Abstract
Malachite green is a common environmental pollutant that poses a great threat to non-target organisms, including humans. This study reports the characterization of a bacterial strain, Pseudomonas veronii JW3-6, which was isolated from a malachite green enrichment culture. This strain degraded malachite green efficiently in a wide range of temperature and pH levels. Under optimal degradation conditions (32.4 °C, pH 7.1, and inoculum amount of 2.5 × 107 cfu/mL), P. veronii JW3-6 could degrade 93.5% of 50 mg/L malachite green within seven days. Five intermediate products from the degradation of malachite green were identified: leucomalachite green, 4-(dimethylamino) benzophenone, 4-dimethylaminophenol, benzaldehyde, and hydroquinone. We propose a possible degradation pathway based on these findings. The present study is the first to report the degradation of malachite green by P. veronii and the identification of hydroquinone as a metabolite in the degradation pathway.
Subject terms: Environmental microbiology, Microbiology techniques, Biotechnology, Microbiology
Introduction
Malachite green is a common triphenylmethane dye that is widely used in the textile and dyeing industries. This compound has also long been applied as a fishery medicine in China’s aquaculture industry because of its ability to control certain diseases, such as saprolegniasis1. However, malachite green residues in aquaculture products and the environment may threaten human health, and the potential teratogenic, carcinogenic, and mutagenic effects of malachite green have been reported since the 1970s2,3. Based on its hazardous properties, the Ministry of Agriculture and Rural Affairs of China banned the use of malachite green in aquaculture in 2002. Nevertheless, malachite green is still used illegally because of its low price and good sterilizing effect4. In addition, thousands of tons of wastewater from triphenylmethane dye production are discharged into rivers and lakes and the residues persist in soils. Triphenylmethane dye residues enter the food chain, thus considerably threatening human health5. Therefore, developing an efficient method for malachite green degradation is crucial.
Although triphenylmethane dyes are stable and difficult to degrade, some environmental microorganisms have evolved the ability to decolorize or degrade these dyes. These microorganisms have become the most effective “weapon” against triphenylmethane dye pollution in the environment6,7. Several strains of bacteria and fungi that can decolorize or degrade triphenylmethane dyes have been isolated from soils, lakes, and liquid waste. In previous studies, quite a few bacterial degradation strains such as Aeromonas hydrophila, Tenacibaculum sp. HMG1, Pseudomonas sp. YB2 have been reported8–14 (Table 1). Recently, some novel malachite green biodegradation strains have been isolated, including Pseudomonas sp. DY1 and Exiguobacterium sp. MG2, and the degradation metabolites and pathways have been studied4,12. Fungi that can degrade malachite green have also been isolated; these include Cunninghamella elegans, Phanerochaete chrysosporium, and Irpex lacteus15–18. These microbial species serve as resources for the biodecolorization of triphenylmethane dyes and the bioremediation of environmental pollution. Nevertheless, further studies are needed, as previous studies mainly isolated decolorizing bacteria through traditional cultivation methods. Thus, new and efficient methods should be explored. Moreover, only the upstream decolorization pathway of triphenylmethane dyes, such as malachite green has been reported. The complete pathway for microbial degradation and triphenylmethane dye metabolism remains unknown19.
Table 1.
Reported microorganisms for degradation of triphenylmethane dyes.
| Name of strain | kingdom of strain | Triphenylmethane dye type | Reference |
|---|---|---|---|
| Aeromonas hydrophila | Bacteria | Malachite green, crystal violet | 8 |
| Arthrobacter sp. M6 | Bacteria | Malachite green | 9 |
| Bacillus subtilis sp. | Bacteria | Malachite green | 10 |
| Citrobacter sp. | Bacteria | Malachite green, crystal violet, brilliant green | 11 |
| Exiguobacterium sp. MG2 | Bacteria | Malachite green, crystal violet, brilliant green | 12 |
| Pseudomonas sp. YB2 | Bacteria | Malachite green | 13 |
| Pseudomonas sp. strain DY1 | Bacteria | Malachite green | 4 |
| Tenacibaculum sp. HMG1 | Bacteria | Malachite green | 14 |
| Cunninghamella elegans | Fungi | Malachite green | 15 |
| Irpex lacteus F17 | Fungi | Malachite green, crystal violet | 16 |
| Phanerochaete chrysosporium | Fungi | Malachite green | 17 |
| Pleurotus ostreatus | Fungi | brilliant blue R | 18 |
The objectives of the present study were: (1) to isolate a promising bacterial strain for the treatment of malachite green-contaminated environments; (2) to determine the kinetic parameters for the biodegradation of malachite green and other triphenylmethane dyes; and (3) to detect the metabolites and deduce the possible downstream degradation pathway of this strain, and examine the mechanism underlying malachite green degradation by P. veronii JW3-6.
