Abstract
During thymic development, mouse γδ T cells commit to either an IFN-γ- or an IL-17-producing phenotype through mechanisms that remain unclear. Here, we investigated the extent to which the SLAM/SAP signaling pathway regulates the functional programming of γδ T cells. Characterization of SLAM family receptor expression revealed that thymic γδ T cell subsets were each marked by distinct co-expression profiles of SLAMF1, SLAMF4, and SLAMF6. In the thymus, Vγ1 and Vγ4 T cells that exhibited a SLAMF1+SLAMF6+ double positive (DP) phenotype were largely contained within immature CD24+CD73− and CD24+CD73+ subsets, while SLAMF1 single positive (SP), SLAMF6 SP, or SLAMF1SLAMF6 double negative (DN) cells were found within mature CD24−CD73+ and CD24−CD73− subsets. In the periphery, SLAMF1 and SLAMF6 expression distinguished IL-17- and IFN-γ-producing γδ T cells, respectively. Disruption of SLAM family receptor signaling through deletion of SAP resulted in impaired thymic Vγ1 and Vγ4 T cell maturation at the CD24+CD73−SLAMF1+SLAMF6+ DP stage that was associated with a decreased frequency of CD44+RORγt+ γδ T cells. Impaired development was in turn associated with decreased γδ T cell IL-17 and IFN-γ production in the thymus as well as in peripheral tissues. The role for SAP was subset-specific, as Vγ1Vδ6.3, Vγ4, Vγ5, but not Vγ6 subsets were SAP-dependent. Together, these data suggest that the SLAM/SAP signaling pathway plays a larger role in γδ T cell development than previously appreciated, and represents a critical checkpoint in the functional programming of both IL-17 and IFN-γ-producing γδ T cell subsets.
Introduction:
Innate-like γδ T cells are unusual T cells that are highly enriched in skin and mucosal tissues where they constitute a prominent source of cytokines and chemokines (1). In mice, most γδ T cells produce either IL-17 or IFN-γ, and IL-17 production is primarily restricted to the CD27−CCR6+ Vγ4 and Vγ6 subsets (Heilig and Tonegawa nomenclature (2)), whereas IFN-γ production is observed in CD27+CCR6− Vγ1, Vγ4, Vγ5, and Vγ7 subsets (3-6). Accumulating data suggest that the relative balance of these IL-17 or IFN-γ-producing subsets can have significant consequences on disease outcome. IFN-γ-producing γδ T cells, for example, contribute to anti-tumor immunity (7) and inflammation in a model of cerebral malaria (3, 8). IL-17-producing γδ T cells, in contrast, have been associated with protection against bacterial and fungal infections (9-11), and promotion of tumor metastasis (12, 13) and immunopathologies in settings of autoimmunity (14-19).
Like their innate-like T cell counterparts, iNKT cells and MAIT cells, the functional programming of innate-like γδ T cells occurs during thymic development. Unlike iNKT and MAIT cells, however, a significant portion of γδ T cell developmental programming is restricted to a discrete developmental window (20), as it has been demonstrated that most natural IL-17-producing γδ T cells (γδT17) are generated only during the embryonic/early post-natal stage (21). Although the mechanisms that regulate the differentiation of γδ T cells into IFN-γ or IL-17-producing subsets remain unclear, a significant body of data supports a model in which strong or weak TCR signals skew the developing γδ T cell toward IFN-γ (strong signal) or IL-17 (weak signal) production (22-26). In contrast, there is also evidence supporting a TCR-independent model in which the development of some γδT17 subsets is dependent on pre-existing hardwired transcriptional programs (27-29). Exactly how these seemingly independent mechanisms converge to regulate the developmental programming of innate-like γδ T cell subsets remains an open question.
Slam receptors comprise a family of nine cell surface receptors, SLAMF1 (SLAM; CD150), SLAMF2 (CD48), SLAMF3 (Ly9; CD229), SLAMF4 (2B4; CD244), SLAMF5 (CD84), SLAMF6 (Ly108; CD352, SLAMF7 (CRACC; CD319), SLAMF8 (BLAME), and SLAMF9 (SF2001; CD84H), that are expressed primarily on hematopoietic cells and which play diverse roles in immune development and function (30). Most SLAM family receptors interact in a homophilic manner and therefore serve as self-ligands, the exception to this rule being the heterophilic interaction between SLAMF4 and SLAMF2. SLAM receptor signals can be both activating or inhibitory, depending on balanced recruitment of the cytosolic signaling adapter proteins SAP (31) and EAT-2 (32), and inhibitory SHP-1 or SHIP phosphatases to immunoreceptor tyrosine switch motifs in the SLAM family receptor cytoplasmic domain.
Inactivation of the gene encoding SAP (Sh2d1a) disrupts the signaling pathway of the majority of SLAM family receptors and results in the development of the primary immunodeficiency X-linked lymphoproliferative disease (XLP) which is characterized by broad immunological defects, among which is a failure in iNKT (33, 34) and MAIT (35) cell development. Investigations into the identity of the relevant SLAM family receptors involved in NKT cell development have revealed a complex interplay of individual receptors, including SLAMF1, SLAMF3, and SLAMF6 (34, 36-40). Interestingly, within the γδ T cell compartment, it was previously demonstrated that development of the Vγ1Vδ6.3 (γδNKT) T cell subset is dependent on SAP (41), indicating that the requirement for SLAM/SAP signaling during development is shared by NKT cells, MAIT cells, and at least one γδ T cell subset. Yet, very little is known regarding SLAM family receptor expression on γδ T cells in either the periphery or the thymus, or the extent to which this receptor system is involved in γδ T cell development. In this study, therefore, we explored the extent to which the SLAM/SAP signaling pathway regulates the developmental programming of thymic γδ T cells that determines their effector function.
Materials and Methods
Mice
C57BL/6J, BALB/cJ, and B6.Sh2d1a−/− (SAP−/−) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were housed and bred in a specific pathogen-free barrier room at the AAALAC-approved animal facility of the University of Vermont. Age and sex matched mice between 8 and 12 weeks of age were used for experiments involving adult naïve mice as indicated in the figure legends. To analyze γδ T cells in embryonic thymus, timed-pregnant matings were used. The presence of a vaginal plug indicated day 0.5 of pregnancy. No attempt of sex matching was done for fetuses and newborn mice. All procedures involving animals were approved by the University of Vermont Institutional Animal Care and Use Committee.
Cell preparation and stimulation
Single cell suspensions of splenocytes, lymph nodes, and thymocytes were obtained by gently pressing tissue through nylon mesh. RBCs were lysed using Gey’s solution. Lung cell preparations were conducted as described (42). Lungs were dissected away from the trachea and finely minced in enzymatic digestion buffer (Dulbecco’s modified Eagle’s medium (DMEM), 1 mg/ml collagenase type IV (Invitrogen), 0.2 mg/ml DNase (Sigma)). Lung tissue was resuspended in additional enzymatic digestion buffer and shaken at 200 rpm at 37°C for 20 min. The samples were triturated with a 16-gauge needle and allowed to digest for an additional 20 min, after which they were triturated one last time. After RBC lysis using Gey’s solution, the digested tissue was passed through a 70-μm filter and washed in PBS/2% FCS.
Single cell suspensions from skin were prepared as described (43). Briefly, depilated skin was digested in Trypsin/GNK buffer (0.3% bovine pancreatic trypsin (Type XI) (Sigma), 5 mM KCl, 150 mM NaCl, and 0.1% glucose, pH 7.6) for 2 h at 37 °C., after which the epidermis was separated from the dermis. Epidermis was then placed in Trypsin/GNK buffer containing 0.5% DNase I and incubated for 15 min at 37 °C. After incubation, samples were passed through 70 μm mesh, washed with DMEM + 10% FBS, and resuspended in RPMI 1640 supplemented with 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 50 U/ml penicillin, 50 ug/ml streptomycin + 10 U/ml IL-2 and incubated overnight at 37 °C in a 5% CO2 incubator prior to staining.
Liver intrahepatic leukocytes (IHLs) were isolated as previously described (44). Briefly, livers were perfused through the portal vein with PBS, subsequently removed, minced, and gently pressed through nylon mesh. The resulting cell suspension was washed twice and then resuspended in isotonic 33.8% Percoll (GE Healthcare). After centrifugation, the IHL cell pellet was resuspended, washed with PBS, and red blood cells were lysed using Gey’s solution. Single cell suspensions were resuspended in flow cytometry staining buffer.
Intestinal Intraepithelial Lymphocytes (IEL) were isolated as previously described (45). Briefly, mouse small intestines were flushed with cold CMF buffer (1 mM HEPES, 2.5 mM NaHCO3, 2% FBS in Ca2+/Mg2+-free HBSS pH 7.2). Surrounding fat tissue was removed, intestines opened with a longitudinal incision, and excess mucous removed by gentle scraping. Tissue was cut into small pieces, washed 3 times with CMF, followed by shaking in CMF+10% FBS/1 mM dithioerythritol (DTE) for 20 minutes at 37 °C. This step was repeated and the cells in the supernatants of both extractions were combined. Cells were incubated for 20 minutes at 4 °C, before transferring cell supernatants into a fresh tube. Cells were centrifuged, washed once with RPMI, before resuspending in 44% Percoll solution. Cells were overlaid on a 67% Percoll solution, spun for 20 minutes at room temperature. Cells at the interface were collected, washed twice with RPMI, prior to counting and further analysis.
For all tissues, cell numbers were determined on a MACSQuant VYB (Miltenyi) using propidium iodide (1 ug/ml in PBS) for dead cell exclusion. Cell stimulations were conducted in Click’s medium (Sigma) supplemented with 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 50 μg/ml gentamycin, 50 U/ml penicillin, 50 ug/ml streptomycin, and 5% FBS with 25 ng/ml PMA and 500 ng/ml ionomycin for 4 hours at 37 °C and 5% CO2. After 2 hours, 2 μM monensin was added to the culture.
Flow Cytometry
Cells were stained with Live Dead Fixable Blue staining reagent (ThermoFisher, Grand Island, NY) at 4°C in PBS for 30 min, after which they were washed and resuspended in PBS/2% FCS containing 0.1% sodium azide and Fc-Block (Biolegend) to block non-specific antibody binding prior to the addition of antibodies. Brilliant Violet Staining buffer (BD Biosciences) was used in experiments containing more than one Brilliant Violet or Brilliant Ultraviolet fluorophore conjugated antibody. To identify the Vγ6 subset, the staining procedure described in Roark et al. was followed (46).
Cells were washed twice with PBS/2% FCS/0.1% sodium azide solution and fixed in PBS/1% PFA buffer. For intracellular cytokine staining, cells were permeabilized with PBS/1% FCS/0.1% sodium azide/0.1% saponin after surface staining and fixation. 25 μg/ml total rat IgG were added to reduce unspecific binding prior to cytokine staining with fluorescent-labeled antibodies. Nuclear transcription factors were analyzed using the Foxp3 transcription factor fixation/permeabilization reagent (ThermoFisher). Briefly, after staining for surface markers cells were resuspended in the fixation/permeabilization reagent and incubated overnight at 4 °C. Cells were washed, incubated with 25 μg/ml total rat IgG prior to adding antibodies against transcription factors. After staining at 4 °C, cells were washed and resuspended in PBS/2% FCS/0.1% sodium azide. Flow cytometry data were collected on a LSRII flow cytometer (BD Bioscience) and analyzed with FlowJo software (Treestar). Representative gating schemes used to identify γδ T cell subsets in thymus and lung are shown in the supplement (Suppl. Fig. 1).
