Abstract
Escherichia coli carrying blaCTX–M–1 mediating resistance to extended-spectrum cephalosporins was recently described as a new genotype in Norwegian broiler production. The aim of this study was to characterize these isolates (n = 31) in order to determine whether the emergence of the genotype was caused by clonal expansion or horizontal dissemination of blaCTX–M–1-carrying plasmids. All included isolates were subjected to whole genome sequencing. Plasmid transferability was determined by conjugation, and plasmid replicons in the transconjugants were described using PCR-based replicon typing. Plasmid sizes were determined using S1 nuclease digestion. Plasmids in a subset of strains were reconstructed and compared to plasmids from broiler production in other European countries. The isolates belonged to nine different sequence types (STs), with the largest group being ST57 (n = 12). The vast majority of blaCTX–M–1-carrying plasmids were conjugative. All transconjugants were positive for the IncI1-Iγ replicon, and several also harbored the IncFIB replicon. Highly similar plasmids were present in different E. coli STs. Additionally, high similarity to previously published plasmids was detected. A reconstructed plasmid from an ST57 isolate harbored both IncI1-Iγ and IncFIB replicons and was considered to be co-integrated. The presence of one large plasmid was confirmed by S1 nuclease digestion. Our results show that dissemination of blaCTX–M–1 in Norwegian broiler production is due to both clonal expansion and horizontal transfer of plasmids carrying blaCTX–M–1. The blaCTX–M–1/IncI1-Iγ plasmids grouped into two main lineages, namely clonal complex (CC)-3 and CC-7. The genetic diversity at both strain and plasmid level indicates multiple introductions to Norway. We also show that the blaCTX–M–1 plasmids circulating in Norwegian broiler production are highly similar to plasmids previously described in other countries.
Keywords: plasmid, IncI1, cephalosporin resistance, Escherichia coli, broiler, blaCTX–M–1
Introduction
Since the first description of an Escherichia coli displaying extended-spectrum cephalosporin (ESC) resistance in broilers in 2000–2001 (Briñas et al., 2003), numerous reports have underlined the global occurrence of these bacteria in the broiler production (Carattoli, 2008; Smet et al., 2008; Dierikx et al., 2010; Doi et al., 2010; Ewers et al., 2012; Hiroi et al., 2012b). Import of breeding animals is believed to be a contributing factor to the wide dissemination. The first detection of E. coli mediating ESC resistance in Norwegian livestock dates back to year 2006. The isolate originated from healthy broilers and harbored the blaTEM–20 gene on an IncI1 plasmid (NORM/NORM-VET, 2007; Sunde et al., 2009). In 2011, a selective method for detection of ESC-resistant E. coli was implemented in the Norwegian monitoring programme for antimicrobial resistance in bacteria from food, feed and animals (NORM-VET) (NORM/NORM-VET, 2012). Since then, all ESC-resistant E. coli isolated from broiler production in the NORM-VET program have displayed an AmpC phenotype due to presence of the blaCMY–2 gene (NORM/NORM-VET, 2012, 2013, 2015, 2017). However, an extensive sampling of broiler- and parent flocks during 2016 revealed an additional presence of E. coli producing extended-spectrum beta-lactamases (ESBLs), with blaCTX–M–1 being the most frequently detected ESBL-encoding gene (Mo et al., 2019 and unpublished data). In other European countries, the blaCTX–M–1 gene has been frequently associated with ESBL-producing E. coli from the broiler production chain. Several studies have shown that the blaCTX–M–1 gene commonly occurs on conjugative IncI1 plasmids (Zurfluh et al., 2014; Smith et al., 2015; Irrgang et al., 2018; Touzain et al., 2018).
The aim of this study was to characterize the blaCTX–M–1 harboring E. coli isolates in order to determine whether the emergence of this genotype was due to clonal expansion of resistant strains or horizontal transmission of blaCTX–M–1-carrying plasmid(s). The blaCTX–M–1-harboring plasmids identified were further investigated, and a selection of plasmids were reconstructed and compared to plasmids originating from broiler production in other European countries and from other sources in order to identify successful plasmids.
Materials and Methods
Bacterial Isolates
The isolates included were retrieved from a previous study. In short; during the period from May to October 2016 all broiler flocks reared in Norway were investigated for ESC-resistant Enterobacteriaceae (n = 2110). Approximately 10% of the flocks were positive, of which the majority contained the blaCMY–2 gene responsible for ESC resistance. A minor proportion (n = 24, 1.1% of total flocks) were positive for blaCTX–M–1, and these isolates were further characterized in the present study. Detailed description of the sampling and detection methods is described previously (Mo et al., 2019). The isolates originated from 24 unique broiler flocks reared at 15 different farms. Nine farms had one positive flock, and four farms had two positive flocks, while two farms had three and four positive flocks, respectively. In addition, seven E. coli with blaCTX–M–1 isolated from parent flocks were included (unpublished data). These isolates originated from three different parent flocks sampled at multiple times during a 4 week period in November–December 2016. One parent flock was positive at one sampling, while the two remaining flocks were positive at three samplings each.
Susceptibility Testing
The minimum inhibitory concentration (MIC) values of the isolates were determined by broth microdilution (EUVSEC, Sensititre®, TREK Diagnostic Systems Ltd., Thermo Fisher Scientific). For quality control, E. coli ATCC25922 was tested on a regular basis. Epidemiological cut-off values from EUCAST (European Committee on Antimicrobial Susceptibility Testing) were used to categorize the isolates as susceptible or resistant. MIC values for all broiler isolates (n = 24) were available from a previous study (Mo et al., 2019), while isolates originating from parent flocks (n = 7) were tested as a part of this study.
Determination of Phylogroups
Classification of isolates into phylogroups A, B1, B2, or D was performed on all isolates using a previously described multiplex PCR (Doumith et al., 2012). Positive and negative controls were included in each PCR run.
Conjugation Experiments and Characterization of Transconjugants
To determine the transferability of the blaCTX–M–1-carrying plasmids, all isolates were subjected to conjugation experiments in broth as previously described (Mo et al., 2016). E. coli DH5α CCUG32825 was used as recipient. Transconjugants were confirmed on the basis of their characteristic colony morphology on blood agar and on lactose-saccharose-bromthymol blue agar (Mo et al., 2016), and PCR was carried out to confirm the presence of blaCTX–M (Hasman et al., 2005). Further confirmation of transconjugants included PCR-based replicon typing (PBRT) to determine the replicon type(s) of the transferred plasmids in transconjugants. This was done using the PBRT kit (Diatheva, Fano, Italy) as described by the manufacturer. Positive and negative controls were included in each PCR run. To determine the approximate size of selected blaCTX–M–1-carrying plasmids, transconjugants of ST57 isolates (n = 12) were subjected to S1 nuclease digestion (InvitrogenTM, Carlsbad, CA, United States) of DNA embedded in agarose followed by pulsed-field gel electrophoresis (PFGE) as previously described (Barton et al., 1995).
