Summary
Spermatogonial stem cells (SSCs) serve as a resource for producing genetically modified animals. However, genetic manipulation of SSCs has met with limited success. Here, we show efficient gene transfer into SSCs via a lentivirus (FV-LV) using a fusion protein (F), a Sendai virus (SV) envelope protein involved in virion/cell membrane fusion. FV-LVs transduced cultured SSCs more efficiently than conventional LVs. Although SSCs infected with SV failed to produce offspring, those transduced with FV-LVs were fertile. In vivo microinjection showed that FV-LVs could penetrate not only the basement membrane of the seminiferous tubules but also the blood-testis barrier, which resulted in successful transduction of both spermatogenic cells and testicular somatic cells. Cultured SSCs transfected with FV-LVs that express drug-inducible CRISPR/Cas9 against Kit or Sycp3 showed impaired spermatogenesis upon transplantation and drug treatment in vivo. Thus, FV-LVs provide an efficient method for functional analysis of genes involved in SSCs and spermatogenesis.
Keywords: pseudotyping, lentivirus, Sendai virus, spermatogonia, testis
Graphical Abstract

Highlights
-
•
Sendai virus-derived F protein enhances lentiviral infection of male germ cells
-
•
Transfected spermatogonial stem cells undergo germline transmission
-
•
Lentivirus pseudotyped with F protein penetrates the blood-testis barrier
-
•
This method is compatible with in vivo conditional gene editing
In this article, Shinohara and colleagues show that a lentivirus pseudotyped with Sendai virus F protein significantly improves the transduction efficiency of spermatogonial stem cells (SSCs). The virus penetrates the blood-testis barrier and allows conditional gene editing in vivo. This method overcomes problems associated with SSC transfection and provides new possibilities for germline manipulation.
Introduction
Spermatogonial stem cells (SSCs) undergo continuous self-renewal and differentiation, which underlies the lifelong maintenance of male fertility (de Rooij and Russell, 2000, Meistrich and van Beek, 1993). Although embryonic stem cells have been used for germline modification for decades, SSCs represent an alternative resource for producing genetically modified animals because SSCs are the only stem cells in the germline (Kubota and Brinster, 2018, Kanatsu-Shinohara and Shinohara, 2013). However, because SSCs comprise only a small population in testes (0.02%–0.03% of total germ cells) and are surrounded by somatic cells (Meistrich and van Beek, 1993, Tegelenbosch and de Rooij, 1993), gene transduction into SSCs has met with limited success. Nevertheless, development of spermatogonial transplantation techniques has provided the first opportunity for SSC manipulation and transfection. SSCs microinjected into the seminiferous tubules of infertile animals reinitiated spermatogenesis and resulted in fertile sperm (Brinster and Zimmermann, 1994). By transfecting SSCs before transplantation, transgenic offspring were born (Nagano et al., 2001). In vitro SSC culture techniques further improved transfection efficiency and provided an opportunity for genetic selection of transfected clones. Adding fibroblast growth factor 2 and glial cell line-derived neurotrophic factor (GDNF), both of which are SSC self-renewal factors, to testis cultures allowed for long-term in vitro expansion of SSCs, which can proliferate for more than 2 years without losing fertility (Kanatsu-Shinohara et al., 2003). These cells, which were designated as germline stem (GS) cells, allow production of transgenic or knockout (KO) animals after transplantation of drug-selected GS cell clones into seminiferous tubules (Kanatsu-Shinohara et al., 2005, Kanatsu-Shinohara et al., 2006). More recent experiments also demonstrated successful gene editing using similar approaches (Chapman et al., 2015, Sato et al., 2015, Wu et al., 2015). Development of transplantation and culture techniques has greatly improved the utility of SSCs for germline modification.
Despite these successes, there is still a considerable room to improve SSC manipulation techniques. Low gene transduction efficiency has been a major problem in SSC research. Although most of the conventional transfection techniques can be applied to SSCs, difficulties in drug selection and the slow growth of GS cells have hampered efficient clonal selection. Among several transfection methods, SSCs have been most successfully transfected by virus vectors. Retroviruses (RVs) were the first vectors used to transduce SSCs (Nagano et al., 2000). However, because RVs have very low transduction efficiency, lentiviruses (LVs) are more widely used for SSC transduction. Unlike conventional RVs, LVs can transduce non-dividing cells, which makes them useful for transducing tissue stem cells that rarely divide or do not divide at all. Although RVs and LVs integrate into the host genome, adenoviruses (AVs) do not integrate into the genome. Moreover, because AVs can be concentrated at higher titers, AVs transduce SSCs more efficiently than do LVs (Takehashi et al., 2007). However, the major problem with AVs is their toxicity, because continued exposure to AVs induces apoptosis of GS cells. Fortunately, this problem of cell toxicity has recently been overcome by adeno-associated viruses (AAVs) (Watanabe et al., 2017, Watanabe et al., 2018). AAVs have much less toxicity and transduce SSCs without integrating into the host genome. However, application of AAVs is often limited by their relatively small insert size (~4.5 kb).
Although these virus vectors have been used in many SSC studies, we and others recently tested the potential of Sendai virus (SV) for SSC transduction (Shiromoto et al., 2013, Watanabe et al., 2019). SV is a non-segmented negative-strand RNA virus of the Paramyxoviridae family (Lamb and Kolakofsky, 2001, Li et al., 2000, Whelan et al., 2004). SV was discovered in Japan in 1952 when an outbreak of newborn pneumonitis occurred at Tohoku University. SVs was found not to be responsible for the pneumonitis or to be pathogenic to humans, but was subsequently found to have hemagglutinin activity as well as cell fusion activity. More recently, SV has been used as a virus vector (Li et al., 2000). SV has several unique features that make it suitable for gene transduction because it has a broad range of hosts and expresses transgenes at high levels. Because SV does not have a DNA phase in replicative cycles, the virus genome does not integrate into the host genome. Its usefulness was demonstrated in our previous study, in which SV transduced mouse, hamster, rabbit and marmoset SSCs or SSC-like cells for long-term in vivo after xenogeneic transplantation into immunodeficient mice (Watanabe et al., 2019). This was in contrast to other virus vectors, which showed limited transduction. Although these results clearly showed the superiority of SV over the other virus vectors, the molecular mechanism underlying the efficient transduction of SV remains unclear.
In this study, we hypothesized that the surface properties of SV play a critical role in the transduction efficiency of SSCs. SV has two envelope proteins, HN and F (Kobayashi et al., 2003). HN protein binds to sialic acids on host cells and is required for interaction between SV and host cells. F protein is responsible for the fusion of SV with host cells and is essential for virus entry. These proteins appear to influence transfection efficiency, because several studies have demonstrated that pseudotyping of LVs or simian immunodeficiency viruses (SIVs) with both F and HN improved transduction efficiency to human hepatocytes, respiratory epithelium and several types of cultured cells compared with those pseudotyped with vesicular stomatitis virus G (VSVG) protein (Kowolik and Yee, 2002, Mitomo et al., 2010, Murakami et al., 2010). Based on this hypothesis, we produced several hybrid LVs containing SV-derived envelope proteins and examined their transduction efficiencies as well as their usefulness in gene editing of GS cells.
