Abstract
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked handicapping disease due to the loss of an essential muscle protein dystrophin. Dystrophin-null animals have been extensively used to study disease mechanisms and to develop experimental therapeutics. Despite decades of research, however, treatment options for DMD remain very limited.
Areas covered
High-throughput high-content screening and precision medicine offer exciting new opportunities. Here, the authors review animal models that are suitable for these studies.
Expert opinion
Nonmammalian models (worm, fruit fly, and zebrafish) are particularly attractive for cost-effective large-scale drug screening. Several promising lead compounds have been discovered using these models. Precision medicine for DMD aims at developing mutation-specific therapies such as exon-skipping and genome editing. To meet these needs, models with patient-like mutations have been established in different species. Models that harbor hotspot mutations are very attractive because the drugs developed in these models can bring mutation-specific therapies to a large population of patients. Humanized hDMD mice carry the entire human dystrophin gene in the mouse genome. Reagents developed in the hDMD mouse-based models are directly translatable to human patients.
Keywords: adeno-associated virus, AAV, Becker muscular dystrophy, BMD, CRISPR, DMD, dystrophin, exon-skipping, genome editing, personized therapy
1. Introduction
Duchenne muscular dystrophy (DMD) is the most common lethal muscle disease affecting boys and young men 1. DMD is caused by mutations in the dystrophin gene 2. The dystrophin gene is one of the largest genes in the genome. The full-length dystrophin gene spans ~2.4 mb at the X chromosome locus Xp21. It contains 79 exons and transcribes into a 14 kb mRNA. The 427 kD full-length dystrophin protein can be divided into four major domains including the N-terminal, rod, cysteine-rich and C-terminal domain. The rod domain contains 24 spectrin-like repeats and four hinges. In DMD patients, dystrophin expression is abolished mostly by frame-shift mutations. Becker muscular dystrophy (BMD) is a mild allelic variant of DMD 3. BMD patients carry in-frame mutations that result in the production of a truncated but partially functional dystrophin protein.
Dystrophin is located immediately underneath the sarcolemma in muscle cells. It interacts with the filamentous cytoskeleton through its N-terminal domain and connects to the extracellular matrix via the interaction between the cysteine-rich domain and transmembrane protein dystroglycan. Dystrophin also interacts with a variety of cytosolic proteins, including syntrophin, dystrobrevin and neuronal nitric oxide synthase (nNOS) via the C-terminal domain and rod domain. In addition to these direct interactions, dystrophin also associates with transmembrane protein sarcospan and sarcoglycans. Together, dystrophin and its associated proteins form the dystrophin glycoprotein complex (DGC) 4, 5. The DGC stabilizes sarcolemma during contraction. The DGC also mediates muscle cell signaling. Loss of dystrophin compromises both mechanical and signaling function. As a consequence, muscle cells undergo a cascade of pathogenic events which eventually lead to degeneration, necrosis, inflammation and replacement of muscle by fibrofatty tissues 6.
Two different strategies have been used to develop drugs for DMD. One aims at restoring dystrophin expression using genetic approaches. The other aims at ameliorating downstream pathological changes with pharmaceutical drugs. Irrespective of the approach, rigorous preclinical testing in animal models is the critical first step. Below we review animal models used in DMD drug discovery with an emphasis on models that are suitable for high throughput and high content screening and precision medicine.
2. Naturally occurring DMD models have laid the foundation for DMD drug discovery
About one-third of DMD patients have no family history 7, 8. The disease in these patients are due to spontaneous mutations. Similar to humans, naturally-occurring mutations also present in animals. The mdx mouse and golden retriever muscular dystrophy (GRMD) dog were discovered in 1984 and 1981, respectively 9, 10. Both models show elevated serum creatine kinase (CK) and muscle pathology. However, only the GRMD dog displays clinical signs similar to that of human patients. The mdx mouse and GRMD dog were later confirmed to carry spontaneous null mutations in the dystrophin gene 11, 12. In the mdx mouse, a C to T transition in exon 23 creates a nonsense mutation that stops dystrophin protein translation 11. In the GRMD dog, an A to G transition near the end of intron 6 disrupts the conserved splicing acceptor site. This results in erroneous splicing of exon 6 to exon 8 and subsequent frame shift 13.
Since mdx mice and GRMD dogs are the first confirmed dystrophin deficient animals, they become the most widely used DMD models in pre-clinical studies. Mdx mice are especially popular in research arena because they are fertile, have a near normal lifespan, easy to maintain, readily available, and importantly, affordable to most research laboratories 14. The use of mdx mice has also benefited from ample reagents, tools and outcome measurement protocols that are already developed for murine studies. Finally, research in mdx mice is promoted by the extensive knowledge on the disease natural history in this model and the establishment of the standard operating procedures to study mdx mice 15, 16. Indeed, mdx mice have been used to establish preliminary efficacy data for essentially every pharmaceutical drug that has been or is being tested in human patients, as well as many that are still in the early phase of the drug development pipeline (reviewed in 17–22). Besides drug therapy, mdx mice have also served as the testbed to establish the proof-of-principle for various gene and cell therapies that are intended to restore dystrophin expression (reviewed in 23–33). An excellent example in this regard is systemic micro-dystrophin gene therapy with vectors derived from adeno-associated virus (AAV) (reviewed in 22, 34–36).
Mild clinical presentation is a major drawback for the mdx model 37. This is largely due to the presence of various compensatory mechanisms. To overcome this hurdle, a number of double-knock out mice have been created by genetic inactivation of compensatory genes (such as utrophin, integrin, MyoD, telomerase RNA) in mdx mice (reviewed in 14, 38). Alternatively, clinically more severe mice are obtained by breeding mdx mice from their original C57Bl/10 background to other backgrounds (such as DBA/2 and Cmah) (reviewed in 14, 38, 39). Although these phenotypic mouse models resemble patients better, they have not become the mainstream model in DMD research yet. There are several reasons. Most of these models lack comprehensive natural history data. Some are difficult to establish a colony. In the case of double knock out mice, there are always concerns on data interpretation because human patients do not carry mutation in the second gene that is inactivated in mice.
In contrast to mdx mice, GRMD dogs show a clinical course much more similar to that of human patients (reviewed in 10, 14, 40). Further the body size of the dog is much larger than that of the mouse. The larger body size is extremely appealing for scaling up gene therapy and cell therapy. Despite these advantages, the GRMD dog has only been used in a limited number of studies for testing drug, gene and cell therapy (reviewed in 10, 41, 42). This is largely because of the difficulties in the generation and caring of affected dogs, the high cost-associated with the colony maintenance, the large dose of vectors needed for systemic delivery, and the lack of expertise and dedicated personnel in most laboratories. Nevertheless, studies conducted in the canine model are considered much more informative in shaping the design of clinical trials. A carefully planned and sufficiently powered study in dystrophic dogs should be encouraged at the late stage of preclinical drug development.
3. High-throughput high-content screening in nonmammalian DMD modes opens the door to novel therapeutic targets
Our understanding on disease mechanisms has constituted the foundation for traditional DMD drug discovery. With the recognition of dystrophin deficiency as the molecular culprit 2, a great deal of effort has been placed on dystrophin restoration with gene therapy. On the other side, pharmaceutic approaches for DMD have centered on downstream secondary pathogenic events such as inflammation, fibrosis, calcium dysregulation, oxidative stress, mitochondria dysfunction, ischemia, leaky sarcolemma, and muscle atrophy 19, 21. While these target-defined strategies have been very fruitful in establishing the current landscape of DMD drug discovery, there are also important limitations. For example, we still do not have a crystal-clear understanding of all the molecular and cellular changes in DMD. In a small but significant group of patients, clinical manifestations remain unexplainable by the reading-frame rule 43–50. For patients with genotype-phenotype disconnection, the lack of mechanistic insights poses an unsurmountable challenge for the traditional drug discovery approaches because there is no clear druggable target.
Current DMD drug discovery heavily depends on the mouse model. This is doable when dealing with one or two drug candidates. However, it becomes challenging or even impossible if one wants to test a broad range of doses of a particular drug or to find the most effective drug from dozens or hundreds of chemical variants. The time-consuming (often months) and labor-intensive (often multiple assays by multiple people) nature of the mouse study also eliminates the possibility of exhaustively evaluating all known drugs that belong to the same category of a lead drug.
The development of nonmammalian DMD models has opened the door for high-throughput, high-content screening of large libraries of chemical compounds with either known or unknown function (Table 1). Three most commonly used nonmammalian models are Caenorhabditis elegans (C. elegans), Drosophila melanogaster and zebrafish. These models carry many excellent features that make them highly desirable for high throughput, high content screening. These include but not limit to their easy maintenance, short life cycle, large progeny size, genetic tractability, and visible phenotype (Figure 1). The dystrophin gene is highly conserved between mammals and nonmammals 51–55. The homolog of the human dystrophin gene has been identified in chromosome 1, 3, and 1 in C. elegans, Drosophila, and zebrafish, respectively (https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=173038) 55, 56. DMD models have also been generated in C. elegans, Drosophila, and zebrafish (reviewed in 14, 57–62).
Table 1.
DMD drug discovery with nonmammalian models
| C. elegans | Drosophila | Zebrafish | ||
|---|---|---|---|---|
| Compound screened | >1000 compounds | No library screened yet | 3 candidate drugs | >4500 compounds |
| Screening Method | Phalloidin staining. Staining is continuous and uninterrupted in WT but discontinuous/disrupted in DMD. | Wing posterior cross vein. The vein is present in WT but absent in DMD. | Projectin immunostaining. Staining is strong and continuous in WT but weak and/or fragmented in DMD. | Birefringence assay. WT shows continuous birefringence but DMD shows fragmented birefringence. |
| Results | 20 candidate drugs have been identified including prednisone, serotonin, carbonic anhydrous inhibitors, cyclosporin A | THI, THI-oxime, FTY720 | 25 lead compounds have been identified including epirizole, homochlorcyclizine dihydrochloride, conessine, aminophylline, equilin, pentetic acid, proscillaridin A, ergotamine, pergolide, fluoxetine, flunarizine, ropinirole, homochlorcyclizine, pentetic acid, nitromide, propantheline bromide, androsterone acetate, crassin acetate, pomiferin, sildenafil citrate, cerulenin, 9a-11b-prostaglandin F2 | |
| References | 64 to 68 | 79 | 62,77 | 61, 100 to 102 |
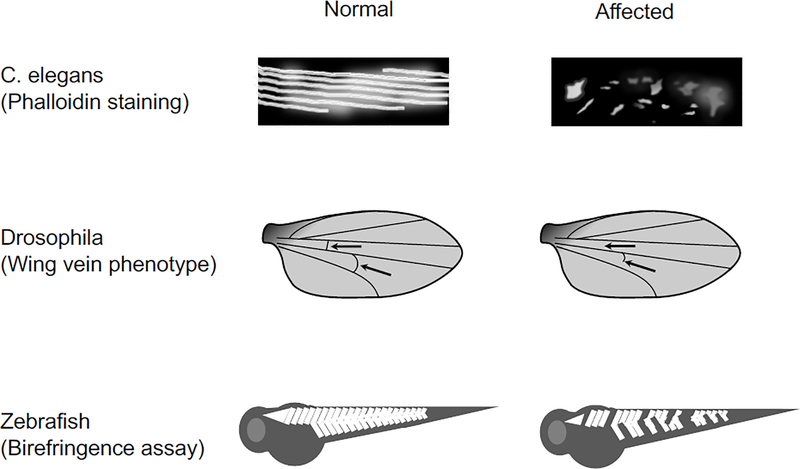
Figure 1. Methods for high-throughput phenotype screening in nonmammalian DMD models.
