Abstract
N6-methyladenosine (m6A) is the most prevalent eukaryotic messenger RNA modification. Diabetic cataract (DC) is caused by high glucose (HG) in diabetes mellitus. However, the regulatory mechanism of m6A in the DC pathogenesis is poorly understood. In present research, we performed the m6A-RNA immunoprecipitation sequencing (MeRIP-Seq) analysis and detected the m6A modification profile in the HG- or normal glucose (NG)-induced human lens epithelial cells (HLECs). Results revealed that methyltransferase-like 3 (METTL3) was upregulated in the DC tissue specimens and HG-induced HLECs. Besides, total m6A modification level was higher in the HG-induced HLECs. Functionally, METTL3 knockdown promoted the proliferation and repressed the apoptosis of HLECs induced by HG. MeRIP-Seq analysis revealed that ICAM-1 might act as the target of METTL3. Mechanistically, METTL3 targets the 3′ UTR of ICAM-1 to stabilize mRNA stability. In conclusion, this research identified the regulation of METTL3 in the HG-induced HLECs, providing a potential insight of the m6A modification for DC.
Keywords: N6-methyladenosine, human lens epithelial cells, diabetic cataract, METTL3, ICAM-1
Introduction
Diabetic cataract (DC) is characterized by the disorder and nubecula of human lens epithelial cells (HLECs) caused by abnormal blood supply of terminal circulation in diabetes mellitus.1,2 In addition, other complications may be aroused by the diabetes mellitus, such as microcirculation abnormity and metabolic disorders.3 In the DC pathogenesis, high glucose (HG) could give rise to the lens apoptosis and metabolism disorder.
N6-methyladenosine (m6A) is the most common internal modification of eukaryotic mRNA, participating in the protein-coding transcripts, exports, translation, and decay.4,5 For the m6A installation, methyltransferase protein complex could deposit the methyl to the adenosine, including methyltransferase-like 3 (METTL3), METTL14, Wilms’ tumor 1-associating protein (WTAP), and Virilizer homolog (KIAA1429).5 For the uninstallation, demethylases remove the methyl from adenosine, including AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated (FTO). Besides, the reader proteins are responsible for the recognition, including YTH family domain of YT521-B homology (YTHDF1-3, YTHDC1-2).6
Approximately, 0.1%–0.4% of adenosines in total RNAs are modified by m6A methylation.7 The consensus motif of m6A is identified as RRACH (R: G, A, U; R: G, A; H: U, A, C).8 According to the high-throughput m6A profiling, results revealed that m6A sites are mainly enriched in 3′ untranslated regions (3′ UTRs) and near stop codons. METTL3 is a critical m6A writer and functions as an essential initiating factor in multiple pathogenesis. For instance, METTL3 associates with ribosomes and enhances mRNA translation through an interaction with the translation initiation machinery to promote the translation in the cytoplasm.9
In present research, we performed the m6A-RNA immunoprecipitation sequencing (MeRIP-Seq) analysis and found that the m6A peaks distribution was distributed near the stop codon, covering the coding sequence (CDS) and 3′ UTR. METTL3 silencing could promote the proliferation and repress the apoptosis of HLECs induced by HG. Further experiments revealed that METTL3 targets the 3′ UTR of ICAM-1 to stabilize its protein expression. In conclusion, this research identified the regulation of METTL3 in the HG-induced HLECs, providing a potential insight of the m6A modification for DC.