Results
Failure to Produce Offspring Using SV-Transfected GS Cells
In our previous study, we showed that SVs transduce GS cells with high efficiency (Watanabe et al., 2019). Transfected GS cells reinitiated normal spermatogenesis after spermatogonial transplantation. Therefore, we hypothesized that SV would be a useful vehicle for genetic modification of the male germline. Because SV is an RNA virus, we hypothesized that it would induce genetic modification without being transmitted to the next generation. To test this possibility, we transfected GS cells with Azami Green (AG) fluorescent protein-expressing SV (Figure 1A), and the transfected GS cells were transplanted them into infertile mice. At least 4 weeks after transplantation, the recipients were mated with wild-type females to produce offspring.
Figure 1.
Transduction of GS Cells by Pseudotyped V-LVs
(A) Schematic diagrams of an SV and pseudotyped V-LVs by F and/or HN proteins.
(B) Histological appearance of a recipient testis transplanted with SV-infected GS cells.
(C) Appearance of GS cells 3 days after infection by transduction of pseudotyped LVs without VSVG.
(D) Appearance of GS cells 3 days after infection by transduction of pseudotyped LVs with VSVG.
(E) Flow cytometric analysis of GS cells 3 days after infection (n = 3). Cells were infected with the same MOI (= 30). V-LVs without PB were used as a control.
(F and G) Mean fluorescence intensity (MFI) of VENUS expression (F) and the percentage of cells expressing VENUS (G) were determined. Results of three independent experiments (n = 3; F and G).
(H) Failure to improve FV-LV infection efficiency by PB.
Scale bars, 50 μm (B–D and H). See also Figure S1. Results are mean ± SEM.
Although more than 12 recipients were mated for more than 6 months, none of the recipient animals produced offspring. GS cell recipients usually produce offspring within 3–4 months (Kanatsu-Shinohara et al., 2016), so these results implied that the SV-infected GS cells had spermatogenic defects. To determine the reason for infertility, we sacrificed the recipient mice and analyzed their testes. Histological analysis revealed that spermatogenesis was severely impaired (Figure 1B). This was in contrast to the results in our previous study, which showed normal spermatogenesis at 3 months after transplantation (Watanabe et al., 2019). Therefore, the SV-transfected GS cells gradually lost spermatogenic potential despite colonization and successful differentiation.
Improved Transduction Efficiency of GS Cells by LVs Pseudotyped with F
Although the results detailed in the preceding section revealed the potential toxicity of SV, the mechanism was not clear. There are many structural and functional characteristics that are unique to SV, making it difficult to determine which part of the virus or infection process is responsible for impairing spermatogenic potential. However, because the high infectivity of SVs can be conferred by pseudotyping of LV using F and HN proteins (Kowolik and Yee, 2002, Mitomo et al., 2010, Murakami et al., 2010), we reasoned that a similar pseudotyping strategy might be applicable to SSCs to improve transduction efficiency. We generated three types of LVs pseudotyped with F, HN, or both F and HN (F/HN) proteins (Figure 1A). These LVs expressed mNeonGreen (mNG), a monomeric GFP derived from Branchiostoma lanceolatum, under the Eif1a promoter. We also used conventional LVs using VSVG as a control. We added polybrene (PB) for V-LV because it enhances the transfection efficiency and is commonly added to SSC infection experiments (Nagano et al., 2000). PB was not added to LVs pseudotyped with F or HN proteins. When the GS cells were examined 3 days after transfection, none of the three LVs with SV-derived envelopes showed fluorescence, while HEK293T cells were infected successfully by the three types of LVs (Figure 1C).
Based on these observations, we carried out another set of experiments by adding VSVG to F or HN proteins. In these experiments, we used VENUS as a fluorescent marker. Contrary to the initial experiments, we were able to detect strong fluorescence in GS cells without PB (Figure 1D). When compared with each other, VENUS expression level was highest in FV-LV-infected GS cells. To quantify this result, we used flow cytometry (Figures 1E–1G), which confirmed that the strongest fluorescence was by FV-LVs. The infection efficiency was approximately four times higher than that of V-LVs. HNV-LVs also increased the infection efficiency, albeit at lesser degree. F/HNV-LVs did not improve the infection efficiency and there was no significant difference between HN-LV and F-LV. Adding PB to FV-LV did not increase the infection efficiency (Figure 1H). These results implied that a combination of F protein and VSVG is important to confer infectivity to GS cells.
To examine the optimal MOI of FV-LVs, we transfected GS cells with FV-LVs expressing Venus at an MOI of 1, 5, 15, 30, or 60, and fluorescence levels were analyzed 3 days post-infection by fluorescence microscopy and flow cytometry (Figures S1A and S1B). Fluorescence levels were increased in a dose-dependent manner and >90% of GS cells showed VENUS expression at an MOI of 30–60 (Figure S1C). We next evaluated the effect of FV-LV on cell proliferation. The numbers of GS cells recovered after FV-LV or LV infection were examined at an MOI of 30. Cell recovery after FV-LV infection increased significantly compared with that after V-LV infection (Figure S1D). This difference in cell recovery was likely due to increased apoptosis of V-LV-infected cells, because Terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) staining showed that the number of TUNEL+ cells increased by 5-fold after V-LV infection (Figure S1E).
Because PB is potentially toxic (Cornetta and Anderson, 1989), we checked the toxicity of PB to GS cells. Adding PB decreased cell recovery in a dose-dependent manner 3 days after the treatment (Figure S1F), while the number of TUNEL+ cells increased in a reciprocal manner (Figure S1G). An almost 10-fold increase in TUNEL+ cells was noted at 10 μg/mL compared with cells cultured without PB. Taken together, these results imply that pseudotyping of LVs with F and VSVG proteins dramatically increases the SSC transduction efficiency with significantly reduced cell toxicity.
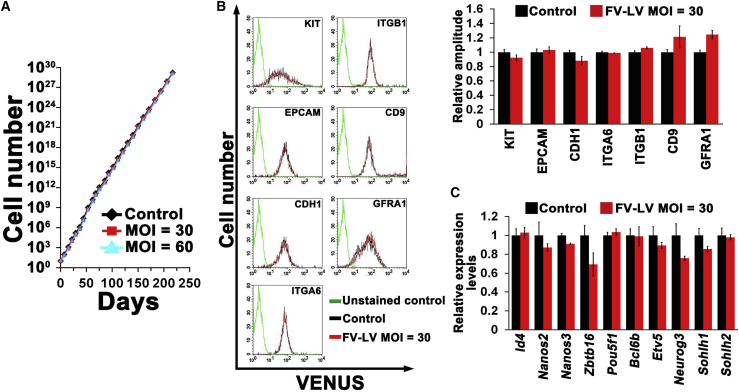
Phenotypic Analysis of GS Cells Stably Infected with FV-LVs
GS cells transfected with FV-LVs continued to proliferate normally for more than 7 months (Figure 2A). Because SV-transfected GS cells exhibited a significantly altered phenotype and several downregulated spermatogonia markers, we examined the phenotype of FV-LV-infected cells. We first examined whether FV-LV infection influenced the expression levels of spermatogonia cell surface markers by flow cytometry (KIT, EPCAM, CDH1, ITGA6, ITGB1, CD9, and GFRA1) (Figure 2B). In contrast to the results of SV infection, which showed downregulation of all examined cell surface markers other than EPCAM (Watanabe et al., 2019), no significant changes were observed in any tested markers.
