Abstract
Hypoxic-ischemic brain damage (HIBD) is a major cause of fatality and morbidity in neonates. However, current treatment approaches to alleviate HIBD are not effective. Various studies have highlighted the role of microRNAs (miRNAs) in various biological functions in multiple diseases. This study investigated the role of miR-339-5p in HIBD progression. Neonatal HIBD mouse model was induced by ligation of the right common carotid artery. Neuronal cell model exposed to oxygen-glucose deprivation (OGD) was also established. The miR-339-5p expression in mouse brain tissues and neuronal cells was quantified, and the effects of miR-339-5p on neuronal cell activity and apoptosis induced by hypoxia-ischemia were explored. The overexpression or knockdown of long non-coding RNA (lncRNA) nuclear-enriched abundant transcript 1 (NEAT1) in hippocampal neurons was used to determine the effect of lncRNA NEAT1 on the expression of miR-339-5p and homeobox A1 (HOXA1) and apoptosis. Short hairpin RNA targeting lncRNA NEAT1 and miR-339-5p antagomir were used in neonatal HIBD mice to identify their roles in HIBD. Our results revealed that miR-339-5p was downregulated in neonatal HIBD mice and neuronal cells exposed to OGD. Downregulated miR-339-5p promoted neuronal cell viability and suppressed apoptosis during hypoxia-ischemia. Moreover, lncRNA NEAT1 competitively bound to miR-339-5p to increase HOXA1 expression and inhibited neuronal cell apoptosis under hypoxic-ischemic conditions. The key observations of the current study present evidence demonstrating that lncRNA NEAT1 upregulated HOXA1 to alleviate HIBD in mice by binding to miR-339-5p.
Keywords: ischemic brain damage, hypoxic-ischemic brain damage, HOXA1, NEAT1, long non-coding RNA, microRNA, miR-339-5p, neonatal mice
Introduction
Neonatal hypoxia-ischemia has been widely documented to be characterized by a severe inflammatory response, which exerts further damage on brain tissues.1 Perinatal brain damage is a crucial factor regarding long-term neurological disorders with a multifactorial etiology, including that of hypoxia-ischemia injury.2 Hypoxic-ischemic brain damage (HIBD) is a major cause of fatality and morbidity.3,4 Studies have revealed that approximately 7 per 1,000 preterm newborns and 3 per 1,000 term newborns are reported to suffer from HIBD.5,6 However, only about 60% of patients survive during the neonatal phase, whereas approximately 30% fall victim to subsequent long-term neurological disabilities, including visual difficulties and learning deficiencies caused by impaired brain development.1,7 Although therapeutic hypothermia is currently regarded as the gold standard treatment approach for HIBD, the development of immunomodulatory therapeutic strategies has been discussed as a novel avenue for the future of HIBD management.8,9 Owing to HIBD’s notoriously high rates of morbidity and mortality, the swift development of more effective alternative treatments is of urgent need.10
MicroRNAs (miRNAs) represent a family of small non-coding RNAs with a length of about 22 nt that have been widely reported to participate in normal brain development.11 Furthermore, studies have implicated miRNAs in the development of HIBD, with reports highlighting the poor expression exhibited by miR-182 in pineal gland in cases of neonatal HIBD.12,13 Another miRNA, miR-339-5p, has been found to be downregulated in the brain of patients with Alzheimer disease.14 Moreover, miR-339-5p has been implicated in the inhibitory effects of neuronal apoptosis induced by glycine-histidine-lysine.15 A previously conducted functional study has highlighted the enforced expression of miR-339-5p following ischemic insult to the brain.16 Given the highly suggestive neuroprotective effects of miR-339-5p, the objective of the current study was to determine the poorly understood role of miR-339-5p in HIBD. Homeobox A1 (HOXA1) belongs to the HOX family of transcription proteins that regulate diverse biological processes through gene expression, including neurogenesis.17 Many HOX genes function in the development of embryonic brain.18 The HOXA1 gene has been linked to brainstem and cerebellum development.19,20 Furthermore, miR-339-5p has been predicted to be a downstream miRNA of long non-coding RNA (lncRNA) nuclear-enriched abundant transcript 1 (NEAT1) through in silico analysis. lncRNAs, incapable of encoding proteins, have over 200 nt, and they have been reported to participate in neurodevelopment.21 The relationship between lncRNAs and HIBD has been emphasized in previous studies based on the differential expression of multiple lncRNAs in brains of neonatal rats suffering from HIBD.21,22 Elevated expression of lncRNA NEAT1 has been demonstrated to repress cell apoptosis and inflammation, which ultimately contributes to traumatic brain injury recovery.23 The role of lncRNA NEAT1 in the recovery of HIBD remains unclear. Thus, lncRNAs have recently been extensively reported to interact with miRNA to exert post-transcriptional regulatory effects as competing endogenous RNAs (ceRNAs).24 In light of the aforementioned studies, we hypothesized that lncRNA NEAT1 could serve as a ceRNA, bind to miR-339-5p, regulate the expression of HOXA1, and participate in the development of HIBD.
Results
miR-339-5p Expression Is Reduced in Mouse and Cell Models of HIBD
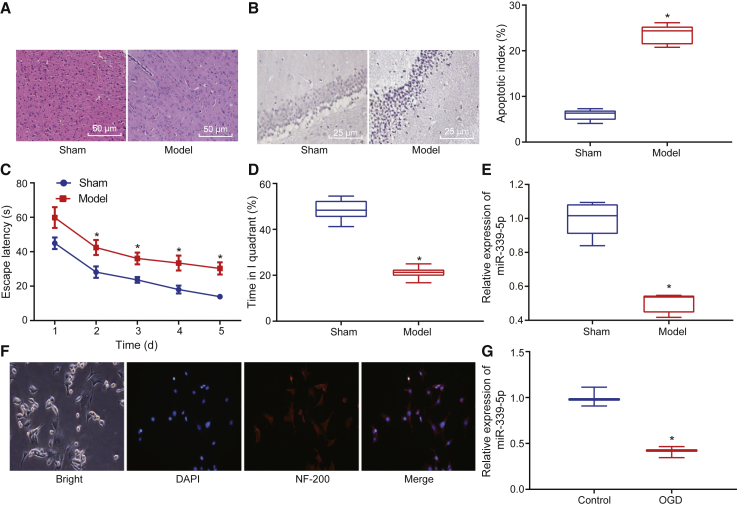
Recently, miRNAs have been found to play essential roles in the development of HIBD.25 Hence, in the current study, we set out to elucidate the role of miR-339-3p in HIBD. HIBD mice had distinct brain damage when compared with sham-operated mice (Figure 1A). Terminal deoxynucleotidyl transferase-mediated 2′-deoxyuridine 5′-triphosphate (dUTP)-biotin nick end labeling (TUNEL) staining revealed that cell apoptosis in the neonatal HIBD increased (Figure 1B).
Figure 1.