Top panel, Schematic cartoon illustration of phalloidin staining of the body wall muscle in normal and affected worms. In normal, phalloidin staining reveals a well-organized pattern of actin in muscle. In affected, phalloidin staining shows a patchy fragmented pattern. Middle panel, Schematic cartoon illustration of wing vein phenotype in fruit fly. Posterior cross veins are intact in a normal fly but is absent or defective in an affected fly. Arrow, location of the posterior cross vein. Bottom panel, Schematic cartoon illustration of birefringence assay in zebrafish. Normal zebrafish has a bright ordered birefringence pattern. Affected zebrafish has random pocketed region with reduced birefringence.
3.1. The C. elegans DMD model
The first C. elegans DMD model [dys-1(cx18), hlh-1(cc561ts)] was developed in 2000 in the Segalat lab 63. This model carries a null mutation in the dystrophin gene [dys-1(cx18)] and a thermosensitive mutation in the MyoD gene [hlh-1(cc561ts)] 63. The double mutant worms show a time-dependent locomotion impairment and muscle degeneration. Mobility change can be visualized by eyeballing. Muscle degeneration can be examined with phalloidin staining (Figure 1). Using these assays, the Segalat lab blindly screened ~1,000 approved drugs 64–68. They identified more than 20 candidate drugs that are capable of blocking muscle degeneration and/or improving mobility. Interestingly, these drugs not only include the ones that were known previously (such as prednisone, cyclosporin A and calcium antagonist Nifedipine) 64, 67, 68, but also include many new drugs such as antidepressants (serotonin and tricyclic compounds) 65, 67 and carbonic anhydrase inhibitors (methazolamide and dichlorphenamide) (Table 1) 66. Subsequent validation studies in mdx mice confirmed muscle protection effect of antidepressants and carbonic anhydrase inhibitors 66, 67.
Recently, Hewitt et al reported a new worm DMD model only deficient for dystrophin [dys-1(cx33)] 69. The dys-1(cx33) mutant carries a different dystrophin null mutation and shows a more severe phenotype than the dys-1(cx18) mutant. Using an automatic force measurement system called NemaFlex, the authors found strength reduction in the dys-1(cx33), but not dys-1(cx18) mutant. The author further validated this new model as a drug screening platform using drugs that are known to improve muscle force in the DMD model (prednisone and melatonin). In conclusion, the authors suggest that future drug screening should use the dys-1(cx33) model 69.
3.2. The Drosophila DMD model
A large collection of Drosophila DMD models have been developed (reviewed in 59, 70–73). According to the FlyBase website (https://flybase.org/reports/FBhh0000191.html#do_annotation_sub), at least 17 knockdown models have been generated using RNA interference 74, 75. Besides the knockdown model, at least 7 genetic loss-of-function mutants are available including Dys8−2, DysE17, Dysdet−1, DysEP3397, DysDf, Dyskx43, and DysExel618474, 76, 77. The fly DMD models show progressive climbing deficits and muscle degeneration. Interestingly, dystrophin plays an important role in wing vein development in Drosophila 78. As a result, morphological changes in the wing vein can be used as a visible phenotype for screening (Figure 1) 79, 80.
Dilated cardiomyopathy is a major health threat in DMD patients 81. Two compound mutants Dyskx43/DysExel6184 and Dyskx43/Dys8−2 display cardiac dysfunction resembling dilated cardiomyopathy 76. Surprisingly, heart disease in the fly DMD model can be rescued by Dp116, a naturally occurring truncated dystrophin isoform containing only spectrin-like repeats 22, 23, 24, hinge 4, the cysteine-rich domain and the C-terminal domain. Considering the fact that Dp116 worsens skeletal muscle disease in dystrophin-null mdx4cv mice 82, cautions should be taken when using Dyskx43/DysExel6184 and Dyskx43/Dys8−2 as a DMD cardiomyopathy model.
Although the fly DMD model has been proposed for large-scale, high-throughput small molecular screening (Table 1) 62, 70, no blind screening has been conducted using a large chemical compound library yet. Nevertheless, using a candidate drug approach, Pantoja et al found that drugs that can increase the level or signaling of sphingosine 1-phosphate, including 2-acetyl-4(5)-tetrahydroxybutyl imidazole (THI), THI-oxime (a derivative of THI), and FTY720 (gilenya), can suppress dystrophic phenotypes in several fly DMD models 77. A subsequent study in mdx mice validated findings obtained from the fly model 83.
3.3. The zebrafish DMD model
The zebrafish model is the most frequently used DMD model for high-throughput, high-content drug screening (reviewed in 60, 61, 84, 85). Similar to the fly model, both knockdown and genetic null DMD models are available in zebrafish. In the knockdown model, dystrophin expression was reduced by inhibiting mRNA translation with a morpholino oligomer 86. A total of five dystrophin null zebrafish DMD models have been identified in the genetic screen following ethylnitrosourea (ENU) mutagenesis 87–90. These include sapje (dmdta222a), sapje-like (sapcl100), dmdpc1, dmd pc2, and dmdtm90c87, 88, 91. Four of these contains nonsense mutation in exons 4 (sapje), 21 (dmdpc1), 32 (dmd pc2) and 53 (dmdtm90c), respectively 88, 91. The sapje-like model carries a mutation in the donor splice junction of exon 62 87.
Dystrophin deficient zebrafish share many characteristic features of human patients. They die prematurely before reaching the reproductive age. Their muscles not only show histological lesions (degeneration/regeneration, necrosis, inflammation and fibrosis) but also are highly susceptible to mechanical strain 92. Further, both absolute and specific muscle force are reduced 92, 93. Importantly, the extent of force deficit is similar to what was reported in GRMD dogs 92.
Due to premature death, the zebrafish model is maintained as heterozygous. Dystrophin-null zebrafish are generated by crossing heterozygous animals. Zebrafish do not have defined X and Y chromosomes 94. Hence, 25% of the offspring will be dystrophin deficient due to recessive inherence of the mutated dystrophin gene. Dystrophic signs can be readily identified using a birefringence assay at 3 to 4 days post-fertilization in affected zebrafishes (Figure 1). The birefringence assay is a sensitive, inexpensive noninvasive assay to quickly evaluate muscle integrity in translucent zebrafish larvae 95, 96. Normal larval zebrafish show a bright, highly ordered array of birefringence under polarized light. In contrast, affected larvae display reduced birefringence in areas corresponding to damaged muscle (Figure 1). The recently developed automated system and software have further streamline the assay for screening chemically treated larvae in 96 and even 384-well plates 97–99. Besides the birefringence assay, the dystrophic phenotype can also be evaluated by quantifying survival, muscle force, and the touch-evoked escape response 96.
High throughput, high content small molecule screening in the zebrafish DMD model has greatly benefited from the birefringence assay (Figure 1). To date, at least five chemical libraries have been screened in dystrophic zebrafish 61. These include Prestwick collection 1 (1,280 chemicals), Prestwick collection 2 (1,120 chemicals), NINDS 2 compound library (1,040 chemicals), ICCB Known Bioactives 2012 (480 chemicals), and FDA Approved Drugs (640 chemicals) (Table 1) 61. These screens have yielded at least 25 lead compounds 100–102. The details of these 25 compounds have been reported elsewhere and will not be discussed here 102.
4. Developing personalized therapy using hotspot mutation mammalian models
More than 7,000 mutations have been reported in DMD patients 46, 103, 104. Among these, ~68% are large deletions (≥ 1 exon), ~11% are large duplications (≥ 1 exon), and ~20% are small mutations (small deletions, small duplications, splice site mutations, nonsense and missense mutations) 103. About 67 to 75% of all deletion mutations occur between exons 45–55 50, 105. This region is termed the major mutation hotspot (Figure 2A). Approximately 20% of all deletion mutations are found between exons 2 to 20 105. This region is termed the minor mutation hotspot (Figure 2B). Duplication mutations are clustered between exons 2 to 20 45, 46, 103.
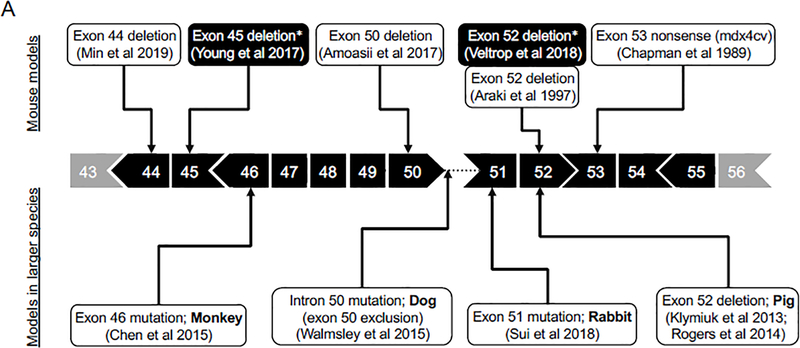
Figure 2. DMD animal models with mutations in regions of exon 45–55 and exon 2–20.
In human patients, about two-third of deletion mutations occur between exons 45 and 55 (the major mutation hotspot) and about one-fifth of deletion mutations occur between exons 2 and 20 (the minor mutation hotspot). A variety of animal models with mutations found in the regions corresponding to the major and minor hotspots of the human DMD gene have been reported. A, Models with mutations in the major mutation hotspot region. B, Models with mutations in the minor mutation hotspot region.
The great mutation spectrum creates a significant challenge for sequence-specific therapies, such as exon-skipping and genome editing. It is unrealistic to make a model for each mutation in light of the huge number of mutations found in patients (several thousands) 46, 47, 103. However, if models can be generated to harbor mutations in hotspot regions, majority of patients will benefit from sequence-specific drugs developed in these models. For example, 13% patients can benefit from CRISPR-mediated exon 51 deletion or antisense oligonucleotide (AON)-mediated exon 51 skipping 103, 106–108. Removing or skipping exons 45, 53, 44, 46, 52, 50, and 43 can benefit 8.1%, 7.7%, 6.2%, 4.3%, 4.1%, 4.0% and 3.8% DMD patients, respectively 103, 106–108. Patients with large in-frame deletion of exons 45–55 often show very mild clinical disease 106, 109, 110. About two-third of DMD patients can benefit from a variant dystrophin protein that lacks exons 45 to 55 103, 106–108 (Figure 3A).