Results
MeRIP-Seq Analysis Reveals the m6A Modification in HLECs
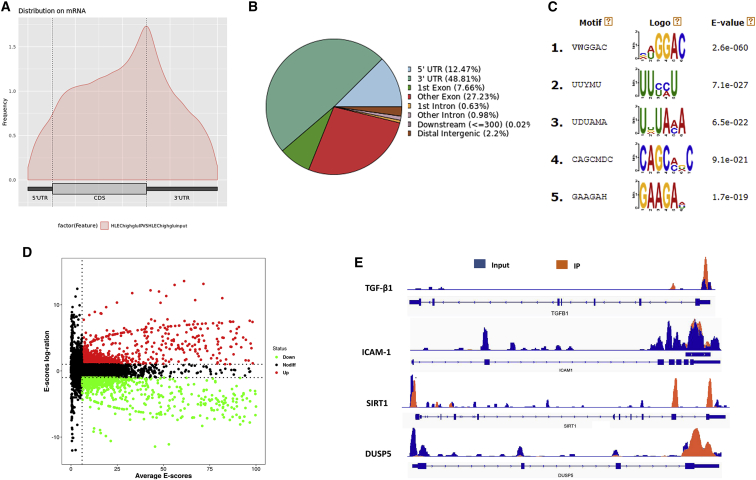
In order to investigate the m6A modification of DC, MeRIP-Seq analysis was performed in the HG-induced HLECs. Density distribution of m6A peaks across the mRNA transcripts revealed that the main components of m6A peaks were concentrated on the stop codon, covering CDS and 3′ UTR (Figure 1A). Proportion of m6A peaks distribution illustrated that m6A peaks were distributed in 3′ UTR, 5′ UTR, exon, intron, downstream region, and distal intergenic region (Figure 1B). Based on the MeRIP-Seq analysis, several potential consensus m6A motifs were identified (Figure 1C). Among these candidate motifs, the consensus sequence “GGAC” occupied a significant portion, which is in accord with the previous reports.10,11 Volcano plot of the MeRIP-Seq analysis showed the expression difference in the m6A-tagged or un-tagged transcripts (Figure 1D). The Integrative Genomics Viewer (IGV) tool revealed the m6A peaks distribution in several candidate target genes, including transforming growth factor-β1 (TGF-β1), ICAM-1, SIRT1, and DUSP5 (Figure 1E). Overall, these data revealed the m6A modification in the HLECs detected by MeRIP-Seq analysis.
Figure 1.
MeRIP-Seq Analysis Reveals the m6A Modification in HLECs
(A) Density distribution of m6A peaks across the mRNA transcripts, including 5′ untranslated regions (3′ UTR), covering coding sequence (CDS) and 3′ UTR. (B) Representative chart illustrated the proportion of m6A peaks distribution in 3′ UTR, 5′ UTR, exon, intron, downstream region, and distal intergenic region. (C) Several potential consensus m6A motif were identified from the MeRIP-Seq analysis. (D) Volcano plot showed the expression difference in the m6A-tagged transcripts of the MeRIP-Seq analysis. (E) The Integrative Genomics Viewer (IGV) tool revealed the m6A peaks distribution in several candidate target genes, including TGF-β1, ICAM-1, SIRT1, and DUSP5.
METTL3 Was Upregulated in DC Tissue and HG-Induced HLECs
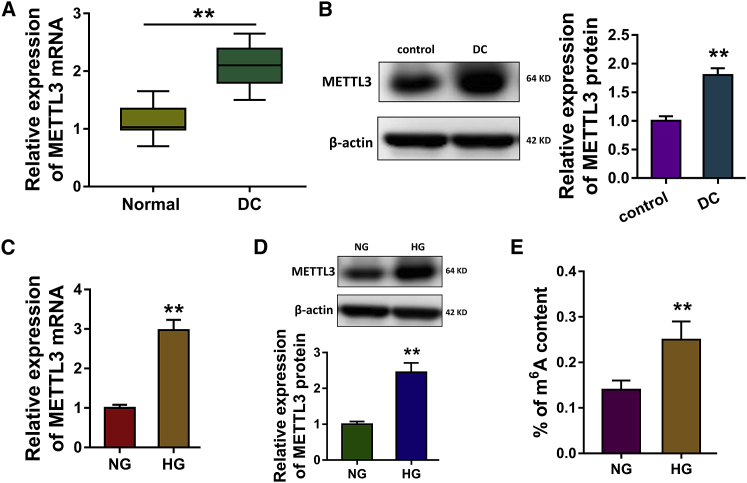
In the enrolled DC tissue samples and lens anterior capsules samples, RT-PCR revealed that METTL3 expression was upregulated in the DC tissue samples (Figure 2A). Western blot analysis revealed that METTL3 protein was upregulated in the DC tissue samples (Figure 2B). In the HG-induced HLECs, RT-PCR revealed that METTL3 mRNA was upregulated as comparing to the normal glucose (NG) induced HLECs (Figure 2C). Moreover, western blot analysis showed that the METTL3 protein was upregulated in HG-induced HLECs (Figure 2D). The m6A quantitative analysis unveiled that the percentage of m6A content in the total RNA was increased in the HG-induced HLECs (Figure 2E). Overall, these findings concluded that METTL3 was upregulated in DC tissue and HG-induced HLECs, which was also correlated with the m6A content.
