Summary
The design and development of specific recognition and sensing systems for biologically important anionic species has received growing attention in recent years, as they play significant roles in biology, pharmacy, and environmental sciences. Herein, a new supramolecular sensing probe L1 was developed for highly selective differentiation of nucleotides. L1 displayed extremely marked absorption and emission differentiation upon binding with nucleotide homologs of AMP, ADP, and ATP, due to the divergent spatial orientations of guests upon binding, which allowed for a naked-eye colorimetric differentiation for nucleotides. A differentiating mechanism was unambiguously rationalized by using various spectroscopic studies and theoretical calculations. Furthermore, we successfully demonstrated that L1 can be applied to the real-time monitoring of the enzyme-catalyzed phosphorylation/dephosphorylation processes and thus demonstrated an unprecedented visualizable strategy for selectively differentiating the structurally similar nucleotides and real-time monitoring of biological processes via fluorescent and colorimetric changes.
Subject Areas: Analytical Chemistry, Analytical Chemistry Applications, Chemistry
Graphical Abstract

Highlights
-
•
Bis-quinoline CDXs as sensing probes to differentiate the nucleotide homologs in water
-
•
Its responses in the visible range allowed for a naked-eye colorimetric differentiation
-
•
The mechanism is proposed based on divergent spatial orientations of guests binding
-
•
Differentiation is realized by in situ enzymatic ATP hydrolysis and AMP phosphorylation
Analytical Chemistry; Analytical Chemistry Applications; Chemistry
Introduction
Ubiquitously present enzymatic active sites, where target substrates are selectively extracted from a complex mixture (e.g., biological fluid) and transformed with efficient rate accelerations, predetermined geometry (beneficial orientation and conformation), and superb stereocontrol under physiological conditions (at ca 36°C, in aqueous media at pH 7), represent the paradigmatic ideal for supramolecular chemistry (Kirby, 1996, Cacciapaglia et al., 2004, Ringe and Petsko, 2008). The construction of various artificial supramolecular receptors that mimic enzymes with highly selective binding of biologically important molecules in aqueous media has received substantial attention in recent years (Hembury et al., 2008, Liu and Bonizzoni, 2014, Zhou et al., 2011, Hargrove et al., 2011, You et al., 2015, Busschaert et al., 2015, O'Neil and Smith, 2006, Martinez-Manez and Sancenon, 2003, Molina et al., 2017). Cyclodextrins (CDXs), which are prominent naturally occurring host molecules that possess tunable hydrophobic cavities capable of selectively binding specific substrates in water via non-covalent interactions, are of particular interest in this regard (Szejtli, 1998, Rekharsky and Inoue, 1998). These fascinating properties of CDXs can be further enhanced by selective modifications that enable them to be used in various applications such as catalysis, enzyme mimetic, and molecular sensing (You et al., 2015, Busschaert et al., 2015, O'Neil and Smith, 2006, Martinez-Manez and Sancenon, 2003, Molina et al., 2017, Ogoshi and Harada, 2008, Szente and Szeman, 2013, Hapiot et al., 2012). This CDX-based supramolecular sensing approach, in which selectivity and sensitivity can be tuned to the desired analyte by choosing the specific cavity size, shape, and charge complementarity, has apparent advantages over conventional analytical chemistry techniques and instruments, such as ionic conductivity, mass spectrometry, electrochemical sensing, bio-polymer-based sensing, and indicator displacement methods (You et al., 2015). Taking advantage of multiple weak non-covalent interactions involved in the formation of supramolecular complexes, CDX-based systems are able to produce novel and important practical functions.
Nucleotides are composed of a five-carbon sugar (ribose or deoxyribose) unit connected to a hydrophobic nitrogenous base at the anomeric carbon and one to three negatively charged phosphate groups on the primary hydroxyl moiety. Among all nucleotides, adenosine 5′-monophosphate (AMP), adenosine 5′-diphosphate (ADP), and adenosine 5′-triphosphate (ATP) are particularly important since these nucleotides play a decisive and fundamental role in metabolism at the cellular level and they are involved in several processes including cellular signal transmission and transduction, modulation of ion channels, bioenergetics, and the transfer of genetic information (Knowles, 1980, Berg et al., 2002). ATP is a universal energy supplier in living cells, with the required energy for the cell accomplished by metabolic conversion of the nucleotide back into its precursors ADP and/or AMP (Knowles, 1980, Berg et al., 2002). Therefore, effective differentiation of ATP from ADP and AMP by artificial sensors is highly important to quantify the amount of each of these nucleotides in a cell. Based on the ATP/AMP ratio, one can understand and control the accompanying metabolic pathways in the cell that generate and consume ATP. Indeed, this is one of the most efficient methods to determine the amount of energy required for a cell. Furthermore, as ADP and ATP have different biochemical roles in metabolic processes, their interaction with proteins and other biomolecules are varied. Remarkably, PII proteins are one of the most widely distributed families of signal transduction proteins in nature and their competitive interaction with ATP and ADP serves as a sensor for the “energy charge” of bacteria (Wolfe et al., 2007, Jiang and Ninfa, 2007, Huergo et al., 2013). Therefore, sensing and distinguishing these nucleotides using supramolecular fluorescent probes (in aqueous medium) is an urgently needed yet challenging task for researchers, and the development of such a sensor is in high demand. It is important to note that these specially designed supramolecular receptors should distinguish the structurally similar nucleotides based on their different total anionic charge densities via differential binding affinities.
Many selective fluorescent and/or colorimetric chemosensing probes for adenosine nucleotides have been developed (Zhou et al., 2011, Martinez-Manez and Sancenon, 2003, Ramaiah et al., 2010, Zhao and Huang, 2010, García-Espana et al., 2012, Wu et al., 2017, Agafontsev et al., 2019, Ojida et al., 2006, McCleskey et al., 2003, Xu et al., 2009, Huang et al., 2015, Kumar et al., 2014a, Kumar et al., 2014b, Shi et al., 2013, Das et al., 2017, Sancenon et al., 2001, Jose et al., 2007, Li et al., 2005, Wang et al., 2016). A singular sensing probe with a differential signaling response to the three phosphorylation states of adenosine, however, has not been reported. Indeed, the design of most previously reported receptors is mainly focused on targeting the negatively charged phosphate group via interactions with a metal or cationic unit tethered to a fluorophore. Moreover, such sensors are typically employed in organic or aqueous-organic media (You et al., 2015, Busschaert et al., 2015, O'Neil and Smith, 2006, Martinez-Manez and Sancenon, 2003, Molina et al., 2017, Ramaiah et al., 2010, Zhao and Huang, 2010, García-Espana et al., 2012, Wu et al., 2017, Agafontsev et al., 2019, Ojida et al., 2006, McCleskey et al., 2003, Xu et al., 2009, Huang et al., 2015, Kumar et al., 2014a, Kumar et al., 2014b, Shi et al., 2013, Das et al., 2017, Sancenon et al., 2001, Jose et al., 2007); while only a few systems are able to function in a complex aqueous media though being not suited for practical applications (Ramaiah et al., 2010, Zhao and Huang, 2010, García-Espana et al., 2012, Wu et al., 2017, Agafontsev et al., 2019, Kumar et al., 2014a, Kumar et al., 2014b, Shi et al., 2013, Das et al., 2017, Sancenon et al., 2001). Furthermore, when designing an aqueous media nucleotide sensor system, the selectivity for nucleotides over related inorganic phosphate analogues with similar anionic charge densities, such as phosphate (Pi), diphosphate (PPi), and triphosphate (triPi, Scheme 1), must be taken into account. Many studies have collectively reported on selective binding and diverse recognition interactions between positively charged CDXs and various nucleotide phosphates (Hargrove et al., 2011, Eliseev and Schneider, 1993, Eliseev and Schneider, 1994, Schwinte et al., 1998, Vizitiu and Thatcher, 1999, Cotner and Smith, 1998, Hauser et al., 2000, Mourtzis et al., 2007, Yuan et al., 2007, Aggelidou et al., 2009, Yang et al., 2015, Formoso, 1973, Formoso, 1974). Nevertheless, such highly adaptable systems have yet to yield selective chemosensors for real-time applications. Recently, Hayashita and co-workers disclosed a guest-targeting azo/coumarin-modified CDX-copper complex as a probe for distinguishing ATP from AMP and ADP (Fujita et al., 2017, Yamada et al., 2018). The majority of known sensors, however, suffer from a low selectivity and sensitivity toward structurally similar nucleotides, and they demonstrate significantly reduced binding affinity in complex aqueous media, thus making them inappropriate for applications under biologically relevant conditions (Ramaiah et al., 2010, Zhao and Huang, 2010, García-Espana et al., 2012, Wu et al., 2017, Agafontsev et al., 2019). There are only a few examples of selective nucleotide sensors available (You et al., 2015, Busschaert et al., 2015, O'Neil and Smith, 2006, Martinez-Manez and Sancenon, 2003, Molina et al., 2017, Ramaiah et al., 2010, Zhao and Huang, 2010, García-Espana et al., 2012, Wu et al., 2017, Agafontsev et al., 2019, Ojida et al., 2006, McCleskey et al., 2003, Xu et al., 2009, Huang et al., 2015, Kumar et al., 2014a, Kumar et al., 2014b, Shi et al., 2013, Das et al., 2017, Sancenon et al., 2001, Jose et al., 2007), and these examples still are not well suited to the requirements of chemosensing probes, owing to poor solubility in pH-buffered solutions, competitive interference from similar anionic analytes, inefficient detecting ability in competitive aqueous media, poor bio-compatibility, unacceptably low bio-sensing responses in cells and biological fluids, or response in high analyte concentrations. Therefore, the design and development of a single supramolecular sensor that is able to distinguish between structurally similar nucleotides based on their different total anionic charge densities is a fascinating goal for many researchers working in biological and supramolecular chemistry.