DNA Isolation and Sequencing
For Illumina sequencing, DNA was extracted using the QIAamp DSP DNA mini kit (Qiagen, Hilden, Germany) with the QiaSymphony automated extractor system (Qiagen). The DNA yield and purity was measured on a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and on a Qubit fluorometer (Invitrogen) using Qubit dsDNA BR Assay kit (Invitrogen). Whole genome sequencing (WGS) was performed on all isolates using the Nextera XT library preparation kit (Illumina, San Diego, CA, United States) and a NextSeq 500 Illumina platform resulting in 150 bp paired-end reads.
To facilitate reconstruction and in-depth characterization of blaCTX–M–1-carrying plasmids, nine isolates representing each multilocus sequence type (MLST)/core genome (cg)MLST cluster identified were selected for Pacific Biosciences (PacBio, Menlo Park, CA, United States) long-read sequencing (Table 1). Total genomic DNA was extracted using a Qiagen Genomic-tip 100/G kit (Qiagen). The quality and quantity of DNA was assessed as described in the previous paragraph. The DNA library was prepared using a protocol for single molecule real-time (SMRT) bellTM libraries (PacBio). The library was sequenced on a PacBio Sequel platform using sequel polymerase v3.0, a 10 kb insert library, SMRT cells v3 LR and Sequencing chemistry v3.0 (PacBio).
TABLE 1.
Overview of characteristics associated with 31 Escherichia coli isolates with blaCTX–M–1 originating from broiler- (n = 24) and parent flocks (n = 7, isolates from three flocks) in Norway in 2016.
| Isolate | Origin | Parent flock | Broiler farm | Phylo-group | MLST | Pathogroup (diarrhoeagenic) | Plasmid replicon type(s) | IncI1 pMLST (clonal complex) | Other AMR genes | Transferable plasmid | Plasmid replicon type(s) in transconjugant | PacBio |
| 2016-40-14272 | Broiler | B | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-16262 | Broiler | B | D | 57 | I1, FIB, B/O/K/Z, I2 | 42 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-17200 | Broiler | B | D | 57 | I1, FIB, B/O/K/Z | 42 (CC-3) | Sul2 | + | I1,FIB | |||
| 2016-40-19738 | Broiler | B | D | 57 | I1, FIB, B/O/K/Z | 42 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-14497 | Broiler | C | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-17437 | Broiler | C | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | + | ||
| 2016-40-19583 | Broiler | C | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-16990 | Broiler | D | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-17074 | Broiler | D | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-16344 | Broiler | G | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-17091 | Broiler | H | D | 57 | I1, FIB | 3 (CC-3) | sul2 | + | I1,FIB | |||
| 2016-40-20481 | Broiler | M | D | 57 | I1, FIB, B/O/K/Z | 42 (CC-3) | sul2 | + | I1 | |||
| 2016-40-19016 | Broiler | E | B1 | 297 | I1, FIB, FIC/FII, FIA, B/O/K/Z | IC | sul2 | + | I1 | |||
| 2016-40-19138 | Broiler | F | B1 | 297 | I1, FIB, FIC/FII, FIA, B/O/K/Z | 3 (CC-3) | sul2 | + | I1 | + | ||
| 2016-40-17381 | Broiler | I | B1 | 297 | I1, FIB, FIC/FII, FIA, B/O/K/Z, X1 | IC | sul2 | + | I1,B/O/K/Z | |||
| 2016-40-19148 | Broiler | J | B1 | 297 | I1, FIB, FIC/FII, FIA, B/O/K/Z | IC | sul2 | + | I1,B/O/K/Z | |||
| 2016-40-19970 | Broiler | K | B1 | 297 | I1, FIB, FIC/FII, FIA, B/O/K/Z | IC | sul2 | + | I1,B/O/K/Z | |||
| 2016-40-21210 | Broiler | N | B1 | 297 | I1, FIB, FIC/FII, FIA, B/O/K/Z | 3 (CC-3) | sul2 | + | I1 | |||
| 2016-40-21270 | Broiler | O | B1 | 297 | I1, FIB, FIC/FII, FIA, B/O/K/Z | IC | sul2 | + | I1 | |||
| 2016-40-14263 | Broiler | A | A | 752 | EPEC | I1, FIB, FIC/FII | 7 (CC-7) | aph | - | NA | + | |
| 2016-40-20703 | Broiler | A | A | 752 | EPEC | I1, FIB, FII | 7 (CC-7) | aph | - | NA | ||
| 2016-40-23575 | Parent | 3 | A | 752 | EPEC | I1, FIB, FII, FIC, I2 | 7 (CC-7) | aph | - | NA | ||
| 2016-40-22440 | Parent | 1 | A | 48 | I1, FIB, FII, HI1B | 3 (CC-3) | sul2, tetA | + | I1 | + | ||
| 2016-40-23572 | Parent | 1 | A | 48 | I1, FIB, FII, HI1B | 3 (CC-3) | sul2, tetA | + | I1 | |||
| 2016-40-24053 | Parent | 1 | A | 48 | I1, FIB, FII, HI1B, X4 | 3 (CC-3) | sul2, tetA | + | I1 | |||
| 2016-40-22638 | Parent | 2 | A | 1638 | I1, FIB, FII | 3 (CC-3) | aadA, dfrA, sul1, sul2, tetA | + | I1 | + | ||
| 2016-40-23574 | Parent | 2 | A | 1638 | I1, FIB, FII | 3 (CC-3) | aadA, dfrA, sul1, sul2, tetA | + | I1 | |||
| 2016-40-21249 | Broiler | F | B1 | 162 | I1, FIB, FII, X1 | 3 (CC-3) | sul2 | + | I1 | + | ||
| 2016-40-21254 | Broiler | E | A | 10 | I1 | 3 (CC-3) | sul2 | + | I1 | + | ||
| 2016-40-24003 | Parent | 2 | A | 1251 | I1, FIB, FII, HI1B | 3 (CC-3) | aadA, dfrA, sul1, sul2, tetA | + | I1 | + | ||
| 2016-40-20426 | Broiler | L | D | 641 | I1, FIB, FII, FIA | 3 (CC-3) | sul2 | + | I1 | + |
EPEC, enteropathogenic Escherichia coli; IC, incomplete sequence type; AMR, antimicrobial resistance; NA, not applicable.
The sequencing was performed by the Norwegian Sequencing Centre.