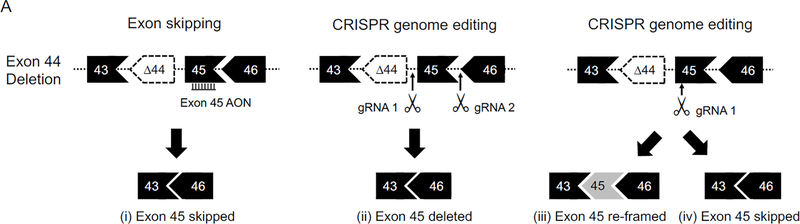
Figure 3. Strategies for treating hotspot mutation with personalized medicine.
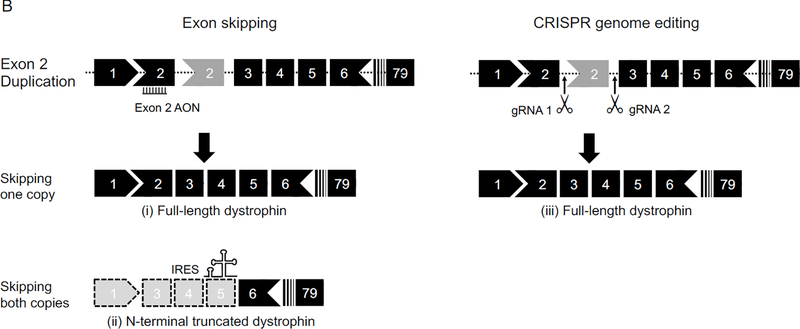
A, Exon 44 deletion can be treated with (i) exon 45 skipping using antisense oligonucleotides, (ii) exon 45 deletion using CRISPR, (iii) exon 45 re-framing using CRISPR, and (iv) exon 45 skipping using CRISPR. B, Exon 2 duplication can be treated with (i) skipping one copy of exon 2 using antisense oligonucleotides, (ii) skipping both copies of exon 2 using antisense oligonucleotides to induce IRES-mediated translation of a N-terminal partially truncated dystrophin, and (iii) deleting one copy of exon 2 using CRISPR. AON, antisense oligonucleotides; gRNA, guide RNA; IRES, internal ribosome entry site.
4.1. Major hotspot mutation mouse models
The major hotspot is located between exons 45 to 55 (Figure 2A) 50, 105. The mdx4cv mouse is the first model harboring a mutation in the major hotspot. This mouse is generated by ENU mutagenesis in 1989 and it carries a nonsense mutation in exon 53 111. This model has been used to test CRISPR-mediated deletion of exons 52 and 53 from the genome 112, or AON-mediated skipping of exons 52 and 53 from the transcript 113. In both cases, the reading frame was restored. The next major hotspot model is the mdx52 mouse. This model was created by targeted deletion of exon 52 using traditional gene knockout technique 114. In this model, not only the full-length dystrophin protein (Dp427) but also two smaller isoforms (Dp260 and Dp140) are eliminated 114. The mdx52 mouse has been used to demonstrate the proof-of-principle for exon 51 skipping 115 and exon 45 to 55 multi-exon skipping 116–118.
Two more major hotspot mouse models, the ΔEx44 and ΔEx50 model, were developed using the CRISPR technology recently 119, 120. In ΔEx44 and ΔEx50 mice, dystrophin expression is aborted by exon 44 deletion and exon 50 deletion, respectively 119, 120. These two models have been used to test CRISPR-mediated gene repair therapy 119, 120. Amoasii et al designed a single guider RNA (gRNA) to target an exonic splicing enhancer (ESE) near the 5’-end of exon 51. They then delivered the gRNA and SpCas9 to ΔEx50 mice using AAV vectors. SpCas9 created a double strand break at the ESE of exon 51. Subsequent repair by nonhomologous end joining (NHEJ) should disrupt the ESE and prevent inclusion of exon 51 in the transcript. Interestingly, only ~10% of the transcript showed the expected exon 51 skipping. On the other hand, ~70% of the transcript were re-framed due to insertion of an adenosine at the double strand break site in exon 51 119. A similar strategy was tested in the ΔEx44 model by Min et al 120. This time, the authors co-delivered SpCa9 and a single gRNA targeting exon 45 with AAV vectors. They observed exon 45 skipping and re-framing in ~7% and ~42% of the transcript (Figure 3A) 120.
Above mentioned models carry mutations in the mouse genome. These models are good for proof-of-principle studies. However, due to species-specific differences in the genome sequence, reagents developed in these models cannot be directly used in human patients. To overcome this hurdle, a series of new hotspot mouse models were developed in the hDMD mouse, a transgenic strain carrying the full-length human dystrophin gene 121. These include a human exon 45 deletion model and two human exon 52 deletion models 122–124. The application of these models in DMD drug discovery is discussed in section 4.6.
4.2. Major hotspot mutation models in larger species
Besides mice, major hotspot mutation models have also been created in other species (Figure 2A). These include a rabbit model in which exon 51 is disrupted by partial deletion 125, a dog model with a point mutation at the 5’-end of intron 50 which leads to the exclusion of exon 50 in the mRNA, two independent pig models in which exon 52 is deleted 126, 127, and a monkey model in which exon 46 (or both exons 4 and 46) is disrupted by small deletions and insertions 128. As dystrophic manifestations seem to be proportional to the size of the animal, it is likely that these newly developed models will show more severe clinical disease than mouse models do 129, 130.
The rabbit model was characterized by Sui et al recently 125. Affected rabbits showed characteristic serological, histological and physiological features resembling those of human patients, including significant elevation of serum CK, aspartate aminotransferase, and alanine aminotransferase, variable myofiber size, centrally localized myonuclei (~30%), muscle inflammation, fibrosis, muscle atrophy, growth delay, mobility reduction, and premature death (~20% die within 2 weeks after birth and ~43% die within the first six months) 125. A unique feature of the rabbit model is the early onset of cardiomyopathy. The left ventricular ejection fraction (EF) and fraction shortening (FS) were significantly reduced by 4 months of the age. Consistently, the authors observed myocardial inflammation, fibrosis, and fatty cell infiltration 125. The rabbit model will be very useful for testing sequence-specific therapies, for example removing or skipping of exons 45–55.
The dog model was initially discovered in a Cavalier King Charles Spaniel and named CKCS-MD 131. Subsequently, the dog was crossed to the beagle background and renamed as deltaE50-MD 129, 132. The CKCS-MD dogs are characterized by marked elevation of serum CK, early onset dysphagia (2 to 3 months of age), exercise intolerance, and characteristic pathology changes in skeletal muscle (inflammation, variable fiber size, degeneration and regeneration) 131. However, myocardial pathology appeared mild in a 2-year-old CKCS-MD dog 131. The beagle background deltaE50-MD dogs are currently being characterized 133–135. Preliminary conference reports revealed skeletal muscle atrophy, inflammation and fibrosis 134. The deltaE50-MD dogs were also more susceptible to eccentric contraction-induced force drop 133. In 6-minute walk test, affected dogs significantly underperformed normal dogs. The distance walked by affected dogs plateaued at ~ 6 months. By 18 months, the averaged distance walked by normal and affected dogs were 359 and 97 meters, respectively 135. The deltaEx50-MD dog was recently used to test local and systemic CRISPR gene editing therapy 132, 136, 137. Removing or skipping exon 51 should restore the reading frame in the deltaEx50-MD dog. To achieve this goal, Amoasii et al targeted SpCas9 to an ESE adjacent to the exon 51 splice acceptor with AAV vectors. This strategy will not remove exon 51 from the genome. However, it should result in exon 51 exclusion in the mRNA via exon-skipping. Alternatively, it may result in re-framing by adding one nucleotide (adenine) to exon 51. AAV vectors were delivered to four 1-m-old deltaEx50-MD dogs via intramuscular (2 dogs) or intravenous (2 dogs) injection. Robust dystrophin restoration was observed at 6 to 8 weeks after injection. Similar to what was found in ΔEx44 and ΔEx50 mice 119, 120, the authors observed much more re-framing events than exon-skipping events in deltaEx50-MD dogs 132.
Two independent exon 52-deleted pig models have been generated 126, 127. In the model reported by Klymiuk et al, affected pigs showed CK elevation, characteristic skeletal muscle histopathology (such as excessive fiber size variation, myofiber degeneration and necrosis, centrally localized myonuclei, interstitial fibrosis, fatty replacement, and inflammation), muscle weakness, reduced mobility, and premature death by 3 months of age due to respiratory muscle failure 126. The other pig model has not been published in a peer reviewed journal yet 127.
A non-human primate DMD model was reported in 2015 by Chen et al 128. The authors co-delivered the Cas9 mRNA and two gRNAs (one targeting exon 4 and the other targeting exon 46) to the fertilized rhesus monkey eggs and obtained 14 live monkeys and four stillborn. Among live birth, frame-shifting mutations were identified in eight monkeys including two males and six females. Four had mutations in exon 4, two had mutations in exon 46, and two had mutations in both exons. At the time of publication, these monkeys were between 4 to 6 months of age and none developed obvious clinical signs of muscular dystrophy. For the remaining live birth, five did not have mutation, one animal had a small in-frame deletion in exon 4. Among four stillborn monkeys, two had normal dystrophin expression, one had greatly reduced dystrophin expression, and one more had no dystrophin expression. In two dystrophin-deficient stillborn monkeys, the authors found mosaic frame-disrupting mutations in 87% of the dystrophin gene alleles. They also found histological evidence of muscle disease such as the presence of hypertrophic myofibers with centrally localized myonuclei.
4.3. Minor hotspot mutation mouse models
The minor hotspot is located between exons 2 to 20 (Figure 2B) 105. The mdx5cv mouse is the first model harboring a mutation in the minor hotspot. This mouse is generated by ENU mutagenesis in 1989 111. In mdx5cv mice, an A to T transversion in the middle of exon 10 creates a new splice donor. Aberrant splicing results in a 53 bp deletion in the mRNA and reading frame shift 138. Interestingly, compared to that of mdx mice, mdx5cv mice show more severe muscle function deficits although both strains display similar histopathology 139.
Base editing is a newly developed genome editing tool. It replaces a single nucleotide without breaking the genome 140. Kim et al created an exon 20 nonsense mutation mouse model using the cytidine base editor 141. Specifically, a stop codon (TAG) was created by converting cytidine in a glutamine codon (CAG) in exon 20 to thymidine. This model was subsequently used to test the adenine base editor as a tool to treat nonsense mutation in DMD 142. In this case, adenine in the antisense strand of the TAG stop codon (CTA) was substituted with guanine to become CTG. As a result, the TAG stop codon in the sense strand was reverted to the original glutamine codon (CAG) in exon 20 142.
The first minor hotspot region deletion mouse model was reported by Egorova et al 143. Using the CRISPR technology, the authors removed a 430 kb fragment of the mouse dystrophin gene between introns 7 and 34. The resulting DMDdel8−34 model lost ~12% of the dystrophin coding sequence spanning exons 8 to 34 and displayed a phenotype similar to that of mdx mice. The reading frame of the DMDdel8−34 model can be restored by skipping or deleting exons 6 and 7. Removing exons 6 and 7 is predicted to treat ~3% DMD patients 107. Interestingly, variable clinical manifestations, from the mild BMD type to the severe DMD type, have been reported in patients with in-frame deletions containing exons 6 and 7 143. The DMDdel8−34 model offers an excellent platform to experimentally test exons 6/7 removal therapy.