Scheme 1.
Structures of the Supramolecular Receptors and Analytes.
Structure of the (A) designed supramolecular A/B/GCDX-QUI-2 receptors and (B) nucleotides (AMP, ADP, and ATP) and inorganic phosphates (Pi, PPi, and triPi).
Indeed, introduction of fluorophores with functional groups to the primary or secondary side arms of CDXs gives two advantages for the development of selective nucleotide probes: an external additional binding site and a fluorogenic signaling unit for guest molecules. Quinoline motifs frequently are used as sensing probes (Meng et al., 2012, Tang et al., 2016, Mikata et al., 2017, Pramanik and Das, 2009, Cai et al., 2013) for various analytes owing to their desirable structure and unique photophysical properties such as relatively high photostability, long-wavelength absorption, and moderate fluorescence emission. Therefore, many mono-quinoline-appended CDX derivatives have been used as sensing probes for cations and neutral analytes (Liu et al., 2003, Liu et al., 2004, Liu et al., 2007a, Liu et al., 2007b, Liu et al., 2009, Liu and Bonizzoni, 2014, Yu et al., 2010, Chen et al., 2007, Zhai et al., 2013, Li et al., 2014, Cheng and Dong, 2016, Cheng et al., 2015). Herein, for the first time, we designed bis-quinoline receptors, A/B/GCDX-QUI-2 (Scheme 1), which contain two main recognition sites: the CDX cavity as a hydrophobic guest binding site and the appended mono-N-bis-(8-methylaminoquinolyl) unit acting simultaneously as a fluorophore and metal-binding unit. This combination is indispensable to the substrate/analyte-specific response function. In the present study, the copper-coordinated BCDX-QUI-2 receptor (probe L1) was used as a differentiating fluorescence and colorimetric sensor for detecting adenosine-based nucleotides (AMP, ADP, and ATP), and it demonstrated significant selectivity toward AMP and ATP in aqueous media. A unique binding mechanism is proposed on the basis of UV-vis absorption, fluorescence measurements, NMR experiments, and theoretical simulations.
Results and Discussion
Conformational Analysis of Sensing Receptors
In general, the cavity of a CDX host provides a chiral microenvironment to the tethered achiral chromophoric moiety and generates a corresponding induced circular dichroism (ICD) signal (Harata and Uedaira, 1975, Kajtar et al., 1982, Kodaka, 1993) that depends on the mutual spatial arrangement of the CDX axis and chromophore position. It is important to note that the geometry of molecules with two chromophores linked to a single nitrogen atom of amine as coordinating functional group attaching to the chiral carbon could be controlled by oxidation state of complexing metal ion, hence resulting in the opposite sign of exciton-coupled circular dichroism (ECCD) spectra, which was dependent upon the cationic oxidation form (Harada and Nakanishi, 1972, Zahn and Canary, 1999, Zahn and Canary, 2000, Canary et al., 1998). We herein synthesized the A/B/GCDX-QUI-2 receptors with different cavity sizes and analyzed the effect of the CDX cavity size and solvent used on the conformation of the tethered mono-N-bis-(8-methylquinolyl) chromophoric moiety.
The supramolecular receptors, A/B/GCDX-QUI-2 (Scheme 1), were synthesized by reacting two equivalents of 8-(bromomethyl)quinoline with the respective mono-6-amino-α/β/γ-CDXs (see Scheme S3). The synthetic A/B/GCDX-QUI-2 derivatives were fully characterized by using various analytical techniques (see Figures S4–S19), and their conformations in water and methanol were analyzed by CD (Harata and Uedaira, 1975, Kajtar et al., 1982, Kodaka, 1993) and 1H and 2D NMR spectroscopic studies. The UV-vis absorption spectra of A/B/GCDX-QUI-2 have similar absorption maxima at 206, 226 (shoulder), 284, and 315 nm and a single emission band observed at 427 nm upon excitation (λexc) at 315 nm in both water and methanol (Figures 1 and S23). The absorption bands at 226, 284, and 315 nm are attributed to the long (1Bb) and short (1La,b)-axis polarized π-π∗ transitions of the quinoline chromophore (Meng et al., 2012, Tang et al., 2016, Mikata et al., 2017, Pramanik and Das, 2009, Cai et al., 2013, Liu et al., 2004, Liu et al., 2007a, Liu et al., 2007b, Liu et al., 2009, Chen et al., 2007, Zhai et al., 2013, Li et al., 2014, Cheng and Dong, 2016, Cheng et al., 2015). As shown in Figure 1, the chiroptical behaviors in water and methanol of the three CDX-QUI-2 sensors depend significantly upon the CDX cavity size.
Figure 1.
Conformational Analysis of Sensing Receptors
Circular dichroism and UV-vis absorption spectra of receptors (A) ACDX-QUI-2 (3.95 × 10−5 M), (B) BCDX-QUI-2 (3.68 × 10−5 M), and (C) GCDX-QUI-2 (3.17 × 10−5 M) in water and methanol at 25°C. (D) Schematic representation of the quinoline chromophore conformations proposed by applying the empirical rule on the ICD phenomena of CDX complexes (Harata and Uedaira, 1975, Kajtar et al., 1982, Kodaka, 1993) and the exciton chirality theory (Harada and Nakanishi, 1972, Zahn and Canary, 1999, Zahn and Canary, 2000, Canary et al., 1998) to the CD spectra of A/B/GCDX-QUI-2.
In the CD spectra of A/B/GCDX-QUI-2 derivatives (Figure 1), the ICD signals of the two quinoline chromophores appeared at ∼230 and 290 nm and are attributed to the corresponding 1Bb and 1La,b electronic transitions. ACDX-QUI-2 has almost the same CD profile in both water and methanol, and this result is indicative of conformational similarities of the quinoline moieties in either solvent (Figure 1A). BCDX-QUI-2 and GCDX-QUI-2, on the other hand, demonstrate different chiroptical behavior in these two solvents, apparently as a result of solvent-dependent conformational variability. Specifically, the CD spectra of BCDX-QUI-2 and GCDX-QUI-2 in water consist of a negative couplet (a weak negative Cotton effect [CE] signal located at low energy of the spectrum followed by a strong positive CE signal located at high energy of the spectrum) appearing around 234 and 233 nm, respectively (Figures 1B and 1C). According to the empirical rule on the ICD phenomena of CDX complexes (Harata and Uedaira, 1975, Kajtar et al., 1982, Kodaka, 1993), the observed CD signals of BCDX-QUI-2 and GCDX-QUI-2 indicate that the anchored quinoline chromophores locate inside the CDX cavities with the long 1Bb transition band being parallel to the CDX axis and the short 1La,b transition band being almost vertical to the CDX axis. Both the quinoline chromophores (in GCDX-QUI-2) or only one quinoline chromophore (in BCDX-QUI-2) accommodated inside the CDX cavity (see below) resulted in the counterclockwise orientation of its electronic transitions that correspond to the negative exciton chirality ECCD signals (a negative CE followed by positive CE, reading from a longer to shorter wavelength) according to the exciton chirality method (Harada and Nakanishi, 1972, Zahn and Canary, 1999, Zahn and Canary, 2000, Canary et al., 1998). In methanol, BCDX-QUI-2 shows a relatively strong negative couplet, hence indicating that both of the quinoline chromophores are outside the CDX cavity and are relatively in proximity to each other. This conformational mode is a result of the high solvation and its location at the primary hydroxyl rim with the long 1Bb transitions being slightly tilted to the CDX axis. Similarly, because of its smaller cavity size, ACDX-QUI-2 gives a strong negative exciton couplet in both water and methanol, implying that both quinoline chromophores locate outside of the CDX cavity (Figure 1A). The weak positive CE peak at around 239 nm observed for GCDX-QUI-2 in methanol indicates that both of the quinoline chromophores are not completely excluded from the CDX cavity. This is because the large accommodating cavity size of the γ-CDX host results in the parallel orientation of the 1Bb electronic transitions to the CDX axis, hence producing a positive CD signal (Figure 1C).
The solvent-dependent conformational change of the three CDX-QUI-2 derivatives when transitioning from deuterated water to deuterated methanol was further analyzed by NMR measurements (Figures 2 and S24–S30). The 1H NMR spectra of the CDX-QUI-2 derivatives show complicated patterns with multiple overlapped signals from the CDX protons appearing between 3.0 and 4.2 ppm and well-resolved quinoline protons observed further downfield (7.0–9.0 ppm, Figures S24–S30). The CDX protons of both BCDX-QUI-2 and GCDX-QUI-2 in D2O experienced corresponding up-field shifts observed at around 3.0–3.25 ppm in comparison with those in CD3OD. This clearly indicates that the appended quinoline chromophore is self-included into the CDX cavity as a predominant conformer (Figure 1D). The self-included pattern of BCDX-QUI-2 was also confirmed by the appearance of cross peaks between the quinoline protons and adjacent CDX protons in the 2D ROESY spectrum (Figures 2 and S28A). In CD3OD, the absence of these up-field shifted protons from CDX and the broadening of the quinoline signals indicate that both the quinoline moieties are located outside the cavity. This proposal was supported by the absence of the corresponding cross contour peaks in the 2D ROESY spectrum (Figure S28B). ACDX-QUI-2 also shows the absence of the up-field shift for the CDX protons. This feature, along with the strong negative exciton couplet signals in both water and in methanol, clearly indicate that ACDX-QUI-2 exists as a single conformer with both the quinoline chromophores located outside the CDX cavity (Figure 1D).