Results
Diversity and community changes in enrichment cultures
High-throughput sequencing yielded 63,153, 80,192, 79,079, and 78,197 16 S rRNA gene sequences of the distinct V4–V5 regions of the four sequential enrichment cultures JWY, JW1, JW2, and JW3, respectively. After statistical analysis and annotation, 63,153 sequences in the initial JWY culture were clustered into 1,568 OTUs. The highest abundance of OTUs belonged to Flavobacterium, Pseudomonas, Arenimonas, Methylotenera, and Dechloromonas (Fig. 1). The diversity of bacteria in the first transfer culture JW1 was significantly lower, as 80,192 sequences were clustered into 967 OTUs. The main genera included Pseudomonas, Methylotenera, Methylobacillus, Acinetobacter, and Labrys. Pseudomonas replaced Flavobacterium as the genus with the highest abundance. The bacterial diversity after the second passage in culture JW2 also decreased, as 63,153 sequences were clustered into 576 OTUs. The main genera included Pseudomonas, Labrys, Acinetobacter, Achromobacter, and Cupriavidus. Finally, 78,197 sequences were clustered into 396 OTUs in culture JW3 after the third passage. The main microbiota belonged to Pseudomonas, Labrys, Acinetobacter, Cupriavidus, and Achromobacter. The above results suggest that some bacteria could not use the malachite green to grow and thus died out or were outcompeted with sequential passages of the enrichment culture. As a result, the population diversity in the microbial enrichment gradually declined with the increase of passage generations. This decline was accompanied by pronounced changes in the community composition. The dominant genus changed from Flavobacterium in the initial enrichment culture, to Pseudomonas after the first passage, and the proportion of Pseudomonas reached 67.8% after the third passage (Fig. 1). This result provides strong evidence that Pseudomonas were involved in malachite green degradation. In contrast, the rapid reduction in the proportion of Flavobacterium to less than 0.1% after one passage seems to indicate that Flavobacterium did not contribute to malachite green degradation. The increased proportion of Labrys in the total OTUs from 0.16% to 14.6% suggests that Labrys was also a major bacterial genus that participated in malachite green degradation. In addition, the relative proportions of Acinetobacter, Cupriavidus, and Achromobacter increased after the second passage, suggesting that these bacteria may also contribute to malachite green degradation.
Figure 1.

Diversity and community changes in enrichment cultures JWY, JW1, JW2 and JW3 based on the relative abundance of the Illumina sequences. The histogram was constructed by using R software.
Isolation and identification of malachite green degraders
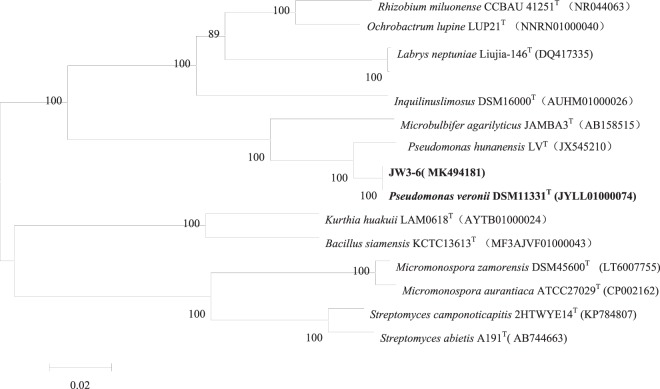
Thirteen bacterial strains able to degrade malachite green were isolated from enrichment JW3(Table 2). JW3-6 was the best malachite green degrader among the isolated strains. The degradation efficiency of JW3-6 reached 92.2%, and was significantly higher than those of other strains. JW3-6 colonies on TSA plates were milk white in color and exhibited smooth surfaces and clear edges. JW3-6 cells were Gram-negative, aerobic, nonmotile, and featured short rods that were 0.5–0.8 mm in length and 0.1 mm in width. Phylogenetic analysis indicated that strain JW3-6 belonged to the genus Pseudomonas and clustered strongly with Pseudomonas veronii DSM11331T (99.69%, accession number JYLL01000074). On the basis of the results of morphological and 16 S rRNA sequence evaluation, JW3-6 was identified as a member of genus Pseudomonas and named as Pseudomonas veronii JW3-6 (Fig. 2). Pseudomonas veronii JW3-6 has been deposited in the China General Microbiological Culture Collection Center (CGMCC) with the accession number 17523.
Table 2.
The taxonomic status, 16 S rRNA gene accession numbers, degradation efficiency and the growth media of the malachite green degrading strains isolated from JW3.
| Name of strain | Species | Accession number of 16S rRNA gene | Degradation efficiency | Grwothmedium |
|---|---|---|---|---|
| JW3-6 | Pseudomonas veronii | MK494181 | 91.2% | MSM |
| JW3-5 | Labrys neptuniae | MN945372 | 59.9% | MSM |
| JW3-2 | Pseudomonas plecoglossicida | MN946618 | 49.9% | MSM |
| JW3-9 | Pseudomonas umsongensis | MN946621 | 28.8% | MSM |
| JW3-12 | Tenacibaculum mesophilum | MN946619 | 23.5% | TSA |
| JW3-15 | Pseudomonas hunanensis | MN946620 | 48.6% | TSA |
| JW3-18 | Rhizobium miluonense | MN946624 | 35.7% | TSA |
| JW3-21 | Pseudomonas granadensis | MN946622 | 39.2% | TSB |
| JW3-22 | Kurthia gibsonii | MN946626 | 29.9% | TSB |
| JW3-34 | Inquilinus limosus | MN946628 | 29.2% | LB |
| JW3-35 | Citrobacter sedlakii | MN946625 | 35.5% | LB |
| JW3-39 | Pseudomonas lactis | MN946627 | 25.1% | LB |
Figure 2.
Neighbour-joining phylogenetic tree based on a comparison of the 16 S rRNA gene sequences of P. veronii JW3-6 and its closest relatives and several out-group strains. The numbers at the nodes indicate the percentages of bootstrap sampling derived from 1000 replications. GenBank accession numbers are given in parentheses. Bar, 0.02 nucleotide substitution per nucleotide position.