Mouse and Cell Model of HIBD
(A) The representative micrographs showing morphological changes in brain tissues stained by H&E (original magnification ×200). (B) The representative micrographs showing hippocampal apoptosis measured by TUNEL staining (original magnification ×400). (C) The escape latency in Morris water maze. (D) The time spent in platform quadrant in Morris water maze. (E) The expression of miR-339-5p in mouse brain tissues determined by qRT-PCR. (F) The representative micrographs showing expression of NF-200 in hippocampal neuronal cells detected using immunofluorescence assay (original magnification ×200). (G) The expression of miR-339-5p in hippocampal neuronal cells after exposure to OGD. *p < 0.05 versus sham-operated mice or untreated hippocampal neuronal cells. The measurement data were expressed as mean ± standard deviation, and comparison of data between two groups was performed using unpaired t test. Data in Morris water maze task were analyzed using repeated-measures ANOVA, followed by Bonferroni’s post hoc test. Cell experiments were repeated three times independently.
In the Morris water maze test, escape latency in HIBD mice was longer than that in the sham-operated mice (p < 0.05; Figure 1C). In the spatial probe test, the sham-operated mice spent the majority of the time in the platform quadrant, whereas the neonatal HIBD mice spent a considerably shorter time at the platform quadrant (p < 0.05; Figure 1D). The aforementioned results confirmed that the neonatal HIBD mouse model had been successfully established.
The expression of miR-339-5p was lower in the brain tissues of the HIBD mice when compared with that of the sham-operated mice (p < 0.05; Figure 1E). Moreover, positive expression of NF-200 was detected in the primary hippocampal neurons (Figure 1F). The expression of miR-339-5p was decreased in the hippocampal neuronal cells following exposure to oxygen-glucose deprivation (OGD) (p < 0.05; Figure 1G). Taken together, downregulated miR-339-5p was consistently detected in both the HIBD mouse models and the HIBD cell models.
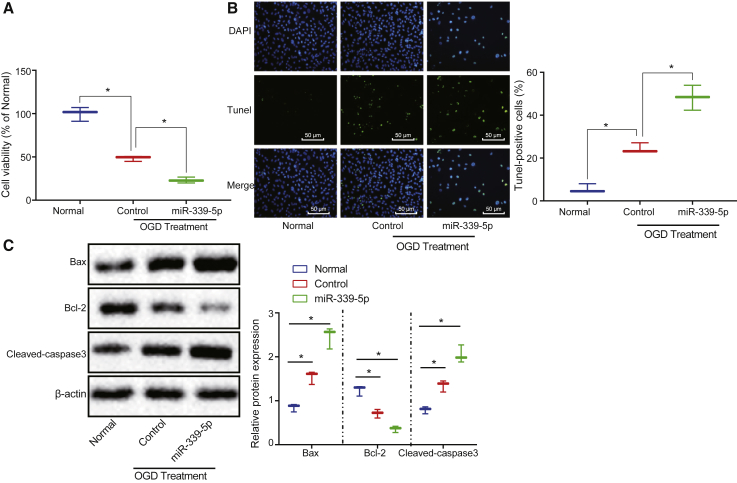
miR-339-5p Overexpression Inhibits Neuronal Cell Viability and Promotes Apoptosis Induced by Hypoxia-Ischemia
The effects of miR-339-5p on viability and apoptosis of the neuronal cells during OGD were determined when miR-339-5p was overexpressed. Our results revealed that overexpressed miR-339-5p inhibited the neuronal cell viability induced by OGD (p < 0.05; Figure 2A). Additionally, the overexpression of miR-339-5p potentiated hypoxia-ischemia-induced neuronal cell apoptosis (p < 0.05; Figure 2B). Besides, miR-339-5p overexpression was observed to elevate the expression of Bax and cleaved caspase-3, but downregulated Bcl-2 expression (p < 0.05; Figure 2C). Taken together, the overexpression of miR-339-5p inhibited neuronal cell viability and enhanced apoptosis during hypoxia-ischemia events.
Figure 2.
Neuronal Cell Viability Is Potentiated and Neuronal Cell Apoptosis Induced by Hypoxia-Ischemia Is Inhibited with miR-339-5p Reduced under Hypoxia-Ischemia
(A) The neuronal cell viability detected using CCK-8 assay. (B) The neuronal cell apoptosis determined by TUNEL staining (original magnification ×200). (C) The expression of apoptosis-related proteins normalized to β-actin determined by western blot analysis. *p < 0.05 versus neuronal cells without treatment. The data were measurement data and are expressed as mean ± standard deviation. Data among multiple groups were compared using one-way ANOVA, followed by Tukey’s post hoc test. The experiments were repeated three times independently.
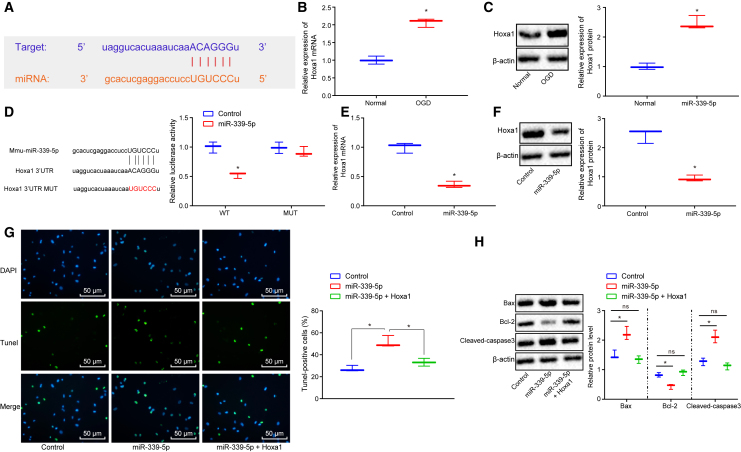
Depletion of miR-339-5p Upregulates HOXA1 to Repress Hypoxia-Ischemia-Induced Neuronal Cell Apoptosis
The starBase database, as well as the Gene Expression Omnibus database, GEO: GSE37777, provided data indicating that miR-339-5p targets HOXA1 (Figure 3A). In addition, HOXA1 mRNA (Figure 3B) and protein (Figure 3C) expression exhibited elevated levels in the OGD-treated cells when compared with control (p < 0.05).
Figure 3.
Binding Relationship between miR-339-5p and HOXA1 and Their Effects on Neuronal Cell Apoptosis during Hypoxia-Ischemia
(A) The binding site between miR-339-5p and HOXA1 predicted by starBase database. (B) HOXA1 mRNA expression. (C) HOXA1 protein expression. (D) Binding of miR-339-5p to HOXA1 evaluated by dual-luciferase reporter gene assay. (E) HOXA1 mRNA expression when miR-339-5p was overexpressed. (F) HOXA1 protein expression when miR-339-5p was overexpressed. (G) The neuronal cell apoptosis assessed by TUNEL staining (original magnification ×200). (H) The expression of neuronal cell apoptosis-related proteins normalized to β-actin determined by western blot analysis. *p < 0.05 versus untreated neuronal cells or neuronal cells treated with overexpressed miR-339-5p during OGD. The measurement data were expressed as mean ± standard deviation. Data among multiple groups were compared using one-way ANOVA, followed by Tukey’s post hoc test. The experiment was repeated three times independently.
Additionally, dual-luciferase reporter gene assay was conducted in order to confirm the binding relationship between miR-339-5p and HOXA1. The results illustrated that miR-339-5p bound to HOXA1-wild-type (WT) but failed to bind to HOXA1-mutant (MUT; p < 0.05; Figure 3D). Moreover, HOXA1 mRNA (Figure 3E) and protein (Figure 3F) expression was decreased in the neuronal cells in the presence of overexpressed miR-339-5p. These results indicated that miR-339-5p could bind to and diminish the expression of HOXA1.
