Summary
Naive and primed human pluripotent stem cells (hPSCs) have provided useful insights into the regulation of pluripotency. However, the molecular mechanisms regulating naive conversion remain elusive. Here, we report intermediate naive conversion induced by overexpressing nuclear receptor 5A1 (NR5A1) in hPSCs. The cells displayed some naive features, such as clonogenicity, glycogen synthase kinase 3β, and mitogen-activated protein kinase (MAPK) independence, expression of naive-associated genes, and two activated X chromosomes, but lacked others, such as KLF17 expression, transforming growth factor β independence, and imprinted gene demethylation. Notably, NR5A1 negated MAPK activation by fibroblast growth factor 2, leading to cell-autonomous self-renewal independent of MAPK inhibition. These phenotypes may be associated with naive conversion, and were regulated by a DPPA2/4-dependent pathway that activates the selective expression of naive-associated genes. This study increases our understanding of the mechanisms regulating the conversion from primed to naive pluripotency.
Keywords: human pluripotent stem cells, primed pluripotency, naive pluripotency, intermediate naive conversion, nuclear receptor 5A1
Graphical Abstract
Highlights
-
•
hPSCs in an intermediate state between primed and naive pluripotency are generated
-
•
Conversion to the state is induced by nuclear receptor 5A1 overexpression
-
•
The state displays phenotypic similarities and differences from primed and naive cells
-
•
DPPA2/4 regulate gene expression in conversion
Human stem cells exist in two different states, primed and naive. However, the mechanisms regulating conversion between them remain unknown. Here, Hirofumi Suemori and colleagues report a stable intermediate conversion stage, induced by overexpressing nuclear receptor 5A1. This state is phenotypically distinct from primed and naive cells and provides a model to elucidate the molecular events involved in naive conversion.
Introduction
Human embryonic stem cells (hESCs) are derived from the inner cell mass (ICM) of blastocysts at the preimplantation stage (Thomson et al., 1998). Initially, hESCs were thought to be similar to ICM cells, such as mouse ESCs (mESCs), which are derived from the ICMs of mouse blastocysts (Evans and Kaufman, 1981). However, significant differences have been observed between hESCs and mESCs; for example, maintenance of the hESC undifferentiated state depends on fibroblast growth factor 2 (FGF2) and the transforming growth factor β (TGF-β) family (Vallier et al., 2005) but does not require leukemia inhibitory factor (LIF). In mESCs, dual inhibition of the glycogen synthase kinase 3 (GSK3) and the mitogen-activated protein kinase (MAPK)-extracellular signal-regulated kinase (ERK) pathways (termed 2i) (Ying et al., 2008) together with LIF induces mESCs into a state more closely resembling ICM cells (Boroviak et al., 2014, Huang et al., 2014). These mESCs are considered to be in a “ground” or “naive” state. In parallel with these studies, a pluripotent state of epiblast stem cells (mEpiSCs) established from post-implantation mouse embryos (Brons et al., 2007, Tesar et al., 2007) is designated as a “primed” state, which shares many common features with hESCs. Importantly, they can be converted into the naive state by reprogramming factors, including Oct3/4 (also known as Pou5f1), Nanog, Prdm14, Klf2, Klf4, Esrrb, Nr5a1, and Nr5a2, under the 2i/LIF condition (Festuccia et al., 2012, Gillich et al., 2012, Guo et al., 2009, Hall et al., 2009, Hanna et al., 2009, Silva et al., 2009). Efforts to induce a naive state of human pluripotency identified OCT3/4, KLF2, and KLF4 as important inducing factors (Hanna et al., 2010). It was also shown that simultaneous expression of NANOG and KLF2 can generate two kinds of naive cells, termed 5i/L/A and reset cells, that are considered closer to in vivo peri-implantation pluripotent cells in some aspects than other established naive-like cells (Chan et al., 2013, Chen et al., 2015, Gafni et al., 2013, Liu et al., 2017, Qin et al., 2016, Takashima et al., 2014, Theunissen et al., 2014, Ware et al., 2014). Comparison analyses of the cell lines unveiled significant differences between primed and naive cells in terms of cellular responses to 2i conditions, metabolism, transcriptional and epigenetic profiles, and X chromosome status. Such findings provide criteria for the definitions of human naive pluripotency, but the molecular mechanisms regulating induction of the naive state from the primed state remain unclear. In this study, we examine the effects of various transcription factors on naive state induction in hPSCs.
Results
NR5A1 Overexpression Induces Naive-Associated Gene Expression in Primed hPSCs
To understand the molecular mechanisms controlling naive conversion in hPSCs, we examined whether exogenous expression of various transcription factors could promote expression of the naive-associated genes NANOG and KLF4 (Figures S1A and S1B). We selected ten genes that are either known inducers of mouse naive pluripotency or play important roles in mouse pluripotency (Festuccia et al., 2012, Gillich et al., 2012, Guo et al., 2009, Hall et al., 2009, Hanna et al., 2009, Martello et al., 2012, Niwa et al., 2009, Silva et al., 2009). A lentiviral system was used to transfect each gene into the hESC line H9, which was cultured under feeder-free conditions with mouse embryonic fibroblast (MEF)-conditioned medium to maintain a primed state (Figure S1A). The MEF-conditioned medium was used for initial screening because we routinely cultivate hESCs on MEFs with KnockOut Serum Replacement-based medium whose components are similar to those of the MEF-conditioned medium. Real-time PCR analysis revealed that expression of both NANOG and KLF4 was upregulated by NR5A1 overexpression (Figure S1B). This result was unexpected as NR5A1 overexpression in mEpiSCs does not induce expression of naive-associated genes under primed conditions (Guo and Smith, 2010), prompting us to further investigate the ability of NR5A1 as an inducer of human naive pluripotency.