Optimization of the degradation conditions for JW3-6
The influences of multiple environmental factors on malachite green degradation by P. veronii JW3-6 were analyzed. This bacterial strain exhibited good malachite green degradation efficiency over a wide temperature range from 20 °C–40 °C (Fig. 3a). The degradation efficiency of P. veronii JW3-6 peaked (92.9%) at 30 °C and decreased to 70.1% at 40 °C. The strain can also degrade malachite green over a pH range of 4–9. The degradation efficiency of P. veronii JW3-6 over the pH range of 5–7 exceeded 80% and peaked (93.1%) at pH 7 (Fig. 3b). The degradation efficiency of strain under alkaline conditions was low. Degradation decreased to 55.8% when the pH was increased to 8 and further decreased to 43.1% at pH 9. This behavior indicates that P. veronii JW-6 exhibits good degradation under neutral or acid conditions. On the other hand, the influence of initial inoculum size on malachite green degradation by P. veronii JW3-6 was also studied. The degradation efficiency of P. veronii JW3-6 exceeded 80% when the initial inoculum was 1–5 × 107 cfu/mL and peaked (92.8%) at an initial inoculum of 3 × 107 cfu/mL (Fig. 3c). The initial concentration of malachite green also influenced the degradation efficiency of JW3-6. The degradation efficiency of P. veronii JW3-6 with 20 mg/L malachite green reached 95.1% and decreased to 81.7% when the malachite green concentration reached 100 mg/L (Fig. 3d). When the initial concentration of malachite green exceeded 200 mg/L, the degradation efficiency of P. veronii JW3-6 decreased to less than 70%. These results indicate that high concentrations of malachite green inhibit the growth of JW3-6.
Figure 3.
Degradation kinetics of malachite green under different conditions (a) temperature(°C), (b) pH level, (c) inoculum amount(×107 cfu/mL), and (d) initial concentration(mg/L) of malachite green. The symbols represent averages of triplicate experiments and the error bars indicate their corresponding standard deviations.
Degradation conditions were further designed through the response surface method in accordance with the results of single-factor experiments, and 17 degradation tests were conducted (Table 3). The following second-degree polynomial equation was obtained through the statistical analysis of data to explain malachite green biodegradation by P. veronii JW3-6:
| 1 |
where R1 denotes the predicted malachite green degradation efficiency, and A, B, and C are the coded values for temperature, pH level, and inoculum amount, respectively. Analysis of variance (ANOVA) for the fitted quadratic polynomial model is shown in Table 4. The model was significant (P < 0.05) with R2 = 0.9618 and Adj R2 = 0.9411. The results of regression analysis showed that temperature and pH level were significant terms (P < 0.05), whereas the inoculum amount was a nonsignificant term (P > 0.05). The response surface was used to illustrate the effects of temperature, pH level and inoculum amount on malachite green biodegradation. At the theoretical maximum point, the optimal conditions for malachite green degradation by JW3-6 were 32.4 °C and pH 7.1 and inoculum amount 2.5 × 107 cfu/mL (Fig. 4).
Table 3.
Box-Behnken experimental design with three independent variables.
| Run | X1 | X2 | X3 | Responses degradation efficiency(%) |
|---|---|---|---|---|
| 1 | 35.0 | 6.5 | 1.0 | 79.8 |
| 2 | 30.0 | 8.0 | 5.0 | 80.4 |
| 3 | 30.0 | 6.5 | 3.0 | 93.6 |
| 4 | 35.0 | 8.0 | 3.0 | 75.8 |
| 5 | 35.0 | 6.5 | 5.0 | 79.7 |
| 6 | 25.0 | 6.5 | 1.0 | 73.4 |
| 7 | 30.0 | 8.0 | 1.0 | 86.5 |
| 8 | 25.0 | 8.0 | 3.0 | 78.9 |
| 9 | 25.0 | 5.0 | 3.0 | 79.8 |
| 10 | 30.0 | 6.5 | 3.0 | 93.3 |
| 11 | 25.0 | 6.5 | 5.0 | 81.3 |
| 12 | 30.0 | 6.5 | 3.0 | 93.1 |
| 13 | 30.0 | 5.0 | 1.0 | 86.5 |
| 14 | 30.0 | 6.5 | 3.0 | 92.8 |
| 15 | 30.0 | 5.0 | 5.0 | 86.5 |
| 16 | 35.0 | 5.0 | 3.0 | 82.6 |
| 17 | 30.0 | 6.5 | 3.0 | 93.4 |
X1: temperature (°C), X2: pH level, X3: inoculum amount (×107 cfu/mL).
Table 4.
Analysis of variance (ANOVA) for the fitted quadratic polynomial model.
| Source | Sum of squares | DF | Mean Square | F Value | P Value |
|---|---|---|---|---|---|
| X1 | 2.53 | 1 | 2.53 | 0.63 | 0.4532 |
| X2 | 23.80 | 1 | 23.80 | 5.93 | 0.0451 |
| X3 | 0.36 | 1 | 0.36 | 0.090 | 0.7729 |
| X1X2 | 8.70 | 1 | 8.70 | 2.17 | 0.1844 |
| X1X3 | 16.00 | 1 | 16.00 | 3.99 | 0.0860 |
| X2X3 | 9.30 | 1 | 9.30 | 2.32 | 0.1717 |
| X1X1 | 437.63 | 1 | 437.63 | 109.04 | 0.0001 |
| X2X2 | 59.84 | 1 | 59.84 | 14.91 | 0.0062 |
| X3X3 | 85.07 | 1 | 85.07 | 21.20 | 0.0025 |
| Model | 694.31 | 9 | 77.15 | 19.22 | 0.0004 |
| Error | 0.37 | 4 | 0.093 | ||
| Total | 722.40 | 16 |
P Value < 0.05 indicates the model terms are significant.
Figure 4.