Figure 2.
1H–1H ROESY Spectrum of BCDX-QUI-2 in D2O at 25°C with a Mixing Time of 250 ms (400 MHz) and Proposed Conformation
The Q1 proton of quinoline moieties proved to be the best conformational indicator. Hence, in all CDX-QUI-2 derivatives the proton provided a sharp and intense signal in D2O, whereas in CD3OD, the resulting signal was less intense. This is evidence that the quinoline chromophores are expelled from the CDX cavity via strong solvation in the organic solvent. The expelled quinoline subunits can interchange their positions at a rate substantially faster than the NMR timescale, hence resulting in less intense and broadened NMR signals for the Q1 proton. According to the aforementioned discussion about conformational switching, in aqueous solution, the hydrophobic quinoline chromophores are expected to be immersed in the chiral microenvironment of appropriately sized CDX cavities, but this is not necessarily when using less polar methanol as solvent. Combining the results from CD and NMR experiments provides strong support for the existence of the proposed A/B/GCDX-QUI-2 conformers in both water and methanol (Figure 1D).
The relationship between solvent and conformation was further investigated for BCDX-QUI-2. In organic solvents such as CDCl3, DMSO-d6, CD3OD, and acetone-d6, the primary conformation of BCDX-QUI-2 appears to be one in which the quinoline chromophores are expelled from the chiral hydrophobic CDX cavity (Figure S31). It should be noted that the presence of small organic molecules such as isopropanol and ethyl acetoacetate do not affect the predominant self-included conformer of BCDX-QUI-2 in D2O, with the corresponding co-inclusion complexes forming instead (Figure S32).
Application of Adenosine Nucleotide Sensing Probe
Although positively charged CDXs with mono/per-aminomethyl and guanidine moieties have been reported to strongly bind nucleotides like AMP, ADP, and ATP (Eliseev and Schneider, 1993, Eliseev and Schneider, 1994, Schwinte et al., 1998, Vizitiu and Thatcher, 1999, Cotner and Smith, 1998, Hauser et al., 2000, Mourtzis et al., 2007, Yuan et al., 2007, Aggelidou et al., 2009, Yang et al., 2015, Formoso, 1973, Formoso, 1974), the design and construction of such supramolecular systems as chemical sensors for nucleotides under practical applications remains a challenging task. Introduction of fluorophores to the primary or secondary side arms of CDXs may provide additional binding sites and fluorogenic signaling units, which can be used as artificial sensing receptors for biologically important small molecules and anions (Zhong et al., 2009, Ngo et al., 2012, Kumar et al., 2012, Kumar et al., 2014a, Kumar et al., 2014b). The fluorogenic bis-quinoline motifs in A/B/GCDX-QUI-2 are able to form the corresponding chelated complexes with Cu(II) and Zn(II) ions (Schemes 2 and S5, Figures S33–S36), which can be used as sensing and differentiating receptors for nucleotides and inorganic phosphates. As will be detailed below, the selective response of the BCDX-QUI-2:Cu(II) complex L1 toward various adenosine-based nucleotides in an aqueous medium was clearly demonstrated via UV-vis absorption and fluorescence spectroscopic studies and further supported by control experiments, 1H and 31P NMR spectroscopy, mass spectrometry analysis, and theoretical simulations.
Scheme 2.
Structures of Probes L1-L4
Schematic structures for probes L1 and L1(Zn), as well as control compounds L2-L4 (green cone, CDX moiety; purple, quinoline groups; dark blue, chelating nitrogen atoms; light blue, Cu(II) or Zn(II) cation).
Selective Response of Probe L1 in the Presence of Various Nucleotides and Inorganic Phosphates
The selective response of β-CDX-derived probe L1 toward various adenosine nucleotides and inorganic phosphates was investigated by using UV-vis and fluorescence titrations (Figures 3 and 4). The measurements were carried out by adding five equivalents of nucleotides and inorganic phosphates to a fixed amount of probe L1. Remarkably, among the analytes tested, only AMP exhibited an enhancement of the absorption maxima at 303 and 315 nm (Figure 3A), accompanied by a blue-shifted (from 427 to 377 nm, Δλem = 50 nm) and quenched fluorescence emission (Figure 4A). ADP and ATP, on the other hand, both showed a small hypochromic effect in the absorption spectra and enhanced fluorescence emission at 427 nm (λexc. = 315 nm). Inorganic phosphates exhibited a relatively negligible change in the absorption and the fluorescence emission of the probe L1. Most importantly, it was only upon addition of AMP to L1 that the aqueous solution turned from colorless to light blue with corresponding absorption maxima appearing at 303 and 315 nm and a less intense ligand-to-metal charge transfer (LMCT) band around >325 nm. The relative absorption and fluorescence changes for selective sensing and the differentiating response of the probe L1 in the presence of various nucleotides and inorganic phosphates are given in Figures 3B and 4B. This unique change and differential binding behavior of L1 with AMP, ADP, and ATP allows the molecular probe to be used as an effective sensor for discriminating between structurally similar nucleotides using either spectral means or even the naked eye.
Figure 3.
Absorption based Sensing Response of Probe L1 towards Various Analytes
(A) UV-vis absorption spectra of probe L1 (37 μM) in the absence (black) and presence of nucleotides (5 equiv; AMP [red], ADP [blue], ATP [magenta]) and inorganic phosphates (5 equiv; Pi [green], PPi [navy], and triPi [violet]) in water at room temperature.
(B) UV-vis absorption change (at 315 nm) of probe L1 (37 μM) toward the nucleotides (5 equiv) AMP, ADP, ATP and inorganic phosphates (5 equiv) Pi, PPi, and triPi in water at room temperature (green bars represents the sole absorbance of probe L1 and nucleotides at 315 nm; red bars represents the observed absorbance changes [at 315 nm] of probe L1 upon addition of nucleotides and inorganic phosphates, respectively).
Figure 4.
Fluorescence based Sensing Response of Probe L1 towards Various Analytes
(A) Fluorescence spectra of probe L1 (37 μM) in the absence (black) and presence of nucleotides (10 equiv; AMP [red], ADP [blue], ATP [magenta]) and inorganic phosphates (10 equiv; Pi [green], PPi [navy], and triPi [violet]) in water at room temperature (λexc = 315 nm).
(B) Fluorescence response (at 427 nm) of probe L1 (37 μM) toward the nucleotides (10 equiv) AMP, ADP, ATP and inorganic phosphates (10 equiv) Pi, PPi, and triPi in water at room temperature (green bars represents the sole fluorescence of probe L1 and nucleotides at 427 nm; red bars represents the observed fluorescence changes (at 427 nm) of probe L1 upon addition of nucleotides and inorganic phosphates, respectively).
Investigation of Sensing and Differentiating Binding Response of Probe L1 with AMP, ADP, and ATP
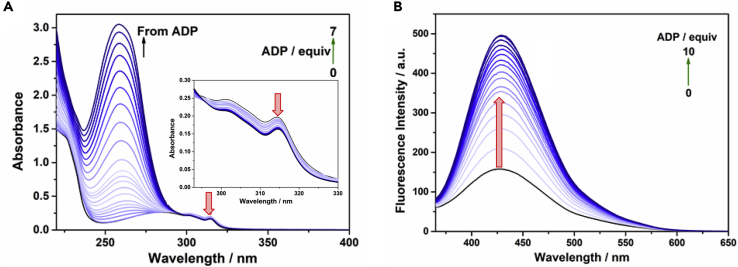
In order to verify the differentiating binding response of probe L1 with nucleotides AMP, ADP, and ATP, the corresponding absorption and emission titrations were carried out separately in water (Figures 5, 6, and 7). It was found that L1 shows a moderate fluorescence emission observed at 427 nm (λexc. = 315 nm) in water, which was blue shifted (from 427 to 377 nm, Δλem = 50 nm) and considerably quenched upon addition of 0–10 equivalents of AMP (Figure 5B), which is most probably due to the charge interaction between the adenine moiety and the quinoline chromophore when L1 binds with AMP (see “Theoretical Studies” for detail). In the UV-vis spectra there is a noticeable enhancement of the absorption maxima at 303 and 315 nm with small red shifts (∼2–5 nm) and new long wavelength band (>325 nm) upon addition of AMP (Figure 5A). Moreover, the color of the solution turned from colorless to light blue upon increasing the AMP concentration (Figure 5C). Indeed, Cu(II) ions having empty d-orbitals are able to form an LMCT ternary complex with the probe L1 and electron-rich phosphate group of AMP, hence exhibiting a longer wavelength absorption band and changing the solution color to light blue (Kim et al., 2009, Kim and Quang, 2007, Santis et al., 1995, de Silva et al., 2009, Guo et al., 2016, Wang et al., 2017). In contrast, addition of ADP and ATP to the probe L1 resulted in a significant enhancement of the fluorescence emission at 427 nm (λexc. = 315 nm) (Figures 6B and 7B), and they demonstrated hypochromic effects of absorption maxima at 303 and 315 nm in the absorption spectra (Figures 6A and 7A).
Figure 5.
