Abstract
Objective: Cancer is expected to be the leading cause of death worldwide within the 21st century and is the single most important obstacle to extending life expectancy. Unfortunately, the most effective approach to combating cancers remains a complex and unsolved problem. Siglec-15 is a member of the Siglec family and plays a conserved regulatory role in the immune system of vertebrates. Previous studies on Siglec-15 have focused on its function in osteoclast regulation. The purpose of this study was to explore the significance of Siglec-15 mRNA in human cancer mainly based on information obtained from online databases.
Method: Data were collected from several online databases. Serial analysis of gene expression (SAGE) and Virtual Northern, UALCAN Database Analysis, Catalog of Somatic Mutations in Cancer (COSMIC) analysis, the cBio cancer genomics portal, Cancer Regulome tools and data, Kaplan-Meier Plotter Analysis and the UCSC Xena website were used to analyze the data.
Results: Compared with normal tissues, Siglec-15 up-regulation was widely observed in tuomrs. Differences in Siglec-15 expression were associated with different prognoses. Siglec-15 mutations are widely observed in tumors and interact with different genes in different cancer types.
Conclusion: Siglec-15 is a potential target for the expansion of cancer immunotherapy.
Keywords: Siglec-15, cancer, immunotherapy, biomarker
Introduction
With the development of medical science, novel methods for the diagnosis and treatment of many diseases are being discovered and bring greater opportunities and increased hope to patients worldwide. However, the most effective approach to combating cancer remains a complex and unsolved problem for not only doctors but also scientists. According to the Global Cancer Statistics 2018 report, there were an estimated 18.1 million new cancer cases (17.0 million excluding nonmelanoma skin cancer) and 9.6 million cancer deaths (9.5 million excluding nonmelanoma skin cancer) in 2018 1. The report also stated that cancer is expected to be the leading cause of death worldwide within the 21st century and is the single most important obstacle to extending life expectancy 1. Traditional treatments for cancer include surgery, chemotherapy and radiotherapy. Immunotherapy and targeted therapies have also been applied gradually in the clinic, but limitations related to cancer diagnosis and drug resistance remain. To solve these problems, potential biomarkers are needed for the development of novel diagnostic and treatment approaches.
The sialic acid-binding immunoglobulin-like lectin (Siglec) family is the known largest group of vertebrate lectins that can recognize sialylated glycans. Originally, Siglec family members were found on immune cells and determined to be involved in coupling glycan recognition to immunological regulation 2. Siglec-15 is a member of the Siglec family and is considered to play a conserved, regulatory role in the immune system of vertebrates 2. The extracellular domain of Siglec-15 consists of an immunoglobulin variable region and a type 2 constant region (IgC2), which has more than 30% homology with the B7 gene family. These results indicate that Siglec-15 may have similar immunomodulatory functions with the B7 family 3, which includes the renowned immunological therapy biomarker PD-L1. Siglec-15 has been reported to activate the AKT pathway through DAP12 4. The activation of AKT could promote tumor cell proliferation 5-7. Previous studies on Siglec-15 have focused on its function in osteoclast regulation 4, 8, 9. However, some recent studies have demonstrated that Siglec-15 is highly expressed in some cancers and might play important roles in tumor immunity 10, 11.
Few studies have focused on the role of Siglec-15 in tumors, and thus, its expression in tumors remains unclear. To elucidate the role of Siglec-15 in tumors, in this study, database tools were used to obtain information from well-known online databases, such as TCGA, GEO and EGA, to examine Siglec-15 mRNA expression in different types of solid tumors.
Material & Methods
Serial Analysis of Gene Expression (SAGE) and Virtual Northern
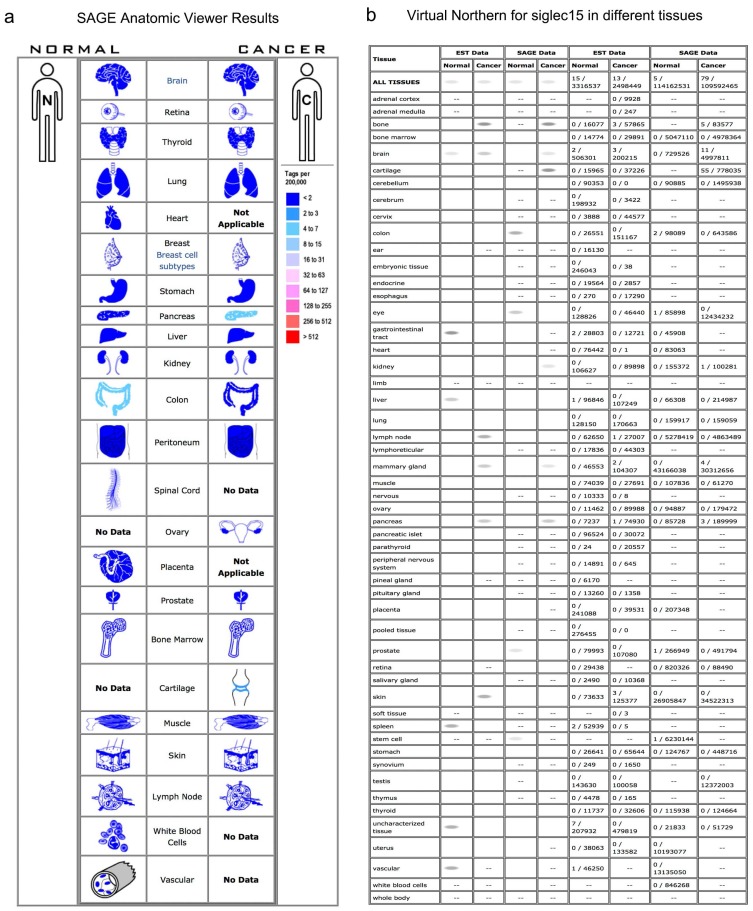
SAGE is a useful technique developed for the genome-wide analysis of gene expression. It utilizes efficient computational tools to acquire and analyze tag sequences from raw sequence files to compare tag abundances between different libraries 12. Monochromatic SAGE/cDNA Virtual Northern displays the expression of genes in all human organs. In this study, both SAGE and Monochromatic SAGE/cDNA Virtual Northern tools from the Cancer Genome Anatomy Project (https://cgap.nci.nih.gov/Genes) were used. The SAGE results revealed the expression of Siglec-15 in cancer and normal tissues, and the Virtual Northern results displayed the levels of Siglec-15 in human tissues.