Antibodies used in these experiments were: CD11b (M1/70), CD19 (1D3), CD24 (M1/69), CD27 (LG.3A10), CD44 (IM7), CD45 (30-F11), CD48 (HM48-1), CD73 (TY/11.8), CD84 (mCD84.7), CD150 (TC15-12F12.2), CD229 (Ly9ab3), CD352 (330-AJ), IFN-γ (XMG1.2), IL-17A (TC11-18H10.1), TCRβ (H57-597), TCRδ (GL3), Vγ1 (2.11), Vγ5 (536), Vδ6.3 (C504.17c) from Biolegend; CD196 (140706), CD244 (2B4), RORγt (Q31-378) from BD Biosciences; CD244.1 (C9.1), CD352 (13G3-19D), Eomes (Dan11mag), Vγ4 (UC3-10A6) from eBiosciences
t-SNE analysis
Dimensionality reduction of flow cytometry data was conducted using t-stochastic neighbor embedding (t-SNE) in FlowJo. First, traditional gating was used to identify γδ T cells, after which the γδ T cell node data from 5 B6 and 5 BALB/c mice was concatenated for analysis. t-SNE was conducted on the SLAMF1, SLAMF4, SLAMF6, Vγ1, Vγ4, Vγ6, CD44, and CD27 parameters on the concatenated B6/BALB/c data using 1000 iterations, perplexity = 30, learning rate = 957, after which the B6 and BALB/c data were extracted for independent visualization.
Transcriptional profiling
Leukocytes from lung cell preparations were isolated using anti-CD45 Microbeads and LS columns according to the manufacturer’s instruction (Miltenyi). Cells were stained with Fixable Viability Dye eFluor780 (eBioscience), washed, and surface stained with Abs against SLAMF1, SLAMF6, NK1.1, TCRδ, TCR-Vγ1, TCR-Vγ4, TCRβ, CD11b, CD19, and CD45 in sterile PBS/2% FBS. Cells were sorted on a FACS Aria III (BD Bioscience) into culture medium, pelleted, washed, and snap frozen until later use. RNA was isolated using a RNeasy Micro kit (Qiagen). RNA quality was assessed using an Agilent Bioanalyzer 2100, and RNA concentration was calculated using a Qubit fluorometer (ThermoFisher Scientific). Amplified cDNA was prepared using the Ovation Pico WTA System V2 (Nugen), and biotinylated using the Encore biotin module (Nugen). Hybridization with GeneChip Mouse Gene 2.0 ST arrays (Affymetrix) was conducted overnight at 45 °C. at 60 rpm. CEL files were analyzed using Transcriptome Analysis Console (ThermoFisher) software. Genes that demonstrated a greater than or less than 2-fold difference in expression and a false-discovery rate of <0.2 were identified. This resulted in 316 SLAMF1-enriched genes and 229 SLAMF6-enriched genes. These gene lists were filtered by removing unannotated transcripts, predicted transcripts, and by removing all but 1 of the 28 multiple transcript isoforms of Syne1, yielding 244 SLAMF1-enriched and 181 SLAMF6-enriched transcripts. All differentially expressed genes in this list exhibited a p-value < 0.004. The raw data files for this analysis were deposited in NCBI GEO (GSE128544), https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128544.
Data were analyzed for gene ontological and pathway enrichment using NIH DAVID analysis and IPA (filtered to include only those with a p-value and FDR <0.05). Heatmaps were generated using prettyheatmap with row-scaling. Upstream regulator prediction using IPA analysis was conducted on the 425 differentially expressed genes based on fold-change (SLAMF1+SLAMF6−/SLAMF1−SLAMF6+). The result was filtered to include only those predicted regulators with p-value of overlap < 1 X 10−5, ranked according to activation z-score, after which the ten highest (predicted to be active in SLAMF1+SLAMF6−) and lowest (predicted to be active in SLAMF1−SLAMF6+) were chosen for display.
Statistical analysis
Statistical analysis was performed using Prism (GraphPad Software, San Diego, CA). Unpaired Student t-tests, one-way ANOVA, or two-way ANOVA was used where appropriate. For ANOVA posttest comparisons, the Dunnett, Tukey, or Sidak multiple comparison tests were used where appropriate. In all cases, tests were considered significant when p < 0.05. Unless otherwise indicated, error bars represent standard deviation.
Results:
Thymic γδ T cell developmental stages are associated with distinct SLAM family receptor expression profiles.
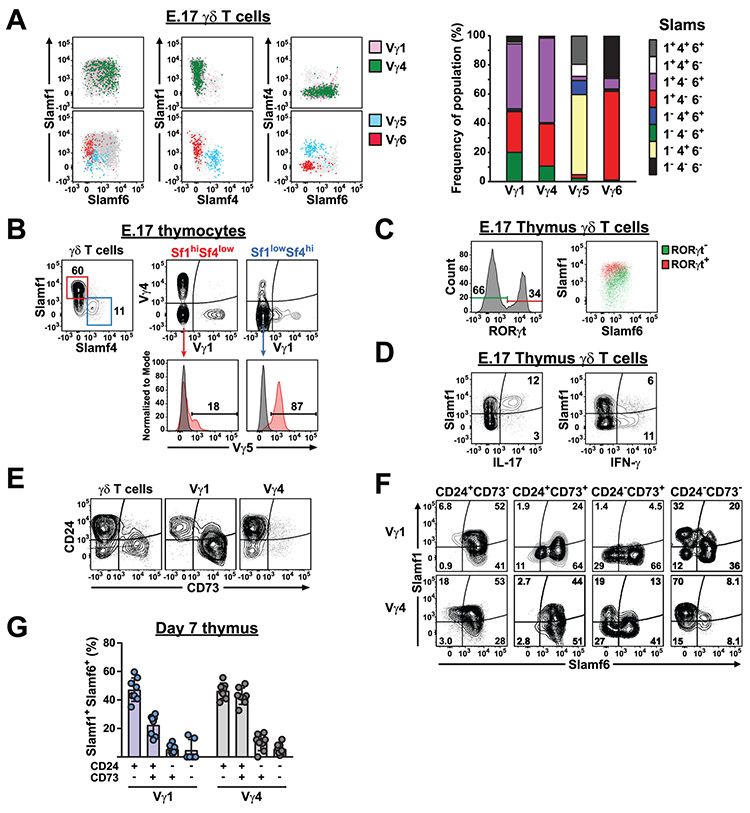
γδ T cell effector functions are largely programmed during thymic development, and IL-17 functional programming is thought to occur primarily during embryonic and neonatal thymic development (21). Therefore, we examined the co-expression of SLAMF1 through SLAMF6 receptors on γδ T cell subsets in C57BL/6 embryonic day 17 (E.17) thymus. This analysis revealed that SLAMF5 was undetectable on γδ T cells and that the expression of SLAMF2 and SLAMF3 was relatively uniform among γδ T cell subsets (Suppl. Fig. 2). In contrast, SLAMF1, SLAMF4, and SLAMF6 exhibited a surprising degree of heterogeneity in their co-expression (Fig. 1A). For example, a significant fraction of E.17 thymic γδ T cells were SLAMF1−SLAM4+SLAMF6− and this population consisted primarily of Vγ5 T cells (Fig. 1B). In the skin, mature Vγ5 T cells exhibit the same SLAMF1−SLAMF4+SLAMF6− phenotype (Suppl. Fig. 2), suggesting that the Vγ5 SLAM family receptor expression profile is established during embryonic thymic development (47). Similarly, we observed that Vγ6 T cells were predominantly SLAMF1+SLAMF4−SLAMF6− in the E.17 thymus (Fig. 1A), similar to the SLAM receptor expression profile that they exhibit in the adult lung (see below).
Fig. 1. Slam receptor expression profiles on different γδ T cell subsets are established during thymic development.
A) SLAMF1, SLAMF4, and SLAMF6 expression on B6 E.17 γδ T cells subsets. The position of Vγ1+ (pink) and Vγ4+ (green) γδ T cells on plots of SLAMF1, SLAMF4, and SLAMF6 is shown above. The position of Vγ5+ (blue) and Vγ6+ (red) γδ T cells is shown below. Total γδ T cells are indicated in grey. Right, cumulative frequencies of SLAM receptor expression profiles on the thymic γδ T cell subsets are shown as bar graph. Data are representative of 2 independent experiments, 6 mice each, sex was not determined. B) SLAMF1 and SLAMF4 expression on E.17 thymic Vγ1, Vγ4, and Vγ5 subsets. Representative contour plots of SLAMF1 and SLAMF4 staining on thymic γδ T cells is shown at left. SLAMF1hiSLAMF4lo and SLAMF1loSLAMF4hi γδ T cells are gated in red and blue, respectively. Vγ1, Vγ4, and Vγ5 staining on SLAMF1hiSLAMF4lo and SLAMF1loSLAMF4hi subsets is shown at right. Data is representative of 2 independent experiments, 6-7 mice each, sex was not determined. C) SLAMF1 and SLAMF6 expression on RORγt+ and RORγt− E.17 thymic γδ T cells. The position of RORγt+ (red) and RORγt− (green) cells are shown on a dot plot of SLAMF1 and SLAMF6 expression. Data are representative of 3 independent experiments with 3-8 mice each experiment, sex was not determined. D) SLAMF1 expression on IL-17 and IFN-γ-producing E.17 thymic γδ T cells. Representative contour plots are shown. Data are representative of 2 independent experiments with 4 and 6 mice each experiment, sex was not determined. E) CD24 and CD73 expression on day 7 thymus γδ T cell subsets. Data shown are concatenated data from 8 male and female B6 mice and are representative of 2 independent experiments. F) SLAMF1 and SLAMF6 expression on day 7 thymus Vγ1 and Vγ4 T cell subsets at different stages of development. Data shown are concatenated data from 8 B6 mice and are representative of 2 independent experiments. G) Frequency of SLAMF1SLAMF6 DP at different stages of γδ T cell maturation. Day 7 thymus Vγ1 and Vγ4 T cells were gated according to CD24 and CD73 expression, and the frequency of SLAMF1SLAMF6 DP cells was calculated. Each circle represents an individual B6 male or female mouse and are representative of 2 independent experiments.