Sequence Analysis and Characterization
The Bifrost pipeline (Lagesen, 2020)1 was used for initial analysis of fastq data from the Illumina sequencing. Quality control of the raw reads was done using FastQC2, and multiQC (Ewels et al., 2016) was used to merge the results. Furthermore, Trimmomatic (Bolger et al., 2014) was used to trim the reads, and bbduk3 removed PhiX. Thereafter, de novo assembly was done using SPAdes (Bankevich et al., 2012). Only contigs longer than 500 bp were withheld in the assemblies. Assemblies were polished and improved using Pilon (Walker et al., 2014), and QUAST (Gurevich et al., 2013) was used to evaluate the assemblies using E. coli K-12 substr. MG1655 (accession number NZ_CP027060.1) as reference. The sequence types (STs) were predicted by the ARIBA (antimicrobial resistance identification by assembly) tool (Hunt et al., 2017) using the Achtman scheme (Wirth et al., 2006). ARIBA was also used to predict the acquired antimicrobial resistance (AMR) genes present using the ResFinder database (Zankari et al., 2012), and virulence genes using the VirulenceFinder database (Joensen et al., 2014). Furthermore, we used chewbacca (Silva et al., 2018) to perform cgMLST using the scheme available from Enterobase4. Vizualisation of cgMLST data was done in R version 3.5.2 (R Core Team, 2019). Plasmid replicons and pMLST profiles were determined using PlasmidFinder (Carattoli et al., 2014) and pMLST 2.0 (Carattoli et al., 2014).
Long reads were demultiplexed using the barcoding pipeline on SMRT Link (v6.0.0.47841, SMRT Link Analysis Services and GUI v6.0.0.47836) with 40 as minimum barcode score. Read mapping was carried out to assess the quality of data and coverage using Minimap2 (Li, 2018). Illumina reads were used as reference in the filtlong tool5 to produce a better subset of PacBio reads. Hybrid assemblies were generated using Unicycler version v0.4.7 (Wick et al., 2017). Bandage (Wick et al., 2015) was used to assess the quality of assemblies. Circularized plasmid contigs were investigated using BLASTn6, 15th August 2019, and the curated complete plasmid sequences were annotated using Prokka version 1.13.7 (Seemann, 2014). Annotations were analyzed and curated using Artemis (Carver et al., 2005) and compared using Easyfig (Sullivan et al., 2011). Sequence differences in the genes encoding replication (inc), partition (parAB), and entry exclusion (traY-excA) can be used to distinguish between plasmids of the IncI1 and IncIγ groups (Takahashi et al., 2011). Thus, the nucleotide sequence of the inc gene, and amino acid sequence of parAB and ExcA were compared to the corresponding regions in R64 (accession number AP005147) and R621a (accession number AP011954.1) to further analyze the plasmid replicon types.
Reconstructed plasmids were compared to IncI plasmids from broilers in the Netherlands, Switzerland, France, and Denmark available from GenBank.
Complete plasmid sequences are available in GenBank; MN419430 (p17437), MN419431 (p19138), MN419432 (p21254), MN419433 (p20426), MN419434 (21249), MN419435 (p14263), MN419436 (p22440), MN419437 (p22638), and MN419438 (p24003). Raw reads for the 31 E. coli isolates are available from ENA (bioproject number PRJEB45077).
Results
Strain Characterization
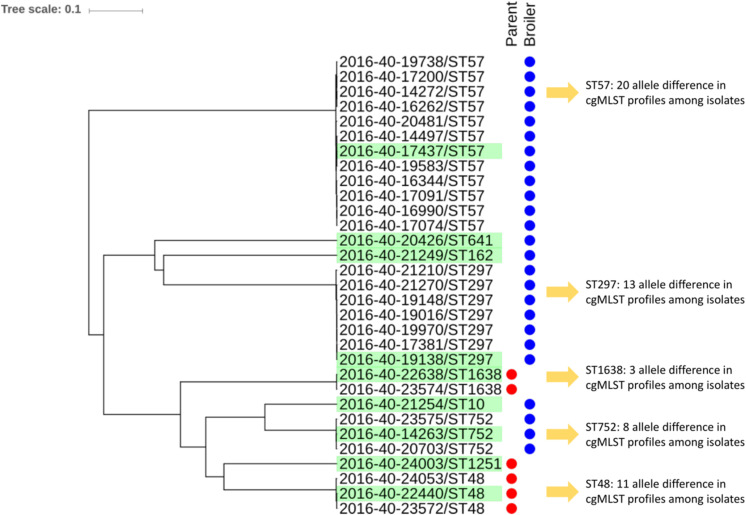
An overview of the strain characteristics is presented in Table 1. Most isolates belonged to phylogroup D (n = 13), followed by A (n = 10), and B1 (n = 8). None of the isolates were characterized as B2 (Table 1). The 31 E. coli isolates belonged to nine different STs, with ST57 (n = 12) and ST297 (n = 7) being the most common. Each MLST group represented one cluster in the cgMLST analysis (Figure 1). On broiler farm A, E. coli ST752 was detected twice, differing by eight alleles. On broiler farm B, ST57 isolates were detected four times, two isolates with identical cgMLST profiles, and only three allele differences between all four isolates. Two ST57 isolates with identical cgMLST profiles and a third isolate differing by only one allele were detected from farm C. All ST297 isolates originated from different farms. Among these, two shared identical cgMLST patterns, and the maximum allele difference was 13.
FIGURE 1.
Clustering of 31 Escherichia coli isolates with blaCTX–M–1 isolated from Norwegian broiler production in 2016 based on core genome MLST using the scheme available from Enterobase. Isolates selected for long read sequencing are highlighted in green.
Table 2 presents the results from the antimicrobial susceptibility testing. Six isolates displayed multidrug resistance (MDR, resistance to three or more antimicrobial classes, Table 1). All isolates were cephalosporin-resistant, and most were co-resistant to at least one antimicrobial class. The most commonly detected co-resistance was to sulfonamides (n = 27, 87%), followed by tetracycline (n = 6, 19%), and trimethoprim (n = 3, 10%). Resistance to the critically important antimicrobials carbapenem and colistin was not observed. WGS data confirmed the presence of blaCTX–M–1 in all isolates. Additional AMR genes detected in the isolates are shown in Table 1. All three ST752 isolates harbored the eae gene, and were categorized as enteropathogenic E. coli (EPEC) (Table 1; Kaper et al., 2004). The remaining isolates did not harbor virulence genes associated with an E. coli pathogroup. An overview of all virulence genes detected is available in Supplementary Table 1.
TABLE 2.
Minimum inhibitory concentrations (MICs) and antimicrobial resistance in Escherichia coli harboring blaCTX–M–1 (n = 31) isolated from broiler- and parent flocks in Norway during 2016.
