Abstract
Purpose
Compared to breast cancer risk genes such as BRCA2, ATM, PALB2, and NBN, no defined phenotype is currently associated with biallelic pathogenic variants (PVs) in CHEK2. This study compared the prevalence of breast and other cancers in women with monoallelic and biallelic CHEK2 PVs.
Methods
CHEK2 PV carriers were identified through commercial hereditary cancer panel testing (09/2013–07/2019). We compared cancer histories of 6473 monoallelic carriers to 31 biallelic carriers. Breast cancer risks were estimated using multivariate logistic regression and are reported as odds ratios (OR) with 95% confidence intervals (CI).
Results
Breast cancer frequency was higher among biallelic CHEK2 PV carriers (80.6%, 25/31) than monoallelic carriers (41.2%, 2668/6473; p < 0.0001). Biallelic carriers were more likely to be diagnosed at or before age 50 (61.3%, 19/31) and to have a second breast cancer diagnosis (22.6%, 7/31) compared to monoallelic carriers (23.9%, 1548/6473; p < 0.0001 and 8.1%, 523/6473; p = 0.0107, respectively). Proportionally more biallelic carriers also had any cancer diagnosis and > 1 primary diagnosis. Compared to women with no PVs, biallelic PV carriers had a higher risk of developing ductal invasive breast cancer (OR 8.69, 95% CI 3.69–20.47) and ductal carcinoma in situ (OR 4.98, 95% CI 2.00–12.35) than monoallelic carriers (OR 2.02, 95% CI 1.90–2.15 and OR 1.82, 95% CI 1.66–2.00, respectively).
Conclusions
These data suggest that biallelic CHEK2 PV carriers have a higher risk for breast cancer, are more likely to be diagnosed younger, and to have multiple primary breast cancers compared to monoallelic carriers. Biallelic carriers also appear to have a higher risk of cancer overall. Therefore, more aggressive management may be appropriate for women with biallelic PVs in CHEK2 compared with current recommendations for monoallelic carriers.
Keywords: CHEK2, Breast cancer, Germline, Biallelic
Introduction
CHEK2 is considered a moderate risk breast cancer gene, with estimates of the relative risk for women carrying a single pathogenic variant (PV) ranging from 2.0 to 4.8 for a first breast cancer. The risk for a second primary breast cancer following an initial diagnosis is estimated to be increased 2.8- to 3.5-fold over individuals with breast cancer without pathogenic variants (PVs) in breast cancer risk genes [1–4]. An increased risk of colorectal cancer has also been reported for CHEK2 PV carriers, however the evidence for this association is not well established [5–7]. More recent evidence supports a possible association with other cancers such as testicular germ cell tumors [8], renal cell cancer [9], and lethal prostate cancer [10]. Despite known and possible cancer associations for CHEK2 PV carriers, phenotypic differences between monoallelic and biallelic carriers are not yet understood. Biallelic carriers of PVs in other, dominant breast cancer susceptibility genes such as BRCA2, ATM, PALB2, and NBN are known to have more severe cancer phenotypes than monoallelic carriers [11–13]. Similar patterns for CHEK2 PV carriers have yet to be adequately established.
A single CHEK2 founder mutation, c.1100del, is present in 1.1% of individuals of Northern and Eastern European ancestry [14]. Assuming Hardy–Weinberg equilibrium, roughly 3/10000 of Northern and Eastern Europeans will be homozygous for c.1100del, with the possibility of additional biallelic individuals if other CHEK2 PVs are included. Homozygous CHEK2 c.1100del has, in fact, been reported in 14 female breast cancer cases in the Dutch population. In one study of 8 female homozygous carriers, the risk of breast cancer was estimated to be twice the risk in heterozygous carriers [15]. Because the number of homozygous carriers in this study was small, the calculated confidence intervals (CI) for breast cancer risk were wide, making the interpretation of these findings challenging. Similar results were observed in another study of six homozygous cases [16].
