Abstract
A lack of tools to precisely control gene expression has limited our ability to evaluate relationships between expression levels and phenotypes. Here, we describe an approach to titrate expression of human genes using CRISPR interference and series of single guide RNAs (sgRNAs) with systematically modulated activities. We used large-scale measurements across multiple cell models to characterize activities of sgRNAs containing mismatches to their target sites and derived rules governing mismatched sgRNA activity using deep learning. These rules enabled us to synthesize a compact sgRNA library to titrate expression of ~2,400 genes essential for robust cell growth and to construct an in silico sgRNA library spanning the human genome. Staging cells along a continuum of gene expression levels combined with single-cell RNA-seq readout revealed sharp transitions in cellular behaviors at gene-specific expression thresholds. Our work provides a general tool to control gene expression, with applications ranging from tuning biochemical pathways to identifying suppressors for diseases of dysregulated gene expression.
The complexity of biological processes arises not only from the set of expressed genes but also from quantitative differences in their expression levels. As a classic example, some genes are haploinsufficient and thus sensitive to a 50% decrease in expression, whereas other genes are permissive to far stronger depletion1. Enabled by tools to titrate gene expression levels such as series of promoters or hypomorphic mutants, the underlying expression-phenotype relationships have been explored systematically in yeast2–4 and bacteria5–8. These efforts have revealed gene- and environment-specific effects of changes in expression levels4 and yielded insight into the opposing evolutionary forces that determine gene expression levels including the cost of protein synthesis and the need for robustness against random fluctuations3,6,8.
The availability of equivalent tools in mammalian systems would enable similar efforts to probe expression-phenotype relationships in more complex models. In addition, such tools could be used to identify the functionally sufficient levels of gene products, which can serve as targets for rescue by gene therapy or chemical treatment, or as targets of inhibition for anti-cancer drugs. It is possible to titrate the expression of individual genes in mammalian systems by incorporating microRNA binding sites of varied strength into the 3′-UTR of the endogenous locus9 or using synthetic promoters and regulators10, but these approaches require engineering of the endogenous locus for each target, limiting scalability and transferability across models. The development of artificial transcription factors, such as TALEs11 or the CRISPR-based effectors underlying CRISPR interference (CRISPRi) and activation (CRISPRa)12, has now provided tools to systematically knock down or overexpress genes in mammalian models. CRISPR/Cas9 based systems in particular have attracted considerable attention due to the exquisite programmability of targeting a locus via sequence complementarity to an associated single guide RNA (sgRNA)13. Thus far, however, these tools have been primarily optimized for strong knockdown or overexpression14,15 and do not afford nuanced control over gene expression levels.
Studies of the targeting mechanisms of Cas9 and its nuclease-dead variants (dCas9) have established that both activity and binding can be modulated by introducing mismatches into the sgRNA targeting region, modifying the sgRNA constant region, or adding hairpin extensions13,16–20. In addition, (d)Cas9 activity can be controlled using small molecules, degrons, or anti-CRISPRs (e.g. 21–24), but these approaches generally have not been optimized to afford precise control over activity levels and can be challenging to transfer across models. Here, we report a systematic approach to control DNA binding of dCas9 effectors through modified sgRNAs as a general method to titrate gene expression in mammalian cells. We describe both an empirically validated compact sgRNA library to titrate the expression of essential genes and a genome-wide in silico library derived from deep learning analysis of the empirical data. As a starting point for analyses of expression-phenotype relationships in mammalian cells, we examined transcriptional phenotypes derived from single-cell RNA-seq at various expression levels of 25 essential genes. Our data reveal gene-specific expression-phenotype relationships and expression level-dependent cell responses at single-cell resolution, highlighting the utility of systematically attenuated sgRNAs in staging cells along a continuum of expression levels in order to explore fundamental biological questions.
Results
Mismatched sgRNAs mediate diverse intermediate phenotypes
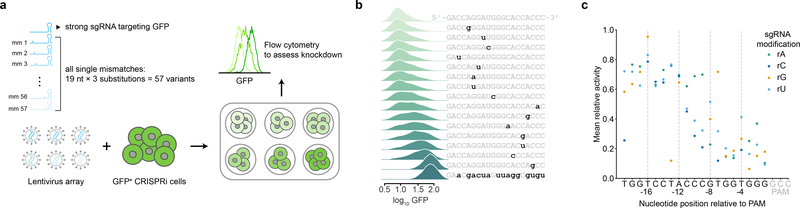
To comprehensively characterize the activities of mismatched sgRNAs in CRISPRi-mediated knockdown, we measured the knockdowns mediated by all 57 singly mismatched variants of a GFP-targeting sgRNA25 (Fig. 1a). K562 cells harboring mismatched sgRNAs experienced knockdown levels between those of cells with the perfectly matched sgRNA (94%) and cells with a non-targeting control sgRNA (Fig. 1b, S1a–c, Table S1). As expected, sgRNAs with mismatches in the PAM-proximal seed region13,16 had strongly attenuated activity. By contrast, sgRNAs with mismatches in the PAM-distal region mediated GFP knockdown to an extent similar to that of the unmodified sgRNA, albeit with substantial variability depending on the type of mismatch (Fig. 1b–c). The distributions of GFP levels with mismatched sgRNAs were largely unimodal, although the distributions were typically broader than those with the perfectly matched sgRNA or the control sgRNA (Fig. 1b, S1c). These results suggest that series of mismatched sgRNAs can be used to titrate gene expression at the single-cell level, but that mismatched sgRNA activity is modulated by complex factors.
Figure 1.
Mismatched sgRNAs titrate GFP expression at the single-cell level. (a) Experimental design to test knockdown conferred by all mismatched variants of a GFP-targeting sgRNA. (b) Distributions of GFP levels in cells with a perfectly matched sgRNA (top), mismatched sgRNAs (middle), and a non-targeting control sgRNA (bottom). Sequences of sgRNAs are indicated on the right (without the PAM). (c) Relative activities of all mismatched sgRNAs, defined as the ratio of fold-knockdown conferred by a mismatched sgRNA to fold-knockdown conferred by the perfectly matched sgRNA. Data represent mean relative activities obtained from two replicate transductions.
Rules of mismatched sgRNA activity derived from a large-scale screen
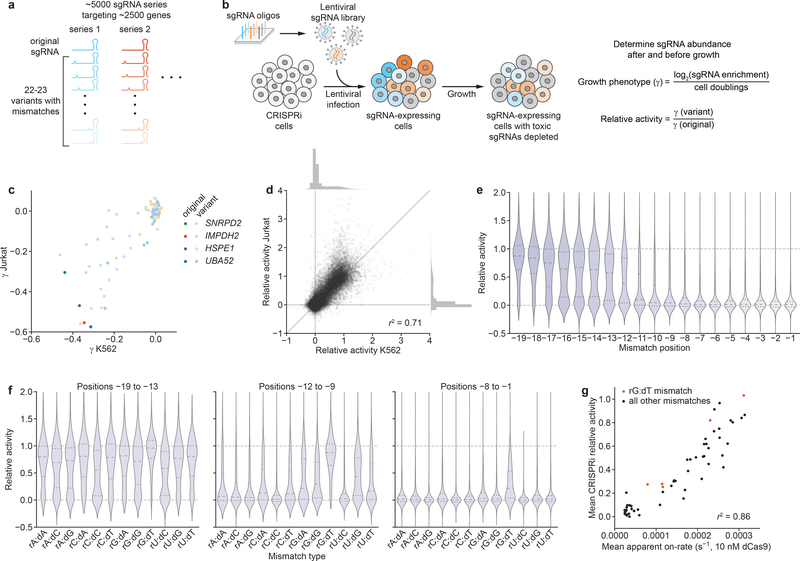
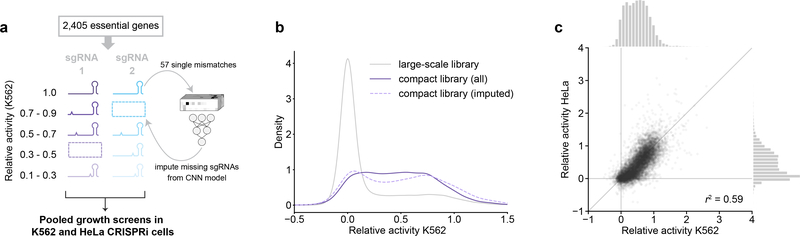
We reasoned that we could empirically derive the factors governing the influence of mismatches on sgRNA activity by measuring growth phenotypes imparted by a large number of mismatched sgRNAs in a pooled screen. For this purpose, we generated a ~120,000-element library comprising series of variants for 4,898 sgRNAs targeting 2,449 genes with growth phenotypes in K562 cells14. Each individual series, herein referred to as an allelic series, contains the original, perfectly matched sgRNA and 22–23 variants harboring one or two mismatches (Fig. 2a, Table S2, the first nucleotide of the sgRNA was held as a G regardless of its match in the genome, Methods). We then measured CRISPRi growth phenotypes (γ; a more negative value indicates a stronger growth defect) for each sgRNA in both K562 and Jurkat cells using pooled screens18,26 (Fig. 2b, S2a–b, Methods). Growth phenotypes of targeting sgRNAs were well-correlated in replicate screens (Fig. S2a–b, Tables S3–S4) and recapitulated previously reported phenotypes14 (Fig. S2c).
Figure 2.
A large-scale CRISPRi screen identifies factors governing mismatched sgRNA activity. (a) Design of large-scale mismatched sgRNA library. (b) Schematic of pooled CRISPRi screen to determine activities of mismatched-sgRNAs. (c) Growth phenotypes (γ) in K562 and Jurkat cells for four sgRNA series, with the perfectly matched sgRNAs shown in darker colors and mismatched sgRNAs shown in corresponding lighter colors. Phenotypes represent the mean of two replicate screens. Differences in absolute phenotypes likely reflect cell type-specific essentiality. A γ of 0 is equivalent to the average phenotype of non-targeting control sgRNAs. (d) Comparison of mismatched sgRNA relative activities in K562 and Jurkat cells. Marginal histograms depict distributions of relative activities along the corresponding axes. n = 41,512 sgRNAs; r2 = squared Pearson correlation coefficient. (e) Distribution of mismatched sgRNA relative activities stratified by position of the mismatch. Position –1 is immediately adjacent to the PAM. n = 1372–3374 sgRNAs. (f) Distribution of mismatched sgRNA relative activities stratified by type of mismatch, grouped by mismatches located in positions –19 to –13 (PAM-distal region), positions –12 to –9 (intermediate region), and positions –8 to –1 (PAM-proximal/seed region). Division into these regions was based on previous work13,16 and the patterns in panel e. n = 437–2342 sgRNAs. (g) Comparison of mean apparent on-rates measured in vitro for mismatched variants of a single sgRNA29 and mean relative activities from large-scale screen. Values are compared for identical combinations of mismatch type and mismatch position; mean relative activities were calculated by averaging relative activities for all mismatched sgRNAs with a given combination. Data are from n = 57 unique combinations of mismatch type and position; r2 = squared Pearson correlation coefficient. Lines in violin plots (e, f) denote distribution quartiles.
