in fibers located in the mid-myocardial and sub-endocardial regions [104]. In addition, cardiac magnetic resonance imaging modality revealed that cMyBPC’s absence severely compromises in vivo mechanical function by significantly depressing the rates and magnitudes of LV wall strain and torsion [101]. Thus, differences between fiber effects and in vivo function in KO mice likely arise from compromised cardiac mechanics [101], modified LV architecture [104], altered EC coupling, and structural rearrangements like myocyte disarray and fibrosis, which are consistent with cellular hypertrophy [102] at the whole-heart level –changes that are difficult to replicate in fiber settings. Therefore, a certain level of caution should be exerted when drug effects in skinned fibers to whole heart function.
5. Whole Heart and Animal Model Techniques
The intact heart is composed of cardiac myocytes that are electrically coupled by gap junctions. This unique arrangement allows for the efficient and rapid spread of action potentials that trigger synchronous contraction during each heartbeat, a mechanism that is absent in skinned cardiac preparations. Thus, in conjunction with all the techniques reviewed above, it is essential to test drug candidates in animal models before planning human clinical trials because animal models can capture major aspects of the pathophysiology seen clinically. Animal models are indispensable tools in identifying drug effects at the whole-organ or whole-animal level and determining the chronic impacts of therapy on disease progression.
There are several available tools for studying cardiovascular physiology and pharmacodynamics in animal models. These included artificially perfused isolated hearts (e.g., Langendorff model), invasive pressure-volume (PV) hemodynamic analysis, ultrasound echocardiogram, and cardiac magnetic resonance (CMR) imaging. One advantage of perfused isolated heart models is the ability to easily manipulate perfusion parameters such as coronary flow rate/pressure, preload, afterload while isolating the heart from the effects of animal stress and sympatho-adrenergic activation [105]. One can also easily study the electrical properties of the heart by direct surface electrical conductance recording, voltage-sensitive dye optical mapping, and surface electrode stimulation/pacing. These advantages make the perfused isolated heart an important tool in assessing the effects of pharmacologic intervention on cardiac electrophysiology and EC coupling.
There are some important disadvantages of the perfused isolated heart model to consider. This model most frequently uses the murine heart and technical expertise to quickly isolate and prepare hearts for reproducible experiments [106]. Furthermore, classical Langendorff models rely on retrograde flow to perfuse the coronary arteries but do not provide LV inflow, which hinders the assessment of contractility parameters under the most physiological conditions. The modified “ejecting heart” model addresses this by adding a cannulation into the left atrium (LA), providing inflow, and thereby giving the LV physiologic preload and afterload [107]. However, this complicated setup is technically challenging and not frequently used. Overall, the perfused isolated heart offers a flexible platform capable of obtaining electrical and mechanical data in the study of physiology and acute pharmacologic response in one experiment, bridging in vitro and in vivo assays in cardiovascular research.
By comparison, whole-heart in vivo studies using invasive PV catheter hemodynamic measurement is the gold standard in monitoring ventricular contractile mechanics and energetics. PV loop analysis is especially useful in characterizing end-systolic PV relationships (ESPVR) and end-diastolic PV relationships (EDPVR), which are surrogates for LV contractility and LV refill compliance, respectively. Additionally, PV loops can also be used to estimate stroke work (SW) by taking the total area of the PV loop, which is calculated clinically as a product of stroke volume and mean arterial pressure. The SW can thus be used as a correlate to examine myocardial energetics due to changes in XB cycling. The product of SW and heart rate (HR) is the total power output of the heart, which can be used as the readout for the effects of different pharmacologic interventions [108]. The major disadvantages of invasive PV loop catheterization are the need for surgical expertise to obtain consistent catheter placement directly in the LV chamber, and carefully calibrated instruments are required to obtain reproducible pressure and volume measurements. Experiments in small animal models are also often limited to single time-points.
Non-invasive imaging tools are indispensable in pre-clinical studies assessing changes in myocardial contractility over multiple time points and have the highest potential for functional comparisons with contemporary clinical imaging modalities. Both CMR and speckle-tracking echocardiography (STE) based imaging analysis have seen significant advancements in the past decade in preclinical studies. Traditionally, CMR has been used to assess ventricular ejection fraction and measure global function. Recent advancements now allow the study of localized deformation or strain as well. In addition, CMR is a versatile technique that can be combined with other techniques to be used for ex vivo pig heart perfusion [109], non-invasive quantification of PV relationships [110], and functional imaging using positron emission tomography [111].
2D and 3D STE are other promising methods that can provide important insights into both acute and chronic effects of pharmacologic interventions during preclinical screening [112]. STE has been shown to be more sensitive in detecting subclinical disease states in the absence of ventricular hypertrophy [113]. Furthermore, regional strain analysis has been especially useful in the study of regional contractile inhomogeneity and chamber geometry in the presence of ischemic disease, valvular diseases, and cardiomyopathies. Using strain to understand the pathophysiology of these diseases and the effects of interventions has helped identify early patterns of subclinical disease that can predict long term cardiac function [114]. It is important to also consider the need to optimize each of these techniques further. A key limitation in the use of mouse models in CMR and STE preclinical studies is the need for high frame rate imaging, due to high basal HR that limits the temporal resolution of each contractile cycle. Comparably, 2D STE offers higher frame rates than CMR but is limited by the quality of the acoustic window. The higher relative cost and lower availability of small animal CMR are additional factors that limit its widespread use. Similar to CMR, 3D STE offers the capability to study complex rotational mechanics. However, CMR is limited by low temporal and spatial resolution. Lastly, there are significant differences in vendor software that hinder the establishment of standardized methods of functional and disease characterization.