Based on the evidence of increased breast cancer risk in women with monoallelic CHEK2 PVs, current NCCN guidelines recommend annual mammography and consideration of breast MRI beginning at age 40 [12]. It is important to establish if women who are homozygous for CHEK2 c.1100del or are biallelic carriers of any CHEK2 PV have higher breast cancer risks than monoallelic carriers, since this could impact management recommendations. In order to better assess the cancer risks in rare female carriers of biallelic CHEK2 PVs, we identified CHEK2 PV carriers in a large series of patients undergoing hereditary cancer panel testing. We then compared the prevalence of breast cancer and other cancers in women with biallelic and monoallelic CHEK2 PVs.
Methods
Patients and hereditary cancer testing
Monoallelic and biallelic female carriers of CHEK2 PVs were identified through clinical pan-hereditary cancer panel testing at a Clinical Laboratory Improvement Amendments and College of American Pathologists-approved commercial laboratory (Myriad Genetic Laboratories, Inc., Salt Lake City, UT) between September 2013 and July 2019. The panel test included at least 25 genes (APC, ATM, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A, CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD51C, RAD51D, SMAD4, STK11, and TP53) or up to 35 genes (with the step-wise addition of GREM1, POLD1, and POLE, followed by HOXB13, and then AXIN2, GALNT12, MSH3, NTHL1, RNF43, and RPS20). All patients provided informed consent for clinical testing. De-identified clinical information was obtained from provider-completed test request forms (TRFs). Data was included only for women residing in states with no legal prohibitions on the use of de-identified data for research purposes.
Variant analysis, phase determination, and statistical methods
Variants were classified according to the American College of Medical Genetics and Genomics Guidelines supplemented with additional statistical algorithms [17, 18]. The CHEK2 founder mutations c.470C > T (p.Ile157Thr) and c.1283C > T (p.Ser428Phe) were not included in this analysis, as these were classified as variants of uncertain significance by the laboratory during the study time period and this analysis was restricted to PVs [19]. CHEK2 PV carriers who had a PV in another gene were also excluded. Phase in biallelic carriers was determined by allelic haplotypes or by confirming carrier status in relatives. Biallelic carriers with first-degree relatives confirmed to have only one PV were considered confirmed to carry PVs in trans. Fisher’s exact tests for difference in proportions were used to determine differences between monoallelic and biallelic CHEK2 carriers. p-values < 0.05 were considered significant. Adjustments for multiple comparisons were not performed as this analysis was hypothesis-driven.
A previously-published multivariable logistic regression model was used to estimate breast cancer risks [4]. A separate analysis was performed for breast cancer according to subtype with CHEK2 PV carrier status (monoallelic and biallelic) as a dependent variable. Independent variables included personal (binary flags) and family (numeric counts weighted by degree of relative) cancer histories of breast (lobular invasive, ductal invasive, ductal carcinoma in situ [DCIS], and male), ovarian, colorectal, melanoma, gastric, pancreatic, prostate, endometrial, and colon (polyps) as well as age at testing, sex and ancestry. 95% confidence intervals (CIs) were calculated using Wald statistics.
Results
Cohort description
In this study, we identified 6473 monoallelic and 42 biallelic carriers of CHEK2 PVs. Of those 42 biallelic CHEK2 PV carriers, phase was confirmed to be in trans for 31 cases, 16 of which were homozygous for CHEK2 c.1100del. In this cohort, the majority of CHEK2 PV carriers were of White/Non-Hispanic origin, both in monoallelic and biallelic carriers (69.8% and 71.0%, respectively; Table 1). Among biallelic PV carriers, there appeared to be a possible over-representation of patients with Hispanic/Latino ancestry compared to among monoallelic carriers (12.9% and 5.9%, respectively). On average, biallelic carriers of CHEK2 PVs were diagnosed with breast and other cancers at younger ages than monoallelic carriers.
Table 1.