Mismatched sgRNAs mediated a range of phenotypes, spanning from that of the corresponding perfectly matched sgRNA to those of negative control sgRNAs (Fig. 2c). To account for differences in absolute growth phenotypes, we normalized the phenotype of each mismatched sgRNA to that of its corresponding perfectly matched sgRNA (relative activity, Fig. 2b) and filtered for series in which the perfectly matched sgRNA had a strong growth phenotype (Methods). Relative activities measured in K562 and Jurkat cells were well-correlated (Fig. 2d), regardless of differences in absolute phenotype of the perfectly matched sgRNAs (Fig. S2d–e). We therefore averaged relative activities from both cell lines for further analysis. Although the majority of mismatched sgRNAs were inactive (Fig. 2d), particularly if they contained two mismatches (Fig. S2f), approximately 25% of mismatched sgRNAs exhibited intermediate activity (relative activity 0.1–0.9).
To understand the rules governing the impacts of mismatches on activity, we stratified the relative activities of singly-mismatched sgRNAs by properties of the mismatch. As expected, mismatch position was a strong determinant of activity, with mismatches closer to the PAM leading to lower relative activity (Fig. 2e). In agreement with patterns of Cas9 off-target activity27,28, sgRNAs with rG:dT mismatches (A to G mutations in the sgRNA) retained substantial activity even for mismatches close to the PAM (Fig. 2f). Other factors had smaller effects on activity and were more context-dependent. For example, sgRNAs with higher GC content or for which the first, invariant G matched the genome retained higher activity for mismatches located 9 or more bases upstream of the PAM (positions –9 to –19), and mismatch-surrounding G nucleotides were associated with marginally higher activity for mismatches in the intermediate region (Fig. S2g–i). CRISPRi activities of mismatched sgRNAs were moderately correlated with Cas9 cutting scores in the presence of mismatches (CFD scores27), but Cas9 cutting appears to be less sensitive to many types of mismatches (Fig. S2j). By contrast, the CRISPRi activities of mismatched sgRNAs were well-correlated with previous in vitro measurements of dCas9 binding on-rates in the presence of mismatches29 (Fig. 2g). The activities of mismatched sgRNAs in CRISPRi thus appear to be determined by general biophysical rules; a premise further supported by the high correlation of relative activities obtained in different cell lines (Fig. 2d).
Overall, 86.7% of sgRNA series contained at least 2 sgRNAs with intermediate activity (relative activity 0.1–0.9, Fig. S2k). As we explored only ~20% of possible single mismatches and <1% of possible double mismatches, it is likely that intermediate-activity sgRNAs also exist for the remaining series. Altogether, these results suggest that systematically mismatched sgRNAs provide a general method to titrate CRISPRi activity and, consequently, target gene expression.
Controlling sgRNA activity with modified constant regions
We also explored the orthogonal approach of generating intermediate-activity sgRNAs through modifications to the sgRNA constant region, which is required for binding to Cas9. Although previous work has established that such modifications can lead to varied effects on Cas9 activity19,30–34, the mutational landscape of the constant region has only been sparsely explored, and largely with the goal of preserving sgRNA activity.
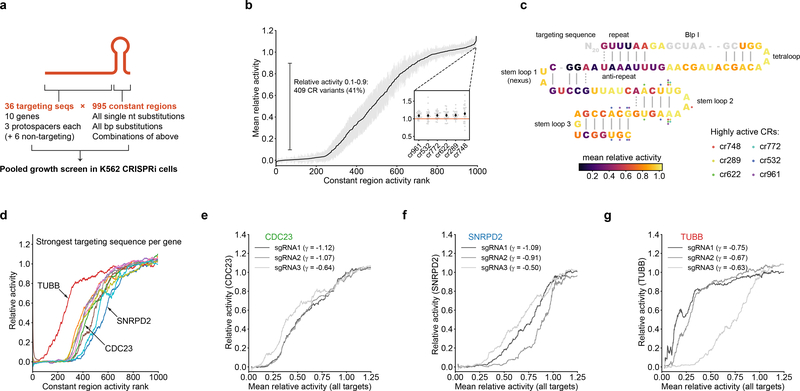
To comprehensively assess the activities of modified sgRNA constant regions, we designed 995 constant region variants comprising all single nucleotide substitutions, base pair substitutions, and combinations of these changes (Methods, Table S5) and determined the growth phenotypes for each variant paired with 30 targeting sequences against 10 essential genes in pooled CRISPRi screens in K562 cells (Fig. 3a, S3a; Tables S1, S6, S7). We calculated relative activities for each targeting sequence:constant region pair by normalizing its phenotype to that of the targeting sequence paired with the unmodified constant region, identifying 409 constant region variants that on average conferred intermediate activity (0.1–0.9, Fig. 3b). Ten variants selected for individual evaluation mediated intermediate mRNA knockdown (Fig. S3b). Mapping the activities of constant region variants with single base substitutions onto the structure recapitulated known relationships between constant region structure and function: mutation of bases in the first stem loop or the nexus that mediate contacts19 with Cas9 reduced activity, whereas mutations in regions not contacted by Cas9 (e.g. the hairpin region of stem loop 2) were well-tolerated (Fig. 3c). Notably, several variants carrying mutations in stem loop 2 had consistently increased activities (Fig. 3b–c).
Figure 3.
Identification and characterization of intermediate-activity constant regions. (a) Design of constant region variant library. (b) Mean relative activities of constant region variants, calculated by averaging relative activities for all targeting sequences; n = 995 constant region variants, gray margins denote 95% confidence interval of 30 targeting sequences. Inset: Focus on 6 constant region variants with higher activity than the original constant region. Black diamonds denote mean relative activity, gray dots denote relative activities of individual targeting sequences. (c) Mapping of constant region variant relative activities onto the constant region structure. Each constant region base is colored by the average relative activity of the three constant region variants carrying a single mutation at that position. Positions mutated in 6 highly active constant regions (inset in panel b) are indicated by colored dots. The BlpI site (gray) is used for cloning and was not mutated. (d) Constant region activities by targeting sequence, plotted against ranked mean constant region activity. For each gene, the activities with the strongest targeting sequence are shown as rolling means with a window size of 50. (e-g) Constant region activities by targeting sequence for all three targeting sequences against the indicated genes. Growth phenotypes (γ) of each targeting sequence paired with the unmodified constant region are indicated in the legend.
Evaluating the relative activities of constant region variants across the 30 targeting sequences revealed consistent rank ordering but substantial variation in the actual values (Fig. 3d, S3c). For example, a targeting sequence against TUBB retained high activity with ~100 constant region variants with which other targeting sequences lost activity, whereas a targeting sequence against SNRPD2 lost activity with ~50 variants that otherwise conferred intermediate activity (Fig. 3d). In some but not all cases (Fig. 3e), this heterogeneity extended to different targeting sequences against the same gene, both at the level of growth phenotype (Fig. 3f–g, S3d–e) and mRNA knockdown (Fig. S3b). This heterogeneous behavior could be a consequence of structural interactions between specific targeting sequences and constant regions or of differences in basal sgRNA expression levels such that lowly expressed sgRNAs are more susceptible to constant region modifications. Thus, although modified constant regions can be used to titrate gene expression, the activity of a given constant region:targeting sequence pair is difficult to predict. We therefore focused on sgRNAs with mismatches in the targeting region for the remainder of our work, given that the activities of these sgRNAs appeared to be governed directly by more readily discernible biophysical principles.
A neural network predicts mismatched sgRNA activities with high accuracy
We next sought to leverage our large-scale dataset of mismatched sgRNA activities to learn the underlying rules in a principled manner and enable predictions of intermediate-activity sgRNAs against other genes. We reasoned that a convolutional neural network (CNN) would be well-suited to uncovering these rules due to the ability of CNNs to learn complex global and local dependencies on spatially-ordered features such as nucleotide sequences35, including factors governing CRISPR guide RNA activity36,37.
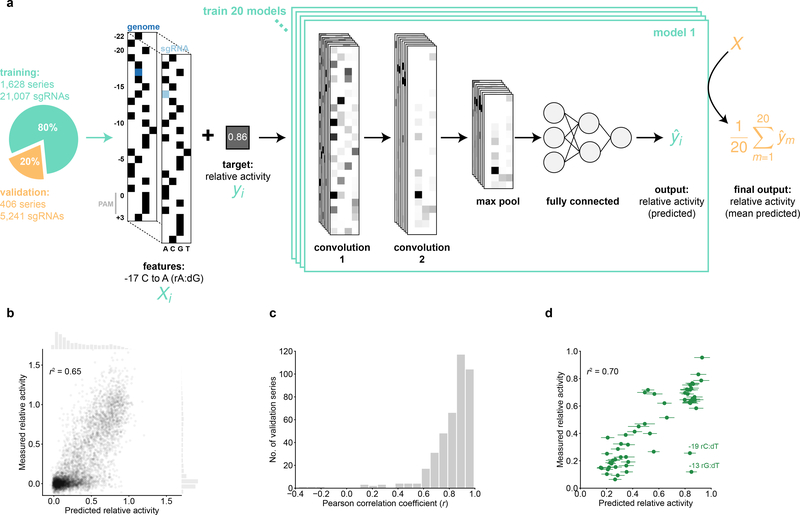
We constructed our CNN model using two convolution steps, a pooling step, and a 3-layer fully connected neural network (Fig. 4a, S4a). As inputs, the model received sgRNA relative activities paired with nucleotide sequences represented by binarized 3D arrays denoting the genomic sequence of the target and the associated sgRNA mismatch (Fig. 4a, Table S8). After optimizing hyperparameters using a cross-validated randomized grid search on the training dataset (80% of randomly selected sgRNA series, Fig. S4b–d, Methods), we trained 20 independent, equivalently initialized models for 8 epochs, which minimized loss without extensive over-fitting (Fig. S4e). Predicted and measured sgRNA relative activities for the validation sgRNA set (i.e. the remaining 20% of series that were not used to optimize parameters or train the model) were well-correlated (r2 = 0.65), with mean predictions of the 20-model ensemble outperforming all individual models (Fig. 4b, S4f). The correlation coefficients for individual sgRNA series were unimodally distributed (25th-75th percentile range: 0.77–0.93), indicating that the model performed comparably well for most series (Fig. 4c). Model accuracy varied by mismatch position and type, with the highest accuracies corresponding to mismatches in the PAM-proximal seed region (Fig. S4g–h). The accuracy of CNN predictions showed no correlation with off-target specificity scores, suggesting that off-target effects did not substantially contribute to the phenotypes we measured (Fig. S4i). Despite the fact that the model was trained on relative growth phenotypes, it accurately predicted relative fluorescence values measured in the GFP experiment (Fig. 4d), further supporting the hypothesis that relative growth phenotypes report on biophysical attributes of sgRNA:DNA interactions.