Figure 2.
Phenotypic Analysis of GS Cells Infected with FV-LVs
(A) In vitro expansion of GS cells infected with FV-LV at MOI = 30 and 60.
(B) Flow cytometric analysis of surface markers for spermatogonia in FV-LV-infected GS cells. Wild-type GS cells without infection were used as a control. Results of three independent experiments (n = 3).
(C) Real-time PCR analysis of spermatogonial markers in FV-LV infected GS cells (n = 3). Results of three independent experiments (n = 3).
We then used real-time PCR to examine the expression of several spermatogonial transcription factors (Id4, Nanos2, Nanos3, Zbtb16, Pou5f1, Bcl6b, Neurog3, Sohlh1, and Sohlh2). Although SV-infected GS cells have been shown to downregulate Id4, Nanos2, Zbtb16, Pou5f1, Bcl6b, and Etv5 (Watanabe et al., 2019), none of these factors was affected by FV-LV infection (Figure 2C). In summary, FV-LV infection did not disturb cell growth or spermatogonia marker expression.
Transduction Kinetics of Non-integrating FV-LVs
Although FV-LV efficiently infected GS cells without changing the phenotype, LVs integrate into the genome of the infected cells, which potentially alters the expression of neighboring genes. To overcome this problem, we generated integration-deficient FV-LVs (FV-IDLVs) whose integrase had a D64V point mutation leading to the inactivation of integration (Certo et al., 2011). We examined the transduction kinetics of FV-IDLVs in GS cells at an MOI of 120 using fluorescence microscopy and flow cytometry (Figures 3A and 3B). GS cells with FV-LVs were used as a control (MOI = 30). We used a lower MOI for FV-LVs because preliminary experiments showed weaker fluorescence of FV-IDLVs.
Figure 3.
Transduction Kinetics of Non-integrating FV-LV
(A) Appearance of GS cells after FV-LV (top) or FV-IDLV (bottom) infection. Cells were infected at MOI = 30 (FV-LV) or 120 (F-IDLV).
(B) Quantification of MFI by flow cytometric analysis. Results of three independent experiments (n = 3).
(D) Flow cytometric analysis of EYFP expression in R26R-Eyfp GS cells following infection of Cre-expressing F-IDLVs (MOI = 120). Cells were analyzed 5 days after infection. Results of three independent experiments (n = 3).
(E) PCR analysis of Cre-mediated deletion 3 days after transfection.
(F) PCR analysis of Cre cDNA after transfection. Cells were passaged three times before analysis at 30 days. Scale bar,= 50 μm (A).
GS cells transfected with FV-IDLV showed mNG fluorescence as early as 1 day after transfection (Figure S2A). Almost all cells exhibited fluorescence in a manner similar to FV-IDLV infection. Expression levels of the FV-IDLV-transfected GS cells peaked at 3 days and gradually declined thereafter. By 10 days after infection, flow cytometry detected few mNG+ cells. Even at its peak, the level of mNG fluorescence was less than 2-fold that seen at 1 day after transfection. By contrast, FV-LV-infected GS cells showed mNG expression on the next day after infection and mNG expression gradually increased until 14 days after transfection. Compared with 1 day post-infection, GS cells at this point showed an approximately 30-fold increase in mNG expression.
We examined the utility of FV-IDLVs by infecting GS cells established from R26R-Efyp reporter mice, which have a Pgk-Neo cassette flanked by loxP before Eyfp cDNA and express Eyfp upon Cre transfection (Figure 3C). Although simple Cre overexpression in GS cells induces apoptosis (Kanatsu-Shinohara et al., 2008), AAVs or AVs that transiently express Cre can delete the target gene and induce the expression of Eyfp. Four days after infection with FV-IDLV-Cre (Figure S2B), EYFP expression was observed in most GS cell from Gt(ROSA)26Sortm1(EYFP)Cos mice (designated R26R-Eyfp); flow cytometry showed that 84% expressed EYFP (Figure 3D). PCR analysis confirmed the CRE-mediated recombination (Figure 3E). We also confirmed the lack of Cre cDNA 30 days after transfection (Figure 3F), which suggested that the gradual decline in mNG fluorescence in FV-IDLV-infected GS cells was most likely due to a lack of genome integration. This infection efficiency was comparable with those of AV-Cre and AAV1-Cre (Watanabe et al., 2017), implying that FV-IDLV is useful for functional analysis of genes by conditional KO in GS cells.
In Vivo Transduction into Germ Cells and Sertoli Cells by FV-LVs
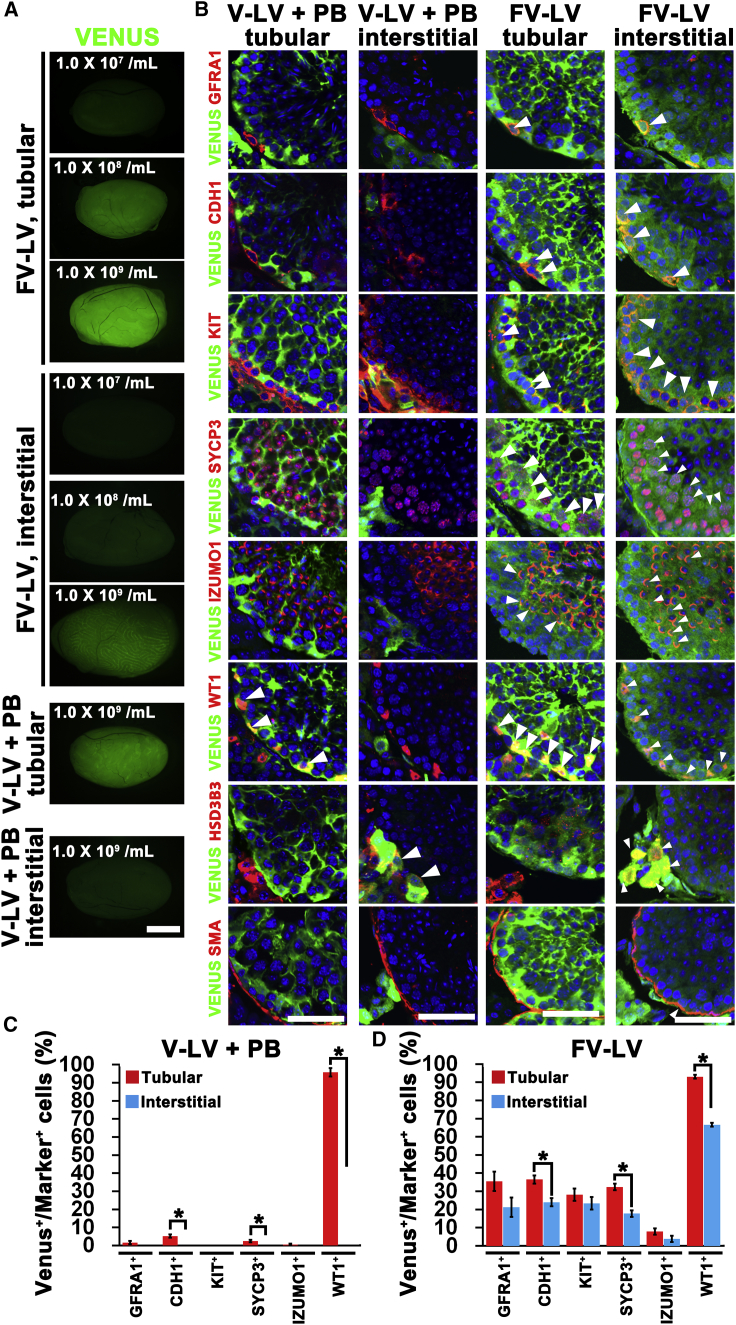
Because we found that FV-LVs can efficiently transduced GS cells in vitro, we examined the feasibility of transducing germ cells in vivo. We microinjected FV-LVs expressing VENUS into adult mouse testes at three different doses (1.0 × 107/mL, 1.0 × 108/mL and 1.0 × 109/mL). We used V-LVs as a positive control. The same amount of virus particles was microinjected into the seminiferous tubules or interstitial tissues. When the testes samples were recovered 7 days after microinjection of FV-LVs, VENUS fluorescence was detected regardless of the injection route (Figure 4A). Although we found VENUS fluorescence in testes with tubular injection of V-LV, no apparent signals were found in testes after interstitial injection.