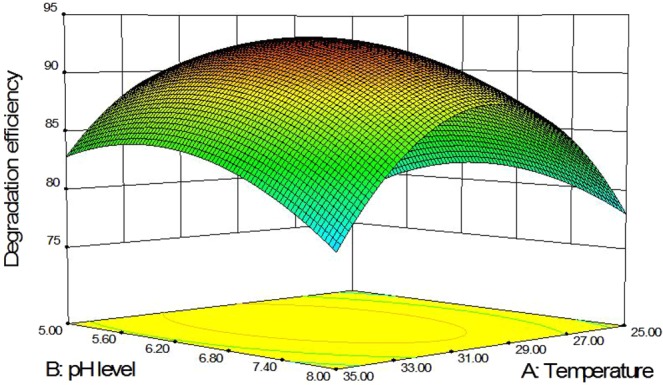
Response surface curves demonstrating the effects of medium temperature (°C) and pH level on malachite green biodegradation efficiency (%) with an inoculum amount of P. veronii JW3-6 at 2.5 × 107cfu/mL.
Degradation of various triphenylmethane dyes
The degradation of malachite green and triphenylmethane dyes by P. veronii JW3-6 were studied under the optimal conditions. The results showed that P. veronii JW3-6 degraded most of the malachite green within the first 4 days, and its degradation efficiency exceeded 80% on day 4 (Fig. 5). Meanwhile, the cell density of strain JW3-6 increased to approximately 8.92 × 108 cfu/mL during this period of malachite green degradation. The degradation efficiency decreased after 4 days, and was accompanied by no further increase in JW3-6 cell density. Under the optimal conditions, the degradation efficiency of P. veronii JW3-6 reached 93.5% on day 7 when the initial concentration of malachite green was 50 mg/L. P. veronii JW3-6 was also capable of degrading other triphenylmethane dyes, most likely because of their similar chemical structures. The degradation efficiencies of strain JW3-6 for ethyl violet, crystal violet, fuchsin basic, brilliant green, and victoria blue B were 91.5%, 86.4%, 75.8%, 62.2%, and 57.2% (Fig. 6). These results show that P. veronii JW3-6 has a broad specificity for the degradation of triphenylmethane dyes and has considerable potential for processing triphenylmethane dye pollution in the environment.
Figure 5.
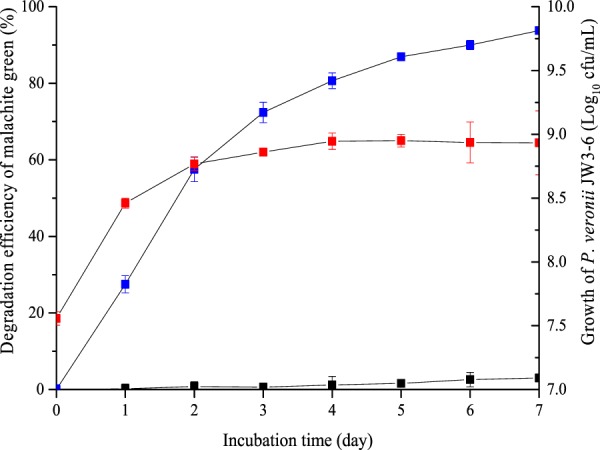
Growth of P. veronii JW3-6 along time and degradation efficiency of malachite green. Symbol: blue square, malachite green degradation by P. veronii JW3-6; black square, malachite green degradation by killed P. veronii JW3-6 cells as negative control; red square, the cell growth of P. veronii JW3-6. Data represent mean values of three replicates with standard deviation.
Figure 6.
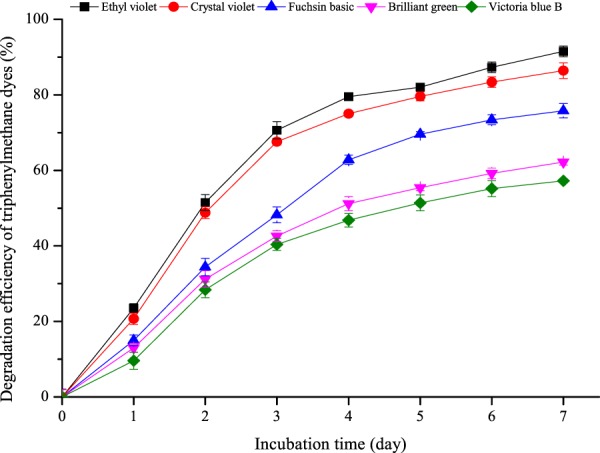
Degradation kinetics of various triphenylmethane dyes by P. veronii JW3-6. The symbols represent averages of triplicate experiments and the error bars indicate their corresponding standard deviations.