Figure 4.
Neural network predictions of sgRNA activity. (a) Schematic of a singly-mismatched sgRNA feature array (Xi) and the convolutional neural network architecture trained on pairs of such arrays and their corresponding relative activities (yi). Black squares in Xi represent the value 1 (the presence of a base at the indicated position); white represents 0. The mean prediction from 20 independently trained models was used to assign a final prediction (ŷ) to each sgRNA in the hold-out validation set (orange). (b) Comparison of measured relative growth phenotypes from the large-scale screen and predicted activities assigned by the neural network. Marginal histograms show distributions of relative activities along the corresponding axes. n = 5,241 sgRNAs; r2 = squared Pearson correlation coefficient. (c) Distribution of Pearson r values (predicted vs. measured relative activity) for each sgRNA series in the validation set. n = 406 series. (d) Comparison of measured relative activity (i.e. relative knockdown) in the GFP experiment and predicted relative sgRNA activity. Two outliers with lower-than-predicted activity are annotated with their respective mismatch position and type. Predictions are shown as mean ± S.D. from the 20-model ensemble. n = 57 sgRNAs; r2 = squared Pearson correlation coefficient.
To derive intermediate-activity sgRNAs for all human genes, we used the CNN ensemble to predict relative activities for all 57 singly-mismatched sgRNAs for the top 5 sgRNAs against each gene in the hCRISPRi-v2.1 library14 (Table S9). Based on the accuracy of predictions for the validation set, we estimate that for any given gene, sampling 3 sgRNAs with predicted relative activity between 0.37 and 0.63 will yield at least one sgRNA of intermediate activity (0.1–0.9) over 95% of the time (Fig. S4j–m). This resource should therefore enable the titration of any gene(s) of interest.
To further understand the features of mismatched sgRNAs that contribute most to their activity, which is difficult to assess directly with a deep learning model, we also trained an elastic net linear regression model on the same data using a curated set of features (Methods). This linear model explained less variance in relative activities than the CNN model (r2 = 0.52, Fig. S5a–b), implying that our feature set was incomplete and/or sgRNA activity is partly determined by non-linear combinations of features; nonetheless, the relative activities predicted by the different models were well-correlated (r2 = 0.74, Fig. S5c). Consistent with our earlier observations, mismatch position and type were assigned the largest weights in the model, although other features such as GC content and the identities of flanking bases up to 3 nucleotides from the mismatch were weighted as well (Fig. S5d–e). For any given position the type of mismatch contributed differentially to the prediction, which was especially pronounced in the sgRNA intermediate region (Fig. S5f). Taken together, these data demonstrate that the activities of mismatch-containing sgRNAs are determined by multiple factors which can be captured using supervised machine learning approaches.
A compact mismatched sgRNA library conferring intermediate phenotypes
We next set out to design a more compact version of our large-scale library to titrate essential genes with a limited number of sgRNAs. We selected 2,405 genes which we had found to be essential for robust growth of K562 cells in our large-scale screen, divided the relative activity space into six bins, and attempted to select mismatched variants from each of the center four bins (relative activities 0.1–0.9) for two sgRNA series targeting each gene. If a bin did not contain a previously measured sgRNA, we selected one from the CNN model ensemble predictions, filtered to exclude sgRNAs with off-target binding potential (Fig. 5a, S6a–c; Table S10).
Figure 5.
Compact mismatched sgRNA library targeting essential genes. (a) Design of library. For activity bins lacking a previously measured sgRNA, novel mismatched sgRNAs were included according to predicted activity. (b) Distribution of relative activities from the large-scale library (gray) and the compact library (purple) in K562 cells. The dashed line represents sgRNAs that were selected based on predicted activity from the deep learning model. (c) Comparison of relative activities of mismatched sgRNAs in HeLa and K562 cells. Marginal histograms show the distributions of relative activities along the corresponding axes. n = 9,514 sgRNAs; r2 = squared Pearson correlation coefficient.
We evaluated the relative activities of sgRNAs in this compact library using pooled CRISPRi growth screens in K562 and HeLa cells (Fig. S6d–f, Tables S11, S12). The correlation of measured and predicted relative activities of the imputed sgRNAs was lower than that observed for the validation set in our CNN model (r2 = 0.24; Fig. S6g), although the imputed sgRNAs were selected from predicted activity bins that are associated with higher model errors, and indeed the per-bin errors were consistent between the imputed sgRNAs and the CNN model validation set (Fig. S6h). Whereas the majority of mismatched sgRNAs in the large-scale screen were inactive, relative activities in the compact library were evenly distributed (Fig. 5b, S6i). Relative sgRNA activities measured in K562 cells were well-correlated with those measured in the large-scale screen (r2 = 0.7), and relative activities were also well-correlated between K562 and HeLa cells (r2 = 0.59, Fig. 5c). In addition, in a chemical-genetic screen in K562 cells for sensitivity to lovastatin, a potent HMG-CoA reductase inhibitor, even moderate-activity sgRNAs targeting HMGCR strongly reduced growth in the presence of lovastatin, suggesting that our approach could be used to probe drug-gene interactions (Fig. S6j–k, Tables S11, S12). Altogether, these data demonstrate that our library reproducibly provides access to intermediate phenotypes for this core gene set in multiple cell types.
Exploring expression-phenotype relationships with sgRNA series
Finally, we sought to use sgRNA series to explore expression-phenotype relationships for a diverse set of genes. To simultaneously measure gene expression levels and the resulting cellular phenotypes for multiple series, we used Perturb-seq, which enables matched capture of the transcriptome and the identity of an expressed sgRNA for each individual cell in pools of cells34,38–40 (Fig. S7a). We targeted 25 genes involved in essential cell biological processes (Table S13) with series of 5–6 sgRNAs (138 sgRNAs total including 10 non-targeting controls, Table S1). We then subjected pooled K562 CRISPRi cells expressing these sgRNAs from a modified CROP-seq vector40,41 to single-cell RNA-seq (scRNA-seq), using the sgRNA barcodes to assign unique sgRNA identities to ~19,600 cells (median 122 cells per sgRNA, Fig. S7b–c, Table S14). In addition to the single-cell transcriptomes, we measured the bulk growth phenotypes conferred by the sgRNAs in these cells, which were well-correlated with those from the large-scale screen and were used to assign sgRNA relative activities for further analysis (Methods, Fig. S7d–e, Tables S15, S16).
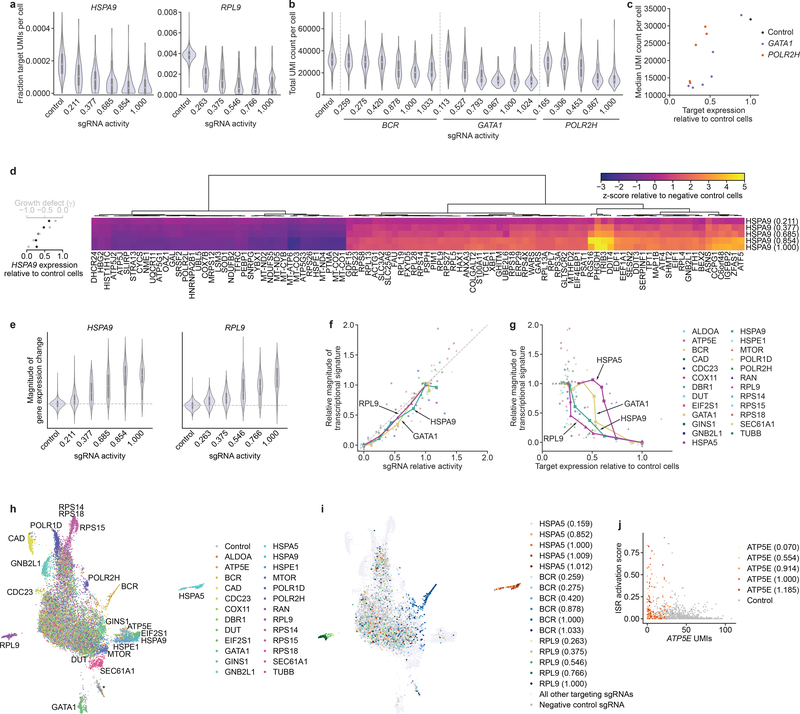
We first used the scRNA-seq data to assess the expression levels of each targeted gene. To account for cell-to-cell variability in transcript capture efficiency, we quantified target gene unique molecular identifier (UMI) counts as a fraction of total UMI count in a given cell (Fig. S8a), although analyzing raw UMI counts yielded similar results (Fig. S9). For approximately half of the genes targeted we were able to directly assess expression levels on the single-cell level (median >10 UMIs per cell, Fig. 6a, S8a). These expression levels were largely unimodally distributed, with medians shifting downwards with increasing sgRNA activity (Fig. 6a). For some genes, however, two populations with different knockdown levels were apparent (Fig. 6a, S8a). These populations were present both with intermediate-activity sgRNAs and the perfectly matched sgRNAs, suggesting that they did not result from limited knockdown penetrance for intermediate-activity sgRNAs. For genes with intermediate to low expression we typically observed 0–4 UMIs per cell, rendering the quantification of single-cell expression levels more difficult. We nonetheless observed a shift of the distribution to lower UMI numbers with increasing sgRNA activity (Fig. S8a, S9) as well as a decrease in mean expression levels when averaging expression across all cells with the same sgRNA (Fig. S8b).
Figure 6.
Rich phenotyping of cells with intermediate-activity sgRNAs by Perturb-seq. (a) Distributions of HSPA9 and RPL9 expression in cells with indicated perturbations. Expression is quantified as target gene UMI count normalized to total UMI count per cell. sgRNA activity is calculated using relative γ measurements from the Perturb-seq cell pool after 5 days of growth. (b) Distributions of total UMI counts in cells with indicated perturbations. (c) Comparison of median UMI count per cell and target gene expression in cells with GATA1- or POLR2H- targeting sgRNAs. (d) Right: Expression profiles of 100 genes in populations with HSPA9-targeting sgRNAs. Genes were selected by lowest FDR-corrected p-values in cells with the strongest sgRNA from a two-sided Kolmogorov-Smirnov test (Methods). Expression is quantified as z-score relative to population of cells with non-targeting sgRNAs. Left: Growth phenotype and knockdown for each sgRNA. (e) Distribution of gene expression changes in populations with indicated sgRNAs. Magnitude of gene expression change is calculated as sum of z-scores of genes differentially expressed in the series (FDR-corrected p < 0.05 with any sgRNA in the series, two-sided Kolmogorov-Smirnov test, Methods), with z-scores of individual genes signed by the direction of change in cells with the perfectly matched sgRNA. Distribution for negative control sgRNAs is centered around 0 (dashed line).