In vivo animal models provide the opportunity to model human disease across the lifespan of the organism. For instance, the cMyBPC−/− mouse model of hypertrophic cardiomyopathy (HCM) demonstrates characteristic features of systolic and diastolic dysfunction, which closely resemble the clinical presentation of late-stage HCM patients [102,115,116]. Hundreds of other sarcomeric mutations have been identified in humans that cause cardiomyopathies, and many of these mutant genes have been incorporated into animal models in order to understand the molecular derangements driving the disease processes [117]. In particular, rodent models have been widely used to recapitulate human diseases as a means of studying major forms of dilated, hypertrophic, and arrhythmogenic cardiomyopathies [118]. Preclinical studies have also involved a diverse array of small and large animal species in modeling human disease conditions. While the advantages and disadvantages of various animal models are beyond the scope of the present review, they have been reviewed elsewhere [119,120] while we highlight the major roles of different models in the drug validation process.
Small animal models are the mainstay of in vivo studies; they provide a highly accessible and cost-effective platform for studying drug effects at the whole organ and organism levels. Early studies of mouse models of cardiomyopathies demonstrated the cardioprotective benefits of restoring homeostatic cardiomyocyte contractile function [121–123]. These studies relied on a purely transgenic approach, where mice expressing one pathologic mutation are crossed with mice expressing a mutation of disparate effect. Normalized heart function, as reflected by improved LV function and cardiac morphology, can thus be achieved when the net effect of the double mutant normalizes cardiomyocyte contractility [123–125]. These studies in small animal models have provided a proof of concept for the efficacy of contractility modulation. As mentioned above, novel therapies are being developed in the form of the new classes of pharmacologic sarcomeric contractility modulators. We previously found differential effects of OM in vitro that was dependent on the presence or absence of cMyBPC [61]. This is clinically relevant since MYBPC3 mutations are the most common cause of inherited HCM, with the majority of these being nonsense mutations leading to haploinsufficiency of cMyBPC protein expression [126,127]. Thus it is prudent to test whether the differential effect of OM observed in skinned fiber experiments translate to functional differences at whole-heart level using animal models that express reductions in cMyBPC in the heart. Such testing would help reinforce a personalized medicine approach in dose optimization in patients with truncating MYBPC3 mutations that reduce the cMyBPC content.
For all their benefits, small animal models have significant limitations in terms of physiological similarity to humans. These may include important differences in electrophysiology, HR, metabolism, energetic reserve, vasculature, disease progression, etc. For example, unlike humans, mice lack native collateral cardiac circulation. However, following an ischemic injury to the heart, mice develop collateral cardiac circulation, the extent of which is genetically influenced [128]. This may be an important consideration when using small animal models of HF.
Large animal models of HF overcome many of these physiological limitations. For drug studies, porcine and canine models are common choices because of their close similarities with human physiology in terms of HR, contractility, protein isoform expression, cardiomyocyte progenitor population, and vascular anatomy [119]. These key factors highlight the need to further validate the results of drug studies in small animal models. For example, pigs have been routinely utilized in studies of ischemic and non-ischemic HF models [129,130]. One drug study of the myosin activator OM in a pig model of HF found that in addition to improving cardiac contractility, OM significantly increased cardiac energetic demand under load [131]. Such studies rely heavily on the cardiovascular similarities of these large animal models to best approximate a human response to pharmacological intervention in the context of energetically compromised myocardium. While large animal models provide one of the best available platforms to mimic human disease, simulating inherited diseases to study long term effects of human diseases can be technically and logistically challenging [132]. Because large animals requires more test compounds to be synthesized based on the animal’s weight, this can also pose a practical limitation in preclinical research.
While each type of preclinical animal model comes with limitations, utilizing these models is undoubtedly a vital step in identifying the efficacy and mechanism of action of novel cardiac sarcomeric drugs. Although it is not possible to perfectly mimic the chronic and multifactorial nature of human cardiovascular disease in animals, advancements in genomics and techniques to study whole-heart function allow for a better understanding of the pathophysiology of cardiac diseases. These models are also suitable testbeds for determining the subset of patients that could receive maximal benefit from drug-mediated cardiomyocyte contractility modulation.
6. Conclusion
In the sections above, we describe some of the various preclinical techniques available to better support clinical trials. The bottom-up approach that we suggest addresses the challenges posed by the complex pathophysiology of HF by first considering drug effects in simple but less physiologically relevant systems to elucidate later results in more complex and realistic settings. Of course, there remain logistical challenges in implementing a bottom-up approach: science rarely follows such idealized trajectories. However, by understanding the advantages and limitations of various preclinical techniques, basic scientists, clinicians, and drug companies can collaborate to better prepare for cardiovascular clinical trials.