In contrast to the Vγ5 and Vγ6 populations, E.17 Vγ1 and Vγ4 T cells showed broad expression of SLAMF1 and SLAMF6 and they rarely expressed SLAMF4 (Fig. 1A). We identified prominent SLAMF1 and SLAMF6 single positive (SP) Vγ1 and Vγ4 T cell populations that represented the predominant phenotypes detectable in peripheral tissues (Fig. 1A). Consistent with a previous report that IL-17-producing γδ T cells preferentially express SLAMF1 (48), RORγt+ E.17 γδ T cells were largely found within a SLAMF1hiSLAMF6low γδ T cell subset (Fig. 1C) and the majority of E.17 IL-17-producing γδ T cells were SLAMF1+, while the majority of IFN-γ-producing γδ T cells were SLAMF1− (Fig. 1D).
In addition to the SLAMF1 and SLAMF6 SP Vγ1 and Vγ4 T cell populations, we detected prominent SLAMF1SLAMF6 double positive (DP) Vγ1 and Vγ4 T cell populations (Fig. 1A) that were rarely observed in the periphery (see below). To characterize this SLAMF1SLAMF6 DP population in more detail, we evaluated neonate thymus Vγ1 and Vγ4 SLAMF1 and SLAMF6 expression in the context of CD24 and CD73 expression which can be used to track γδ T cell development (49, 50). Recently, two distinct pathways of Vγ1 and Vγ4 maturation have been defined using these markers (51). In pathway 1, utilized by both Vγ1 and Vγ4 T cells, immature CD24+CD73− γδ T cells upregulate CD73 (CD24+CD73+) prior to CD24 downregulation and maturation (CD24−CD73+). In pathway 2, utilized by Vγ4 and Vγ6 T cells, immature CD24+CD73− γδ T cells downregulate CD24 (CD24−CD73−) prior to CD73 upregulation. Consistent with a previous report (51), we found that day 7 thymus Vγ1 and Vγ4 T cells exhibited distinct patterns of CD24 and CD73 expression and that Vγ1 cells exhibited prominent immature CD24+CD73− and mature CD24−CD73+ populations, while Vγ4 cells were primarily CD24+CD73− and CD24−CD73− (Fig. 1E).
Examination of SLAMF1 and SLAMF6 expression on these subsets revealed that in both Vγ1 and Vγ4 T cells, the SLAMF1SLAMF6 DP populations largely resided within the immature CD24+CD73− population, while SLAMF1 SP and SLAMF6 SP and SLAMF1SLAMF6 DN cells predominated in the mature CD24−CD73+ and CD24−CD73− populations (Fig. 1F). In Vγ1 cells, which develop using pathway 1, there was a significant decrease in the frequency of SLAMF1SLAMF6 DP cells as cells progressed through the CD24+CD73− to CD24+CD73+ to CD24−CD73+ stages (Fig. 1G), and the prominent mature CD24−CD73+ Vγ1 population was largely dominated by SLAMF6 SP Vγ1 cells (Fig. 1F). In Vγ4 cells, which develop primarily using pathway 2 during neonatal development (51), there was a significant decrease in the frequency of SLAMF1SLAMF6 DP cells as they progressed from CD24+CD73− to CD24−CD73− (Fig. 1G). Interestingly, mature CD24−CD73− Vγ4 cells were predominantly SLAMF1 SP, while the small number of mature CD24−CD73+ Vγ4 cells were predominantly SLAMF6 SP and SLAMF1SLAMF6 DN (Fig. 1F). Similar results were observed in adult thymus, with the exception that in Vγ4 cells, which develop primarily using pathway 1 during adult development (51), there was a decrease in the frequency of SLAMF1SLAMF6 DP cells as they progressed from CD24+CD73− to CD24+CD73+ (Suppl. Fig. 2). In both Vγ1 and Vγ4 subsets, we noted the presence of SLAMF1 SP and/or SLAMF6 SP γδ T cells in the immature CD24+CD73− populations (Fig. 1F), suggesting that the alterations in SLAM family receptor expression are already occurring at this developmental stage. Taken together, these data indicated that the co-expression of SLAMF1, SLAMF4, and SLAMF6 distinguished 3 groups of γδ T cell subsets in the thymus: Vγ1/Vγ4, Vγ5, and Vγ6. Our findings were consistent with a developmental scheme in which immature CD24+CD73− thymic Vγ1 and Vγ4 T cells initially possess a SLAMF1SLAMF6 DP profile that resolves into either mature SLAMF1, SLAMF6 SP, or SLAMF1SLAMF6 DN phenotypes upon maturation and acquisition of function.
Impaired γδ T development in SAP-deficient mice
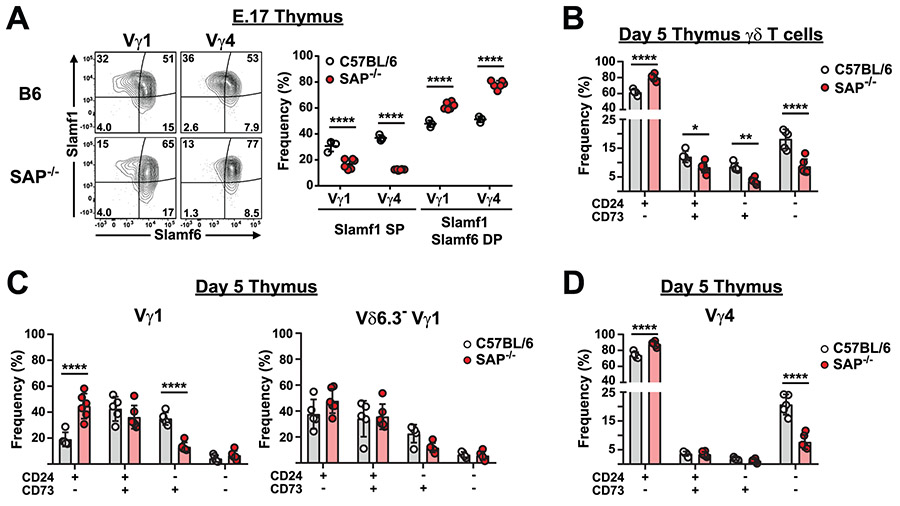
SLAMF1, SLAMF4, and SLAMF6 transduce signals by utilizing the signaling adapter proteins SAP and EAT-2, which bind to immunoreceptor tyrosine switch motifs (ITSMs) located in the cytoplasmic tails of SLAM family receptors (30). Sh2d1a (encoding SAP) is the most widely expressed of these adapter proteins in γδ T cells and SAP-deficient mice have previously been reported to be deficient in the IFN-γ-producing Vγ1Vδ6.3 (γδNKT) subset (52). To determine whether the SLAM/SAP signaling pathway played any additional roles in the functional programming that γδ T cells undergo during thymic development, we examined γδ T cell development in SAP-deficient mice during embryonic and neonatal stages of development.
A comparison of thymic γδ T cells between B6 and B6.SAP-deficient mice revealed no significant change in the overall frequencies of Vγ4, Vγ5, or Vγ6 T cell subsets, although we did confirm the critical role of SAP in the development of the Vγ1Vδ6.3 subset on day 5 (52)(Suppl. Fig. 3). An examination of the SLAM family receptor expression profiles on E.17 γδ T cells revealed a significant decrease in the frequency of SLAMF1 SP Vγ1 and Vγ4 T cells (Fig. 2A). The reduction in the SLAMF1 SP subset coincided with a significant increase in the frequency of SLAMF1SLAMF6 DP subsets in both E.17 (Fig. 2A) and neonate (Suppl. Fig. 3) B6.SAP−/− thymus. In accordance with our observations that SLAMF1SLAMF6 DP largely resided within the CD24+CD73− population, while SLAMF1 and SLAMF6 SP cells largely resided within the CD24−CD73− and CD24−CD73+ populations (Fig. 1F), we observed a significant increase in the frequency of CD24+CD73− thymic γδ T cells in SAP-deficient mice (Fig. 2B). Concomitant with this increase was a significant decrease in CD24+CD73+, CD24−CD73+ and CD24−CD73− γδ T cells (Fig. 2B).
Fig. 2. Altered γδ T cell developmental progression in SAP-deficient mice.
A) SAP-dependent alteration in SLAMF1 SP and SLAMF1SLAMF6 DP γδ T cells. Representative contour plots depicting SLAMF1 and SLAMF6 expression on E.17 and day 6 thymic Vγ4 T cells is shown at left. The frequencies of SLAMF1 and SLAMF6-expressing subsets is shown at right, ****p < 0.0001, as determined by two-way ANOVA followed by Sidak’s multiple comparisons test. Sex of the mice was not determined. B) The effect of SAP deficiency on the developmental progression of thymus γδ T cell. Flow cytometry was used to calculate the frequency of CD24 and CD73-expressing γδ T cells from B6 and B6.SAP−/− day 5 thymus. Data are representative of 2 independent experiments, 8-9 male and female mice per group. *p < 0.05, **p < 0.01, ****p < 0.0001, as determined by t-tests corrected for multiple comparisons using the Holm-Sidak method. C-D) The effect of SAP deficiency on the developmental progression of thymus C) Vγ1 and D) Vγ4 T cells. Flow cytometry was used to calculate the frequency of CD24 and CD73-expressing γδ T cells from B6 and B6.SAP−/− day 5 thymus. Data are representative of 2 independent experiments, 8-9 male and female mice per group. ****p < 0.0001, as determined by t-tests corrected for multiple comparisons using the Holm-Sidak method.
Examination of the individual γδ T cell subsets revealed that among Vγ1 T cells, SAP deficiency resulted in increased CD24+CD73− and CD24+CD73+ populations and a decreased frequency of CD24−CD73+ cells (Fig. 2C). This was due in large part to the near complete absence of the Vγ1Vδ6.3+ subset, which was primarily CD24+CD73+ and CD24−CD73+ (Suppl. Fig. 3), as we observed no significant SAP-dependent changes in maturation among the Vγ1Vδ6.3− T cells (Fig. 2C). Therefore, among neonate Vγ1 T cells, the major effect of SAP deletion was on the Vγ1Vδ6.3+ subset. Surprisingly, however, we also observed that among Vγ4 T cells, SAP deficiency resulted in significantly more CD24+CD73−, and significantly fewer CD24−CD73− Vγ4 cells, suggesting that SAP was critical in the CD24+CD73− to CD24−CD73− (i.e., Pathway 2) transition of a Vγ4 subset (Fig. 2D). No effect of SAP deficiency on the CD24 expression of the Vγ6 subset was observed (not shown). Together, these data suggested that SAP-mediated signaling is integral to the developmental progression of Vγ4 as well as Vγ1 γδ T cells during thymic development and that it functions to promote γδ T cell maturation.