UALCAN Database
UALCAN (http://ualcan.path.uab.edu) is a comprehensive, user-friendly, and interactive web resource for analyzing cancer OMICS data. The database uses 31 types of cancer RNA-seq and clinical information from TCGA to complete in-depth analyses of TCGA gene expression data, including analysis of the relative expression of a query gene across tumor and normal samples 13. In this study, the UALCAN database was used to obtain data from the TCGA database and compare the expression of Siglec-15 mRNA in tumors and normal tissues.
Catalog of Somatic Mutations in Cancer (COSMIC)
COSMIC (https://cancer.sanger.ac.uk/cosmic/) is the world's largest and most comprehensive resource for exploring the impact of somatic mutations in human cancer. It has a collection of data on millions of coding mutations, noncoding mutations, genomic rearrangements, fusion genes, copy number abnormalities and gene expression variants in the human genome all in one database so that researchers can explore these data more easily14. In this study, COSMIC was used to investigate the mutations of Siglec-15 in human cancers, and the results are depicted in pie charts.
Mutation Rate and Distribution of Different Exons
The cBio cancer genomics portal (http://cbioportal.org) is an open-access resource that simplifies the molecular analysis of cancer tissues and cell lines into easy-to-understand genetic, epigenetic, gene expression, and proteomic events15, 16. We used cBioPortal to analyze Siglec-15 in TCGA pan-cancer data to identify coexpressed genes.
Integrative Data Visualization
The Cancer Regulome tools and data (http://explorer.cancerregulome.org/) from the TCGA database were used to create circus plots to display the expression of Siglec-15 and its correlation with other genes in tumors. Spearman correlation was used to show the pairwise correlation between two genes. Only genes with P values > - log10 are shown in the circus plots.
Kaplan-Meier Plotter
The Kaplan-Meier Plotter (http://kmplot.com/analysis/) is an online tool that is typically used to draw survival curves. The system includes gene chip and RNA-seq data, and sources for the databases include GEO, EGA, and TCGA 17. In this study, the mRNA expression levels of Siglec-15 in each cancer were stratified into high and low groups by the web, and then survival curves were drawn. Hazard ratios with 95% confidence intervals and log-rank P values were also calculated.
Genomic Data and Views
The UCSC Xena website (http://xena.ucsc.edu/) offers tools for the visualization and exploration of TCGA genomic data 18. According to the coexpression results from Oncomine and an interesting study 11, we performed a search on Siglec-15, SMAD7, ATP5A1 and CD274 (PD-L1) using the Genes viewing mode.
Results
Siglec-15 mRNA in Normal and Tumor Tissues
Normally, Siglec-15 has low expression in nonimmune organs. As shown in the SAGE results in Figure 1a, a higher expression of Siglec-15 was observed in pancreatic cancer, cartilage tumors and normal colon tissues. In the Virtual Northern results, compared with normal tissues, Siglec-15 was upregulated in bone, breast and pancreatic cancer tissues (Figure 1b).
Figure 1.
The SAGE Anatomic Viewer. a. Expression profile for Siglec-15 in human cancers displayed by the SAGE Digital Gene Expression Displayer (DGED). b Monochromatic SAGE/cDNA Virtual Northern results for Siglec-15 in different tissues.