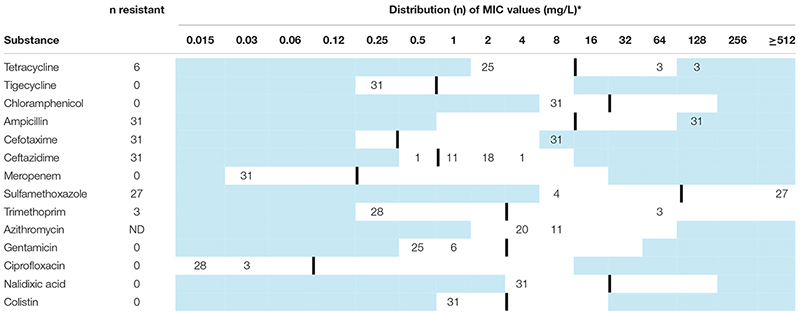 |
*Bold vertical lines denote epidemiological cut-off values for resistance. ND, cut-off not defined by EUCAST. White fields denote range of dilutions tested for each antimicrobial agent. MIC values higher than the highest concentration tested are given as the lowest MIC value above the range. MIC values equal to or lower than the lowest concentration tested are given as the lowest concentration tested.
Plasmid Characterization
The blaCTX–M–1 gene was located on conjugative plasmids in 28 of the 31 isolates (90%) (Table 1). This was confirmed by the presence of a single replicon type in most transconjugants (Table 1), and the presence of a complete transfer region on all fully sequenced pMLST3 plasmids (Figure 2; Carattoli et al., 2018). The blaCTX–M–1-carrying plasmids in the three remaining isolates were not able to conjugate into E. coli DH5α.
FIGURE 2.
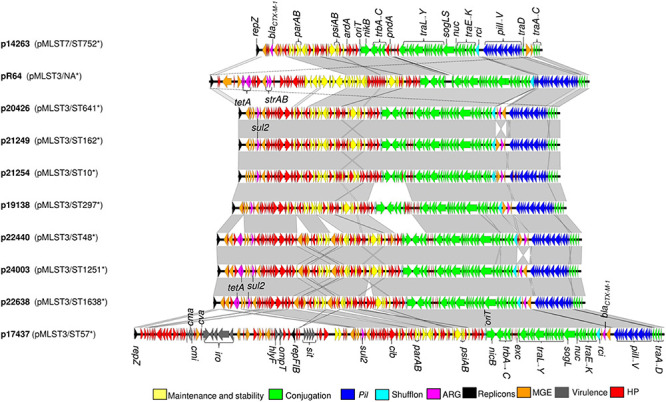
Genetic organization of plasmids p14263, p20426, p21249, p21254, p19138, p22440, p24003, p22638, and p17437 including comparison to the IncI1 reference plasmid R64 (accession number AP005147). Areas shaded gray indicate homologus regions, nucleotide identity threshold >95%. ARG, antimicrobial resistance genes; MGE, mobile genetic elements; HP, hypothetical proteins; NA, not available; *, sequence type of host strain.
PlasmidFinder confirmed the presence of IncI1-Iγ replicons in all isolates. Additional replicons like FIB and B/O/K/Z were also present in subsets of the isolates (Table 1). pMLST revealed that 19 of the IncI1-Iγ plasmids were ST3, three were ST7 and four were ST42. ST3 and ST42 group into the same clonal complex (CC), namely CC-3. Five IncI1-Iγ plasmids had an incomplete ST (IC) (Table 1). PBRT showed that all transconjugants harbored IncI1 replicons, and WGS data confirmed that blaCTX–M–1 was located on IncI1-Iγ plasmids in all isolates. Some transconjugants also harbored FIB and B/O/K/Z replicons as shown in Table 1.
The genetic organization of the nine reconstructed IncI1-Iγ plasmids is visualized in Figure 2. The backbone structures of the plasmids were similar to R64. The traABCD regulatory gene cluster was located at the distal end of the type IV pili locus region in ST3 plasmids (Figure 2). In the re-constructed ST7 plasmid, the traC gene was disrupted by insertion of the ISSoEn3 transposase. The shufflon consisted of a number of invertible DNA segments (Table 3), and was located at the proximal end of the type IV pili locus region. Genes encoding addiction systems, namely relBE, phd/doc, and pndAC were present on all plasmids (Table 3). Some differences were observed in the regions encoding trbABC and nikB in p19138 compared to the other ST3 plasmids (Figure 2). In most plasmids, the nucleotide sequence of the inc gene was identical to that found in the R64 plasmid, while three deletions in the 3’ end was present in one plasmid (p17437) (Supplementary Figure 1). The ParAB amino acid sequences were also identical to that of R64 in all plasmids (Supplementary Figures 2, 3). In seven of our plasmids, the amino acid sequence of ExcA had 64% sequence identity compared to R64, and 38% sequence identity compared to R621a. p17437 differed by eight amino acids at the N-terminal compared to the rest of our plasmids. The amino acid sequence of ExcA in the ST7 plasmid (p14263) had 99.5% sequence identity to the one in R621a (Figure 3). The results show that our plasmids cannot be unambiguously grouped as belonging to the I1 or Iγ, as they show intermediate evolutionary levels between these replicon types. Thus, we refer to them as IncI1-Iγ.
TABLE 3.
Overview of characteristics associated with IncI1-Iγ plasmids carrying blaCTX–M–1 in nine Escherichia coli isolated from Norwegian broilers and parent flocks in 2016.
| Isolate | MLST | Plasmid | pMLST | Estimated plasmid size (bp) | Other AMR genes | Shufflon regions | Virulence determinants |
| 2016-40-14263 | ST752 | p14263 | ST7 | 91030 | ND | CC’B’BA’A | ND |
| 2016-40-20426 | ST641 | p20426 | ST3 | 102183 | sul2 | B’BC’CA’A | cib |
| 2016-40-21249 | ST162 | p21249 | ST3 | 102184 | sul2 | BB’C’CA’A | cib |
| 2016-40-21254 | ST10 | p21254 | ST3 | 102373 | sul2 | BB’CC’A’A | cib |
| 2016-40-19138 | ST297 | p19138 | ST3 | 106298 | sul2 | BB’CC’A’A | cib |
| 2016-40-22440 | ST48 | p22440 | ST3 | 115586 | sul2, tetA | B’BCC’A’A | cib |
| 2016-40-24003 | ST1251 | p24003 | ST3 | 116056 | sul2, tetA | CC’BB’A’A | cib |
| 2016-40-22638 | ST1638 | p22638 | ST3 | 118243 | sul2, tetA | B’BCC’A’A | cib |
| 2016-40-17437 | ST57 | p17437 | ST3 | 168655 | sul2 | B’BC’CA’A | cib, cma, cva, sit, iro, hlyF, ompT |
AMR, antimicrobial resistance; ND, not detected.
FIGURE 3.