The Dup2 mouse is one more mouse model with mutation located between exons 2 to 20 144. This mouse carries an exon 2 duplication mutation and is discussed in section 4.5.
4.4. Minor hotspot mutation models in larger species
A rat model with mutations in the minor hotspot was created in 2014 (Figure 2B) 145. The CRISPR technology was used to induce reading-frame disrupting indels in exons 3 and/or 16. The resulting founder rats and their first-generation progeny displayed motor function deficiency and characteristic histological lesions in muscle. It is worth pointing out that the rat model does not have the acute necrotic phase, a unique phase only seen in the mouse model 14. Further, DMD-mutated rats showed early onset cardiac disease, an observation confirmed in another rat model that carries a small deletion in exon 23 146.
Several canine DMD models carry mutations between exons 2 and 20 (Figure 2B). These include the GRMD (point mutation in intron 6) 10, beagle-background canine X-linked muscular dystrophy in Japan (CXMDJ) (point mutation in intron 6 as in GRMD dogs) 147, Welsh corgi muscular dystrophy dog (repetitive element insertion in intron 13) 148, Labrador retriever muscular dystrophy dog (repetitive element insertion in intron 19) 14, and border collie muscular dystrophy dog (a single nucleotide deletion in exon 20) 149. GRMD dogs have been used to test multi-exon skipping with the AAV U7 approach 150, 151. CXMDJ dogs have been used to test multi-exon skipping with AON 152. The Welsh corgi and Labrador retriever models are unique because a full-length dystrophin protein can be produced following genomic deletion of the inserted repetitive element with the CRISPR or other genome editing technology. Repetitive element insertion has been seen in introns in the minor hotspot in human patients 153. The Welsh corgi and Labrador retriever models are thus ideal to develop personalized therapy for these patients. The border collie model can be used to test exon 19/20 or 20/21 skipping and removing therapies. Such treatments will meet the need of 1.6% DMD patients 107.
Several dystrophin-deficient monkeys carry indel mutations in exon 4 128. Although exon 4 skipping/removing therapies can restore the reading frame in 0.2% DMD patients 107, it is uncertain whether testing such therapies is the best use of the precious non-human primate model.
4.5. Animal models for duplication mutation
Duplication is the second most common mutation type in DMD patients 103. However, there is only one duplication model. Using a targeted knock-in strategy, the Flanigan laboratory inserted exon 2 and its flanking sequence into intron 2 at a location corresponding to the duplication mutation hotspot in human patients 144. The resulting Dup 2 mouse carries a duplicated copy of exon 2 (Figure 2B and 3B). This disrupts the open reading frame in the transcript and aborts dystrophin expression in muscle. The Dup 2 mouse exhibits a phenotype similar to that of the mdx mouse except that (i) the Dup 2 mouse rarely has revertant fibers, (ii) the Dup 2 mouse has a significantly less pronounced CK elevation in the first 12 weeks, and (iii) the Dup 2 mouse has trace amount (≤ 2%) of dystrophin 144, 154.
Exon 2 duplication is the most common duplication mutation in DMD patients 155. Removing exon 2 is expected to treat 12.7% of single exon duplication patients (1.9% of all DMD patients) 107. Deletion of one copy of exon 2 restores the full-length protein (Figure 3B). However, deletion of both copies of exon 2 leads to reading frame shift (Figure 3B). Surprisingly, patients without exon 2 display a very mild BMD phenotype instead of the predicted severe DMD phenotype 154. Subsequent mechanistic studies revealed activation of an internal ribosome entry site in exon 5 and production of a slightly truncated dystrophin protein missing only the first half of the N-terminal actin-binding domain. Approximately 6% of DMD patients can benefit from this N-terminal shortened dystrophin protein. To test exon 2 deletion as a personalized therapy for patients who carry mutations at the 5’-end of the dystrophin gene, Wein et al treated the Dup 2 mouse with an AAV-U7 vector to skip both copies of exon 2 in the Dup 2 mouse. Although the treatment created a frame-shift mutation, it induced the expression of the N-terminal shortened dystrophin protein (Figure 3B). This counter-intuitive therapy significantly improved muscle force and attenuated muscle pathology 154.
4.6. Humanized mouse models for precision medicine
The ultimate goal of preclinical evaluation of a personalized therapy is to apply the animal-validated medication to human patients. Despite the highly conserved nature of the dystrophin gene, there exist significant differences in the DNA sequence among different species. As a result, custom-designed genetic medication for one species often cannot treat the same mutation in a different species. Ideally, sequence-specific DMD therapy should be developed in muscles that carry the human dystrophin gene.
To address this unmet need, ‘t Hoen et al generated the hDMD mouse 121. Specifically, they introduced the full-length human dystrophin gene to murine embryonic stem cells with a yeast artificial chromosome 121. The hDMD mouse generated from these cells contained a single copy of the stably integrated human dystrophin gene in mouse chromosome 5. The hDMD mouse maintained the tissue-specific expression pattern of the smaller isoforms of human dystrophin (Dp427m, Dp427c, Dp427p, Dp260, and Dp71). Both human dystrophin and mouse dystrophin were expressed in the hDMD mouse. Subsequent crossing with the mdx mouse resulted in the hDMD/mdx mouse which only expressed human dystrophin 121. Although the expression of mouse dystrophin is abolished in the hDMD/mdx mouse, this mouse still carries the full-length mouse dystrophin gene. Given the high homology between the human and mouse dystrophin gene sequence, there is a possibility that a therapeutic reagent designed for the human sequence may cross-react with the mouse sequence. To overcome this hurdle, Echigoya et al crossed hDMD mice with DMD-null mice 156. In DMD-null mice, the entire mouse dystrophin gene is deleted 157. The resulting hDMD/Dmd-null mice only carry the human dystrophin gene 156.
Although the hDMD mouse, hDMD/mdx mouse, and hDMD/Dmd-null mouse are useful to determine whether human sequence-specific drugs can result in expected changes in the DNA sequence and/or the RNA transcript of the human dystrophin gene, these models are not suitable for studying therapeutic efficacy because they do not have muscle disease. To truly capitalize these models for preclinical development of patient-ready precision therapies, several groups introduced deletions in the major mutation hotspot region of the human dystrophin gene in the hDMD mouse or hDMD/mdx mouse 122–124. Using the CRISPR technology, Young et al removed exon 45 from the human dystrophin gene in the hDMD mouse and created the hDMD del45 mouse 122. The hDMD del45 mouse was subsequently crossed with the mdx mouse and the DBA/2-background mdx mouse to generate the hDMD del45 mdx mouse and hDMD del45 mdxD2 mouse, respectively. Both human and mouse dystrophin were absent in these two newly generated strains. Consequently, these mice exhibited dystrophic pathology. The hDMD del45 mdx mouse and hDMD del45 mdxD2 mouse are now ready for in vivo evaluation of personalized genetic interventions that are designed to treat human patients. Such personalized therapies may include but not limit to skipping/removal of exon 44 (treat 6.2% DMD patients), exon 46 (treat 4.3% patients), or exons 45 to 55 (treat 60% DMD patients) 107, 158. As a proof-of-principle, the authors showed restoration of human dystrophin after CRISPR-mediated deletion of exons 46 to 55 from the hDMD del45 mdxD2 mouse 122, 158.
Using the transcription activator ligand effector nuclease (TALEN) technology, Veltrop et al deleted exon 52 from the human dystrophin gene in embryonic stem cells derived from the hDMD/mdx mouse 159. The hDMD del52/mdx mouse derived from these embryonic stem cells showed dystrophic muscle pathology and functional impairment 123. Similar to the humanized models generated by Young et al 122, the hDMD del52/mdx mouse is also ready for preclinical evaluation of personalized therapies developed for treating human patients. Examples of such therapies include skipping/removal of exon 51 (treat 13% DMD patients), exon 53 (treat 7.7% patients), or exons 45 to 55 (treat 60% DMD patients) 107. To validate the utility of the model, the authors tested human sequence-specific AONs for exons 51 and 53 skipping in the hDMD del52/mdx mouse 159. The exon 51 AON had two mismatches with the mouse sequence and the exon 53 AON had four mismatches with the mouse sequence. Direct muscle injection resulted in pronounced skipping of the human dystrophin transcript and restoration of human dystrophin. While the exon 53 AON only induced human-specific skipping, the exon 51 AON also induced low-level skipping of the mouse dystrophin transcript. These results suggest that species-specific sequence differences indeed greatly influence the outcome of precision medicine.
5. Conclusion
DMD is the most common inherited muscle-wasting disease. Currently, there is no cure. Animal models are essential for therapy development. Naturally occurring mdx mice and GRMD dogs have been the primary animal models in DMD drug discovery in the last three decades. High-throughput high-content screening provides a means to rapidly evaluate large collections of chemicals and small molecules for potential drug hits without prior knowledge of the mechanism of action. A series of nonmammalian DMD models have been developed in nematode, fruit fly and zebrafish. Easy to use methods that are based on dystrophin-deficiency related phenotype are now available for conducting large-scale screening. Many interesting hits have been discovered in these screens. Some of these hits may eventually become marketed drugs after preclinical validation in mammalian models and clinical evaluation in DMD patients.
Precision medicine is an emerging approach for developing customized therapy to meet the need of individual patient. DMD is caused by mutations that disrupt the open reading frame. Sequence-specific therapy such as exon skipping and genome editing restores the reading frame at the transcript and gene level, respectively. In DMD, most mutations are deletions and duplications clustered at two hotspots. Many hotspot mutation models were created in the mouse, rat, rabbit, pig and monkey in last several years. Several naturally occurring hotspot mutation models have also been discovered in dogs. Together, 21 hotspot mutation DMD models are now available. Many of these have been used in proof-of-principle studies to establish the feasibility of novel exon-skipping therapies and CRISPR therapies. Importantly, several mouse models were generated on the background of transgenic mice that carry the full-length human dystrophin gene. These humanized mouse models are especially valuable for in vivo evaluation of drugs that are intended for exon-skipping and genome editing in human patients.
6. Expert opinion
DMD drug discovery has followed two independent but intervening pathways. One pathway aims at restoring dystrophin expression in muscle using gene and/or cell therapies. The other pathway aims at ameliorating secondary and/or downstream defects such as inflammation and fibrosis via pharmaceutical interventions. In either case, animal models are essential in identifying the leads, demonstrating the proof-of-principle, and establishing preclinical efficacy and safety data for investigative new drug (IND) application before moving to human trials.
Naturally occurring mdx mice and GRMD dogs have served as the primary workhorse in DMD drug discovery in last several decades. Although these models are still very useful, they cannot fully meet all the needs of DMD drug discovery due to their inherent limitations. For example, the cost and workload have prevented the use of mammalian models in large-scale chemical screening. Most common mutations in DMD patients are large exon deletions and duplications. However, both mdx mice and GRMD dogs carry point mutation and hence are not ideal for precision medicine which aims at developing patient-tailored therapies.