Absorption and Fluorescence Spectral Changes of Probe L1 towards AMP
(A) Absorption and (B) fluorescence spectral changes (λexc. = 315 nm) of L1 (37 μM) upon addition of AMP (0–10 equiv) in water at room temperature; (C) expansion of Figure 5A in the region of >280 nm. Inset: Color change of probe L1 upon addition of AMP.
Figure 6.
Absorption and Fluorescence Spectral Changes of Probe L1 towards ADP
(A) Absorption and (B) fluorescence spectral changes (λexc. = 315 nm) of probe L1 (37 μM) upon addition of ADP (0–10 equiv) in water at room temperature. Inset: Expansion of Figure 6A in the region of 293–330 nm.
Figure 7.
Absorption and Fluorescence Spectral Changes of Probe L1 towards ATP
(A) Absorption and (B) fluorescence spectral changes (λexc = 315 nm) of probe L1 (37 μM) upon addition of ATP (0–10 equiv) in water at room temperature. Inset: Expansion of Figure 7A in the region of 293–330 nm.
It is important to emphasize that poor binding efficiency has been observed for nucleotides upon interaction with native CDXs. For example, the association constant (ka) of β-CDX with AMP is 41 M−1, through non-specific hydrophobic interactions (Formoso, 1973, Formoso, 1974). Nucleotide binding to CDX can be enhanced considerably, however, upon functionalization with a respective number of cationic amino groups on the primary side of the cyclic oligosaccharide framework (ka of per-methylamino-β-CDX with AMP and ATP are 1.26 × 105 M−1 and 3.24 × 106 M−1, respectively, whereas per-guanidino-β-CD with ADP or ATP form irreversible complexes due to the ionic/columbic interaction and salt bridge formation) (Hargrove et al., 2011, Eliseev and Schneider, 1993, Yuan et al., 2007). These results demonstrate that the sensing and binding response of probe L1 with adenosine nucleotides differed based on the length of anionic phosphate esters attached to the hydrophobic adenine moiety. This response depends mainly on the coordinating ability of the phosphoric anion group to the metal ion and the nature of host:guest (H:G) interaction of the adenine moiety with the CDX cavity. The calculated ka values of BCDX-QUI-2 with AMP, ADP, and ATP are 5.55 × 103, 443, and 308 M−1, respectively (Table S1 and Figure S38), which are considerably less in comparison with the ka values of probe L1 with the same nucleotides (4.24 × 104, 4.78 × 104, and 5.15 × 104 M−1, respectively; Table 1 and Figures S40–S42). This observation clearly indicates that the metal ion plays a key role in sensing and differentiating the binding response. It is interesting to find that the binding constants of nucleotides with L1 are very much similar, despite their different charge in nucleotides in water. This is most probably due to the unique binding mode of L1 with nucleotides, as only one phosphate group of nucleotides could bind the metal center of L1, whereas the hydrophobic part of the nucleotides that complexes with the CD cavity is exactly same.
Table 1.
Association Constants (ka) Determined for Probe L1 and Nucleotides in Aqueous Medium at Room Temperature
| Host | Guest | ka/M−1 |
|---|---|---|
| Probe L1 | AMP | 4.24 × 104 |
| Probe L1 | ADP | 4.78 × 104 |
| Probe L1 | ATP | 5.15 × 104 |
| Probe L1 | Other anionsa | – |
All of the nucleotides were added as sodium salts. ka was determined by fitting the absorption change in the UV-vis titration spectra of probe L1 upon addition of nucleotides.
Other anions—not determined.
Supramolecular Complex Stoichiometry
The stoichiometry of the supramolecular complexes are mainly found from the Job's method, which is a common method and powerful tool for the determination of the complex stoichiometry of an equilibria (Job, 1928). Job's plots based on absorption titrations suggest the formation of a 1:1 complex (Figure S43) between the probe L1 and nucleotides AMP, ADP, or ATP.
NMR Studies
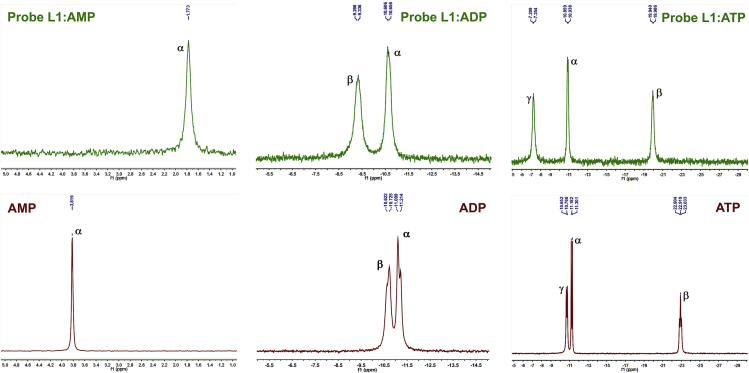
The origin behind the observed remarkable sensing and differentiating response of probe L1 with the nucleotides was investigated by using 1H NMR and 31P NMR spectroscopy. Since the paramagnetic Cu(II) ion of L1 is not suitable for the NMR studies, the analogous zinc complex L1(Zn) was used to examine the mode of binding of the formed complexes. It was found that the aromatic protons of quinoline are slightly shifted upfield and became broader upon sequential addition of AMP, ADP, and ATP. Additionally, the CDX protons (3.0–4.2 ppm) were completely broadened and also experienced a large upfield shift upon addition of the nucleotides (Figures S44–S46). This is due to the ring current effect from adenine aromatic ring strongly affecting the CDX protons. Upon complexation with AMP, the aromatic adenine signals of the included nucleotide base underwent upfield shifts. In the case of ADP complexation, signals for protons in the pyrimidine ring also showed an upfield shift, whereas signals from the imidazole ring experienced a downfield shift. For ATP complexation, the proton resonances of the aromatic adenine moiety were shifted downfield as a result of the interaction mode, in which the adenine moiety is located outside the CDX cavity and thus experienced the downfield shift. These observations clearly support the suggestion that the existence of significantly variable binding patterns of the adenine moiety and the involved H:G interactions between L1(Zn) and nucleotides are responsible for the differentiating and sensing response of probe L1(Zn) toward AMP, ADP, and ATP.
The 31P NMR spectra of the complexes of the probe L1(Zn) with AMP, ADP, and ATP were recorded separately (Figure 8). It was revealed that the respective α-, β-, and γ-phosphorus atoms of AMP, ADP, and ATP (Scheme 1) showed different chemical shifts with respect to their distance from the metal center and the H:G interaction with the CDX cavity of L1(Zn). In the case of AMP bound to L1(Zn), there is a relatively large upfield shift (Δδ = +2.046 ppm) of the α-P signal from −3.819 to −1.773 ppm, indicating that the phosphate group strongly coordinates to the metal ion. On the other hand, the P signals of ADP (two signals) and ATP (three signals) show downfield shifts upon binding to L1(Zn), which varied in extent with respect to the binding of terminal phosphate group to the Zn cation (Figure 8). For ADP, the α-P and β-P signals experienced a downfield shift (Δδ = −0.526 and −1.373 ppm, respectively). For both ADP and ATP, the shift of α-P resonance was relatively smaller than that of β-P resonance, hence indicating that the β-P atom of ADP strongly interacts with the Zn ion. Similarly, the α-, β-, and γ-P signals of ATP show downfield shifts (Δδ = −0.343, −2.956, and −3.478 ppm, respectively) upon complexation with the probe L1(Zn), also suggesting that the γ-P atom of ATP strongly interacts with the metal center. These observations indicate that only the more negatively charged terminal phosphate groups (owing to the presence of two negative charges) play a key role in binding to the Zn ion. On the basis of literature data (Eliseev and Schneider, 1993, Eliseev and Schneider, 1994, Schwinte et al., 1998, Vizitiu and Thatcher, 1999, Cotner and Smith, 1998, Hauser et al., 2000, Mourtzis et al., 2007, Yuan et al., 2007, Aggelidou et al., 2009, Yang et al., 2015, Formoso, 1973, Formoso, 1974, Zhong et al., 2009, Ngo et al., 2012) and the observed results, the following differentiating models for binding of L1 to AMP, ADP, and ATP can be illustrated schematically, as demonstrated in Scheme 3.
Figure 8.
31P NMR Spectra of Nucleotides and its Complexes with Probe L1(Zn)
31P NMR spectra of AMP, ADP, and ATP (lower panel) and its complexes with the probe L1(Zn) (upper panel)
Scheme 3.
Proposed Sense and Differentiating Mechanism
Proposed sense and differentiating binding patterns of L1 with AMP, ADP, and ATP in water.
Importance of the CDX Cavity and Its Size
The observed differential binding responses between L1 and the three different nucleotides, such as absorption changes, blue-shifted fluorescence quenching by AMP (Figure 5), hypochromic effects in absorption, and enhanced fluorescence emission at 427 nm by ADP and ATP (Figures 6 and 7), unambiguously indicate that the cavity of the β-CDX group in L1 plays a crucial role in this structural selectivity. To rationalize this result, a reference molecule without the CDX cavity (Bu-QUI-2) was synthesized (Schemes 2 and S4) and its Cu(II) complex (L4) was used to study the corresponding sensing and differentiating response. These control experiments were carried out in the presence and absence of native (non-modified) β-CDX serving as an external host cavity. Upon addition of the nucleotides to an aqueous solution of L4, we found a negligible variation in the absorption spectra (Figure S48), both with and without β-CDX (Figure S49). This result indicates a synergistic contribution between the hydrophobic cavity of β-CDX and the covalently linked bis-quinoline moiety in L1 toward generating the differentiating binding pattern and spectral response of the molecular probe. Additionally, control experiments with the complex between L1 and the strongly binding guest molecule 1-adamantanecarboxylic acid (1-AdCA) (Chen et al., 2007) were carried out so as to validate the importance of β-CDX cavity in the probe L1 for efficient binding of nucleotides. The L1:1-AdAC complex was titrated against nucleotides and showed no discernible spectral change (Figure S50). This result demonstrates that the strongly bound 1-AdCA effectively blocks access of the nucleotide to the CDX cavity, hence preventing the corresponding nucleotide binding.