Cohort demographics
| Characteristic | Variable | Monoallelic (N = 6473) | Biallelica (N = 31) |
|---|---|---|---|
| Age at testing (years) | Range | 16, ≥ 90 | 27, 74 |
| Median (IQR) | 48 (38, 58) | 51 (40, 63) | |
| ≤ 50 | 56.3% | 48.4% | |
| Age at cancer diagnosis (any cancer)b | Range | 1, ≥ 90 | 24, 66 |
| Median (IQR) | 47 (40, 56) | 44 (37.5, 49.5) | |
| Age at breast cancer diagnosisc | Range | 16, ≥ 90 | 30, 66 |
| Median (IQR) | 48 (42, 57) | 44 (38, 49) | |
| Ancestry | White/Non-Hispanic | 4521 (69.8%) | 22 (71.0%) |
| Hispanic/Latino | 384 (5.9%) | 4 (12.9%) | |
| Black/African | 87 (1.3%) | 0 | |
| Ashkenazi Jewish | 68 (1.1%) | 0 | |
| Middle Eastern | 34 (0.5%) | 1 (3.2%) | |
| Asian | 33 (0.5%) | 0 | |
| Otherd | 65 (1.0%) | 0 | |
| Multiple | 352 (5.4%) | 2 (6.5%) | |
| Not provided | 929 (14.4%) | 2 (6.5%) | |
| Personal cancer historye | Breast | 2668 (41.2%) | 25 (80.6%) |
| Ovarian | 230 (3.6%) | 2 (6.5%) | |
| Endometrial | 117 (1.8%) | 0 | |
| Colorectal | 97 (1.5%) | 0 | |
| Melanoma | 98 (1.5%) | 0 | |
| Pancreatic | 10 (0.2%) | 1 (3.2%) | |
| Gastric/stomach | 2 (< 0.1%) | 0 | |
| Colorectal polyps | 201 (3.1%) | 0 | |
| Non-colorectal polyps | 1 (< 0.1%) | 0 | |
| Other | 441 (6.8%) | 3 (9.7%) | |
| No cancer history | 3234 (50.0%) | 3 (9.7%) |
aConfirmed in trans
bOnly includes patients with a cancer diagnosis (28 biallelic PV carriers, 3239 monoallelic PV carriers)
cOnly includes patients with a breast cancer diagnosis (25 biallelic PV carriers, 2668 monoallelic PV carriers)
dOther includes Native American, Pacific Islander, and all other ancestries
eRows are not exclusive; patients could have a personal history of multiple cancers
Cancer prevalence in monoallelic and biallelic CHEK2 PV carriers
Only 9.7% (3/31) of biallelic CHEK2 PV carriers were reported with no history of cancer, compared to 50.0% (3234/6473) of monoallelic CHEK2 PV carriers (Table 1). Breast cancer was the most prevalent cancer reported in both monoallelic and biallelic carriers of CHEK2 PVs, with ovarian cancer being the second most prevalent single cancer in both. The frequency of at least one primary breast cancer at any age was significantly (p < 0.0001) higher in biallelic carriers (80.6%, 25/31) than in monoallelic carriers (41.2%, 2668/6473; Table 2). Biallelic CHEK2 PV carriers were significantly more likely to be diagnosed with breast cancer at or before age 50 (61.3%, 19/31), as well as to have a second breast cancer diagnosis (22.6%, 7/31), as compared to monoallelic carriers (23.9%, 1548/6473; p < 0.0001 and 8.1%, 523/6473; p = 0.0107, respectively). The increased prevalence of breast cancer, breast cancer at age 50 or younger, and second primary breast cancer was also observed when the comparisons were restricted to monoallelic and homozygous carriers of CHEK2 c.1100del (Table 2). No excess of any specific non-breast cancer was clearly identified in the biallelic CHEK2 PV carriers overall, though pancreatic cancer did appear to be more common among biallelic and CHEK2 c.1100del homozygous carriers (3.2% and 6.2%, respectively) as compared to monoallelic and CHEK2 c.1100del monoallelic carriers (0.2%; p = 0.0512 and 0.1%; p = 0.0277, respectively).