High-throughput high-content library screening offers a great opportunity for new drug discovery. It also provides an economical and expedite way to repurpose medications that have already been approved for other indications. In this regard, simple nonmammalian animal models such as worm, fruit fly and zebrafish are particularly useful. These animals have a small body size, large progeny number, short life cycle, and are easy to breed and maintain. Importantly, the dystrophic phenotype in these models can be easily quantified using tools compatible with high-throughput high-content screening (Figure 1). The availability of nonmammalian DMD models enables unbiased phenotypic screening beyond what can be offered from in vitro models such as patient cells, induced pluripotent stem cells, engineered tissues, and organoids. Fruitful results have been obtained from screens performed in the C. elegans and zebrafish DMD models. Of interest, these screening have identified drugs that are currently in use in DMD patients (such as prednisone) and drugs that have been tested in clinical trials in DMD patients (such as cyclosporin A, nifedipine and sildenafil citrate). These findings suggest that high throughput screening in nonmammalian models may represent a useful approach in DMD drug discovery. Despite immense potential of the nonmammalian models in speeding up new drug discovery, it should be recognized that these animals are evolutionarily very distant from humans, the dose response and physiological consequences could be significantly different between non-mammals and mammals. The effect observed in nonmammalian models could be irrelevant for humans. For this reason, the hit identified in nonmammalian model screening has to be carefully validated in mammalian DMD models before clinical translation.
One of the most important goals of animal model development is to generate models that are suitable for testing novel therapies that can be directly applied to human patients. Hotspot deletion mutation is a characteristic feature in DMD. Unfortunately, mutation in most of the existing DMD models does not fit this profile. New animal models are in urgent need for developing therapeutic interventions that can treat hotspot deletion mutation. To fill the gap, a series of new mammalian DMD models were established in last several years. The currently available major hotspot models include the exon 44 deletion mouse model 120, exon 45 deletion mouse model 122, exon 46 mutation monkey model 128, exon 50 deletion mouse model 119, exon 50 exclusion dog model 131, exon 51 mutation rabbit model 125, exon 52 deletion mouse model 123, and exon 52 deletion pig models 126, 127 (Figure 2A). The currently available minor hotspot models include the exon 2 duplication mouse model 144, exon 3 and/or 16 mutation rat model 145, exon 4 mutation monkey model 128, exon 8 to 34 deletion mouse model 143, intron 13 and 19 mutation dog models 14, 148, exon 20 nonsense mutation mouse model 141, and exon 20 mutation dog model 149 (Figure 2B). Among these models, the ones with deletion mutation in the human dystrophin gene are particularly useful for evaluating sequence-specific precision medicine such exon-skipping and genome editing 122, 123. The humanized mouse model provides a unique opportunity to study interactions between the candidate drug and the human sequence in a dystrophic mammal. The pharmacokinetic and pharmacodynamic information learnt from these studies would be of great value for clinical trial design.
Technology advances, in particular targeted genome editing with CRISPR, have significantly simplified the process of animal model creation. Before 2000, there are only two major hotspot models (exon 52 deletion mice 114 and exon 53 nonsense mutation mdx4cv mice 111) and two minor hotspot models (exon 10 mutation mdx5cv mice 111 and exon 7 exclusion GRMD dogs 12). Since 2010, the number of major and minor hotspot models has increased to 11 and 10, respectively. However, model characterization has fallen behind model development. Research colonies remain to be established for many new models (e.g. the rabbit, pig and non-human primate models) in order to establish the natural history. A better understanding of onset and progression of skeletal muscle disease and cardiomyopathy in these new models will help us select the most appropriate models for a particular study. For example, one may want to use the rat and rabbit models in studies designed to treat DMD-associated cardiomyopathy because these two models display early onset heart disease 125–127. An alternative option for cardiomyopathy related studies is the recently established Australian Labradoodle dog model because this model also shows prominent cardiac disease 160. If the survival is the primary readout, one may want to use the pig (affected pigs die prematurely before 3 months of age) or rabbit (~43% affected rabbit die before 6 months of age) model 125, 126.
It is worth pointing out that the newly developed animal models can also be used to investigate some puzzling clinical observations. Results from such studies may have important implications for personalized therapy in DMD. A good example is the use of the Dup 2 mouse to elucidate the molecular mechanism underlying the paradox genotype-phenotype miscorrelation seen in exon 2 deletion patients 144, 154. The discovery of the novel ribosome entry site in exon 5 reveals a highly promising new approach to treat patients that have mutations at the 5’-end of the dystrophin gene 154. By the same token, the hDMD del 45/mdx mouse will be very useful to determine whether in-frame exon 45–47 or 45–48 deletion can indeed yield better outcome than in-frame exon 45–46 deletion 161.
In summary, with newly developed nonmammalian models and hotspot mutation models, investigators can now conduct studies that are not possible with traditional mdx and GRMD models, for example, high-throughput high-content screening, in vivo evaluation of human sequence-specific AONs for exon skipping and human sequence-specific guide RNAs for CRISPR editing. These new studies will no doubt accelerate DMD drug discovery. However, it does not mean we have solved all animal model-related issues. Several important models are still to be generated such as the hDMD/mdx mouse with human exon duplication mutation and the hDMD/mdx mouse with deletion mutation in the minor hotspot region of the human dystrophin gene. Nearly all current therapies aim to covert the severe DMD phenotype to a relatively mild BMD phenotype. But BMD is not disease-free. Currently, very few models exist for BMD 162, 163. If we were to further improve DMD therapy, we also need to develop more BMD models in preparation for BMD drug discovery in the future.
Last but not least, we should always bear in mind that no model is perfect. Depending on the question asked and the stage of drug discovery, different models should be used. Nonmammalian models should only be reserved for library screening. Large animal models should be used for late-stage scale-up studies after the candidate drug has been validated in the murine model. The hDMD mouse-based models are excellent for on-target effect evaluation when developing human-ready personalized therapy. However, these models cannot be used to study off-target effect because the hDMD mouse does not carry the entire human genome, it only carries the human dystrophin gene. In this case, human cells may be a better platform to study off-target editing in the human genome.
Article Highlights.
Duchenne muscular dystrophy (DMD) is a lethal muscle disease with limited therapeutic options.
Dystrophin-deficient mdx mice and golden retriever muscular dystrophy dogs are the most widely used animal models in the last three decades.
Nonmammalian DMD models in worms, flies and zebrafishes open the door to high-throughput drug discovery.
Hotspot mutation models are excellent platform for developing personalized therapy for a large population of DMD patients.
The human dystrophin gene-containing mouse models allow accelerated translation of sequence-specific therapies to DMD patients.
Funding
The authors thank the support from the National Institutes of Health (NIH) (AR-69085, AR-70517 and NS-90634 to D Duan, GM-063732 and GM-117059 to S Chen), the Department of Defense (MD150133 to D Duan) and the Jackson Freel DMD Research Fund (to D Duan).
Footnotes
Declaration of Interest
D Duan is a member of the scientific advisory board for Solid Biosciences and an equity holder of Solid Biosciences. The Duan lab has received research supports unrelated to CRISPR editing from Solid Biosciences and Edgewise Therapeutics. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
- 1.Rondot PGBA Duchenne de Boulogne (1806–1875). Journal of neurology 2005. July;252(7):866–7. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel LM. 2004. William Allan award address. cloning of the DMD gene. Am J Hum Genet 2005 Feb;76(2):205–14.* An excellent review on the discovery of dystrophin gene and protein.
- 3.Zeidman LA, Kondziella D. Peter Becker and his Nazi past: the man behind Becker muscular dystrophy and Becker myotonia. J Child Neurol 2014. April;29(4):514–9. [DOI] [PubMed] [Google Scholar]
- 4.Ervasti JM, Sonnemann KJ. Biology of the striated muscle dystrophin-glycoprotein complex. Int Rev Cytol 2008;265:191–225. [DOI] [PubMed] [Google Scholar]
- 5.Gao QQ, McNally EM. The dystrophin complex: structure, function, and implications for therapy. Comprehensive Physiology 2015. July 1;5(3):1223–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, van Ommen GJ, Kunkel LM. The pathogenesis and therapy of muscular dystrophies. Annual review of genomics and human genetics 2015. August 24;16:281–308. [DOI] [PubMed] [Google Scholar]
- 7.Prior TW, Bridgeman SJ. Experience and strategy for the molecular testing of Duchenne muscular dystrophy. J Mol Diagn 2005. August;7(3):317–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caskey CT, Nussbaum RL, Cohan LC, Pollack L. Sporadic occurrence of Duchenne muscular dystrophy: evidence for new mutation. Clin Genet 1980. November;18(5):329–41. [DOI] [PubMed] [Google Scholar]
- 9.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A 1984;81(4):1189–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornegay JN. The golden retriever model of Duchenne muscular dystrophy. Skelet Muscle 2017. May 19;7(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science 1989;244(4912):1578–80. [DOI] [PubMed] [Google Scholar]
- 12.Cooper BJ, Winand NJ, Stedman H, Valentine BA, Hoffman EP, Kunkel LM, et al. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature 1988;334(6178):154–6. [DOI] [PubMed] [Google Scholar]
- 13.Sharp NJ, Kornegay JN, Van Camp SD, Herbstreith MH, Secore SL, Kettle S, et al. An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy. Genomics 1992. May;13(1):115–21. [DOI] [PubMed] [Google Scholar]
- 14.McGreevy JW, Hakim CH, McIntosh MA, Duan D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis Model Mech 2015. March;8(3):195–213.**A comprehensive review of mammlian DMD models and their use in gene therapy research
- 15.van Putten M, Aartsma-Rus A, Grounds MD, Kornegay JN, Mayhew A, Gillingwater TH, et al. Update on Standard Operating Procedures in Preclinical Research for DMD and SMA Report of TREAT-NMD Alliance Workshop, Schiphol Airport, 26 April 2015, The Netherlands. Journal of neuromuscular diseases 2018;5(1):29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan D, Rafael-Fortney JA, Blain A, Kass DA, McNally EM, Metzger JM, et al. Standard operating procedures (SOPs) for evaluating the heart in preclinical studies of Duchenne muscular dystrophy. Journal of cardiovascular translational research 2016. February;9(1):85–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurana TS, Davies KE. Pharmacological strategies for muscular dystrophy. Nat Rev Drug Discov 2003. May;2(5):379–90. [DOI] [PubMed] [Google Scholar]
- 18.De Luca A Pre-clinical drug tests in the mdx mouse as a model of dystrophinopathies: an overview. Acta myologica : myopathies and cardiomyopathies : official journal of the Mediterranean Society of Myology / edited by the Gaetano Conte Academy for the study of striated muscle diseases 2012. May;31(1):40–7. [PMC free article] [PubMed] [Google Scholar]
- 19.Spinazzola JM, Kunkel LM. Pharmacological therapeutics targeting the secondary defects and downstream pathology of Duchenne muscular dystrophy. Expert opinion on orphan drugs 2016;4(11):1179–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blat Y, Blat S. Drug Discovery of Therapies for Duchenne Muscular Dystrophy. J Biomol Screen 2015. December;20(10):1189–203. [DOI] [PubMed] [Google Scholar]
- 21.Guiraud S, Davies KE. Pharmacological advances for treatment in Duchenne muscular dystrophy. Current opinion in pharmacology 2017. June;34:36–48. [DOI] [PubMed] [Google Scholar]
- 22.Verhaart IEC, Aartsma-Rus A. Therapeutic developments for Duchenne muscular dystrophy. Nature reviews Neurology 2019. July;15(7):373–86. [DOI] [PubMed] [Google Scholar]
- 23.Chamberlain JR, Chamberlain JS. Progress toward gene therapy for Duchenne muscular dystrophy. Mol Ther 2017. May 03;25(5):1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nance ME, Hakim CH, Yang NN, Duan D. Nanotherapy for Duchenne muscular dystrophy. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2018. April 11;10(2):e1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan D Dystrophin gene replacement and gene repair therapy for Duchenne muscular dystrophy in 2016. Human gene therapy Clinical development 2016. March;27(1):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barthelemy F, Wein N. Personalized gene and cell therapy for Duchenne Muscular Dystrophy. Neuromuscul Disord 2018. October;28(10):803–24. [DOI] [PubMed] [Google Scholar]
- 27.Nelson CE, Robinson-Hamm JN, Gersbach CA. Genome engineering: a new approach to gene therapy for neuromuscular disorders. Nature reviews Neurology 2017. November;13(11):647–61. [DOI] [PubMed] [Google Scholar]
- 28.Wong TWY, Cohn RD. Therapeutic Applications of CRISPR/Cas for Duchenne Muscular Dystrophy. Current gene therapy 2017;17(4):301–08. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Long C, Bassel-Duby R, Olson EN. Myoediting: Toward Prevention of Muscular Dystrophy by Therapeutic Genome Editing. Physiological reviews 2018. July 1;98(3):1205–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Zaidy S, Rodino-Klapac L, Mendell JR. Gene therapy for muscular dystrophy: moving the field forward. Pediatric neurology 2014. November;51(5):607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdul-Razak H, Malerba A, Dickson G. Advances in gene therapy for muscular dystrophies. F1000Res 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skuk D, Tremblay JP. Cell therapy in muscular dystrophies: many promises in mice and dogs, few facts in patients. Expert opinion on biological therapy 2015;15(9):1307–19. [DOI] [PubMed] [Google Scholar]
- 33.Aartsma-Rus A, Straub V, Hemmings R, Haas M, Schlosser-Weber G, Stoyanova-Beninska V, et al. Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic Acid Ther 2017. October;27(5):251–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan D Micro-dystrophin gene therapy goes systemic in Duchenne muscular dystrophy patients. Hum Gene Ther 2018. February 20;29(7):733–6.** A comprehensive review on the history and current status of AAV micro-dystrophin gene therapy.
