Abstract
Human preimplantation development is a complex process involving dramatic changes in transcriptional architecture. For a better understanding of their time-spatial development, it is indispensable to identify key genes. Although the single-cell RNA sequencing (RNA-seq) techniques could provide detailed clustering signatures, the identification of decisive factors remains difficult. Additionally, it requires high experimental cost and a long experimental period. Thus, it is highly desired to develop computational methods for identifying effective genes of development signature. In this study, we first developed a predictor called EmPredictor to identify developmental stages of human preimplantation embryogenesis. First, we compared the F-score of feature selection algorithms with differential gene expression (DGE) analysis to find specific signatures of the development stage. In addition, by training the support vector machine (SVM), four types of signature subsets were comprehensively discussed. The prediction results showed that a feature subset with 1,881 genes from the F-score algorithm obtained the best predictive performance, which achieved the highest accuracy of 93.3% on the cross-validation set. Further function enrichment demonstrated that the gene set selected by the feature selection method was involved in more development-related pathways and cell fate determination biomarkers. This indicates that the F-score algorithm should be preferentially proposed for detecting key genes of multi-period data in mammalian early development.
Keywords: developmental mRNA signature, single-cell transcriptome, machine learning, feature selection, prediction, differential gene expression, DGE
Introduction
Human preimplantation embryo development refers to the first 7 days of fertilization, which proceeds through stages of the two-cell stage, four-cell stage, eight-cell stage, morula, blastocyst, and late hatched blastocyst.1,2 The first process of zygote development is zygotic genome activation (ZGA) when the embryo gradually stops depending on maternally inherited transcripts and proteins and initiates zygotic genome transcription.3,4 After a small transcriptional activation wave from oocytes to the four-cell stage, major ZGA genes are upregulated between the four-cell and eight-cell stage and start to regulate the biological development of the embryo.5,6 Then, the differences between embryonic cells begin to appear, and three different blastocyst cell lineages are formed.7 The formation of the trophectoderm (TE) reflects the first lineage segregation, followed by the next lineage segregation when the inner cell mass (ICM) is divided into primitive endoderm (PE) and epiblast (EPI) cells.8 In fact, there have been numerous studies of mRNA identification for embryo development. For example, it has been found that pioneering factors (ARGFX, CPHX1, LEUTX, and DUX4) activate the ZGA program by an overexpression experiment and transcriptional analysis.9,10 In mice, CDX2 represses OCT4 expression in the outer cells, leading to TE-ICM lineage segregation,11 but in human CDX2-OCT4 antagonism may not be necessary.12 DPPA2 and DPPA4 regulate expression of Dux and LINE-1 in mouse embryonic stem cells, suggesting that they are an upstream factor of ZGA.13,14 Moreover, Yan et al.15 have identified 2,733 potential novel long non-coding RNAs (lncRNAs) that were involved in preimplantation. However, potential molecular events of embryo development are not fully understood.
Recently, the single-cell RNA sequencing (RNA-seq) techniques are the main method for detecting developmental trajectories and cellular heterogeneity in early preimplantation embryos;16, 17, 18, 19 however, such techniques could only provide detailed clustering signatures, and the identification of decisive factors remains difficult and requires high experimental cost and a long experimental period. As good complements to experimental techniques, computational methods play high potential roles for cancer diagnosis and sequence classification.20, 21, 22, 23, 24, 25, 26, 27 For example, Capper et al.28 proposed random forest (RF) to classify approximately 100 known tumor types of the central nervous system based on DNA methylation data. Based on a single-cell transcriptome, single-cell variational inference (scVI) aggregates information across similar cells and genes by stochastic optimization and deep neural networks,29 and Scialdone et al.30 constructed a predictor for identifying cell-cycle stage. In addition, feature selection methods are independent of prior knowledge of biological dependencies, having been applied in bioinformatics, including protein prediction and biomarker discovery.31,32 The QSPred-FL tool is based on the fact that quorum-sensing peptides in large-scale proteomic data can be detected by feature representation learning and machine learning algorithms,31 and the GF-ICF (gene frequency-inverse cell frequency) pipeline can provide an effective and simple workflow for feature selection and subsequent analyses. However, many studies have advanced new computational methods to interpret single-cell RNA-seq data,29,30,33,34 but most existing methods cannot build predictive models of development. To the best of our knowledge, so far there is no computational tool available for identifying signature genes of development.