7. Expert Opinion
In the sections above, we have proposed a bottom-up approach to preclinical cardiovascular studies as a means to account for the complex physiology underlying heart function and HF. The bottom-up approach would also foster hypothesis-driven drug discovery where novel therapies are designed to alter specific physiological parameters known to play functional roles in disease. While the idea of hypothesis-driven drug discovery has been discussed before some in the contexts of single-molecule and fiber studies, a larger discussion between clinicians and basic scientists will be required to extend these concepts to encompass a wider gamut of preclinical techniques [47,50,87]. An important part of the discussion revolves around the current clinical definitions of HF. Currently, HF is clinically characterized by global heart function parameters that gloss over the specific molecular etiology [3,133]. While conventional cardiovascular therapies that target downstream mechanisms may be agnostic to the varying molecular origins of HF, precision therapies targeting the sarcomere are not - similar to how cancer treatment is not general [9,134]. More precise HF definitions may also be needed to define better target subpopulations of HF patients that are most likely to benefit from a particular therapy [3,9]. Continuing advances in the accessibility of genomic sequencing and further development and deployment of highly specific, minimally-invasive phenotyping methods will help in this regard [135]. From the basic scientist’s perspective, more specific HF definitions would also help guide what biological systems and mechanisms researchers study, thus maximizing the translatability and clinical relevance of preclinical studies.
Perhaps a larger obstacle facing the adoption of hypothesis-driven drug discovery involves the current culture surrounding cardiovascular drug discovery and clinical trials. As others have identified, drug companies often choose to pursue a drug target based on its drugability and commercial potential rather than its hypothesized role in a disease mechanism [136]. Additionally, drug searches frequently rely on high throughput screenings of massive libraries of random small molecules to find a drug candidate that is then refined [5]. While the previous successes of such a shotgun approach have been well documented, its limitations as a general paradigm are becoming more apparent [4,5]. The shotgun paradigm may also be partly responsible for the high failure rate of cardiovascular clinical trials. As discussed previously, high throughput assays for sarcomere targeting drugs, most commonly based on the ATPase assay, struggle to faithfully capture many crucial aspects of the sarcomere and whole heart function but are used because they are inexpensive and scalable. The financial incentives to be first to market worsen the problem. From the perspective of the pharma industry, only one HF drug needs to succeed to justify the R&D investment due to the over 25 million global cardiovascular patients spending over 100 billion USD on treatments annually [2,137] As a result of the potential payout, drug candidates are rushed to clinical trials once identified, often forgoing detailed preclinical studies at the fiber and whole heart level that require longer time periods to complete properly [138].
Consequently, one cannot easily learn from the failures of clinical trials. Without the mechanistic granularity afforded by preclinical studies, researchers and clinicians cannot properly analyze the physiological reasons leading to a trial’s outcome [139]. Thus, the same costly mistakes are bound to repeat. The bottom-up approach to preclinical trials that we have proposed would greatly help in this respect as well. By first studying the drug effects at the atomic, then molecular, fiber, and whole organ level, preclinical studies can construct a solid foundation of mechanistic insights that drug companies can use to intelligently pick their drug targets. In this way, we can leverage hypothesis-driven drug-discovery techniques to reduce the number of clinical trials launched while maximizing the success rate.
Article Highlights.
Cardiovascular preclinical studies must often trade between mechanistic granularity and physiological fidelity.
Computer atomic models and techniques allow for detailed analysis of ligand interactions and can assist with efficient structure-based drug optimization but are limited by computational complexity and approximations.
Solution-based and single-molecule techniques characterize sarcomeric drug effects at the molecular level but struggle to capture drug-induced changes in ensemble behaviors and other important sarcomeric mechanisms.
Muscle fiber-based experiments can test drug candidates on functional sarcomeres but are difficult to scale and do not consider the heart’s gross architecture and dynamic regulation.
In vivo animal studies offer a complete picture of whole heart function but can pose considerable logistical challenges and suffer key physiological differences from humans.
Acknowledgments
Funding:
This work was supported by the National Institutes of Health (NIH) and National Heart, Lung, and Blood Institute (NHLBI) under grant 1R01HL146676-01.
Footnotes
Declaration of Interest:
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures:
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose
References
- [1].Desai AS, Stevenson LW. Rehospitalization for Heart Failure. Circulation. 2012;126:501–506. [DOI] [PubMed] [Google Scholar]
- [2].Cook C, Cole G, Asaria P, et al. The annual global economic burden of heart failure. Int. J. Cardiol 2014;171:368–376. [DOI] [PubMed] [Google Scholar]
- [3].Ahmad T, Miller PE, McCullough M, et al. Why has positive inotropy failed in chronic heart failure? Lessons from prior inotrope trials. Eur. J. Heart Fail 2019;21:1064–1078.** Discusses the challenges faced by clinical trials of inotropic cardiac drugs and serves as the main motivation for this review.