For a-e, the cell numbers for each perturbation are listed in Table S14. Box plots inside violin plots denote quartile ranges (box), median (center mark), and 1.5 × interquartile range (whiskers).
(f) Comparison of relative growth phenotype and magnitude of gene expression change for all individual sgRNAs. Growth phenotype and magnitude of gene expression change are normalized in each series to those of the sgRNA with the strongest knockdown. (g) Comparison of magnitude of gene expression change and target gene knockdown, as in f. (h) UMAP projection of all single cells with assigned sgRNA identity in the experiment, colored by targeted gene. Clusters clearly assignable to a genetic perturbation are labeled. Cluster labeled * contains a small number of cells with residual stress response activation and could represent apoptotic cells. Note that ~5% cells appear to have confidently but incorrectly assigned sgRNA identities (Methods). Given the strong trends in the other results, we concluded that such misassignment did not substantially affect our ability to identify trends within cell populations and in the future may be avoided by approaches to directly capture the expressed sgRNA41. n = 19,587 cells. (i) UMAP projection, as in h, with selected series colored by sgRNA activity. n = 19,587 cells. (j) Comparison of extent of ISR activation to ATP5E UMI count in cells with knockdown of ATP5E or control cells.
Titration is also apparent at the level of the transcriptional responses, which provide a robust single-cell measurement of the phenotype induced by depletion of the targeted gene. In the simplest cases, knockdown led to substantial reductions in cellular UMI counts, consistent with large-scale inhibition of mRNA transcription (Fig. 6b, Fig. S10a). Examples include GATA1, a central myeloid lineage transcription factor, POLR2H, a core subunit of RNA polymerase II (and RNA polymerases I and III), or to a lesser extent BCR, which is fused to the driver oncogene ABL1 in K562 cells. Notably, the reduction in UMI counts correlated linearly with growth phenotype within sgRNA series (Fig. 6b, Fig. S10b) but exhibited non-linear relationships with target gene knockdown at least in the cases of GATA1 and POLR2H (Fig. 6c, S10b, BCR levels are difficult to quantify accurately). Both relationships appeared to be sigmoidal but with different thresholds: whereas cellular UMI counts dropped sharply once GATA1 mRNA levels were reduced by 50%, a larger reduction of POLR2H mRNA levels was required to achieve a similarly sized effect.
Knockdown of most other targeted genes did not perturb total UMI counts to the same extent (Fig. S10a) but resulted in other transcriptional responses. Knockdown of CAD, for example, triggered cell cycle stalling during S-phase, consistent with previous observations34, with a higher frequency of stalling with increasing sgRNA activity (Fig. S10c, S10d). Knockdown of HSPA9, the mitochondrial Hsp70 isoform, induced the expected transcriptional signature corresponding to activation of the integrated stress response (ISR) including upregulation of DDIT3 (CHOP), DDIT4, ATF5, and ASNS34,42. The magnitude of this transcriptional signature increased with increasing sgRNA activity on both the population (Fig. 6d) and the single-cell level (Fig. 6e), although populations with intermediate-activity sgRNAs had larger cell-to-cell variation in response magnitude. Similarly, the transcriptional responses to knockdown of other genes scaled with sgRNA activity and exhibited larger variance for intermediate-activity sgRNAs (Fig. 6e).
We next compared the expression levels of the targeted gene to the magnitudes of the resulting phenotypes. Within each series, two metrics of phenotype, bulk population growth phenotype and transcriptional response, were well-correlated, despite substantial differences in the absolute magnitudes of the transcriptional responses with different series (Fig. 6f, S10e–g). By contrast, the relationships between either metric of phenotype and target gene expression were strongly gene-specific (Fig. 6g, Fig. S10h–j). For HSPA5 and GATA1, for example, a reduction in mRNA levels by ~50% was sufficient to induce a near-maximal transcriptional response and growth defect, whereas for most other genes a larger reduction was required. These results suggest that K562 cells are intolerant to moderate decreases in expression of GATA1 and HSPA5, with sharp transitions from growth to death once expression levels drop below a threshold. More broadly, these results highlight the utility of titrating gene expression to map expression-phenotype relationships and quantitatively define gene expression sufficiency.
Following single-cell trajectories along a continuum of gene expression levels
To gain further insight into the diversity of responses induced by depletion of essential genes, we compared the transcriptional profiles induced by each individual sgRNA. Averaging transcriptional profiles across all cells with the same sgRNA and clustering the resulting mean profiles revealed multiple groups segregated by biological function, including a cluster of ribosomal proteins and POLR1D, a subunit of the rRNA-transcribing RNA polymerase I (and of RNA polymerase III), and a cluster of perturbations that activate the ISR (HSPA9, HSPE1, and EIF2S1/eIF2α, Fig. S11a). To further visualize the space of transcriptional states, we performed dimensionality reduction on the single-cell transcriptomes using UMAP43. The resulting projection recapitulated the clustering, as indicated for example by the close proximity of cells with perturbations of HSPA9, HSPE1, and EIF2S1 (Fig. 6h). Within individual series, cells projected further outward in UMAP space with increasing sgRNA activity, further highlighting the titration of gene expression levels on the single-cell level (Fig. 6i).
Closer examination of the UMAP projection revealed more granular structure, including the grouping of a subset of cells with knockdown of ATP5E, a subunit of ATP synthase, with cells with ISR-activating perturbations (Fig. 6h). This subset of cells indeed exhibited classical features of ISR activation (Fig. S11b). The frequency of ISR activation increased with lower ATP5E mRNA levels, but even at the lowest levels some cells did not exhibit ISR activation (Fig. 6j, S11b). These results suggest that depletion of ATP synthase under these conditions predisposes cells to ISR activation, perhaps by exacerbating transient phases of mitochondrial stress in a manner that is proportional to ATP synthase levels. More broadly, these results highlight the utility of titrating gene expression in probing cell biological phenotypes, especially in conjunction with rich phenotyping methods such as scRNA-seq.
Discussion
Here we describe the development of an approach to systematically titrate gene expression in human cells using allelic series of attenuated sgRNAs. These series, either individually or as a pool, have a broad range of applications across basic and biomedical research. We highlight the utility of the approach by mapping gene expression levels to phenotypes with single-cell resolution, enabling identification of gene-specific viability thresholds and expression level-dependent cell fates.
Our approach builds on in vitro work describing the biophysical principles by which modifications to the sgRNA modulate (d)Cas9 binding on-rates and activity16,28,29,44,45. In cells, modifications to the sgRNA constant region were affected by specific interactions with targeting sequences, rendering sgRNA activities difficult to predict. By contrast, the effects of targeting sequence mismatches on sgRNA activity followed readily discernible biophysical principles, enabling us to apply machine learning approaches to derive the underlying rules and predict series for arbitrary sgRNAs. The resulting genome-wide in silico library (Table S9) enables titration of any expressed gene. We also describe a compact (~25,000-element) library that enables titration of ~2,400 essential genes (Table S10), with potential applications for example in focused screens for sensitization to chemical or genetic perturbations. Our approach yields intermediate-activity sgRNAs in a predictable manner, is readily scalable to target any number of genes, by contrast to approaches that titrate gene expression using microRNAs or synthetic biology tools, and provides access to many expression levels of each gene in a single pooled experiment, by contrast to approaches that rely on small molecules to control (d)Cas9 activity. The sgRNA activities also hold across different cell models, suggesting that the approach should be widely applicable to models in which CRISPRi is available, including primary cell models such as iPSCs or organoids23,46,47. In these settings, combining sgRNA series with single-cell readout can circumvent limitations such as small cell numbers and low transduction efficiency, as meaningful phenotypes can be extracted from far fewer cells than typically needed for bulk readouts.
These sgRNA series now enable systematic mapping of expression-phenotype relationships directly in mammalian systems, with implications for human genetics, evolutionary biology, and disease biology. As an example, we highlight how the minimal expression levels that sustain cell growth vary for different genes, with K562 cells being particularly sensitive to depletion of GATA1 and HSPA5. This variability suggests gene-specific buffering capacities, in line with findings in yeast4, but the logic by which these buffering capacities are determined in mammalian systems remains unclear. Comprehensive efforts to generate such dose-response curves across cell models could begin to reveal the underlying principles that have shaped gene expression levels. Analogous efforts to map dose-response curves in cancer cells could identify specific vulnerabilities as targets for therapeutics and, vice versa, mapping these curves for cancer driver genes or genes underlying specific diseases could enable defining the corresponding therapeutic windows as goals for drug development.
Our intermediate-activity sgRNAs also provide access to diverse cell states including loss-of-function phenotypes that otherwise may be obscured by cell death or neomorphic behavior. Thus, our approach enables positioning cells at states of interest to record chemical-gene or gene-gene interactions or to characterize transcriptional trajectories near phenotypic transitions. These sgRNA series will also facilitate recapitulating gene expression levels of disease-relevant states such as haploinsufficiency or partial loss-of-function, enabling efforts to identify suppressors or modifiers, or modeling quantitative trait loci associated with multigenic traits in conjunction with rich phenotyping to identify the mechanisms by which they interact and contribute to such traits. Finally, mismatched sgRNAs can be used to titrate dCas9 occupancy and activity in other applications such as CRISPRa or other dCas9-based epigenetic modifiers.
In summary, our allelic series approach provides a scalable tool to titrate gene expression and evaluate dose-response relationships in mammalian systems. This resource should be equally enabling to systematic large-scale efforts and detailed single-gene investigations in basic cell biology, drug development, and functional genomics.
Online methods
Reagents and cell lines
K562 and Jurkat cells were grown in RPMI 1640 medium (Gibco) with 25 mM HEPES, 2 mM l-glutamine, 2 g/L NaHCO3 supplemented with 10% (v/v) standard fetal bovine serum (FBS, HyClone or VWR), 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (Gibco). HEK293T and HeLa cells were grown in Dulbecco’s modified eagle medium (DMEM, Gibco) with 25 mM d-glucose, 3.7 g/L NaHCO3, 4 mM l-glutamine and supplemented with 10% (v/v) FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine. K562 (chronic myelogenous leukemia) and HeLa (cervical carcinoma) cells are derived from female patients. Jurkat (acute T-cell lymphocytic leukemia) cells are derived from a male patient. HEK293T (embryonic kidney) cells are derived from a female fetus. The K562 and HeLa CRISPRi cell lines and the GFP+ K562 CRISPRi cell line were previously published18,25,34. Jurkat CRISPRi cells (Clone NH7) were obtained from the Berkeley Cell Culture Facility. All cell lines were grown at 37 °C in the presence of 5% CO2. All cell lines were periodically tested for Mycoplasma contamination using the MycoAlert Plus Mycoplasma detection kit (Lonza).