Upregulation of Siglec-15 mRNA in Cancer
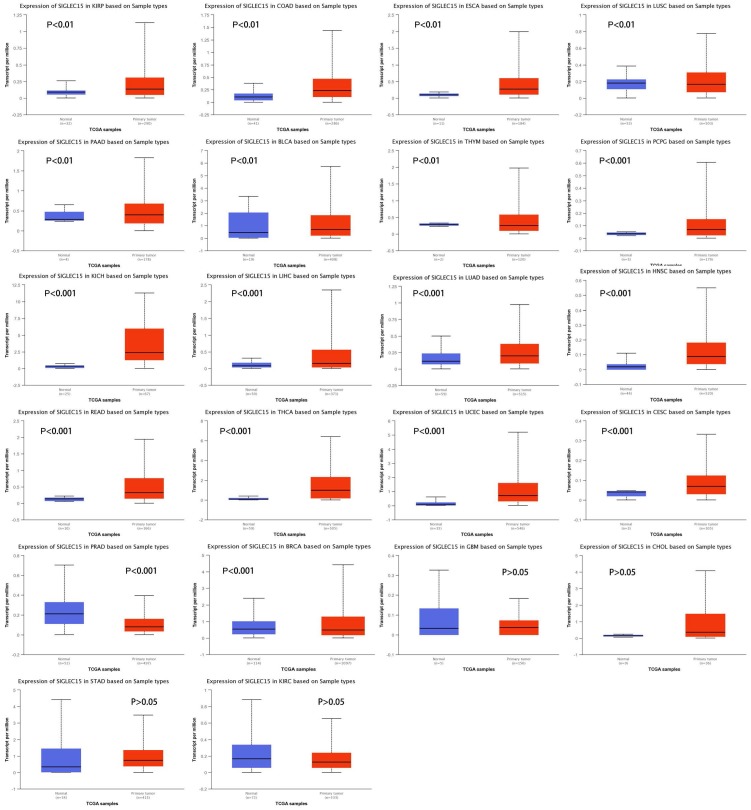
To examine Siglec-15 mRNA levels across different cancer types, data from the UALCAN database were used. We chose TCGA data as the data source and compared the Siglec-15 mRNA levels in cancer tissues with those in normal tissues. As shown in Figure 2, the expression of Siglec-15 mRNA in most cancers was obviously different from that in normal tissues. Higher Siglec-15 mRNA levels were found in kidney renal papillary cell carcinoma (KIRP), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), pancreatic adenocarcinoma (PAAD), bladder urothelial carcinoma (BLCA), pheochromocytoma and paraganglioma (PCPG), kidney chromophobe (KICH), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), head and neck squamous cell carcinoma (HNSC), rectum adenocarcinoma (READ), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC) and cervical squamous cell carcinoma (CESC),while downregulation could be observed in thymoma (THYM), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD) and breast invasive carcinoma (BRCA). However, no significant differences were observed in glioblastoma multiforme (GBM), cholangiocarcinoma (CHOL), stomach adenocarcinoma (STAD) and kidney renal clear cell carcinoma (KIRC).
Figure 2.
Siglec-15 mRNA was evaluated in human cancers by UALCAN database analysis.
Siglec-15 Mutations in Cancer
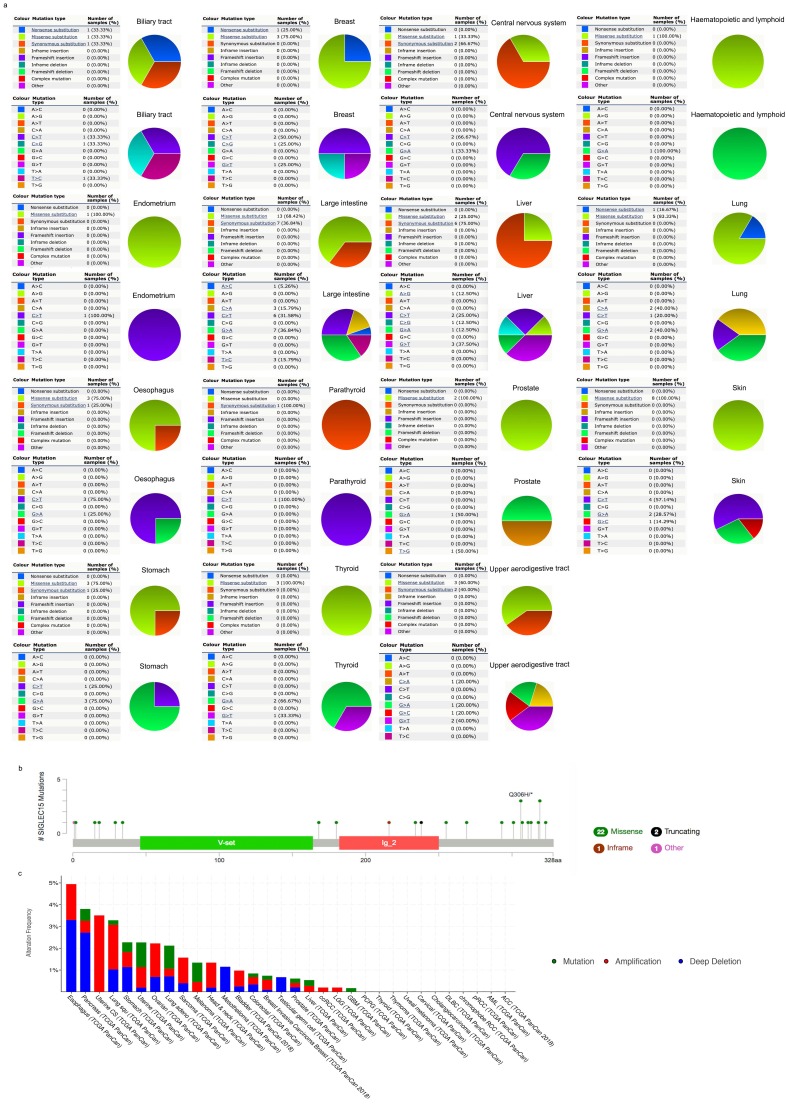
COSMIC provided information on Siglec-15 mutations in different cancers, which included substitution missense, nonsense and synonymous mutations, and the results are depicted in pie charts. Nonsense substitutions were found in biliary tract cancer (33.33%), breast cancer (25%) and lung cancer (16.67%), while substitution missense mutations were observed in biliary tract cancer (33.33%), breast cancer (75%), central nervous system cancer (33.33%), hematopoietic and lymphoid cancer (100%), endometrial cancer (100%), large intestine cancer (68.42%), liver cancer (25%), lung cancer (83.33%), esophageal cancer (75%), prostate cancer (100%), skin cancer (100%), stomach cancer (75%), thyroid cancer (100%) and upper aerodigestive tract cancer (60%). Additionally, synonymous substitution mutations appeared in biliary tract cancer (33.33%), central nervous system cancer (66.67%), large intestine cancer (36.84%), liver cancer (75%), esophageal cancer (25%), parathyroid cancer (100%), stomach cancer (25%) and upper aerodigestive tract cancer (40%). C>T and G>A mutations were most common in the Siglec-15 coding strand, both of which were observed in eleven cancer types. A>T and T>A mutations in Siglec-15 were not found in the TCGA cancer samples. Other types of base mutations occurred sporadically in different cancers (Figure 3a). With the help of cBioPortal, we found that a total of 22 mutation sites were detected and located between amino acids 0 and 328 (Figure 3b). Figure 3c shows the mutation results from cBioPortal, which shows the Siglec-15 mutation level in the TCGA cancer database. Pancreatic cancer, lung cancer, stomach cancer, uterine cancer, melanoma cancer, colorectal cancer, breast cancer, prostate cancer, liver cancer and glioblastoma multiforme all had high mutation levels.