- [4].Hay M, Thomas DW, Craighead JL, et al. Clinical development success rates for investigational drugs. Nat. Biotechnol 2014;32:40–51. [DOI] [PubMed] [Google Scholar]
- [5].Harrer S, Shah P, Antony B, et al. Artificial Intelligence for Clinical Trial Design. Trends Pharmacol. Sci 2019;40:577–591. [DOI] [PubMed] [Google Scholar]
- [6].Berndt ER, Cockburn IM. Price Indexes for Clinica Trial Research: A Feasibility Study. 2013. [Google Scholar]
- [7].Gordon AM, Homsher E, Regnier M. Regulation of Contraction in Striated Muscle. Physiol. Rev 2000;80:853–924. [DOI] [PubMed] [Google Scholar]
- [8].Maack C, Eschenhagen T, Hamdani N, et al. Treatments targeting inotropy. Eur. Heart J 2019;40:3626–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].MacRae CA. Closing the ‘phenotype gap’ in precision medicine: improving what we measure to understand complex disease mechanisms. Mamm. Genome 2019;30:201–211. [DOI] [PubMed] [Google Scholar]
- [10].Usaj MM, Styles EB, Verster AJ, et al. High-Content Screening for Quantitative Cell Biology. Trends Cell Biol. 2016;26:598–611. [DOI] [PubMed] [Google Scholar]
- [11].Szymański P, Markowicz M, Mikiciuk-Olasik E. Adaptation of High-Throughput Screening in Drug Discovery—Toxicological Screening Tests. Int. J. Mol. Sci 2011;13:427–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Haasen D, Schopfer U, Antczak C, et al. How Phenotypic Screening Influenced Drug Discovery: Lessons from Five Years of Practice. Assay Drug Dev. Technol 2017;15:239–246. [DOI] [PubMed] [Google Scholar]
- [13].Park CY, Zhou EH, Tambe D, et al. High-throughput screening for modulators of cellular contractile force. Integr. Biol 2015;7:1318–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hwang H, Barnes DE, Matsunaga Y, et al. Muscle contraction phenotypic analysis enabled by optogenetics reveals functional relationships of sarcomere components in Caenorhabditis elegans. Sci. Rep 2016;6:19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Young J, Margaron Y, Fernandes M, et al. MyoScreen, a High-Throughput Phenotypic Screening Platform Enabling Muscle Drug Discovery. SLAS Discov. Adv. Life Sci. R&D 2018;23:790–806. [DOI] [PubMed] [Google Scholar]
- [16].Guhathakurta P, Prochniewicz E, Grant BD, et al. High-throughput screen, using time-resolved FRET, yields actin-binding compounds that modulate actin–myosin structure and function. J. Biol. Chem 2018;293:12288–12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reid BG, Stratton MS, Bowers S, et al. Discovery of novel small molecule inhibitors of cardiac hypertrophy using high throughput, high content imaging. J. Mol. Cell. Cardiol 2016;97:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].del Álamo JC, Lemons D, Serrano R, et al. High throughput physiological screening of iPSC-derived cardiomyocytes for drug development. Biochim. Biophys. Acta - Mol. Cell Res 2016;1863:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee J, Razu ME, Wang X, et al. Biomimetic Cardiac Microsystems for Pathophysiological Studies and Drug Screens. J. Lab. Autom 2015;20:96–106. [DOI] [PubMed] [Google Scholar]
- [20].Blundell TL, Jhoti H, Abell C. High-throughput crystallography for lead discovery in drug design. Nat. Rev. Drug Discov 2002;1:45–54. [DOI] [PubMed] [Google Scholar]
- [21].Myers S, Baker A. Drug discovery—an operating model for a new era. Nat. Biotechnol 2001;19:727–730. [DOI] [PubMed] [Google Scholar]
- [22].DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J. Health Econ 2003;22:151–185. [DOI] [PubMed] [Google Scholar]
- [23].MacRae CA, Roden DM, Loscalzo J. The Future of Cardiovascular Therapeutics. Circulation. 2016;133:2610–2617. [DOI] [PubMed] [Google Scholar]
- [24].Caraus I, Alsuwailem AA, Nadon R, et al. Detecting and overcoming systematic bias in high-throughput screening technologies: a comprehensive review of practical issues and methodological solutions. Brief. Bioinform 2015;16:974–986. [DOI] [PubMed] [Google Scholar]
- [25].Dar KB, Bhat AH, Amin S, et al. Modern Computational Strategies for Designing Drugs to Curb Human Diseases: A Prospect. Curr. Top. Med. Chem 2018;18:2702–2719. [DOI] [PubMed] [Google Scholar]
- [26].Toepfer CN, Sharma A, Cicconet M, et al. SarcTrack. Circ. Res 2019;124:1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bajorath J Integration of virtual and high-throughput screening. Nat. Rev. Drug Discov 2002;1:882–894. [DOI] [PubMed] [Google Scholar]
- [28].Kitchen DB, Decornez H, Furr JR, et al. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov 2004;3:935–949. [DOI] [PubMed] [Google Scholar]
- [29].Planelles-Herrero VJ, Hartman JJ, Robert-Paganin J, et al. Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun 2017;8:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem 2009;30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dewan S, McCabe KJ, Regnier M, et al. Insights and Challenges of Multi-Scale Modeling of Sarcomere Mechanics in cTn and Tm DCM Mutants—Genotype to Cellular Phenotype. Front. Physiol 2017;8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Campbell KS, Yengo CM, Lee L-C, et al. Closing the therapeutic loop. Arch. Biochem. Biophys 2019;663:129–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Campbell SG, McCulloch AD. Multi-scale computational models of familial hypertrophic cardiomyopathy: genotype to phenotype. J. R. Soc. Interface 2011;8:1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Szatkowski L, Lynn ML, Holeman T, et al. Proof of Principle that Molecular Modeling Followed by a Biophysical Experiment Can Develop Small Molecules that Restore Function to the Cardiac Thin Filament in the Presence of Cardiomyopathic Mutations. ACS Omega. 2019;4:6492–6501.* A key study that combines biophysical experiments, computational modeling, and simulations to identify drug effects in cardiac disease.