In order to examine the effect of cavity size on the differential binding behavior of probe L1 to nucleotides, we repeated the nucleotide binding studies with α-CDX-based L2 and γ-CDX-derived probe L3, which possess smaller and larger cavity sizes, respectively, in comparison with L1 (Figures 9 and S47). The titration of L2 with the corresponding nucleotides showed an insignificant change in the absorption spectra and a small enhancement of the fluorescence emission upon addition of AMP, ADP, or ATP (Figures 9 and S47). Similar titration studies were also carried out with L3 showing even less variation in the absorption spectra and relatively small changes in the fluorescence spectra (Figures 9 and S47). These results demonstrate the requirements of the CDX cavity for effective nucleotide binding, which in turn depends upon the size of the cavity acting as a portal for the sensing and differentiating response.
Figure 9.
Absorption and Fluorescence Spectral Changes of Probe L1-L3 upon Binding with Analytes
Comparison of (A) absorption (ΔAbs.) and (B) fluorescence spectral changes (ΔEmis.; λexc. = 315 nm) of probe L1-3 upon addition of nucleotides (AMP, ADP, and ATP) and inorganic phosphates in aqueous medium at room temperature.
Theoretical Studies
The proposed differential binding mechanism is strongly supported by corresponding theoretical calculations. On the basis of these experimental results and in accordance with the literature data (Eliseev and Schneider, 1993, Eliseev and Schneider, 1994, Schwinte et al., 1998, Vizitiu and Thatcher, 1999, Cotner and Smith, 1998, Hauser et al., 2000, Mourtzis et al., 2007, Yuan et al., 2007, Aggelidou et al., 2009, Yang et al., 2015, Formoso, 1973, Formoso, 1974, Fujita et al., 2017, Yamada et al., 2018, Kim et al., 2009, Kim and Quang, 2007, Santis et al., 1995, de Silva et al., 2009, Guo et al., 2016, Wang et al., 2017, Butler, 2014, Hewitt et al., 2017, Wongkongkatep et al., 2017), the sensor probe L1 and the corresponding nucleotide complexes (L1:AMP/ADP/ATP) were examined by density functional theory (DFT) and time-dependent density functional theory (TD-DFT) calculations at the B3LYP/6-31G(d) level by using the Gaussian 09 program (Figures 10, S51, and S52) (Frisch et al., 2010). For BCDX-QUI-2, both the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) are distributed on the tethered N-bis-8-methylamino part and quinolyl scaffolds, respectively, with an energy difference (ΔE) of 4.93 eV that decreased down to 4.60 eV upon coordination with Cu2+ in the probe L1. For comparison, the HOMO and LUMO of AMP, ADP, and ATP are located at the adenine moiety with ΔE values decreasing in the following order: 5.43, 5.42, and 5.35 eV, respectively, which correlate with increasing the number of phosphate groups. For gaining more insights into the binding pattern between the probe L1 and these three nucleotides, the corresponding calculations were performed separately with the results shown in Figure 10.
Figure 10.
DFT Study of Probe L1: nucleotide Complexes
DFT optimized structures, HOMO-LUMO energy levels, and interfacial plots of the orbitals for the three proposed L1:nucleotide complexes
The energy minimized structures of the complexes were acquired from the design strategy and the experimental results discussed above via the coordination of the terminal phosphate group of the nucleotide to the metal ion of L1. The calculated ΔE values of the L1:AMP/ADP/ATP complexes (6.3764, 5.2567, and 4.8731 eV, respectively) decreased considerably upon the increases in the number of phosphate groups in the nucleotides. Furthermore, the HOMO electron clouds of these complexes were only distributed on the adenine moiety, whereas the LUMO electron cloud distributions varied with the differential binding patterns of the different nucleotides with the probe L1. The LUMO level electron clouds for the complex L1:AMP is distributed on the N-bis-8-methylamino part only, whereas for the L1:ADP/ATP complexes, the LUMO electron clouds distributed on both the N-bis-8-methylamino part and quinoline scaffolds. This indicates a good match between the molecular size of AMP and probe L1, hence allowing the adenine base moiety in AMP to occupy the hydrophobic cavity region of β-CDX via corresponding non-covalent interactions (ka = 4.24 × 104 M−1). In turn, the CDX walls restrict the electron density shift process from the adenine moiety to the quinoline chromophore, which is consistent with the observed results (quenched fluorescence emission, enhanced absorption maxima at 303 and 315 nm, and new long wavelength absorption band at >325 nm with blue coloration upon addition of AMP to the probe L1). Binding of the terminal phosphate groups of ADP and ATP to the metal center of probe L1 forces the adenine chromophore to sit outside of the CDX cavity, which simultaneously allows the electron density to be shifted from the adenine moiety to both the N-bis-8-methylamino part and the quinolyl chromophore. This shift in electron density also is consistent with the obtained experimental data (enhanced fluorescence emission, small hypochromic effect in the absorption maxima at 303 and 315 nm, and no optical color change upon addition of ADP or ATP to the probe L1). In addition, the TD-DFT ground and excited state calculations of L1 and its nucleotide complexes provide the corresponding simulated absorption (303 and 315 nm) and emission (for AMP and ADP/ATP, 377 and 427 nm, respectively) bands, which are in full agreement with the experimentally observed results. Therefore, the present study establishes an effective design strategy for the differentiation of nucleotides by using the probe L1.
Enzyme and Stimuli Responsive In Situ Monitoring of Optical Signaling, Enzymatic ATP Hydrolysis, and AMP Phosphorylation
As nucleotides play a vital role in most fundamental biological reactions catalyzed by enzymes (Schlattner et al., 2006, Zimmermann, 1999, Chen and Urban, 2016), monitoring such enzymatic activity in real time has a significant practical importance to biological and medical science. Thus, on the basis of the established differentiating ability of the probe L1 toward AMP, ATP, and ADP, a real-time monitoring of the enzymatic activity of the enzymes creatine kinase (CK, AMP/ADP → ATP) and apyrase (ATP → ADP → AMP) was tested (Scheme 4) (Kumar et al., 2012, Kumar et al., 2014a, Kumar et al., 2014b, Schlattner et al., 2006, Zimmermann, 1999, Chen and Urban, 2016).
Scheme 4.
Schematic Representation of Enzyme Catalyzed AMP/ADP Phosphorylation and ATP/ADP Hydrolysis Process
Two enzymatic reactions involving adenosine nucleotides and their products detected by using the differentiating probe L1, (A) creatine kinase (CK)-catalyzed phosphorylation of AMP and/or ADP using creatine phosphate (CP) to produce ATP and creatine, (B) hydrolytic enzyme apyrase-catalyzed dephosphorylation of ATP → ADP → AMP and Pi.
The enzyme-catalyzed phosphorylation and dephosphorylation reactions were carried out with and without the probe L1 and monitored by two different methods (Phosphorylation Method I/II (PM-I/II) and Dephosphorylation Method I/II (dPM-I/II); see Transparent Methods section in Supplemental Information and Figure S53). In both methods, under the catalysis by CK, ADP phosphorylated with creatine phosphate (CP) to produce ATP and creatine, and the fluorescence intensity of the probe L1 thus increased over time due to the formation of the L1: ATP complex (Figures 11A and 11B). CK-catalyzed AMP phosphorylation with CP is comparatively less feasible in comparison with ADP (Figures S54 and S55 in SI). The fluorescence spectra of L1 in the presence of substrate (ADP + CP) and product (ATP + creatine) were measured in water (Figure 11A). The observed fluorescence enhancements over time during the CK catalysis are in good agreement with the separate fluorescence measurements of L1 in the presence of substrate and product. Similarly, the hydrolytic apyrase enzyme catalyzed the dephosphorylation of ATP and ADP and the reaction was also followed by monitoring the fluorescence of dissolved L1 at different time intervals. Such studies showed a corresponding fluorescence quenching over time (Figures 11D and S57). The obtained results are comparable with the fluorescence emissions of L1:AMP/ADP/ATP and Pi complexes (Figures 11C and S56). In both cases, the fluorescence kinetic data indicate that the enzymatic conversion reaction rates of ADP → ATP and ATP →→ AMP were accelerated upon increasing the relative concentration of the enzymes CK or apyrase, respectively (Figures 11B and 11D). The real-time monitoring of the enzymatic activity of apyrase (ATP → ADP → AMP and Pi) also was verified by 31P NMR spectroscopy measurements at different time intervals with EDTA as a catalyst quencher (Figure S58). The observed 31P NMR spectral peaks of a reaction mixture containing the probe L1(Zn):ATP + apyrase revealed the ratio of hydrolyzed products (ADP and AMP) at different time intervals.
Figure 11.