Next, we determined the causal relationship between miR-339-5p and HOXA1 on neuronal cell apoptosis. The number of apoptotic neuronal cells was increased when miR-339-5p was overexpressed during OGD but decreased when both miR-339-5p and HOXA1 were overexpressed (p < 0.05; Figure 3G). The apoptosis-related protein expression of Bax and cleaved caspase-3 was elevated, whereas Bcl-2 was diminished in response to upregulated miR-339-5p during OGD (Figure 3H). These results were opposite when both miR-339-5p and HOXA1 were overexpressed (p < 0.05). The findings demonstrated that HOXA1 overexpression reversed the apoptotic effects of miR-339-5p during OGD.
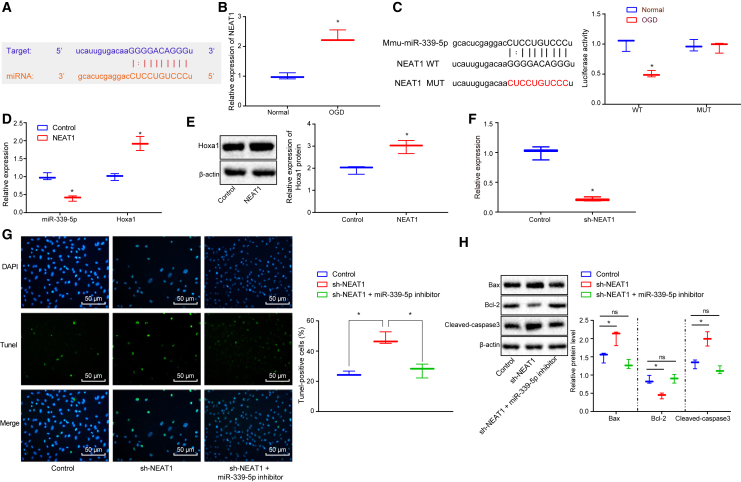
lncRNA NEAT1 Overexpression Downregulates miR-339-5p to Inhibit Neuronal Cell Apoptosis in Hypoxia-Ischemia
It was hypothesized that the expression of miR-339-5p in HIBD was regulated by an upstream lncRNA. Hence the starBase database was used to predict the potential lncRNA binding miR-339-5p. It was predicted that there was a complementary fragment between lncRNA NEAT1 and miR-339-5p (Figure 4A). In addition, lncRNA NEAT1 was found to be elevated during OGD (p < 0.05; Figure 4B), which was consistent with the hypothesis that lncRNA NEAT1 regulates the expression of miR-339-5p.
Figure 4.
Elevation of lncRNA NEAT1 Downregulates miR-339-5p to Increase the Expression of HOXA1 and Neuronal Cell Apoptosis during Hypoxia-Ischemia
(A) The binding site between lncRNA NEAT1 and miR-339-5p predicted by starBase database. (B) The mRNA expression of HOXA1 in neuronal cells measured by qRT-PCR. (C) The binding between miR-339-5p and lncRNA NEAT1 by dual-luciferase reporter gene assay. (D) The expression of miR-339-5p and HOXA1 in neuronal cells measured by qRT-PCR. (E) The protein expression of HOXA1 in neuronal cells normalized to β-actin determined by western blot analysis. (F) The expression of lncRNA NEAT1 in OGD-treated neuronal cells in response to lncRNA NEAT1 knockdown. (G) The apoptosis of OGD-treated neuronal cells detected by TUNEL staining (original magnification ×200). (H) The expression of neuronal cell apoptosis-related proteins normalized to β-actin as determined by western blot analysis. *p < 0.05 versus untreated neuronal cells or neuronal cells treated with sh-NEAT1. The measurement data were expressed as mean ± standard deviation. Data between two groups were analyzed using unpaired t test, and data among multiple groups were tested using one-way ANOVA, followed by Tukey’s post hoc test. The experiment was repeated three times independently.
Dual-luciferase reporter gene assay was then performed in order to confirm the binding relationship between lncRNA NEAT1 and miR-339-5p. The results demonstrated that NEAT1-WT, but not NEAT1-MUT, bound to miR-339-5p (p < 0.05; Figure 4C). Next, the overexpression of lncRNA NEAT1 was found to diminish the expression of miR-339-5p to elevate HOXA1 mRNA (Figure 4D) and protein (Figure 4E) expression in neuronal cells (p < 0.05), indicating that overexpressed lncRNA NEAT1 depleted miR-339-5p and upregulated HOXA1.
Moreover, lncRNA NEAT1 was knocked down in the OGD model to investigate the effects of lncRNA NEAT1 on hypoxia-ischemia-induced neuronal cell apoptosis. The results revealed that the knockdown efficiency of lncRNA NEAT1 was approximately 78.36%, which was qualified for the initiation of the following experiments (p < 0.05; Figure 4F). lncRNA NEAT1 knockdown enhanced neuronal cell apoptosis (Figure 4G). In contrast, short hairpin (sh)-NEAT1 or miR-339-5p inhibitor attenuated neuronal cell apoptosis (p < 0.05; Figure 4G). Moreover, Bax and cleaved caspase-3 were elevated, whereas Bcl-2 reduced in response to lncRNA NEAT1 knockdown (Figure 4H). These results were reversed by combining with treatment of miR-339-5p inhibitor (p < 0.05; Figure 4H). Collectively, lncRNA NEAT1 bound to and downregulated miR-339-5p, leading to reduced neuronal cell apoptosis during hypoxia-ischemia.
lncRNA NEAT1 Downregulates miR-339-5p to Increase HOXA1 and Alleviates HIBD Progression
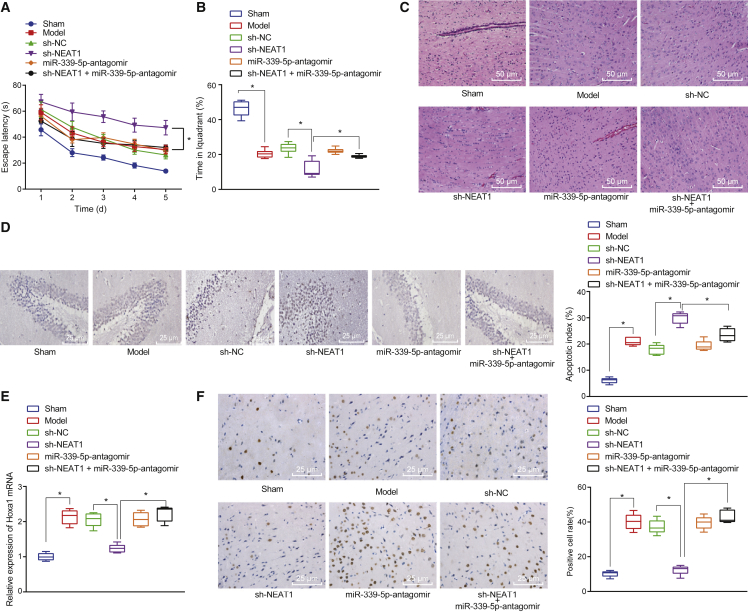
Next, we sought to clarify the role of the lncRNA NEAT1/miR-339-5p axis in a neonatal HIBD mouse model in vivo. The neonatal HIBD mice were treated with sh-NEAT1 and miR-339-5p-antagomir to inhibit lncRNA NEAT1 and miR-339-5p, respectively. During the navigation test, sh-NEAT1 was found to significantly increase the escape latency when compared with that of the controls (Figure 5A). However, the escape latency was normalized when the mice were treated with combined sh-NEAT1 and miR-339-5p-antagomir (p < 0.05; Figure 5A). During the spatial probe test, sh-NEAT1 increased time spent in the platform quadrant when compared with control, whereas the addition of miR-339-5p-antagomir normalized the amount of time spent at the platform quadrant (p < 0.05; Figure 5B). The results suggested that lncRNA NEAT1 could enhance learning and memory in neonatal HIBD mice.