Figure 3.
(a) Pie chart showing the percentage of the different mutation types of Siglec-15 in human cancers according to the COSMIC database. (b) Mutation diagram of Siglec-15 in different cancer types across protein domains. (c) Siglec-15 mutation level in the TCGA cancer database.
Genome-wide Association of Siglec-15 mRNA in Cancer
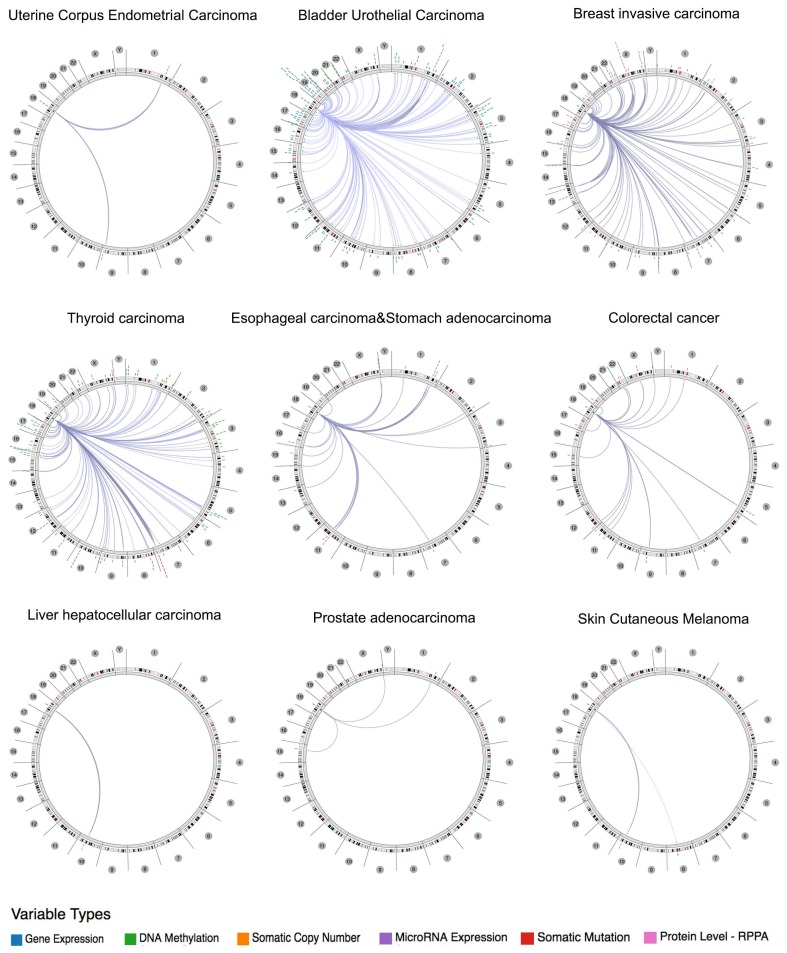
Using the Regulome Explorer, we further analyzed the relevant human genome location and the correlation between certain genes and Siglec-15 in human cancer. Based on the association among genes, DNA methylation, somatic copy number, somatic mutation and protein level, circus plots were drawn to display the interrelation between Siglec-15 and other genes. According to the data from TCGA, Siglec-15 was associated with other genes that could be detected in colorectal cancer, bladder urothelial carcinoma, breast invasive carcinoma, esophageal carcinoma and stomach adenocarcinoma, uterine corpus endometrial carcinoma, thyroid carcinoma, liver hepatocellular carcinoma, prostate adenocarcinoma and skin cutaneous melanoma (Figure 4). Detailed data can be found in Supplementary Tables 1 to 9.
Figure 4.
The correlation between Siglec-15 and other genes from the TCGA database (Regulome program).