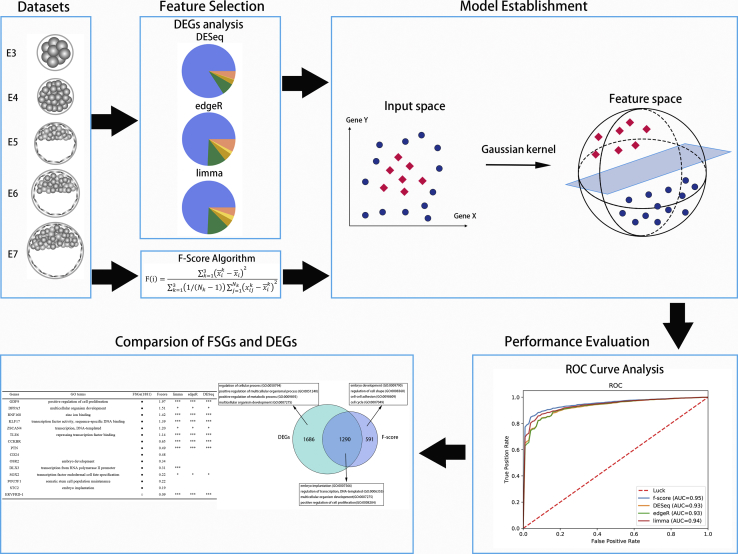
Here, we develop the EmPredicitor, a novel machine learning-based tool for predicting stages of human embryonic development. In this predictor, we compared three traditional differential gene expression (DGE) analyses with a feature selection method based on a single-cell RNA-seq dataset of preimplantation embryos. Figure 1 shows a schematic diagram of the model establishment workflow. The dataset was first integrated and removed genes with no expression in all cells. Then, we applied three DGE methods (edgeR, limma, and DESeq) and a feature selection method (F-score algorithm) to obtain signature genes. By comparing these method performances based on support vector machine (SVM) and functional enrichment analysis, the F-score algorithm had the highest performance and obtained an area under the receiver operating characteristic (ROC) curve (AUC) of 0.95. Our results also suggested that DGE analysis relied on pairwise comparison and overlap, inducing the loss of some key genes that were highly expressed at multiple stages, and the F-score algorithm considered gene expression at all stages and ignored low expression of transcripts.
Figure 1.
The Workflow of Construction and Validation of the Embryo Development Signatures
The best signature from the F-score method consisted of 1,881 genes related to embryo development that was constructed and validated using gene expression from publicly available datasets.
Results
Global Expression Profiles of Human Embryos
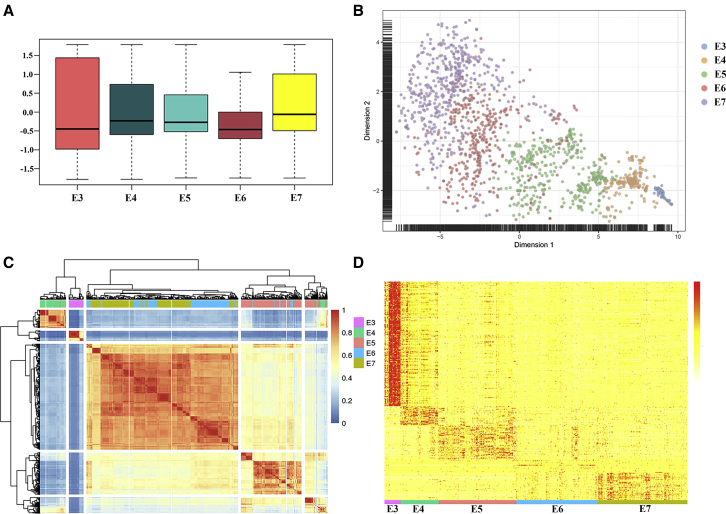
Global transcriptome profiles were first analyzed based on a dataset of early human embryos (Figure 2A, reads per kilobase transcript per million mapped reads [RPKM] > 0). The gene expression level of E3 cells was higher than that for other stages, indicating that the zygote genome was activated and began to identify the genetic program that may control this process.15 Also, the gene expression level of the cells began to decrease during the E3–E6 stage but increased during the E7 stage, suggesting that E7 embryos may initiate new transcriptional activation to begin embryo implantation. To determine whether embryos at the same stage showed a high correlation, we analyzed RNA-seq data of the E3–E7 embryos using the SC3 package (Figure 2C).35 Most of the cells from the same stage were clustered into one cluster. E3 cells and E4 cells have a high correlation. However, E5 cells have two clusters, because after ZGA, differences of embryos begin to emerge and E5 cells appear to segregate ICM and TE.12 Interestingly, the embryos at the E6 and E7 stages were clustered together and divided into three clusters, suggesting that preimplantation of the early embryo resolved in the formation of three distinct cell lineages of blastocysts.36
Figure 2.
Global Transcriptome Profiles of Human Embryos
(A) Expression-level boxplots for the expressed gene in E3–E7 embryos normalized to the Z score. (B) Heatmap of E3–E7 cells and the top 1,881 genes based on the F-score using unsupervised clustering. (C) Two-dimensional t-SNE representation of 1,529 single-cell preimplantation transcriptomes using the top 1,881 genes according to the F-score feature selection. (D) Heatmap of E3–E7 cells and 4,876 differentially expressed genes based on DESeq.