Figure 2.
METTL3 Was Upregulated in DC Tissue and HG-Induced HLECs
(A) RT-PCR revealed the METTL3 expression in the DC tissue samples and lens anterior capsules samples. (B) Western blot analysis revealed the METTL3 protein in the DC tissue samples and lens anterior capsules samples. (C) RT-PCR revealed that METTL3 mRNA in the high glucose (HG) or normal glucose (NG) induced HLECs. (D) Western blot analysis showed the METTL3 protein in HG- or NG-induced HLECs. (E) The m6A quantitative analysis unveiled the percentage of m6A content in the total RNA. **p < 0.01 indicates the significant difference.
METTL3 Regulated the HG-Induced HLECs Proliferation and Apoptosis
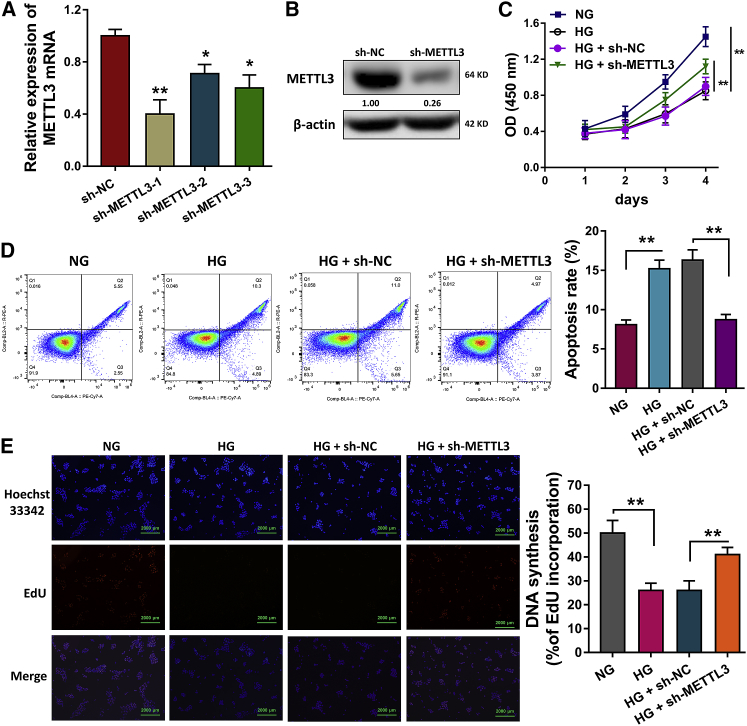
In order to investigate the biological role of METTL3, short hairpin RNA (shRNA) for METTL3 was constructed and transfected into HLECs to silence the METTL3 mRNA (Figure 3A). Western blot analysis revealed that the METTL3 protein was decreased after the sh-METTL3 transfection (Figure 3B). Cell counting kit-8 (CCK-8) assay revealed that the HG administration reduced the proliferation of HLECs, and sh-METTL3 transfection rescued the proliferation (Figure 3C). Flow cytometry revealed that the HG administration promoted the apoptosis and HG administration alleviated the apoptosis of HLECs (Figure 3D). 5-Ethynyl-2′-deoxyuridine (EdU) assay illustrated that HG administration repressed the proliferation of HLE B-3 cells and sh-METTL3 transfection promoted the proliferation (Figure 3E). These data indicated that METTL3 participated in the HG-induced HLECs proliferation and apoptosis.
Figure 3.
METTL3 Regulated the HG-Induced HLECs Proliferation and Apoptosis
(A) Short hairpin shRNA (shRNA) for METTL3 were constructed and transfected into HLECs (HLE B-3) to silence the METTL3 mRNA. The relative level was detected using RT-PCR. (B) Western blot analysis revealed the METTL3 protein after the sh-METTL3 transfection. (C) CCK-8 assay revealed the proliferation of HLECs. (D) Flow cytometry revealed the apoptosis of HLECs with the NG or HG administration and sh-METTL3 or control transfection. (E) EdU assay illustrated the proliferation of HLECs with the NG or HG administration and sh-METTL3 or control transfection. **p < 0.01 indicates the significant difference.