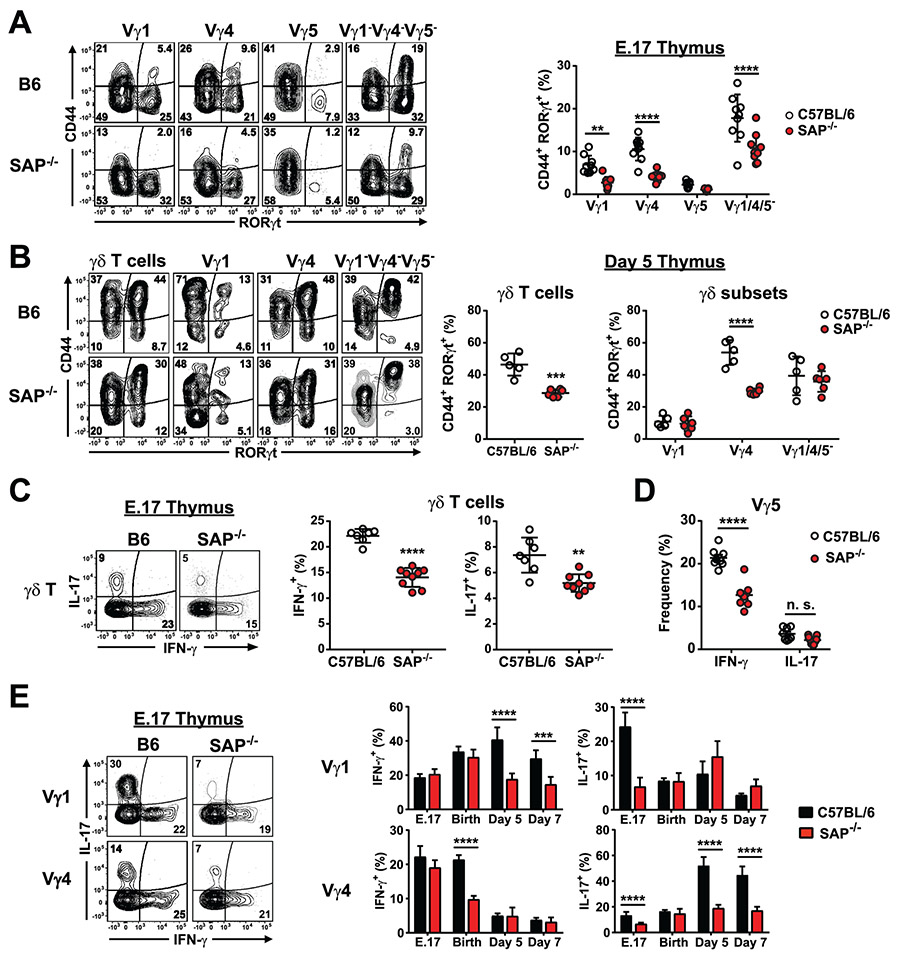
Next, we assessed whether SAP-deficient thymic γδ T cells exhibited any functional defects. Examination of the E.17 CD44+RORγt+ γδ T cell population revealed a significant decrease in the frequency of CD44+RORγt+ γδ T cell subsets in SAP-deficient mice (Fig. 3A). Significant decreases were observed in the Vγ1, Vγ4, and Vγ1−4−5− subsets, but not in Vγ5, very few of which were CD44+RORγt+ (Fig. 3A). Examination of neonatal day 5 thymocytes revealed that the SAP-dependent decrease in CD44+RORγt+ γδ T cells was not restricted to embryonic thymic development, as there was a significant decrease in the frequency of CD44+RORγt+ Vγ4 T cells, which comprise the majority of thymic γδ T cells at this stage of development (Fig. 3B). However, the SAP-dependent decrease in CD44+RORγt+ Vγ1 and Vγ1−4−5− populations was not observed in the neonatal thymus (Fig. 3B).
Fig. 3. Impaired γδ T cell functional programming in SAP-deficient mice.
A) Significant decrease in CD44+RORγt+ thymic γδ T cells in E.17 B6.SAP−/− mice. Thymocytes from E.17 B6 and B6.SAP−/− mice were stained for surface markers and nuclear staining was used to assess RORγt expression. Representative contour plots depicting CD44 and RORγt expression are shown at left. Cumulative frequencies of CD44+RORγt+ E.17 thymic γδ T cells is shown at right, **p < 0.01, ****p < 0.0001, as determined by two-way ANOVA followed by Sidak’s multiple comparisons test. Data are representative of 2 independent experiments, 7-9 mice per group, sex was not determined. B) SAP-dependent decrease in CD44+RORγt+ γδ T cells in day 5 neonate thymus. Representative contour plots are shown at left. Cumulative frequencies of CD44+RORγt+ are shown at right, ***p < 0.001, ****p < 0.0001, as determined by two-way ANOVA followed by Sidak’s multiple comparisons test. Data are representative of 2 independent experiments, 5-6 mice per group, sex was not determined. C) Intracellular IFN-γ and IL-17 production in E.17 thymic γδ T cells. Representative contour plots are shown at left. Numbers indicate the percentage of cells. Cumulative data are shown at right, **p < 0.01, ****p < 0.0001 as determined by an unpaired two-tailed Student’s t-test. Sex of the mice was not determined. D) Decreased E.17 Vγ5 IFN-γ production in SAP-deficient mice. ****p < 0.0001, as determined by Student’s t-test, sex was not determined. E) IFN-γ and IL-17 production in thymic γδ T cell subsets from B6 and B6.SAP−/− mice at different developmental stages as indicated. Cumulative data represent the combined results of 4 independent experiments, 5 – 9 mice per group, sex was not determined. ***p < 0.001, ****p < 0.0001, as determined by two-way ANOVA followed by Sidak’s multiple comparisons test.
Next, we investigated whether SAP played a role in the acquisition of γδ T cell function during thymic development. A comparison of E.17 γδ T cell cytokine production between B6 and B6.SAP−/− mice revealed a significant decrease in both IL-17 and IFN-γ production in SAP-deficient mice (Fig. 3C). Subset analysis revealed that the reduction in IFN-γ production was due primarily to an effect on Vγ5 γδ T cells (Fig. 3D), while the reduction in IL-17 production was instead observed in both Vγ1 and Vγ4 γδ T cell subsets (Fig. 3E). No significant decrease in the IL-17-producing Vγ1−Vγ4−Vγ5− population was observed at this time, consistent with SAP-independent development of the Vγ6 population (data not shown).
Analysis of later stages of development revealed that the effect of SAP deficiency was dependent on both the subset and developmental stage. A significant effect of SAP deficiency was observed on thymic Vγ4 IL-17 production at all stages examined except for day 0. Conversely, we found that Vγ4 IFN-γ production was significantly decreased in SAP-deficient mice on day 0, but not any of the other timepoints examined. Similarly, we noted decreased thymic Vγ1 IL-17 in B6.SAP−/− mice only at E.17, and decreased thymic Vγ1 IFN-γ only on days 5 and 7 (Fig. 3E). Together, these data suggested that the SLAM/SAP signaling pathway played a significant role in the developmental programming of thymic Vγ1, Vγ4, and Vγ5 IL-17 and IFN-γ production during embryonic and neonatal thymic development and that SAP coordinately regulated γδ T cell maturation and the acquisition of γδ T cell function.
Tissue-specific heterogeneity in SLAM receptor expression profiles on γδ T cell subsets in the periphery.
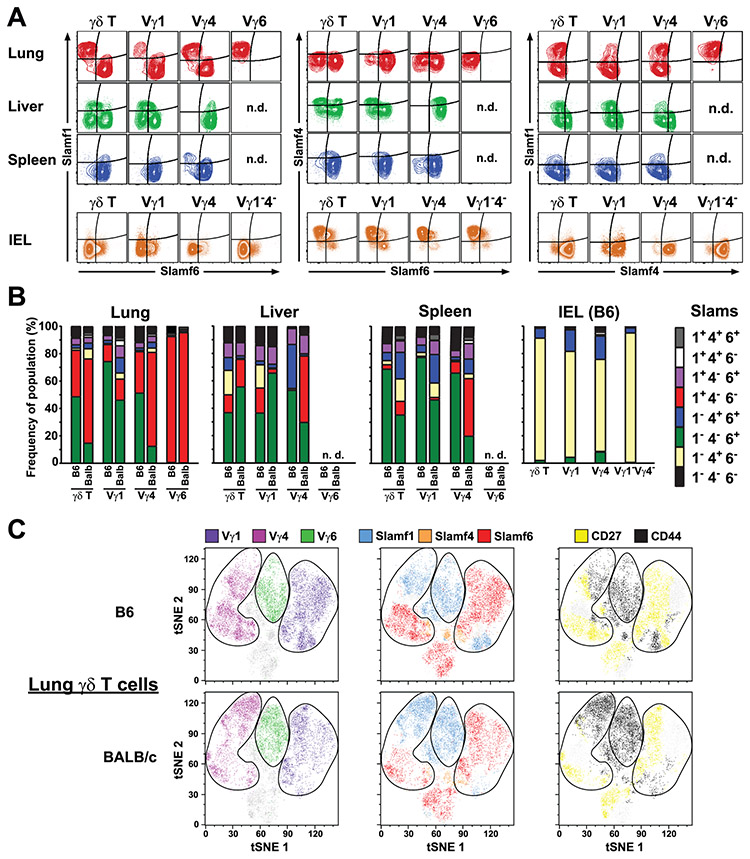
Next, we examined the extent to which SAP-dependent thymic γδ T cell development regulated γδ T cell function in peripheral tissues. We first assessed the co-expression of SLAMF1 through SLAMF6 on γδ T cell subsets from different tissues. Similar to our results in the thymus, we found little evidence of SLAMF5 expression, and we found uniform expression SLAMF2 and SLAMF3 on γδ T cells and from the lung, spleen, and lymph node (Suppl. Fig. 4). In contrast, our analysis revealed significant heterogeneity in SLAMF1, SLAMF4, and SLAMF6 co-expression on γδ T cell subsets obtained from different tissues (Fig. 4A). In the lung, we found two distinct populations of Vγ1 and Vγ4 γδ T cells which were either SLAMF1 SP or SLAMF6 SP. In contrast, Vγ6 γδ T cells were uniformly SLAMF1 SP with no detectable SLAMF6 expression (Fig. 4A). A similar SLAM receptor expression pattern was observed in the spleen although the frequency of the SLAMF1 SP Vγ4 T cell population was markedly reduced compared with the lung and essentially absent in the Vγ1 subset (Fig. 4A). In contrast to lung and spleen, a significant proportion of SLAMF4-expressing γδ T cell subsets were detected in the liver, while gut IEL γδ T cells only expressed SLAMF4, and not SLAMF1 or SLAMF6 (Fig. 4A). Heterogeneity in SLAM family receptor co-expression was also observed in BALB/cJ γδ T cells, which possess a different Slam haplotype (53) (Fig. 4B). We did note significant differences between the strains indicating that SLAM receptor co-expression profiles were influenced by genetic background. Notably, we observed a significantly greater proportion of SLAMF1+SLAMF6− lung and spleen Vγ4 T cells in BALB/c compared to their B6 counterparts (Fig. 4B).
Figure 4. Tissue-dependent heterogeneity in γδ T cell SLAM family receptor expression profiles.