Table 2.
Breast cancer frequency in CHEK2 PV carriers
| Variable | Monoallelic | Biallelic | p-value | c.1100del Monoallelic | c.1100del Homozygous | p-value |
|---|---|---|---|---|---|---|
| Breast cancer (any age) | 2668 (41.2%) | 25 (80.6%) | < 0.0001 | 1403 (41.1%) | 13 (81.2%) | 0.0015 |
| Breast cancer (≤ 50 years) | 1548 (23.9%) | 19 (61.3%) | < 0.0001 | 799 (23.4%) | 9 (56.2%) | 0.0048 |
| Breast cancer (any age) + 2nd primary breast cancer | 523 (8.1%) | 7 (22.6%) | 0.0107 | 280 (8.2%) | 5 (31.2%) | 0.0078 |
| Breast cancer (any age) + any non-breast cancer | 324 (5.0%) | 3 (9.7%) | 0.2027 | 188 (5.5%) | 3 (18.8%) | 0.0558 |
The prevalence of multiple cancers among monoallelic and biallelic CHEK2 PV carriers was also compared. Biallelic CHEK2 PV carriers were significantly more likely to be affected with at least one primary cancer as compared to monoallelic PV carriers (90.3% vs. 50.0%, p < 0.0001; Fig. 1). Similarly, biallelic CHEK2 PV carriers were significantly more likely to have at least two primary cancers when compared to monoallelic PV carriers (32.3% vs. 13.5%, p = 0.0061). These differences were also significant for carriers of homozygous CHEK2 c.1100del PVs as compared to monoallelic CHEK2 c.1100del carriers. The converse comparison of unaffected monoallelic and biallelic CHEK2 PV carriers revealed that biallelic carriers were significantly less likely to be unaffected with cancer as compared to monoallelic carriers (p < 0.0001). Similarly, 50.4% of CHEK2 c.1100del monoallelic carriers were unaffected while only 12.5% of CHEK2 c.1100del homozygous carriers were unaffected by any cancer (p = 0.0022).
Fig. 1.
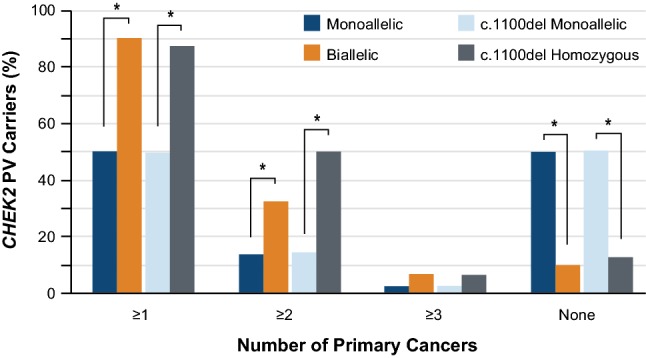
Distribution of women with one or two in trans PVs in CHEK2 according to number of primary cancers of any type. *p < 0.05
Age-related cancer penetrance in monoallelic and biallelic CHEK2 PV carriers
The median age of first cancer diagnosis was compared in monoallelic and biallelic CHEK2 PV carriers for any cancer, and for breast cancer specifically. A trend toward younger median age at cancer diagnosis was observed in biallelic CHEK2 PV carriers as compared to monoallelic carriers for all evaluated categories (≥ 1, ≥ 2, or ≥ 3 cancer diagnoses of any kind; 1 or ≥ 2 breast cancer diagnosis), though this trend was generally not statistically significant (Fig. 2). This trend held true when the analysis was restricted only to carriers of the CHEK2 c.1100del founder mutation. Notably, biallelic CHEK2 PV carriers with at least one primary cancer were significantly younger at the time of diagnosis compared to monoallelic carriers (44 and 47 years, respectively; p = 0.0268). Similarly, homozygous carriers of CHEK2 c.1100del with at least two cancers were significantly younger than monoallelic carriers in this category (47 and 36 years, respectively; p = 0.0476).