- 35.Duan D Systemic AAV Micro-dystrophin Gene Therapy for Duchenne Muscular Dystrophy. Mol Ther 2018;26(10):2337–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies KE, Guiraud S. Micro-dystrophin Genes Bring Hope of an Effective Therapy for Duchenne Muscular Dystrophy. Mol Ther 2019. March 6;27(3):486–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan D Considerations on preclinical neuromuscular disease gene therapy design In: Duan D, Mendell JR, eds. Muscle gene therapy. Second ed. Switzerland: Springer Nature Switzerland AG; 2019:291–326. [Google Scholar]
- 38.Rodrigues M, Echigoya Y, Fukada SI, Yokota T. Current Translational Research and Murine Models For Duchenne Muscular Dystrophy. Journal of neuromuscular diseases 2016. March 3;3(1):29–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordish-Dressman H, Willmann R, Dalle Pazze L, Kreibich A, van Putten M, Heydemann A, et al. “Of Mice and Measures”: A Project to Improve How We Advance Duchenne Muscular Dystrophy Therapies to the Clinic. Journal of neuromuscular diseases 2018;5(4):407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shrader SM, Wrighten R, Smith BF. Animal models for muscle disease and muscle gene therapy In: Duan D, Mendell JR, eds. Muscle gene therapy. Second ed. Switzerland: Springer Nature Switzerland AG; 2019:41–64. [Google Scholar]
- 41.Duan D Duchenne muscular dystrophy gene therapy in the canine model. Hum Gene Ther Clin Dev 2015. March;26(1):57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X, Bao B, Echigoya Y, Yokota T. Dystrophin-deficient large animal models: translational research and exon skipping. American journal of translational research 2015;7(8):1314–31. [PMC free article] [PubMed] [Google Scholar]
- 43.Vengalil S, Preethish-Kumar V, Polavarapu K, Mahadevappa M, Sekar D, Purushottam M, et al. Duchenne Muscular Dystrophy and Becker Muscular Dystrophy Confirmed by Multiplex Ligation-Dependent Probe Amplification: Genotype-Phenotype Correlation in a Large Cohort. J Clin Neurol 2017. January;13(1):91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan R, Yi J, Xie Z, Zheng Y, Han M, Hou Y, et al. Genotype-phenotype correlation in Becker muscular dystrophy in Chinese patients. Journal of human genetics 2018. October;63(10):1041–48. [DOI] [PubMed] [Google Scholar]
- 45.Juan-Mateu J, Gonzalez-Quereda L, Rodriguez MJ, Baena M, Verdura E, Nascimento A, et al. DMD Mutations in 576 Dystrophinopathy Families: A Step Forward in Genotype-Phenotype Correlations. PLoS One 2015;10(8):e0135189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 2006. August;34(2):135–44. [DOI] [PubMed] [Google Scholar]
- 47.Tuffery-Giraud S, Beroud C, Leturcq F, Yaou RB, Hamroun D, Michel-Calemard L, et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat 2009. June;30(6):934–45. [DOI] [PubMed] [Google Scholar]
- 48.Magri F, Govoni A, D’Angelo MG, Del Bo R, Ghezzi S, Sandra G, et al. Genotype and phenotype characterization in a large dystrophinopathic cohort with extended follow-up. Journal of neurology 2011. September;258(9):1610–23. [DOI] [PubMed] [Google Scholar]
- 49.Mori-Yoshimura M, Mitsuhashi S, Nakamura H, Komaki H, Goto K, Yonemoto N, et al. Characteristics of Japanese Patients with Becker Muscular Dystrophy and Intermediate Muscular Dystrophy in a Japanese National Registry of Muscular Dystrophy (Remudy): Heterogeneity and Clinical Variation. Journal of neuromuscular diseases 2018;5(2):193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Echigoya Y, Lim KRQ, Nakamura A, Yokota T. Multiple Exon Skipping in the Duchenne Muscular Dystrophy Hot Spots: Prospects and Challenges. J Pers Med 2018. December 7;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts RG, Bobrow M. Dystrophins in vertebrates and invertebrates. Hum Mol Genet 1998;7(4):589–95. [DOI] [PubMed] [Google Scholar]
- 52.Bolanos-Jimenez F, Bordais A, Behra M, Strahle U, Mornet D, Sahel J, et al. Molecular cloning and characterization of dystrophin and Dp71, two products of the Duchenne Muscular Dystrophy gene, in zebrafish. Gene 2001. August 22;274(1–2):217–26. [DOI] [PubMed] [Google Scholar]
- 53.Greener MJ, Roberts RG. Conservation of components of the dystrophin complex in Drosophila. FEBS Lett 2000;482(1–2):13–8. [DOI] [PubMed] [Google Scholar]
- 54.Segalat L. Dystrophin and functionally related proteins in the nematode Caenorhabditis elegans. Neuromuscul Disord 2002. October;12 Suppl 1:S105–9. [DOI] [PubMed] [Google Scholar]
- 55.Steffen LS, Guyon JR, Vogel ED, Beltre R, Pusack TJ, Zhou Y, et al. Zebrafish orthologs of human muscular dystrophy genes. BMC genomics 2007. March 20;8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neuman S, Kaban A, Volk T, Yaffe D, Nudel U. The dystrophin / utrophin homologues in Drosophila and in sea urchin. Gene 2001. January 24;263(1–2):17–29. [DOI] [PubMed] [Google Scholar]
- 57.Sparrow J, Hughes SM, Segalat L. Other model organisms for sarcomeric muscle diseases. Adv Exp Med Biol 2008;642:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wells DJ, Wells KE. What do animal models have to tell us regarding Duchenne muscular dystrophy? Acta myologica : myopathies and cardiomyopathies : official journal of the Mediterranean Society of Myology / edited by the Gaetano Conte Academy for the study of striated muscle diseases 2005. December;24(3):172–80. [PubMed] [Google Scholar]
- 59.Plantie E, Migocka-Patrzalek M, Daczewska M, Jagla K. Model organisms in the fight against muscular dystrophy: lessons from drosophila and Zebrafish. Molecules 2015. April 9;20(4):6237–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M, Hromowyk KJ, Amacher SL, Currie PD. Muscular dystrophy modeling in zebrafish. Methods Cell Biol 2017;138:347–80. [DOI] [PubMed] [Google Scholar]
- 61.Widrick JJ, Kawahara G, Alexander MS, Beggs AH, Kunkel LM. Discovery of Novel Therapeutics for Muscular Dystrophies using Zebrafish Phenotypic Screens. Journal of neuromuscular diseases 2019. July 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pantoja M, Ruohola-Baker H. Drosophila as a starting point for developing therapeutics for the rare disease Duchenne Muscular Dystrophy. Rare Dis 2013;1:e24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gieseler K, Grisoni K, Segalat L. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Current biology : CB 2000. September 21;10(18):1092–7. [DOI] [PubMed] [Google Scholar]
- 64.Gaud A, Simon JM, Witzel T, Carre-Pierrat M, Wermuth CG, Segalat L. Prednisone reduces muscle degeneration in dystrophin-deficient Caenorhabditis elegans. Neuromuscul Disord 2004. June;14(6):365–70. [DOI] [PubMed] [Google Scholar]
- 65.Carre-Pierrat M, Mariol MC, Chambonnier L, Laugraud A, Heskia F, Giacomotto J, et al. Blocking of striated muscle degeneration by serotonin in C. elegans. J Muscle Res Cell Motil 2006;27(3–4):253–8. [DOI] [PubMed] [Google Scholar]
- 66.Giacomotto J, Pertl C, Borrel C, Walter MC, Bulst S, Johnsen B, et al. Evaluation of the therapeutic potential of carbonic anhydrase inhibitors in two animal models of dystrophin deficient muscular dystrophy. Hum Mol Genet 2009. November 1;18(21):4089–101. [DOI] [PubMed] [Google Scholar]
- 67.Carre-Pierrat M, Lafoux A, Tanniou G, Chambonnier L, Divet A, Fougerousse F, et al. Pre-clinical study of 21 approved drugs in the mdx mouse. Neuromuscul Disord 2011. May;21(5):313–27. [DOI] [PubMed] [Google Scholar]
- 68.Giacomotto J, Brouilly N, Walter L, Mariol MC, Berger J, Segalat L, et al. Chemical genetics unveils a key role of mitochondrial dynamics, cytochrome c release and IP3R activity in muscular dystrophy. Hum Mol Genet 2013. November 15;22(22):4562–78. [DOI] [PubMed] [Google Scholar]
- 69.Hewitt JE, Pollard AK, Lesanpezeshki L, Deane CS, Gaffney CJ, Etheridge T, et al. Muscle strength deficiency and mitochondrial dysfunction in a muscular dystrophy model of Caenorhabditis elegans and its functional response to drugs. Dis Model Mech 2018. December 4;11(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lloyd TE, Taylor JP. Flightless flies: Drosophila models of neuromuscular disease. Annals of the New York Academy of Sciences 2010. January;1184:e1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jagla K, Kalman B, Boudou T, Henon S, Batonnet-Pichon S. Beyond mice: Emerging and transdisciplinary models for the study of early-onset myopathies. Semin Cell Dev Biol 2017. April;64:171–80. [DOI] [PubMed] [Google Scholar]
- 72.Kreipke RE, Kwon YV, Shcherbata HR, Ruohola-Baker H. Drosophila melanogaster as a Model of Muscle Degeneration Disorders. Curr Top Dev Biol 2017;121:83–109. [DOI] [PubMed] [Google Scholar]
- 73.Potikanond S, Nimlamool W, Noordermeer J, Fradkin LG. Muscular Dystrophy Model. Adv Exp Med Biol 2018;1076:147–72. [DOI] [PubMed] [Google Scholar]
- 74.Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, et al. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. The EMBO journal 2007. January 24;26(2):481–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Plas MC, Pilgram GS, de Jong AW, Bansraj MR, Fradkin LG, Noordermeer JN. Drosophila Dystrophin is required for integrity of the musculature. Mechanisms of development 2007. August;124(7–8):617–30. [DOI] [PubMed] [Google Scholar]
- 76.Taghli-Lamallem O, Akasaka T, Hogg G, Nudel U, Yaffe D, Chamberlain JS, et al. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging cell 2008. March;7(2):237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pantoja M, Fischer KA, Ieronimakis N, Reyes M, Ruohola-Baker H. Genetic elevation of sphingosine 1-phosphate suppresses dystrophic muscle phenotypes in Drosophila. Development 2013. January 1;140(1):136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christoforou CP, Greer CE, Challoner BR, Charizanos D, Ray RP. The detached locus encodes Drosophila Dystrophin, which acts with other components of the Dystrophin Associated Protein Complex to influence intercellular signalling in developing wing veins. Dev Biol 2008. January 15;313(2):519–32. [DOI] [PubMed] [Google Scholar]