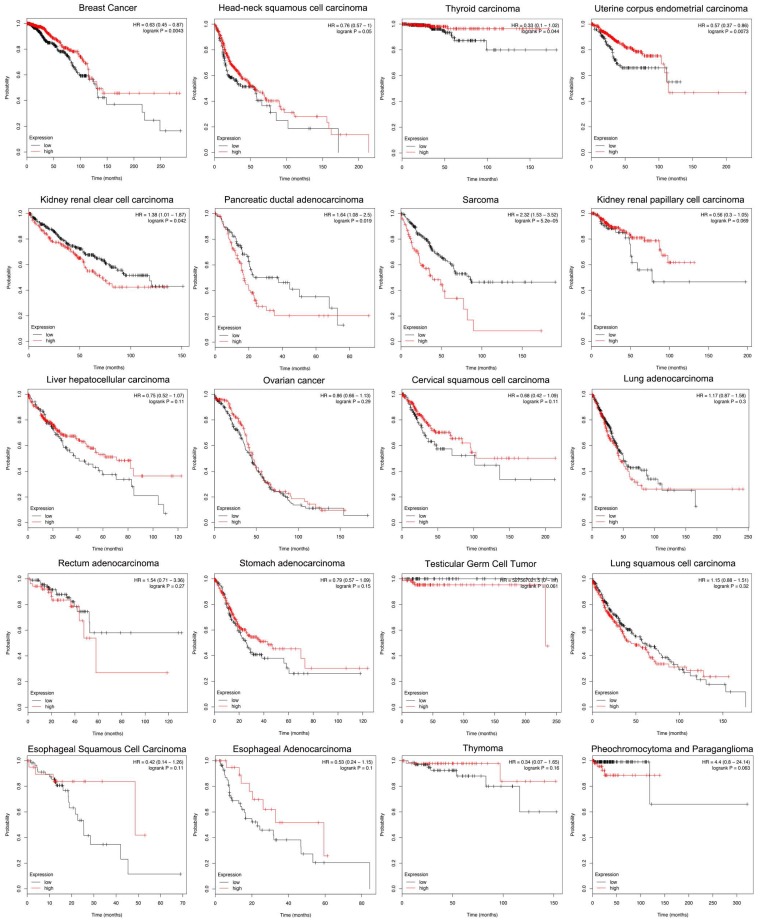
Siglec-15 and the Survival Rate of Cancers
According to the Kaplan-Meier analysis results, higher levels of Siglec-15 mRNA indicated worse overall survival in pancreatic ductal adenocarcinoma (P=0.019), sarcoma (P=5.2E-05) and kidney renal clear cell carcinoma (P=0.042). However, the opposite result was observed in breast cancer (P=0.0043), head and neck squamous cell carcinoma (P=0.05), thyroid carcinoma (P=0.044), and uterine corpus endometrial carcinoma (P=0.0073). The expression of Siglec-15 mRNA level have no significant influence in kidney renal papillary cell carcinoma (P=0.069), liver hepatocellular carcinoma (P=0.11), ovarian cancer (P=0.29), cervical squamous cell carcinoma (P=0.11), lung adenocarcinoma (P=0.3), rectum adenocarcinoma (P=0.27), stomach adenocarcinoma (P=0.15), testicular germ cell tumor (P=0.061), lung squamous cell carcinoma (P=0.32), esophageal squamous cell carcinoma (P=0.11), pheochromocytoma and paraganglioma (P=0.063), esophageal adenocarcinoma (P=0.1) and thymoma (P=0.16) (Figure 5).
Figure 5.
Different expression levels of Siglec-15 mRNA will result in different overall survival rates in cancers (Kaplan-Meier analysis).
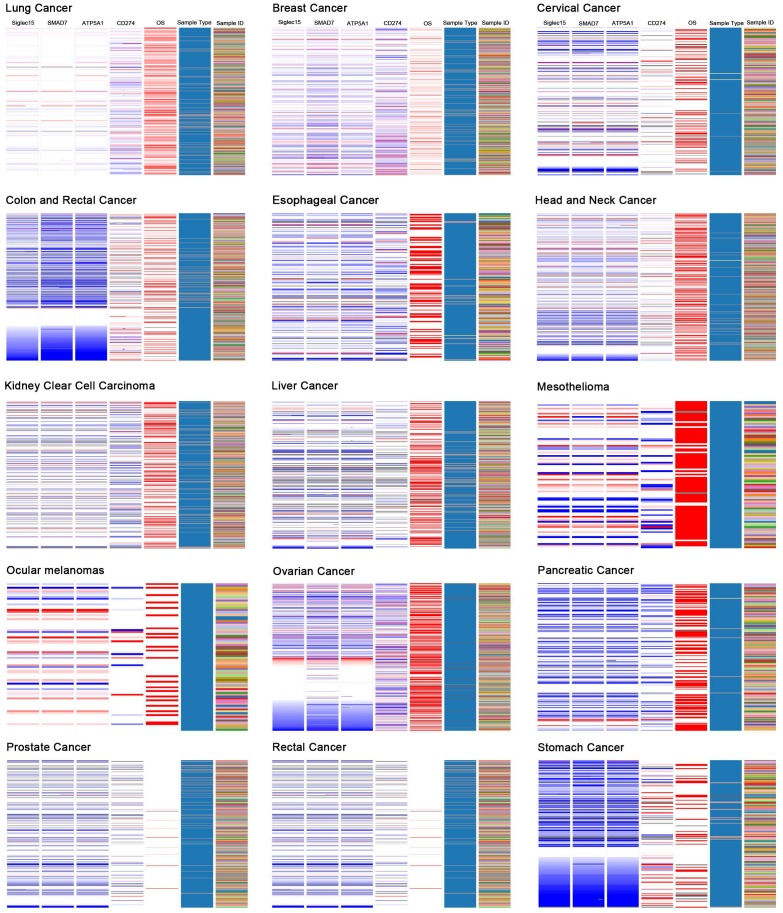
Correlations among Siglec-15, SMAD7, ATP5A1, and CD274 (PD-L1)
Based on the results of the Oncomine coexpression analysis and a newly published study 11, we chose SMAD family member 7 (SMAD7), ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit 1, cardiac muscle (ATP5A1), and CD274 (PD-L1) in this study. According to the image created by UCSC Xena, in the TCGA tumor samples, the DNA copy numbers of Siglec-15, SMAD7 and ATP5A1 showed a similar trend, while Siglec-15 and CD274 (PD-L1) showed the opposite trend.