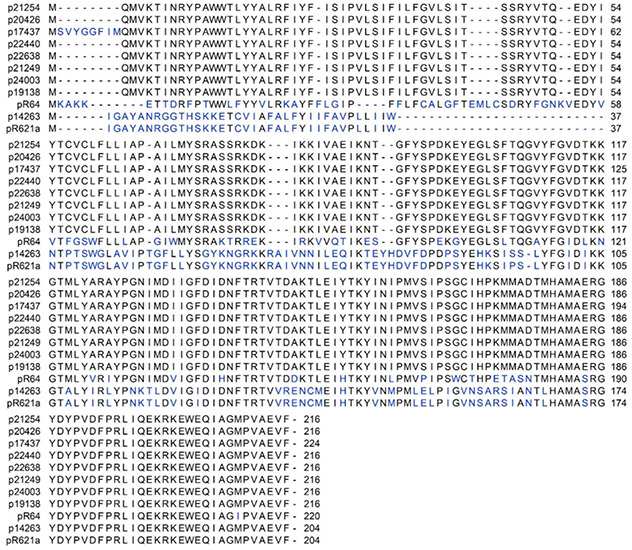
Alignment of the amino acid sequences of ExcA of the nine IncI1-Iγ plasmids reconstructed in this study, IncI1 prototype R64 (accession number AP005147) and IncIγ prototype R621a (accession number AP011954.1). Gaps are marked by dashes, and amino acid differences are shown in blue.
Highly similar plasmids belonging to ST3 were detected in E. coli ST1638, ST1251 and ST48. These plasmids ranged in size from 115586-118243 bp and carried the sul2 and tetA genes inserted as a part of the accessory module. In addition, somewhat smaller IncI1-Iγ/ST3 plasmids, ranging from 102183-102373 bp and carrying only sul2 in addition to blaCTX–M–1, were present in ST641, ST162, and ST10 (Table 2). On all IncI1-Iγ/ST3 plasmids, blaCTX–M–1 was linked with ISEcp1, and located in close proximity to the shufflon. In contrast, the ISEcp1-blaCTX–M–1 region was inserted in the accessory module in the IncI1-Iγ/ST7 plasmid. All IncI1-Iγ/ST3 plasmids harbored the cib gene encoding a colicin (Table 2).
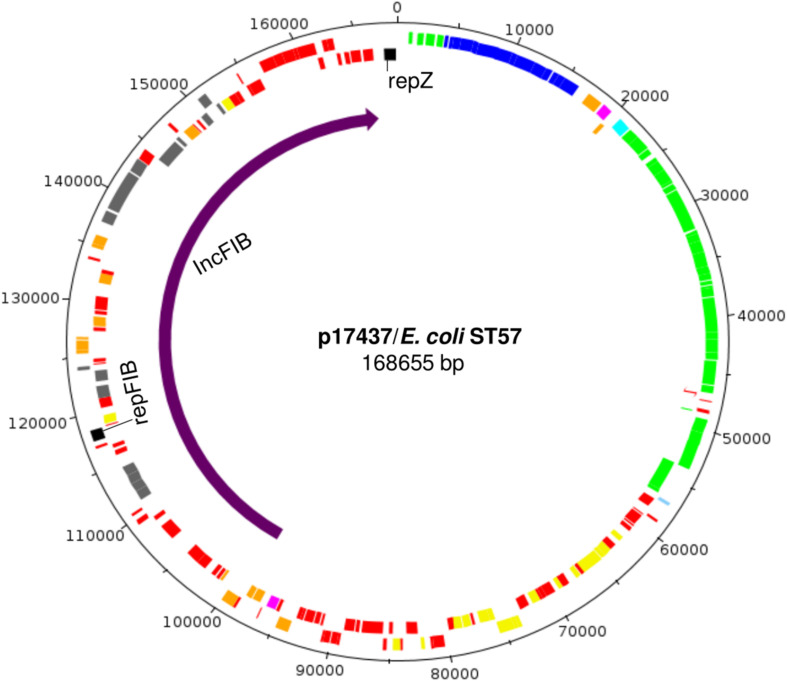
In one reconstructed ST3 plasmid, two replicons were detected, namely IncI1-Iγ and IncFIB (p17437, Table 3). Further characterization implied that this IncI1-Iγ/IncFIB co-integrated plasmid consisted of a complete IncI1-Iγ plasmid and a fraction of an IncFIB plasmid (Figures 2, 4). Several virulence determinants, including sit, iroN and hlyF, were encoded on the IncFIB fraction of the plasmid. The total length of the co-integrated plasmid was 168655 bp (Figures 2, 4 and Table 3). S1 nuclease digestion confirmed the presence of the plasmid in all ST57 transconjugants that were positive for both the IncI1-Iγ and IncFIB replicons. One ST57 transconjugant had only the IncI1-Iγ replicon, and harbored a smaller plasmid (Supplementary Figure 4).
FIGURE 4.
Genomic map of the co-integrated plasmid p17437 harboring both IncI1-Iγ and IncFIB replicons. The region indicated by the purple arrow is the IncFIB specific region, while the remaining part is the IncI1-Iγ specific region. Positions of genes are indicated by color, and their meaning is as follows; yellow, maintenance and stability cluster; green, transfer locus; blue, pilus; black, plasmid replicons; aqua, shufflon; pink, antimicrobial resistance genes; orange, transposome; gray, virulence genes; red, hypothetical proteins.
Comparison with IncI1-Iγ plasmids originating from broiler production in other European countries revealed close similarity to plasmids characterized in this study (Supplementary Figures 5–7). Our data show clonal dissemination of E. coli STs carrying blaCTX–M–1, but also horizontal transfer of highly similar blaCTX–M–1 carrying plasmids in Norwegian broiler production is illustrated in Supplementary Figure 8.
Discussion
The isolates investigated represent a unique collection including all blaCTX–M–1 isolates originating from Norwegian broilers sampled during a six month period (May–October) and Norwegian parent flocks sampled during a four week period (medio November–medio December) in 2016. Thus, it gives us substantial knowledge regarding different E. coli STs and plasmids circulating, and enables evaluation of plasmid- and ST flux and epidemiology over a prolonged time period in the broiler/parent population at national level.
Nine different E. coli STs were detected among the 31 isolates included, underlining some genetic diversity among the isolates. However, ST57 and ST297 were detected from several flocks and farms indicating possible clonal dissemination of certain E. coli STs with blaCTX–M–1 in Norwegian broiler production. This is supported by identical or highly similar cgMLST profiles among the isolates. All three isolates from parent flock 1 belonged to ST48 and clustered together in the cgMLST analysis, differing by 11 alleles. On the contrary, one of the three isolates from parent flock 2 belonged to ST1251, while the other two belonged to ST1638. However, the blaCTX–M–1 carrying plasmids in these three isolates were highly similar, possibly indicating horizontal plasmid transfer between different E. coli STs. On six of the 15 broiler farms two or more flocks were positive for blaCTX–M–1. This could be due to on-farm persistence of ESC-resistant bacteria as previously suggested (Hiroi et al., 2012a; Dierikx C. et al., 2013; Laube et al., 2013; Agersø et al., 2014; Nilsson et al., 2014).