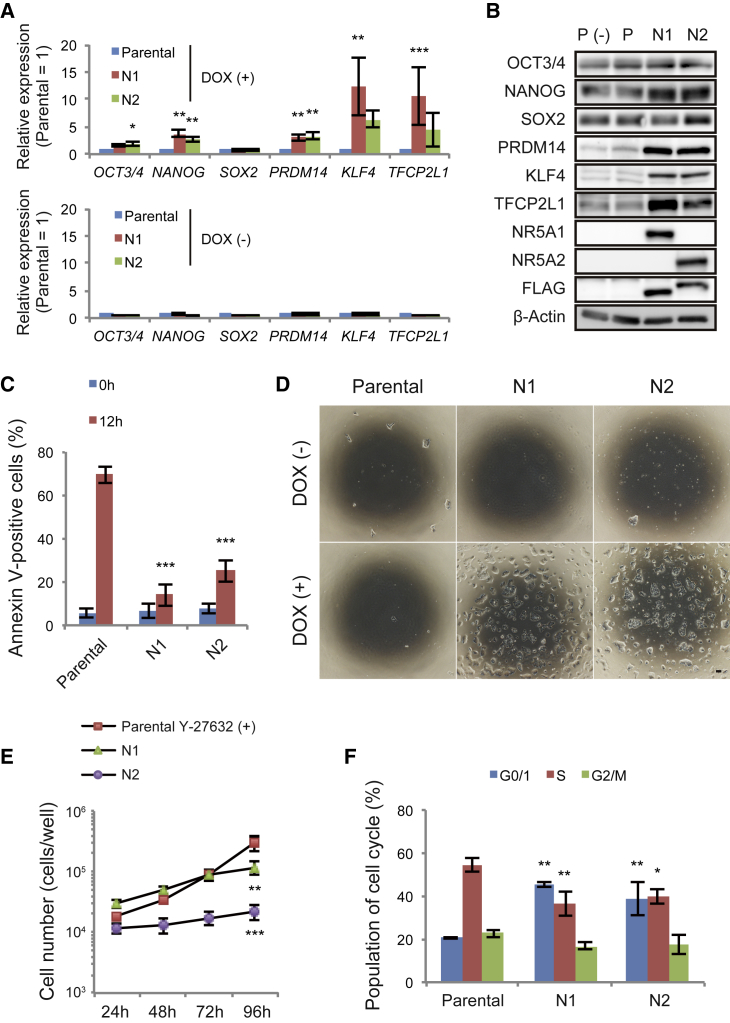
To facilitate our analyses, we established a doxycycline (DOX)-inducible expression system for NR5A1 (Figures S1C and S1D). An NR5A2 expression system was generated in parallel because of its high sequence similarity with NR5A1. DOX activated transgene expression in transfectants carrying NR5A1 or NR5A2 (referred to as N1 and N2 cells unless specified otherwise) cultured in mTeSR1 (referred to as the TGF-β and FGF2 [TF] condition for primed cells; Figures S1E and S1F). Because the MEF-conditioned medium used for initial screening contains undefined factors from feeder cells, which hampers detailed molecular analysis, we used the defined medium mTeSR1 instead. Naive-associated genes, such as PRDM14 and TFCP2L1 (Dunn et al., 2014, Takashima et al., 2014, Theunissen et al., 2014) as well as NANOG and KLF4 were upregulated at the mRNA and protein levels in the transfectants compared with parental cells cultured with DOX (referred to as parental cells unless specified otherwise; Figures 1A and 1B). We next investigated whether NR5A transfectants can survive after single-cell dissociation, because, unlike cells in the primed state, naive-state hESCs are resistant to cell death caused by dissociation. Flow cytometric analysis revealed a dramatic reduction in the proportion of annexin V-positive cells (indicating dying or dead cells) in NR5A transfectants compared with parental cells 12 h after dissociation (Figures 1C and S1G). Both N1 and N2 cells were maintained by single-cell passaging for over ten passages with sustained expression of OCT3/4, NANOG, and SOX2 (Figures 1D and S1H). However, the growth rates of both transfectants were decreased compared with parental cells (Figure 1E). Cell-cycle analysis revealed an increased G0/1 phase population and a decreased S phase population (Figure 1F). These results suggest that NR5A-induced cells lost a subset of the features of primed pluripotency. The changes in gene expression and tolerance to single-cell passaging were no longer apparent by day 10 after DOX withdrawal, indicating their dependence on NR5A expression (Figures S1I–S1K).
Figure 1.
Overexpression of NR5A Receptors Induces Naive-Associated Gene Expression in hPSCs
(A and B) Expression of genes associated with the naive pluripotent state. Data from parental (P), NR5A1-expressing (N1), and NR5A2-expressing (N2) cells were analyzed by the comparative ΔCt method. DOX-treated and untreated (A, top and bottom, respectively) samples are shown. In (A), expression in parental cells under each condition was set to 1. β-Actin was used as an internal control. P (−) represents parental cells cultured without DOX.
(C) Percentages of annexin V-positive cells before (0 h) and 12 h after single-cell dissociation. ∗∗∗p < 0.001 versus parental cells at 0 or 12 h (Dunnett's test).
(D) Representative images of parental cells and NR5A transfectants in the presence or absence of DOX at day 3 after single-cell dissociation. Scale bar, 100 μm.
(E) Growth rates of parental cells and NR5A transfectants. Cells were counted every 24 h. Parental cells were treated with Y-27632 for the first 24 h of culture.
(F) Cell proliferation analysis by Click-iT 5-ethynyl-2′-deoxyuridine (EdU) assay. After EdU treatment for 1 h, the cells were co-stained with FxCycle Violet and analyzed by flow cytometry.
Data are represented as mean ± SD (A and E) or SEM (C and F) of three biological replicates. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus parental cells (Dunnett's test). See also Figure S1.
NR5A1-Induced Cells Are Stably Maintained in Culture with GSK3 Inhibition
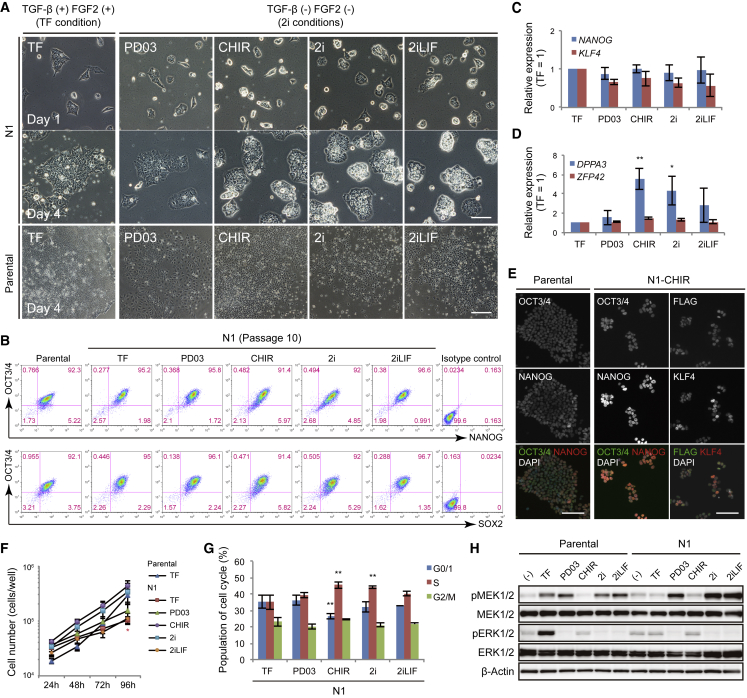
NR5A-induced cells displayed a cell proliferation defect in the primed (TF) condition, suggesting that the naive (2i/LIF) condition is more suitable for cell growth while sustaining pluripotency. We investigated whether N1 and N2 cells can be maintained in the presence of the mitogen-activated protein kinase (MEK)-ERK inhibitor PD0325901 (PD03), the GSK3 inhibitor CHIR99021 (CHIR), both inhibitors (2i), or 2i/LIF in custom mTeSR1 lacking TGF-β and FGF2, which are not required for maintaining cells in the naive state. Under 2i and 2i/LIF conditions, parental cells differentiated soon after culturing, whereas all transfectants were stably maintained over ten passages by single-cell passaging with sustained expression of OCT3/4, NANOG, and SOX2 (Figures 2A, 2B, S2A, and S2B). Interestingly, NR5A transfectants also maintained their pluripotency with PD03 or CHIR alone, with sustained expression of NANOG and KLF4 (Figure 2C). However, when we examined naive pluripotency marker expression, including DPPA3 and ZFP42 (Takashima et al., 2014, Theunissen et al., 2014), N1 cells treated with CHIR alone (N1-CHIR cells) had significantly increased expression of DPPA3, but not ZFP42 (Figure 2D). N1-CHIR cells were maintained as packed colonies, similar to the 2i and 2i/LIF conditions (Figure 2A), and expressed NANOG and KLF4 (Figure 2E). Both N1- and N2-CHIR cells formed teratomas containing the three germ layers of ectoderm, mesoderm, and endoderm when transplanted into immunodeficient mice (Figure S2C). Furthermore, the CHIR condition increased cell proliferation and enhanced entry into S phase of the cell cycle (Figures 2F and 2G). In contrast, PD03 had little or no effect, and LIF also had little effect. These results suggested that CHIR treatment alone was sufficient to maintain pluripotency and facilitate the proliferation of NR5A-induced cells.