DNA transfections and virus production
Lentivirus was generated by transfecting HEK239T cells with four packaging plasmids (for expression of VSV-G, Gag/Pol, Rev, and Tat, respectively) as well as the transfer plasmid using TransIT-LT1 Transfection Reagent (Mirus Bio). Viral supernatant was harvested two days after transfection and filtered through 0.44 μm PVDF filters and/or frozen prior to transduction.
Cloning of individual sgRNAs
Individual perfectly matched or mismatched sgRNAs were cloned essentially as described previously18. Briefly, two complementary oligonucleotides (Integrated DNA Technologies), containing the targeting region as well as overhangs matching those left by restriction digest of the backbone with BstXI and BlpI, were annealed and ligated into an sgRNA expression vector digested with BstXI (NEB or Thermo Fisher Scientific) and BlpI (NEB) or Bpu1102I (Thermo Fisher Scientific). The ligation product was transformed into Stellar chemically competent E. coli cells (Takara Bio) and plasmid was prepared following standard protocols.
Individual evaluation of sgRNA phenotypes for GFP knockdown
For individual evaluation of GFP knockdown phenotypes, sgRNAs were individually cloned as described above, ligated into a version of pU6-sgCXCR4–2 (marked with a puromycin resistance cassette and mCherry, Addgene #46917)25, modified to include a BlpI site. Sequences used for individual evaluation are listed in Table S1. The sgRNA expression vectors were individually packaged into lentivirus and transduced into GFP+ K562 CRISPRi cells34 at MOI < 1 (15 – 40% infected cells) by centrifugation at 1000 × g and 33 °C for 0.5–2 h. GFP levels were recorded 10 d after transduction by flow cytometry using a FACSCelesta flow cytometer (BD Biosciences), gating for sgRNA-expressing cells (mCherry+). Experiments were performed in duplicate from the transduction step. Relative activities were defined as the fold-knockdown of each mismatched variant (GFPsgRNA[non-targeting] / GFPsgRNA[variant]) divided by the fold-knockdown of the perfectly-matched sgRNA. The background fluorescence of a GFP– strain was subtracted from all GFP values prior to other calculations. Data were analyzed in Python 2.7 using the FlowCytometryTools package (v0.5.0). The distributions of GFP values in Fig. 1B were plotted following the example in https://seaborn.pydata.org/examples/kde_ridgeplot.
Design of large-scale mismatched sgRNA library
To generate the list of targeting sgRNAs for the large-scale mismatched sgRNA library, hit genes from a growth screen performed in K562 cells with the CRISPRi v2 library14 were selected by calculating a discriminant score (phenotype z-score × –log10(Mann-Whitney P)). Discriminant scores for negative control genes (randomly sampled groups of 10 non-targeting sgRNAs) were calculated as well, and hit genes were selected above a threshold such that 5% of the hits would be negative control genes (i.e. an estimated empirical 5% FDR). This procedure resulted in the selection of 2477 genes. Of these genes, 28 genes for which the second strongest sgRNA by absolute value had a positive growth phenotype were filtered out as these were likely to be scored as hits solely due to a single sgRNA. For the remaining 2,449 genes, the two sgRNAs with the strongest growth phenotype were selected, for a total of 4,898 perfectly matched sgRNAs.
For each of these sgRNAs, a set of 23 variant sgRNAs with mismatches was designed: 5 with a single randomly chosen mismatch within 7 bases of the PAM, 5 with a single randomly chosen mismatch 8–12 bases from the PAM, and 3 with a single randomly chosen mismatch 13–19 bases from the PAM (the first base of the targeting region was never selected for this purpose as it is an invariant G in all sgRNAs to enable transcription from the U6 promoter). The remaining 10 variants had 2 randomly chosen mismatches selected from positions –1 to –19. The compiled sgRNA sequences were then filtered for sgRNAs containing BstXI, BlpI, and SbfI sites, which are used during library cloning and sequencing library preparation, and 2,500 negative controls (randomly generated to match the base composition of our hCRISPRi-v2 library) were added. Note that the first base of all sgRNAs was fixed as a G, regardless of whether or not it matched the genome, consistent with the design of our hCRISPRi-v2 library14. Sequences of sgRNAs and descriptions of mismatches are listed in Table S2.
Assessment of off-target potential
To assess the off-target potential of mismatched sgRNAs, we first extended our previous strategy to estimate sgRNA off-target effects14,18. Briefly, for each target in the genome, a FASTQ entry was created for the 23 bases of the target including the PAM, with the accompanying empirical Phred score indicating an estimate of the anticipated importance of a mismatch in that base position. Bowtie (http://bowtie-bio.sourceforge.net)48 was then used to align each designed sgRNA back to the genome (or a subset of the genome solely encompassing annotated transcription start sites flanked by 500 base pairs), parameterized so that sgRNAs were considered to mutually align if and only if: a) no more than 3 mismatches existed in the PAM-proximal 12 bases and the PAM, b) the summed Phred score of all mismatched positions across the 23 bases was less than a threshold. This alignment was done iteratively with decreasing thresholds, and any sgRNAs which aligned successfully to no other site in the genome at a particular threshold were then deemed to have a specificity at said threshold.
Subsequently, the empirical measurements of relative activities of CRISPRi sgRNAs in the presence of mismatches from our large-scale screen afforded the opportunity to calculate the off-target potential in a more nuanced manner, akin to the methods used to measure off-target potential for CRISPR cutting as implemented for example in GuideScan49. Briefly, we used Cas-OFFinder50 to first find all potential off-target sites up to 3 mismatches away for each sgRNA. We then aggregated these off-target sites into a specificity score for each sgRNA:
Where n represents the number of sites with up to 3 mismatches, RA the empirically measured relative CRISPRi activity of each sgRNA at this target site given the positions and types of mismatches, and q the number of times the ith site occurs in the genome. In particular, RA was calculated as follows:
Where m represents the number of mismatches between the sgRNA and the target site and RAj represents the mean relative activity of sgRNAs with mismatch j (given mismatch type at given sgRNA position). An equivalent methodology was previously used to assess off-target potential of sgRNAs in CRISPR cutting27,49. If the “mismatched site” was the intended on-target site (because many of our sgRNAs contained mismatches to the intended on-target site), we instead assigned it RA = 1 to keep specificity scores on a scale of 0 to 1. A specificity score of 1 indicates that there are no off-target sites with up to 3 mismatches in the genome.
We also calculated equivalent specificity scores using the empirically measured CFD scores, which were determined by measuring cutting frequency at mismatched sites27. Note that CRISPR cutting appears to be less sensitive to mismatches (see also Fig. S2j), and thus specificity scores calculated using CFD scores are frequently lower than those calculated using relative CRISPRi activities.
We also note that the off-target potential calculated in this manner is likely overestimated, as binding of CRISPRi sgRNAs in most regions of the genome outside of promoters/TSSs, enhancers, or similar regions is relatively innocuous. Nonetheless, these off-target specificity scores can serve as guidelines in sgRNA selection. All four off-target scoring metrics (Bowtie threshold genome-wide, Bowtie threshold near TSSs only, off-target specificity score calculated using CRISPRi relative activities, off-target specificity score calculated using CFDs) are included in Table S2 as well as in Tables S9 and S10.
Pooled cloning of mismatched sgRNA libraries
Pooled sgRNA libraries were cloned largely as described previously18,26,51. Briefly, oligonucleotide pools containing the desired elements with flanking restriction sites and PCR adapters were obtained from Agilent Technologies. The oligonucleotide pools were amplified by 15 cycles of PCR using Phusion polymerase (NEB). The PCR product was digested with BstXI (Thermo Fisher Scientific) and Bpu1102I (Thermo Fisher Scientific), purified, and ligated into BstXI/Bpu1102I-digested pCRISPRia-v2 (marked with a puromycin resistance cassette and BFP, Addgene #84832)14 at 16 °C for 16 h. The ligation product was purified by isopropanol precipitation and then transformed into MegaX DH10B electrocompetent cells (Thermo Fisher Scientific) by electroporation using the Gene Pulser Xcell system (Bio-Rad), transforming ~100 ng purified ligation product per 100 μL cells. The cells were allowed to recover in 3–6 mL SOC medium for 2 h. At that point, a small 1–5 μL aliquot was removed and plated in three serial dilutions on LB plates with selective antibiotic (carbenicillin). The remainder of the culture was inoculated into 0.5 to 1 L LB supplemented with 100 μg/mL carbenicillin, grown at 37 °C with shaking at 220 rpm for 16 h and harvested by centrifugation. Colonies on the plates were counted to confirm a transformation efficiency greater than 100-fold over the number of elements (>100x coverage). The pooled sgRNA plasmid library was extracted from the cells by GigaPrep (Qiagen or Zymo Research). Even coverage of library elements was confirmed by sequencing a small aliquot on a HiSeq 4000 (Illumina).
Large-scale mismatched sgRNA screen and sequencing library preparation
Large-scale screens were conducted similarly to previously described screens14,18,26. The large-scale library was transduced in duplicate into K562 CRISPRi and Jurkat CRISPRi cells at MOI <1 (percentage of transduced cells 2 days after transduction: 20–40%) by centrifugation at 1000 × g and 33 °C for 2 h. Replicates were maintained separately in 0.5 L to 1 L of RPMI-1640 in 1 L spinner flasks for the course of the screen. 2 days after transduction, the cells were selected with puromycin for 2 days (K562: 2 days of 1 μg/mL; Jurkat: 1 day of 1 μg/mL and 1 day of 0.5 μg/mL), at which point transduced cells accounted for 80–95% of the population, as measured by flow cytometry using an LSR-II flow cytometer (BD Biosciences). Cells were allowed to recover for 1 day in the absence of puromycin. At this point, t0 samples with a 3000x library coverage (400 × 106 cells) were harvested and the remaining cells were cultured further. The cells were maintained in spinner flasks by daily dilution to 0.5 × 106 cells/mL at an average coverage of greater than 2000 cells per sgRNA with daily measurements of cell numbers and viability on an Accuri bench-top flow cytometer (BD BioSciences) for 11 days, at which point endpoint samples were harvested by centrifugation with 3000x library coverage.