Figure 4.
In Vivo Transduction of Mouse Testes by FV-LVs
(A) Macroscopic appearance of testes 7 days after tubular or interstitial injection of Venus-expressing FV-LV or V-LVs (1.0 × 109/mL).
(B) Immunohistochemistry of FV-LV- and V-LV-infected testes using antibodies against markers for undifferentiated spermatogonia (GFRA1, CDH1), differentiating spermatogonia (KIT), spermatocytes (SYCP3), spermatids (IZUMO1), Sertoli cells (WT1), Leydig cells (HSD3B3), and peritubular myoid cells (SMA).
(C and D) Quantification of VENUS expression in each cell type by V-LV (C) and FV-LV (D). Arrowheads indicate cells expressing both VENUS and lineage markers. At least 180 cells in ten tubules were counted. For GFRA1+ and CDH1+ spermatogonia, 15 cells in ten tubules were counted because there were fewer of them than other cell types. Asterisk indicates statistical significance (p < 0.05). Results of three independent experiments (n = 3).
Scale bars, 1 mm (A), 50 μm (B). See also Table S1. Results are mean ± SEM.
To investigate which cell types of testicular cells were transduced by FV-LVs and V-LVs, we performed immunohistochemistry of testes 7 days post-infection using markers for germ cells (GFRA1: Asingle, Apaired spermatogonia and some Aaligned spermatogonia; CDH1: undifferentiated spermatogonia; KIT: differentiating spermatogonia; SYCP3: spermatocytes; IZUMO1 for spermatids developing into sperm) and somatic cells (WT1: Sertoli cells: HSD3B3: Leydig cells; SMA: peritubular myoid cells) (Figure 4B). The number of VENUS+ cells expressing each antigen was counted for quantification (Figures 4C and 4D).
LVs efficiently infected WT1+ Sertoli cells in tubular injection, but VENUS expression was barely detectable in germ cells (Figures 4B and 4C), which confirmed the result of previous studies (Ikawa et al., 2002). With interstitial injection of V-LVs, germ cells and WT1+ Sertoli cells were not infected at all and VENUS signals were detected only in HSD3B3+ Leydig cells (Figures 4B and 4C). By contrast, tubular injection of FV-LVs successfully transduced both germ cells and Sertoli cells. Germ cells were transduced not only in the adluminal compartment but also those in the basal compartment. Similar results were obtained with interstitial injection of FV-LVs (Figures 4B and 4D). The infection efficiency of germ cells of all germ cells except for IZUMO+ cells was ~20%–40% for both infection routes. WT1+ Sertoli cells were more efficiently transduced by tubular injection than by interstitial injection (93% versus 66%). Interstitial injection of FV-LVs efficiently transduced HSD3B3+ Leydig cells and SMA+ peritubular myoid cells, although these cell types were not transduced by tubular injection. Taken together, FV-LVs enabled in vivo gene transfer into both undifferentiated spermatogonia and differentiating germ cells regardless of injection route, implying that FV-LVs acquired the ability to penetrate the blood-testis barrier (BTB) and basement membrane of the seminiferous tubules.
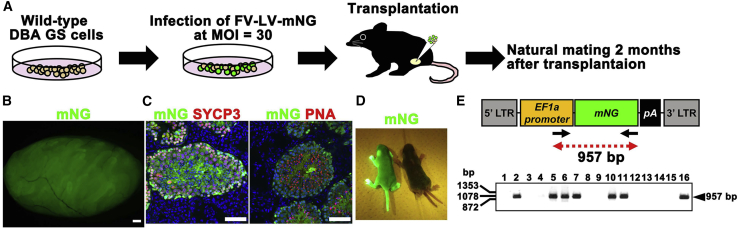
Generation of Transgenic Mice Derived from FV-LV Transduced GS Cells
Although these results showed the improved transduction efficiency of FV-LVs, it was possible that F protein influenced spermatogenesis and impaired fertility. To test this possibility, wild-type GS cells were infected with FV-LVs expressing mNG at an MOI of 30 (Figure 5A). The transfected cells were then cultured for about 2 weeks for in vitro expansion before transplantation into busulfan-treated C57BL6/J (B6) × DBA/2 F1 (BDF1) mouse testes. The recipient males were mated with wild-type females to produce offspring. Two months after transplantation, we confirmed extensive colonization of donor GS cells (Figure 5B). Development of SYCP3+ spermatocytes and peanut agglutinin (PNA)+ haploid spermatids were confirmed by immunostaining of the recipient testes (Figure 5C).
Figure 5.
Transgenic Mice Produced by Transfection of GS Cells Using FV-LV Expressing mNG
(B) Macroscopic appearance of a recipient testis transplanted with GS cells infected with mNG-expressing FV-LV.
(C) Immunostaining of recipient testis by spermatocyte (SYCP3) and haploid (PNA) markers.
(D) Offspring born after spermatogonial transplantation showing donor cell-derived mNG fluorescence under UV light.
(E) PCR analysis of mNG transgene in tail DNA.
Offspring with normal appearance were obtained at 4 months after spermatogonial transplantation. These mice showed mNG fluorescence under UV light, which confirmed their donor cell origin (Figure 5D). After genotyping of 16 offspring by PCR to specifically detect transgenes of FV-LVs, seven offspring were found to have transgenes derived from FV-LV infected GS cells (Figure 5E). These results show that GS cells transduced with FV-LVs retain fertility.
Genome Editing in GS Cells by FV-LVs Expressing CRISPR/Cas9
To study the utility of FV-LVs, we used FV-LVs and GS cells to examine the feasibility of gene editing. We constructed two types of FV-LVs (Figure 6A). The first vector expressed spCas9 under the control of Eif1α promoter. Another vector expressed guide RNA (gRNA) against Cldn11 or Fgf10 exons under the control of human U6 promoter. We also generated V-LVs containing the same transgenes for control experiments. These viruses were transduced into GS cells, and mutations were analyzed by DNA sequencing 7 and 21 days after infection without drug selection. Although V-LVs scarcely induced mutations, FV-LVs successfully induced mutations in both Cldn11 and Fgf10 gene loci (Figures 6B and 6C). We also quantified genome editing efficiency by Tide analysis (Figure 6D) (Brinkman et al., 2014). At day 7 after infection, <5% of the genomes of V-LV-treated cells were edited, whereas 20%–30% of FV-LV-treated cells were successfully edited. Similarly, at day 21, approximately 70% of the genome of FV-LV-treated cells were mutated, while ~10% of V-LV-treated cells showed evidence of gene editing (Figure 6C). These results show that FV-LV-mediated gene transfer of CRISPR/Cas9 efficiently induces mutations in GS cells.