Fig. 2.
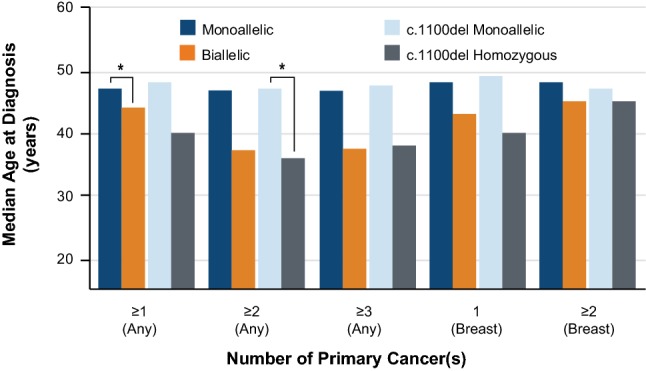
Median age at cancer diagnosis in women with one or two in trans CHEK2 PVs. *p < 0.05
Breast cancer risk estimates
Multivariate logistic regression analysis was used to quantify the risk of breast cancer in biallelic CHEK2 PV carriers and monoallelic carriers. As there were no cases of lobular invasive breast cancer among the 31 biallelic CHEK2 PV carriers, it was not possible to determine an overall odds ratio (OR) for developing any type of breast cancer (lobular invasive, ductal invasive, and DCIS). Therefore, ORs were determined separately for the breast cancer types for which there were sufficient numbers in both biallelic and monoallelic CHEK2 PV carriers: ductal invasive and DCIS (Fig. 3). Biallelic CHEK2 PV carriers had an OR of 8.69 (95% CI 3.69–20.47) for ductal invasive breast cancer compared to 2.02 for monoallelic CHEK2 PV carriers (95% CI 1.90–2.15). Similarly, biallelic CHEK2 PV carriers had a higher risk of developing DCIS than did monoallelic carriers, with biallelic carriers having an OR of 4.98 (95% CI 2.00–12.35) and monoallelic carriers having an OR of 1.82 (95% CI 1.66–2.00). While the CI for biallelic CHEK2 PV carriers are wide for both ductal invasive breast cancer and DCIS, the lower end of the CI range for both show at least a twofold higher risk of cancer compared to non-carriers.
Fig. 3.

OR of developing breast cancer according to subtype and CHEK2 PV carrier status (biallelic or monoallelic), shown with 95% CI
Discussion
This study compared the cancer phenotypes of women with biallelic PVs in CHEK2 to monoallelic carriers identified by hereditary cancer panel testing. Using a similar cohort of patients who underwent clinical hereditary cancer testing, our laboratory has previously shown that women with any PV in CHEK2 have approximately twofold greater risk of developing any breast cancer than PV-negative women [4]. This is consistent with our present findings, where the ORs for invasive breast cancer and DCIS were 2.02 and 1.82, respectively. In this cohort, biallelic CHEK2 PV carriers had a significantly higher risk for breast cancer, were more likely to be diagnosed at or before age 50, and were more likely to have multiple primary breast cancers compared to monoallelic CHEK2 PV carriers. Biallelic CHEK2 PV carriers also appeared to have a higher risk of cancer overall, although we did not observe a statistically significant excess of any individual cancer other than breast. The ORs calculated for women with biallelic PVs (8.69 for invasive breast cancer and 4.98 for DCIS) suggest that biallelic CHEK2 PV carriers could have breast cancer risks higher than those associated with PVs in BRCA1 and BRCA2. It may therefore be reasonable to consider biallelic findings in CHEK2 as high, rather than moderate, penetrance for the purposes of management.