Metabolic pathways of malachite green degradation by P. veronii JW3-6
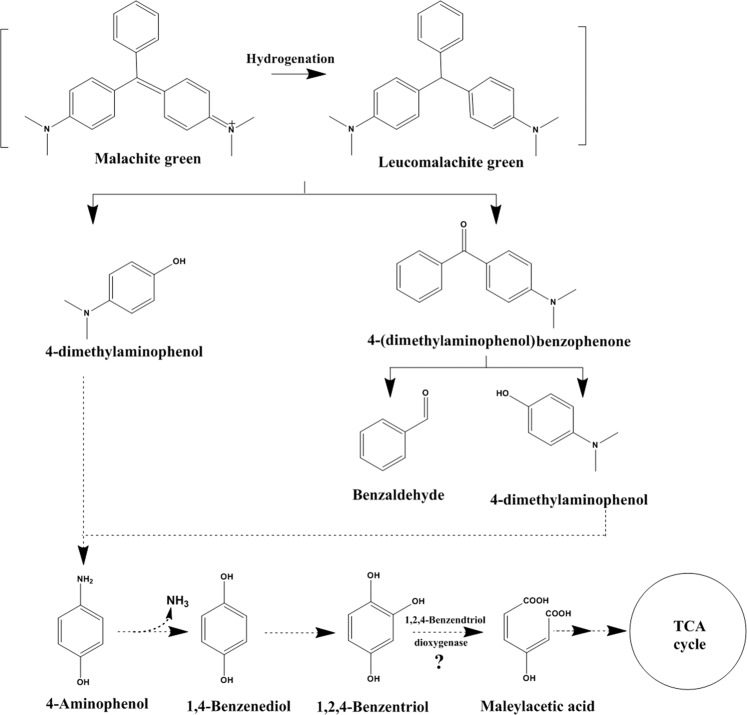
LC–MS detected five intermediates of malachite green degradation by P. veronii JW3-6. The candidate products were identified in accordance with their m/z values. These products were leucomalachite green (330 m/z), 4-(dimethylamino) benzophenone, 4-dimethylaminophenol, benzaldehyde, and hydroquinone. A possible metabolic pathway for malachite green biodegradation by P. veronii JW3-6 was proposed in accordance with the chemical properties of malachite green and the metabolites (Fig. 7). In this proposed pathway, malachite green is first transformed into leucomalachite green through the hydrogenation of the central carbon. Then, leucomalachite green is degraded via dibenzocyclization into two metabolites: 4-(dimethylamino) benzophenone and 4-dimethylamino aminophenol. These metabolites were further degraded by JW3-6; 4-(dimethylamino) benzophenone was decomposed into 4-diaminophenol and benzaldehyde through C–C bond cleavage, and 4-diaminophenol might be further degraded into hydroquinone by the action of esterases and deamination. Although the ring-opening products of hydroquinone were not detected in the present study, hydroquinone could be further degraded by phenyldiphenyl-degrading microorganisms until CO2 and water are produced.
Figure 7.
Proposed pathway for the degradation of malachite green by P. veronii JW3-6.
Discussion
P. veronii JW3-6 was isolated from an enrichment culture by means of culture dependent techniques and also retrieved by high-throughput sequencing methods. The strain is highly effective in degrading malachite green and other triphenylmethane dyes. P. veronii is well known for its ability as a biological control agent; it exhibits antagonistic activity against nematodes of agronomic importance20. However, the use of P. veronii in bioremediation of environmental pollutants has received inadequate attention. Several reported species of Pseudomonas can degrade xenobiotic compounds, such as sulfonylurea herbicides and pyrethroid pesticides21,22. Thus far, however, the biodegradation of malachite green and other triphenylmethane dyes by P. veronii strains has not been reported. This study is the first to report the isolation of a P. veronii strain that efficiently degrades a wide range of triphenylmethane dyes.
In the present study, P. veronii JW3-6 utilized malachite green as the sole carbon source, and it could degrade 93.5% of 50 mg/L malachite green within seven days. Although several malachite green-degrading bacterial strains have been isolated, most of them degrade malachite green in a nutrient-rich medium or require an additional carbon or nitrogen source in the medium. For example, Qu et al.13 reported that Tenacibaculum spp. HMG1 can decolorize 98.8% of 20 mg/L malachite green in 12 h in LB medium. Tao et al.14 reported that Pseudomonas spp. YB2 can almost completely decolorize 1000 mg/L malachite green within 12 h in LB medium. However, achieving similar degradation efficiencies in the natural environment presents difficulties. Another important property of P. veronii JW3-6 is that it can degrade malachite green and other triphenylmethane dyes over a wide range of temperatures (20 °C–40 °C) and pH levels (4–9). These results demonstrate the potential of this microorganism for use in biodegradation and open up new opportunities for its future applications.
Several bacteria and fungi have been isolated from various environmental samples, including soils, lakes, and liquid waste, and most of these studies focused on malachite green decolorization. However, some of the decolorization products of malachite green, such as leucomalachite green, are more toxic than the dye itself. Therefore, studying the metabolites and complete catabolic pathways of microorganisms that degrade malachite green is important. According to existing studies, different bacteria might have similar pathways of triphenylmethane dye decolorization. Ioth et al. identified that Michler’s ketone and diaminophenol are the main metabolites of crystal violet degradation by Bacillus subtilis IF0 13719 and N. coralline10. Wang et al. detected six metabolites from malachite green degradation by Exiguobacterium sp. MG2 and proposed a possible metabolic pathway12. In this pathway, malachite green produces leucomalachite through hydrogenation. Then, leucomalachite is cleaved into (4-dimethylamino-phenyl)-phenyl-methano through the de-benzene ring reaction. Subsequently, (4-dimethylamino-phenyl)-phenyl-methano is transformed into 3-dimethylamino-phenol and benzaldehyde through further cleavage of C–C bonds. The upstream metabolites and metabolic pathways identified in the present study are similar to those identified in previous works. Although ring-opening products were absent, 4-dimethylaminobenzene and hydroquinone were detected as metabolites. Hydroquinone could be transformed into 1,2,4-benzentriol through catalysis by phenol hydroxylase, which has been purified from certain bacteria, such as Pseudomonas stutzeri OX123,24. In addition, 1,2,4-benzentriol can be converted into maleylacetate through catalysis by 1,2 dioxygenase. Then, the further degradation of maleylacetate produces CO2 and water. A new possible degradation pathway is proposed for the microbial degradation of malachite green.