Figure 2.
NR5A1-Induced Cells Are Stably Maintained in Culture with a GSK3 Inhibitor
(A) Representative images of parental at day 4 and N1 cells at passage 10 after culturing with combined GSK3 and MEK-ERK inhibition.
(B) Flow cytometric analysis of pluripotency markers in parental and N1 cells at passage 10 cultured under each condition.
(C and D) Expression of the indicated genes in N1 cells. RNA was extracted from cells cultured for >10 passages under each condition. Data were analyzed by the comparative ΔCt method. The TF condition was set to 1.
(E) Immunocytochemical staining of pluripotency markers and FLAG in parental and CHIR-treated N1 (N1-CHIR) cells at passage 6.
(F) Growth rates of N1 cells. Cells after switching from TF to each condition were counted every 24 h. Parental cell data from Figure 1E are shown here for comparison.
(G) Cell proliferation analysis of N1 cells cultured for >3 passages under inhibitor conditions by EdU assay.
(H) Phosphorylated MEK1/2 and ERK1/2 in parental and N1 cells. The cells were transferred from the TF condition (mTeSR1) to custom mTeSR1 lacking TGF-β and FGF2. After 23 h of treatment, inhibitors were added to each culture for 1 h. Under TF conditions, custom mTeSR1 was changed to the TF condition. Under (−) conditions, the cells were treated with DMSO. β-Actin was used as a loading control.
Scale bars, 100 μm. Data are represented as mean ± SD (C, D, and F) or SEM (G) of three biological replicates. ∗p < 0.05; ∗∗p < 0.01 versus TF condition (C, D, and G) or versus parental cells (F); Dunnett's test. See also Figures S2 and S3.
In line with the result under the TF condition (Figures S1I–S1K), we confirmed that the pluripotency of N1-CHIR cells was dependent on NR5A1 expression, because DOX withdrawal induced differentiation even under the CHIR condition (Figure S2D). To ensure that the cellular response to CHIR was independent of the hPSC lines used, we established N1- or N2-CHIR cells derived from the hESC line KhES-1 and the human induced pluripotent stem cell (hiPSC) line 253G1, in which we constitutively overexpressed the NR5A receptors (Figure S2E). Transgene expression significantly reduced the proportions of annexin V-positive cells (Figure S2F), and the transfectants maintained their pluripotency for more than ten passages, except for N2-CHIR cells derived from KhES1 cells, which displayed >40% differentiation (Figure S2G). This result may reflect the lower NANOG, KLF4, and TFCP2L1 expression in N2-CHIR cells compared with N1-CHIR cells (Figures S2H and S2I), suggesting that NR5A2 is a weaker inducer of naive pluripotency than NR5A1.
In primed cells, suppression of MEK-ERK signaling by PD03 decreased OCT3/4 and NANOG expression soon after treatment (Figure S2J). In naive cells, reactivation of MEK-ERK signaling by PD03 withdrawal induces downregulation of NANOG and KLF4 (Theunissen et al., 2014). Conversely, our results revealed that the presence or absence of PD03 had little impact on pluripotency in NR5A-induced cells. To understand this, we investigated whether PD03 suppressed ERK1/2 phosphorylation by MEK (Figure 2H). Phosphorylated ERK1/2 was not detected in PD03-treated control or N1 cells, indicating that PD03 acted as expected. PD03 treatment induced phosphorylation of MEK1/2 in both parental and N1 cells, probably because the suppression of ERK phosphorylation by PD03 reduced negative feedback regulation, as reported previously (Sturm et al., 2010). Interestingly, activation of MEK1/2 and ERK1/2 by FGF2, a known activator of MEK-ERK signaling, was abrogated in N1 cells. We, therefore, examined whether NR5A1 overexpression decreased the expression of FGFR1, a major receptor for FGF2 (Ornitz et al., 1996). FGFR1 was significantly downregulated at the mRNA and protein levels after NR5A1 overexpression, while the low levels of FGFR2, 3, and 4 remained unchanged (Figures S2K and S2L). These results suggest that decreased FGFR1 expression diminished the response to FGF2, suppressing MEK/ERK activation in N1 cells. Taken together, we can conclude that the pluripotent state induced by NR5A1 is independent of MEK-ERK signaling, making treatment with PD03 unnecessary. Therefore, we focused on investigating the characteristics of N1 cells cultured with CHIR.
As the self-renewal of primed hESCs is dependent on TGF-β/activin/nodal signaling (Weinberger et al., 2016), we next cultured N1 cells with the activin receptor-like kinase inhibitors A83-01 (A83) and SB431542 (SB43). Their dependency on TGF-β/activin/nodal signaling became pronounced as the passage number increased (Figure S3A), indicating that NR5A1-induced cells were dependent on these pathways.
In the primed state, hESCs maintain a glycolytic metabolic state with a low mitochondrial respiration capacity, which changes to a low glycolysis, highly aerobic respiration-dependent state in the naive state (Guo et al., 2016, Sperber et al., 2015, Takashima et al., 2014, Zhou et al., 2012). Therefore, we analyzed the metabolic state of N1-CHIR cells in comparison with parental cells, N1-DOX (−), and N1-DOX (+) (−) cells, which were transferred from the CHIR condition with DOX to the TF condition without DOX (Figures S3B and S3C). Although the basal oxygen consumption rate revealed minimal changes between the cell lines, maximal respiration was significantly increased in N1-CHIR cells compared with the other lines.
Previous reports have demonstrated a higher frequency of genomic instability in naive cells subjected to continuous passaging by single-cell dissociation (Pastor et al., 2016, Theunissen et al., 2014). In this study, while NR5A transfectants derived from H9 cells displayed normal karyotypes for >10 passages, chromosomal abnormalities were observed at 25 and 19 passages in N1 cells derived from KhES-1 and 253G1 cells, respectively (Figures S3D and S3E). This indicates that the risk of genomic instability increased by single-cell dissociation, even though the cells retained their pluripotent state.