Figure 6.
Genome Editing of GS Cells by FV-LVs Using the CRISPR/Cas9 System
(A) Schematic diagrams of FV-LVs expressing the CRISPR/Cas9 system. gRNAs against Cldn11 and Fgf10 loci in the mouse genome were tested.
(B and C) Representative results of sequence analysis in Cldn11 and Fgf10 loci at 7 days (B) and 21 days post-infection (C).
(D) Quantification of genome editing efficiency in each tested locus by Tide analysis (https://tide.nki.nl/). Results of three independent experiments (n = 3).
See also Table S2. Results are mean ± SEM.
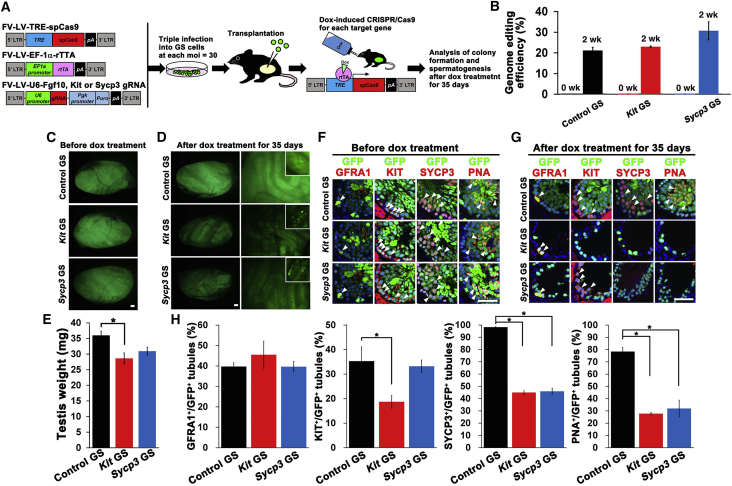
In Vivo Analysis of Gene Functions Involved in Spermatogenesis by Inducible Gene Editing of GS Cells
In the final set of experiments, FV-LVs were used for in vivo analysis of genes involved in spermatogenesis. We used the Tet-On system to induce CRISPR/Cas9 genome editing (Figure 7A). The Tet-On system is a drug-inducible gene expression system. rtTA is a transcription factor that can be activated in the presence of dox, and dox-rtTA binds to tetracycline responsive element and induces expression of a gene of interest. We first tested the efficiency of this system in vitro. In this experiment, we used gRNAs targeting Kit and Sycp3 exons. Fgf10 is not expressed in mouse testes and was used as a control (https://www.ncbi.nlm.nih.gov/gene/14165). Previous reports on Kit mutants showed that Kit inactivation abrogates development of differentiating spermatogonia and does not affect SSC self-renewal (Kubota et al., 2009, Ohta et al., 2003, Yoshinaga et al., 1991). In Sycp3 KO cells, spermatogonia proliferated normally but meiosis was abrogated (Yuan et al., 2000). We transfected these constructs into EGFP-expressing GS cells and produced GS cells with Kit (Kit GS) or Sycp3 (Sycp3 GS) gRNA. One month after transfection, mutation analysis was carried out in GS cells without dox treatment. However, no mutation was found in control, Kit and Sycp3 GS cells (Figure 7B). To test the efficiency of the Tet-On system, we added dox to GS cells in vitro and found that 20%–30% of GS cells were mutated without drug selection (Figure 7B).
Figure 7.
In Vivo Functional Analysis of Spermatogenic Genes Using FV-LVs by Inducible Gene Editing
(A) Experimental Procedures and schematic diagrams of FV-LVs expressing the Tet-On-dependent CRISPR/Cas9 system.
(B) Evaluation of mutation levels in dox-treated GS cells by Tide analysis. Results of three independent experiments (n = 3).
(C and D) Macroscopic appearance of testes transplanted with GS cells transduced with FV-LV expressing Tet-On CRISPR/Cas9 genes before (C) and after dox treatment (D).
(E) Testis weight of transplanted testes after dox treatment (n = 9 for control gRNA; n = 4 for Sycp3 and Kit gRNA). Results of three independent experiments (n = 3).
(F and G) Immunostaining of recipient testes before (F) and after (G) dox treatment.
(H) Quantification of the number of seminiferous tubules showing EGFP fluorescence and germ cell markers. Results of three independent experiments (n = 3). Asterisks indicates statistical significance (p < 0.05).
Scale bars, 1 mm (C, D), 50 μm (F, G). See also Tables S1 and S2. Results are mean ± SEM.
To test the usefulness of this system in vivo, GS cells were infected by each virus and the transfected cells were transplanted into busulfan-treated mouse testes more than 45 days after infection (Figure 7A). The recipients received dox via drinking water 2 months after transplantation to induce gene editing in vivo. The testes of these mice were analyzed at this point because donor-derived spermatogenesis completes around 2 months after transplantation (Nagano et al., 1999). We observed normal colonization of GS cells infected with FV-LV-Tet-CRISPR for each gRNA (Figure 7C). After administering dox for 35 days, the intensity of donor cell-derived fluorescence was weaker in testes transplanted with Kit or Sycp3 GS cells (Figure 7D). Consistent with this observation, the testis weight of the Kit GS cell sample was significantly decreased (Figure 7E).
To assess the degree of spermatogenic defect quantitatively, we performed immunostaining of dox-treated testes using antibodies against GFRA1, KIT, and SYCP3 (Figures 7F and 7G). The testes were also stained with PNA to identify haploid cells. We quantified the number of EGFP+ tubules that expressed each marker to confirm donor-derived spermatogenesis (Figure 7H). The number of tubules expressing GFRA1 did not change. However, the number of tubules with KIT+ cells was significantly decreased in testes transplanted with Kit GS cells. In recipients with Sycp3 and Kit GS cells, SYCP3+ and KIT+ tubules were also significantly reduced (Figure 7G), which was consistent with previous studies. These results suggest that in vivo gene editing using FV-LVs with GS cell transplantation is useful for functional analysis of genes associated with spermatogenesis.
Discussion
We initiated this study to overcome the male infertility problem caused by SV infection of GS cells. Although we did not notice abnormalities in initial experiments, the recipients gradually lost spermatogenesis and many empty seminiferous tubules were found in the long-term. Although the exact mechanism underlying this phenomenon is unknown, toxicity of SV has been reported by several groups (Heylbroeck et al., 2000, Tanaka et al., 2007, Waddington et al., 2004). Based on previous reports that showed increased transfection efficiency of LVs pseudotyped with F and HN proteins, we reasoned that these envelope proteins are primarily responsible for the enhanced transduction efficiency of SV and sought to develop a similar LV vector that exhibits increased transduction efficiency without impairing fertility. Although we initially produced LVs with F and HN proteins, these LVs did not infect GS cells. However, because VSVG is often used to increase virus transduction efficiency (Burns et al., 1993), we included VSVG in LV production, which dramatically improved GS cell transfection efficiency.