In previous studies, malachite green degrading bacteria were mainly isolated through traditional methods, and the direct enrichment and isolation of degradative microorganisms were implemented. In this study, samples were first subjected to high-throughput sequencing to identify the primary members of the microbial community. Microbial changes during the enrichment of different generations of malachite green-degrading bacteria were analyzed to identify possible strains involved in malachite green degradation. The increased proportions of Pseudomonas and Labrys in from 9.39% to 67.8% and from 0.16% to 14.6%, respectively, after continuous enrichment passage indicate that these strains are likely to participate in malachite green degradation. Subsequently, 13 strains, including P. veronii and Labrys neptuniae, with malachite green degrading ability were isolated on different media, such as MSM, LB, TSA and TSB. Among them, P. veronii JW3-6 was found to be capable of utilizing malachite green and other triphenylmethane dyes as the sole carbon source for growth over a wide range of temperature and pH levels. On the other hand, microbial composition could be characterized through high-throughput sequencing. Moreover, the bacterial genera involved in malachite green degradation can be predicted from these data and then specific strains can try to be isolated and screened for the desired degradation ability. Overall, this work also provides a combined strategy for the isolation of microorganisms in the degradation of other environmental pollutants.
Methods
Chemicals and media
Malachite green (analytical grade, >98.5%) was purchased from Aladdin Industrial Shanghai Co., Ltd., China. Analytical-grade acetonitrile was purchased from Sigma–Aldrich, USA. All other reagents used were of analytical grade. The microbial community was enriched using modified mineral salt medium (MSM) modified with glucose (1.0 g/L) and supplemented with 20 µL trace element solution25. LB (Difco), tryptic soy agar medium (TSA, Difco), and tryptic soy broth (TSB, Difco) were used to isolate degradative strains.
Microbial consortium construction and high-throughput sequencing
Activated sludge was collected from a purification tank for liquid waste in a dyestuff factory in Hangzhou City, Zhejiang Province, China. Samples were collected in sterile bottles and then transported back to the laboratory for subsequent tests. In the laboratory, 5 g of activated sludge was added to a 250 mL triangular flask with 100 mL MSM supplemented with 100 mg/L malachite green as the sole carbon source. The culture was then incubated on a rotary shaker (150 rpm) at 30 °C in the dark. After incubation for 7 days, portions of the culture (10%, v/v) were transferred to fresh MSM supplemented with 200 mg/L malachite green and incubated for another 7 days. A third passage was carried out in MSM containing 300 mg/L malachite green26. The sludge sample was designated as JWY, and the three generations of enrichment cultures were designated as JW1, JW2, and JW3. Total DNA from each of the samples was extracted by using a total soil DNA kit (Omega, USA). The 16 S rRNA genes of distinct regions V4–V5 were amplified with the following primers: 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′)27. After the purification of polymerase chain reaction (PCR) products, a library was constructed by utilizing the DNA Library Prep Kit for Illumina (NEB, USA). The amplicon library was submitted to the Maggi Technologies Company for PE2500 sequencing. Sequence analyses were performed by using Arch software (V10, http://www.drive5.com/usearch/)28. Sequences with ≥97% identity were assigned to the same operational taxonomic unit (OTU), and each OTU was considered to represent a species. R software(V2.15.3, https://www.R-project.org) was used for statistical analysis and histogram construction based on the relative abundance of OTUs in the samples29. The raw data of JWY, 1–3 is deposited at NCBI SRA database under the project PRJNA601309.
Isolation and identification of malachite green-degrading bacteria
The JW3 enrichment culture was serially diluted and spread on LB, TSA, TSB, and MSM solid plates supplemented with 50 mg/L malachite green12. Bacterial colonies with different morphologies were selected and purified by repeated streaking. The malachite green-degrading ability of the isolates was detected by using a high-performance liquid chromatography (LC) system (1290, Agilent, USA) equipped with an eclipse Plus C18 column (4.6 mm × 150 mm, 5 µm). First, 10 mL samples were collected and an equal volume of methanol was added to each. The mixture was processed for 1 min. Then, the tube was shaken at 220 rpm for 30 min in the dark prior to centrifugation for 5 min at 5,000 rpm. The upper organic phase was collected and filtered through a 0.22 µm membrane for HPLC analysis. The elution mobile phase was comprised of a mixture of acetonitrile and distilled water (85/15, v/v) running at a flow rate of 0.6 mL/min. The injection volume was 5 µL. The photodiode array detector was operated at a wavelength of 600 nm, and the column temperature was 30 °C30. The analytical curve employed for malachite green quantification is shown in Figure S1. The strains with the malachite green degradation capacity were identified on the basis of their 16 S rRNA sequences. The 16 S rRNA gene fragments was amplified as described by Ruan et al., and purified PCR products (approximately 1.5 kb) were sequenced by Maggi Technologies Company31. The obtained sequences were deposited in GenBank, the accession numbers are shown in Table 2. For further identifying the strain with the highest degradation capacity, multiple sequence alignment was conducted by using Clustal X software and phylogenetic relationships were analyzed via the neighbor-joining method with MEGA 6 software32.
Inoculum preparation
Strain activation was conducted before each experiment as follows. Strain JW3-6 was inoculated into MSM, incubated at 30 °C on a rotary shaker at 150 rpm in the dark. Bacterial cells in the late exponential growth phase were harvested (6 min, 8,000 rpm) at 4 °C. The supernatant was removed, and the bacterial cells were washed three times with 0.9% NaCl for subsequent studies.
Optimization of the degradation conditions for JW3-6
The effects of temperature (20 °C, 25 °C, 30 °C, 35 °C and 40 °C), pH (4, 5, 6, 7, 8, and 9), inoculum size (1 × 107, 2 × 107, 3 × 107, 5 × 107, 8 × 107, and 10 × 107 cfu/mL), and initial concentration (20 mg/L, 50 mg/L, 100 mg/L, 200 mg/L, and 300 mg/L) on the degradation of malachite green by JW3-6 were investigated through a single-environmental-factor experiment. The conditions during this experiment were the same as those during activation except that the factors were selected as independent variables. To account for evaporation loss, sterile distilled water was supplemented daily according to the loss of weight.