In order to investigate whether these gene expression profiles were related to developmental stages, we conducted t-distributed stochastic neighbor embedding (t-SNE) on all individual embryos (Figure 2B)37,38 and found that embryos at the same developmental stage were clustered together, and the primary segregating factor was developmental time. With the development of embryos, the heterogeneity of embryos increased gradually. In addition, we used differentially expressed genes (DEGs) to plot a heatmap by DESeq, which reflected that embryo cells segregated into five groups, that E6 cells were less different from cells in adjacent stages, and that E3 cells have more DEGs than do other stages (Figure 2D).
Identification of the Developmental Signature by Comparing with F-Score and Differential Expression Analysis
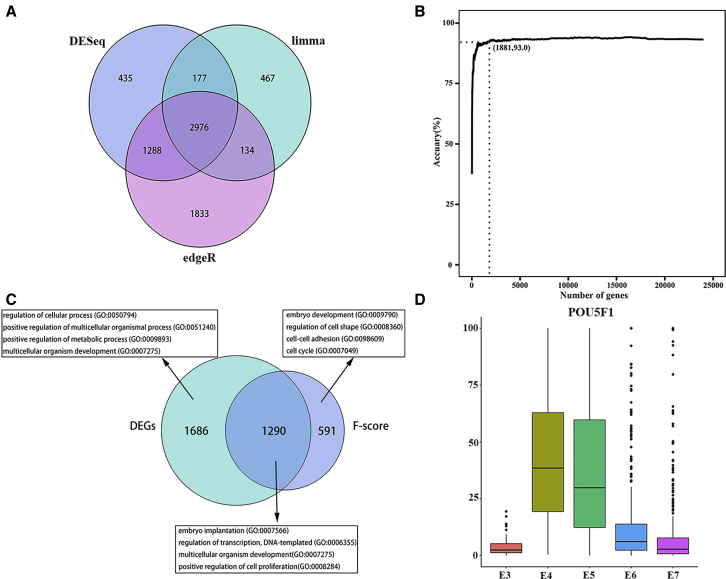
To identify the best signature genes related to embryonic development, we obtained 24,444 gene expression profiles of 1,529 individual cells (81 E3, 190 E4, 377 E5, 415 E6, and 466 E7 cells) from a public database. As DGE analysis usually applied sequence count data, count data were analyzed by comparing three DGE analyses and the F-score algorithm. By using the same parameter (fold change > 2, p < 0.05), limma, DESeq, and edgeR identified 3,754, 4,876 and 6,231 DEGs, respectively, and the number of overlapping genes based on these methods was 2,976 (Figure 3A; Figure S1A). E3 cells had the highest number of DEGs compared to other stages (Figure S1A), and edgeR had more differential genes than did other methods (Figure 3A). The F-score algorithm calculated and ranked each gene score, but we still did not know how many genes should finally be selected. To optimize signature gene selection, we tried the number of signature genes (ranging from 10 to 24,444 genes) for training the support vector machine model and calculated their prediction performance, which applied incremental feature selection (IFS) (Figure 3B).39,40 More genes may bring the best performance but lead to more hardware and time loss, so we chose 1,881 of the top genes by considering gene number and their performance, the accuracy of which was 0.93, and this cost low memory consumption and took only 43 min to obtain the best model. Then, we plotted a heatmap of 1,881 genes based on F-score algorithms (FSGs) and using the SC3 package (Figure S1B). Interestingly, E5 cells were separated into two clusters, suggesting that embryo lineage separation showed the formation of TE and ICM.41,42
Figure 3.
Identification of Development Signature by Comparing the F-Score with Differential Expression Analysis
(A) Venn diagram of DEGs from limma, edgeR, and DESeq. The number of overlapping genes from the three DGE methods is 2,976. (B) The IFS curve with the number of genes and the performance of classifiers. The x axis is the number of genes used for SVM classifier construction and the y axis is the accuracy of the SVM classifier evaluated with 5-fold cross-validation. (C) Venn diagram showing a comparison of DEGs and FSGs. Representative gene ontology (GO) terms are listed. (D) Boxplots of POU5F1 relative expression level at the E3–E7 stages.
We also compared DEGs and FSGs by gene function enrichment (Figure 3C; Table S1). Unique DEGs were enriched for regulation of the cellular process, positive regulation of the multicellular organismal process, positive regulation of the metabolic process, and multicellular organism development. Unique FSGs were enriched for embryo development, regulation of cell shape, cell-cell adhesion, and the cell cycle, which are most relevant to embryonic development. The genes overlapped by DEGs and FSGs were enriched for embryo implantation, regulation of transcription, DNA-templated, multicellular organism development, and positive regulation of cell proliferation, which related to transcription and development. Briefly, unique FSGs related more to embryonic development compared to unique DEGs.