- [35].Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov 2005;4:649–663. [DOI] [PubMed] [Google Scholar]
- [36].Kirchmair J, Williamson MJ, Tyzack JD, et al. Computational Prediction of Metabolism: Sites, Products, SAR, P450 Enzyme Dynamics, and Mechanisms. J. Chem. Inf. Model 2012;52:617–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sorsa T, Pollesello P, Solaro RJ. The contractile apparatus as a target for drugs against heart failure: Interaction of levosimendan, a calcium sensitiser, with cardiac troponin c. Mol. Cell. Biochem 2004;266:87–107. [DOI] [PubMed] [Google Scholar]
- [38].Lounnas V, Ritschel T, Kelder J, et al. Current progress in Structure-Based Rational Drug Design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J 2013;5:e201302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kairys V, Baranauskiene L, Kazlauskiene M, et al. Binding affinity in drug design: experimental and computational techniques. Expert Opin. Drug Discov 2019;14:755–768. [DOI] [PubMed] [Google Scholar]
- [40].Sliwoski G, Kothiwale S, Meiler J, et al. Computational methods in drug discovery. Pharmacol. Rev 2014;66:334–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Niederer SA, Lumens J, Trayanova NA. Computational models in cardiology. Nat. Rev. Cardiol 2019;16:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Williams MR, Lehman SJ, Tardiff JC, et al. Atomic resolution probe for allostery in the regulatory thin filament. Proc. Natl. Acad. Sci 2016;113:3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gräter F, Shen J, Jiang H, et al. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J 2005;88:790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yuan Y, Bai X, Luo C, et al. The virtual heart as a platform for screening drug cardiotoxicity. Br. J. Pharmacol 2015;172:5531–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chan HCS, Shan H, Dahoun T, et al. Advancing Drug Discovery via Artificial Intelligence. Trends Pharmacol. Sci 2019;40:592–604. [DOI] [PubMed] [Google Scholar]
- [46].Mamoshina P, Vieira A, Putin E, et al. Applications of Deep Learning in Biomedicine. Mol. Pharm 2016;13:1445–1454. [DOI] [PubMed] [Google Scholar]
- [47].Liu C, Kawana M, Song D, et al. Controlling load-dependent kinetics of β-cardiac myosin at the single-molecule level. Nat. Struct. Mol. Biol 2018;25:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tardiff JC. Thin filament mutations: Developing an integrative approach to a complex disorder. Circ. Res 2011;108:765–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sommese RF, Sung J, Nag S, et al. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human -cardiac myosin motor function. Proc. Natl. Acad. Sci 2013;110:12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Spudich JA. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys J. 2014;106:1236–1249.* A detailed review of single-molecule techniques and how they can be used to study inotropic cardiac drug interactions with myosin.
- [51].Scapin G Structural Biology and Drug Discovery. Curr. Pharm. Des 2006;12:2087–2097. [DOI] [PubMed] [Google Scholar]
- [52].Ghosh AK, Gemma S. Structure-based Design of Drugs and Other Bioactive Molecules: Tools and Strategies. Struct. Des. Drugs Other Bioact. Mol. Tools Strateg. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2015. [Google Scholar]
- [53].Malik FI, Hartman JJ, Elias KA, et al. Cardiac Myosin Activation: A Potential Therapeutic Approach for Systolic Heart Failure. Science. 2011;331:1439–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].De La Cruz EM, Michael Ostap E. Chapter 6 Kinetic and Equilibrium Analysis of the Myosin ATPase 1st ed. Methods Enzymol. Elsevier Inc.; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Morgan BP, Muci A, Lu PP, et al. Discovery of omecamtiv mecarbil the first, selective, small molecule activator of cardiac myosin. ACS Med. Chem. Lett 2010;1:472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Green EM, Wakimoto H, Anderson RL, et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kovács M, Tóth J, Hetényi C, et al. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem 2004;279:35557–35563. [DOI] [PubMed] [Google Scholar]
- [58].Herrmann C, Travers F, Barman T, et al. Effect of 2, 3-Butanedione Monoxime on Myosin and Myofibrillar ATPases. An Example of an Uncompetitive Inhibitor. Biochemistry. 1992;31:12227–12232. [DOI] [PubMed] [Google Scholar]