NR5A1-Induced Cells Have Transcriptional Features of Both Primed and Naive Pluripotency
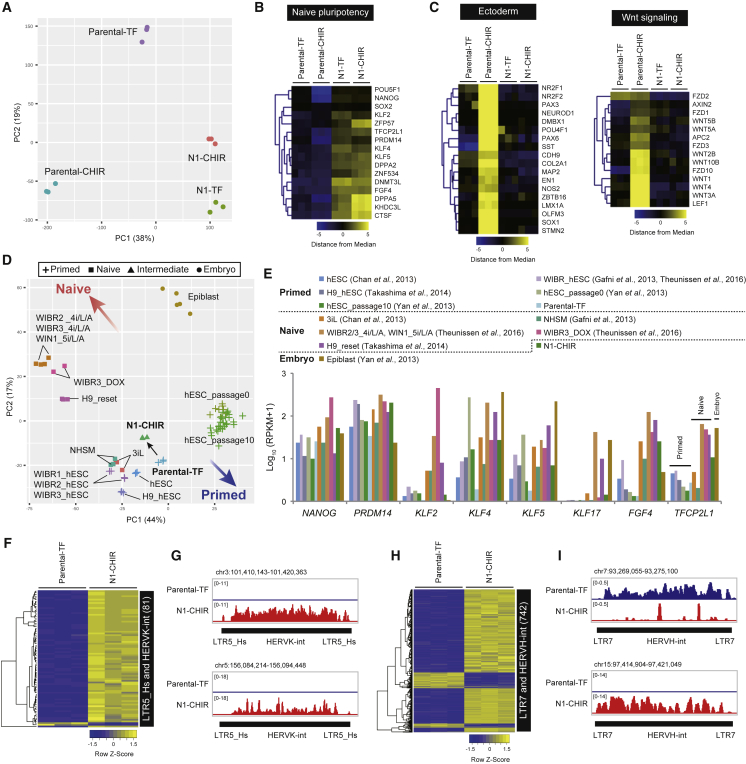
To examine the global transcriptional state of NR5A1-induced cells grown under the CHIR condition, we first performed microarray analysis of N1-CHIR cells, comparing them with parental cells cultured under TF or CHIR conditions (parental-TF and parental-CHIR cells, respectively) and N1 cells cultured under the TF condition (N1-TF cells; Figures 3A–3C). Principal-component analysis and heatmaps showed similar transcriptional states for N1 cells under both culture conditions, but these patterns were completely different from those of parental-TF or parental-CHIR cells (Figures 3A and 3B). However, a set of genes associated with naive pluripotency, including ZFP57, FGF4, KHDC3L, and CTSF was upregulated in N1-CHIR cells compared with N1-TF cells (Figure 3B; Table S3), suggesting that the CHIR condition is more suitable for the promotion of naive-associated gene expression than the TF condition. CHIR treatment of parental cells induced a marked increase in the expression of gene sets associated with ectoderm differentiation and Wnt signaling (Figures 3C and S4A), whereas upregulation of these genes was not observed in N1-CHIR cells, indicating that the response of NR5A1-induced cells to GSK3 blockade was altered at the transcriptional level. We next performed RNA sequencing (RNA-seq) analysis of parental-TF and N1-CHIR cells (Figures 3D–3I); comparative analysis of RNA-seq datasets from this study and of human naive-like cell lines (Chan et al., 2013, Gafni et al., 2013, Takashima et al., 2014, Theunissen et al., 2014) indicated that N1-CHIR cells were closer to 3iL and NHSM cells than to 4i/L/A, 5i/L/A, WIBR3-DOX, or reset cells (Figure 3D; Table S4). Among the naive-associated genes examined, KLF2 and KLF17 expression was lower in N1-CHIR cells than in 4i/L/A, 5i/L/A, WIBR3-DOX, or reset cells (Figures 3E and S4B). Overexpression of NR5A1 markedly increased DNA methyltransferase 3 beta (DNMT3B) expression at the mRNA and protein levels (Figures S4B and S4C). We next examined the expression of transposable elements (TEs), such as human endogenous retroviruses (HERVs), HERV-associated long terminal repeats (LTRs), and SINE-VNTR-Alu (SVA) elements, as diminished transcription of LTR7 and HERVH-int elements and elevated transcription of LTR5_Hs, HERVK-int, and SVA elements was reported in 5i/L/A and reset cells (Guo et al., 2017, Theunissen et al., 2016). LTR5_Hs and HERVK-int elements were listed in the top 100 TEs (log2 fold change > 1.5, adjusted p < 0.05; Table S5). The heatmap revealed significantly increased expression of these transcripts in N1 cells (Figures 3F and 3G). However, while expression of some LTR7 and HERVH-int elements was reduced, a large number of elements showed increased expression (Figures 3H and 3I). SVA elements were low and unchanged between parental-TF and N1-CHIR cells (Figure 4D, right panel). These results demonstrate that NR5A1-induced cells have features of both primed and naive pluripotency, suggesting that they represent an intermediate state of conversion between these states.
Figure 3.
NR5A1-Induced Cells Bear Transcriptional Features of Both Primed and Naive Pluripotency
(A) Principal-component analysis (PCA) of microarray data from parental and N1 cells cultured under TF or CHIR conditions.
(B and C) Heatmaps of genes selected from microarray data under each condition.
(D) PCA of RNA-seq data from this study and previously published human naive-like cell lines (3iL, NHSM, WIBR2_4i/L/A, WIBR3_4i/L/A, WIN1_5i/L/A, and WIBR3_DOX, H9_reset), human primed cell lines (hESC_passage0 and 10), and human embryos.
(E) Expression of individual genes in the RNA-seq data in (D). Values are shown as the log10 (RPKM+1). RPKM, reads per kilobase of exon per million mapped reads.
(F–I) Heatmaps (adjusted p < 0.05, log2 FC > 1.5; F and H) and expression patterns (G and I) of selected TEs under each condition.
All microarray and RNA-seq data in this study were obtained from three biological replicates. See also Figure S4 and Tables S3, S4, and S5.
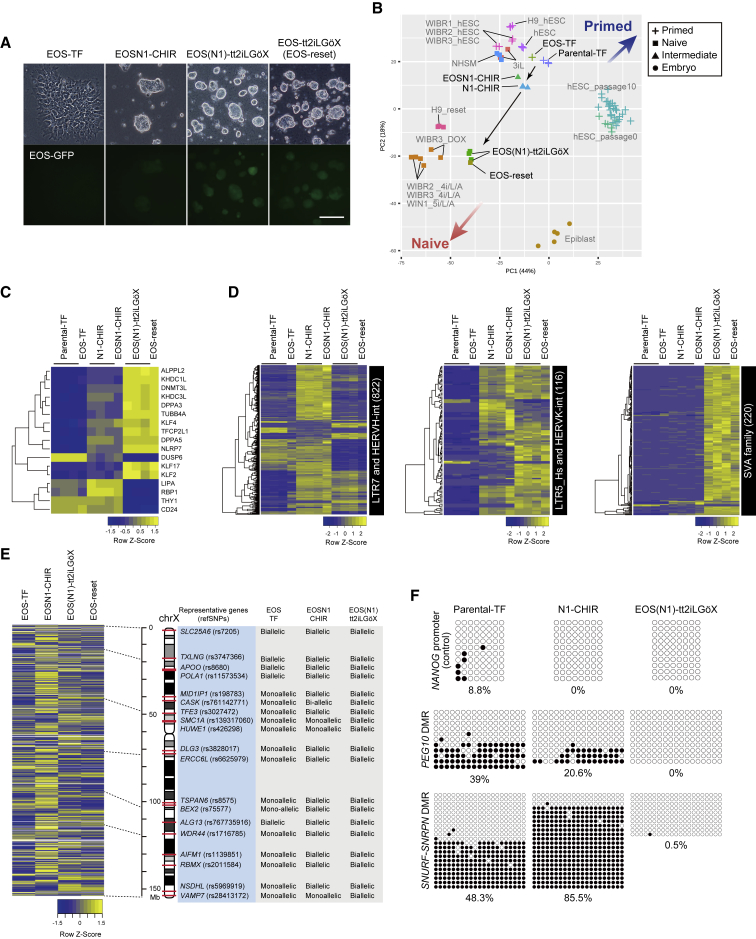
Figure 4.
Induction of the Full Naive Pluripotent State in NR5A1-Induced Cells
(A) Representative images of cells cultivated under TF, CHIR, and tt2iLGöX conditions. (N1) represents the absence of NR5A1 expression by DOX withdrawal. EOS-reset cells were established as a control for naive cells. Scale bar, 100 μm.
(B) PCA of RNA-seq data from this study and previously published human naive-like cell lines (3iL, NHSM, WIBR2_4i/L/A, WIBR3_4i/L/A, WIN1_5i/L/A, and WIBR3_DOX, H9_reset), human primed cell lines (hESC_passage0 and 10), and human embryos.
(C and D) Heatmaps of naive-associated genes (C) and TEs (D) FDR < 0.05 among parental, N1, and EOS(N1) cells under each condition.