These newly produced LVs are more useful than conventional SVs for several reasons. First, SVs have immunogenicity and relatively slow clearance of SV RNA (Yoshizaki et al., 2006), which might have caused infertility in GS cell recipients. Second, SVs perturb expression of several spermatogonial genes involved in self-renewal and adhesion (Watanabe et al., 2019). For example, GFRA1, which is a component of GDNF receptor, was significantly downregulated. This might have caused problems in vivo because the concentration of GDNF is probably lower in vivo. Therefore, although the transfected GS cells retained SSC activity, the decreased GFRA1 expression levels might have interfered with normal SSC self-renewal and contributed to the loss of spermatogenesis. Third, because SVs do not have a DNA phase, gene expression levels and patterns cannot be controlled using conventional DNA-based promoters. In addition to these viral properties, SVs require higher biosafety containment measures, which particularly hampers in vivo studies. Moreover, SVs have more restrictions imposed by licensors or vendors (Schlaeger et al., 2015). In this context, pseudotyped LVs are more attractive because they can be readily produced and show significantly improved infectivity.
Pseudotyping of LVs was not as simple as we originally assumed. Because other groups have already demonstrated that LVs or SIVs pseudotyped with both F and HN exhibit improved transduction efficiency in human hepatocytes, respiratory epithelium, and several types of cultured cells (Kowolik and Yee, 2002, Mitomo et al., 2010, Murakami et al., 2010), we expected that pseudotyping with SV envelope proteins alone would be sufficient to improve transfection efficiency. The mechanism of the SV infection process has been well studied. It is thought that SV binds to host cells through interaction between HN proteins and sialic acids of the host cell. Then, SV enters the inside of host cell mediated by the fusion activity of F proteins (Kobayashi et al., 2003). Because sialic acids on GS cells are responsible for SV infection (Markwell and Paulson, 1980, Watanabe et al., 2019), expressing both F and HN proteins appeared to be sufficient for GS cell transfection. However, contrary to our expectations, LVs pseudotyped with a combination of F and HN proteins failed to transduce GS cells. The key molecule that led to successful infection was VSVG, because VSVG allowed transduction of GS cells with either F or HN protein. Considering that we were able to infect GS cells with SV in our previous study (Watanabe et al., 2019), this result implies that additional molecules, with a function similar to VSVG, work cooperatively with F and HN proteins to transduce GS cells during SV infection.
We further showed that pseudotyping with VSVG and F proteins gave the best infection efficiency. This result suggested that HN protein is dispensable for infection. Although we expected HN protein to increase the transduction efficiency, co-transfection of all of these proteins significantly reduced the transduction efficiency. This occurred despite HN protein promoting infection efficiency when it was expressed with VSVG. It is thought that binding of HN protein to the receptor induces a conformational change in the F protein that exposes a hydrophobic region to trigger virion/cell membrane fusion. Given our results, VSVG might have performed the same function as HN to promote initial cell fusion and trigger this conformational change. VSVG was initially used by Burns et al. (1993) for lentivirus production and it also acts as a fusion-inducing molecule by itself (Yao et al., 2003). It has been suggested that low-density lipoprotein receptor and its family members serve as cellular receptors for VSVG (Finkelshtein et al., 2013). Although we have no direct evidence of interaction between F and VSVG proteins, HN protein might have interfered with this interaction and decreased the fusion efficiency. Because several mutant F proteins that lack fusion activity are available (Paterson and Lamb, 1987, Rapaport et al., 1995), future analysis using those mutants will clarify the molecular mechanism by which VSVG and F proteins cooperate for efficient fusion.
In addition to the increased transduction efficiency, pseudotyping with the F protein made it possible for F-LVs to penetrate the BTB as well as the basement membrane of the seminiferous tubules. Although LVs have been used to transduce SSCs in vitro and have the ability to disrupt the tight junctions in the blood-brain barrier (BBB) (Nagano et al., 2002, Spindler and Hsu, 2012), it has not been possible to transduce spermatogenic cells in vivo. Although the mechanism underlying these phenomena is currently unknown, increased infectivity must play a role because neither SVs nor LVs alone could infect germ cells in vivo. To penetrate the basement membrane, it is possible that viruses that have high transduction efficiency infect spermatogonia and early spermatocytes, because exogenous antibodies can reach these cell populations after intravenous administration (Yoshinaga et al., 1991). By contrast, the penetration of the BTB is currently inexplicable. We recently showed that AAVs can similarly transduce spermatogenic cells in vivo by penetrating the BTB (Watanabe et al., 2018). Because AAVs are thought to pass through the BBB by transcytosis (Di Pasquale and Chiorini, 2006), it seems reasonable to speculate that LVs penetrated the BTB by transcytosis in the same manner. Indeed, transcytosis of LVs has been reported (Gonzalez and Sagar, 2016).
Unlike SVs, FV-LV did not alter spermatogonial marker gene expression, and normal offspring were produced by natural mating of GS cell recipients. Therefore, FL-LV can overcome the most critical problem associated with SV infection. We further demonstrated the utility of FV-LVs by making FV-IDLVs with inducible gene editing. Although the expression levels of transgenes by FV-IDLVs was lower than those achieved by FV-LVs, they did successfully transfect GS cells and induce Cre-loxP recombination in R26R-Eyfp GS cells in a manner similar to AVs and AAVs (Takehashi et al., 2007, Watanabe et al., 2017). By contrast, gene editing and subsequent transplantation of GS cells have created a new opportunity to analyze spermatogenesis in a functional manner. Genome editing in GS cells is a laborious process because of the relatively low infection efficiency and slow proliferation of GS cells. However, conventional methods of gene transfer, such as electroporation and subsequent drug selection have demonstrated the feasibility of gene editing in these cells (Chapman et al., 2015, Sato et al., 2015, Wu et al., 2015). Because we achieved genome editing of more than 20% of GS cells by FV-LVs without drug selection, we could directly analyze the function of the target gene in less time. Currently, a range of applications of CRISPR/Cas9 is available, including epigenome editing, chromatin imaging plus manipulation and base editing (Adli, 2018). Combining FV-LV-mediated gene transfer with these tools will greatly facilitate functional analysis of SSC self-renewal and spermatogenic differentiation.
Our study showed that pseudotyping of LVs with F proteins not only overcomes the problems associated with SVs but also improves the transduction efficiency and changes the transduction patterns of original LVs. It will be interesting to examine whether these newly generated LVs can transduce SSCs from other animal species that are resistant to conventional LV transfection. The ability to penetrate the BTB and the basement membrane of the seminiferous tubules will also improve the efficiency of genetic modification of male germ cells in vivo. Theoretically, almost all membrane proteins can be used for LV pseudotyping. They are not limited to virus envelope proteins and species of organisms, and include membrane-associated antibodies and artificially engineered membrane proteins. Therefore, it is likely that even better molecules may become available for improving the transduction efficiency. Thus, our study provides a basis for developing optimized protocols for SSC transduction, and future studies will overcome the problems associated with genetic manipulation of the male germline.