Figure 5.
Effects of lncRNA NEAT1 and miR-339-5p Inhibition on HOXA1 Expression and Neonatal HIBD Mice
(A) The escape latency in the navigation test of Morris water maze. (B) The time spent in platform quadrant in the spatial probe test of Morris water maze. (C) The representative micrographs showing morphological changes in hippocampal tissues after H&E staining (original magnification ×200). (D) The neuronal cell apoptosis determined by TUNEL staining (original magnification ×400). (E) The mRNA expression of HOXA1 in hippocampal tissues detected by qRT-PCR. (F) HOXA1 protein expression in hippocampal tissues determined by immunohistochemistry (original magnification ×400). *p < 0.05 versus sham-operated mice or neonatal HIBD mice treated with sh-NEAT1. The measurement data were expressed as mean ± standard deviation. Data among multiple groups were compared using one-way ANOVA, followed by Tukey’s post hoc test. Data in Morris water maze task were analyzed using repeated-measures ANOVA, followed by Bonferroni’s post hoc test.
Next, the hippocampal tissues were collected from HIBD mice to observe and analyze the morphological changes via hematoxylin and eosin (H&E) staining. Treatment with sh-NEAT1 was found to aggravate brain damage, whereas the addition of miR-339-5p-antagomir attenuated brain damage (Figure 5C). The number of apoptotic neuronal cells was elevated following treatment with sh-NEAT1 but normalized in response to both sh-NEAT1 and miR-339-5p-antagomir (p < 0.05; Figure 5D). These findings suggested that lncRNA NEAT1 could attenuate HIBD via miR-339-5p downregulation.
The effects of the lncRNA NEAT1/miR-339-5 axis on HOXA1 were then explored. HOXA1 mRNA was downregulated following sh-NEAT1 treatment but normalized following combined treatment of sh-NEAT1 and miR-339-5p-antagomir (p < 0.05; Figure 5E). Similar results were obtained in HOXA1 protein expression as evidenced by immunohistochemistry results (Figure 5F). Taken together, the results suggested that lncRNA NEAT1 absorbed miR-339-5p to upregulate HOXA1 and inhibit the development of HIBD.
Discussion
Neonatal HIBD is a devastating condition often accompanied by high mortality and long-term disabilities. Despite its severity, current treatment approaches are inadequate.26 For example, although hypothermia has been used to treat HIBD, many newborns fail to survive, and many subsequently experience neurological disabilities.3 It is important to understand the underlying mechanism in which neonatal HIBD progresses, in order to identify new treatments approaches. The current study aimed to explore the role of the lncRNA NEAT1/miR-339-5p/HOXA1 axis on the progression of HIBD using mice and neuronal cell models. Our findings provided evidence indicating that lncRNA NEAT1 functions as a ceRNA of miR-339-5p, leading to increased HOXA1 expression and attenuated HIBD progression.
Numerous miRNAs have been reported to play a protective role against various diseases, many of which lead to brain damage. For instance, miR-21 has been reported to suppress apoptosis and potentiate angiogenesis following traumatic brain damage, and therefore may protect against traumatic brain damage.27 However, the effect of miR-339-5p in HIBD remains less studied. Intriguingly, our results revealed that miR-339-5p was downregulated in both neonatal HIBD mice and the neuronal cell OGD model. More importantly, we discovered that downregulated miR-339-5p promoted neuronal cell viability and suppressed apoptosis. Likewise, re-oxygenation after OGD brought about a reduction in the expression of miR-135b-5p in the hippocampal neurons.28 Existing literature has indicated that miR-130a downregulation promotes cell viability and attenuates apoptosis in myocardial hypoxia/reoxygenation injury.29 Bcl-2, Bax, and caspase-3 are all crucial regulators of neuronal cell apoptosis in ischemia/reperfusion brain injury.30 Consistent with the findings of the current study, decreased levels of caspase-3 and Bax, and increased Bcl-2 level have been identified to promote apoptosis, which was evidenced in our results when miR-339-5p was overexpressed.25 Collectively, the results demonstrate that miR-339-5p downregulation could potentially confer protection against HIBD.
Another key finding of our study revealed that lncRNA NEAT1 competitively absorbs miR-339-5p to upregulate HOXA1. Similarly, a previous study concluded that lncRNA H19 is highly expressed following hypoxic injury and functions as a ceRNA to sponge miR-139 and increase expression of Sox8.31 A previous study asserted that miR-525-5p is downregulated in the ischemic brain and highlighted its inhibitory role against neuronal cell death through its target gene, ADAMTS13.32 In addition, miR-144-3p expression was reported to be reduced in the OGD-reoxygenation-treated neurons, ultimately suppressing apoptosis and attenuating neuronal injury by directly targeting Brahma-related gene 1.33 HOXA1 has been reported to be directly targeted by miR-577 and miR-99a, respectively, in addition to studies highlighting its role in the biological processes of hepatocellular carcinoma and breast cancer.34,35 Taken together, our results added new information for the understanding of the neuronal apoptotic pathway that involved miRs in HIBD.
A notable observation of the current study revealed that lncRNA NEAT1 was upregulated in neuronal cells treated with OGD. Moreover, lncRNA NEAT1 absorbed miR-339-5p to upregulate HOXA1 to inhibit neuronal cell apoptosis during hypoxia-ischemia. Apoptosis is a particularly important factor in HIBD because it has been shown to be a significant trigger of neonatal HIBD.36 Consistent with our findings, upregulation of lncRNA NEAT1 has also been detected in the midbrain of mice with Parkinson’s disease (PD).37 Besides, lncRNA NEAT1 has been reported to confer neuroprotective effects in PD in human brain tissues and mouse and cellular models.37,38 Furthermore, lncRNA NEAT1 is highly expressed in the brains of transgenic mice with Huntington’s disease (HD).39 The postmortem brains from patients with HD exhibit elevated expression of lncRNA NEAT1, which has been shown to protect against neuronal injury.39 Likewise, a previous study has reported that brain microvascular endothelial cells (BMECs) exposed to OGD have elevated expression of lncRNA NEAT1 and reduced expression of miR-377, leading to decreased cell apoptosis in BMECs.40 Mice with a targeted disruption of Hoxa1 were observed to die shortly after birth from breathing defects, which are thought to result from mispatterning of the hindbrain.41 Moreover, it has been revealed that downregulation of miR-24 suppresses hippocampal neuronal apoptosis, and also downregulated miR-124 can upregulate the expression of Ku70 to attenuate neuronal cell death, thus protecting against brain injury induced by ischemia and reperfusion.42,43
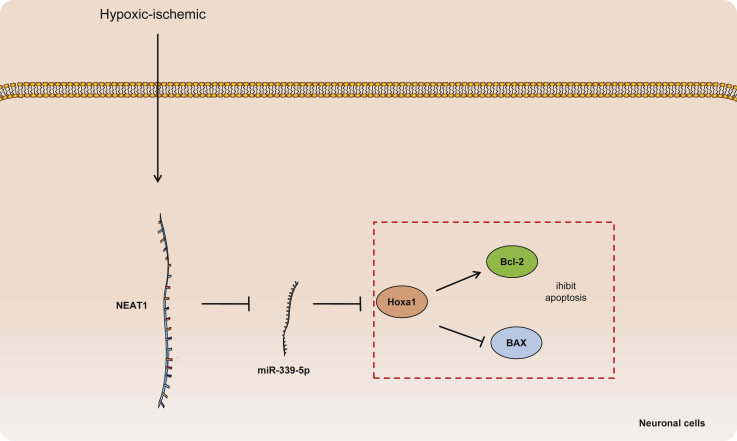
Taken together, our findings provided evidence that lncRNA NEAT1 increases the expression of HOXA1 to prevent neuronal cell apoptosis by functioning as a ceRNA, thereby absorbing miR-339-5p (for a detailed pathway, see Figure 6). This effect may be beneficial to HIBD and serve as a potential novel therapeutic target that deserves further investigation.