As shown in Table 1, some key genes were selected from FSGs and DEGs. DPPA5, ZSCAN4, and SOX2 were selected by using these methods (rank 9, 22, and 1,520, respectively). DPPA5 stabilizes NANOG and supports human pluripotent stem cell (hPSC) self-renewal and cell reprogramming in feeder-free conditions.43 ZSCAN4 is a unique gene highly expressed at the zygotic genome activation stage.44,45 The POU5F1 gene is vital for PSC maintenance in the mammalian embryo.46,47 Interestingly, POU5F1 (rank 1,470) was not selected by the DGE method but was obtained by F-score. Therefore, we analyzed the expression of POU5F1 among E3–E7 stages, and POU5F1 was highly expressed in E4 and E5 cells (Figure 3D). Therefore, DGE analysis mainly relies on pairwise comparison and overlap, so if a gene is highly expressed at two or more stages, differential expression analyses may lose the gene, suggesting that DGE analysis only considered DEGs highly expressed at a stage. The F-score algorithm showed the importance of a gene in all stages. If the expression level of a gene was too low, the F-score algorithm would give a low score for this gene, especially similar to ERVFRD-1 (Figure S1C; Table S2). Although transcripts of low expression may be important, most of these are outliers, suggesting that transcripts with low expression levels were preprocessed, in line with previous studies.48
Table 1.
Comparison between Key Genes of FSGs and DEGs
| Genes | Gene Ontology Terms | FSGs (1,881) | F-Score | limma | edgeR | DESeq |
|---|---|---|---|---|---|---|
| GDF9 | positive regulation of cell proliferation | ● | 1.97 | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| DPPA5 | multicellular organism development | ● | 1.51 | ∗ | ∗ | ∗ |
| RNF168 | zinc ion binding | ● | 1.42 | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| KLF17 | transcription factor activity, sequence-specific DNA binding | ● | 1.39 | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| ZSCAN4 | transcription, DNA templated | ● | 1.20 | ∗ | ∗ | ∗ |
| TLE6 | repressing transcription factor binding | ● | 1.14 | ∗∗∗ | ∗∗∗ | ∗∗∗ |
| CCKBR | ● | 0.65 | ∗∗∗ | ∗∗∗ | ∗∗∗ | |
| PTN | ● | 0.49 | ∗∗∗ | ∗∗∗ | ∗∗∗ | |
| CD24 | ● | 0.48 | ||||
| OSR2 | embryo development | ● | 0.34 | |||
| DLX3 | transcription from RNA polymerase II promoter | ● | 0.31 | ∗∗∗ | ||
| SOX2 | transcription factor endodermal cell fate specification | ● | 0.22 | ∗ | ∗ | ∗ |
| POU5F1 | somatic stem cell population maintenance | ● | 0.22 | |||
| STC2 | embryo implantation | ● | 0.19 | |||
| ERVFRD-1 | ○ | 0.09 | ∗∗∗ | ∗∗∗ | ∗∗∗ |
If a gene belongs to the dataset, replace it with ●; otherwise, replace it with ○. F-score shows the importance of features selected by the F-score algorithm. ∗p < 0.05, ∗∗∗p < 0.001.
In addition to known markers, several less described markers were identified, such as RNF168, CCKBR, PTN, CD24 and STC2 (rank 12, 151, 332, 349, and 1,851). CCKBR, a cholecystokinin B receptor, has been found in a diverse range of cancers.49 We found that in the late blastocyst, E6 and E7 cells expressed high levels of CCKBR, indicating that CCKBR may be involved in the ICM segregation of EPI and PE cells. PTN-encoded protein has significant roles in cell growth, migration, and tumorigenesis,50,51 and it was expressed in the late blastocyst, suggesting that PTN may be involved in embryonic cell migration. Then, we found that most of the 500 top-ranked genes were high relative expression genes of E3 stages (Table S2), similar to what has been previously reported.15
Predictor of Human Preimplantation Development and the Web Server
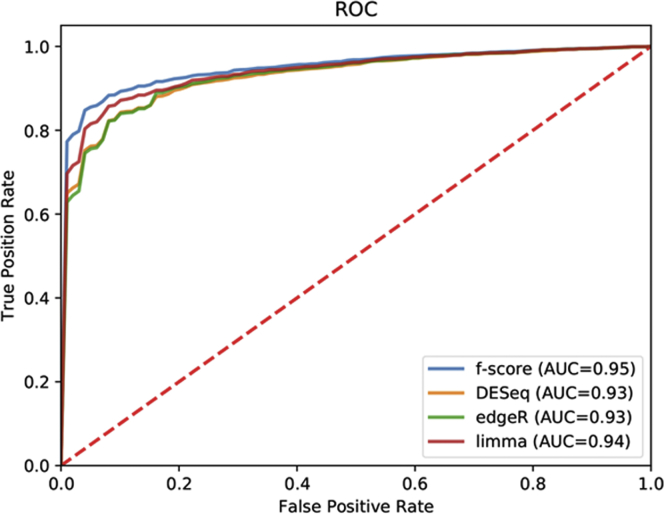
To develop a predictor to identify developmental stages of human preimplantation embryogenesis, we applied the support vector machine classifier to train models based on three DGE analyses and F-score algorithm in 5-fold cross-validation, and we obtained the performance of the four methods (Table 2). The models of the four method showed high performances; however, FSGs achieved precision, recall, accuracy, and F1 measure values of 0.933, 0.929, 0.930, and 0.930, respectively, and the number of FSGs had the fewest (Table 2). In addition, the classifier using the F-score algorithm also showed high performance, with an AUC greater than 95% (Figure 4).