Experimental Procedures
Lentivirus Construction and Infection
In experiments to determine the transfection properties in GS cells, we used CSII-Eif1a-IRES2-Venus or CSII-Eif1a-mNG. mNG cDNA was purchased from Allele Biotechnology (San Diego, CA) and cloned into a CSII-Eif1a vector. For production of LV with VSVG (V-LV), PB (Sigma) was added at a final concentration of 10 μg/mL. LV particles were prepared as described previously (Morimoto et al., 2013). To prepare 1.0 × 109/mL of LVs, virus supernatants were concentrated by ultrafiltration (Amicon Ultra-4 4ML - 100 kDa cutoff, UFC810096, Millipore, Darmstadt, Germany) using PBS. To produce pseudotyped LVs, pCAG-F and/or pCAG-HN, pCAG-HIVgp, and pCMV-VSV-G-RSV-Rev were co-transfected with pCSII vector into HEK293T cells. F and HN cDNAs were derived from the Z strain of an SV (a gift from Dr. T. Irie, Hiroshima University, Hiroshima, Japan). To prepare FV-IDLV, we used psPAX2-D64V (Certo et al., 2011), pMD2-G, pCAG-F, and pCSII-Eif1a-Cre. For production of LVs expressing CRISPR/Cas9, we used lentiCas9-Blast (no. 52962; Addgene, Watertown, MA) and lentiGuide-Puro (no. 52963, Addgene). To construct inducible LVs expressing Tet-On CRISPR/Cas9, spCas9 cDNA was amplified by PCR using pX330-U6-Chimeric-BB-CBh-hSpCas9 (Cong et al., 2013) as a template, and cloned into pTetO-FUW (Brambrink et al., 2008). We also cloned rtTA cDNA (a gift from Dr. K. Yagita, Kyoto Prefectural University of Medicine, Kyoto, Japan) into pCSII-Eif1a vector. The following gRNA sequences were used in this study; Fgf10 gRNA (5′-TGTTTGGATCGTCATGGGG-3′), Cldn11 gRNA (5′-ATGGGCCACGAGCCTGGAG-3′), Sycp3 gRNA (5′-TTTTAGGCTGATCAACCAA-3′), and Kit gRNA (5′-TCAGCCATCTGCAAGTCCA-3′). The titers of all types of viruses used in this study were measured using a Lenti-X p24 Rapid Titer Kit (Clontech, Mountain View, CA) according to the manufacturer's instructions. All types of LVs were infected at the indicated MOI, which ranged from 0 to 60.
Statistical Analyses
Results are presented as the means ± SEM. Data were analyzed using Student's t-test. Multiple comparison analyses were performed using ANOVA followed by Tukey's HSD test.
Author Contributions
T.S. carried out the experiments and analyzed the data. T.S. and M.K.-S. wrote the manuscript.
Acknowledgments
We thank Ms. S. Ikeda and Mr. S. Watanabe for technical assistance and Dr. T. Irie (Hiroshima University) for providing a plasmid encoding SV envelope genes. Financial support for this research was provided by AMED (17933225), and Grants-in-aid for Scientific Research on Innovative Areas (19H05750, 19H04906, 18H04882) from The Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Published: March 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.02.001.
Supplemental Information
References
- Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H., Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R.L., Zimmermann J.W. Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. U S A. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J.C., Friedmann T., Driever W., Burrascano M., Yee J.K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. U S A. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo M.T., Ryu B.Y., Annis J.E., Garibov M., Jarjour J., Rawlings D.J., Scharenberg A.M. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K.M., Medrano G.A., Jaichander P., Chaudhary J., Waits A.E., Nobrega M.A., Hotaling J.M., Ober C., Hamra F.K. Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Rep. 2015;10:1828–1835. doi: 10.1016/j.celrep.2015.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornetta K., Anderson W.F. Protamine sulfate as an effective alternative to polybrene in retroviral-mediated gene-transfer: implications for human gene therapy. J. Virol. Methods. 1989;23:187–194. doi: 10.1016/0166-0934(89)90132-8. [DOI] [PubMed] [Google Scholar]
- de Rooij D.G., Russell L.D. All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 2000;21:776–798. [PubMed] [Google Scholar]
- Di Pasquale G., Chiorini J.A. AAV transcytosis through barrier epithelia and endothelium. Mol. Ther. 2006;13:506–516. doi: 10.1016/j.ymthe.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. U S A. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez O.A., Sagar M. Antibodies and acidic environment do not enhance HIV-1 transcytosis. J. Infect. Dis. 2016;214:1221–1224. doi: 10.1093/infdis/jiw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heylbroeck C., Balachandran S., Servant M.J., DeLuca C., Barber G.N., Lin R., Hiscott J. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 2000;74:3781–3792. doi: 10.1128/jvi.74.8.3781-3792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa M., Tergaonkar V., Ogura A., Ogonuki N., Inoue K., Verma I.M. Restoration of spermatogenesis by lentiviral gene transfer: offspring from infertile mice. Proc. Natl. Acad. Sci. U S A. 2002;99:7524–7529. doi: 10.1073/pnas.072207299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S., Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Toyokuni S., Shinohara T. Genetic selection of mouse male germline stem cells in vitro: offspring from single stem cells. Biol. Reprod. 2005;72:236–240. doi: 10.1095/biolreprod.104.035659. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ikawa M., Takehashi M., Ogonuki N., Miki H., Inoue K., Kazuki Y., Lee J., Toyokuni S., Oshimura M. Production of knockout mice by random or targeted mutagenesis in spermatogonial stem cells. Proc. Natl. Acad. Sci. U S A. 2006;103:8018–8021. doi: 10.1073/pnas.0601139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Takehashi M., Shinohara T. Brief history, pitfalls, and prospects of mammalian spermatogonial stem cell research. Cold Spring Harb. Symp. Quant. Biol. 2008;73:17–23. doi: 10.1101/sqb.2008.73.033. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Shinohara T. Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell Dev. Biol. 2013;29:163–187. doi: 10.1146/annurev-cellbio-101512-122353. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Morimoto H., Shinohara T. Fertility of male germline stem cells following spermatogonial transplantation in infertile mouse models. Biol. Reprod. 2016;94:1–11. doi: 10.1095/biolreprod.115.137869. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Iida A., Ueda Y., Hasegawa M. Pseudotyped lentivirus vectors derived from simian immunodeficiency virus SIVagm with envelope glycoproteins from paramyxovirus. J. Virol. 2003;77:2607–2614. doi: 10.1128/JVI.77.4.2607-2614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowolik C.M., Yee J.K. Preferential transduction of human hepatocytes with lentiviral vectors pseudotyped by Sendai virus F protein. Mol. Ther. 2002;5:762–769. doi: 10.1006/mthe.2002.0603. [DOI] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Schmidt J.A., Brinster R.L. Spermatogonial stem cells derived from infertile Wv/Wv mice self-renew in vitro and generate progeny following transplantation. Biol. Reprod. 2009;81:293–301. doi: 10.1095/biolreprod.109.075960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Brinster R.L. Spermatogonial stem cells. Biol. Reprod. 2018;99:52–74. doi: 10.1093/biolre/ioy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A., Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fundamental Virology. Fourth Edition. Lippincott Williams &Wilkins; 2001. pp. 1305–1340. [Google Scholar]