- [59].Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci 1992;89:4884–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kawas RF, Anderson RL, Ingle SRB, et al. A small-molecule modulator of cardiac myosin acts on multiple stages of the myosin chemomechanical cycle. J. Biol. Chem 2017;292:16571–16577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mamidi R, Li J, Doh CY, et al. Impact of the Myosin Modulator Mavacamten on Force Generation and Cross‐Bridge Behavior in a Murine Model of Hypercontractility. J. Am. Heart Assoc 2018;7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Muretta JM, Rohde JA, Johnsrud DO, et al. Direct real-time detection of the structural and biochemical events in the myosin power stroke. Proc. Natl. Acad. Sci 2015;112:14272–14277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rohde JA, Thomas DD, Muretta JM. Heart failure drug changes the mechanoenzymology of the cardiac myosin powerstroke. Proc. Natl. Acad. Sci 2017;114:E1796–E1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Rohde JA, Roopnarine O, Thomas DD, et al. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc. Natl. Acad. Sci 2018;115:E7486–E7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci 1986;83:6272–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Greenberg MJ, Moore JR. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton. 2010;67:273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Capitanio M, Canepari M, Maffei M, et al. Ultrafast force-clamp spectroscopy of single molecules reveals load dependence of myosin working stroke. Nat. Methods 2012;9:1013–1019. [DOI] [PubMed] [Google Scholar]
- [68].Sung J, Nag S, Mortensen KI, et al. Harmonic force spectroscopy measures load-dependent kinetics of individual human β-cardiac myosin molecules. Nat. Commun 2015;6:7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Woody MS, Greenberg MJ, Barua B, et al. Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat. Commun 2018;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tang W, Unrath WC, Desetty R, et al. Dilated cardiomyopathy mutation in the converter domain of human cardiac myosin alters motor activity and response to omecamtiv mecarbil. J. Biol. Chem 2019;294:17314–17325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Liu Y, White HD, Belknap B, et al. Omecamtiv Mecarbil Modulates the Kinetic and Motile Properties of Porcine β-Cardiac Myosin. Biochemistry. 2015;54:1963–1975. [DOI] [PubMed] [Google Scholar]
- [72].Millman BM. The Filament Lattice of Striated Muscle. Physiol. Rev 1998;78:359–391. [DOI] [PubMed] [Google Scholar]
- [73].Tardiff JC, Carrier L, Bers DM, et al. Targets for therapy in sarcomeric cardiomyopathies. Cardiovasc. Res 2015;105:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Coulton AT, Stelzer JE. Cardiac Myosin Binding Protein C and Its Phosphorylation Regulate Multiple Steps in the Cross-Bridge Cycle of Muscle Contraction. Biochemistry. 2012;51:3292–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mamidi R, Li J, Gresham KS, et al. Cardiac myosin binding protein-C: a novel sarcomeric target for gene therapy. Pflügers Arch. - Eur. J. Physiol 2014;466:225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mamidi R, Gresham KS, Li A, et al. Molecular effects of the myosin activator omecamtiv mecarbil on contractile properties of skinned myocardium lacking cardiac myosin binding protein-C. J. Mol. Cell. Cardiol 2015;85:262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Stehle R, Tesi C. Kinetic coupling of phosphate release, force generation and rate-limiting steps in the cross-bridge cycle. J. Muscle Res. Cell Motil 2017;38:275–289. [DOI] [PubMed] [Google Scholar]
- [78].Stehle R, Iorga B. Kinetics of cardiac sarcomeric processes and rate-limiting steps in contraction and relaxation. J. Mol. Cell. Cardiol 2010;48:843–850. [DOI] [PubMed] [Google Scholar]
- [79].Caremani M, Melli L, Dolfi M, et al. The working stroke of the myosin II motor in muscle is not tightly coupled to release of orthophosphate from its active site. J. Physiol 2013;591:5187–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Walcott S, Warshaw DM, Debold EP. Mechanical Coupling between Myosin Molecules Causes Differences between Ensemble and Single-Molecule Measurements. Biophys. J 2012;103:501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Månsson A, Ušaj M, Moretto L, et al. Do Actomyosin Single-Molecule Mechanics Data Predict Mechanics of Contracting Muscle? Int. J. Mol. Sci 2018;19:1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Best PM. Cardiac muscle function: results from skinned fiber preparations. Am. J. Physiol. Circ. Physiol 1983;244:H167–H177. [DOI] [PubMed] [Google Scholar]
- [83].Mamidi R, Gresham KS, Li J, et al. Cardiac myosin binding protein-C Ser302 phosphorylation regulates cardiac b-adrenergic reserve. Sci. Adv 2017;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mamidi R, Michael JJ, Muthuchamy M, et al. Interplay between the overlapping ends of tropomyosin and the N terminus of cardiac troponin T affects tropomyosin states on actin. FASEB J. 2013;27:3848–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Konhilas JP, Irving TC, de Tombe PP. Length-dependent activation in three striated muscle types of the rat. J. Physiol 2002;544:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mamidi R, Gresham KS, Verma S, et al. Cardiac myosin binding protein-C phosphorylation modulates myofilament length-dependent activation. Front. Physiol 2016;7:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mamidi R, Li J, Doh CY, et al. Lost in translation: Interpreting cardiac muscle mechanics data in clinical practice. Arch. Biochem. Biophys 2019;662:213–218.* A review of common fiber-based experiments and assays and how they may relate to in vivo cardiac function.