ICAM-1 Acted as a Target Protein of METTL3 in the HLECs
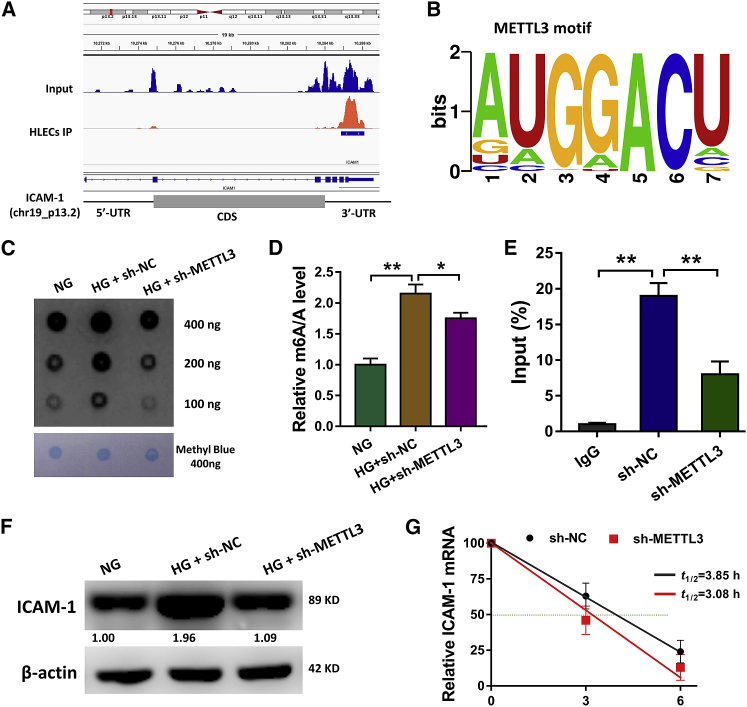
The IGV tool revealed that, comparing with the Input group, the m6A peaks distribution IP group was located in the 5′ UTR and 3′ UTR (Figure 4A). Moreover, we found that the major m6A peaks were distributed at the 3′ UTR. With the help of online bioinformatics tools (http://www.cuilab.cn/sramp), we found that the METTL3 motif in the HLECs is the consensus sequence (Figure 4B). Dot blot analysis revealed that the m6A quantity is upregulated in the HG-induced HLECs, while the quantity is reduced after the METTL3 silencing (Figure 4C). Analogously, m6A quantitative analysis revealed that METTL3 silencing could reduce the m6A level as comparing to the control group (Figure 4D). RIP-qPCR showed that the METTL3 silencing remarkedly decreased the ICAM-1 mRNA enrichment precipitated by antibody (Figure 4E). Western blot illustrated that the HG administration could enhance the ICAM-1 protein expression as comparing to NG, however, the METTL3 silencing repressed the ICAM-1 protein expression (Figure 4F). RNA stability assay showed that METTL3 silencing decreased the mRNA half-life (t1/2) as compared to the control (Figure 4G). In conclusion, these data supported that ICAM-1 acted as a target protein of METTL3 in the HLECs.
Figure 4.
ICAM-1 Acted as a Target Protein of METTL3 in the HLECs
(A) The Integrative Genomics Viewer (IGV) tool revealed the m6A peaks distribution in Input group and IP group. (B) METTL3 motif in the HLECs is the consensus sequence. (C) Dot blot analysis revealed the m6A quantity in the HG-induced HLECs. (D) m6A quantitative analysis illustrated the m6A level with or without METTL3 silencing. (E) RIP-qPCR (RNA immunoprecipitation following qPCR) showed the ICAM-1 mRNA enrichment precipitated by METTL3 antibody. (F) Western blot illustrated the ICAM-1 protein expression with HG administration and METTL3 silencing transfection. (G) RNA stability assay showed the ICAM-1 mRNA half-life (t1/2) with METTL3 silencing. **p < 0.01 indicates the significant difference.
Discussion
The m6A modification for RNA is a critical regulation in the epigenetic regulation catching more and more researchers’ attention. In multiple human pathophysiological processes, including cancer and embryonic development, m6A has been identified to modify extensive RNA transcription and protein generation.12,13 However, the roles of m6A in the DC pathogenesis are still unclear.
The m6A for RNA is a dynamic and reversible process catalyzed by methyltransferase complex, including METTL3, METTL14, and WTAP. In our present research, we performed the MeRIP-Seq and found that the m6A peaks distribution was located near the stop codon, covering CDS and 3′ UTR. Moreover, the MeRIP-Seq analysis unveiled several potential consensus m6A motifs. Volcano plot of the MeRIP-Seq analysis showed the ectopically expressed m6A-tagged or un-tagged transcripts within HG-induced or NG-induced HLECs. Besides, we selected several candidate target genes with remarkable m6A peaks, including ICAM-1, TGF-β1, and DUSP5.