Table 2.
Performance of Stage Predicting Models with 5-Fold Cross-Validation
| Method | Gene No. | Precision (%) | Recall (%) | Accuracy (%) | F1 Measure (%) |
|---|---|---|---|---|---|
| DESeq | 4,876 | 90.23 | 89.81 | 89.85 | 89.73 |
| limma | 3,754 | 91.5 | 91.23 | 91.24 | 91.24 |
| edgeR | 6,231 | 90.82 | 90.36 | 90.42 | 90.31 |
| F-score | 1,881 | 93.3 | 92.91 | 93.01 | 93.2 |
Underlined text represents the maximum value of every performance evaluation criterion.
Figure 4.
The Predictor of Human Preimplantation Development Based on Machine Learning
ROC curve and AUC showing that the F-score method obtained high performance.
Based on our proposed model, a user-friendly and publicly accessible web server for EmPredictor was established (available at http://bioinfor.imu.edu.cn/empredictor), where users can upload or paste a dataset of the eight key genes to predict the stage of their samples. The home page of EmPredictor is shown in Figure 5. We also considered that users may want to know the relative expression trend of a gene, so the server provides the function of searching for a gene on a single-cell dataset from E-MTAB-3929. The user guide is available on the web page.
Figure 5.
A Semi-screenshot Showing the Home Page of the EmPredictor Web Server
Discussion
Herein, we have proposed the first EmPredicitor, a novel machine learning-based tool for predicting stages of human embryonic development. Based on three DGE analyses (limma, edgeR, DESeq) and F-score algorithm, the single-cell transcriptomes data obtain 3,754, 4,876, 6,231, and 1,881 signature genes, respectively. Then, supervised machine learning is used to estimate the contribution of embryonic development to these signature genes. Toward the application of 5-fold cross-validation on a benchmark dataset, the F-score algorithm can achieve the highest accuracy of 0.93 and AUC of 0.95. Furthermore, functional enrichment analysis showed that the F-score algorithm can obtain key signaling pathways related to embryo development. Based on prior biological knowledge, some key genes were used to estimate the assessment of F-score and DGE analyses. DGE analyses rely on pairwise comparison and overlap to obtain differentially expressed genes. F-score detected key genes of multi-period data that contributed to identifying early embryo stages. In addition, we constructed a user-friendly and publicly accessible web server where users can upload or paste a dataset of the eight key genes to predict the stage of their samples.
There are still some disadvantages of this work. Here, we investigated only predicting embryonic days. However, embryonic development is a complex process involving lineage specification and X chromosome dosage compensation.7,12,46 Integrating genetic and epigenetic data with gene expression may provide a more comprehensive view of embryonic development. In addition, feature selection methods have irreplaceable advantages in processing single-cell transcriptome data and are independent of prior knowledge of biological dependencies, which extend the development analysis pipeline. In the future, we will use advanced feature selection methods to study embryonic development based on more accurate molecular events and multi-omics data.
Materials and Methods
Data and Preprocessing
We downloaded a single-cell transcriptome dataset of human preimplantation embryos from ArrayExpress under accession E-MTAB-3929,12 including 1,529 samples. The dataset has five different cell stages, which are embryonic day (E)3, E4, E5, E6, and E7. The E3 stage has 81 cells, the E4 stage has 190 cells, the E5 stage has 377 cells, the E6 stage has 415 cells, and the E7 stage has 466 cells.
The data were processed using TrueSeq dual-index sequencing primers (Illumina) according to the manufacturer’s recommendations on an Illumina HiSeq 2000.12 The data quality was checked and reads were mapped to the human genome (hg19) using STAR with default settings.52 RPKM were calculated using rpkmforgenes53 by the uniquely mapped read counts. Genes were filtered, keeping 24,444 out of 26,178 genes that were expressed in at least 1 out of 1,529 cells (count > 0).
Feature Selection
Linear Model
During the past decade, the limma package54 has been a popular choice for gene discovery through differential expression analyses of microarrays. Recently, limma has also provided differential expression and differential splicing analyses of RNA-seq data. limma uses the voom function by converting mean variance to precision weights and using a linear model,
| (Equation 1) |
where Xi is a vector of covariates and βg is a vector of unknown coefficients representing log2 fold changes between experimental conditions. In matrix terms
| (Equation 2) |
where yg is the vector of log cpm values for gene g, and X is the design matrix with the Xi as rows. The limma package is available at https://bioconductor.org/packages/release/bioc/html/limma.html.