Figure 6.
Schematic Diagram Showing Potential Mechanism in which the lncRNA NEAT1/miR-339-5/HOXA1 Axis Is Involved in HIBD
lncRNA NEAT1 sponges miR-339-5p, reduces inhibition to HOXA1, and inhibits neuronal cell apoptosis during hypoxia-ischemia in neonatal HIBD mice.
Materials and Methods
Ethics Statement
All experimental protocols were approved by the Experimental Animal Ethics Committee in Affiliated Hospital of North Sichuan Medical College (approval number: 201805001). Extensive efforts were made to minimize the number of animals used and pain suffered by the animals.
Animal HIBD Model and Treatments
Seven-day-old neonatal female C57BL/6 mice (n = 108, 4.1 ± 0.5 g; license number of animal use: SYXK [Shanghai] 2018-0005; Shanghai Lingchang Biotechnology, Shanghai, China) were housed in an animal facility under a controlled 12-h light/dark cycle. The mice were subsequently subjected to a sham operation (n = 18) or HIBD model establishment (n = 90). The neonatal HIBD mouse model was established via right common carotid artery occlusion based on the Rice-Vannucci method. Under isoflurane anesthesia (2.5%–3% for induction and 1.5%–2% for maintenance), an incision was made to the midline of the mouse neck under a dissecting microscope, after which the right common carotid artery was isolated, followed by double ligation at distal and proximal ends using a 6-0 suture. After 2 h, the mice were moved to an airtight hypoxic container at constant temperature with 8% oxygen and 92% nitrogen for 1 h. The neonatal mice were then returned to their mothers. The sham-operated mice were subjected to only right common carotid artery isolation without ligation and hypoxia treatment. Next, short hairpin RNA targeting lncRNA NEAT1 (shRNA-NEAT1), negative control shRNA sequence (shRNA-NC), or miR-339-5p-antagomir (Guangzhou RiboBio, Guangzhou, Guangdong, China) was continuously administered into the lateral ventricle of the HIBD mice for 7 days using an osmotic pump (n = 18 in each group).44 Approximately zero to three mice failed to survive the surgery or treatment procedures in each group. Five mice from each group were randomly selected for tissue collection. The remaining 10 mice in each group were used for Morris water maze.
Hippocampal Primary Cell Culture and Treatment
The 18-day-old C57BL/6 mice were euthanized via isoflurane anesthesia in order to separate the hippocampal neuronal cells. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO, Grand Island, NY, USA) containing 20% fetal bovine serum and filtered by a 40-mm filter. The cells were then inoculated on an eight-well poly-L-lysine-treated slide or culture dish (30,000 cells/well) and cultured in DMEM (4.5 g/L glucose, 100 U/mL penicillin, 100 mg/mL streptomycin, 2 mM glutamine, and B27) at 37°C with 5% CO2.
The packaging plasmids (pVSVG, pREV, pMDL, and PIK) and expression plasmids including pLKO.1 (Amp+) with lncRNA NEAT1 knockdown, LV3 (Amp+) with miR-339-5p overexpression, pBABE (Puromycin) with lncRNA NEAT1 overexpression, and pBABE (Puromycin) with HOXA1 overexpression were transferred to the DH5α-competent cells. All plasmids were purchased from Cyagen Biosciences (Guangzhou, Guangdong, China). The corresponding plasmids were extracted using plasmid extraction kits (DP103-03; TIANGEN Biotechnology, Beijing, China). Afterward, the four types of plasmids were delivered into the 293T cells using Turbofect reagent (R0531; Thermo Fisher Scientific, Waltham, MA, USA). The culture medium was changed after 12 h. The culture liquid was collected at 24 and 48 h, respectively, and filtered using a 0.22-μm filter to collect viruses. Virus titer was measured by QuickTiter Lentivirus Titer Kit (VPK-107; Cell Biolabs, San Diego, USA). A total of 1 × 106 primary hippocampal neurons were infected with 1 mL viruses and cultured for 24 h. After 48 h, the cells with stably knocked down lncRNA NEAT1, overexpressed miR-339-5p, as well as HOXA1 were all screened by puromycin (1 μg/mL).
Cell OGD Model
The primary mouse hippocampal neurons that had been cultured for 7 days were washed three times with phosphate-buffered saline (PBS). The culture medium was replaced with glucose-free DMEM. The neurons were cultured in an anaerobic culture system on hypoxic culture dishes with 95% N2 and 5% CO2 at 37°C for 3 h. Next, the glucose-free DMEM was renewed with neuron culture medium. The hippocampal neurons were subsequently cultured with normal oxygen concentration at 37°C with 5% CO2 for 6–24 h. The neuronal cells that had not been subject to OGD treatment were set as controls.
NF-200 Expression Determined by Immunofluorescence Assay
The hippocampal neuronal cells were inoculated on poly-L-lysine-treated slides. When cell confluence reached 50%, the cells were immersed in 4% polyformaldehyde for 30 min, permeabilized using 2% Triton X-100 for 15 min, cleared by 2 M hydrochloric acid (HCl) for 20 min, and blocked with 2% bovine serum albumin for 45 min. The cells were then incubated overnight at 4°C with NF-200 antibody (N4142, Rabbit; 1:200; Sigma-Aldrich Chemical Company, St. Louis, MO, USA), followed by a further period of incubation with fluorescent secondary goat anti-rabbit immunoglobulin-G H&L (ab150080; Rabbit; 1:400; Abcam, Shanghai, China) for 2 h. The slides were stained with 2 μg/mL 4′,6-diamidino-2-phenylindole and then mounted. The expression of NF-200 was detected under a fluorescence microscope. The fluorescence intensity was quantified using ImageJ software (NIH, Bethesda, MD, USA).