- 79.Kucherenko MM, Pantoja M, Yatsenko AS, Shcherbata HR, Fischer KA, Maksymiv DV, et al. Genetic Modifier Screens Reveal New Components that Interact with the Drosophila Dystroglycan-Dystrophin Complex. Plos One 2008. June 11;3(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kucherenko MM, Marrone AK, Rishko VM, Magliarelli Hde F, Shcherbata HR. Stress and muscular dystrophy: a genetic screen for dystroglycan and dystrophin interactors in Drosophila identifies cellular stress response components. Dev Biol 2011. April 15;352(2):228–42. [DOI] [PubMed] [Google Scholar]
- 81.McNally EM, Kaltman JR, Benson DW, Canter CE, Cripe LH, Duan D, et al. Contemporary cardiac issues in Duchenne muscular dystrophy. Circulation 2015. May 5;131(18):1590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Judge LM, Haraguchi M, Chamberlain JS. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J Cell Sci 2006. April 15;119(Pt 8):1537–46. [DOI] [PubMed] [Google Scholar]
- 83.Ieronimakis N, Pantoja M, Hays AL, Dosey TL, Qi J, Fischer KA, et al. Increased sphingosine-1-phosphate improves muscle regeneration in acutely injured mdx mice. Skelet Muscle 2013. August 1;3(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berger J, Currie PD. Zebrafish models flex their muscles to shed light on muscular dystrophies. Dis Model Mech 2012. November;5(6):726–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawahara G, Kunkel LM. Zebrafish based small molecule screens for novel DMD drugs. Drug Discov Today Technol 2013 Spring;10(1):e91–e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guyon JR, Mosley AN, Zhou Y, O’Brien KF, Sheng X, Chiang K, et al. The dystrophin associated protein complex in zebrafish. Hum Mol Genet 2003. March 15;12(6):601–15. [PubMed] [Google Scholar]
- 87.Guyon JR, Goswami J, Jun SJ, Thorne M, Howell M, Pusack T, et al. Genetic isolation and characterization of a splicing mutant of zebrafish dystrophin. Hum Mol Genet 2009. January 1;18(1):202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berger J, Berger S, Jacoby AS, Wilton SD, Currie PD. Evaluation of exon-skipping strategies for Duchenne muscular dystrophy utilizing dystrophin-deficient zebrafish. J Cell Mol Med 2011. December;15(12):2643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 1996. December;123:1–36. [DOI] [PubMed] [Google Scholar]
- 90.Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 1996. December;123:399–413. [DOI] [PubMed] [Google Scholar]
- 91.Bassett DI, Bryson-Richardson RJ, Daggett DF, Gautier P, Keenan DG, Currie PD. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development 2003. December;130(23):5851–60. [DOI] [PubMed] [Google Scholar]
- 92.Widrick JJ, Alexander MS, Sanchez B, Gibbs DE, Kawahara G, Beggs AH, et al. Muscle dysfunction in a zebrafish model of Duchenne muscular dystrophy. Physiological genomics 2016. November 1;48(11):850–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li M, Andersson-Lendahl M, Sejersen T, Arner A. Muscle dysfunction and structural defects of dystrophin-null sapje mutant zebrafish larvae are rescued by ataluren treatment. FASEB J 2014. April;28(4):1593–9. [DOI] [PubMed] [Google Scholar]
- 94.Nagabhushana A, Mishra RK. Finding clues to the riddle of sex determination in zebrafish. J Biosci 2016. March;41(1):145–55. [DOI] [PubMed] [Google Scholar]
- 95.Berger J, Sztal T, Currie PD. Quantification of birefringence readily measures the level of muscle damage in zebrafish. Biochem Biophys Res Commun 2012. July 13;423(4):785–8. [DOI] [PubMed] [Google Scholar]
- 96.Smith LL, Beggs AH, Gupta VA. Analysis of skeletal muscle defects in larval zebrafish by birefringence and touch-evoke escape response assays. J Vis Exp 2013. December 13(82):e50925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leet JK, Lindberg CD, Bassett LA, Isales GM, Yozzo KL, Raftery TD, et al. High-content screening in zebrafish embryos identifies butafenacil as a potent inducer of anemia. PLoS One 2014;9(8):e104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raftery TD, Isales GM, Yozzo KL, Volz DC. High-content screening assay for identification of chemicals impacting spontaneous activity in zebrafish embryos. Environ Sci Technol 2014;48(1):804–10. [DOI] [PubMed] [Google Scholar]
- 99.Yozzo KL, Isales GM, Raftery TD, Volz DC. High-content screening assay for identification of chemicals impacting cardiovascular function in zebrafish embryos. Environ Sci Technol 2013. October 1;47(19):11302–10. [DOI] [PubMed] [Google Scholar]
- 100.Kawahara G, Karpf JA, Myers JA, Alexander MS, Guyon JR, Kunkel LM. Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 2011. March 29;108(13):5331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kawahara G, Gasperini MJ, Myers JA, Widrick JJ, Eran A, Serafini PR, et al. Dystrophic muscle improvement in zebrafish via increased heme oxygenase signaling. Hum Mol Genet 2014. April 1;23(7):1869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Waugh TA, Horstick E, Hur J, Jackson SW, Davidson AE, Li X, et al. Fluoxetine prevents dystrophic changes in a zebrafish model of Duchenne muscular dystrophy. Hum Mol Genet 2014. September 1;23(17):4651–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, et al. The TREAT-NMD DMD global database: analysis of more than 7000 Duchenne muscular dystrophy mutations. Hum Mutat 2015. January 21;36(4):395–402.* This paper report a comprehesive database of all known mutations in DMD patients.
- 104.Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat 2009. December;30(12):1657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aartsma-Rus A, Van Ommen GJ. Antisense-mediated exon skipping for Duchenne muscular dystrophy In: Duan D, ed. Muscle gene therapy. New York: Springer Science + Business Media, LLC; 2010:69–84. [Google Scholar]
- 106.Nakamura A, Shiba N, Miyazaki D, Nishizawa H, Inaba Y, Fueki N, et al. Comparison of the phenotypes of patients harboring in-frame deletions starting at exon 45 in the Duchenne muscular dystrophy gene indicates potential for the development of exon skipping therapy. Journal of human genetics 2017. April;62(4):459–63. [DOI] [PubMed] [Google Scholar]
- 107.Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat 2009. March;30(3):293–9.** This paper shows the propotion of DMD patients that can be treated with different mutation-specific medications
- 108.Beroud C, Tuffery-Giraud S, Matsuo M, Hamroun D, Humbertclaude V, Monnier N, et al. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum Mutat 2007. February;28(2):196–202. [DOI] [PubMed] [Google Scholar]
- 109.Nakamura A, Yoshida K, Fukushima K, Ueda H, Urasawa N, Koyama J, et al. Follow-up of three patients with a large in-frame deletion of exons 45–55 in the Duchenne muscular dystrophy (DMD) gene. J Clin Neurosci 2008. July;15(7):757–63. [DOI] [PubMed] [Google Scholar]
- 110.Taglia A, Petillo R, D’Ambrosio P, Picillo E, Torella A, Orsini C, et al. Clinical features of patients with dystrophinopathy sharing the 45–55 exon deletion of DMD gene. Acta myologica : myopathies and cardiomyopathies : official journal of the Mediterranean Society of Myology / edited by the Gaetano Conte Academy for the study of striated muscle diseases 2015. May;34(1):9–13. [PMC free article] [PubMed] [Google Scholar]
- 111.Chapman VM, Miller DR, Armstrong D, Caskey CT. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc Natl Acad Sci U S A 1989;86(4):1292–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 2017. February 14;8:14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mitrpant C, Fletcher S, Iversen PL, Wilton SD. By-passing the nonsense mutation in the 4 CV mouse model of muscular dystrophy by induced exon skipping. J Gene Med 2009. January;11(1):46–56. [DOI] [PubMed] [Google Scholar]
- 114.Araki E, Nakamura K, Nakao K, Kameya S, Kobayashi O, Nonaka I, et al. Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in Duchenne muscular dystrophy. Biochem Biophys Res Commun 1997;238(2):492–7. [DOI] [PubMed] [Google Scholar]
- 115.Aoki Y, Nakamura A, Yokota T, Saito T, Okazawa H, Nagata T, et al. In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol Ther 2010. November;18(11):1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aoki Y, Yokota T, Nagata T, Nakamura A, Tanihata J, Saito T, et al. Bodywide skipping of exons 45–55 in dystrophic mdx52 mice by systemic antisense delivery. Proc Natl Acad Sci U S A 2012. August 21;109(34):13763–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Echigoya Y, Aoki Y, Miskew B, Panesar D, Touznik A, Nagata T, et al. Long-term efficacy of systemic multiexon skipping targeting dystrophin exons 45–55 with a cocktail of vivo-morpholinos in mdx52 mice. Molecular therapy Nucleic acids 2015;4:e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Echigoya Y, Lim KRQ, Melo D, Bao B, Trieu N, Mizobe Y, et al. Exons 45–55 Skipping Using Mutation-Tailored Cocktails of Antisense Morpholinos in the DMD Gene. Mol Ther 2019. November 6;27(11):2005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Amoasii L, Long C, Li H, Mireault AA, Shelton JM, Sanchez-Ortiz E, et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med 2017. November 29;9(418):pii: eaan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv 2019. March;5(3):eaav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.‘t Hoen PA, de Meijer EJ, Boer JM, Vossen RH, Turk R, Maatman RG, et al. Generation and characterization of transgenic mice with the full-length human DMD gene. J Biol Chem 2008. February 29;283(9):5899–907. [DOI] [PubMed] [Google Scholar]
- 122.Young CS, Mokhonova E, Quinonez M, Pyle AD, Spencer MJ. Creation of a Novel Humanized Dystrophic Mouse Model of Duchenne Muscular Dystrophy and Application of a CRISPR/Cas9 Gene Editing Therapy. Journal of neuromuscular diseases 2017;4(2):139–45.*This paper reports a humanized exon 44 deletion DMD model that can be used to develop precision medicine for human patients.