Other breast cancer susceptibility genes have recessive phenotypes associated with biallelic PVs, including Fanconi Anemia for BRCA2 and PALB2, Ataxia Telangiectasia for ATM, and Nijmegen Breakage Syndrome for NBN [13, 20–22]. CHEK2 is distinguished by not having a defined recessive phenotype [23]. Ascertainment of cancer phenotypes relied on provider-completed TRFs, which focus on cancers for which patients meet guideline-based testing criteria. For this reason, it is possible that this study did not identify rare cancers or cancers that are not recognized for current HBOC or hereditary colorectal cancer syndrome testing criteria in North America. Therefore, this study does not rule out the possibility of other rare or common non-breast cancer phenotypes associated with biallelic inheritance of CHEK2 PVs. Additionally, clinical information unrelated to cancers and precancerous findings were not documented on the TRF, and this study could not identify an association between biallelic CHEK2 PVs and other phenotypes.
A possible limitation of the current study is that cancer histories were extracted from provider-completed TRFs and the reported diagnoses were not verified. Studies evaluating the accuracy of reporting of family cancer histories have shown that accuracy decreases with increasing distance in degree of relationship to the proband [24, 25]. However, self-reporting of common cancers by probands has been shown to be highly accurate [26] and the majority of the analyses presented here are based solely on the probands clinical data. Although the logistic regression model did incorporate family history information, the previously reported sensitivity analyses show that the ORs calculated using this methodology were not substantially impacted when assuming differential family history reporting [4]. Similarly, ascertainment of this sample was from a population of patients who had clinical hereditary cancer testing and this population is therefore likely to be higher risk than the general population. However, all comparisons were done within the same testing population, making the differences between groups valid regardless of baseline risk. The small sample size of the biallelic CHEK2 PV carriers also limits this study, as certain analyses would be underpowered and could therefore not be performed. While this does not weaken the conclusions drawn from the statistically-powered analyses presented, it limits the types of analyses that could be performed.
Current breast cancer screening guidelines for women who have inherited CHEK2 PVs include breast mammography with consideration of tomosynthesis and consideration of breast MRI starting by age 40 [12]. The findings from this study suggest that biallelic carriers of CHEK PVs have a considerably higher breast cancer risk than monoallelic carriers, and therefore may benefit from more aggressive management, possibly beginning at younger ages. The observed high risk for more than one primary breast cancer diagnosis in women with biallelic PVs may also have significant implications for treatment of an initial malignancy. The findings of this study add evidence to support germline testing in affected women for informing future cancer risk.
Funding
This study was supported my Myriad Genetics, Inc.
Compliance with ethical standards
Conflict of interest
All authors are employed by and have stock options in Myriad Genetics, Inc.
Ethical approval and informed consent
All analyses were performed using de-identified data obtained during the course of routine healthcare operations. All patients provided informed consent for clinical testing. De-identified clinical information was obtained from provider-completed test request forms (TRFs). Data were included only for women residing in states with no legal prohibitions on the use of de-identified data for research purposes. Only aggregate data are presented in the manuscript. Therefore, this analysis did not meet the U.S. Health and Human Services definition of research on human subjects (HHS 46.102) and did not require Institutional Review Board approval.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weischer M, Bojesen SE, Ellervik C, Tybjærg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008;26(4):542–548. doi: 10.1200/JCO.2007.12.5922. [DOI] [PubMed] [Google Scholar]