Genomic DNA was isolated from frozen cell samples and the sgRNA-encoding region was enriched, amplified, and processed for sequencing essentially as described previously14. Briefly, genomic DNA was isolated using a NucleoSpin Blood XL kit (Macherey-Nagel), using 1 column per 100 × 106 cells. The isolated genomic DNA was digested with 400 U SbfI-HF (NEB) per mg DNA at 37 °C for 16 h. To isolate the ~500 bp fragment containing the sgRNA expression cassette liberated by this digest, size separation was performed using large-scale gel electrophoresis with 0.8% agarose gels. The region containing DNA fragments between 200 and 800 bp was excised and DNA was purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). The isolated DNA was quantified using a Qubit Fluorometer (Thermo Fisher Scientific) and then amplified by 23 cycles of PCR using Phusion polymerase (NEB), appending Illumina adapter and unique sample indices in the process. Each DNA sample was divided into 5–50 individual 100 μL reactions, each with 500 ng DNA as input. To ensure base diversity during sequencing, the samples were divided into two sets, with all samples for a given replicate always being assigned to the same set. The two sets had the Illumina adapters appended in opposite orientations, such that samples in set A were sequenced from the 5′ end of the sgRNA sequence in the first 20 cycles of sequencing and samples in set B were sequenced from the 3′ end of the sgRNA sequence in the next 20 cycles of sequencing. With updates to Illumina chemistry and software, this strategy is no longer required to ensure high sequencing quality, and all samples are amplified in the same orientation. Following the PCR, all reactions for a given DNA sample were combined and a small aliquot (100–300 μL) was purified using AMPure XP beads (Beckman-Coulter) with a two-sided selection (0.65x followed by 1x). Sequencing libraries from all samples were combined and sequencing was performed on a HiSeq 4000 (Illumina) using single-read 50 runs and with two custom sequencing primers (oCRISPRi_seq_V5 and oCRISPRi_seq_V4_3′, Table S17). For samples that were amplified in the same orientation, only a single custom sequencing primer was added (oCRISPRi_seq_V5), and the samples were supplemented with a 5% PhiX spike-in.
Sequencing reads were aligned to the library sequences, counted, and quantified using the Python-based ScreenProcessing pipeline (https://github.com/mhorlbeck/ScreenProcessing). Calculation of phenotypes was performed as described previously14,18,26. Untreated growth phenotypes (γ) were derived by calculating the log2 change in enrichment of an sgRNA in the endpoint and t0 samples, subtracting the equivalent median value for all non-targeting sgRNAs, and dividing by the number of doublings of the population18,26. For sgRNAs with a read count of 0, a pseudocount of 1 was added. sgRNAs with <50 reads in both the endpoint and t0 samples in a given replicate were excluded from analysis. Read counts and phenotypes for individual sgRNAs are available in Table S3 and Table S4, respectively. To calculate relative activities, phenotypes of mismatched sgRNAs were divided by those for the corresponding perfectly matched sgRNA. Relative activities were filtered for series in which the perfectly matched sgRNA had a growth phenotype greater than 5 z-scores outside the distribution of negative control sgRNAs for all further analysis (3,147 and 2,029 sgRNA series for K562 and Jurkat cells, respectively). Relative activities from both cell lines were averaged if the series passed the z-score filter in both. All analyses were performed in Python 2.7 using a combination of Numpy (v1.14.0), Pandas (v0.23.4), and Scipy (v1.1.0).
Design and pooled cloning of constant region variants library
The sequences in the library of modified constant regions were derived from the sgRNA (F+E) optimized sequence30 modified to include a BlpI site18. Each modified constant region was paired with 36 sgRNA targeting sequences (3 sgRNAs targeting each of 10 essential genes and six non-targeting negative control sgRNAs). The cloning strategy (described below) allowed the mutation of most positions in the sgRNA constant region. A variety of modifications were made, including substitutions of all single bases not in the BlpI restriction site (which is used for cloning), double substitutions including all substitutions at base-paired position pairs not before or in the BlpI site, and a variety of triple, quadruple, and sextuple substitutions, including base-pair-preserving substitutions at adjacent base-pairs.
The library was ordered and cloned in two parts. One part consisted of ~100 modifications to the eight bases upstream of the BlpI restriction site. Constant region variants with mutations in this section were paired with each of the 36 targeting sequences, ordered as a pooled oligonucleotide library (Twist Biosciences), and cloned into pCRISPRia-v2 as described above. The second part consisted of ~900 modifications to the 71 bases downstream of the BlpI restriction site. This part was cloned in two steps. First, all 36 targeting sequences were individually cloned into pCRISPRia-v2 as described above. The vectors were then pooled at an equimolar ratio and digested with BlpI (NEB) and XhoI (NEB). The modified constant region variants were ordered as a pooled oligonucleotide library (Twist Biosciences), PCR amplified with Phusion polymerase (NEB), digested with BlpI (NEB) and XhoI (NEB), and ligated into the digested vector pool, in a manner identical to previously published protocols and as described above, except for the different restriction enzymes.
Compact mismatched sgRNA library and constant region library screens
Screens with the compact mismatched sgRNA library and the constant region library were conducted largely as described above, with smaller modifications during the screening procedure and an updated sequencing library preparation protocol. Briefly, the libraries were transduced in duplicate into K562 CRISPRi (both libraries) or HeLa CRISPRi cells (compact mismatched sgRNA library) as described above. K562 replicates were maintained separately in 0.15 to 0.3 L of RPMI-1640 in 0.3 L spinner flasks for the course of the screen. HeLa replicates were maintained in sets of ten 15-cm plates. Cells were selected with puromycin as described above (K562: 1 day of 0.75 μg/mL and 1 day of 0.85 μg/mL; HeLa: 2 days of 0.8 μg/mL and 1 day of 1 μg/mL). The remainder of the screen was carried out at >1000x library coverage (K562 compact mismatched sgRNA library: >2000x; HeLa compact mismatched sgRNA library: >1000x; K562 constant region library: >2000x). For the drug screen, 10 μM lovastatin (ApexBio) or an equivalent volume of DMSO (vehicle) was added to flasks at t=0, and 3 days later cells were pelleted and re-suspended in fresh medium. Lovastatin (12 μM) or DMSO was again added after 5 and 9 days of growth, with media exchanges 3 days after drug supplementation. Multiple samples were harvested after 4 to 8 days for the K562 and HeLa growth screens. Both drug-treated and vehicle-treated samples were harvested after 12 days for the drug screen, which allowed for a difference of 3.5 to 4.1 cell population doublings between drug- and vehicle-treated groups.
Genomic DNA was isolated from frozen cell samples as described above. The subsequent sequencing library preparation was simplified to omit the enrichment step by gel extraction. In particular, following the genomic DNA extraction, DNA was quantified by absorbance at 260 nm using a NanoDrop One spectrophotometer (Thermo Fisher Scientific) and then directly amplified by 22–23 cycles of PCR using NEBNext Ultra II Q5 PCR MasterMix (NEB), appending Illumina adapter and unique sample indices in the process. Each DNA sample was divided into 50–200 individual 100 μL reactions, each with 10 μg DNA as input. All samples were amplified using the same strategy and in the same orientation. The PCR products were purified as described above and sequencing libraries from all samples were combined. For the compact mismatched library screens, sequencing was performed on a HiSeq 4000 (Illumina) using single-read 50 runs with a 5% PhiX spike-in and a custom sequencing primer (oCRISPRi_seq_V5, Table S17). For the constant region screens, the PCR primers were adapted to allow for amplification of the entire constant region and to append a standard Illumina read 2 primer binding site (Table S17). Sequencing was then performed in the same manner including the custom sequencing primer (oCRISPRi_seq_v5) and a 5% PhiX spike-in, but using paired-read 150 runs.
Sequencing reads were processed as described above, except that sgRNAs with <50 reads (compact mismatched sgRNA library) or <25 reads (constant region library) in both the endpoint and t0 samples in a given replicate or with a read count of 0 in either sample were excluded from analysis. Read counts and phenotypes for individual sgRNAs are available in Tables S6–S7 (constant region screen) and Tables S11–S12 (compact mismatched sgRNA library screen).
Generation and evaluation of individual constant region variants by RT-qPCR
Constant region variants were evaluated in the background of a constant region with an additional base pair substitution in the first stem loop (fourth base pair changed from AT to GC32). Ten constant region variants with average relative activities between 0.2 and 0.8 from the screen and carrying substitutions after the BlpI site were selected (Table S17). Cloning of individual constant regions was performed essentially as the cloning of sgRNA targeting regions, described above, except that the BlpI and XhoI restriction sites were used for cloning (the XhoI site is immediately downstream of the constant region) and that cloning was performed with a variant of pCRISPRia-v2 with the modified stem loop. For each of the ten constant region variants as well as the constant region carrying only the stem loop substitution, two different targeting regions against DPH2 were then cloned as described above (Table S1). These 22 vectors as well as a vector with a non-targeting negative control sgRNA (Table S1) were individually packaged into lentivirus and transduced into K562 CRISPRi cells at MOI < 1 (10 – 50% infected cells) by centrifugation at 1000 × g and 33 °C for 2 h. Cells were allowed to recover for 2 days and then selected to purity with puromycin (1.5 – 3 μg/mL), as assessed by measuring the fraction of BFP-positive cells by flow cytometry on an LSR-II (BD Biosciences), allowed to recover for 1 day, and harvested in aliquots of 0.5 – 2 × 106 cells for RNA extraction. RNA was extracted using the RNeasy Mini kit (Qiagen) with on-column DNase digestion (Qiagen) and reverse-transcribed using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific) with oligo(dT) primers in the presence of RNaseOUT Recombinant Ribonuclease Inhibitor (Thermo Fisher Scientific). Quantitative PCR (qPCR) reactions were performed in 22 μL reactions by adding 20 μL master mix containing 1.1x Colorless GoTaq Reaction Buffer (Promega), 0.7 mM MgCl2, dNTPs (0.2 mM each), primers (0.75 μM each), and 0.1x SYBR Green with GoTaq DNA polymerase (Promega) to 2 μL cDNA or water. Reactions were run on a LightCycler 480 Instrument (Roche). For each cDNA sample, reactions were set up with qPCR primers against DPH2 and ACTB (sequences listed in Table S17). Experiments were performed in technical triplicates.
Machine learning
In order to establish a subset of highly active sgRNAs with which to train a machine learning model, we filtered for perfectly matched sgRNAs with a growth phenotype greater than 10 z-scores outside the distribution of negative control sgRNAs in the K562 and/or Jurkat pooled screens (K562 γ < –0.21; Jurkat γ < –0.35). All singly-mismatched variants derived from sgRNAs passing the filter were then included, and relative activities were calculated as described previously, averaging the replicate measurements for each sgRNA. In cases where a perfectly matched sgRNA passed the filter in the K562 and Jurkat screen, the average relative activity across both cell types was calculated for each mismatched variant; otherwise the relative activities for only one cell type were considered. This filtering scheme resulted in 26,248 mismatched sgRNAs comprising 2,034 series targeting 1,292 genes, with approximately 40% of relative activity values averaged from K562 and Jurkat cells.