In present research, we established the DC and control models using the HG-induced or NG-induced HLECs. In the HG-induced HLECs, m6A methyltransferase METTL3 was upregulated as comparing to the NG-induced HLECs. Meanwhile, the m6A level was upregulated in the HG-induced HLECs, indicating that the HG could stimulate the m6A level in the HLECs. These data supported that HG stimulation could enhance the methylation level in total transcripts, illustrating that the RNA methylation might regulate the DC pathogenesis. Therefore, the investigation of m6A modification in the DC could shed light on its underlying mechanism.
In the functional experiments, we found that METTL3 silencing promoted the proliferation and alleviated the apoptosis of HG-induced HLECs. Therefore, the finding clued that METTL3 indeed could regulate the biological phenotype of HLECs. The roles of METTL3 in the human diseases have been wildly illustrated. For example, in the colorectal carcinoma, METTL3 specifically methylated the SOX2 transcripts via targeting the CDS regions through specific m6A “reader” IGF2BP2.14 In bladder cancer, the increased METTL3 induced the upregulation of ITGA6, and METTL3 could increase m6A methylations of the ITGA6 mRNA 3′ UTR to promote the translation of ITGA6 mRNA via readers (YTHDF1, YTHDF3).15
In present research, we found that METTL3 specifically targeted the ICAM-1 3′ UTR and promoted its protein expression. The m6A peak targeted by the METTL3 is found to be located in the 3′ UTR, which is approximate to the stop codon. These findings in our research are in accord with previous literature that m6A sites are intensively distributed near the stop codon.6,16 In our previous research, we investigated the roles of long noncoding RNAs (lncRNAs) in the HLECs and identified the functions of lncRNAs.17,18 In further research, we would perform more investigation to unveil the interaction within lncRNA and m6A.19, 20, 21
In conclusion, present research found that METTL3 was upregulated in the HG-induced HLECs, and METTL3 installed the higher methylation level. Mechanistically, METTL3 targets the 3′ UTR of ICAM-1 to stabilize its protein expression. Overall, the finding could provide a novel insight for the m6A modification for DC.
Materials and Methods
Tissue Samples Collection
Anterior lens capsule tissues were collected from DC patients and normal patient individuals without diabetes mellitus. The clinical assay was approved by the Ethics Committee of Eye Hospital of Tianjin Medical University. Informed consents were signed by every volunteer before the surgery.
Cells and Culture
Human LEC line (HLE B-3) was purchased from ATCC (CRL-11421) and cultured in the DMEM medium supplemented with 10% FBS (fetal bovine serum) and 15 mM HEPES as previously described,22 and then maintained at 37°C with 5% CO2.
MeRIP-Seq
Total RNA was extracted from the HLEC cells administrated with HG and NG. MeRIP-Seq analysis was performed by Jiayin Biotechnology (Shanghai, China).
Oligonucleotides Transfection
The shRNA and blank control targeting METTL3 were synthesized by the GENEWIZ (Suzhou, China). The transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s specifications.
RNA Isolation and qRT-PCR
Total RNA was isolated from HLECs and DC tissue specimens using QIAGEN RNeasy Mini kit (Hilden, Germany). cDNA was reverse-transcribed using the iScript cDNA Synthesis Kit (BioRad) according to the manufacturer’s protocol. qRT-PCR was performed using the SYBR Premix Ex Taq (Takara, Otsu, Japan) using Applied Biosystems StepOne Plus Real-Time Thermal Cycling. GAPDH was used as an internal control for mRNA. The relative level of RNA was calculated using the 2−ΔΔCt method. The primer sequences were shown in Table S1.