(E) Transcriptional status of the two X chromosomes under each condition. A heatmap of X-linked genes and SNP-based allelic expression patterns in the RNA-seq data were analyzed. Allelic expression patterns of representative genes (blue box) are shown with their reference SNP IDs (rs), locations on the X chromosome (red bars), and type of expression (“monoallelic” or “biallelic”; gray box). RNA-seq data were obtained from one (EOS-TF, EOSN1-CHIR, and EOS-reset) and three (EOS(N1)-tt2iLGöX) biological samples.
(F) Bisulfite sequencing analysis of DMRs in imprinted genes. Open and closed circles represent unmethylated and methylated CpG sites, respectively. See also Figure S5, Tables S4 and S6.
NR5A1-Induced Cells Can Progress to the Full Naive Pluripotent State without NR5A1 Overexpression
To examine whether NR5A1-induced cells exist in a state where they can be converted to naive pluripotency, we next investigated whether N1 cells can progress to the full naive state. Analyses of N1 cells revealed that 2i and 2i/LIF conditions had little effect on DPPA3 expression and proliferation (Figures 2D and 2F), leading us to test a tt2iLGöX condition containing titrated CHIR, PD03, LIF, the protein kinase inhibitor Gö6983, and the tankyrase inhibitor XAV939, which was recently reported to be a more efficient protocol for naive induction (Guo et al., 2017). Because naive induction under the tt2iLGöX condition is accompanied by differentiation and cell death (Guo et al., 2017), the EOS-C(3+)-GFP-IRES-Puro (EOS-GFP) reporter system, which monitors the activity of a multimerized CR4 element derived from the mouse Oct3/4 distal enhancer driven by an early transposon promoter (Guo et al., 2017, Hotta et al., 2009, Takashima et al., 2014), was used to detect and purify naive cells. GFP expression in DOX-treated parental cells carrying EOS-GFP (EOS-TF cells) was lower than in cells with NR5A1 expression and CHIR treatment (referred to as EOSN1-CHIR cells; Figures 4A and S4D). After switching from CHIR to tt2iLGöX medium, colonies cultured with DOX gradually collapsed and displayed slower proliferation (Figure S4E), whereas DOX-untreated cells (referred to as EOS(N1)-tt2iLGöX cells) formed dome-shaped colonies with robust GFP expression (Figures 4A and S4D). These results suggest that NR5A1 overexpression is dispensable upon conversion, as previously reported in mice (Guo and Smith, 2010). We next performed RNA-seq analysis of EOS(N1)-tt2iLGöX cells for direct comparison with the reset cells (Guo et al., 2017) as well as cells in formative pluripotency, which is an intermediate state of the conversion from naive to primed pluripotency (Rostovskaya et al., 2019). The results demonstrated that EOS(N1)-tt2iLGöX cells shared features with reset cells in terms of both gene and TE expression (Figures 4B–4D; Tables S4 and S6). Notably, N1 cells were clustered closely together with cells at days 7 and 10 of formative transition when expression of formative pluripotency-associated genes was compared (Rostovskaya et al., 2019) (Figure S4F). These findings suggest that NR5A1-induced cells are in an intermediate state between naive and primed states in terms of their gene expression profile and were converted to the full naive state. Moreover, the findings suggest that naive pluripotency can be induced from the intermediate cell state, which is stably maintained by NR5A1 overexpression and CHIR treatment.
X chromosome status is an important hallmark used to define the state of hPSCs (Sahakyan et al., 2017, Theunissen et al., 2016). We therefore investigated this in NR5A1-induced intermediate cells by RNA-fluorescence in situ hybridization (FISH) and SNP-based allelic expression analysis of X-linked genes using RNA-seq data. RNA-FISH of XACT and HUWE1, which are subjected to X chromosome reactivation (Sahakyan et al., 2017), and SNP analyses revealed eroded X inactivation (XaXe) of EOS-TF cells (Figures 4E, S5A, and S5B), as reported previously (Mekhoubad et al., 2012). In EOSN1-CHIR cells, no XIST coating was observed (Figure S5B). Almost all examined X-linked genes were biallelically expressed in the cells, although monoallelic expression of HUWE1 was strongly enforced (Figures S5A and S5B). This result is consistent with the increased transcriptional states of X-linked genes in EOSN1-CHIR cells compared with EOS-TF cells (Figure 4E). In tt2iLGöX medium, the proportion of cells expressing HUWE1 biallelically was markedly increased with the appearance of monoallelic XIST coating, but the majority of cells bore no XIST clouds (Figures S5B and S5C), and expression of X-linked genes was decreased (Figure 4E). Similar phenotypes were observed in our reset cells (Figures 4E and S5A–S5C) and in previously established reset cells (Guo et al., 2017, Sahakyan et al., 2017). These results suggest that both X chromosomes are highly activated in the NR5A1-induced intermediate cell state.
Because naive cells are hypomethylated at differentially methylated regions (DMRs) in their imprinted genes (Theunissen et al., 2016), we performed bisulfite sequencing analysis to examine the methylation levels of the DMRs of PEG10 and SNURF-SNRPN (Figure 4F). The DMRs of parental-TF cells were partially methylated, as previously demonstrated in hPSCs (Kim et al., 2007). DNA methylation was maintained on the DMRs of EOSN1-CHIR cells but absent in EOS(N1)-tt2iLGöX cells. These results strongly suggest that hypomethylation of imprinted DMRs is not established at the intermediate cell state, probably because of the sustained high expression of DNMT3A and DNMT3B in N1 cells (Figure S4C).
NR5A1 Binds Regions of DPPA2 and DPPA4 that Regulate the Expression of TFCP2L1 and FGF4 but Not KLF4
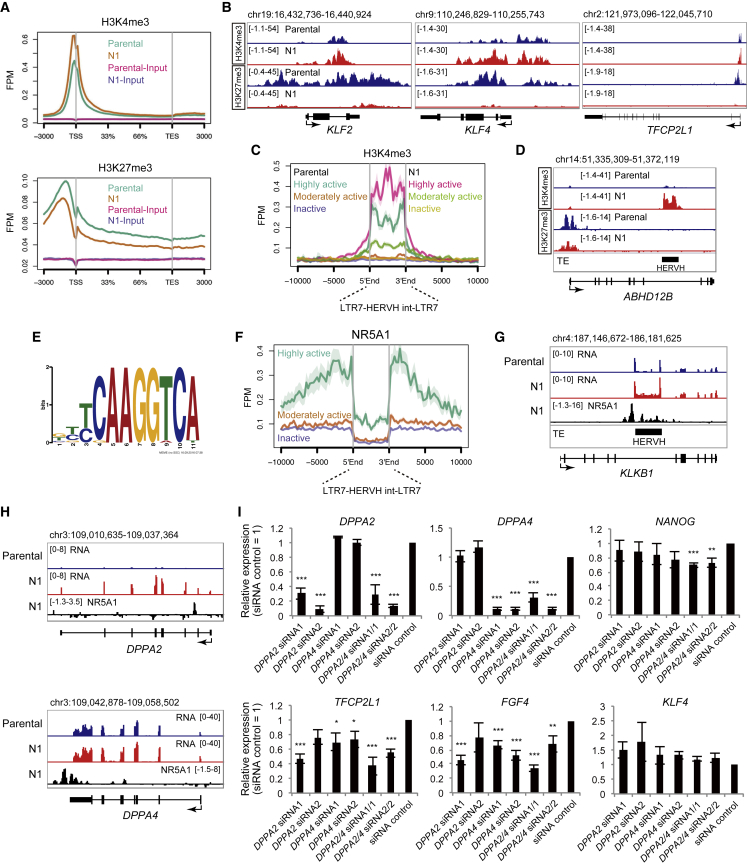
To investigate the epigenetic status of NR5A1-induced intermediate cells, the active histone mark histone 3 lysine 4 trimethylation (H3K4me3) and the repressive mark histone 3 lysine 27 trimethylation (H3K27me3) were analyzed by chromatin immunoprecipitation sequencing (ChIP-seq). A sharp H3K4me3 peak observed in parental cells at the transcription start site (TSS) was increased in N1-CHIR cells (Figure 5A). In contrast, H3K27me3 peaks upstream of the TSS to downstream of the transcriptional end site were decreased in these cells. Naive pluripotency genes, including KLF2, KLF4, and TFCP2L1, which harbored bivalent H3K4me3 and H3K27me3 marks in parental cells, exhibited a loss of H3K27me3 in the intermediate state (Figure 5B). However, a clear reduction in H3K27me3 was detected at the TSSs of only a few development-associated genes, such as NKX2-5, in N1-CHIR cells (Figures S6A and S6B), whose phenotypes are different from those of previously established naive cells (Theunissen et al., 2014). This analysis also revealed that H3K4me3 signals were increased on transcriptionally activated HERVHs in NR5A1-induced intermediate cells (Figures 5C and 5D).