For each sgRNA, a set of features was defined based on the sequences of the genomic target and the mismatched sgRNA. First, the genomic sequence extending from 22 bases 5′ of the beginning of the PAM to 1 base 3′ of the end of the PAM (26 bases in all) was binarized into a 2D array of shape (4, 26), with 0s and 1s indicating the absence or presence of a particular nucleotide at each position, respectively. Next, a similar array was constructed representing the mismatch imparted by the sgRNA, with an additional potential mismatch at the 5′ terminus of the sgRNA (position –20), which invariably begins with G in our libraries due to the U6 promoter. Thus, the mismatched sequence array was identical to the genomic sequence array except for 1 or 2 positions. Finally, the arrays were stacked into a 3D volume of shape (4, 26, 2), which served as the feature set for that particular sgRNA.
The training set of sgRNAs was established by randomly selecting 80% of sgRNA series, with the remaining 20% set aside for model validation. A convolutional neural network (CNN) regression model was then designed using Keras (https://keras.io/) with a TensorFlow backend engine, consisting of two sequential convolution layers, a max pooling layer, a flattening layer, and finally a three-layer fully connected network terminating in a single neuron. Additional regularization was achieved by adding dropout layers after the pooling step and between each fully connected layer. To penalize the model for ignoring under-represented sgRNA classes (e.g. those with intermediate relative activity), training sgRNAs were binned according to relative activity, and sample weights inversely proportional to the population in each bin were assigned. Hyperparameters were optimized using a randomized grid search with 3-fold cross-validation with the training set as input. Parameters included the size, shape, stride, and number of convolution filters, the pooling strategy, the number of neurons and layers in the dense network, the extent of dropout applied at each regularization step, the activation functions in each layer, the loss function, and the model optimizer. Ultimately, 20 CNN models with identical starting parameters were individually trained for 8 epochs in batches of 32 sgRNAs. Performance was assessed by computing the average prediction of the 20-model ensemble for each validation sgRNA and comparing it to the measured value.
A linear regression model was trained on the same set of sgRNAs, albeit with modified features more suited for this approach. These features include the identities of bases in and around the PAM, whether the invariant G at the 5′ end of the sgRNA is base paired, the GC content of the sgRNA, the change in GC content due to the point mutation, the location of the protospacer relative to the annotated transcription start site, the identities of the 3 RNA bases on either side of the mismatch, and the location and type of each mismatch. All features were binarized except for GC and delta GC content. In total, each sgRNA was represented by a vector of 270 features, 228 of which describe the mismatch position and type (19 possible positions by 12 possible types). Prior to training, feature vectors were z-normalized to set the mean to 0 and variance to 1. Finally, an elastic net linear regression model was created using the scikit-learn Python package (https://scikit-learn.org), and key hyperparameters (alpha and L1 ratio) were optimized using a grid search with 3-fold cross validation during training.
Design of compact library
Genes targeted by the compact allelic series library were required to have at least one perfectly matched sgRNA with a growth phenotype greater than 2 z-scores outside the distribution of negative control sgRNAs (γ < –0.04) in a single replicate of a K562 pooled screen (this work or Horlbeck et al.14). By this metric, 4,722 unique sgRNAs targeting 2,405 essential genes were included. Next, for each perfectly matched sgRNA, variants containing all 57 single mismatches in the targeting sequence (positions –19 to –1) were generated in silico, and sequences with off-target binding potential in the human genome were filtered out as described previously14. Remaining variant sgRNAs were whitelisted for potential selection in subsequent steps.
For each gene being targeted, if both of the perfectly matched sgRNAs imparted growth phenotypes greater than 3 z-scores outside the distribution of negative controls (γ < –0.06) in this work’s large-scale K562 screen, then one series of 4 variant sgRNAs was generated from each. Otherwise, one series of 8 variants was generated from the sgRNA with the stronger phenotype. Both perfectly matched sgRNAs were included regardless of their growth phenotype, for a total of 2 perfectly matched and 8 mismatched sgRNAs per gene.
In order to select mismatched sgRNAs, we first divided the relative activity space into 6 bins with edges at 0.1, 0.3, 0.5, 0.7, and 0.9. For each series, we attempted to select sgRNAs from each of the middle 4 bins (centers at 0.2, 0.4, 0.6, and 0.8 relative activity) as measured in this work’s K562 screen. If multiple sgRNAs were available in a particular bin, they were prioritized based on distance to the center of the bin and variance between replicate measurements. If no previously measured sgRNA was available in a given bin, then the CNN model was run on all whitelisted (novel) mismatched sgRNAs belonging to that series, and sgRNAs were selected based on predicted activity as needed. In total, the compact library was composed of 4,722 unique perfectly matched sgRNAs, 19,210 unique mismatched sgRNAs, and 1,202 non-targeting control sgRNAs. Approximately 68% of mismatched sgRNAs were evaluated in previous screens (72% single mismatches, 28% double mismatches), with the remaining 32% imputed from the CNN model (all single mismatches). Sequences of sgRNAs and descriptions of mismatches are listed in Table S10.
Availability of sgRNA libraries
The large-scale and compact mismatched sgRNA libraries are available at Addgene under catalog numbers 136478 (large-scale) and 136479 (compact).
Perturb-seq
The Perturb-seq experiment targeted 25 genes involved in a diverse range of essential functions (Table S13). For each target gene, the original sgRNAs and 4–5 mismatched sgRNAs covering the range from full to low activity were chosen from the large-scale screen. These 128 targeting sgRNAs as well as 10 non-targeting negative control sgRNAs (Table S1) were individually cloned into a modified variant of the CROP-seq vector40,41 as described above, except into the different vector. Lentivirus was individually packaged for each of the 138 sgRNAs and was harvested and frozen in array. To determine viral titers, each virus was individually transduced into K562 CRISPRi cells by centrifugation at 1000 × g and 33 °C for 2 h, and the fraction of transduced cells was quantified as BFP+ cells using an LSR-II flow cytometer (BD Biosciences) 48 h after transduction.
To generate transduced cells for single-cell RNA-seq analysis, virus for all 138 sgRNAs was pooled immediately before transduction and then transduced into K562 CRISPRi cells by centrifugation at 1000 × g and 33 °C for 2 h. To achieve even representation at the intended time of single-cell analysis, the virus pooling was adjusted both for titer and expected growth-rate defects. 3 d after transduction, transduced (BFP+) cells were selected using FACS on a FACSAria2 (BD Biosciences) and then resuspended in conditioned media (RPMI formulated as described above except supplemented with 20% FBS and 20% supernatant of an exponentially growing K562 culture). 2 d after sorting, the cells were loaded onto three lanes of a Chromium Single Cell 3′ V2 chip (10x Genomics) at 1000 cells/μL and processed according to the manufacturer’s instructions.
The CROP-seq sgRNA barcode was PCR amplified from the final single cell RNA-seq libraries with a primer specific to the sgRNA expression cassette (oBA503, Table S17) and a standard P5 primer (Table S17), purified on a Blue Pippin 1.5% agarose cassette (Sage Science) with size selection range 436–534 bp, and pooled with the single cell RNA-seq libraries at a ratio of 1:100. The libraries were sequenced on a HiSeq 4000 (Illumina) according to the manufacturer’s instructions (10x Genomics).
To measure the growth rate defects conferred by each sgRNA for comparison with the transcriptional phenotypes, samples of ~500,000 transduced cells were taken from the same transduced cell population used in the Perturb-seq experiment 2, 7, and 12 days after transduction. Genomic DNA was extracted using the Nucleospin Blood kit (Macherey-Nagel) and sgRNA amplicons were prepared as described previously and above14, albeit with no genomic DNA digestion or gel purification, and sequenced on HiSeq 4000 as described above for the other screens. Growth phenotypes were calculated by comparing normalized sgRNA abundances at day seven and twelve to those at day two, as described above. Read counts and growth phenotypes (γ and relative activity) for individual sgRNAs are available in Table S15 and Table S16, respectively. Relative sgRNA activities measured at day 7 (5 days of growth) were used to assign sgRNA activities in further analysis.
Perturb-seq data analysis
i) Cell barcode and UMI calling, assignment of perturbations
UMI count tables with UMI counts for all genes in each individual cell were calculated from the raw sequencing data using CellRanger 2.1.1 (10x Genomics) with default settings. Perturbation calling was performed as described previously34. Briefly, reads from the specifically amplified sgRNA barcode libraries were aligned to a list of expected sgRNA barcode sequences using bowtie (flags: -v3 -q -m1). Reads with common UMI and barcode identity were then collapsed to counts for each cell barcode, producing a list of possible perturbation identities contained by that cell. A proposed perturbation identity was identified as “confident” if it met thresholds derived by examining the distributions of reads and UMIs across all cells and candidate identities: (1) reads > 50, (2) UMIs > 3, and (3) coverage (reads/UMI) in the upper mode of the observed distribution across all candidate identities. As described previously52, perturbation identities were called for any cell barcode with greater than 2,000 UMIs to enable capture of cells with strong growth defects. Any cell barcode containing two or more confident identities was deemed a “multiplet”, and may arise from either multiple infection or simultaneous encapsulation of more than one cell in a droplet during single-cell RNA sequencing. Cell barcodes passing the 2,000 UMI threshold and bearing a single, unambiguous perturbation barcode were included in all subsequent analyses. Cell counts for each perturbation are summarized in Table S14.
ii). Expression normalization
Some portions of analysis use normalized expression data. We used a relative normalization procedure based on comparison to the gene expression observed in control cells bearing non-targeting sgRNAs, as described previously34:
Total UMI counts for each cell barcode are normalized to have the median number of UMIs observed in control cells.
- For each gene x, expression across all cell barcodes is z-normalized with respect to the mean (μx) and standard deviation (σx) observed in control cells:
Following this normalization, control cells have average expression 0 (and standard deviation 1) for all genes. Negative/positive values therefore represent under/overexpression relative to control.
iii). Target gene quantification
Expression levels of genes targeted by a given sgRNA were quantified by normalizing UMI counts of the targeted gene to the total UMI count for each individual cell (Fig. S8). Considering raw UMI counts of the targeted gene (Fig. S9) or z-normalized target gene expression as described above yielded similar results. Note that the sgRNA targeting BCR is toxic due to knockdown of the BCR-ABL1 fusion present in K562 cells. Knockdown was apparent both in BCR and ABL1 expression, but we used BCR expression for further analysis as there are likely additional copies of ABL1 that are not fused to BCR (and thus would not be affected by the BCR-targeting sgRNA) contributing to ABL1 expression.
iv). Cell cycle analysis
Calling of cell cycle stages was performed using a similar approach to Macosko et al.53 and largely as described in Adamson and Norman et al.34. Briefly, lists of marker genes showing specific expression in different cell cycle stages from the literature were first adapted to K562 cells by restricting to those that showed highly correlated expression within our experiment. The total (log2-normalized) expression of each set of marker genes was used to create scores for each cell cycle stage within each cell, and these scores were then z-normalized across all cells. Each cell was assigned to the cell cycle stage with the highest score.
v). Differential gene expression analysis
We took two approaches to differential expression, as described previously52. For both approaches, we only considered genes with expression greater than 0.25 UMIs per cell on average across all cells. First, for a given gene, we could assess the changes in the expression distribution of that gene induced by a given genetic perturbation by comparing to the expression distribution observed in control cells bearing non-targeting sgRNAs. We performed this comparison using a two-sample Kolmogorov-Smirnov test and corrected for multiple hypothesis testing at an FDR of 0.001 using the Benjamini-Yekutieli procedure.