CCK-8 Assay
The hippocampal neuronal cells (1 × 105 cells/well) were inoculated into a 96-well plate, with 100 μL culture liquid in each well. After 7 days of culture, the cells were exposed to OGD and subsequently incubated for 2 h with 10 μL Cell Counting Kit-8 (CCK-8) solution (C0038; Beyotime Biotechnology, Shanghai, China) in each well. The optical density value was measured at 450 nm.
Learning and Memory Evaluated by Morris Water Maze Task
Learning and memory capacities were evaluated by Morris water maze 5 weeks after surgery in 18 mice from each group. A pool (diameter: 150 cm; height: 60 cm) was divided into four quadrants with a hidden platform placed 1 cm below the water surface in one quadrant. The mice then underwent a navigation test and a spatial probe test over 5 consecutive days. In the navigation test, the mice were allowed to swim freely in the pool for 2 min 1 day prior to the test. During the test, the mice were randomly placed in the pool at different quadrants with the time that each mouse required to find the platform recorded. The process was repeated four times per day. In the event the mouse was unable to find the platform within 60 s, the mouse was removed and the time was recorded as 60 s. During the spatial probe test, the hidden platform was removed. The mice were placed in the pool for 60 s. Time spent in each quadrant was then recorded. The percentage of time duration in the platform quadrant in relation to the entire time spent in the pool was calculated using Morris water maze motion measurement software (Actimetrics software, Evanston, IL, USA).
Brain Morphology Determined by H&E Staining
Five mice from each group were anaesthetized by isoflurane for thoracotomy. Normal saline at 4°C was infused through the heart, followed by the addition of 4% paraformaldehyde. The mice were subsequently decapitated and after which their brains were collected. The brain tissues were fixed with 10% neutral formaldehyde overnight, paraffin embedded, and cut into serial sections. The tissue sections were then deparaffinized using xylene, hydrated with gradient alcohol, and stained with H&E. The morphological variations in the brain tissues were observed and analyzed under a high-power microscope.
qRT-PCR
Total RNA was extracted using TRIzol kits (15596-018; Beijing Solarbio Science & Technology, Beijing, China). The primer sequences are depicted in Table 1 (Takara Biotechnology, Dalian, Liaoning, China). Complementary DNA (cDNA) of miRNA was obtained using PolyA RNA-tailing detection kits (B532451; Sangon Biotech, Shanghai, China). The cDNA of the non-miRNA was obtained using cDNA RT kits (K1622; Beijing Reanta Biotechnology, Beijing, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was regarded as the internal control for lncRNA NEAT1 and HOXA1. Relative transcription levels of genes to be tested were calculated by the 2−ΔΔCt method.45
Table 1.
Primer Sequence of qRT-PCR
| Gene | Primer Sequence |
|---|---|
| NEAT1 | F: 5′-TACATGGGGGCAGTGTCCTA-3′ |
| R: 5′-ACCACAGAAGAGGAAGCACG-3′ | |
| miR-339-5p | F: 5′-GGGTCCCTGTCCTCCA-3′ |
| R: Universal PCR Primer R | |
| HOXA1 | F: 5′-CCCAGACGGCTACTTACCAG-3′ |
| R: 5′-CCCACCACTTACGTCTGCTT-3′ | |
| GAPDH | F: 5′-AGGCCGGTGCTGAGTATGTC-3′ |
| R: 5′-TGCCTGCTTCACCACCTTCT-3′ | |
| U6 | F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
F, forward; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HOXA1, homeobox A1; miR-339-5p, microRNA-339-5p; NEAT1, nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription polymerase chain reaction; R, reverse.
Western Blot Analysis
Total protein from hippocampal neuronal cells was obtained by adding lysis buffer containing phenylmethylsulfonyl and phosphatase inhibitor. The protein concentration was measured using bicinchoninic acid protein assay kits. A total of 50 μg protein was dissolved in 2× sodium dodecyl sulfate (SDS) followed by the application of 10% SDS-polyacrylamide gel electrophoresis. The proteins were then transferred onto the polyvinylidene fluoride membrane and subsequently blocked with 5% skimmed milk for 1 h, followed by incubation with primary antibodies against HOXA1 (ab230513; 1:1,000, rabbit), cleaved caspase-3 (#9661; 1:1,000, rabbit), Bcl-2-associated X (BAX; ab232479; 1:1,000, rabbit), B cell CLL/Lymphoma 2 (Bcl2; ab32124; 1:1,000, rabbit), and β-actin (ab8226; 1:1,000, mouse). Among the aforementioned antibodies, cleaved caspase-3 was purchased from Cell Signaling Technologies (Beverly, MA, USA), whereas the rest were purchased from Abcam (Cambridge, MA, USA). The membrane was further incubated for 1 h with horseradish peroxidase (HRP)-labeled secondary goat anti-mouse or goat anti-rabbit antibody (Beijing TransGen Biotech, Beijing, China). The membrane was then developed using enhanced chemiluminescence followed by imaging with a gel imager. After the images had been captured using Bio-Rad image analysis system (Bio-Rad, Richmond, CA, USA), further analysis was conducted using Quantity One v.4.6.2. The relative expression of the proteins was expressed as the gray value ratio between the band of protein to be assessed relative to that of β-actin.
TUNEL Staining
Hippocampal tissues were collected from five mice in each group for TUNEL staining (Roche, Nutley, NJ, USA). The tissue sections were deparaffinized using xylene, hydrated using gradient ethanol, and treated with Proteinase K. After that, the tissues were incubated with TUNEL reaction solution at 37°C for 1 h, fixed with formaldehyde solution containing 3% H2O2, and then incubated with streptavidin-peroxidase at 37°C for 30 min and developed with diaminobenzidine. The cell nucleus was counterstained with hematoxylin, dehydrated, permeabilized, and mounted with neutral balsam. TUNEL-positive cells were stained in brown under an optical microscope. Cell apoptosis percentage = number of TUNEL-positive cells/number of total cells × 100%.
The hippocampal neuronal cells were fixed with 4% polyformaldehyde for 30 min, as well as with formaldehyde solution containing 0.3% H2O2 for 30 min, and subsequently reacted with 0.3% Triton X-100 for 2 min. TUNEL reaction solution was prepared as per the instructions of the TUNEL cell apoptosis detection kits (Promega, Madison, WI, USA). The cells were incubated with either dUTP labeled by 50 μL TdT + 450 μL luciferase or dUTP labeled by 50 μL luciferase at 37°C for 60 min. The cells were observed and analyzed under a fluorescence microscope (Bio-Rad, Richmond, CA, USA).
Dual-Luciferase Reporter Gene Assay
The interaction among lncRNA NEAT1, HOXA1, and miR-339-5p was analyzed in the biological prediction website starBase and further determined using dual-luciferase reporter gene assay. Dual-luciferase reporter gene vectors of lncRNA NEAT1 and HOXA1, as well as the MUT of their respective binding sites in miR-339-5p, were constructed as PGLO-NEAT1-WT, PGLO-HOXA1-WT, PGLO-NEAT1-MUT, and PGLO-HOXA1-MUT. The above-mentioned vectors were co-treated with plasmids overexpressing miR-339-5p or NC plasmids in 293T cells. After 24 h of treatment, the cells were lysed and centrifuged at 12,000 rpm for 1 min to collect supernatant. The luciferase activity was detected using a Dual-Luciferase Reporter Assay System (E1910; Promega, Madison, WI, USA). Firefly (100 μL) and Renilla (100 μL) luciferase reagents were subsequently added. The relative luciferase activity was expressed as the ratio between Firefly and Renilla luciferase.