Western Blot Analysis
Protein was extracted from HLECs and DC tissue specimens using radioimmunoprecipitation assay (RIPA) lysis buffer with 1% protease inhibitor (Solarbio, Beijing, China). Proteins were separated by sodium dodecylsulphate-polyacrylamide gel electrophoresis (10% SDS-PAGE) and transferred to polyvinylidene difluoride member (PVDF, Millipore, MA, USA). Primary antibody (anti-METTL3, Abcam, ab195352, 1:1,000, anti-ICAM-1, ab53013, 1:2,000) was incubated with PVDF member overnight at 4°C. Then, the PVDF members were incubated with secondary antibodies (anti-β-actin, 1:1,000). The signals were visualized using enhanced chemiluminescence (Thermo Scientific, Rockford, IL, USA) and graphed using Gel Doc 2000 imaging scanner (Bio-Rad).
m6A Quantification
Total RNA was isolated using TRIzol (Invitrogen, CA) according to the manufacturer’s instruction. After the quality of RNA validation by NanoDrop, the m6A content in the total RNAs was assessed using m6A RNA methylation quantification kit (ab185912, Abcam).
HLECs Proliferative Assay
The proliferative ability was performed using CCK-8 and EdU assay. For the CCK-8 assay, HLECs were seeded in 96-well culture plates and administrated with 10 μL of CCK-8 assay kit (Dojindo Japan). 24 h later, cells were measured for the absorbance at 450 nm. For the EdU assay, HLECs were seeded in 24-well plates at 2 × 104 cells using the EdU labeling/detection kit (Ribobio, Guangzhou, China). EdU labeling medium was added and fixed in 4% paraformaldehyde (pH 7.4) for 30 min. After counterstaining with 250 mL DAPI (Invitrogen, Molecular Probes, Eugene, OR, USA) for 25 min, EdU-positive cells were imaged under a fluorescence microscope (Nikon Corporation, Tokyo, Japan).
Flow Cytometry
The apoptosis of HLECs were detected using flow cytometry assay. In brief, HLECs were washed with PBS and resuspended in 100 μL of 1 × binding buffer. Fluorescein isothiocyanate (FITC) Annexin V (5 μL) and propidium iodide (PI, 5 μL) were added for 15 min at room temperature in the dark. The cells were analyzed by FACS Canto II flow cytometry (BD Biosciences).
Dot Blot Assay
Dot blots were performed as previously described.23 RNA (400 ng) was spotted onto a nylon membrane (GE Healthcare, Fairfield, CT, USA). Nylon membranes were UV crosslinked and then incubated with m6A antibody (anti-m6A, ab208577, Abcam, 1:100) and horseradish peroxidase (HRP) conjugate anti-rabbit immunoglobulin G (IgG). The final images were incubated by HRP-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology was added to the blots for 1 h at room temperature. The same RNA (400 ng) were spotted on the nylon membrane, stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2) for 2 h, and washed with ribonuclease-free water for 5 h.
RNA Stability
The ICAM-1 mRNA stability was detected as previously described.24 RNA was extracted using Trizol reagent (Invitrogen, Grand Island, NY, USA) from HLECs at different time points, mRNAs were treated with actinomycin D (1 μg/mL). Reverse transcription was performed using oligo (dT) primers and ICAM-1 mRNA level was detected using qRT-PCR.
RNA Immunoprecipitation PCR (RIP-qPCR)
Total RNA was isolated from HLECs using Trizol. Anti-METTL3 antibody (Abcam, ab195352) and anti-IgG (Abcam, ab172730) were conjugated to protein A/G magnetic beads in IP buffer (140 mM NaCl, 1% NP-40, 2 mM EDTA, 20 mM Tris pH 7.5) for overnight at 4°C. The total RNA was incubated with the antibody in IP buffer and then precipitated RNA was eluted from the beads. Finally, the precipitated RNA and input total RNA were eluated and reverse-transcribed for qRT-PCR. The relative fold enrichment was calculated using 2-ΔΔCt methods.
Statistical Analysis
All statistical analysis was calculated by SPSS software version 19.0 (Chicago, IL, USA) and the GraphPad Prism 5.0 software (La Jolla, CA, USA). Data are displayed as mean ± SD. Group pairs were calculated using Student’s t test. p < 0.05 was considered statistically significant.
Author Contributions
Jun Yang acts as the major performer. Jingshu Liu acts as the assitance. Shaozhen Zhao and Fang Tian act as the project designer.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the Tianjin Clinical Key Discipline Project (grant number: TJLCZDXKQ007 and TJLCZDXKM003), Science & Technology Development Fund of Tianjin Education Commission for Higher Education (grant number: 2019YD06).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.02.002.
Contributor Information
Shaozhen Zhao, Email: zhaosz1997@sina.com.
Fang Tian, Email: tianfang06@tmu.edu.cn.