Figure 5.
ChIP-Seq Analysis of Histone Modifications and NR5A1 in NR5A1-Induced Cells
(A) Average ChIP-seq signals for H3K4me3 (top) and H3K27me3 (bottom) concentrated around transcription start sites (TSSs) in parental and N1 cells.
(B) H3K4me3 and H3K27me3 signal distributions on individual genes.
(C) Signal enrichment of H3K4me3 marks on moderately and highly active types of HERVHs, characterized based on their expression in primed hPSCs, parental, and N1 cells. FPM, fragments per million mapped fragments.
(D) Distributions of H3K4me3 and H3K27me3 signals on the ABHD12B gene in N1 cells.
(E) NR5A1 consensus motif detected by the de novo motif discovery algorithm MEME.
(F) NR5A1 signal distributions on the three types of HERVHs in N1 cells.
(G) NR5A1 signal distribution on the KLKB1 gene in N1 cells.
(H) NR5A1 signal distribution on the DPPA2 and DPPA4 genes in N1 cells.
(I) Knockdown of DPPA2 and DPPA4 in N1 cells. Data were analyzed by the comparative ΔCt method. Cells treated with non-targeted siRNA (siRNA control) were set to 1. Data are represented as mean ± SD of three biological replicates. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student's t test). See also Figure S6.
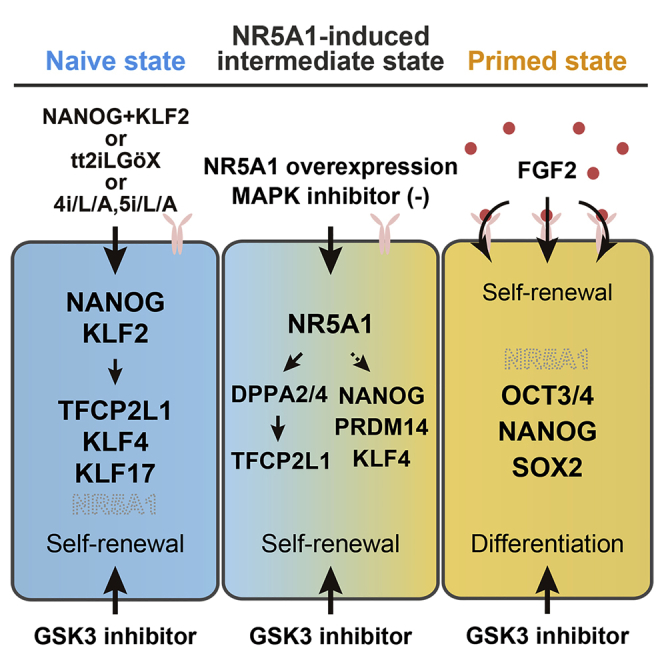
In mice, the Oct3/4, Nanog, and Klf2 genes are direct targets of NR5A2 during somatic cell reprograming into iPSCs (Heng et al., 2010). To examine whether these genes are targets of NR5A1, we performed ChIP followed by deep sequencing. Motif analysis of NR5A1 revealed a previously reported consensus motif (Figure 5E) (Martin and Tremblay, 2010). ChIP signals were distributed near the TSSs of some genes, such as NPAS4 (Figure S6C), which were reported previously in the human adrenal cortical carcinoma cell line H295R (Doghman et al., 2013) suggesting that NR5A1 binding to these genes is independent of cell type, and confirming the accuracy of our ChIP-seq analysis. The analysis also revealed strong NR5A1 binding to nearby HERVH regions that are activated in primed hPSCs (Wang et al., 2014) (Figures 5F and 5G), suggesting that NR5A1 is involved in the transcriptional activation of HERVHs, resulting in the stall at the intermediate conversion state. Significant enrichments were not observed on the OCT3/4, NANOG, or KLF2 genes, but were observed in regions of DPPA2 and DPPA4 (Figure 5H). As both genes are key reprogramming factors in mice (Hernandez et al., 2018), we performed knockdowns using short interfering RNAs (siRNAs) against DPPA2 and DPPA4 to confirm that these genes were functional in the NR5A1-mediated pluripotent state (Figures 5I and S6D). Gene expression analysis of N1 cells after knockdown of both genes revealed downregulation of naive-associated genes, such as NANOG, TFCP2L1, and FGF4, although KLF4 expression was maintained. Furthermore, PAX6 and FOXF1 were upregulated. These results suggest that the pluripotent state of N1 cells shifts toward differentiation rather than reverting to the primed state, suggesting that DPPA2 and DPPA4 may be involved in the transcriptional regulation of a subset of naive-associated genes in the NR5A1-induced intermediate state between prime and naive pluripotency (Figure 6A).
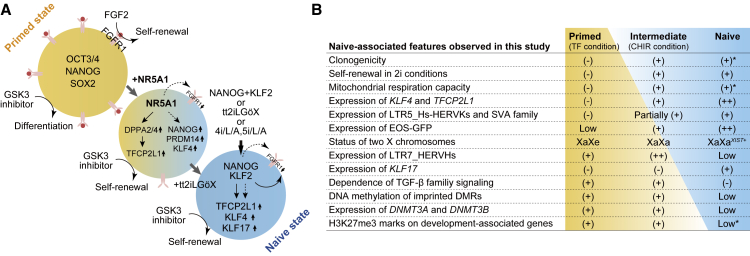
Figure 6.
Graphical Summary of This Study
(A) Transcription factors induced by NR5A1 overexpression. Arrows and dotted arrows represent direct and indirect potential targets of the indicated genes, respectively.
(B) Representative features of hPSCs in the primed, intermediate, and naive states observed in this study. Asterisks indicate results from previous studies.
Discussion
Accumulating studies on human naive pluripotency have provided ample criteria for evaluating whether established cells are distinct from primed pluripotent cells (Chan et al., 2013, Chen et al., 2015, Duggal et al., 2015, Gafni et al., 2013, Qin et al., 2016, Takashima et al., 2014, Theunissen et al., 2014, Ware et al., 2014, Yang et al., 2015). Based on these criteria, we concluded that NR5A1-induced cells exist in an intermediate state between the primed and naive states (Figure 6B). These cells bear a subset of features of naive pluripotency, such as clonogenicity, GSK3 and MEK-ERK independence, increased mitochondrial respiration capacity, naive-specific gene and LTR5-HERVK expression profiles, and two activated X chromosomes. The results suggest that these phenotypic changes occur early in the process of naive conversion. Features, such as TGF-β/activin/nodal and FGF independence, naive-specific expression profiles of TEs, DNA demethylation of imprinted genes, and XIST coating of the active X chromosome were observed under the tt2iLGöX condition, suggesting that these features are acquired during later stages of conversion. In addition to these results, KLF17 was expressed at low levels in N1-CHIR cells, but was upregulated after switching to the tt2iLGöX condition. Previous studies have revealed that KLF17 is detectable in cells in the later stages of naive conversion (Kilens et al., 2018), supporting our definition of the pluripotent state of the NR5A1-induced cells. Collectively, our results suggest that naive conversion progresses in a stepwise fashion.