HOXA1 Expression and Localization by Immunohistochemistry
The hippocampal tissues obtained from five mice in each group were fixed with 4% polyformaldehyde, paraffin embedded, and sliced into 4-μm serial sections. The sections were then deparaffinized, followed by antigen retrieval. The sections were stained using Histostain SP-9000 immunohistochemistry reagent (Zymed, San Francisco, CA, USA) and incubated overnight at 4°C with primary rabbit antibody against HOXA1 (ab230513; 1:500; Abcam, Cambridge, UK) after which they were subjected to incubation at 37°C for 30 min with secondary antibody (ab6728; 1:1,000; Abcam, Cambridge, UK). After PBS washing and incubation with HRP-labeled working solution, the sections were developed using diaminobenzidine, counterstained with hematoxylin, and mounted. The tissues that were stained in brown around the cytoplasm viewed using an optical microscope (Nikon, Tokyo, Japan) were considered to be positive. The sham-operated mice were regarded as the NC.
Statistical Analysis
All statistical analyses were conducted using SPSS 21.0 software (IBM, Armonk, NY, USA). Data were expressed as mean ± standard deviation. The comparison of data between two groups was performed by unpaired t test. Comparison of data among multiple groups was conducted using one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. Data generated from Morris water maze were analyzed by repeated measures of ANOVA, followed by Bonferroni’s post hoc test. Significant difference was regarded as p < 0.05.
Author Contributions
J.Z. wrote the paper and conceived and designed the experiments; L.H. analyzed the data; L.Y. collected and provided the sample for this study.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81300528), the Sichuan Science and Technology Program (No. 2017JY0115), the Popularization Project of Health and Family Planning Commission of Sichuan Province (No. 18PJ041), and the School Cooperation Research Fund of Nanchong City (No. NSMC20170425).
References
- 1.Rocha-Ferreira E., Hristova M. Antimicrobial peptides and complement in neonatal hypoxia-ischemia induced brain damage. Front. Immunol. 2015;6:56. doi: 10.3389/fimmu.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thornton C., Rousset C.I., Kichev A., Miyakuni Y., Vontell R., Baburamani A.A., Fleiss B., Gressens P., Hagberg H. Molecular mechanisms of neonatal brain injury. Neurol. Res. Int. 2012;2012:506320. doi: 10.1155/2012/506320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Q., Chen W., Sinha B., Tu Y., Manning S., Thomas N., Zhou S., Jiang H., Ma H., Kroessler D.A. Neuroprotective agents for neonatal hypoxic-ischemic brain injury. Drug Discov. Today. 2015;20:1372–1381. doi: 10.1016/j.drudis.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Arteaga O., Álvarez A., Revuelta M., Santaolalla F., Urtasun A., Hilario E. Role of Antioxidants in Neonatal Hypoxic-Ischemic Brain Injury: New Therapeutic Approaches. Int. J. Mol. Sci. 2017;18:e265. doi: 10.3390/ijms18020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagberg H., Mallard C., Ferriero D.M., Vannucci S.J., Levison S.W., Vexler Z.S., Gressens P. The role of inflammation in perinatal brain injury. Nat. Rev. Neurol. 2015;11:192–208. doi: 10.1038/nrneurol.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalak L.F., Rollins N., Morriss M.C., Brion L.P., Heyne R., Sánchez P.J. Perinatal acidosis and hypoxic-ischemic encephalopathy in preterm infants of 33 to 35 weeks’ gestation. J. Pediatr. 2012;160:388–394. doi: 10.1016/j.jpeds.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins R.D., Raju T., Edwards A.D., Azzopardi D.V., Bose C.L., Clark R.H., Ferriero D.M., Guillet R., Gunn A.J., Hagberg H. Hypothermia and other treatment options for neonatal encephalopathy: an executive summary of the Eunice Kennedy Shriver NICHD workshop. J. Pediatr. 2011;159:851–858.e1. doi: 10.1016/j.jpeds.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B., Concepcion K., Meng X., Zhang L. Brain-immune interactions in perinatal hypoxic-ischemic brain injury. Prog. Neurobiol. 2017;159:50–68. doi: 10.1016/j.pneurobio.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas-Escobar M., Weiss M.D. Hypoxic-ischemic encephalopathy: a review for the clinician. JAMA Pediatr. 2015;169:397–403. doi: 10.1001/jamapediatrics.2014.3269. [DOI] [PubMed] [Google Scholar]
- 10.Meloni B.P. Pathophysiology and Neuroprotective Strategies in Hypoxic-Ischemic Brain Injury and Stroke. Brain Sci. 2017;7:e110. doi: 10.3390/brainsci7080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponnusamy V., Yip P.K. The role of microRNAs in newborn brain development and hypoxic ischaemic encephalopathy. Neuropharmacology. 2019;149:55–65. doi: 10.1016/j.neuropharm.2018.11.041. [DOI] [PubMed] [Google Scholar]
- 12.Cui H., Yang L. Analysis of microRNA expression detected by microarray of the cerebral cortex after hypoxic-ischemic brain injury. J. Craniofac. Surg. 2013;24:2147–2152. doi: 10.1097/SCS.0b013e3182a243f3. [DOI] [PubMed] [Google Scholar]
- 13.Ding X., Sun B., Huang J., Xu L., Pan J., Fang C., Tao Y., Hu S., Li R., Han X. The role of miR-182 in regulating pineal CLOCK expression after hypoxia-ischemia brain injury in neonatal rats. Neurosci. Lett. 2015;591:75–80. doi: 10.1016/j.neulet.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 14.Long J.M., Ray B., Lahiri D.K. MicroRNA-339-5p down-regulates protein expression of β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J. Biol. Chem. 2014;289:5184–5198. doi: 10.1074/jbc.M113.518241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., Wang Y., He Z. Glycine-Histidine-Lysine (GHK) Alleviates Neuronal Apoptosis Due to Intracerebral Hemorrhage via the miR-339-5p/VEGFA Pathway. Front. Neurosci. 2018;12:644. doi: 10.3389/fnins.2018.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhiraj D.K., Chrysanthou E., Mallucci G.R., Bushell M. miRNAs-19b, -29b-2* and -339-5p show an early and sustained up-regulation in ischemic models of stroke. PLoS ONE. 2013;8:e83717. doi: 10.1371/journal.pone.0083717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makki N., Capecchi M.R. Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev. Biol. 2011;357:295–304. doi: 10.1016/j.ydbio.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland P.W., Takahashi T. The evolution of homeobox genes: Implications for the study of brain development. Brain Res. Bull. 2005;66:484–490. doi: 10.1016/j.brainresbull.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Canu E., Boccardi M., Ghidoni R., Benussi L., Duchesne S., Testa C., Binetti G., Frisoni G.B. HOXA1 A218G polymorphism is associated with smaller cerebellar volume in healthy humans. J. Neuroimaging. 2009;19:353–358. doi: 10.1111/j.1552-6569.2008.00326.x. [DOI] [PubMed] [Google Scholar]