- [88].Stelzer JE, Dunning SB, Moss RL. Ablation of Cardiac Myosin-Binding Protein-C Accelerates Stretch Activation in Murine Skinned Myocardium. Circ. Res. 2006;98:1212–1218. [DOI] [PubMed] [Google Scholar]
- [89].Ford SJ, Chandra M, Mamidi R, et al. Model representation of the nonlinear step response in cardiac muscle. J. Gen. Physiol 2010;136:159–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Hanft LM, Cornell TD, McDonald CA, et al. Molecule specific effects of PKA-mediated phosphorylation on rat isolated heart and cardiac myofibrillar function. Arch. Biochem. Biophys 2016;601:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Colson BA, Bekyarova T, Fitzsimons DP, et al. Radial Displacement of Myosin Cross-bridges in Mouse Myocardium due to Ablation of Myosin Binding Protein-C. J. Mol. Biol. 2007;367:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gresham KS, Mamidi R, Stelzer JE. The contribution of cardiac myosin binding protein-c Ser282 phosphorylation to the rate of force generation and in vivo cardiac contractility. J. Physiol 2014;592:3747–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mamidi R, Li J, Gresham KS, et al. Dose-Dependent Effects of the Myosin Activator Omecamtiv Mecarbil on Cross-Bridge Behavior and Force Generation in Failing Human Myocardium. Circ. Hear. Fail 2017;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Swenson AM, Tang W, Blair CA, et al. Omecamtiv Mecarbil Enhances the Duty Ratio of Human β-Cardiac Myosin Resulting in Increased Calcium Sensitivity and Slowed Force Development in Cardiac Muscle. J. Biol. Chem 2017;292:3768–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Chandra M, Mamidi R, Ford S, et al. Nebulin Alters Cross-bridge Cycling Kinetics and Increases Thin Filament Activation. J. Biol. Chem 2009;284:30889–30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mamidi R, Mallampalli SL, Wieczorek DF, et al. Identification of two new regions in the N-terminus of cardiac troponin T that have divergent effects on cardiac contractile function. J. Physiol 2013;591:1217–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hwang PM, Sykes BD. Targeting the sarcomere to correct muscle function. Nat. Rev. Drug Discov. 2015;14:313–328. [DOI] [PubMed] [Google Scholar]
- [98].Cleland JGF, Teerlink JR, Senior R, et al. The effects of the cardiac myosin activator, omecamtiv mecarbil, on cardiac function in systolic heart failure: a double-blind, placebo-controlled, crossover, dose-ranging phase 2 trial. Lancet. 2011;378:676–683. [DOI] [PubMed] [Google Scholar]
- [99].Teerlink JR, Clarke CP, Saikali KG, et al. Dose-dependent augmentation of cardiac systolic function with the selective cardiac myosin activator, omecamtiv mecarbil: a first-in-man study. Lancet. 2011;378:667–675. [DOI] [PubMed] [Google Scholar]
- [100].Stehle R, Solzin J, Iorga B, et al. Insights into the kinetics of Ca2+-regulated contraction and relaxation from myofibril studies. Pflügers Arch. - Eur. J. Physiol 2009;458:337–357. [DOI] [PubMed] [Google Scholar]
- [101].Desjardins CL, Chen Y, Coulton AT, et al. Cardiac Myosin Binding Protein C Insufficiency Leads to Early Onset of Mechanical Dysfunction. Circ. Cardiovasc. Imaging 2012;5:127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Harris SP, Bartley CR, Hacker TA, et al. Hypertrophic Cardiomyopathy in Cardiac Myosin Binding Protein-C Knockout Mice. Circ. Res. 2002;90:594–601. [DOI] [PubMed] [Google Scholar]
- [103].Tong CW, Stelzer JE, Greaser ML, et al. Acceleration of Crossbridge Kinetics by Protein Kinase A Phosphorylation of Cardiac Myosin Binding Protein C Modulates Cardiac Function. Circ. Res. 2008;103:974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wang TT, Kwon HS, Dai G, et al. Resolving Myoarchitectural Disarray in the Mouse Ventricular Wall with Diffusion Spectrum Magnetic Resonance Imaging. Ann. Biomed. Eng 2010;38:2841–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bell RM, Mocanu MM, Yellon DM. Retrograde heart perfusion: The Langendorff technique of isolated heart perfusion. J. Mol. Cell. Cardiol. Elsevier; 2011. p. 940–950. [DOI] [PubMed] [Google Scholar]
- [106].Olejnickova V, Novakova M, Provaznik I. Isolated heart models: cardiovascular system studies and technological advances. Med. Biol. Eng. Comput 2015. p. 669–678. [DOI] [PubMed] [Google Scholar]
- [107].Liao R, Podesser BK, Lim CC. The continuing evolution of the Langendorff and ejecting murine heart: New advances in cardiac phenotyping. Am. J. Physiol. - Hear. Circ. Physiol 2012. p. H156–H167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Bastos MB, Burkhoff D, Maly J, et al. Invasive left ventricle pressure–volume analysis: overview and practical clinical implications. Eur. Heart J. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Schuster A, Grünwald I, Chiribiri A, et al. An isolated perfused pig heart model for the development, validation and translation of novel cardiovascular magnetic resonance techniques. J. Cardiovasc. Magn. Reson 2010;12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Seemann F, Arvidsson P, Nordlund D, et al. Noninvasive Quantification of Pressure-Volume Loops From Brachial Pressure and Cardiovascular Magnetic Resonance. Circ. Cardiovasc. Imaging 2019;12:e008493. [DOI] [PubMed] [Google Scholar]
- [111].Lau JMC, Raptis DA, Laforest R, et al. Cardiac Positron Emission Tomography-Magnetic Resonance Imaging. J. Thorac. Imaging. 2018. p. 139–146. [DOI] [PubMed] [Google Scholar]
- [112].Satriano A, Heydari B, Guron N, et al. 3-Dimensional regional and global strain abnormalities in hypertrophic cardiomyopathy. Int. J. Cardiovasc. Imaging 2019; [DOI] [PubMed] [Google Scholar]
- [113].Nagueh SF, Bachinski LL, Meyer D, et al. Tissue Doppler imaging consistently detects myocardial abnormalities in patients with hypertrophic cardiomyopathy and provides a novel means for an early diagnosis before and independently of hypertrophy. Circulation. 2001;104:128–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Voigt J-U, Cvijic M. 2- and 3-Dimensional Myocardial Strain in Cardiac Health and Disease. JACC Cardiovasc. Imaging. 2019;12:1849–1863. [DOI] [PubMed] [Google Scholar]
- [115].Lekanne Deprez RH, Muurling-Vlietman JJ, Hruda J, et al. Two cases of severe neonatal hypertrophic cardiomyopathy caused by compound heterozygous mutations in the MYBPC3 gene. J Med Genet. 2006;43:829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Alders M. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. Eur. Heart J 2003;24:1848–1853. [DOI] [PubMed] [Google Scholar]
- [117].Tayal U, Prasad S, Cook SA. Genetics and genomics of dilated cardiomyopathy and systolic heart failure. Genome Med. 2017;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Li C-J, Chen C-S, Yiang G-T, et al. Advanced Evolution of Pathogenesis Concepts in Cardiomyopathies. J. Clin. Med. 2019;8:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Milani-Nejad N, Janssen PML. Small and large animal models in cardiac contraction research: Advantages and disadvantages. Pharmacol. Ther. 2014;141:235–249.* A comprehensive review of the advantages and disadvantages of various animal models of heart failure.