NR5A1 is an orphan nuclear receptor belonging to the nuclear receptor superfamily. In mice, the protein plays central roles in endocrine organ development and steroidogenesis by regulating the transcription of steroidogenic enzyme and glycolytic genes (Lala et al., 1992, Schimmer and White, 2010). Notably, Nr5a1 is not expressed in the blastocyst (Gu et al., 2005, Ikeda et al., 1994, Pare et al., 2004). Nevertheless, Nr5a1 can replace Oct3/4 as a reprogramming factor in the generation of iPSCs from somatic cells (Heng et al., 2010). Furthermore, it can convert mEpiSCs to a naive pluripotent state (Guo and Smith, 2010). These studies led us to investigate the ability of NR5A1 in inducing human naive pluripotency. Consistent with the previous report on Nr5a1 function in mEpiSCs, continuous NR5A1 expression was dispensable after completing the conversion process in humans. However, NR5A1 overexpression induced only a subset of naive-associated features in hPSCs. The cellular response to suppressed GSK3 and MEK-ERK signaling appears to differ between mice and humans, as extrinsic suppression by 2i is required to establish naive cells from mEpiSCs with Nr5a expression (Guo and Smith, 2010), which is in contrast to our results. Exogenous expression of Nr5a1 enhances the initial expression of Klf2, suggesting that Klf2 could be direct target of Nr5a1 in mEpiSCs (Guo and Smith, 2010), but no significant binding of NR5A1 was detected on KLF2 in hPSCs. More importantly, whether Nr5a1 is involved is the activation of murine ERVs remains unknown. Our results suggest that the ability of NR5A1 to induce naive pluripotency differs slightly in mice and humans, and careful examination will be necessary to understand its molecular functions in different species.
Overexpression of NANOG and KLF2 induces the naive state in hPSCs (Takashima et al., 2014, Theunissen et al., 2014), and overexpression of KLF4 accelerates naive conversion in the t2iLGö condition (Liu et al., 2017). These reports prompted us to investigate whether NANOG and KLF4 are potential targets of NR5A1, but ChIP-seq analysis showed no significant binding to these genes. However, we found that NR5A1 bound to DPPA2 and DPPA4, whose expression was upregulated in NR5A1-induced cells. Knockdown of both genes indicated that a DPPA2/4-dependent pathway regulated the expression of naive-associated genes, such as TFCP2L1 and FGF4, but not KLF4. In addition, the siRNA-mediated depletion of DPPA2 and DPPA4 increased the expression of differentiation-associated genes. These observations suggest that these genes play a vital role in maintaining the proper balance between pluripotency and differentiation in the NR5A1-induced state.
It was unexpected that overexpression of NR5A1 elicited a suppressed response to MEK-ERK signaling by extrinsic FGF2 stimuli in a cell-autonomous manner, as such a response has not been reported in humans or mice. In general, FGF-MEK-ERK signaling has diverse functions, including cell-cycle control (Roovers and Assoian, 2000); thus, it is highly possible that in this study the suppressed FGF2 response led to reduced proliferation and G1-phase cell accumulation in NR5A1-induced cells. Inhibition of ERK phosphorylation by PD03 is required for naive conversion, although FGFR1 expression is downregulated in reset and 5iLA cells (Takashima et al., 2014, Theunissen et al., 2014). Notably, expression of other FGFRs, including FGFR3, is upregulated in these cells, but remained low in N1 cells (Figure S2K). MEK-ERK signaling can be activated through FGFR3 as well as FGFR1 (Ornitz et al., 1996), which might explain the necessity for PD03 in the naive state. Direct binding of NR5A1 to FGFR1 was not observed in our ChIP-seq analysis, suggesting that NR5A1 indirectly regulates FGFR1 expression.
We expected that human NR5A2 would have a similar ability as NR5A1 because mouse Nr5a2 also induces the naive state of mEpiSCs (Guo and Smith, 2010), and human NR5A2 can function as an iPSC reprogramming factor in humans (Wang et al., 2011). However, despite high sequence similarity between the NR5A genes in their DNA-binding domains, NR5A2 only partially mimicked the effects of NR5A1. Primed hPSCs express NR5A2, but show lower expression of PRDM14, KLF4, and TFCP2L1, and unlike NR5A1, NR5A2 overexpression did not promote expression of these genes. These results suggest that NR5A2-specific molecular mechanisms exist in hPSCs.
In summary, NR5A1 has the ability to induce and maintain cells at an intermediate state of naive conversion that has until now been inaccessible for study. Our data suggest that the pluripotent state of N1 cells may be comparable with the state of formative pluripotency, warranting further investigations on the relationship between the NR5A1-induced state and formative transition. Using NR5A1 overexpression to induce the naive state will be advantageous in elucidating the processes that regulate the conversion between naive and primed pluripotency. We cannot exclude the possibility that the pluripotent state induced by NR5A1 does not reflect a particular stage of normal development, as NR5A1 is not expressed in the human ICM or in primed or naive cells (Yan et al., 2013). However, this model will nonetheless contribute to our understanding of the molecular mechanisms that regulate pluripotency in humans.
Experimental Procedures
To establish a DOX-inducible lentiviral system, the pLV-tTRKRAB and pLVCT-tTR-KRAB vectors were purchased from Addgene (plasmids 12249 and 11643, respectively). To construct the pLV-tTR-KRAB-IRES-Neomycin (Neo) vector, an IRES-Neo cassette was inserted into pLV-tTRKRAB. To construct the pLVCT-NR5A-IRES-Puromycin (Puro) and pLVCT-NR5A-IRES-Zeocin (Zeo) vectors, the backbone of pLVCT carrying the CAG promoter was used after removing the DNA fragment (EGFP-IRES-tTR-KRAB). IRES-Puro and IRES-Zeo cassettes were subcloned into the vectors. FLAG-tagged NR5A1 and NR5A2 were amplified by PCR and each inserted into pLVCT-IRES-Puro. The pLVCT-3×FLAG-tagged NR5A1-IRES-Puro vector was generated for ChIP with an anti-FLAG antibody. Primer sequences are listed in Table S1.
Author Contributions
K.Y. and H.S. conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. K.Y. performed the experiments. T.I. analyzed the microarray, RNA-seq, and ChIP-seq data. M.H. and S.C. developed the RNA-seq and ChIP-seq methods and provided technical assistance. E.K., S.C., and K.H. assisted with data analysis and writing the manuscript. N.N. and H.S. supervised the project.
Acknowledgments
This study was partially supported by the Research Project for Practical Applications of Regenerative Medicine from the Japan Agency for Medical Research and Development (project number, JP17bk0104043) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technologies of Japan (project number, JP19K08539). We thank Takamichi Miyazaki and Kei Takada for helpful discussions, Mari Hamao for karyotype analysis, Fumiyuki Nakagawa for plasmid construction, and Emiko Moribe for sequencing. We are grateful to Masako Tada at Toho University (Japan) and Hiroyuki Kugoh at Tottori University (Japan) for advice and support with RNA-FISH, and the RIKEN BRC DNA Bank at the National BioResource Research Center for providing the NR5A1 plasmid.