A) Representative contour plots of SLAMF1, SLAMF4, and SLAMF6 expression on lung, liver, spleen, and IEL γδ T cells. Data represent concatenated files from 4 - 5 female B6 mice 11 weeks of age and are representative of 2-3 independent experiments. n.d., not detected. B) Cumulative frequencies of SLAM family receptor expression profiles on γδ T cell subsets from B6 and BALB/c mice. Data represent groups of 4 - 5 B6 mice (as in A) and 4 female BALB/cJ mice, 12 weeks of age. IEL γδ T cells from BALB/cJ mice were not analyzed. C) SLAM family receptor co-expression segregates with surrogate markers of γδ T cell function. Flow cytometric data concatenated from 5 female C57BL/6J and 5 female BALB/cJ mice, all 10 weeks of age were analyzed using t-SNE. Clusters representing Vγ1, Vγ4 and Vγ6 subsets are circled, and SLAMF1, SLAMF4, and SLAMF6-expressing cells, as well as CD44- and CD27-expressing cells are colored as indicated.
Given the association of SLAMF1 expression with γδ T cell IL-17 production in the thymus (Fig. 1D), we assessed whether distinct SLAM family receptor expression profiles coincided with CD44 and CD27 expression which are canonical markers of IL-17 and IFN-γ-producing γδ T cells, respectively. We focused our attention on lung γδ T cells, since they exhibited a diversity of SLAM family receptor expression profiles. This analysis revealed that in both B6 and BALB/c mice, SLAMF1+SLAMF6− γδ T cells were primarily CD44+CD27− consistent with an IL-17-producing phenotype (48), while SLAMF1−SLAMF6+ γδ T cells were primarily CD44−CD27+ suggestive of an IFN-γ-producing phenotype (Fig. 4C). SLAMF4-expressing γδ T cells, in contrast were distributed among both the CD44+CD27− and CD44−CD27+ populations (Fig. 4C). Together, these data indicated that the co-expression of SLAM family receptors on γδ T cells constituted unique SLAM receptor expression profiles that varied in both a tissue-dependent and Vγ subset-dependent manner. The data also suggested that the SLAM receptor expression profiles of γδ T cell subsets in the periphery largely reflected those established upon maturation during thymic development, the exception being SLAMF4 whose expression on some γδ T cell subsets appeared to be modulated in the periphery. Finally, the data indicated that these SLAM receptor expression profiles were present in strains representative of Slam haplotypes 1 and 2, and that in each haplotype SLAMF1 and SLAMF6 expression exhibited a strong association with canonical markers of γδ T cell function.
SLAM receptor expression profiles distinguish γδ T cell functional subsets in the periphery.
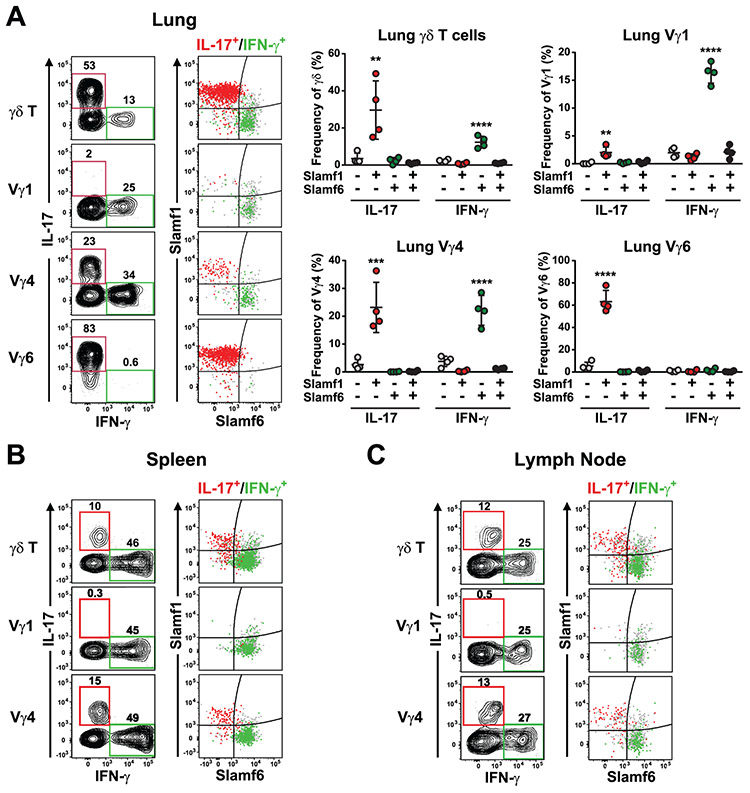
To determine whether SLAM receptor expression profiles marked functional subsets of γδ T cells, we assessed lung γδ T cell cytokine production after stimulation with PMA/ionomycin. These data revealed that lung γδ T cell IFN-γ production was associated with SLAMF1−SLAMF6+ γδ T cells, while IL-17 production was associated with SLAMF1+SLAMF6− γδ T cells (Fig. 5A). Similarly, stimulation of both spleen (Fig. 5B) and lymph node (Fig. 5C) γδ T cells revealed a strong association of SLAMF1 and SLAMF6 expression with γδ T cell IL-17 and IFN-γ production, respectively. Together, these data suggested that the mutually exclusive expression of SLAMF1 and SLAMF6 marked IL-17- and IFN-γ-producing Vγ1, Vγ4, and Vγ6 γδ T cell subsets in the periphery.
Figure 5. SLAM family receptor expression profiles distinguish distinct functional subsets of γδ T cells in the periphery.
SLAMF1 and SLAMF6 expression distinguish IL-17 and IFN-γ-producing γδ T cells in lung (A), spleen (B), and lymph node (C). Cell suspensions were stimulated with PMA/ionomycin for 4 h at 37 °C. in the presence of monensin, after which they were stained for surface markers and intracellularly stained for IFN-γ and IL-17. Representative contour plots of γδ T cell cytokine production after stimulation are shown at left. IL-17 and IFN-γ-producing cells are indicated by red and green gates, respectively. Dot plots depicting SLAMF1 and SLAMF6 expression are shown at right and the position of IL-17-gated and IFN-γ-gated cells are shown in red and green, respectively. All other cells are shown in grey. Cumulative data of lung γδ T cell cytokine production is shown at far right. Data are representative of 3 independent experiments, 4 female C57BL/6J mice per group, 9.5 weeks of age,**p < 0.01, ***p < 0.001, ****p < 0.0001 as determined by one-way ANOVA followed by Tukey’s multiple comparison test.
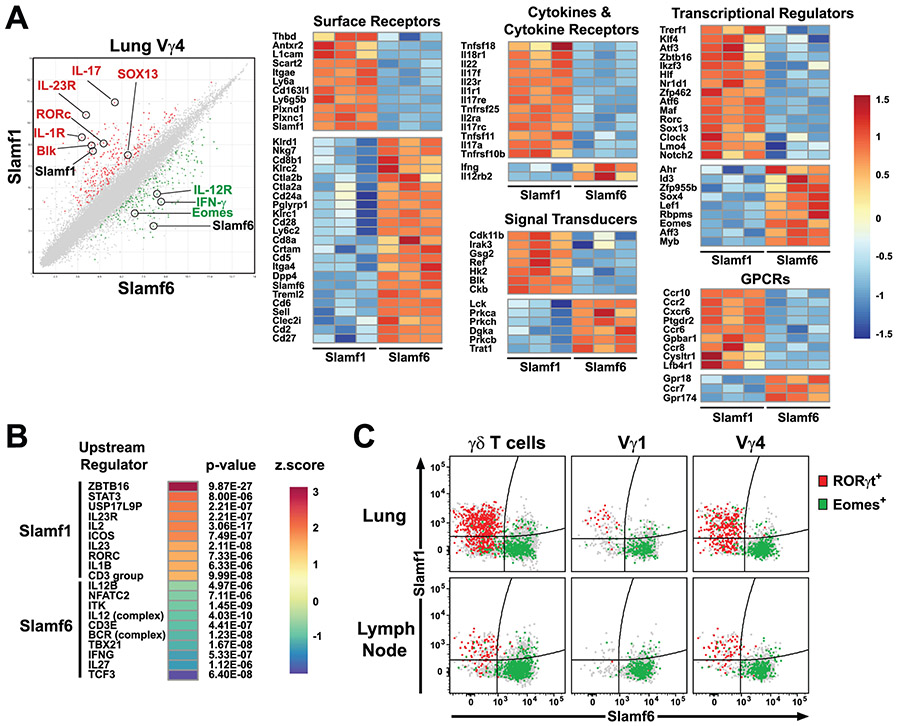
To explore this association further, we compared the ex vivo transcriptional profiles of sorted lung SLAMF1+SLAMF6− and SLAMF1−SLAMF6+ lung Vγ4 from naïve C57BL/6J mice. This analysis revealed the differential expression of 244 genes in SLAMF1+SLAMF6− and 181 genes in SLAMF1−SLAMF6+ lung Vγ4 (Fig. 6A). In accordance with our functional analysis, we found that SLAMF1+SLAMF6− lung Vγ4 γδ T cells exhibited a transcriptional profile highly suggestive of IL-17 production. We observed differential expression of Il23r, Il17a, Il17f, Il1r1, Blk, Rorc, Sox13, Maf, Ccr6, Ccr2, and Scart2 all of which have been previously associated with a γδT17 phenotype (27, 28, 54) (Fig. 6A). In contrast, Ifng, Eomes, IL12rb, and Crtam were preferentially expressed in SLAMF1−SLAMF6+ Vγ4 γδ T cells, suggestive of an IFN-γ-producing phenotype (Fig. 6A). These findings were consistent with a recent report demonstrating that EOMES expression marks IFN-γ-producing γδ T cells (55).
Figure 6. SLAMF1 and SLAMF6 expression distinguishes lung Vγ4 subsets with distinct transcriptional profiles.
SLAMF1+SLAMF6− and SLAMF1−SLAMF6+ lung Vγ4 cells were FACS-sorted from lung cell suspensions pooled from 12 female C57BL/6J mice each (n = 3 pools). RNA from sorted cells was used in a microarray to assess differential gene expression. A) Scatter plot of average sample signals (log2 transformed) observed in SLAMF1+SLAMF6− and SLAMF1−SLAMF6+ cells is shown at left. Genes exhibiting a fold-change of greater or less than 2 and a false discovery rate < 0.2 are colored red (differentially expressed in SLAMF1+SLAMF6−) or green (differentially expressed in SLAMF1−SLAMF6+). Row-normalized heatmaps of differentially expressed genes are shown at right. Receptors, signal transducers, transcriptional regulators, cytokine/cytokine receptors, and GPCRs that exhibited a fold-change of greater or less than 2.5 are shown. Gene expression data are from a single experiment. B) Predicted upstream regulators of differentially expressed genes. Predicted regulators were ranked according to their activation z-score. Positive z-scores suggest activity in SLAMF1+SLAMF6− and negative z-scores suggest activity in SLAMF1−SLAMF6+ subsets. C) SLAMF1 and SLAMF6 distinguish RORγt- and EOMES-expressing γδ T cell subsets. Dot plots depict SLAMF1 and SLAMF6 expression on lung and lymph node γδ T cell subsets. RORγt- and EOMES-expressing γδ T cells are highlighted in red and green, respectively. All other cells are shown in grey. Data are representative of two separate experiments, 4-5 male C57BL/6J mice, 13 weeks of age per group.