- 2.Weischer M, Nordestgaard BG, Pharoah P, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30(35):4308–4316. doi: 10.1200/JCO.2012.42.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Easton D. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9065 controls from 10 studies. Am J Hum Genet. 2004;74(6):1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurian AW, Hughes E, Handorf EA, et al. Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. JCO Precis Oncol. 2017 doi: 10.1200/po.16.00066. [DOI] [PubMed] [Google Scholar]
- 5.Xiang HP, Geng XP, Ge WW, Li H. Meta-analysis of CHEK2 1100delC variant and colorectal cancer susceptibility. Eur J Cancer. 2011;47(17):2546–2551. doi: 10.1016/j.ejca.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 6.Katona BW, Yang Y-X. Colorectal cancer risk associated with the CHEK2 1100delC variant. Eur J Cancer. 2017;83:103–105. doi: 10.1016/j.ejca.2017.05.045. [DOI] [PubMed] [Google Scholar]
- 7.NCCN clinical practice guidelines in oncology, genetic/familial high-risk assessment: colorectal (version 1. 2018). https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf. Accessed 1 Aug 2018
- 8.Aldubayan SH, Pyle LC, Gamulin M, et al. Association of inherited pathogenic variants in checkpoint kinase 2 (CHEK2) with susceptibility to testicular germ cell tumors. JAMA Oncol. 2019;5(4):514–522. doi: 10.1001/jamaoncol.2018.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zlowocka-Perlowska E, Narod SA, Cybulski C. CHEK2 alleles predispose to renal cancer in Poland. JAMA Oncol. 2019;5(4):576–576. doi: 10.1001/jamaoncol.2019.0022. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Yu H, Zheng SL, et al. A comprehensive evaluation of CHEK2 germline mutations in men with prostate cancer. Prostate. 2018;78(8):607–615. doi: 10.1002/pros.23505. [DOI] [PubMed] [Google Scholar]
- 11.Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44(1):1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NCCN clinical practice guidelines in oncology, genetic/familial high-risk assessment: breast and ovarian (version 3. 2019). https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf. Accessed 20 Feb 2019
- 13.Salewsky B, Wessendorf P, Hirsch D, Krenzlin H, Digweed M. Nijmegen breakage syndrome: the clearance pathway for mutant nibrin protein is allele specific. Gene. 2013;519(2):217–221. doi: 10.1016/j.gene.2013.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Meijers-Heijboer H, Van Den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2*1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 15.Adank MA, Jonker MA, Kluijt I, et al. CHEK2*1100delC homozygosity is associated with a high breast cancer risk in women. J Med Genet. 2011;48(12):860. doi: 10.1136/jmedgenet-2011-100380. [DOI] [PubMed] [Google Scholar]
- 16.Huijts PEA, Hollestelle A, Balliu B, et al. CHEK2*1100delC homozygosity in the Netherlands—prevalence and risk of breast and lung cancer. Eur J Hum Genet. 2013;22:46. doi: 10.1038/ejhg.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eggington JM, Bowles KR, Moyes K, et al. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet. 2014;86(3):229–237. doi: 10.1111/cge.12315. [DOI] [PubMed] [Google Scholar]
- 19.Mundt E, Nix P, Brown K, Bowles KR, Manley S. Complexities of Variant Classification in Clinical Hereditary Cancer Genetic Testing. J Clin Oncol. 2017;35(34):3796–3799. doi: 10.1200/JCO.2017.74.5182. [DOI] [PubMed] [Google Scholar]
- 20.Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297(5581):606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 21.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 22.Savitsky K, Bar-Shira A, Gilad S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268(5218):1749. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 23.Rahman N. Scott RH (2007) Cancer genes associated with phenotypes in monoallelic and biallelic mutation carriers: new lessons from old players. Hum Mol Genet. 2007;16(R1):R60–R66. doi: 10.1093/hmg/ddm026. [DOI] [PubMed] [Google Scholar]
- 24.Love RR, Evans AM, Josten DM. The accuracy of patient reports of a family history of cancer. J Chronic Dis. 1985;38(4):289–293. doi: 10.1016/0021-9681(85)90074-8. [DOI] [PubMed] [Google Scholar]
- 25.Murff HJ, Spigel DR, Syngal S. Does this patient have a family history of cancer? An evidence-based analysis of the accuracy of family cancer history. JAMA. 2004;292(12):1480–1489. doi: 10.1001/jama.292.12.1480. [DOI] [PubMed] [Google Scholar]
- 26.Sijmons RH, Boonstra AE, Reefhuis J, et al. Accuracy of family history of cancer: clinical genetic implications. Eur J Hum Genet. 2000;8(3):181–186. doi: 10.1038/sj.ejhg.5200441. [DOI] [PubMed] [Google Scholar]