- Li H.O., Zhu Y.F., Asakawa M., Kuma H., Hirata T., Ueda Y., Lee Y.S., Fukumura M., Iida A., Kato A. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 2000;74:6564–6569. doi: 10.1128/jvi.74.14.6564-6569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell M.A., Paulson J.C. Sendai virus utilizes specific sialyloligosaccharides as host cell receptor determinants. Proc. Natl. Acad. Sci. U S A. 1980;77:5693–5697. doi: 10.1073/pnas.77.10.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich M.L., van Beek M.E.A.B. Spermatogonial stem cells. In: Desjardins C.C., Ewing L.L., editors. Cell and Molecular Biology of the Testis. Oxford University Press; 1993. pp. 266–295. [Google Scholar]
- Mitomo K., Griesenbach U., Inoue M., Somerton L., Meng C., Akiba E., Tabata T., Ueda Y., Frankel G.M., Farley R. Toward gene therapy for cystic fibrosis using a lentivirus pseudotyped with Sendai virus envelopes. Mol. Ther. 2010;18:1173–1182. doi: 10.1038/mt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto H., Iwata K., Ogonuki N., Inoue K., Ogura A., Kanatsu-Shinohara M., Morimoto T., Yabe-Nishimura C., Shinohara T. ROS are required for mouse spermatogonial stem cell self-renewal. Cell Stem Cell. 2013;12:774–786. doi: 10.1016/j.stem.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Ikeda Y., Yonemitsu Y., Miyazaki M., Inoue M., Hasegawa M., Sueishi K., Ishibashi T. Inhibition of choroidal neovascularization via brief subretinal exposure to a newly developed lentiviral vector pseudotyped with Sendai viral envelope proteins. Hum. Gene Ther. 2010;21:199–209. doi: 10.1089/hum.2009.102. [DOI] [PubMed] [Google Scholar]
- Nagano M., Avarbock M.R., Brinster R.L. Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol. Reprod. 1999;60:1429–1436. doi: 10.1095/biolreprod60.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Shinohara T., Avarbock M.R., Brinster R.L. Retrovirus-mediated gene delivery into male germ line stem cells. FEBS Lett. 2000;475:7–10. doi: 10.1016/s0014-5793(00)01606-9. [DOI] [PubMed] [Google Scholar]
- Nagano M., Brinster C.J., Orwig K.E., Ryu B.Y., Avarbock M.R., Brinster R.L. Transgenic mice produced by retroviral transduction of male germ-line stem cells. Proc. Natl. Acad. Sci. U S A. 2001;98:13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Watson D.J., Ryu B.Y., Wolfe J.H., Brinster R.L. Lentiviral vector transduction of male germ line stem cells in mice. FEBS Lett. 2002;524:111–115. doi: 10.1016/s0014-5793(02)03010-7. [DOI] [PubMed] [Google Scholar]
- Ohta H., Tohda A., Nishimune Y. Proliferation and differentiation of spermatogonial stem cells in the W/Wv mutant mouse testis. Biol. Reprod. 2003;69:1815–1821. doi: 10.1095/biolreprod.103.019323. [DOI] [PubMed] [Google Scholar]
- Paterson R.G., Lamb R.A. Ability of the hydrophobic fusion-related external domain of a paramyxovirus F protein to act as a membrane anchor. Cell. 1987;48:441–452. doi: 10.1016/0092-8674(87)90195-4. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Ovadia M., Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Sakuma T., Yokonishi T., Katagiri K., Kamimura S., Ogonuki N., Ogura A., Yamamoto T., Ogawa T. Genome editing in mouse spermatogonial stem cell lines using TALEN and double-nicking CRISPR/Cas9. Stem Cell Reports. 2015;5:75–82. doi: 10.1016/j.stemcr.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger T.M., Daheron L., Brickler T.R., Entwisle S., Chan K., Cianci A., DeVine A., Ettenger A., Fitzgerald K., Godfrey M. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiromoto Y., Kuramochi-Miyagawa S., Daiba A., Chuma S., Katanaya A., Katsumata A., Nishimura K., Ohtaka M., Nakanishi M., Nakamura T. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA. 2013;19:803–810. doi: 10.1261/rna.038521.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler K.R., Hsu T.H. Viral disruption of the blood-brain barrier. Trends Microbiol. 2012;20:282–290. doi: 10.1016/j.tim.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehashi M., Kanatsu-Shinohara M., Inoue K., Ogonuki N., Miki H., Toyokuni S., Ogura A., Shinohara T. Adenovirus-mediated gene delivery into mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:2596–2601. doi: 10.1073/pnas.0609282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Yonemitsu Y., Yoshida K., Okano S., Kondo H., Inoue M., Hasegawa M., Masumoto K., Suita S., Taguchi T., Sueishi K. Impact of deletion of envelope-related genes of recombinant Sendai viruses on immune responses following pulmonary gene transfer of neonatal mice. Gene Ther. 2007;14:1017–1028. doi: 10.1038/sj.gt.3302955. [DOI] [PubMed] [Google Scholar]
- Tegelenbosch R.A., de Rooij D.G. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- Waddington S.N., Buckley S.M., Bernloehr C., Bossow S., Ungerechts G., Cook T., Gregory L., Rahim A., Themis M., Neubert W.J. Reduced toxicity of F-deficient Sendai virus vector in the mouse fetus. Gene Ther. 2004;11:599–608. doi: 10.1038/sj.gt.3302205. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Kanatsu-Shinohara M., Ogonuki N., Matoba S., Ogura A., Shinohara T. Adeno-associated virus-mediated delivery of genes to mouse spermatogonial stem cells. Biol. Reprod. 2017;96:221–231. doi: 10.1095/biolreprod.116.143495. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Kanatsu-Shinohara M., Ogonuki N., Matoba S., Ogura A., Shinohara T. In vivo genetic manipulation of spermatogonial stem cells and their microenvironment by adeno-associated viruses. Stem Cell Reports. 2018;10:1551–1564. doi: 10.1016/j.stemcr.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Kanatsu-Shinohara M., Shinohara T. Sendai virus-mediated transduction of mammalian spermatogonial stem cells. Biol. Reprod. 2019;100:523–534. doi: 10.1093/biolre/ioy192. [DOI] [PubMed] [Google Scholar]
- Whelan S.P.J., Barr J.N., Wertz G.W. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhou H., Fan X., Zhang Y., Zhang M., Wang Y., Xie Z., Bai M., Yin Q., Liang D. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Ghosh K., Epand R.F., Epand R.M., Ghosh H.P. Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low pH but not by heat or denaturant. Virology. 2003;310:319–332. doi: 10.1016/s0042-6822(03)00146-6. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K., Nishikawa S., Ogawa M., Hayashi S., Kunisada T., Fujimoto T., Nishikawa S. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development. 1991;113:689–699. doi: 10.1242/dev.113.2.689. [DOI] [PubMed] [Google Scholar]
- Yoshizaki M., Hironaka T., Iwasaki H., Ban H., Tokusumi Y., Iida A., Nagai Y., Hasegawa M., Inoue M. Naked Sendai virus vector lacking all of the envelope-related genes reduced cytopathogenicity and immunogenicity. J. Gene Med. 2006;8:1151–1159. doi: 10.1002/jgm.938. [DOI] [PubMed] [Google Scholar]
- Yuan L., Liu J.G., Zhao J., Brundell E., Daneholt B., Höög C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.