Pathway analysis (IPA) of differentially expressed genes revealed that transcripts enriched in SLAMF1+SLAMF6− lung Vγ4 cells exhibited significant overlap with genes regulated by ZBTB16 (PLZF), IL23R, RORγT, and IL-1B. In contrast, transcripts enriched in SLAMF1−SLAMF6+ lung Vγ4 exhibited significant overlap with genes regulated by IL-12, TBX21 (T-bet), IFN-γ, IL-27, and TCF3 (E2A) (Fig. 6B). These transcriptional profiles were confirmed using flow cytometry which revealed that RORγt expression was largely confined to the SLAMF1+SLAMF6− γδ T cells in both lung and lymph node, while EOMES expression was found in the SLAMF1−SLAMF6+ γδ T cell subsets (Fig. 6C). Together, these data suggested that lung SLAMF1+SLAMF6− and SLAMF1−SLAMF6+ Vγ4 γδ T cells possessed transcriptional profiles consistent with IL-17 and IFN-γ production, respectively.
Impaired γδ T function in SAP-deficient mice
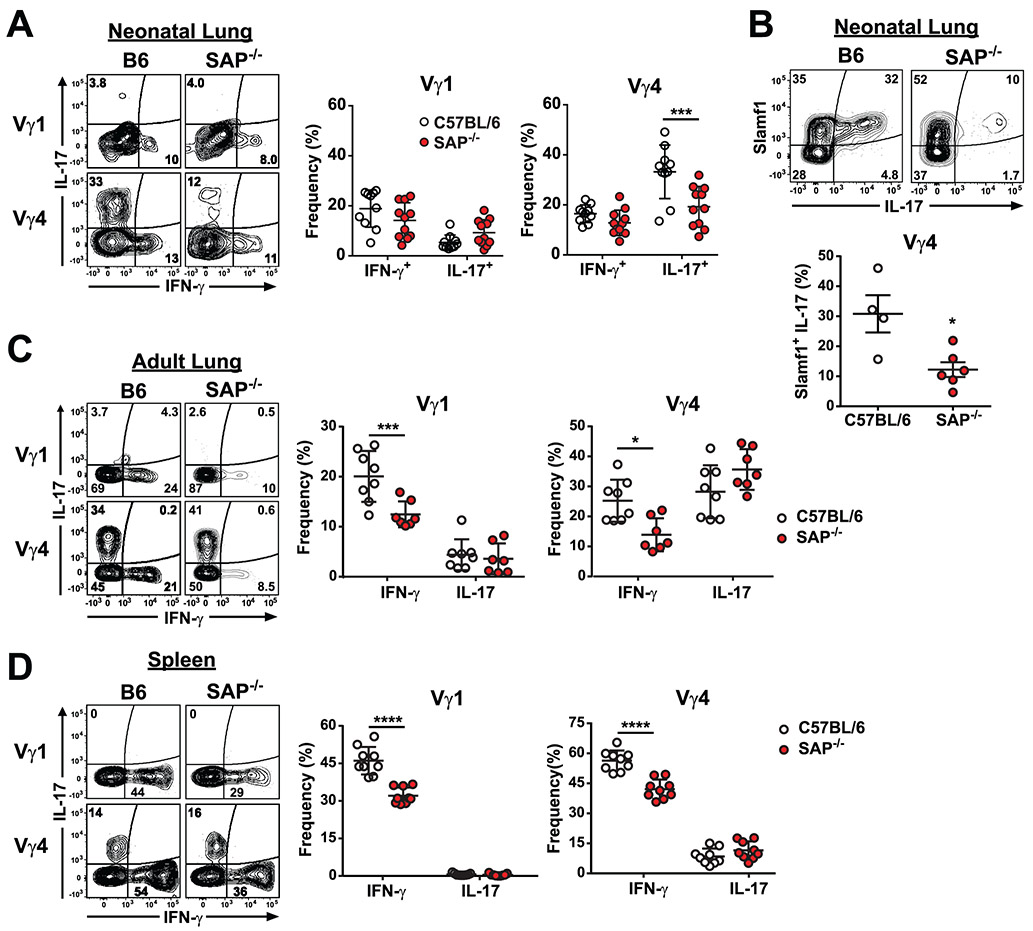
Finally, we examined the extent to which impaired thymic γδ T cell developmental programming in SAP-deficient mice affected γδ T cell function in peripheral lung and lymphoid organs. A comparison of neonate lung γδ T cells between B6 and B6.SAP−/− mice revealed that there was a significant decrease in lung Vγ4 IL-17, but not IFN-γ, production (Fig. 7A) and this decrease corresponded to a significant decrease in SLAMF1+IL-17+ Vγ4 cells (Fig. 7B). No difference was observed in either lung Vγ1 IL-17 or IFN-γ production (Fig. 7A), nor did we observe any SAP-dependent effects on Vγ6 IL-17 production (data not shown). Interestingly, the SAP-dependent decrease in Vγ4 T cell IL-17 production did not persist in the adult lung. Instead, a comparison of adult lung γδ T cell cytokine production between B6 and B6.SAP−/− mice revealed a significant decrease in both lung Vγ1 and Vγ4 IFN-γ production (Fig. 7C). We detected no SAP-dependent effect on Vγ6 IL-17 production (data not shown). A comparison of adult spleen Vγ1 and Vγ4 cytokine production between B6 and B6.SAP−/− mice also revealed significantly less IFN-γ in SAP-deficient Vγ1 and Vγ4 T cells than in their B6 counterparts, indicating that the SAP-dependent decrease was not tissue-specific (Fig. 7D). Together, these data demonstrated that γδ T cell IL-17 and IFN-γ production in the periphery was significantly impaired in SAP-deficient mice. In addition, the data indicated that the SAP-dependent impairment in γδ T cell function in the periphery was both age and subset-dependent.
Fig. 7. Impaired γδ T cell function in the peripheral tissues in SAP-deficient mice.
A) Decreased lung Vγ4 IL-17 production in B6.SAP−/− neonates. Representative contour plots of IL-17 and IFN-γ production from neonate lung Vγ1 and Vγ4 T cells in B6 and B6.SAP−/− mice are shown at left. Numbers indicate the percentage of IL-17 and IFN-γ production in each subset. Cumulative frequencies of IL-17 and IFN-γ-producing cells from γδ T cell subsets are shown at right. Data are the combined results of 2 independent experiments, day 6 to 7 after birth, sex was not determined. ***p < 0.001 as determined by two-way ANOVA followed by Sidak’s multiple comparisons test. B) Decreased SLAMF1+IL-17+ lung Vγ4 in neonate day 7 B6.SAP−/− mice. Representative contour plots of SLAMF1+IL-17+ lung Vγ4 T cells are shown above. The data are representative of 2 independent experiments, *p < 0.05 as determined by an unpaired, two-tailed t-test. The sex of the mice was not determined. C) Decreased Vγ1 and Vγ4 IFN-γ production in adult B6.SAP−/− lung (C) and spleen (D). Representative contour plots of IL-17 and IFN-γ production are shown at left. Frequencies of IL-17 and IFN-γ-producing cells are shown at right. Data representative the combined results of 2 to 3 independent experiments with male and female mice, 10 to 12 weeks of age, ***p < 0.001, ****p < 0.0001 as determined by two-way ANOVA followed by Sidak’s multiple comparisons test.
Discussion
Here, we identified the SLAM/SAP signaling pathway as a critical regulator of the functional programming that generates both IL-17 and IFN-γ-producing γδ T cells during thymic development. We found that maturation of thymic Vγ1 and Vγ4 was associated with the transition from a SLAMF1SLAMF6 DP phenotype to either SLAMF1 or SLAMF6 SP or SLAMF1SLAMF6 DN phenotypes, and that this transition was concomitant with the acquisition of γδ T cell effector function in the thymus. In the periphery, SLAMF1 and SLAMF6 marked IL-17- and IFN-γ-producing γδ T cells, respectively. Importantly, our findings indicated that in addition to its role in Vγ1Vδ6.3 T cell development, SAP also regulated Vγ4 T cell development, and that SAP-deficient Vγ4 T cells exhibited impaired developmental progression in the thymus, and impaired effector function in both the thymus and the periphery. Taken together, these findings suggest that the SAP-dependent signaling through SLAM receptors has a greater influence on γδ T cell development than previously appreciated and that this signaling pathway is a major contributor to the developmental programming that governs γδ T cell effector function.
The exact developmental stage(s) at which thymic γδ T cell functional programming occurs remains an open question. It has previously been shown that the CD24−CD44−CD45RB− γδ T cell population can give rise to both IFN-γ and IL-17 producers (24), but it is unclear whether this population is composed of uncommitted precursors or a mixture of already committed γδ T cells consisting of several functional subsets. Our findings that immature CD24+ thymic γδ T cells are SLAMF1SLAMF6 DP and that mature CD24− γδ T cells are enriched in SLAMF1 and SLAMF6 SP cells suggest that the CD24−CD44−CD45RB− γδ T cell population could be a heterogeneous mixture of SLAMF1 and SLAMF6 SP γδ T cells that have already committed to either IL-17 or IFN-γ production. Our results are consistent with a model in which γδ T cell functional programming occurs within the immature CD24+ population since both RORγt+ and RORγt− γδ T cells are found within the CD24+CD73− population (51), as are SLAMF1 SP γδ T cells which constitute the major subset of IL-17-producing γδ T cells.
A number of studies support a model in which high-affinity binding of γδ TCR during development promotes differentiation toward IFN-γ producing cells, while low-affinity interactions favor programming for IL-17 production (22-26, 56). In light of our findings indicating that both IL-17 and IFN-γ production are altered in SAP-deficient mice, it is interesting to note that SLAM receptor signaling has previously been shown to modulate TCR signaling thresholds in conventional αβ T cells (57). In addition, recent reports indicate that NKT cell differentiation is regulated by TCR signal strength (58, 59), and that the net effect of SLAM receptor signaling is to reduce NKT TCR signal strength (36). Indeed, the co-expression of both SLAMF1 and SLAMF6 on immature CD24+CD73− γδ T cells puts them in an optimal position to influence TCR signal strength and differentiation. Since CD73 expression has been reported to increase as a result of TCR signaling (49), SLAM/SAP-mediated regulation of TCR signal strength might be expected to influence the CD24+CD73− to CD24+CD73+ transition. However, some of our findings are not entirely consistent with such a model. First, SAP deletion did not affect neonatal Vγ4 CD73 expression. Instead, it appeared to block the transition from CD24+CD73− to CD24−CD73− (Pathway 2). Second, we found that the frequency and function of Vγ6 T cells, whose development is regulated by TCR signal strength (25), did not appear to be dependent on SAP. Additional investigation will be required to determine whether the SAP regulates γδ TCR signaling, or a TCR-independent process.