Discussion
The Siglec family is a group of lectins with a structure containing an N-terminal V-set Ig-like domain and a sialic acid-binding domain, followed by variable numbers of C2-set Ig-like domains. Eleven family members were found to bind sialic acid on glycoconjugates 19. Although their structures are similar, their binding specificity differs, and they bind specific sialoglycoconjugates 20. Most Siglec family members are observed on only hematopoietic cells, as they have very low expression in other tissues, but a recent study found that their expression level changed in some tumor cells 21-23. Several lines of evidence have shown that interactions with sialic acid-binding receptors can influence cancer progression; for example, hypersialylation can induce changes in the physical properties of tumor cells and potentiate the evasion of apoptosis in cancer cells 24, 25. As the Siglec family has the ability to bind specific sialoglycoconjugates, it was regarded as a potential targeted therapy biomarker 26.
Siglec-15 is a Siglec gene family member with the ability to bind the sialyl-Tn antigen, and it has been used as a targeted therapy biomarker for osteoporosis 4, 9, 27, 28. Siglec-15 is an I-type transmembrane protein that includes two immunoglobulin-like domains, a transmembrane domain containing lysine residues and a short cytoplasmic tail. It associates with the activating adaptor proteins DNAX activation protein (DAP)12 and DAP10 via its lysine residue in the transmembrane domain, implying that it functions as an activating signaling molecule. Moreover it is the second human Siglec identified to have an activating signaling potential 2. As Figure 1 shows, Siglec-15 mRNA has very low expression in normal human tissues, including most immune cell types; however, it could be detected in macrophages, bone tumors and pancreatic cancer. A study published in Nature proved that Siglec-15 expression was increased in many human cancers; we also obtained the same results, as shown in Figure 2. Compared with normal tissues, the expression of Siglec-15 was upregulated at different levels in KIRP, COAD, ESCA, PAAD, BLCA, PCPG, KICH, LIHC, LUAD, HNSC, READ, THCA, UCEC and CESC. Moreover, a previous study confirmed that Siglec-15 could suppress antigen-specific T cell responses in vitro and in vivo. Additionally, knockdown of Siglec-15 expression did not cause obvious physical abnormalities but did inhibit tumor growth 11. Based on these results, Siglec-15 is believed to be a good potential targeted therapy biomarker.
Since no previous studies have focused on Siglec-15 mutations in human cancers, we explored this topic with the help of COSMIC and cBioPortal. As shown in Figure 3, the results from TCGA demonstrated that Siglec-15 mutations occurred widely in human cancers. The most common type of Siglec-15 mutation was missense substitution, which could be observed in all tumors with mutations. At the base-pair level, C>T and G>A mutations were the most widely observed in tumors. Base substitution can lead to abnormal transcription and translation, which might cause changes in both function and phenotype. Figure 5 shows that a higher level of Siglec-15 expression in pancreatic ductal adenocarcinoma, sarcoma and kidney renal clear cell carcinoma indicates worse overall survival, while the opposite result is true in breast cancer, head and neck squamous cell carcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma. The reason for this phenomenon is still unclear, but the results shown in Figure 5 suggest that patients with different Siglec-15 expression levels have distinct overall survival rates. Nevertheless, Siglec-15 was found to be a useful biomarker for predicting prognosis. Considering the results presented in Figure 4, we postulate that the reason for the opposing results could be that Siglec-15 interacts with entirely different genes in different types of cancers. Alterations in the expression of Siglec-15 may cause a variety of gene changes across different cancers, which could lead to remarkably different results.
Figure 6 displays the coexpression results of Siglec-15 in different cancers. According to the results, SMAD7 and ATP5A1 show similar trends, while Siglec-15 and CD274 (PD-L1) show the opposite trend. SMAD7 is a nuclear protein that binds the E3 ubiquitin ligase SMURF2 and is closely connected with the TGF-β pathway. For cutaneous melanoma patients, high Siglec-15 expression indicates a high risk of tumor aggressiveness and adverse clinical outcomes 29. The same conclusion was drawn for colon, ovarian, and endometrial cancers 30-33. ATP5A1 encodes a subunit of mitochondrial ATP synthase and plays a critical role in catalyzing ATP synthesis, participating in tumor angiogenesis and regulating tumor-related phosphorylation 34. The upregulation of ATP5A1 is widely observed in clear cell renal cell carcinoma, glioblastoma, colorectal cancer and thyroid cancer 34-37. In this study, Siglec-15, SMAD7 and ATP5A1 showed the same expression trends in tumors; thus, they may share some pathways that promote tumor aggressiveness. CD274 (PD-LI) is one of the most well-known biomarkers for tumor immunological therapy. Its inhibitor, atezolizumab, is used in bladder cancer and non-small cell lung cancer, as approved by the FDA 38, 39. In this study, we found that Siglec-15 expression is mutually exclusive from PD-L1. This result is supported by findings reported in a previous study 11. Therefore, targeting Siglec-15 may be an option for patients who are not candidates for PD-L1 immunotherapy.
Figure 6.
The UCSC Cancer Genomics Browser was used to explore the relationships among Siglec-15 and SMAD7, ATP5A1 and PD-L1.
In conclusion, our study suggested that Siglec-15 mRNA is upregulated in many cancers and that the differential expression of Siglec-15 leads to different prognoses. Siglec-15 mutations are widely observed in tumors and interact with different genes in different cancer types, which might explain why high Siglec-15 expression does not necessarily indicate a poor prognosis in all cancers. Siglec-15 represents a potential target for the expansion of cancer immunotherapy in some kinds of cancers.