Supplemental Information
References
- 1.Habib M.S. ILUVIEN® technology in the treatment of center-involving diabetic macular edema: a review of the literature. Ther. Deliv. 2018;9:547–556. doi: 10.4155/tde-2018-0006. [DOI] [PubMed] [Google Scholar]
- 2.Sarao V., Veritti D., Maurutto E., Rassu N., Borrelli E., Loewenstein A., Sadda S., Lanzetta P. Pharmacotherapeutic management of macular edema in diabetic subjects undergoing cataract surgery. Expert Opin. Pharmacother. 2018;19:1551–1563. doi: 10.1080/14656566.2018.1516206. [DOI] [PubMed] [Google Scholar]
- 3.Wong T.Y., Sun J., Kawasaki R., Ruamviboonsuk P., Gupta N., Lansingh V.C., Maia M., Mathenge W., Moreker S., Muqit M.M.K. Guidelines on Diabetic Eye Care: The International Council of Ophthalmology Recommendations for Screening, Follow-up, Referral, and Treatment Based on Resource Settings. Ophthalmology. 2018;125:1608–1622. doi: 10.1016/j.ophtha.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Visvanathan A., Patil V., Arora A., Hegde A.S., Arivazhagan A., Santosh V., Somasundaram K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 5.Yang F., Jin H., Que B., Chao Y., Zhang H., Ying X., Zhou Z., Yuan Z., Su J., Wu B. Dynamic m6A mRNA methylation reveals the role of METTL3-m6A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38:4755–4772. doi: 10.1038/s41388-019-0755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S., Chai P., Jia R., Jia R. Novel insights on m6A RNA methylation in tumorigenesis: a double-edged sword. Mol. Cancer. 2018;17:101. doi: 10.1186/s12943-018-0847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao W., Qi X., Liu L., Liu Z., Ma S., Wu J. Epigenetic Regulation of m6A Modifications in Human Cancer. Mol. Ther. Nucleic Acids. 2019;19:405–412. doi: 10.1016/j.omtn.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brocard M., Ruggieri A., Locker N. m6A RNA methylation, a new hallmark in virus-host interactions. J. Gen. Virol. 2017;98:2207–2214. doi: 10.1099/jgv.0.000910. [DOI] [PubMed] [Google Scholar]
- 11.Wang S., Sun C., Li J., Zhang E., Ma Z., Xu W., Li H., Qiu M., Xu Y., Xia W. Roles of RNA methylation by means of N6-methyladenosine (m6A) in human cancers. Cancer Lett. 2017;408:112–120. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Wu M. Epigenetics in Neurodevelopment: Emerging Role of Circular RNA. Front. Cell. Neurosci. 2019;13:327. doi: 10.3389/fncel.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2020;27:919–933. doi: 10.1038/s41418-019-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T., Hu P.S., Zuo Z., Lin J.F., Li X., Wu Q.N., Chen Z.H., Zeng Z.L., Wang F., Zheng J. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin H., Ying X., Que B., Wang X., Chao Y., Zhang H., Yuan Z., Qi D., Lin S., Min W. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207. doi: 10.1016/j.ebiom.2019.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ianniello Z., Fatica A. N6-Methyladenosine Role in Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2018;19:E2345. doi: 10.3390/ijms19082345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J., Zhao W., Wang Z., Xiang X., Zhang S., Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J. Cell. Mol. Med. 2019;23:680–688. doi: 10.1111/jcmm.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Zhao S., Tian F. SP1-mediated lncRNA PVT1 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract via miR-214-3p/MMP2 axis. J. Cell. Mol. Med. 2020;24:554–561. doi: 10.1111/jcmm.14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai D., Wang H., Zhu L., Jin H., Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124. doi: 10.1038/s41419-017-0129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob R., Zander S., Gutschner T. The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs. Int. J. Mol. Sci. 2017;18:E2387. doi: 10.3390/ijms18112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y., Yang X., Chen Z., Tian L., Jiang G., Chen F., Li J., An P., Lu L., Luo N. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer. 2019;18:87. doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y., Liu S., Yao K. Transcriptome-wide Investigation of mRNA/circRNA in miR-184 and Its r.57c > u Mutant Type Treatment of Human Lens Epithelial Cells. Mol. Ther. Nucleic Acids. 2017;7:71–80. doi: 10.1016/j.omtn.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F., Wang T.T., Xu Q.G., Zhou W.P., Sun S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 24.Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.