ST752 isolates (n = 3) included one isolate from a parent flock and two isolates from broiler flocks. These isolates differed by only eight alleles in their cgMLST profiles. This could indicate clonal dissemination of ESBL-producing E. coli in the production pyramid, which has also been proposed by others (Sunde et al., 2009; Dierikx C.M. et al., 2013; Nilsson et al., 2014; Zurfluh et al., 2014). Unfortunately, we did not have information regarding which parent flocks supplied the broiler flocks included in this study. Thus, we cannot confirm the presence of related isolates in broiler flocks and their supplying parent flock(s) based on our data.
E. coli carrying blaCTX–M–1 on transferable IncI1 plasmids is also commonly occurring in European broiler production (Börjesson et al., 2013a, b; Zurfluh et al., 2014; DANMAP, 2015; Smith et al., 2015; Irrgang et al., 2018; Touzain et al., 2018). Most of the IncI1-Iγ plasmids in our study were identified as ST3 (n = 19), which is also a common ST in broiler production in other countries (Zurfluh et al., 2014; Smith et al., 2015; Baron et al., 2018; Irrgang et al., 2018), and the most commonly reported IncI1-Iγ plasmid associated with blaCTX–M–1 in Europe (Carattoli et al., 2018). We detected highly similar IncI1-Iγ/ST3 plasmids in different E. coli STs, indicating that the plasmids are able to disseminate horizontally in the E. coli population. Further comparison of a selection of IncI1-Iγ/ST3 plasmids from broiler production in Norway and other European countries, including the Netherlands, Switzerland, France, and Denmark (Brouwer et al., 2014; Wang et al., 2014; Zurfluh et al., 2014; Touzain et al., 2018; Valcek et al., 2019), revealed high genetic similarity. Thus, there is reason to believe that highly similar IncI1-Iγ/ST3 plasmids are widespread and successful in the European broiler production. This has also been reported for other plasmids encoding ESC-resistance in broiler production (Hansen et al., 2016; Mo et al., 2016), and is consistent with a common source (Dierikx C.M. et al., 2013; Agersø et al., 2014; Nilsson et al., 2014; Zurfluh et al., 2014). Reconstruction of IncI1-Iγ/ST3 plasmids also revealed some genetic diversity. All shared a common backbone, but some differences were observed, especially in the accessory module. Furthermore, some amino acid differences were observed in regions encoding replication, partitioning, entry exclusion, TrbABC and NikB. This indicates a certain flux of genes at the plasmid level, and further studies are required to determine whether this has an effect on plasmid stability and/or conjugation frequency.
We detected only three IncI1-Iγ plasmids belonging to ST7 in our isolates, all of which were non-transferable. A disruption of the traC gene, known to be essential for conjugation (Kim et al., 1993), was the most likely cause of the non-conjugative phenotype. This could also explain why we only detected them in a limited number of isolates and a single E. coli ST. Thus, it seems that ST7 plasmids are of limited epidemiological importance in Norwegian broiler production, although they have been reported to have high epidemiological relevance internationally (Carattoli et al., 2018). Furthermore, all ST3 plasmids characterized in our study encoded a colicin. Colicin production has been reported to represent a selective advantage, as it can have lethal effect on related bacteria in the gut without colicin production (Cascales et al., 2007; Majeed et al., 2011). It is also plausible that colicin production together with the presence of plasmid-encoded addiction systems, will facilitate persistence of IncI1-Iγ/blaCTX–M–1 plasmids in an E. coli population despite the lack of selection pressure from antimicrobial use, which is the case in Norwegian broiler production (NORM/NORM-VET, 2019).
One of the reconstructed plasmids (p17437) turned out to be a co-integrated plasmid harboring both an IncI1-Iγ and an IncFIB replicon. The IncFIB specific part was inserted into the accessory module. Co-integrated IncI1/IncFIB plasmids have been described in a few previous studies in Salmonella and enterotoxigenic E. coli (ETEC) (Froehlich et al., 2005; Johnson and Nolan, 2009; Crossman et al., 2010; Moreno Switt et al., 2012). It has also been suggested that such human ETEC plasmids might have an animal origin (Johnson et al., 2011). The co-integrated plasmids found in Salmonella (Moreno Switt et al., 2012) had the IncF region inserted in a similar manner as described in this study, adjacent to the IncI replicon repZ. The IncFIB fraction of our co-integrated plasmid harbored several virulence genes. This could represent a selective advantage, as the plasmid will confer both virulence and antimicrobial resistance, but this hypothesis needs to be investigated further. Interestingly, we only detected the co-integrated IncI1-Iγ/IncFIB plasmid in a single ST, namely ST57. This was also the most commonly occurring ST in our material, detected from a total of six different farms. All but one of the ST57 transconjugants were positive for both the IncI1-Iγ and IncFIB replicons. This was confirmed by plasmid profiling, as all but one of the transconjugants from ST57 isolates harbored a large plasmid with a size corresponding to the co-integrative plasmid. One ST3 plasmid (p19138) differed from the others in the regions encoding traABC and nikB. Interestingly, this plasmid was only detected in E. coli ST297. This was the second most common ST in our material, isolated from six different farms. These two examples could indicate successful plasmid-host combinations, and should be further investigated. The systematic and comprehensive sampling behind our dataset can provide support for some trends at the plasmid level. We found that IncI1-Iγ/ST3 plasmids were more common than IncI1-Iγ/ST7 plasmids. A contributing factor could be that the ST7 plasmids were non-conjugative, thus having a lower potential for dissemination in the bacterial population. Furthermore, the clonal dissemination of both ST57 and ST297 can partly explain the dominance of ST3 plasmids in our material. In addition, our dataset enabled comparison of IncI1-Iγ/ST3 plasmids occurring in different E. coli STs. The comparison showed highly conserved backbones, but also variations. This demonstrated dissemination of similar, but distinguishable ST3 plasmids. To a wide extent, the observed variations were associated with the accessory region, which is in concordance with previous findings (Johnson et al., 2011). The observed differences were mainly due to different compositions of AMR genes. In addition, smaller variations were observed on other locations of the plasmid. This further supports the observation by Smith et al. (2015) that accessory genes can integrate on several locations in IncI1-Iγ plasmids.
Conclusion
In conclusion, our results point toward a scenario where both clonal dissemination and horizontal transfer of plasmids have contributed to the dissemination of blaCTX–M–1 in Norway. Our studies revealed circulation of similar plasmids, but some genetic diversity was present among the IncI1-Iγ/blaCTX–M–1 plasmids recovered during the sampling period. The plasmids represented variants grouping into two main lineages, namely CC-3 and CC-7, described from broiler production in several countries. The genetic diversity indicates multiple introductions to the Norwegian broiler production. The close genetic relationship between plasmids originating from broiler production in Norway and other European countries, provides further evidence for the theory of a common source of the ESC-resistant isolates.