Negative Binomial Distribution
edgeR55 is designed for the analysis of replicated count-based expression data. Data are modeled as negative binomial (NB) distributed
| (Equation 3) |
for gene g and sample i. Here, Mi is the library size (total number of reads), φg is the dispersion, and pgi is the relative abundance of gene g in experimental group j to which sample i belongs. The edgeR package is available at https://bioconductor.org/packages/release/bioc/html/edgeR.html.
DESeq56 provides methods to test for differential expression by use of the negative binomial distribution and a shrinkage estimator for the distribution’s variance,
| (Equation 4) |
which has two parameters, the mean μij and the variance . The read counts Kij are non-negative integers. The DESeq package is available at https://bioconductor.org/packages/release/bioc/html/DESeq.html.
F-Score Algorithm
F-score is a simple and basic but effective algorithm for evaluating the importance of each feature in the dataset. F-score is a computed each feature values and
| (Equation 5) |
where are the average of the ith feature of the whole, positive, and negative datasets respectively; is the ith feature of the kth positive instance; and is the ith feature of the kth negative instance. A Python program fselect.py can compute each feature value and rank the feature downloaded from https://www.csie.ntu.edu.tw/∼cjlin/libsvmtools/.57
Machine Learning Model Implementation
The support vector machine (SVM) was proposed by Vapnik et al.58 SVM shows many advantages in solving small sample, nonlinear, and high-dimensional pattern recognition. The idea of SVM is based on transforming the input vector into a high-dimensional Hilbert space and finding a separating hyperplane in this space. Gaussian radial basis function (RBF) kernel function59 is a widely used kernel function because of its high performance in non-line classification:
| (Equation 6) |
We applied LIBSVM as an SVM model with a one-against-one strategy60 and RBF kernel. A grid search strategy with a cross-validation test is always utilized to obtain the best values of the regularization parameter C and kernel parameter g. We used the gird.py file (https://www.csie.ntu.edu.tw/∼cjlin/libsvmtools/) in LIBSVM to search for the best C value and g value (the range of the C parameter is between 2−5 and 210, and the range of the g parameter is between 2−15 and 23).60 Classifier performance was evaluated by 5-fold cross-validation analysis,28,59 where each training dataset was randomly partitioned into four equal parts with one part being used for model training and the remaining part used for testing. We used the cross-validation method to limit overfitting of the classifier. To have a complete measurement of the prediction performance, four statistics, i.e., accuracy, recall, precision, and F1 measure,30,59 were calculated as follows:
| (Equation 7) |
| (Equation 8) |
| (Equation 9) |
| (Equation 10) |
where TP is the true positive correct result, FP is the false unexpected result, FN is the false missing result, and TN is the true correct absence of result.
Code Available
The code for the implementation of the EmPredictor is available on GitHub: https://github.com/liameihao/EmPredictor.
Author Contributions
Y.Z. designed this work. P.L. and W.Y. performed the whole bioinformatics analysis and wrote the manuscript. X.C. and C.L. performed the experiments and helped edit the manuscript. L.Z. and H.L. assisted the experiments.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We are grateful to our laboratory colleagues for their assistance with the bioinformatics analysis. We thank Prof. Fredrik Lanner (Karolinska Universitetssjukhuset) for sharing the single-cell RNA-seq datasets in ArrayExpress database. This work was supported by the National Nature Scientific Foundation of China (61702290, 61861036), the Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (NJYT-18-B01), and by the Fund for Excellent Young Scholars of Inner Mongolia (2017JQ04). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.02.004.