Response surface methodology was used to further optimize the selected parameters and their interactions on the basis of Box–Behnken design33. The second-order polynomial equation is expressed as follows:
| 2 |
where Yi refers to the predicted response, Xi and Xj are variables, bo is a constant, bi denotes the linear coefficient, bii represents the quadratic coefficient, and bij corresponds to the interaction coefficient.
Biodegradation of various triphenylmethane dyes by P. veronii JW3-6
The degradation of 50 mg/L malachite green by strain JW3-6 was conducted under optimal conditions in MSM. A control was set up as described above but was inoculated with killed cells of P. veronii JW3-6. Each treatment was performed in triplicate. The residual malachite green concentration and cell growth were measured every 24 h. The ability of strain JW3-6 to degrade other structurally similar triphenylmethane dyes, including ethyl violet, crystal violet, fuchsin basic, brilliant green, and victoria blue B, were determined under optimal conditions. Analytical methods were the same as previously described34.
Determination of malachite green biodegradation intermediates through LC–mass spectrometry (MS)
Metabolites and intermediates generated during malachite green degradation by strain JW3-6 in MSM cultures were analyzed. The extraction methods were the same as described above. Malachite green was detected by using a Xevo triple-quadrupole mass spectrometer (Waters Corp, Milford, MA, USA) equipped with an electrospray ionization source in the mass range (m/z) of 100–400. The mobile phase consisted of water and acetonitrile. The initial acetonitrile proportion was 10% and was then ramped gradually to 35% for 15 min, then to 60% for 20 min, finally to 95% for 15 min, and then held at 95% for 8 min. The injection volume was 5 µL35.
Supplementary information
Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC, No.31700006, 31670006), Central Public-interest Scientific Institution Basal Research Fund, (CAFS, No. 2017C004).
Author contributions
Y.M., Z.R. and R.P. conceived and designed research. J.S., Y.W., X.J., D.Z. and M.L. conducted experiments. G.H., Z.Y. and Q. M. analyzed and discussed the data. J.S. wrote the manuscript. All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jinlong Song and Gang Han.
Contributor Information
Zhiyong Ruan, Email: ruanzhiyong@caas.cn.
Yingchun Mu, Email: muyc@cafs.ac.cn.
Supplementary information
is available for this paper at 10.1038/s41598-020-61442-z.
References
- 1.Food and Agriculture Organization of the United Nations & World Health Organization. Evaluation of certain veterinary drug residues in food. World Health Organ Tech Rep Ser, 1–134 (2009). [PubMed]
- 2.Allen JL, Hunn JB. Fate and distribution studies of some drugs used in aquaculture. Vet. Hum. Toxicol. 1986;28(Suppl 1):21–24. [PubMed] [Google Scholar]
- 3.Jadhav JP, Govindwar SP. Biotransformation of malachite green by Saccharomyces cerevisiae MTCC 463. Yeast. 2006;23:315–23. doi: 10.1002/yea.1356. [DOI] [PubMed] [Google Scholar]
- 4.Du LN, et al. Biodegradation of malachite green by Pseudomonas sp. strain DY1 under aerobic condition: characteristics, degradation products, enzyme analysis and phytotoxicity. Ecotoxicol. 2011;20:438–446. doi: 10.1007/s10646-011-0595-3. [DOI] [PubMed] [Google Scholar]
- 5.Panandiker A, Fernandes C, Rao TK, Rao KV. Morphological transformation of Syrian hamster embryo cells in primary culture by malachite green correlates well with the evidence for formation of reactive free radicals. Cancer Lett. 1993;1:31–36. doi: 10.1016/0304-3835(93)90040-G. [DOI] [PubMed] [Google Scholar]
- 6.Song J, et al. Biodegradation of nicosulfuron by a Talaromyces flavus LZM1. Bioresour. Technol. 2013;140:243–248. doi: 10.1016/j.biortech.2013.02.086. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, et al. Biodegradation of chloroacetamide herbicides by Paracoccus sp. FLY-8 in vitro. J. Agric. Food Chem. 2001;59:4614–4621. doi: 10.1021/jf104695g. [DOI] [PubMed] [Google Scholar]
- 8.Ren S, Guo J, Zeng G, Sun G. Decolorization of triphenylmethane, azo, and anthraquinone dyes by a newly isolated Aeromonas hydrophila strain. Appl. Microbiol. Biotechnol. 2006;72:1316–1321. doi: 10.1007/s00253-006-0418-2. [DOI] [PubMed] [Google Scholar]
- 9.Fang G, Li L, Li R. Isolation and characterization of a malachite green-degrader Arthrobacter sp. M6. Chin. J. Appl. Environ. Biol. 2010;16:581–584. [Google Scholar]
- 10.Itoh K, Yatome C, Ogawa T. Biodegradation of anthraquinone dyes by Bacillus subtilis. Bull. Env. Contam. Toxicol. 1993;50:522–7. doi: 10.1007/BF00191240. [DOI] [PubMed] [Google Scholar]
- 11.Jang M, et al. Isolation of Citrobacter sp. mutants defective in decolorization of brilliant green by transposon mutagenesis. J. Microbiol. 2004;42:139–142. [PubMed] [Google Scholar]
- 12.Wang J, et al. Pathway and molecular mechanisms for malachite green biodegradation in Exiguobacterium sp. MG2. PLoS One. 2012;7:e51808. doi: 10.1371/journal.pone.0051808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu W, Hong GL, Zhao J. Degradation of malachite green dye by Tenacibaculum sp. HMG1 isolated from Pacific deep-sea sediments. Acta Oceanol. Sin. 2018;37:104–111. doi: 10.1007/s13131-018-1187-3. [DOI] [Google Scholar]