Supplementary Material
Supplementary tables.
Acknowledgments
Data Availability
The datasets analyzed in the current study are available in The Cancer Genome Atlas (http://cancergenome.nih.gov), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) repositories.
Contributions
Zhi-fan Xiong and Sheng-li Yang designed the study; Qiu-ting Li and Zao-zao Huang carried out the research; Qiu-ting Li, Yao-bin Chen, Hong-Yi Yao, Zun-hui Ke, Xiao-xiao He, Meng-jun Qiu and Meng-meng Wang analyzed the data; and Qiu-ting Li wrote the paper.
Funding Statement
This study was supported by The National Key Research and Development Program of China (No: 2018YFC2002000) and the Fundamental Research Funds for the Central Universities: the Independent Innovation Fund of Huazhong University of Science and Technology (No: 2018KFYYXJJ106).
Abbreviations
- TCGA
The Cancer Genome Atlas
- SAGE
Serial analysis of gene expression
- COSMIC
Catalog of Somatic Mutations in Cancer
- Siglec
sialic acid-binding immunoglobulin-like lectin
- SMAD7
SMAD family member 7
- ATP5A1
ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit 1, cardiac muscle
- PD-L1
Programmed cell death 1 ligand 1
- GEO
Gene Expression Omnibus
- EGA
European Genome-phenome Archive
- KIRP
Kidney renal papillary cell carcinoma
- COAD
Colon adenocarcinoma
- ESCA
Esophageal carcinoma
- PAAD
Pancreatic adenocarcinoma
- BLCA
Bladder urothelial carcinoma
- PCPG
Pheochromocytoma, Paraganglioma
- KICH
Kidney chromophobe
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- HNSC
Head and neck squamous cell carcinoma
- READ
Rectum adenocarcinoma
- THCA
Thyroid carcinoma
- UCEC
Uterine corpus endometrial carcinoma
- CESC
Cervical squamous cell carcinoma
- THYM
Thymoma
- LUSC
Lung squamous cell carcinoma
- PRAD
Prostate adenocarcinoma
- BRCA
Breast invasive carcinoma
- GBM
Glioblastoma multiforme
- CHOL
Cholangiocarcinoma
- STAD
Stomach adenocarcinoma
- KIRC
Kidney renal clear cell carcinoma
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology. 2007;17:838–46. doi: 10.1093/glycob/cwm049. [DOI] [PubMed] [Google Scholar]
- 3.Ren X. Immunosuppressive checkpoint Siglec-15: a vital new piece of the cancer immunotherapy jigsaw puzzle. Cancer biology & medicine. 2019;16:205–10. doi: 10.20892/j.issn.2095-3941.2018.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stuible M, Moraitis A, Fortin A, Saragosa S, Kalbakji A, Filion M. et al. Mechanism and function of monoclonal antibodies targeting siglec-15 for therapeutic inhibition of osteoclastic bone resorption. The Journal of biological chemistry. 2014;289:6498–512. doi: 10.1074/jbc.M113.494542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y, Fang L, Wang S, Li Y, Guo Y, Sun YP. Amphiregulin promotes trophoblast invasion and increases MMP9/TIMP1 ratio through ERK1/2 and Akt signal pathways. Life sciences. 2019;236:116899. doi: 10.1016/j.lfs.2019.116899. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Gu L, Li X, Wang J. Silencing of Synuclein-gamma inhibits human cervical cancer through the AKT signaling pathway. Cellular & molecular biology letters. 2019;24:49. doi: 10.1186/s11658-019-0172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Chen X, Hay N. Akt as a target for cancer therapy: more is not always better (lessons from studies in mice) Br J Cancer. 2017;117:159–63. doi: 10.1038/bjc.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kameda Y, Takahata M, Komatsu M, Mikuni S, Hatakeyama S, Shimizu T. et al. Siglec-15 regulates osteoclast differentiation by modulating RANKL-induced phosphatidylinositol 3-kinase/Akt and Erk pathways in association with signaling Adaptor DAP12. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2013;28:2463–75. doi: 10.1002/jbmr.1989. [DOI] [PubMed] [Google Scholar]
- 9.Ishida-Kitagawa N, Tanaka K, Bao X, Kimura T, Miura T, Kitaoka Y. et al. Siglec-15 protein regulates formation of functional osteoclasts in concert with DNAX-activating protein of 12 kDa (DAP12) The Journal of biological chemistry. 2012;287:17493–502. doi: 10.1074/jbc.M111.324194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takamiya R, Ohtsubo K, Takamatsu S, Taniguchi N, Angata T. The interaction between Siglec-15 and tumor-associated sialyl-Tn antigen enhances TGF-beta secretion from monocytes/macrophages through the DAP12-Syk pathway. Glycobiology. 2013;23:178–87. doi: 10.1093/glycob/cws139. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M. et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nature medicine. 2019;25:656–66. doi: 10.1038/s41591-019-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva AP, De Souza JE, Galante PA, Riggins GJ, De Souza SJ, Camargo AA. The impact of SNPs on the interpretation of SAGE and MPSS experimental data. Nucleic acids research. 2004;32:6104–10. doi: 10.1093/nar/gkh937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (New York, NY) 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic acids research. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific reports. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao M, Liu Q, Li B, Liao W, Xie W, Zhang Y. A group of long noncoding RNAs identified by data mining can predict the prognosis of lung adenocarcinoma. Cancer Sci. 2018;109:4033–44. doi: 10.1111/cas.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crocker PR, Varki A. Siglecs, sialic acids and innate immunity. Trends in immunology. 2001;22:337–42. doi: 10.1016/s1471-4906(01)01930-5. [DOI] [PubMed] [Google Scholar]