Data Availability Statement
Plasmid sequences are uploaded to NCBI; MN419430, MN419431, MN419432, MN419433, MN419434, MN419435, MN419436, MN419437, and MN419438. Raw reads from all isolates will be submitted to ENA and accession numbers reported accordingly.
Author Contributions
SM, AT, and MS contributed to the conception and design of the study, while AU was responsible for the conception and design of the study the isolates originated from. SM, AT, KO, HI, and TL performed the laboratory work. SM, AT, KO, JS, CS, and MS performed the in silico analyses. SM wrote the first draft of the manuscript. AT and MS wrote sections of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Håkon Kaspersen is acknowledged for assisting with the bioinformatics analyses. The authors are grateful to the Norwegian Sequencing Centre (www.sequencing.uio.no), a National Technology Platform hosted by the University of Oslo and supported by the Functional Genomics and Infrastructure programs of the Research Council of Norway and the Southeastern Regional Health Authorities, for providing the sequencing service. This work was performed on the Abel Cluster, owned by the University of Oslo and Uninett/Sigma2, and operated by the Department for Research Computing at the University of Oslo IT-department. http://www.hpc.uio.no/.
Funding. This study was funded by the Norwegian Veterinary Institute, the Research Council of Norway (Grant Number 250212/E50), and the European Union’s Horizon 2020 program under grant agreement number 773830, OH-EJP-H2020-JRP-AMR-2-ARDIG.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00333/full#supplementary-material
References
- Agersø Y., Jensen J. D., Hasman H., Pedersen K. (2014). Spread of extended spectrum cephalosporinase-producing Escherichia coli clones and plasmids from parent animals to broilers and to broiler meat in a production without use of cephalosporins. Foodborne Pathog. Dis. 11 740–746. 10.1089/fpd.2014.1742 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S., Le Devendec L., Touzain F., Jouy E., Lucas P., de Boisseson C., et al. (2018). Longitudinal study of Escherichia coli plasmid resistance to extended-spectrum cephalosporins in free-range broilers. Vet. Microbiol. 216 20–24. 10.1016/j.vetmic.2018.01.012 [DOI] [PubMed] [Google Scholar]
- Barton B. M., Harding G. P., Zuccarelli A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226 235–240. 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börjesson S., Bengtsson B., Jernberg C., Englund S. (2013a). Spread of extended-spectrum beta-lactamase producing Escherichia coli isolates in Swedish broilers mediated by an incl plasmid carrying bla(CTX-M-1). Acta Vet. Scand. 55:3. 10.1186/1751-0147-55-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börjesson S., Egervärn M., Lindblad M., Englund S. (2013b). Frequent occurrence of extended-spectrum beta-lactamase- and transferable AmpC beta-lactamase-producing Escherichia coli on domestic chicken meat in Sweden. Appl. Environ. Microbiol. 79 2463–2466. 10.1128/AEM.03893-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briñas L., Moreno M. A., Zarazaga M., Porrero C., Sáenz Y., García M., et al. (2003). Detection of CMY-2, CTX-M-14, and SHV-12 beta-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. Agents Chemother. 47 2056–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer M. S., Bossers A., Harders F., van Essen-Zandbergen A., Mevius D. J., Smith H. E. (2014). Complete genome sequences of IncI1 plasmids carrying extended-spectrum beta-lactamase genes. Genome Announc. 2:e00859-14. 10.1128/genomeA.00859-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2008). Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14(Suppl. 1), 117–123. 10.1111/j.1469-0691.2007.01851.x [DOI] [PubMed] [Google Scholar]
- Carattoli A., Villa L., Fortini D., Garcia-Fernandez A. (2018). Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 10.1016/j.plasmid.2018.12.001 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J. (2005). ACT: the artemis comparison tool. Bioinformatics 21 3422–3423. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- Cascales E., Buchanan S. K., Duche D., Kleanthous C., Lloubes R., Postle K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71 158–229. 10.1128/MMBR.00036-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossman L. C., Chaudhuri R. R., Beatson S. A., Wells T. J., Desvaux M., Cunningham A. F., et al. (2010). A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192 5822–5831. 10.1128/JB.00710-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANMAP (2015). DANMAP 2014. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. Copenhagen: DANMAP. [Google Scholar]
- Dierikx C., van der Goot J., Fabri T., van Essen-Zandbergen A., Smith H., Mevius D. (2013). Extended-spectrum-beta-lactamase- and AmpC-beta-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J. Antimicrob. Chemother. 68 60–67. 10.1093/jac/dks349 [DOI] [PubMed] [Google Scholar]
- Dierikx C. M., van der Goot J. A., Smith H. E., Kant A., Mevius D. J. (2013). Presence of ESBL/AmpC -producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS One 8:e79005. 10.1371/journal.pone.0079005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierikx C., van Essen-Zandbergen A., Veldman K., Smith H., Mevius D. (2010). Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 145 273–278. 10.1016/j.vetmic.2010.03.019 [DOI] [PubMed] [Google Scholar]
- Doi Y., Paterson D. L., Egea P., Pascual A., López-Cerero L., Navarro M. D., et al. (2010). Extended-spectrum and CMY-type beta-lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clin. Microbiol. Infect. 16 33–38. 10.1111/j.1469-0691.2009.03001.x [DOI] [PubMed] [Google Scholar]
- Doumith M., Day M. J., Hope R., Wain J., Woodford N. (2012). Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J. Clin. Microbiol. 50 3108–3110. 10.1128/JCM.01468-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P., Magnusson M., Lundin S., Kaller M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C., Bethe A., Semmler T., Guenther T., Wieler L. H. (2012). Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18 646–655. 10.1111/j.1469-0691.2012.03850.x [DOI] [PubMed] [Google Scholar]
- Froehlich B., Parkhill J., Sanders M., Quail M. A., Scott J. R. (2005). The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J. Bacteriol. 187 6509–6516. 10.1128/JB.187.18.6509-6516.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. H., Bortolaia V., Nielsen C. A., Nielsen J. B., Schønning K., Agersø Y., et al. (2016). Host-specific patterns of genetic diversity among IncI1-Igamma and IncK plasmids encoding CMY-2 beta-lactamase in Escherichia coli isolates from humans, poultry meat, poultry and dogs in Denmark. Appl. Environ. Microbiol. 10.1128/AEM.00495-16 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H., Mevius D., Veldman K., Olesen I., Aarestrup F. M. (2005). beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56 115–121. 10.1093/jac/dki190 [DOI] [PubMed] [Google Scholar]
- Hiroi M., Matsui S., Kubo R., Iida N., Noda Y., Kanda T., et al. (2012a). Factors for occurrence of extended-spectrum beta-lactamase-producing Escherichia coli in broilers. J. Vet. Med. Sci. 74 1635–1637. [DOI] [PubMed] [Google Scholar]