Indeed, accumulating evidence suggests that the development of at least some IL-17-producing γδ T cells can be regulated by TCR-independent mechanisms. γδ T cell development is dependent on a characteristic subset of transcription factors including SOX4, SOX13, Blk, c-maf, HEB, and PLZF, among others (27, 28, 51, 54, 60, 61). HEB was recently demonstrated to be critical for the progression of neonatal IL-17-producing Vγ4 T cells through Pathway 2 (51). Therefore, our finding that SAP also regulates the progression of Vγ4 cells through Pathway 2 provides a point of intersection between HEB and SLAM/SAP signaling that warrants future examination. In addition, it was recently reported that at least some γδT17 programming results from a SOX13-dominated transcriptional hard-wiring of early thymocyte precursors (29). These SOX13 progenitors appear to give rise to IL-17-producing Vγ4, but not Vγ6 cells. Our observation that Vγ4, but not Vγ6 subsets are SAP-dependent are therefore consistent with a role for SLAM receptors and SAP in the initiation of a transcriptional program critical for the acquisition of function. Indeed, because SLAM receptors are expressed on hematopoietic stem cells (62-64), they are in a position to influence γδ T cell programming at the earliest stages of development. Still, we note that because littermate controls were not used in these studies, we cannot exclude the possibility that the SAP-dependent effects on γδ T cell development and function observed here are due to strain-specific differences in microbiota or to developmental differences among the different litters. A future assessment of the role that SLAM/SAP signaling plays in γδ T cell precursor-product relationships using fetal thymic organ culture will be helpful in confirming our observations here, and in elucidating the mechanism(s) through which this pathway regulates γδ development.
Our findings demonstrate the presence of distinct SLAM family receptor expression profiles on γδ T cell subsets in the periphery that are tightly associated with function. These findings not only confirm a previous report that SLAMF1 marks γδT17 cells (48), they reveal the presence of a mutually exclusive expression pattern of SLAMF1 and SLAMF6 on γδ T cells that is tightly associated with distinct transcriptional programs and with IL-17 and IFN-γ production, respectively. Heterogeneous SLAM family receptor co-expression was observed among both B6 and BALB/c γδ T cells which possess different Slam haplotypes (53), although we did note significant strain-dependent differences. Specifically, there was a greater proportion of SLAMF1SP Vγ4 in BALB/c lung, liver, and spleen, which would suggest an increased frequency of IL-17-producing Vγ4. Although we have yet to confirm this prediction, it is interesting to note that NKT cell subsets are also differentially distributed in a strain-dependent manner, and that IL-17-producing NKT cells are also especially prevalent in BALB/c mice (65). The finding that certain SLAM family receptor expression patterns are established during thymic development, and that they are already associated with function as early as embryonic day 17, suggests that they result from a developmental program.
In support of this notion, significant changes in SLAMF1, SLAMF4, SLAMF6, and SLAMF7 expression have been described on B cells as they progress through developmental stages (66), and SLAM receptor expression profiles on hematopoietic stem cell and multipotent progenitor cell populations (62) have recently been demonstrated to mark functionally distinct progenitor populations (64). Together, our findings suggest the possibility that the coordinate signaling of distinct arrays of SLAM family receptors plays a role in the development of γδ T cells in the thymus. This would be consistent with reports indicating that multiple SLAM receptors are involved in NKT cell development (34, 36, 38, 39), and may explain the inability to observe γδT17 defects in mice lacking only a single SLAM family receptor (48).
Our observations indicated that SAP deletion did not completely abolish the development of IL-17-producing Vγ4, but instead reduced their numbers in thymus and neonatal lung by approximately 40%. This finding likely suggests the existence of one or more specific SAP-dependent Vγ4 subsets, as is the case with the Vγ1 T cell population that is composed of the SAP-dependent Vγ1Vδ6.3 subset and other SAP-independent subsets. Recent data demonstrating that the TCR repertoire of IL-17-producing Vγ4 T cells is highly restricted (67, 68), suggests the possibility that the partial decrease in Vγ4 IL-17 production observed here may be due to a complete failure in the development of a specific IL-17-producing Vγ4 subset. Such a model is also in line with the observation that the SAP-dependent decrease in lung γδ T cell IL-17 production observed in neonates was not consistently observed in adult mice. We speculate that the restricted development of IL-17-producing Vγ4 T subsets in embryonic and neonatal thymus (21) results in decreased SAP-dependent Vγ4 T subsets in the neonatal lung, and that over time, homeostatic proliferation of SAP-independent Vγ4 T subsets results in a repopulation of the adult lung. A future analysis of the γδ TCR repertoire in SAP-deficient mice should shed light on this issue.
In summary, our findings demonstrate that the SLAM/SAP signaling pathway plays an important role in shaping the balance of IL-17- and IFN-γ-producing γδ T cell commitment during thymic development. The findings here not only extend the influence of this signaling pathway to γδ T cell subsets other than the Vγ1Vδ6.3 γδNKT cell subset, they also establish a framework of SLAM family receptor co-expression that might be useful in dissecting and understanding γδ T cell development and function. Future studies aimed at understanding the specific mechanisms of SLAM family receptor signaling in this process will be required to integrate this pathway into existing models, and should provide insight into the central mechanisms that regulate γδ T cell functional programming.
Supplementary Material
Key Points:
γδ T cell subsets possess distinct SLAM family receptor co-expression profiles.
SLAMF1 and SLAMF6 identify IL-17- and IFN-γ-producing γδ T cells, respectively.
SAP regulates the development of both IL-17- and IFN-γ-producing γδ T cell subsets.
Acknowledgements
We wish to thank Roxana del Rio Guerra and the Flow Cytometry Cell Sorting Facility for help in cell sorting, Jessica Hoffman and Scott Tighe and the Vermont Integrated Genome Resource for help in microarray, Wendy Havran for providing a protocol for skin γδ T cell isolation, Cynthia Baldwin and Janice Telfer for helpful discussion, and Willi Born and Rebecca O’Brien for the 17D1 hybridoma.
This work was supported by NIHR21AI119974, NIHP30GM118228, and NIHS10OD018175 (JEB).
References
- 1.Chien YH, Meyer C, and Bonneville M. 2014. gammadelta T cells: first line of defense and beyond. Annu. Rev. Immunol 32: 121–155. [DOI] [PubMed] [Google Scholar]
- 2.Heilig JS, and Tonegawa S. 1986. Diversity of murine gamma genes and expression in fetal and adult T lymphocytes. Nature 322: 836–840. [DOI] [PubMed] [Google Scholar]
- 3.Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, Girardi M, Borst J, Hayday AC, Pennington DJ, and Silva-Santos B. 2009. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol 10: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas JD, Gonzalez FH, Schmitz S, Chennupati V, Fohse L, Kremmer E, Forster R, and Prinz I. 2009. CCR6 and NK1.1 distinguish between IL-17A and IFN-gamma-producing gammadelta effector T cells. Eur. J. Immunol 39: 3488–3497. [DOI] [PubMed] [Google Scholar]
- 5.Prinz I, Silva-Santos B, and Pennington DJ. 2013. Functional development of gammadelta T cells. Eur. J. Immunol 43: 1988–1994. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien RL, Roark CL, and Born WK. 2009. IL-17-producing gammadelta T cells. Eur. J. Immunol 39: 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y, Yang W, Pan M, Scully E, Girardi M, Augenlicht LH, Craft J, and Yin Z. 2003. Gamma delta T cells provide an early source of interferon gamma in tumor immunity. J. Exp. Med 198: 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanez DM, Batchelder J, van der Heyde HC, Manning DD, and Weidanz WP. 1999. Gamma delta T-cell function in pathogenesis of cerebral malaria in mice infected with Plasmodium berghei ANKA. Infect. Immun 67: 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami T, Hatano S, Yamada H, Iwakura Y, and Yoshikai Y. 2016. Two Types of Interleukin 17A-Producing gammadelta T Cells in Protection Against Pulmonary Infection With Klebsiella pneumoniae. J. Infect. Dis 214: 1752–1761. [DOI] [PubMed] [Google Scholar]
- 10.Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, Magorien JE, Blauvelt A, Kolls JK, Cheung AL, Cheng G, Modlin RL, and Miller LS. 2010. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Invest 120: 1762–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti HR, Peterson AC, Brane L, Huppler AR, Hernandez-Santos N, Whibley N, Garg AV, Simpson-Abelson MR, Gibson GA, Mamo AJ, Osborne LC, Bishu S, Ghilardi N, Siebenlist U, Watkins SC, Artis D, McGeachy MJ, and Gaffen SL. 2014. Oral-resident natural Th17 cells and gammadelta T cells control opportunistic Candida albicans infections. J. Exp. Med 211: 2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rei M, Goncalves-Sousa N, Lanca T, Thompson RG, Mensurado S, Balkwill FR, Kulbe H, Pennington DJ, and Silva-Santos B. 2014. Murine CD27(−) Vgamma6(+) gammadelta T cells producing IL-17A promote ovarian cancer growth via mobilization of protumor small peritoneal macrophages. Proc. Natl. Acad. Sci. U. S. A 111: E3562–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau CS, Verstegen NJM, Ciampricotti M, Hawinkels L, Jonkers J, and de Visser KE. 2015. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522: 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venken K, Jacques P, Mortier C, Labadia ME, Decruy T, Coudenys J, Hoyt K, Wayne AL, Hughes R, Turner M, Van Gassen S, Martens L, Smith D, Harcken C, Wahle J, Wang CT, Verheugen E, Schryvers N, Varkas G, Cypers H, Wittoek R, Piette Y, Gyselbrecht L, Van Calenbergh S, Van den Bosch F, Saeys Y, Nabozny G, and Elewaut D. 2019. RORgammat inhibition selectively targets IL-17 producing iNKT and gammadelta-T cells enriched in Spondyloarthritis patients. Nat Commun 10: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray EE, Ramirez-Valle F, Xu Y, Wu S, Wu Z, Karjalainen KE, and Cyster JG. 2013. Deficiency in IL-17-committed Vgamma4(+) gammadelta T cells in a spontaneous Sox13-mutant CD45.1(+) congenic mouse substrain provides protection from dermatitis. Nat Immunol 14: 584–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantelyushin S, Haak S, Ingold B, Kulig P, Heppner FL, Navarini AA, and Becher B. 2012. Rorgammat+ innate lymphocytes and gammadelta T cells initiate psoriasiform plaque formation in mice. J. Clin. Invest 122: 2252–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blink SE, Caldis MW, Goings GE, Harp CT, Malissen B, Prinz I, Xu D, and Miller SD. 2014. gammadelta T cell subsets play opposing roles in regulating experimental autoimmune encephalomyelitis. Cell. Immunol 290: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, and Mills KH. 2009. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31: 331–341. [DOI] [PubMed] [Google Scholar]