- 14.Tao Y, et al. Biological decolorization and degradation of malachite green by Pseudomonas sp. YB2: Process optimization and biodegradation pathway. Curr. Microbiol. 2018;74:1210–1215. doi: 10.1007/s00284-017-1306-y. [DOI] [PubMed] [Google Scholar]
- 15.Cha CJ, Doerge DR, Cerniglia CE. Biotransformation of malachite green by the fungus Cunninghamella elegans. Appl. Env. Microbiol. 2001;67:4358–4360. doi: 10.1128/AEM.67.9.4358-4360.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Zheng J, Lu Y, Jia R. Degradation and detoxification of the triphenylmethane dye malachite green catalyzed by crude manganese peroxidase from Irpex lacteus F17. Env. Sci. Pollut. Res. Int. 2016;23:9585–9597. doi: 10.1007/s11356-016-6164-9. [DOI] [PubMed] [Google Scholar]
- 17.Papinutti VL, Forchiassin F. Modification of malachite green by Fomes sclerodermeus and reduction of toxicity to Phanerochaete chrysosporium. FEMS Microbiol. Lett. 2004;231:205–209. doi: 10.1016/S0378-1097(03)00957-1. [DOI] [PubMed] [Google Scholar]
- 18.Vyas B, Molitoris H. Involvement of an extracellular H2O2-dependent ligninolytic activity of the white rot fungus Pleurotus ostreatus in the decolorization of Remazol brilliant blue R. Appl. Env. Microbiol. 1995;61:3919–3927. doi: 10.1128/AEM.61.11.3919-3927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siroosi M, Amoozegar MA, Khajeh K, Dabirmanesh B. Decolorization of dyes by a novel sodium azide-resistant spore laccase from a halotolerant bacterium, Bacillus safensis sp. strain S31. Water Sci. Technol. 2018;77:2867–2875. doi: 10.2166/wst.2018.281. [DOI] [PubMed] [Google Scholar]
- 20.Christian M, et al. A draft genome sequence of Pseudomonas veronii R4: a grapevine (Vitis vinifera L.) root-associated strain with high biocontrol potential. Stand. Genomic Sci. 2016;11:76. doi: 10.1186/s40793-016-0198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H, Zhu J, Liu S, Zhou X. Kinetics study of nicosulfuron degradation by a Pseudomonas nitroreducens strain NSA02. Biodegrad. 2018;29:271–283. doi: 10.1007/s10532-018-9828-y. [DOI] [PubMed] [Google Scholar]
- 22.Saikia N, et al. Biodegradation of beta-cyfluthrin by Pseudomonas stutzeri strain S1. Biodegrad. 2006;16:581–589. doi: 10.1007/s10532-005-0211-4. [DOI] [PubMed] [Google Scholar]
- 23.Sazinsky MH, Bard J, Di Donato A, Lippard SJ. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J. Biol. Chem. 2004;27:30600–30610. doi: 10.1074/jbc.M400710200. [DOI] [PubMed] [Google Scholar]
- 24.Cafaro V, et al. Phenol hydroxylase and toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1: interplay between two enzymes. Appl. Env. Microbiol. 2004;70:2211–2219. doi: 10.1128/AEM.70.4.2211-2219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng W, et al. Hydrolysis of nicosulfuron under acidic environment caused by oxalate secretion of a novel Penicillium oxalicum strain YC-WM1. Sci. Rep. 2017;7:647. doi: 10.1038/s41598-017-00228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thummala VR, Ivaturi MR, Nittala SR. Isolation, identification, and characterization of one degradation product in Ambroxol by HPLC-hyphenated techniques. Sci. Pharmaceutica. 2014;82:247–63. doi: 10.3797/scipharm.1310-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis, C., Desai, D. & LaRoche, J. Influence of 16S rRNA variable region on perceived diversity of marine microbial communities of the Northern North Atlantic. FEMS Microbiol Lett366 (2019). [DOI] [PMC free article] [PubMed]
- 28.Claesson MJ, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellogg CA, Goldsmith DB, Gray MA. Biogeographic Comparison of Lophelia associated bacterial communities in the Western Atlantic Reveals conserved core microbiome. Front. Microbiol. 2017;8:796. doi: 10.3389/fmicb.2017.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayilkan F, et al. Preparation of re-usable photocatalytic filter for degradation of Malachite Green dye under UV and vis-irradiation. J. Hazard. Mater. 2007;148:735–744. doi: 10.1016/j.jhazmat.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 31.Song J, et al. Brevibacillus halotolerans sp. nov., isolated from saline soil of a paddy field. Int. J. Syst. Evol. Microbiol. 2017;67:772–777. doi: 10.1099/ijsem.0.001579. [DOI] [PubMed] [Google Scholar]
- 32.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Vani S, et al. Energy requirement for alkali assisted microwave and high pressure reactor pretreatments of cotton plant residue and its hydrolysis for fermentable sugar production for biofuel application. Bioresour. Technol. 2012;112:300–307. doi: 10.1016/j.biortech.2012.02.076. [DOI] [PubMed] [Google Scholar]
- 34.Chen G, Miao S. HPLC determination and MS confirmation of malachite green, gentian violet, and their leuco metabolite residues in channel catfish muscle. J. Agric. Food Chem. 2010;58:7109–7114. doi: 10.1021/jf9043925. [DOI] [PubMed] [Google Scholar]
- 35.Ruan Z, et al. Isolation and characterization of a novel cinosulfuron degrading Kurthia sp. from a methanogenic microbial consortium. Bioresour. Technol. 2013;147:477–483. doi: 10.1016/j.biortech.2013.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.