- 20.Yamaji T, Nakamura K, Amari S, Suzuki A, Hashimoto Y. Application of a multivalent glycoprobe: characterization of sugar-binding specificity of Siglec family proteins. Methods in enzymology. 2003;363:104–13. doi: 10.1016/S0076-6879(03)01046-2. [DOI] [PubMed] [Google Scholar]
- 21.Ou C, Liu L, Wang J, Dai S, Qu Y, Xiong Y. et al. Enhancement of Siglec-8 expression predicts adverse prognosis in patients with clear cell renal cell carcinoma. Urologic oncology. 2017;35:607.e1–e8. doi: 10.1016/j.urolonc.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, Liu H, Zhang H, Lin C, Li R, Zhang W. et al. Decreased expression of Siglec-8 associates with poor prognosis in patients with gastric cancer after surgical resection. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:10883–91. doi: 10.1007/s13277-016-4859-7. [DOI] [PubMed] [Google Scholar]
- 23.Madge PD, Maggioni A, Pascolutti M, Amin M, Waespy M, Bellette B. et al. Structural characterisation of high affinity Siglec-2 (CD22) ligands in complex with whole Burkitt's lymphoma (BL) Daudi cells by NMR spectroscopy. Scientific reports. 2016;6:36012. doi: 10.1038/srep36012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schauer R. Achievements and challenges of sialic acid research. Glycoconjugate journal. 2000;17:485–99. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz MJ, Swindall AF, Bellis SL. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer metastasis reviews. 2012;31:501–18. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams OJ, Stanczak MA, von Gunten S, Laubli H. Targeting sialic acid-Siglec interactions to reverse immune suppression in cancer. Glycobiology. 2018;28:640–7. doi: 10.1093/glycob/cwx108. [DOI] [PubMed] [Google Scholar]
- 27.Hiruma Y, Hirai T, Tsuda E. Siglec-15, a member of the sialic acid-binding lectin, is a novel regulator for osteoclast differentiation. Biochem Biophys Res Commun. 2011;409:424–9. doi: 10.1016/j.bbrc.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 28.Kameda Y, Takahata M, Mikuni S, Shimizu T, Hamano H, Angata T. et al. Siglec-15 is a potential therapeutic target for postmenopausal osteoporosis. Bone. 2015;71:217–26. doi: 10.1016/j.bone.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 29.Kaczorowski M, Biecek P, Donizy P, Pieniazek M, Matkowski R, Halon A. SMAD7 is a novel independent predictor of survival in patients with cutaneous melanoma. Translational research: the journal of laboratory and clinical medicine. 2019;204:72–81. doi: 10.1016/j.trsl.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J. et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 31.Chen T, Triplett J, Dehner B, Hurst B, Colligan B, Pemberton J. et al. Transforming growth factor-beta receptor type I gene is frequently mutated in ovarian carcinomas. Cancer Res. 2001;61:4679–82. [PubMed] [Google Scholar]
- 32.Lynch MA, Nakashima R, Song H, DeGroff VL, Wang D, Enomoto T. et al. Mutational analysis of the transforming growth factor beta receptor type II gene in human ovarian carcinoma. Cancer Res. 1998;58:4227–32. [PubMed] [Google Scholar]
- 33.Sakaguchi J, Kyo S, Kanaya T, Maida Y, Hashimoto M, Nakamura M. et al. Aberrant expression and mutations of TGF-beta receptor type II gene in endometrial cancer. Gynecologic oncology. 2005;98:427–33. doi: 10.1016/j.ygyno.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Yuan L, Chen L, Qian K, Wang G, Lu M, Qian G. et al. A novel correlation between ATP5A1 gene expression and progression of human clear cell renal cell carcinoma identified by coexpression analysis. Oncology reports. 2018;39:525–36. doi: 10.3892/or.2017.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu G, Li JY. ATP5A1 and ATP5B are highly expressed in glioblastoma tumor cells and endothelial cells of microvascular proliferation. Journal of neuro-oncology. 2016;126:405–13. doi: 10.1007/s11060-015-1984-x. [DOI] [PubMed] [Google Scholar]
- 36.Eldai H, Periyasamy S, Al Qarni S, Al Rodayyan M, Muhammed Mustafa S, Deeb A. et al. Novel genes associated with colorectal cancer are revealed by high resolution cytogenetic analysis in a patient specific manner. PLoS One. 2013;8:e76251. doi: 10.1371/journal.pone.0076251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onda M, Emi M, Yoshida A, Miyamoto S, Akaishi J, Asaka S. et al. Comprehensive gene expression profiling of anaplastic thyroid cancers with cDNA microarray of 25 344 genes. Endocrine-related cancer. 2004;11:843–54. doi: 10.1677/erc.1.00818. [DOI] [PubMed] [Google Scholar]
- 38.Passiglia F, Galvano A, Rizzo S, Incorvaia L, Listi A, Bazan V. et al. Looking for the best immune-checkpoint inhibitor in pre-treated NSCLC patients: An indirect comparison between nivolumab, pembrolizumab and atezolizumab. Int J Cancer. 2018;142:1277–84. doi: 10.1002/ijc.31136. [DOI] [PubMed] [Google Scholar]
- 39.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J. et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–65. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.
Data Availability Statement
The datasets analyzed in the current study are available in The Cancer Genome Atlas (http://cancergenome.nih.gov), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) repositories.