- 19.Roark CL, French JD, Taylor MA, Bendele AM, Born WK, and O’Brien RL. 2007. Exacerbation of collagen-induced arthritis by oligoclonal, IL-17-producing gamma delta T cells. J. Immunol 179: 5576–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jouan Y, Patin EC, Hassane M, Si-Tahar M, Baranek T, and Paget C. 2018. Thymic Program Directing the Functional Development of gammadeltaT17 Cells. Frontiers in immunology 9: 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas JD, Ravens S, Duber S, Sandrock I, Oberdorfer L, Kashani E, Chennupati V, Fohse L, Naumann R, Weiss S, Krueger A, Forster R, and Prinz I. 2012. Development of interleukin-17-producing gammadelta T cells is restricted to a functional embryonic wave. Immunity 37: 48–59. [DOI] [PubMed] [Google Scholar]
- 22.Turchinovich G, and Hayday AC. 2011. Skint-1 identifies a common molecular mechanism for the development of interferon-gamma-secreting versus interleukin-17-secreting gammadelta T cells. Immunity 35: 59–68. [DOI] [PubMed] [Google Scholar]
- 23.Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, Steinman L, Saito T, Locksley RM, Davis MM, Baumgarth N, and Chien YH. 2008. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity 29: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumaria N, Grandjean CL, Silva-Santos B, and Pennington DJ. 2017. Strong TCRgammadelta Signaling Prohibits Thymic Development of IL-17A-Secreting gammadelta T Cells. Cell Rep 19: 2469–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz-Ruiz M, Ribot JC, Grosso AR, Goncalves-Sousa N, Pamplona A, Pennington DJ, Regueiro JR, Fernandez-Malave E, and Silva-Santos B. 2016. TCR signal strength controls thymic differentiation of discrete proinflammatory gammadelta T cell subsets. Nat Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wencker M, Turchinovich G, Di Marco Barros R, Deban L, Jandke A, Cope A, and Hayday AC. 2014. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nat Immunol 15: 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malhotra N, Narayan K, Cho OH, Sylvia KE, Yin C, Melichar H, Rashighi M, Lefebvre V, Harris JE, Berg LJ, Kang J, and C. Immunological Genome Project. 2013. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity 38: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, Kornfeld H, Xiong N, Cohen NR, Brenner MB, Berg LJ, Kang J, and C. Immunological Genome Project. 2012. Intrathymic programming of effector fates in three molecularly distinct gammadelta T cell subtypes. Nat Immunol 13: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spidale NA, Sylvia K, Narayan K, Miu B, Frascoli M, Melichar HJ, Zhihao W, Kisielow J, Palin A, Serwold T, Love P, Kobayashi M, Yoshimoto M, Jain N, and Kang J. 2018. Interleukin-17 Producing gammadelta T Cells Originate from SOX13(+) Progenitors that Are Independent of gammadeltaTCR Signaling. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cannons JL, Tangye SG, and Schwartzberg PL. 2011. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol 29: 665–705. [DOI] [PubMed] [Google Scholar]
- 31.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, and Terhorst C. 1998. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature 395: 462–469. [DOI] [PubMed] [Google Scholar]
- 32.Poy F, Yaffe MB, Sayos J, Saxena K, Morra M, Sumegi J, Cantley LC, Terhorst C, and Eck MJ. 1999. Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol. Cell 4: 555–561. [DOI] [PubMed] [Google Scholar]
- 33.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, and Stein PL. 2005. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med 11: 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, and Bendelac A. 2007. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27: 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, Procopio E, Salou M, Gilet J, Ryffel B, Balvay A, Foussier A, Sarkis M, El Marjou A, Schmidt F, Rabot S, and Lantz O. 2019. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Zhong MC, Qian J, Calderon V, Cruz Tleugabulova M, Mallevaey T, and Veillette A. 2019. SLAM receptors foster iNKT cell development by reducing TCR signal strength after positive selection. Nat Immunol. [DOI] [PubMed] [Google Scholar]
- 37.Cuenca M, Punet-Ortiz J, Ruart M, Terhorst C, and Engel P. 2018. Ly9 (SLAMF3) receptor differentially regulates iNKT cell development and activation in mice. Eur. J. Immunol 48: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, Cai C, Li Z, Liu G, Wang Y, Blonska M, Li D, Du J, Lin X, Yang M, and Dong Z. 2017. Dissection of SAP-dependent and SAP-independent SLAM family signaling in NKT cell development and humoral immunity. J. Exp. Med 214: 475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang B, Gomez-Rodriguez J, Preite S, Garrett LJ, Harper UL, and Schwartzberg PL. 2016. CRISPR-Mediated Triple Knockout of SLAMF1, SLAMF5 and SLAMF6 Supports Positive Signaling Roles in NKT Cell Development. PloS one 11: e0156072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu JK, Crampton JC, Locci M, and Crotty S. 2016. CRISPR-Mediated Slamf1Delta/Delta Slamf5Delta/Delta Slamf6Delta/Delta Triple Gene Disruption Reveals NKT Cell Defects but Not T Follicular Helper Cell Defects. PloS one 11: e0156074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, and Sant’Angelo DB. 2010. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J. Immunol 184: 6746–6755. [DOI] [PubMed] [Google Scholar]
- 42.Benoit P, Sigounas VY, Thompson JL, van Rooijen N, Poynter ME, Wargo MJ, and Boyson JE. 2015. The role of CD1d-restricted NKT cells in the clearance of Pseudomonas aeruginosa from the lung is dependent on the host genetic background. Infect. Immun 83: 2557–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, Boismenu R, and Havran WL. 2002. A role for skin gammadelta T cells in wound repair. Science 296: 747–749. [DOI] [PubMed] [Google Scholar]
- 44.Aktan I, Chant A, Borg ZD, Damby DE, Leenstra PC, Lilley GW, Petty J, Suratt BT, Teuscher C, Wakeland EK, Poynter ME, and Boyson JE. 2010. Slam haplotypes modulate the response to lipopolysaccharide in vivo through control of NKT cell number and function. J. Immunol 185: 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puddington L, Olson S, and Lefrancois L. 1994. Interactions between stem cell factor and c-Kit are required for intestinal immune system homeostasis. Immunity 1: 733–739. [DOI] [PubMed] [Google Scholar]
- 46.Roark CL, Aydintug MK, Lewis J, Yin X, Lahn M, Hahn YS, Born WK, Tigelaar RE, and O’Brien RL. 2004. Subset-specific, uniform activation among V gamma 6/V delta 1+ gamma delta T cells elicited by inflammation. J. Leukoc. Biol 75: 68–75. [DOI] [PubMed] [Google Scholar]
- 47.Schuhmachers G, Ariizumi K, Mathew PA, Bennett M, Kumar V, and Takashima A. 1995. 2B4, a new member of the immunoglobulin gene superfamily, is expressed on murine dendritic epidermal T cells and plays a functional role in their killing of skin tumors. J. Invest. Dermatol 105: 592–596. [DOI] [PubMed] [Google Scholar]
- 48.Schmolka N, Serre K, Grosso AR, Rei M, Pennington DJ, Gomes AQ, and Silva-Santos B. 2013. Epigenetic and transcriptional signatures of stable versus plastic differentiation of proinflammatory gammadelta T cell subsets. Nat Immunol 14: 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coffey F, Lee SY, Buus TB, Lauritsen JP, Wong GW, Joachims ML, Thompson LF, Zuniga-Pflucker JC, Kappes DJ, and Wiest DL. 2014. The TCR ligand-inducible expression of CD73 marks gammadelta lineage commitment and a metastable intermediate in effector specification. J. Exp. Med 211: 329–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buus TB, Geisler C, and Lauritsen JP. 2016. The major diversification of Vgamma1.1(+) and Vgamma2(+) thymocytes in mice occurs after commitment to the gammadelta T-cell lineage. Eur. J. Immunol 46: 2363–2375. [DOI] [PubMed] [Google Scholar]
- 51.In TSH, Trotman-Grant A, Fahl S, Chen ELY, Zarin P, Moore AJ, Wiest DL, Zuniga-Pflucker JC, and Anderson MK. 2017. HEB is required for the specification of fetal IL-17-producing gammadelta T cells. Nat Commun 8: 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, and von Boehmer H. 2009. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. U. S. A 106: 12453–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR Jr., Morel L, and Wakeland EK. 2004. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity 21: 769–780. [DOI] [PubMed] [Google Scholar]
- 54.Zuberbuehler MK, Parker ME, Wheaton JD, Espinosa JR, Salzler HR, Park E, and Ciofani M. 2019. The transcription factor c-Maf is essential for the commitment of IL-17-producing gammadelta T cells. Nat Immunol 20: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lino CNR, Barros-Martins J, Oberdorfer L, Walzer T, and Prinz I. 2017. Eomes expression reports the progressive differentiation of IFN-gamma-producing Th1-like gammadelta T cells. Eur. J. Immunol 47: 970–981. [DOI] [PubMed] [Google Scholar]
- 56.McKenzie DR, Comerford I, Silva-Santos B, and McColl SR. 2018. The Emerging Complexity of gammadeltaT17 Cells. Frontiers in immunology 9: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao F, Cannons JL, Dutta M, Griffiths GM, and Schwartzberg PL. 2012. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity 36: 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tuttle KD, Krovi SH, Zhang J, Bedel R, Harmacek L, Peterson LK, Dragone LL, Lefferts A, Halluszczak C, Riemondy K, Hesselberth JR, Rao A, O’Connor BP, Marrack P, Scott-Browne J, and Gapin L. 2018. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat Commun 9: 2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao M, Svensson MND, Venken K, Chawla A, Liang S, Engel I, Mydel P, Day J, Elewaut D, Bottini N, and Kronenberg M. 2018. Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat Commun 9: 2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laird RM, Laky K, and Hayes SM. 2010. Unexpected role for the B cell-specific Src family kinase B lymphoid kinase in the development of IL-17-producing gammadelta T cells. J. Immunol 185: 6518–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu Y, Cao X, Zhang X, and Kovalovsky D. 2015. PLZF Controls the Development of Fetal-Derived IL-17+Vgamma6+ gammadelta T Cells. J. Immunol 195: 4273–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, and Morrison SJ. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121. [DOI] [PubMed] [Google Scholar]
- 63.Kim I, He S, Yilmaz OH, Kiel MJ, and Morrison SJ. 2006. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood 108: 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oguro H, Ding L, and Morrison SJ. 2013. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 13: 102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, and Hogquist KA. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 14: 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Salort J, Sintes J, Llinas L, Matesanz-Isabel J, and Engel P. 2011. Expression of SLAM (CD150) cell-surface receptors on human B-cell subsets: from pro-B to plasma cells. Immunol. Lett 134: 129–136. [DOI] [PubMed] [Google Scholar]
- 67.Kashani E, Fohse L, Raha S, Sandrock I, Oberdorfer L, Koenecke C, Suerbaum S, Weiss S, and Prinz I. 2015. A clonotypic Vgamma4Jgamma1/Vdelta5Ddelta2Jdelta1 innate gammadelta T-cell population restricted to the CCR6(+)CD27(−) subset. Nat Commun 6: 6477. [DOI] [PubMed] [Google Scholar]
- 68.Wei YL, Han A, Glanville J, Fang F, Zuniga LA, Lee JS, Cua DJ, and Chien YH. 2015. A Highly Focused Antigen Receptor Repertoire Characterizes gammadelta T Cells That are Poised to Make IL-17 Rapidly in Naive Animals. Frontiers in immunology 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.