Results of Enzyme Catalyzed ADP Phosphorylation and ATP Hydrolysis
(A) Fluorescence spectra of the solution of L1 (37 μM) + ATP or ADP (10 equiv), L1 (37 μM) + ATP (10 equiv) + CP (10 equiv), and L1 (37 μM) + ADP (8 equiv) + creatine (10 equiv) in water.
(B) Time-trace plots of CK-catalyzed ADP phosphorylation monitored by emission intensity at 427 nm in two different methods PM-I and PM-II, [ADP] = 300 μM, [CP] = 10 equiv, [CK] = 0, 4, and 7 units, MES buffer (10 mM, pH = 6.5) at 37°C (λexc = 315 nm, slit: 5 nm/5 nm).
(C) Fluorescence spectra of the solution of L1 (37 μM) + ATP or AMP (10 equiv), L1 (37 μM) + ATP (10 equiv) + Pi (10 equiv), and L1 (37 μM) + AMP (10 equiv) + Pi (10 equiv) in water.
(D) Time-trace plots of apyrase-catalyzed ATP dephosphorylation monitored by emission intensity at 427 nm in two different methods PM-I and PM-II, [ATP] = 370 μM, [apyrase] = 0, 3, 6 and 10 units, HEPES buffer solution (10 mM, pH = 7.4) at 37°C (λexc = 315 nm, slit: 5 nm/5 nm).
Conclusions
Herein, we designed and synthesized a series of bis-8-methylquinolyl chromophore-tethered CDX derivatives and studied their conformations in different solvents. The corresponding metal complexes L1–L3 were utilized as efficient sensing and discriminating probes for nucleotide phosphates. β-CDX-based probe L1 was found to serve as a specific supramolecular colorimetric (visible) and fluorescent sensor for discriminating between structurally similar nucleotide phosphates and their related inorganic phosphates in aqueous media. The present study establishes the obvious need of the CDX cavity for nucleotide binding, which in turn depends on the specific cavity size for providing the interaction portals for the corresponding sensing and differentiating response. A mechanism for the signaling and nucleotide differentiation was established from control experiments and theoretical (DFT and TD-DFT) studies. The present sensor also can be applied efficiently for the fluorescent real-time monitoring of nucleotide phosphorylation and dephosphorylation processes catalyzed by enzymes such as creatine kinase and apyrase, respectively. This supramolecular probe-based sensing/differentiating approach and the real-time monitoring of the enzyme-catalyzed phosphorylation/dephosphorylation processes of nucleotides might open up new avenues for many interdisciplinary researchers; this present research results will also be expandable to the detection of other nucleotides and in vivo monitoring applications.
Limitations of the Study
The real-time monitoring of biological processes via fluorescent and colorimetric changes in this study was achieved in vitro; the enzyme-catalyzed phosphorylation and dephosphorylation reactions are surely more complicated in vivo and deserve further investigation but was limited by the present experimental conditions.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors gratefully acknowledge the financial assistance from the National Natural Science Foundation of China (No. 21871194, 21971169, 21572142, 21402129), National Key Research and Development Program of China (No. 2017YFA0505903), and Science and Technology Department of Sichuan Province (2019YJ0160, 2019YJ0090, and 2017SZ0021). V.B. acknowledges the start-up research grant (YZZ16005) from South-Central University for Nationalities. Comprehensive Training Platform of Specialized Laboratory, College of Chemistry, Prof. Peng Wu of Analytical &Testing Center, Sichuan University for characterization and photophysical properties measurement and Ms Jiao Lu, National Engineering Research Center for Biomaterials, Sichuan University for MALDI-TOF-MS measurement are greatly appreciated.
Author Contributions
K.K., C.X., M.R., and C.F. carried out the experiment and analyzed the data; K.K. wrote the manuscript; G.C., D.Z., Z.Z., D.S., X.Y., J.Y., T.H., and J.J.C. analyzed the data and modified the manuscript; and V.B., W.W., and C.Y. conceived of the present idea and modified the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: March 27, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.100927.
Contributor Information
Victor Borovkov, Email: victor.borovkov@scuec.edu.cn.
Wanhua Wu, Email: wuwanhua@scu.edu.cn.
Cheng Yang, Email: yangchengyc@scu.edu.cn.
Supplemental Information
References
- Agafontsev A.M., Ravi A., Shumilova T.A., Oshchepkov A.S., Kataev E.A. Molecular receptors for recognition and sensing of nucleotides. Chemistry. 2019;25:2684–2694. doi: 10.1002/chem.201802978. [DOI] [PubMed] [Google Scholar]
- Aggelidou C., Mavridis I.M., Yannakopoulou K. Binding of nucleotides and nucleosides to per(6-guanidino-6-deoxy)cyclodextrins in solution. Eur. J. Org. Chem. 2009;2009:2299–2305. [Google Scholar]
- Berg J.M., Tymoczko J.L., Stryer L. W.H. Freeman and company; 2002. Biochemistry. [Google Scholar]
- Busschaert N., Caltagirone C., Rossom W.V., Gale P.A. Applications of supramolecular anion recognition. Chem. Rev. 2015;115:8038–8155. doi: 10.1021/acs.chemrev.5b00099. [DOI] [PubMed] [Google Scholar]
- Butler S.J. Ratiometric detection of adenosine triphosphate (ATP) in water and real-time monitoring of apyrase activity with a tripodal zinc complex. Chemistry. 2014;20:15768–15774. doi: 10.1002/chem.201404499. [DOI] [PubMed] [Google Scholar]
- Cacciapaglia R., Di Stefano S., Mandolini L. Effective molarities in supramolecular catalysis of two-substrate reactions. Acc. Chem. Res. 2004;37:113–122. doi: 10.1021/ar020076v. [DOI] [PubMed] [Google Scholar]
- Cai Y., Meng X., Wang S., Zhu M., Pan Z., Guo Q.X. A quinoline based fluorescent probe that can distinguish zinc(II) from cadmium(II) in water. Tetrahedron Lett. 2013;54:1125–1128. [Google Scholar]
- Canary J.W., Allen C.S., Castagnetto J.M., Chiu Y.-H., Toscano P.J., Wang Y. Solid state and solution characterization of chiral, conformationally mobile tripodal ligands. Inorg. Chem. 1998;37:6255–6262. [Google Scholar]
- Cheng Y., Dong X. Preparation of a molecularly imprinted fluorescent chemosensor using quinoline modified vinyl-β-cyclodextrin and acrylamide as monomers for the selective recognition of spermidine. Anal. Methods. 2016;8:5838–5842. [Google Scholar]
- Chen T.-R., Urban P.L. Mass spectrometry-guided refinement of chemical energy buffers. Proc. R. Soc. A. 2016;472:20150812. doi: 10.1098/rspa.2015.0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Han K.-Y., Liu Y. Effective switch-on fluorescence sensing of zinc(II) ion by 8-aminoquinolino-β-cyclodextrin/adamantaneacetic acid system in water. Bioorg. Med. Chem. 2007;15:4537–4542. doi: 10.1016/j.bmc.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Jiang P., Dong X. Molecularly imprinted fluorescent chemosensor synthesized using quinoline-modified-β-cyclodextrin as monomer for spermidine recognition. RSC Adv. 2015;5:55066–55074. [Google Scholar]
- Cotner E.S., Smith P.J. Phosphotyrosine binding by ammonium- and guanidinium-modified cyclodextrins. J. Org. Chem. 1998;63:1737–1739. [Google Scholar]
- Das S., Sarkar H.S., Uddin M.R., Rissanen K., Mandal S., Sahoo P. Differential detection and quantification of cyclic AMP and other adenosine phosphates in live cells. Chem. Commun. (Camb.) 2017;53:7600–7603. doi: 10.1039/c7cc02935g. [DOI] [PubMed] [Google Scholar]
- de Silva A.P., Moody T.S., Wright G.D. Fluorescent PET (Photoinduced Electron Transfer) sensors as potent analytical tools. Analyst. 2009;134:2385–2393. doi: 10.1039/b912527m. [DOI] [PubMed] [Google Scholar]
- Eliseev A.V., Schneider H.J. Aminocyclodextrins as selective hosts with several binding sites for nucleotides. Angew. Chem. Int. Ed. 1993;32:1331–1333. [Google Scholar]
- Eliseev A.V., Schneider H.J. Molecular recognition of nucleotides, nucleosides, and sugars by aminocyclodextrins. J. Am. Chem. Soc. 1994;116:6081–6088. [Google Scholar]
- Formoso C. The interaction of β-cyclodextrin with nucleic acid monomer units. Biochem. Biophys. Res. Commun. 1973;50:999–1005. doi: 10.1016/0006-291x(73)91505-2. [DOI] [PubMed] [Google Scholar]
- Formoso C. The interaction of β-cyclodextrin with dinucleoside phosphates. Biopolymers. 1974;13:909–917. doi: 10.1002/bip.1974.360130507. [DOI] [PubMed] [Google Scholar]
- Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A. Gaussian, Inc.; 2010. Gaussian 09. [Google Scholar]
- Fujita K., Fujiwara S., Yamada T., Tsuchido Y., Hashimoto T., Hayashita T. Design and function of supramolecular recognition systems based on guest-targeting probe-modified cyclodextrin receptors for ATP. J. Org. Chem. 2017;82:976–981. doi: 10.1021/acs.joc.6b02513. [DOI] [PubMed] [Google Scholar]
- Guo A., Zhu R., Ren Y., Dong J., Feng L. A “turn-on” fluorescent chemosensor for aluminum ion and cell imaging application. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016;153:530–534. doi: 10.1016/j.saa.2015.09.009. [DOI] [PubMed] [Google Scholar]
- García-Espana E., Belda R., Gonzalez J., Pitarch J., Bianchi A. Receptors for nucleotides. In: Gale P.A., Steed J.W., editors. Supramolecular Chemistry: From Molecules to Nanomaterials. Wiley-VCH; 2012. pp. 1225–1257. [Google Scholar]
- Harata K., Uedaira H. The circular dichroism spectra of the β-cyclodextrin complex with naphthalene derivatives. Bull. Chem. Soc. Jpn. 1975;48:375–378. [Google Scholar]
- Harada N., Nakanishi K. The exciton chirality method and its application to configurational and conformational studies of natural products. Acc. Chem. Res. 1972;5:257–263. [Google Scholar]
- Hapiot F., Bricout H., Tilloy S., Monflier E. Functionalized cyclodextrins as first and second coordination sphere ligands for aqueous organometallic catalysis. Eur. J. Inorg. Chem. 2012;10:1571–1578. [Google Scholar]
- Hargrove A.E., Nieto S., Zhang T., Sessler J.L., Anslyn E.V. Artificial receptors for the recognition of phosphorylated molecules. Chem. Rev. 2011;111:6603–6782. doi: 10.1021/cr100242s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser S.L., Johanson E.W., Green H.P., Smith P.J. Aryl phosphate complexation by cationic cyclodextrins. an enthalpic advantage for guanidinium over ammonium and unusual enthalpy-entropy compensation. Org. Lett. 2000;2:3575–3578. doi: 10.1021/ol006503r. [DOI] [PubMed] [Google Scholar]
- Hembury G.A., Borovkov V.V., Inoue Y. Chirality-sensing supramolecular systems. Chem. Rev. 2008;108:1–73. doi: 10.1021/cr050005k. [DOI] [PubMed] [Google Scholar]
- Hewitt S.H., Parris J., Mailhot R., Butler S.J. A continuous luminescence assay for monitoring kinase activity: signaling the ADP/ATP ratio using a discrete europium complex. Chem. Commun. (Camb.) 2017;53:12626–12629. doi: 10.1039/c7cc05887j. [DOI] [PubMed] [Google Scholar]
- Huang F., Hao G., Wu F., Feng G. Fluorescence sensing of ADP over ATP and PPi in 100% aqueous solution. Analyst. 2015;140:5873–5876. doi: 10.1039/c5an01291k. [DOI] [PubMed] [Google Scholar]
- Huergo L.F., Chandra G., Merrick M. P(II) signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol. Rev. 2013;37:251–283. doi: 10.1111/j.1574-6976.2012.00351.x. [DOI] [PubMed] [Google Scholar]
- Jiang P., Ninfa A.J. Escherichia coli PII signal transduction protein controlling Nitrogen assimilation acts as a sensor of adenylate energy charge in vitro. Biochemistry. 2007;46:12979–12996. doi: 10.1021/bi701062t. [DOI] [PubMed] [Google Scholar]
- Job P. Formation and stability of inorganic complexes in solution. Ann. Chim. 1928;9:1130–1203. [Google Scholar]
- Jose D.A., Mishra S., Ghosh A., Shrivastav A., Mishra S.K., Das A. Colorimetric sensor for ATP in aqueous solution. Org. Lett. 2007;9:1979–1982. doi: 10.1021/ol0705797. [DOI] [PubMed] [Google Scholar]
- Kajtar M., Horvath-Toro C., Kuthi E., Szejtli J. A simple rule for predicting circular dichroism induced in aromatic guests by cyclodextrin hosts in inclusion complexes. Acta Chim. Acad. Sci. Hung. 1982;110:327–355. [Google Scholar]
- Kim J.S., Quang D.T. Calixarene-derived fluorescent probes. Chem. Rev. 2007;107:3780–3799. doi: 10.1021/cr068046j. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Swamy K.M.K., Lee K.M., Jagdale A.R., Kim Y., Kim S.J., Yoo K.H., Yoon J. Pyrophosphate selective fluorescent chemosensors based on coumarin-DPA-Cu(II) complexes. Chem. Commun. (Camb.) 2009:7215–7217. doi: 10.1039/b913809a. [DOI] [PubMed] [Google Scholar]
- Kirby A.J. Enzyme mechanisms, models, and mimics. Angew. Chem. Int. Ed. 1996;35:706–724. [Google Scholar]
- Kodaka M. A general rule for circular dichroism induced by a chiral macrocycle. J. Am. Chem. Soc. 1993;115:3702–3705. [Google Scholar]
- Knowles J.R. Enzyme-catalyzed phosphoryl transfer reactions. Annu. Rev. Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- Kumar M., Jonnalagadda N., George S.J. Molecular recognition driven self-assembly and chiral induction in naphthalene diimide amphiphiles. Chem. Commun. (Camb.) 2012;48:10948–10950. doi: 10.1039/c2cc35438a. [DOI] [PubMed] [Google Scholar]
- Kumar A., Prasher P., Singh P. A fluorescent probe for estimation of adenosine diphosphate and monitoring of glucose metabolism. Org. Biomol. Chem. 2014;12:3071–3079. doi: 10.1039/c3ob42505c. [DOI] [PubMed] [Google Scholar]
- Kumar M., Brocorens P., Tonnele C., Beljonne D., Surin M., George S.J. A dynamic supramolecular polymer with stimuli-responsive handedness for in situ probing of enzymatic ATP hydrolysis. Nat. Commun. 2014;5:5793. doi: 10.1038/ncomms6793. [DOI] [PubMed] [Google Scholar]
- Li C., Numata M., Takeuchi M., Shinkai S. A sensitive colorimetric and fluorescent probe based on a polythiophene derivative for the detection of ATP. Angew. Chem. Int. Ed. 2005;44:6371–6374. doi: 10.1002/anie.200501823. [DOI] [PubMed] [Google Scholar]
- Li N., Chen Y., Zhang Y.-M., Wang L.-H., Mao W.-Z., Liu Y. Molecular binding thermodynamics of spherical guests by β-cyclodextrins bearing aromatic substituents. Thermochim. Acta. 2014;576:18–26. [Google Scholar]
- Liu Y., Bonizzoni M. A supramolecular sensing array for qualitative and quantitative analysis of organophosphates in water. J. Am. Chem. Soc. 2014;136:14223–14229. doi: 10.1021/ja507905r. [DOI] [PubMed] [Google Scholar]
- Liu Y., Song Y., Wang H., Zhang H.-Y., Wada T., Inoue Y. Selective binding of steroids by 2,2‘-biquinoline-4,4‘-dicarboxamide-bridged bis(β-cyclodextrin): fluorescence enhancement by guest inclusion. J. Org. Chem. 2003;68:3687–3690. doi: 10.1021/jo026908k. [DOI] [PubMed] [Google Scholar]
- Liu Y., Song Y., Chen Y., Li X.-Q., Ding F., Zhong R.-Q. Biquinolin-modified β-cyclodextrin dimers and their metal complexes as efficient fluorescent sensors for the molecular recognition of steroids. Chemistry. 2004;10:3685–3696. doi: 10.1002/chem.200305724. [DOI] [PubMed] [Google Scholar]
- Liu Y., Shi J., Guo D.-S. Novel permethylated β-cyclodextrin derivatives appended with chromophores as efficient fluorescent sensors for the molecular recognition of bile salts. J. Org. Chem. 2007;72:8227–8234. doi: 10.1021/jo071131m. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang N., Chen Y., Wang L.-H. Fluorescence sensing and binding behavior of aminobenzenesulfonamidoquinolino-β-cyclodextrin to Zn2+ Org. Lett. 2007;9:315–318. doi: 10.1021/ol062816w. [DOI] [PubMed] [Google Scholar]
- Liu Y., Yu M., Chen Y., Zhang N. Convenient and highly effective fluorescence sensing for Hg2+ in aqueous solution and thin film. Bioorg. Med. Chem. 2009;17:3887–3891. doi: 10.1016/j.bmc.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Martinez-Manez R., Sancenon F. Fluorogenic and chromogenic chemosensors and reagents for anions. Chem. Rev. 2003;103:4419–4476. doi: 10.1021/cr010421e. [DOI] [PubMed] [Google Scholar]
- McCleskey S.C., Griffin M.J., Schneider S.E., McDevitt J.T., Anslyn E.V. Differential receptors create patterns diagnostic for ATP and GTP. J. Am. Chem. Soc. 2003;125:1114–1115. doi: 10.1021/ja021230b. [DOI] [PubMed] [Google Scholar]
- Meng X., Wang S., Zhu M. Quinoline-based fluorescence sensors. In: Saha S., editor. Molecular Photochemistry - Various Aspects. IntechOpen Limited; 2012. pp. 3–22. [Google Scholar]
- Mikata Y., Takekoshi A., Kaneda M., Konno H., Yasuda K., Aoyama M., Tamotsu S. Replacement of quinolines with isoquinolines affords target metal ion switching from Zn2+ to Cd2+ in the fluorescent sensor TQLN (N, N,N’,N’-tetrakis(2-quinolylmethyl)-2,6-bis(aminomethyl)pyridine) Dalton Trans. 2017;46:632–637. doi: 10.1039/c6dt03948k. [DOI] [PubMed] [Google Scholar]