Supplemental Information
References
- 1.Cockburn K., Rossant J. Making the blastocyst: lessons from the mouse. J. Clin. Invest. 2010;120:995–1003. doi: 10.1172/JCI41229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuo Y., Gao Y., Su G., Bai C., Wei Z., Liu K., Li Q., Bou S., Li G. Irregular transcriptome reprogramming probably causes the developmental failure of embryos produced by interspecies somatic cell nuclear transfer between the Przewalski’s gazelle and the bovine. BMC Genomics. 2014;15:1113. doi: 10.1186/1471-2164-15-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graf A., Krebs S., Heininen-Brown M., Zakhartchenko V., Blum H., Wolf E. Genome activation in bovine embryos: review of the literature and new insights from RNA sequencing experiments. Anim. Reprod. Sci. 2014;149:46–58. doi: 10.1016/j.anireprosci.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Zuo Y., Su G., Cheng L., Liu K., Feng Y., Wei Z., Bai C., Cao G., Li G. Coexpression analysis identifies nuclear reprogramming barriers of somatic cell nuclear transfer embryos. Oncotarget. 2017;8:65847–65859. doi: 10.18632/oncotarget.19504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko M. Zygotic genome activation revisited: looking through the expression and function of Zscan4. Curr. Top. Dev. Biol. 2016;120:103–124. doi: 10.1016/bs.ctdb.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Zuo Y., Su G., Wang S., Yang L., Liao M., Wei Z., Bai C., Li G. Exploring timing activation of functional pathway based on differential co-expression analysis in preimplantation embryogenesis. Oncotarget. 2016;7:74120–74131. doi: 10.18632/oncotarget.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niakan K.K., Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Kwon G.S., Viotti M., Hadjantonakis A.K. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017;49:941–945. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 12.Petropoulos S., Edsgärd D., Reinius B., Deng Q., Panula S.P., Codeluppi S., Plaza Reyes A., Linnarsson S., Sandberg R., Lanner F. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckersley-Maslin M., Alda-Catalinas C., Blotenburg M., Kreibich E., Krueger C., Reik W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019;33:194–208. doi: 10.1101/gad.321174.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Iaco A., Coudray A., Duc J., Trono D. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 2019;20:10. doi: 10.15252/embr.201847382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 16.Farrell J.A., Wang Y., Riesenfeld S.J., Shekhar K., Regev A., Schier A.F. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 2018;360:eaar3131. doi: 10.1126/science.aar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng S., Pei Y., He L., Peng G., Reinius B., Tam P.P.L., Jing N., Deng Q. Single-cell RNA-seq reveals cellular heterogeneity of pluripotency transition and X chromosome dynamics during early mouse development. Cell Rep. 2019;26:2593–2607.e3. doi: 10.1016/j.celrep.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 18.Hu B., Zheng L., Long C., Song M., Li T., Yang L., Zuo Y. EmExplorer: a database for exploring time activation of gene expression in mammalian embryos. Open Biol. 2019;9:190054. doi: 10.1098/rsob.190054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A. mRNA-seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 20.Wong D., Yip S. Machine learning classifies cancer. Nature. 2018;555:446–447. doi: 10.1038/d41586-018-02881-7. [DOI] [PubMed] [Google Scholar]
- 21.Zuo Y., Li Y., Chen Y., Li G., Yan Z., Yang L. PseKRAAC: a flexible web server for generating pseudo K-tuple reduced amino acids composition. Bioinformatics. 2017;33:122–124. doi: 10.1093/bioinformatics/btw564. [DOI] [PubMed] [Google Scholar]
- 22.Liu D., Li G., Zuo Y. Function determinants of TET proteins: the arrangements of sequence motifs with specific codes. Brief. Bioinform. 2019;20:1826–1835. doi: 10.1093/bib/bby053. [DOI] [PubMed] [Google Scholar]
- 23.Feng C.Q., Zhang Z.Y., Zhu X.J., Lin Y., Chen W., Tang H., Lin H. iTerm-PseKNC: a sequence-based tool for predicting bacterial transcriptional terminators. Bioinformatics. 2019;35:1469–1477. doi: 10.1093/bioinformatics/bty827. [DOI] [PubMed] [Google Scholar]
- 24.Chen W., Feng P., Song X., Lv H., Lin H. iRNA-m7G: identifying N7-methylguanosine sites by fusing multiple features. Mol. Ther. Nucleic Acids. 2019;18:269–274. doi: 10.1016/j.omtn.2019.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W., Feng P., Liu T., Jin D. Recent advances in machine learning methods for predicting heat shock proteins. Curr. Drug Metab. 2019;20:224–228. doi: 10.2174/1389200219666181031105916. [DOI] [PubMed] [Google Scholar]
- 26.Zheng L., Huang S., Mu N., Zhang H., Zhang J., Chang Y., Yang L., Zuo Y. RAACBook: a web server of reduced amino acid alphabet for sequence-dependent inference by using Chou’s five-step rule. Database (Oxford) 2019;2019:baz131. doi: 10.1093/database/baz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai H.Y., Zhang Z.Y., Su Z.D., Su W., Ding H., Chen W., Lin H. iProEP: a computational predictor for predicting promoter. Mol. Ther. Nucleic Acids. 2019;17:337–346. doi: 10.1016/j.omtn.2019.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez R., Regier J., Cole M.B., Jordan M.I., Yosef N. Deep generative modeling for single-cell transcriptomics. Nat. Methods. 2018;15:1053–1058. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scialdone A., Natarajan K.N., Saraiva L.R., Proserpio V., Teichmann S.A., Stegle O., Marioni J.C., Buettner F. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 31.Wei L., Hu J., Li F., Song J., Su R., Zou Q. Comparative analysis and prediction of quorum-sensing peptides using feature representation learning and machine learning algorithms. Brief. Bioinform. 2020;21:106–119. doi: 10.1093/bib/bby107. [DOI] [PubMed] [Google Scholar]