- Hiroi M., Yamazaki F., Harada T., Takahashi N., Iida N., Noda Y., et al. (2012b). Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J. Vet. Med. Sci. 74 189–195. [DOI] [PubMed] [Google Scholar]
- Hunt M., Mather A. E., Sanchez-Buso L., Page A. J., Parkhill J., Keane J. A., et al. (2017). ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 3:e000131. 10.1099/mgen.0.000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrgang A., Hammerl J. A., Falgenhauer L., Guiral E., Schmoger S., Imirzalioglu C., et al. (2018). Diversity of CTX-M-1-producing E. coli from German food samples and genetic diversity of the blaCTX-M-1 region on IncI1 ST3 plasmids. Vet. Microbiol. 221 98–104. 10.1016/j.vetmic.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Joensen K. G., Scheutz F., Lund O., Hasman H., Kaas R. S., Nielsen E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52 1501–1510. 10.1128/JCM.03617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Nolan L. K. (2009). Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73 750–774. 10.1128/MMBR.00015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Shepard S. M., Rivet B., Danzeisen J. L., Carattoli A. (2011). Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid 66 144–151. 10.1016/j.plasmid.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Kaper J. B., Nataro J. P., Mobley H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2 123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Kim S. R., Funayama N., Komano T. (1993). Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J. Bacteriol. 175 5035–5042. 10.1128/jb.175.16.5035-5042.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K. (2020). NorwegianVeterinaryInstitute/Bifrost: v1.1 (Version v1.1). Zenodo. 10.5281/zenodo.4020511 [DOI] [Google Scholar]
- Laube H., Friese A., von Salviati C., Guerra B., Käsbohrer A., Kreienbrock L., et al. (2013). Longitudinal monitoring of extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli in German broiler chicken fattening farms. Appl. Environ. Microbiol. 79 4815–4820. 10.1128/AEM.00856-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed H., Gillor O., Kerr B., Riley M. A. (2011). Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J. 5 71–81. 10.1038/ismej.2010.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo S. S., Slettemeås J. S., Berg E. S., Norström M., Sunde M. (2016). Plasmid and host strain characteristics of Escherichia coli resistant to extended-spectrum cephalosporins in the Norwegian broiler production. PLoS One 11:e0154019. 10.1371/journal.pone.0154019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo S. S., Urdahl A. M., Nesse L. L., Slettemeas J. S., Ramstad S. N., Torp M., et al. (2019). Occurrence of and risk factors for extended-spectrum cephalosporin-resistant Enterobacteriaceae determined by sampling of all Norwegian broiler flocks during a six month period. PLoS One 14:e0223074. 10.1371/journal.pone.0223074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Switt A. I., den Bakker H. C., Cummings C. A., Rodriguez-Rivera L. D., Govoni G., Raneiri M. L., et al. (2012). Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PLoS One 7:e41247. 10.1371/journal.pone.0041247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O., Börjesson S., Landén A., Bengtsson B. (2014). Vertical transmission of Escherichia coli carrying plasmid-mediated AmpC (pAmpC) through the broiler production pyramid. J. Antimicrob. Chemother. 69 1497–1500. 10.1093/jac/dku030 [DOI] [PubMed] [Google Scholar]
- NORM/NORM-VET (2007). NORM/NORM-VET 2006. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø: NORM/NORM-VET. [Google Scholar]
- NORM/NORM-VET (2012). NORM/NORM-VET 2011. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø: NORM/NORM-VET. [Google Scholar]
- NORM/NORM-VET (2013). NORM/NORM-VET 2012. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø: NORM/NORM-VET. [Google Scholar]
- NORM/NORM-VET (2015). NORM/NORM-VET 2014. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø: NORM/NORM-VET. [Google Scholar]
- NORM/NORM-VET (2017). NORM/NORM-VET 2016. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø: NORM/NORM-VET. [Google Scholar]
- NORM/NORM-VET (2019). NORM/NORM-VET 2018. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø: NORM/NORM-VET. [Google Scholar]
- R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R foundation for Statistical computing. [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Silva M., Machado M. P., Silva D. N., Rossi M., Moran-Gilad J., Santos S., et al. (2018). chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 4:e000166. 10.1099/mgen.0.000166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smet A., Martel A., Persoons D., Dewulf J., Heyndrickx M., Catry B., et al. (2008). Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob. Agents Chemother. 52 1238–1243. 10.1128/AAC.01285-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Bossers A., Harders F., Wu G., Woodford N., Schwarz S., et al. (2015). Characterization of epidemic IncI1-Igamma plasmids harboring ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob. Agents Chemother. 59 5357–5365. 10.1128/AAC.05006-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M., Tharaldsen H., Slettemeås J. S., Norström M., Carattoli A., Bjorland J. (2009). Escherichia coli of animal origin in Norway contains a blaTEM-20-carrying plasmid closely related to blaTEM-20 and blaTEM-52 plasmids from other European countries. J. Antimicrob. Chemother. 63 215–216. 10.1093/jac/dkn445 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Shao M., Furuya N., Komano T. (2011). The genome sequence of the incompatibility group Igamma plasmid R621a: evolution of IncI plasmids. Plasmid 66 112–121. 10.1016/j.plasmid.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Touzain F., Le Devendec L., de Boisseson C., Baron S., Jouy E., Perrin-Guyomard A., et al. (2018). Characterization of plasmids harboring blaCTX-M and blaCMY genes in E. coli from French broilers. PLoS One 13:e0188768. 10.1371/journal.pone.0188768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcek A., Roer L., Overballe-Petersen S., Hansen F., Bortolaia V., Leekitcharoenphon P., et al. (2019). IncI1 ST3 and IncI1 ST7 plasmids from CTX-M-1-producing Escherichia coli obtained from patients with bloodstream infections are closely related to plasmids from E. coli of animal origin. J. Antimicrob. Chemother. 74 2171–2175. 10.1093/jac/dkz199 [DOI] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Stephan R., Power K., Yan Q., Hachler H., Fanning S. (2014). Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J. Antimicrob. Chemother. 69 2658–2668. 10.1093/jac/dku206 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Schultz M. B., Zobel J., Holt K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31 3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60 1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K., Wang J., Klumpp J., Nuesch-Inderbinen M., Fanning S., Stephan R. (2014). Vertical transmission of highly similar bla CTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front. Microbiol. 5:519. 10.3389/fmicb.2014.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmid sequences are uploaded to NCBI; MN419430, MN419431, MN419432, MN419433, MN419434, MN419435, MN419436, MN419437, and MN419438. Raw reads from all isolates will be submitted to ENA and accession numbers reported accordingly.