We also exploited a machine learning approach that potentially allows correlated expression patterns to be detected and that scales beyond two sample comparisons. Perturbed cells and control cells bearing non-targeting sgRNAs were each used as training data for a random forest classifier that was trained to predict which sgRNA a cell contained from its transcriptional state. As part of the training process the classifier ranks which genes have the most prognostic power in predicting sgRNA identity, which by construction will tend to vary across condition. For most further analysis, the top 100–300 genes by prognostic power were then considered.
To assess the overall magnitude of transcriptional changes in individual cells, z-scores of differentially expressed genes were signed by the direction of change in cells with the perfectly matched sgRNA of a series (such that all z-scores were positive in cells with the perfectly matched sgRNA) and then summed. Conclusions were robust across several metrics used to measure distance in gene expression space and aggregate these distances.
vi). Constructing mean expression profiles
For some analyses, expression profiles were averaged across all cells with the same perturbation. In general, this was done simply by calculating the mean z-normalized expression of all genes with mean expression level of 0.25 UMI or higher across all cells in the experiment or within the specific considered subpopulation (usually all cells with sgRNAs targeting a given gene as well as all control cells with non-targeting sgRNAs).
vii). UMAP Dimensionality reduction
For UMAP dimensionality reduction43 of all cells, the 300 genes with the highest prognostic power in distinguishing cells by targeted gene as ranked by a random forest classifier were selected. Dimensionality reduction was then performed on the z-normalized single-cell expression profiles of these 300 genes using the following parameters: n_neighbors = 40, min_dist = 0.1, metric = ‘euclidean’, spread = 1.0. UMAP dimensionality reduction of subpopulations containing only cells with perturbation of a given gene or control cells was performed analogously but using the expression profiles of the 100 genes with the highest prognostic power and using n_neighbors = 15.
From the UMAP projection, we concluded that ~5% cells had misassigned sgRNA identities, as evident for example by the presence of cells with negative control sgRNAs within the cluster of cells with HSPA5 knockdown. These cells had confidently assigned single perturbations and only expressed the corresponding barcode transcript, suggesting that they did not evade our doublet detection algorithm. We speculate that these cells expressed two different sgRNAs but silenced expression of one of the reporter transcripts. Given the strong trends in the results above, we concluded that this rate of misassignment did not substantially affect our ability to identify trends within cell populations.
viii). ATP5E analysis and ISR scores
Analysis of ISR activation in cells with ATP5E knockdown was confounded by a small subpopulation of cells with residual activation of stress responses (cluster labeled * in Fig. 6h). Cells within this cluster were excluded for analysis of ISR activation to ensure that the measured stress responses were indeed the result of ATP5E knockdown. Magnitude of ISR activation in individual cells was quantified as activation of the PERK (EIF2AK3) regulon from the gene set and activation coefficients determined previously34.
Statistics
Tests for differences in distributions of pairwise correlation coefficients of constant region relative activities within and between gene targets (Fig. S3d) were carried out with a two-tailed Student’s t-test. Tests for differential gene expression in the single-cell RNA-seq data were performed with a two-sample Kolmogorov-Smirnov test and corrected for multiple hypothesis testing at an FDR of 0.001 using the Benjamini-Yekutieli procedure, as described in the Methods section on “Perturb-seq data analysis”. Other methods to analyze the single-cell RNA-seq data are described in the Methods section on “Perturb-seq data analysis”. Correlation coefficients reported are Pearson correlation coefficients unless otherwise indicated. Sample sizes used to calculate statistics are provided in the figure legends.
Reporting Summary
Further information on experimental design and reagents is provided in the Life Sciences Reporting Summary published alongside this paper.
Data Availability Statement
Raw and processed Perturb-seq data are available at GEO under accession code GSE132080. Raw and processed sgRNA read counts from pooled screens are provided as supplementary tables. All other data will be made available by the corresponding author upon request.
Code Availability Statement
Custom scripts in this manuscript largely build on scripts published previously14,34,52. An IPython notebook detailing the initialization of the CNN model and its use to predict mismatched sgRNA activities is included as a supplementary file. All custom scripts will be made available upon request.
Supplementary Material
Acknowledgements
We thank G. Ow and E. Collisson (UCSF) for sharing the mCherry-marked sgRNA expression vector, R. Pak, J. Stern, and A. Xu for help with library cloning and sequencing library preparation, B. Adamson for sharing the modified CROP-seq vector, M. Jones, J. Chen, L. Gilbert, J. Replogle, and all members of the Weissman lab for helpful discussions, and E. Chow, D. Bogdanoff, and K. Chaung from the UCSF Center for Advanced Technology for help with sequencing. This work was funded by National Institutes of Health grants F32 GM116331 and K99 GM130964 (MJ), U01 CA168370, U01 CA217882, and RM1 HG009490 (all to JSW), and R35 GM118061 (CAG) and the Innovative Genomics Institute, UC Berkeley (CAG). JSW is a Howard Hughes Medical Institute Investigator. DAS is supported by NSF Graduate Research Fellowship 1650113 and a Moritz-Heyman Discovery Fellowship. RAS is supported by a Fannie and John Hertz Foundation Fellowship and an NSF Graduate Research Fellowship. MAH is a Byers Family Discovery Fellow and is supported by the UCSF Medical Scientist Training Program and the School of Medicine. TMN is a fellow and JAH is the Rebecca Ridley Kry Fellow of the Damon Runyon Cancer Research Foundation (TMN: DRG-2211-15, JAH: DRG-2262-16).
Footnotes
Competing interests
JSW, MJ, DAS, RAS, MAH, and TMN have filed patent applications related to CRISPRi/a screening, Perturb-seq, and mismatched sgRNAs. JSW consults for and holds equity in KSQ Therapeutics, Maze Therapeutics, and Tenaya Therapeutics. JSW is a venture partner at 5AM Ventures and a member of the Amgen Scientific Advisory Board. MJ, MAH, and TMN consult for Maze Therapeutics.
Editorial summary
Gene expression is precisely modulated with CRISPR interference and mismatched guide RNAs.
References
- 1.Huang N, Lee I, Marcotte EM & Hurles ME Characterising and Predicting Haploinsufficiency in the Human Genome. PLOS Genet. 6, e1001154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rest JS et al. Nonlinear Fitness Consequences of Variation in Expression Level of a Eukaryotic Gene. Mol. Biol. Evol. 30, 448–456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer CR, Li S & Siegal ML Essential gene disruptions reveal complex relationships between phenotypic robustness, pleiotropy, and fitness. Mol. Syst. Biol. 11, 773–773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keren L et al. Massively Parallel Interrogation of the Effects of Gene Expression Levels on Fitness. Cell 166, 1282–1294.e18 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Dykhuizen DE, Dean AM & Hartl DL Metabolic Flux and Fitness. Genetics 115, 25–31 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekel E & Alon U Optimality and evolutionary tuning of the expression level of a protein. Nature 436, 588–592 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Alper H, Fischer C, Nevoigt E & Stephanopoulos G Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. 102, 12678–12683 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perfeito L, Ghozzi S, Berg J, Schnetz K & Lässig M Nonlinear Fitness Landscape of a Molecular Pathway. PLOS Genet. 7, e1002160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaels YS et al. Precise tuning of gene expression levels in mammalian cells. Nat. Commun. 10, 818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patwardhan RP et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 27, 1173–1175 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore R, Chandrahas A & Bleris L Transcription Activator-like Effectors: A Toolkit for Synthetic Biology. ACS Synth. Biol. 3, 708–716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez AA, Lim WA & Qi LS Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 17, 5–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek M et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horlbeck MA et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanson KR et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sternberg SH, Redding S, Jinek M, Greene EC & Doudna JA DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szczelkun MD et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. 111, 9798–9803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert LA et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimasu H et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 156, 935–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocak DD et al. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 37, 657 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maji B et al. A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell 177, 1067–1079.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiarella AM et al. Dose-dependent activation of gene expression is achieved using CRISPR and small molecules that recruit endogenous chromatin machinery. Nat. Biotechnol. 1–6 (2019) doi: 10.1038/s41587-019-0296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian R et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 0, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura M et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert LA et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kampmann M, Bassik MC & Weissman JS Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc. Natl. Acad. Sci. 110, E2317–E2326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu PD et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle EA et al. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl. Acad. Sci. 114, 5461–5466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang Y et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16, 280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grevet JD et al. Domain-focused CRISPR screen identifies HRI as a fetal hemoglobin regulator in human erythroid cells. Science 361, 285–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briner AE et al. Guide RNA Functional Modules Direct Cas9 Activity and Orthogonality. Mol. Cell 56, 333–339 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Adamson B et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eraslan G, Avsec Ž, Gagneur J & Theis FJ Deep learning: new computational modelling techniques for genomics. Nat. Rev. Genet. 20, 389–403 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Kim HK et al. Deep learning improves prediction of CRISPR–Cpf1 guide RNA activity. Nat. Biotechnol. 36, 239–241 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Luo J, Chen W, Xue L & Tang B Prediction of activity and specificity of CRISPR-Cpf1 using convolutional deep learning neural networks. BMC Bioinformatics 20, 332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaitin DA et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Datlinger P et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Replogle JM et al. Direct capture of CRISPR guides enables scalable, multiplexed, and multi-omic Perturb-seq. bioRxiv 503367 (2018) doi: 10.1101/503367. [DOI] [Google Scholar]
- 42.Harding HP et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol. Cell 11, 619–633 (2003). [DOI] [PubMed] [Google Scholar]
- 43.McInnes L, Healy J & Melville J UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv180203426 Cs Stat (2018). [Google Scholar]
- 44.Semenova E et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. 108, 10098–10103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiedenheft B et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. 108, 10092–10097 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandegar MA et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Genga RMJ et al. Single-Cell RNA-Sequencing-Based CRISPRi Screening Resolves Molecular Drivers of Early Human Endoderm Development. Cell Rep. 27, 708–718.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 48.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez AR et al. GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 35, 347–349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bae S, Park J & Kim J-S Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bassik MC et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat. Methods 6, 443–445 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norman TM et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed Perturb-seq data are available at GEO under accession code GSE132080. Raw and processed sgRNA read counts from pooled screens are provided as supplementary tables. All other data will be made available by the corresponding author upon request.