- 20.Paraguison R.C., Higaki K., Yamamoto K., Matsumoto H., Sasaki T., Kato N., Nanba E. Enhanced autophagic cell death in expanded polyhistidine variants of HOXA1 reduces PBX1-coupled transcriptional activity and inhibits neuronal differentiation. J. Neurosci. Res. 2007;85:479–487. doi: 10.1002/jnr.21137. [DOI] [PubMed] [Google Scholar]
- 21.Roberts T.C., Morris K.V., Wood M.J. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao F., Qu Y., Liu J., Liu H., Zhang L., Feng Y., Wang H., Gan J., Lu R., Mu D. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Neonatal Rats Following Hypoxic-ischemic Brain Damage. Sci. Rep. 2015;5:13850. doi: 10.1038/srep13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong J., Jiang L., Huang Z., Zhang H., Cheng C., Liu H., He J., Wu J., Darwazeh R., Wu Y., Sun X. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav. Immun. 2017;65:183–194. doi: 10.1016/j.bbi.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Das S., Ghosal S., Sen R., Chakrabarti J. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS ONE. 2014;9:e98965. doi: 10.1371/journal.pone.0098965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng T., Jia Y.J., Wen Q.Q., Guan W.J., Zhao E.Y., Zhang B.A. [Expression of microRNA in neonatal rats with hypoxic-ischemic brain damage] Zhongguo Dang Dai Er Ke Za Zhi. 2010;12:373–376. [PubMed] [Google Scholar]
- 26.Yang L.J., Wang J., Tian Z.F., Yuan Y.F. Shenfu injection attenuates neonatal hypoxic-ischemic brain damage in rat. Neurol. Sci. 2013;34:1571–1574. doi: 10.1007/s10072-013-1288-x. [DOI] [PubMed] [Google Scholar]
- 27.Ge X.T., Lei P., Wang H.C., Zhang A.L., Han Z.L., Chen X., Li S.H., Jiang R.C., Kang C.S., Zhang J.N. miR-21 improves the neurological outcome after traumatic brain injury in rats. Sci. Rep. 2014;4:6718. doi: 10.1038/srep06718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan Q., Sun W., Yuan H., Mu X. MicroRNA-135b-5p prevents oxygen-glucose deprivation and reoxygenation-induced neuronal injury through regulation of the GSK-3β/Nrf2/ARE signaling pathway. Arch. Med. Sci. 2018;14:735–744. doi: 10.5114/aoms.2017.71076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H., Huan L., Yin J., Qin M., Zhang Z., Zhang Z., Zhang J., Wang S. Role of microRNA-130a in myocardial hypoxia/reoxygenation injury. Exp. Ther. Med. 2017;13:759–765. doi: 10.3892/etm.2016.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G., Wang T., Wang T., Song J., Zhou Z. Effects of apoptosis-related proteins caspase-3, Bax and Bcl-2 on cerebral ischemia rats. Biomed. Rep. 2013;1:861–867. doi: 10.3892/br.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong L.C., Xu H.M., Guo G.L., Zhang T., Shi J.W., Chang C. Long Non-Coding RNA H19 Protects H9c2 Cells against Hypoxia-Induced Injury by Targeting MicroRNA-139. Cell. Physiol. Biochem. 2017;44:857–869. doi: 10.1159/000485354. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L., Hua C., Li Y., Sun Q., Wu W. miR-525-5p inhibits ADAMTS13 and is correlated with Ischemia/reperfusion injury-induced neuronal cell death. Int. J. Clin. Exp. Med. 2015;8:18115–18122. [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Zhao Y., Cheng M., Qiao Y., Wang Y., Xiong W., Yue W. Suppression of microRNA-144-3p attenuates oxygen-glucose deprivation/reoxygenation-induced neuronal injury by promoting Brg1/Nrf2/ARE signaling. J. Biochem. Mol. Toxicol. 2018;32:e22044. doi: 10.1002/jbt.22044. [DOI] [PubMed] [Google Scholar]
- 34.Han S., Liu Z., Wang Y., Wang L., Yao B., Guo C., Song T., Tu K., Liu Q. MicroRNA-577 inhibits the migration and invasion of hepatocellular carcinoma cells by targeting homeobox A1. Oncol. Rep. 2018;39:2987–2995. doi: 10.3892/or.2018.6388. [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Li Y., Qi W., Zhang N., Sun M., Huo Q., Cai C., Lv S., Yang Q. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells. Oncotarget. 2015;6:32737–32747. doi: 10.18632/oncotarget.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilicdag H., Daglioglu Y.K., Erdogan S., Zorludemir S. Effects of caffeine on neuronal apoptosis in neonatal hypoxic-ischemic brain injury. J. Matern. Fetal Neonatal Med. 2014;27:1470–1475. doi: 10.3109/14767058.2013.878694. [DOI] [PubMed] [Google Scholar]
- 37.Yan W., Chen Z.Y., Chen J.Q., Chen H.M. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 2018;496:1019–1024. doi: 10.1016/j.bbrc.2017.12.149. [DOI] [PubMed] [Google Scholar]
- 38.Simchovitz A., Hanan M., Niederhoffer N., Madrer N., Yayon N., Bennett E.R., Greenberg D.S., Kadener S., Soreq H. NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J. 2019;33:11223–11234. doi: 10.1096/fj.201900830R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunwoo J.S., Lee S.T., Im W., Lee M., Byun J.I., Jung K.H., Park K.I., Jung K.Y., Lee S.K., Chu K., Kim M. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington’s Disease. Mol. Neurobiol. 2017;54:1577–1586. doi: 10.1007/s12035-016-9928-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z.W., Zheng L.J., Ren X., Li A.P., Zhou W.S. LncRNA NEAT1 facilitates survival and angiogenesis in oxygen-glucose deprivation (OGD)-induced brain microvascular endothelial cells (BMECs) via targeting miR-377 and upregulating SIRT1, VEGFA, and BCL-XL. Brain Res. 2019;1707:90–98. doi: 10.1016/j.brainres.2018.10.031. [DOI] [PubMed] [Google Scholar]
- 41.Makki N., Capecchi M.R. Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev. Biol. 2010;341:499–509. doi: 10.1016/j.ydbio.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W., Chen X., Zhang Y. Effects of microRNA-21 and microRNA-24 inhibitors on neuronal apoptosis in ischemic stroke. Am. J. Transl. Res. 2016;8:3179–3187. [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu F., Liu J.L., Li J.P., Xiao F., Zhang Z.X., Zhang L. MicroRNA-124 (miR-124) regulates Ku70 expression and is correlated with neuronal death induced by ischemia/reperfusion. J. Mol. Neurosci. 2014;52:148–155. doi: 10.1007/s12031-013-0155-9. [DOI] [PubMed] [Google Scholar]
- 44.Vinciguerra A., Formisano L., Cerullo P., Guida N., Cuomo O., Esposito A., Di Renzo G., Annunziato L., Pignataro G. MicroRNA-103-1 selectively downregulates brain NCX1 and its inhibition by anti-miRNA ameliorates stroke damage and neurological deficits. Mol. Ther. 2014;22:1829–1838. doi: 10.1038/mt.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayuk S.M., Abrahamse H., Houreld N.N. The role of photobiomodulation on gene expression of cell adhesion molecules in diabetic wounded fibroblasts in vitro. J. Photochem. Photobiol. B. 2016;161:368–374. doi: 10.1016/j.jphotobiol.2016.05.027. [DOI] [PubMed] [Google Scholar]