- Molina P., Zapata F., Caballero A. Anion recognition strategies based on combined noncovalent interactions. Chem. Rev. 2017;117:9907–9972. doi: 10.1021/acs.chemrev.6b00814. [DOI] [PubMed] [Google Scholar]
- Mourtzis N., Eliadou K., Aggelidou C., Sophianopoulou V., Mavridis I.M., Yannakopoulou K. Per(6-guanidino-6-deoxy)cyclodextrins: synthesis, characterisation and binding behaviour toward selected small molecules and DNA. Org. Biomol. Chem. 2007;5:125–131. doi: 10.1039/b614899a. [DOI] [PubMed] [Google Scholar]
- Ngo H.T., Liu X., Jolliffe K.A. Anion recognition and sensing with Zn(II)-dipicolylamine complexes. Chem. Soc. Rev. 2012;41:4928–4965. doi: 10.1039/c2cs35087d. [DOI] [PubMed] [Google Scholar]
- Ogoshi T., Harada A. Chemical sensors based on cyclodextrin derivatives. Sensors. 2008;8:4961–4982. doi: 10.3390/s8084961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojida A., Miyahara Y., Wongkongkatep J., Tamaru S.-I., Sada K., Hamachi I. Design of dual-emission chemosensors for ratiometric detection of ATP derivatives. Chem. Asian J. 2006;1:555–563. doi: 10.1002/asia.200600137. [DOI] [PubMed] [Google Scholar]
- O'Neil E.J., Smith B.D. Anion recognition using dimetallic coordination complexes. Coord. Chem. Rev. 2006;250:3068–3080. [Google Scholar]
- Pramanik A., Das G. An efficient phosphate sensor: tripodal quinoline excimer transduction. Tetrahedron. 2009;65:2196–2200. [Google Scholar]
- Ramaiah D., Neelakandan P.P., Nair A.K., Avirah R.R. Functional cyclophanes: promising hosts for optical biomolecular recognition. Chem. Soc. Rev. 2010;39:4158–4168. doi: 10.1039/b920032k. [DOI] [PubMed] [Google Scholar]
- Rekharsky M.V., Inoue Y. Complexation thermodynamics of cyclodextrins. Chem. Rev. 1998;98:1875–1918. doi: 10.1021/cr970015o. [DOI] [PubMed] [Google Scholar]
- Ringe D., Petsko G.A. How enzymes work. Science. 2008;320:1428–1429. doi: 10.1126/science.1159747. [DOI] [PubMed] [Google Scholar]
- Sancenon F., Descalzo A.B., Martinez-Manez R., Miranda M.A., Soto J. A colorimetric ATP sensor based on 1,3,5-Triarylpent-2-en-1,5-diones. Angew. Chem. Int. Ed. 2001;40:2640–2643. doi: 10.1002/1521-3773(20010716)40:14<2640::AID-ANIE2640>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Santis G.D., Fabbrizzi L., Licchelli M., Mangano C., Sacchi D. Redox switching of anthracene fluorescence through the CuII/CuI couple. Inorg. Chem. 1995;34:3581–3582. [Google Scholar]
- Schlattner U., Tokarska-Schlattner M., Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta. 2006;1762:164–180. doi: 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Schwinte P., Darcy R., O'Keeffe F. Ditopic binding of nucleotides by heptakis(6-hydroxyethylamino-6-deoxy)-β-cyclodextrin. J. Chem. Soc. Perkin Trans. 1998;2:805–808. [Google Scholar]
- Shi L., Hu P., Ren Y., Feng G. An unexpected highly selective mononuclear zinc complex for adenosine diphosphate (ADP) Chem. Commun. (Camb.) 2013;49:11704–11706. doi: 10.1039/c3cc47437b. [DOI] [PubMed] [Google Scholar]
- Szejtli J. Introduction and general overview of cyclodextrin chemistry. Chem. Rev. 1998;98:1743–1754. doi: 10.1021/cr970022c. [DOI] [PubMed] [Google Scholar]
- Szente L., Szeman J. Cyclodextrins in analytical chemistry: host-guest type molecular recognition. Anal. Chem. 2013;85:8024–8030. doi: 10.1021/ac400639y. [DOI] [PubMed] [Google Scholar]
- Tang L., Wu D., Huang Z., Bian Y. A fluorescent sensor based on binaphthol-quinoline Schiff base for relay recognition of Zn2+ and oxalate in aqueous media. J. Chem. Sci. 2016;128:1337–1343. [Google Scholar]
- Vizitiu D., Thatcher G.R.J. Binding of phosphates to aminocyclodextrin biomimetics. J. Org. Chem. 1999;64:6235–6238. [Google Scholar]
- Wang L., Yuan L., Zeng X., Peng J., Ni Y., Cheng J., Xu W., Agrawalla B.K., Su D., Kim B., Chang Y.T. A multisite-binding switchable fluorescent probe for monitoring mitochondrial ATP level fluctuation in live cells. Angew. Chem. Int. Ed. 2016;55:1773–1776. doi: 10.1002/anie.201510003. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang H., Meng T., Hao E., Jiao L. Synthetically simple, click-generated quinoline-based Fe3+ sensors. Methods Appl. Fluoresc. 2017;5:024015. doi: 10.1088/2050-6120/aa7170. [DOI] [PubMed] [Google Scholar]
- Wolfe D.M., Zhang Y., Roberts G.P. Specificity and regulation of interaction between the PII and AmtB1 proteins in Rhodospirillum rubrum. J. Bacteriol. 2007;189:6861–6869. doi: 10.1128/JB.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongkongkatep J., Ojida A., Hamachi I. Fluorescence sensing of inorganic phosphate and pyrophosphate using small molecular sensors and their applications. Top. Curr. Chem. 2017;375:30. doi: 10.1007/s41061-017-0120-0. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wen J., Li H., Sun S., Xu Y. Fluorescent probes for recognition of ATP. Chin. Chem. Lett. 2017;28:1916–1924. [Google Scholar]
- Xu Z., Singh N.J., Lim J., Pan J., Kim H.N., Park S., Kim K.S., Yoon J. Unique sandwich stacking of pyrene-adenine-pyrene for selective and ratiometric fluorescent sensing of ATP at physiological pH. J. Am. Chem. Soc. 2009;131:15528–15533. doi: 10.1021/ja906855a. [DOI] [PubMed] [Google Scholar]
- Yamada T., Fujiwara S., Fujita K., Tsuchido Y., Hashimoto T., Hayashita T. Development of dipicolylamine-modified cyclodextrins for the design of selective guest-responsive receptors for ATP. Molecules. 2018;23:635. doi: 10.3390/molecules23030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Wei Y., Kang X., Su Z. Selective recognition and sensing for adenosine triphosphate by label-free electrochemistry based on its inclusion with per-6-ammonium-β-cyclodextrin. Anal. Methods. 2015;7:10129–10135. [Google Scholar]
- You L., Zha D., Anslyn E.V. Recent advances in supramolecular analytical chemistry using optical sensing. Chem. Rev. 2015;115:7840–7892. doi: 10.1021/cr5005524. [DOI] [PubMed] [Google Scholar]
- Yuan D., Izuka A., Fukudome M., Rekharsky M.V., Inoue Y., Fujita K. Heptakis(6-deoxy-6-guanidino)-β-cyclodextrin: an artificial model for mitochondrial ADP/ATP carrier. Tetrahedron Lett. 2007;48:3479–3483. [Google Scholar]
- Yu M., Chen Y., Zhang N., Liu Y. Construction and transmembrane dissociation behavior of supramolecular assembly of quinolinocyclodextrin with porphyrin. Org. Biomol. Chem. 2010;8:4148–4154. doi: 10.1039/c0ob00080a. [DOI] [PubMed] [Google Scholar]
- Zahn S., Canary J.W. Absolute configurations of N,N-dialkyl α-amino acids and β-amino alcohols from exciton-coupled circular dichroism spectra of Cu(II) complexes. Org. Lett. 1999;6:861–864. doi: 10.1021/ol990715a. [DOI] [PubMed] [Google Scholar]
- Zahn S., Canary J.W. Electron-induced inversion of helical chirality in copper complexes of N,N-dialkylmethionines. Science. 2000;288:1404–1407. doi: 10.1126/science.288.5470.1404. [DOI] [PubMed] [Google Scholar]
- Zhai S.-S., Chen Y., Liu Y. Selective binding of bile salts by β-cyclodextrin derivatives with appended quinolyl arms. Chin. Chem. Lett. 2013;24:442–446. [Google Scholar]
- Zhao X.J., Huang C.Z. Small organic molecules as fluorescent probes for nucleotides and their derivatives. TrAC-Trend. Anal. Chem. 2010;29:354–367. [Google Scholar]
- Zhou Y., Xu Z., Yoon Y. Fluorescent and colorimetric chemosensors for detection of nucleotides, FAD and NADH: highlighted research during 2004-2010. Chem. Soc. Rev. 2011;40:2222–2235. doi: 10.1039/c0cs00169d. [DOI] [PubMed] [Google Scholar]
- Zhong C., Mu T., Wang L., Fu E., Qin J. Unexpected fluorescent behavior of a 4-amino-1,8-naphthalimide derived β-cyclodextrin: conformation analysis and sensing properties. Chem. Commun. (Camb.) 2009;2009:4091–4093. doi: 10.1039/b902132a. [DOI] [PubMed] [Google Scholar]
- Zimmermann H. Two novel families of ectonucleotidases: molecular structures, catalytic properties, and a search for function. Trends Pharmacol. Sci. 1999;20:231–236. doi: 10.1016/s0165-6147(99)01293-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.