- [120].Houser SR, Margulies KB, Murphy AM, et al. Animal Models of Heart Failure. Circ. Res 2012;111:131–150. [DOI] [PubMed] [Google Scholar]
- [121].Arber S, Hunter JJ, Ross J, et al. MLP-Deficient Mice Exhibit a Disruption of Cardiac Cytoarchitectural Organization, Dilated Cardiomyopathy, and Heart Failure. Cell. 1997;88:393–403. [DOI] [PubMed] [Google Scholar]
- [122].Li Y, Charles P-YJ, Nan C, et al. Correcting diastolic dysfunction by Ca2+ desensitizing troponin in a transgenic mouse model of restrictive cardiomyopathy. J. Mol. Cell. Cardiol 2010;49:402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Li J, Gresham KS, Mamidi R, et al. Sarcomere-based genetic enhancement of systolic cardiac function in a murine model of dilated cardiomyopathy. Int J Cardiol 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Alves ML, Dias FAL, Gaffin RD, et al. Desensitization of myofilaments to Ca2+ as a therapeutic target for hypertrophic cardiomyopathy with mutations in thin filament proteins. Circ. Cardiovasc. Genet. 2014;7:132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Minamisawa S, Hoshijima M, Chu G, et al. Chronic phospholamban-sarcoplasmic reticulum calcium ATPase interaction is the critical calcium cycling defect in dilated cardiomyopathy. Cell. 1999;99:313–322. [DOI] [PubMed] [Google Scholar]
- [126].Marston S, Copeland O, Jacques A, et al. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency*. Circ. Res. 2009;105:219–222. [DOI] [PubMed] [Google Scholar]
- [127].Cheng Y, Wan X, McElfresh TA, et al. Impaired contractile function due to decreased cardiac myosin binding protein C content in the sarcomere. Am. J. Physiol. Circ. Physiol 2013;305:H52–H65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Zhang H, Faber JE. De-novo collateral formation following acute myocardial infarction: Dependence on CCR2+ bone marrow cells. J. Mol. Cell. Cardiol 2015;87:4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Watanabe S, Ishikawa K, Fish K, et al. Protein Phosphatase Inhibitor-1 Gene Therapy in a Swine Model of Nonischemic Heart Failure. J. Am. Coll. Cardio 2017;70:1744–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Suzuki Y, Lyons JK, Yeung AC, et al. In vivo porcine model of reperfused myocardial infarction: In situ double staining to measure precise infarct area/area at risk. Catheter. Cardiovasc. Interv 2008;71:100–107. [DOI] [PubMed] [Google Scholar]
- [131].Bakkehaug JP, Kildal AB, Engstad ET, et al. Myosin Activator Omecamtiv Mecarbil Increases Myocardial Oxygen Consumption and Impairs Cardiac Efficiency Mediated by Resting Myosin ATPase Activity. Circ. Hear. Fail 2015;8:766–775. [DOI] [PubMed] [Google Scholar]
- [132].Montag J, Petersen B, Flögel AK, et al. Successful knock-in of Hypertrophic Cardiomyopathy-mutation R723G into the MYH7 gene mimics HCM pathology in pigs. Sci. Rep. 2018;8:4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Parikh KS, Sharma K, Fiuzat M, et al. Heart Failure With Preserved Ejection Fraction Expert Panel Report. JACC Hear. Fail 2018;6:619–632. [DOI] [PubMed] [Google Scholar]
- [134].Solomon SD, Pfeffer MA. The Future of Clinical Trials in Cardiovascular Medicine. Circulation. 2016;133:2662–2670. [DOI] [PubMed] [Google Scholar]
- [135].Leopold JA, Loscalzo J. Emerging Role of Precision Medicine in Cardiovascular Disease. Circ. Res. 2018;122:1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].MacRae CA, Loscalzo J. The future of cardiovascular biomedicine. Circulation. 2016;133:2601–2602. [DOI] [PubMed] [Google Scholar]
- [137].Agency for Healthcare and Quality. Medical Expenditure Panel Survey. United States; 2015. [Google Scholar]
- [138].Janssen PML, Elnakish MT. Expert Opinion on Drug Discovery Modeling heart failure in animal models for novel drug discovery and development. Expert Opin. Drug Discov 2019;14:355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].O’Connor CM, Psotka MA, Fiuzat M, et al. Improving Heart Failure Therapeutics Development in the United States. J. Am. Coll. Cardiol 2018;71:443–453. [DOI] [PubMed] [Google Scholar]