Published: February 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.01.012.
Accession Numbers
All microarray and sequencing data have been deposited in the GEO data repository under accession number GSE101089.
Supplemental Information
Raw count data obtained from featureCounts and the results of DESeq2 analysis (adjusted p < 0.05, log2 fold change [FC] > 1.5) are shown.
Raw count data obtained from featureCounts are shown. The DESeq2 analysis was performed in parental-TF, N1-CHIR, and EOS(N1)-tt2iLGöX cells; false discovery rate (FDR) < 0.05.
References
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. The ability of inner cell mass cells to self-renew as embryonic stem cells is acquired upon epiblast specification. Nat. Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chan Y.S., Goke J., Ng J.H., Lu X., Gonzales K.A., Tan C.P., Tng W.Q., Hong Z.Z., Lim Y.S., Ng H.H. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Chen H., Aksoy I., Gonnot F., Osteil P., Aubry M., Hamela C., Rognard C., Hochard A., Voisin S., Fontaine E. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat. Commun. 2015;6:7095. doi: 10.1038/ncomms8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doghman M., Figueiredo B.C., Volante M., Papotti M., Lalli E. Integrative analysis of SF-1 transcription factor dosage impact on genome-wide binding and gene expression regulation. Nucleic Acids Res. 2013;41:8896–8907. doi: 10.1093/nar/gkt658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal G., Warrier S., Ghimire S., Broekaert D., Van der Jeught M., Lierman S., Deroo T., Peelman L., Van Soom A., Cornelissen R. Alternative routes to induce naive pluripotency in human embryonic stem cells. Stem Cells. 2015;33:2686–2698. doi: 10.1002/stem.2071. [DOI] [PubMed] [Google Scholar]
- Dunn S.J., Martello G., Yordanov B., Emmott S., Smith A.G. Defining an essential transcription factor program for naive pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S.R., Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gillich A., Bao S., Grabole N., Hayashi K., Trotter M.W., Pasque V., Magnusdottir E., Surani M.A. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Goodwin B., Chung A.C., Xu X., Wheeler D.A., Price R.R., Galardi C., Peng L., Latour A.M., Koller B.H. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol. Cell. Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W. Epigenetic resetting of human pluripotency. Development. 2017;144:2748–2763. doi: 10.1242/dev.146811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., von Meyenn F., Santos F., Chen Y., Reik W., Bertone P., Smith A., Nichols J. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Hanna J., Cheng A.W., Saha K., Kim J., Lengner C.J., Soldner F., Cassady J.P., Muffat J., Carey B.W., Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Mitalipova M., Cheng A.W., Cassady J.P., Staerk J., Carey B.W., Lengner C.J., Foreman R., Love J. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J.C., Feng B., Han J., Jiang J., Kraus P., Ng J.H., Orlov Y.L., Huss M., Yang L., Lufkin T. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Hernandez C., Wang Z., Ramazanov B., Tang Y., Mehta S., Dambrot C., Lee Y.W., Tessema K., Kumar I., Astudillo M. Dppa2/4 facilitate epigenetic remodeling during reprogramming to pluripotency. Cell Stem Cell. 2018;23:396–411.e8. doi: 10.1016/j.stem.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A., Cheung A.Y., Farra N., Vijayaragavan K., Seguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- Huang K., Maruyama T., Fan G. The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell. 2014;15:410–415. doi: 10.1016/j.stem.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Shen W.H., Ingraham H.A., Parker K.L. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol. Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- Kilens S., Meistermann D., Moreno D., Chariau C., Gaignerie A., Reignier A., Lelievre Y., Casanova M., Vallot C., Nedellec S. Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat. Commun. 2018;9:360. doi: 10.1038/s41467-017-02107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.P., Thurston A., Mummery C., Ward-van Oostwaard D., Priddle H., Allegrucci C., Denning C., Young L. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007;17:1731–1742. doi: 10.1101/gr.6609207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lala D.S., Rice D.A., Parker K.L. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol. Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- Liu X., Nefzger C.M., Rossello F.J., Chen J., Knaupp A.S., Firas J., Ford E., Pflueger J., Paynter J.M., Chy H.S. Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat. Methods. 2017;14:1055–1062. doi: 10.1038/nmeth.4436. [DOI] [PubMed] [Google Scholar]
- Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Gottgens B., Niwa H., Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L.J., Tremblay J.J. Nuclear receptors in Leydig cell gene expression and function. Biol. Reprod. 2010;83:3–14. doi: 10.1095/biolreprod.110.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Ornitz D.M., Xu J., Colvin J.S., McEwen D.G., MacArthur C.A., Coulier F., Gao G., Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Pare J.F., Malenfant D., Courtemanche C., Jacob-Wagner M., Roy S., Allard D., Belanger L. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J. Biol. Chem. 2004;279:21206–21216. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- Pastor W.A., Chen D., Liu W., Kim R., Sahakyan A., Lukianchikov A., Plath K., Jacobsen S.E., Clark A.T. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Hejna M., Liu Y., Percharde M., Wossidlo M., Blouin L., Durruthy-Durruthy J., Wong P., Qi Z., Yu J. YAP induces human naive pluripotency. Cell Rep. 2016;14:2301–2312. doi: 10.1016/j.celrep.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers K., Assoian R.K. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays. 2000;22:818–826. doi: 10.1002/1521-1878(200009)22:9<818::AID-BIES7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Rostovskaya M., Stirparo G.G., Smith A. Capacitation of human naive pluripotent stem cells for multi-lineage differentiation. Development. 2019;146 doi: 10.1242/dev.172916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer B.P., White P.C. Minireview: steroidogenic factor 1: its roles in differentiation, development, and disease. Mol. Endocrinol. 2010;24:1322–1337. doi: 10.1210/me.2009-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., Fischer K.A., Devi A., Detraux D., Gu H. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm O.E., Orton R., Grindlay J., Birtwistle M., Vyshemirsky V., Gilbert D., Calder M., Pitt A., Kholodenko B., Kolch W. The mammalian MAPK/ERK pathway exhibits properties of a negative feedback amplifier. Sci. Signal. 2010;3:ra90. doi: 10.1126/scisignal.2001212. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., Friedli M., He Y., Planet E., O'Neil R.C., Markoulaki S., Pontis J., Wang H., Iouranova A., Imbeault M. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R.A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Wang J., Xie G., Singh M., Ghanbarian A.T., Rasko T., Szvetnik A., Cai H., Besser D., Prigione A., Fuchs N.V. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- Wang W., Yang J., Liu H., Lu D., Chen X., Zenonos Z., Campos L.S., Rad R., Guo G., Zhang S. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl. Acad. Sci. U S A. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C.B., Nelson A.M., Mecham B., Hesson J., Zhou W., Jonlin E.C., Jimenez-Caliani A.J., Deng X., Cavanaugh C., Cook S. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yang Y., Adachi K., Sheridan M.A., Alexenko A.P., Schust D.J., Schulz L.C., Ezashi T., Roberts R.M. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc. Natl. Acad. Sci. U S A. 2015;112:E2337–E2346. doi: 10.1073/pnas.1504778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw count data obtained from featureCounts and the results of DESeq2 analysis (adjusted p < 0.05, log2 fold change [FC] > 1.5) are shown.
Raw count data obtained from featureCounts are shown. The DESeq2 analysis was performed in parental-TF, N1-CHIR, and EOS(N1)-tt2iLGöX cells; false discovery rate (FDR) < 0.05.