- 123.Veltrop M, van Vliet L, Hulsker M, Claassens J, Brouwers C, Breukel C, et al. A dystrophic Duchenne mouse model for testing human antisense oligonucleotides. Plos One 2018. February 21;13(2):e0193289.* This paper reports a humanized exon 52 deletion DMD model that can be used to develop precision medicine for human patients.
- 124.Robinson-Hamm JN, Nelson CE, Gemberling M, Gough V, Castellanos Rivera RM, Aartsma-Rus A, et al. Dystrophin Restoration in a Humanized Mouse Model of Duchenne Muscular Dystrophy by Gene Editing with S. Aureus Cas9. Mol Ther 2018(26):S1. [Google Scholar]
- 125.Sui T, Lau YS, Liu D, Liu T, Xu L, Gao Y, et al. A novel rabbit model of Duchenne muscular dystrophy generated by CRISPR/Cas9. Dis Model Mech 2018. June 4;11(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Klymiuk N, Blutke A, Graf A, Krause S, Burkhardt K, Wuensch A, et al. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Hum Mol Genet 2013. November 1;22(21):4368–82. [DOI] [PubMed] [Google Scholar]
- 127.Rogers CS, Swart JR, inventors; Exemplar Genetics, LLC, assignee. Animal Models of Duchenne Muscular Dystrophy. United States. 2014 8/7/2014.
- 128.Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, Guo X, et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet 2015. July 1;24(13):3764–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wells DJ. Tracking progress: an update on animal models for Duchenne muscular dystrophy. Dis Model Mech 2018. June 13;11(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Valentine BA, Cooper BJ, de Lahunta A, O’Quinn R, Blue JT. Canine X-linked muscular dystrophy--an animal model of Duchenne muscular dystrophy: clinical studies. J Neurol Sci 1988;88(1–3):69–81. [DOI] [PubMed] [Google Scholar]
- 131.Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, Burke MM, Nagel N, Holder A, et al. A Duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient cavalier king charles spaniels is amenable to exon 51 skipping. PLoS One 2010;5(1):e8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018. August 30;362(6410):86–91.* This is the first paper to demonstrate CRISPR editing therapy in a large animal model.
- 133.Riddell D, Harron R, Taylor-Brown F, Wells DJ, Piercy RJ. Progressive age-associated decline in resistance to electrically-induced repetitive eccentric tibiotarsal flexion torque in the deltaE50-MD dog. Neuromuscul Disord 2018;28:S16. [Google Scholar]
- 134.Hildyard JCW, Rawson F, Harron R, Riddell D, Massey C, Taylor-Brown F, et al. Characterising the skeletal muscle histological phenotype of the DeltaE50-MD dog, a preclinical model of Duchenne muscular dystrophy. Neuromuscul Disord 2018;28:S18. [Google Scholar]
- 135.Harron R, Wells DJ, Piercy RJ. Assessment of a 6-minute walk test (6MWT) for non-invasive, phenotypic evaluation of deltaE50-MD dogs, a preclinical model of Duchenne muscular dystrophy. Neuromuscul Disord 2018;28:S15. [Google Scholar]
- 136.Duan D CRISPR alleviates muscular dystrophy in dogs. Nat Biomed Eng 2018. November;2(11):795–96. [DOI] [PubMed] [Google Scholar]
- 137.Wasala NB, Hakim CH, Yang NN, Duan D. Questions answered and unanswered by the first CRISPR editing study in the canine model of Duchenne muscular dystrophy. Hum Gene Ther 2019:in-press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Im WB, Phelps SF, Copen EH, Adams EG, Slightom JL, Chamberlain JS. Differential expression of dystrophin isoforms in strains of mdx mice with different mutations. Hum Mol Genet 1996;5(8):1149–53. [DOI] [PubMed] [Google Scholar]
- 139.Beastrom N, Lu H, Macke A, Canan BD, Johnson EK, Penton CM, et al. Mdx5cv mice manifest more severe muscle dysfunction and diaphragm force deficits than do mdx Mice. Am J Pathol 2011. November;179(5):2464–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nature reviews Genetics 2018. December;19(12):770–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, et al. Highly efficient RNA-guided base editing in mouse embryos. Nature biotechnology 2017. May;35(5):435–37. [DOI] [PubMed] [Google Scholar]
- 142.Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nature biotechnology 2018. June;36(6):536–39. [DOI] [PubMed] [Google Scholar]
- 143.Egorova TV, Zotova ED, Reshetov DA, Polikarpova AV, Vassilieva SG, Vlodavets DV, et al. CRISPR/Cas9-generated mouse model of Duchenne muscular dystrophy recapitulating a newly identified large 430 kb deletion in the human DMD gene. Dis Model Mech 2019. April 25;12(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vulin A, Wein N, Simmons TR, Rutherford AM, Findlay AR, Yurkoski JA, et al. The first exon duplication mouse model of Duchenne muscular dystrophy: A tool for therapeutic development. Neuromuscular Disord 2015. November;25(11):827–34. [DOI] [PubMed] [Google Scholar]
- 145.Nakamura K, Fujii W, Tsuboi M, Tanihata J, Teramoto N, Takeuchi S, et al. Generation of muscular dystrophy model rats with a CRISPR/Cas system. Scientific reports 2014;4:5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Larcher T, Lafoux A, Tesson L, Remy S, Thepenier V, Francois V, et al. Characterization of dystrophin deficient rats: a new model for Duchenne muscular dystrophy. PLoS One 2014;9(10):e110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shimatsu Y, Katagiri K, Furuta T, Nakura M, Tanioka Y, Yuasa K, et al. Canine X-linked muscular dystrophy in Japan (CXMDJ). Exp Anim 2003. April;52(2):93–7. [DOI] [PubMed] [Google Scholar]
- 148.Smith BF, Yue Y, Woods PR, Kornegay JN, Shin J-H, Williams RR, et al. An intronic LINE-1 element insertion in the dystrophin gene aborts dystrophin expression and results in Duchenne-like muscular dystrophy in the corgi breed. Lab Invest 2011. February;91(2):216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mata Lopez S, Hammond JJ, Rigsby MB, Balog-Alvarez CJ, Kornegay JN, Nghiem PP. A novel canine model for Duchenne muscular dystrophy (DMD): single nucleotide deletion in DMD gene exon 20. Skelet Muscle 2018. May 29;8(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Barbash IM, Cecchini S, Faranesh AZ, Virag T, Li L, Yang Y, et al. MRI roadmap-guided transendocardial delivery of exon-skipping recombinant adeno-associated virus restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Gene Ther 2013. March;20(3):274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bish LT, Sleeper MM, Forbes SC, Wang B, Reynolds C, Singletary GE, et al. Long-term restoration of cardiac dystrophin expression in golden retriever muscular dystrophy following rAAV6-mediated exon skipping. Mol Ther 2012. March;20(3):580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S, et al. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol 2009. March 13;65(6):667–76.*This is the first paper to report multi-exon skipping in the canine DMD model.
- 153.Cohen N, Rimessi P, Gualandi F, Ferlini A, Muntoni F. In vivo study of an aberrant dystrophin exon inclusion in X-linked dilated cardiomyopathy. Biochem Biophys Res Commun 2004. May 14;317(4):1215–20. [DOI] [PubMed] [Google Scholar]
- 154.Wein N, Vulin A, Falzarano MS, Szigyarto CA, Maiti B, Findlay A, et al. Translation from a DMD exon 5 IRES results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nature medicine 2014. August 10;20(9):992–1000.*This paper describes the first duplication model and the use of a counterintuitive approach to treat DMD.
- 155.White SJ, Aartsma-Rus A, Flanigan KM, Weiss RB, Kneppers AL, Lalic T, et al. Duplications in the DMD gene. Hum Mutat 2006. September;27(9):938–45. [DOI] [PubMed] [Google Scholar]
- 156.Echigoya Y, Lim KRQ, Trieu N, Bao B, Miskew Nichols B, Vila MC, et al. Quantitative Antisense Screening and Optimization for Exon 51 Skipping in Duchenne Muscular Dystrophy. Mol Ther 2017. November 1;25(11):2561–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kudoh H, Ikeda H, Kakitani M, Ueda A, Hayasaka M, Tomizuka K, et al. A new model mouse for Duchenne muscular dystrophy produced by 2.4 Mb deletion of dystrophin gene using Cre-loxP recombination system. Biochem Biophys Res Commun 2005. March 11;328(2):507–16. [DOI] [PubMed] [Google Scholar]
- 158.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell stem cell 2016. April 7;18(4):533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Veltrop M, van der Kaa J, Claassens J, van Vliet L, Verbeek S, Aartsma-Rus A. Generation of embryonic stem cells and mice for duchenne research. PLoS Curr 2013. September 10;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shrader SM, Jung S, Denney TS, Smith BF. Characterization of Australian Labradoodle dystrophinopathy. Neuromuscular Disord 2018. November;28(11):927–37. [DOI] [PubMed] [Google Scholar]
- 161.Findlay AR, Wein N, Kaminoh Y, Taylor LE, Dunn DM, Mendell JR, et al. Clinical phenotypes as predictors of the outcome of skipping around DMD exon 45. Ann Neurol 2015. April;77(4):668–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Baroncelli AB, Abellonio F, Pagano TB, Esposito I, Peirone B, Papparella S, et al. Muscular dystrophy in a dog resembling human Becker muscular dystrophy. Journal of comparative pathology 2014. May;150(4):429–33. [DOI] [PubMed] [Google Scholar]
- 163.Tanihata J, Nagata T, Ito N, Saito T, Nakamura A, Minamisawa S, et al. Truncated dystrophin ameliorates the dystrophic phenotype of mdx mice by reducing sarcolipin-mediated SERCA inhibition. Biochem Biophys Res Commun 2018. October 20;505(1):51–59. [DOI] [PubMed] [Google Scholar]