- 32.Li J., Lan C.-N., Kong Y., Feng S.S., Huang T. Identification and analysis of blood gene expression signature for osteoarthritis with advanced feature selection methods. Front. Genet. 2018;9:246. doi: 10.3389/fgene.2018.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talwar D., Mongia A., Sengupta D., Majumdar A. AutoImpute: Autoencoder based imputation of single-cell RNA-seq data. Sci. Rep. 2018;8:16329. doi: 10.1038/s41598-018-34688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiselev V.Y., Kirschner K., Schaub M.T., Andrews T., Yiu A., Chandra T., Natarajan K.N., Reik W., Barahona M., Green A.R., Hemberg M. SC3: consensus clustering of single-cell RNA-seq data. Nat. Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blakeley P., Fogarty N.M., del Valle I., Wamaitha S.E., Hu T.X., Elder K., Snell P., Christie L., Robson P., Niakan K.K. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Maaten L., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 38.McCarthy D.J., Campbell K.R., Lun A.T., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang T., Zhang J., Xu Z.P., Hu L.L., Chen L., Shao J.L., Zhang L., Kong X.Y., Cai Y.D., Chou K.C. Deciphering the effects of gene deletion on yeast longevity using network and machine learning approaches. Biochimie. 2012;94:1017–1025. doi: 10.1016/j.biochi.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 40.Chen L., Li J., Zhang Y.H., Feng K., Wang S., Zhang Y., Huang T., Kong X., Cai Y.D. Identification of gene expression signatures across different types of neural stem cells with the Monte-Carlo feature selection method. J. Cell. Biochem. 2018;119:3394–3403. doi: 10.1002/jcb.26507. [DOI] [PubMed] [Google Scholar]
- 41.Rossant J., Tam P.P.L. New insights into early human development: lessons for stem cell derivation and differentiation. Cell Stem Cell. 2017;20:18–28. doi: 10.1016/j.stem.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Ortega N.M., Winblad N., Plaza Reyes A., Lanner F. Functional genetics of early human development. Curr. Opin. Genet. Dev. 2018;52:1–6. doi: 10.1016/j.gde.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Qian X., Kim J.K., Tong W., Villa-Diaz L.G., Krebsbach P.H. DPPA5 supports pluripotency and reprogramming by regulating NANOG turnover. Stem Cells. 2016;34:588–600. doi: 10.1002/stem.2252. [DOI] [PubMed] [Google Scholar]
- 44.Falco G., Lee S.L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long C., Li W., Liang P., Liu S., Zuo Y. Transcriptome comparisons of multi-species identify differential genome activation of mammals embryogenesis. IEEE Access. 2019;7:7794–7802. [Google Scholar]
- 46.Fogarty N.M.E., McCarthy A., Snijders K.E., Powell B.E., Kubikova N., Blakeley P., Lea R., Elder K., Wamaitha S.E., Kim D. Genome editing reveals a role for OCT4 in human embryogenesis. Nature. 2017;550:67–73. doi: 10.1038/nature24033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Ta N., Long C., Zhang Q., Li S., Liu S., Yang L., Zuo Y. The spatial binding model of the pioneer factor Oct4 with its target genes during cell reprogramming. Comput. Struct. Biotechnol. J. 2019;17:1226–1233. doi: 10.1016/j.csbj.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy J., Putt K.S., Coppola D., Leon M.E., Khalil F.K., Centeno B.A., Clark N., Stark V.E., Morse D.L., Low P.S. Assessment of cholecystokinin 2 receptor (CCK2R) in neoplastic tissue. Oncotarget. 2016;7:14605–14615. doi: 10.18632/oncotarget.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai P.S., Xia N., Sun H., Kong Y. Pleiotrophin, a target of miR-384, promotes proliferation, metastasis and lipogenesis in HBV-related hepatocellular carcinoma. J. Cell. Mol. Med. 2017;21:3023–3043. doi: 10.1111/jcmm.13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen D., Podolnikova N.P., Yakubenko V.P., Ardell C.L., Balabiyev A., Ugarova T.P., Wang X. Pleiotrophin, a multifunctional cytokine and growth factor, induces leukocyte responses through the integrin Mac-1. J. Biol. Chem. 2017;292:18848–18861. doi: 10.1074/jbc.M116.773713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramsköld D., Wang E.T., Burge C.B., Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y.W., Lin C.J. Combining SVMs with various feature selection strategies. In: Guyon I., Nikravesh M., Gunn S., Zadeh L.A., editors. vol. 207. Springer; 2006. pp. 315–324. (Feature Extraction: Studies in Fuzziness and Soft Computing). [Google Scholar]
- 58.Vapnik V. Wiley; 1998. Statistical Learning Theory. [DOI] [PubMed] [Google Scholar]
- 59.Dao F.-Y., Yang H., Su Z.D., Yang W., Wu Y., Hui D., Chen W., Tang H., Lin H. Recent advances in conotoxin classification by using machine learning methods. Molecules. 2017;22:1057. doi: 10.3390/molecules22071057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.